Bacteriophages derived from lysogens are abundant in gut microbiomes. Currently, mechanistic knowledge is lacking on the ecological ramifications of prophage carriage yet is essential to explain the abundance of lysogens in the gut. An extensive screen of the bacterial gut symbiont Lactobacillus reuteri revealed that biologically active prophages are widely distributed in this species. L. reuteri 6475 produces phages throughout the mouse intestinal tract, but phage production is associated with reduced fitness of the lysogen. However, phage production provides a competitive advantage in direct competition with a nonlysogenic strain of L. reuteri that is sensitive to these phages. This combination of increased competition with a fitness trade-off provides a potential explanation for the domination of lysogens in gut ecosystem and how lysogens can coexist with sensitive hosts.
KEYWORDS: Lactobacillus reuteri, bacteriophages, intestinal colonization, lysogen, microbial ecology, probiotics, prophage
ABSTRACT
The gut microbiota harbors a diverse phage population that is largely derived from lysogens, which are bacteria that contain dormant phages in their genome. While the diversity of phages in gut ecosystems is getting increasingly well characterized, knowledge is limited on how phages contribute to the evolution and ecology of their host bacteria. Here, we show that biologically active prophages are widely distributed in phylogenetically diverse strains of the gut symbiont Lactobacillus reuteri. Nearly all human- and rodent-derived strains, but less than half of the tested strains of porcine origin, contain active prophages, suggesting different roles of phages in the evolution of host-specific lineages. To gain insight into the ecological role of L. reuteri phages, we developed L. reuteri strain 6475 as a model to study its phages. After administration to mice, L. reuteri 6475 produces active phages throughout the intestinal tract, with the highest number detected in the distal colon. Inactivation of recA abolished in vivo phage production, which suggests that activation of the SOS response drives phage production in the gut. In conventional mice, phage production reduces bacterial fitness as fewer wild-type bacteria survive gut transit compared to the mutant lacking prophages. However, in gnotobiotic mice, phage production provides L. reuteri with a competitive advantage over a sensitive host. Collectively, we uncovered that the presence of prophages, although associated with a fitness trade-off, can be advantageous for a gut symbiont by killing a competitor strain in its intestinal niche.
IMPORTANCE Bacteriophages derived from lysogens are abundant in gut microbiomes. Currently, mechanistic knowledge is lacking on the ecological ramifications of prophage carriage yet is essential to explain the abundance of lysogens in the gut. An extensive screen of the bacterial gut symbiont Lactobacillus reuteri revealed that biologically active prophages are widely distributed in this species. L. reuteri 6475 produces phages throughout the mouse intestinal tract, but phage production is associated with reduced fitness of the lysogen. However, phage production provides a competitive advantage in direct competition with a nonlysogenic strain of L. reuteri that is sensitive to these phages. This combination of increased competition with a fitness trade-off provides a potential explanation for the domination of lysogens in gut ecosystem and how lysogens can coexist with sensitive hosts.
INTRODUCTION
The intestinal tract of vertebrates is dominated by bacteria and their viruses—bacteriophages (1–3), here called phages. Phages may follow one of two life cycles: lytic or lysogenic. Lytic phages infect their host, replicate inside the host cell, and subsequently lyse the bacterium to release their progeny. Experiments in laboratory communities revealed that lytic phages and their host are in a continuous arms race that leads to selection of bacteria that have developed resistance, which in turn leads to selection for phages that have adapted (4,5). This indicates that sensitive and resistant bacterial strains can coexist both with the lytic phage and each other over long periods due to the fitness cost of resistance, especially when resources are limited (6). These interactions provide explanations for the vast diversity of lytic phages in microbial ecosystems and their dynamics observed in nature.
Much less is known about the ecology and evolution of lysogenic phages, especially in gut ecosystems. In human feces, it is estimated there are ∼1011 bacterial cells g−1 and up to 1010 virus-like particles g−1 (7, 8), which include temperate phages (3). Temperate phages infect their host and integrate in the bacterial chromosome, which could lead to a long-lasting relationship (9). However, lysogeny can come with a fitness cost. First, the cell needs to maintain additional genetic material (10–12), which can be as much as 16% of the total genome content (9). Second, activation of lysogenic phages can kill the host (13–15). Despite the fitness burden of prophage carriage, in gut ecosystems half of the identified virus-like particles are derived from lysogens (16–18). Yet, evidence exists that development of phage resistance comes at a fitness cost (19), which is amplified in less productive environments (20). On the other hand, while prophages are dormant, they can provide protection against infection by similar phages (21) and are a source of genetic variation that has been proposed to provide an advantage during the evolution of bacterial species (22–26). Also, prophage carriage has been associated with increased competitiveness. In vitro competition experiments between lysogenic and phage-sensitive Salmonella enterica serovar Typhimurium strains revealed that prophages provide a competitive advantage (25). However, so far only two ecological studies with intestinal bacteria and their prophages have been conducted in the gut environment. In one study, competition experiments in the mouse gut between a nonlysogen and lysogen strain of Enterococcus faecalis revealed that the lysogen strain had outcompeted the nonlysogen by 1.5-fold (27); however, the experiment was limited to 24 h. A more recent study with Escherichia coli and phage λ demonstrated that phage carriage comes at a fitness cost but yields a competitive advantage when in direct competition with E. coli bacteria sensitive to phage λ (14).
The available information points to both beneficial and detrimental effects of prophages in gut microbes. However, it is still unclear to what extent prophages of a mutualistic host-associated microbe impact gut fitness. Currently, experimental evidence is lacking to substantiate claims linking beneficial or detrimental effects to the presence of prophages. This lack of knowledge stems from the absence of mechanistic studies that investigate to what extent prophages impact gut bacterial ecology and fitness in realistic model organisms in relevant experimental settings.
Lactobacillus reuteri is a Gram-positive bacterial gut symbiont that can be found in the intestinal tract of vertebrates, including pigs, mice, rats, birds, and humans (28), and has been established as a model to study the evolution of gut symbionts with their hosts (29). Comparative genome analyses combined with functional experiments in animals revealed host-adapted phylogenetic lineages whose genome content reflects niche characteristics in the respective host species (30–32). In addition, three genome editing tools have been developed for the human isolate L. reuteri 6475, including single-stranded DNA recombineering (33), CRISPR-Cas genome editing (34), and a counterselection marker (35). We applied these tools to develop L. reuteri 6475 as a model to study the functions of prophages. The strain contains two biologically active prophages, LRΦ1 and LRΦ2, which are members of the Siphoviridae family. Each prophage genome is 43 kb and is induced in the gastrointestinal tract (15). Exposure to the short-chain fatty acids acetic acid, propionic acid, and/or butyric acid or metabolism of fructose activates the Ack pathway, which, in turn, activates prophages in a RecA-dependent manner. These findings were not species or strain specific as exposure to short-chain fatty acids also promoted phage production in Lactococcus lactis and in L. reuteri ATCC 55730 (15), a strain that is genetically distinct from L. reuteri 6475 (36). While our previous study unraveled mechanisms by which prophages are induced in gut ecosystems, they have not revealed how prophages evolve with L. reuteri and how they influence its ecology in the gut.
In this study, we show that active prophages are distributed among a broad range of phylogenetically diverse strains within the species Lactobacillus reuteri, which suggests that the gut environment selects for a temperate lifestyle. Using L. reuteri 6475 and its isogenic prophage deletion derivatives, we determined the spatial scale of phage production in the gut and the mechanisms of induction. Also, we established the role of prophages for ecological fitness both in the presence of a complex microbiota and in direct competition with a sensitive strain.
RESULTS
Active prophages are broadly represented in strains of Lactobacillus reuteri.
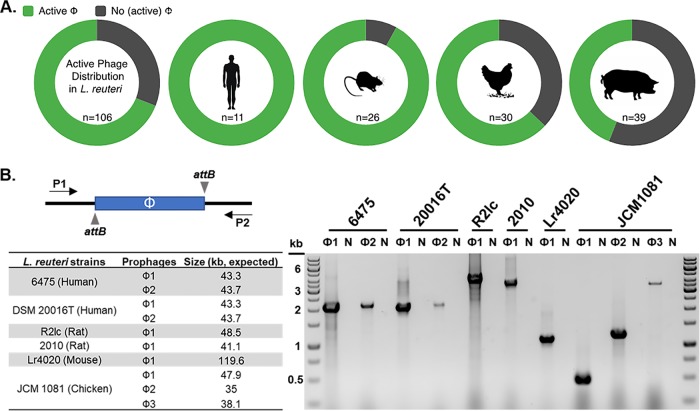
To gain insight into the role of prophages in the evolution of L. reuteri, we first determined the presence of active phages in strains that cover the known phylogenetic diversity of the species. We mapped the prophage distribution in 28 L. reuteri strains of vertebrate origin and identified 17 genomes that are predicted to encode intact prophages, while nearly all genomes contained incomplete prophages or phage remnants (Table 1). To expand on this observation, we assessed the distribution of biologically active prophages in this species by screening an extensive library of L. reuteri strains with mitomycin C (MMC), which activates the bacterial SOS response and cues the prophages to excise and lyse the bacterial cell. We tested 106 strains, including 14 strains for which a genome sequence is available (Table 1; see Fig. S2 in the supplemental material), that originate from the different phylogenetic lineages within the species (32), and are derived from different host origins: humans (n = 11), pigs (n = 39), chickens (n = 30), and rodents (n = 26). Overall, 73 out of 106 strains (69%) were induced by MMC (Fig. 1A; Fig S2). When we analyzed the distribution of active prophages by host origin, we observed that human and rodent isolates have a high distribution of active prophages (100% and 88%, respectively), while fewer active prophages were identified in chicken and pig isolates (63% and 44%, respectively) (Fig. 1A).
TABLE 1.
In silico analysis and mitomycin C induction of prophages present in L. reuteri genomes
| Strain | Genome accession no. | Origin | Status | Size (bp) | No. of prophages |
MMC induction | |||
|---|---|---|---|---|---|---|---|---|---|
| Intact | Questionable | Incomplete | Total | ||||||
| JCM 1112 | AP007281.1 | Human | Complete | 2,039,414 | 2 | 0 | 2 | 4 | Yes |
| DSM 20016 | CP000705.1 | Human | Complete | 1,999,618 | 3 | 1 | 4 | 8 | Yes |
| SD2112 | CP002844.1 | Human | Complete | 2,264,399 | 3 | 1 | 7 | 11 | Yes |
| IRT | NZ_CP011024.1 | Human | Complete | 1,993,967 | 1 | 0 | 2 | 3 | NDb |
| 121 | GCA_001889975.1 | Human | Complete | 2,302,234 | 0 | 1 | 3 | 4 | ND |
| ATCC PTA 6475 | ACGX00000000.2 | Human | Scaffold | 2,067,914 | 2 | 1 | 1 | 4 | Yes |
| CF48-3A | ACHG00000000.1 | Human | Scaffold | 2,107,903 | 2 | 1 | 1 | 4 | ND |
| ATCC PTA-4659 | ACLB00000000.1 | Human | Scaffold | 2,015,721 | 3 | 0 | 2 | 5 | Yes |
| MM34-4A | GCA_002112805.1 | Human | Scaffold | 2,152,944 | 0 | 0 | 0 | 0 | ND |
| M27U15 | GCA_002112195.1 | Human | Scaffold | 2,035,662 | 0 | 2 | 0 | 2 | ND |
| I49 | NZ_CP015408.2 | Mouse | Complete | 2,063,604 | 0 | 1 | 6 | 7 | ND |
| mlc3 | AEAW00000000.1 | Mouse | Scaffold | 2,018,630 | 1 | 0 | 1 | 2 | Yes |
| lpuph | AEAX00000000.1 | Mouse | Scaffold | 2,116,621 | 1 | 0 | 4 | 5 | Yes |
| LR0 | GCA_002156605.1 | Mouse | Contig | 2,148,567 | 0 | 0 | 2 | 2 | ND |
| TD1 | NC_021872.1 | Rat | Complete | 2,145,445 | 1 | 1 | 2 | 4 | ND |
| 100-23 | AAPZ00000000.2 | Rat | Scaffold | 2,305,557 | 3 | 0 | 5 | 8 | Yes |
| ATCC 53608 | CACS00000000.2 | Pig | Complete | 2,091,243 | 0 | 0 | 1 | 1 | No |
| I5007 | CP006011.1 | Pig | Complete | 1,947,706 | 1 | 0 | 2 | 3 | No |
| pg-3b | 2599185334 (IMG)a | Pig | Scaffold | 1,890,545 | 0 | 0 | 0 | 0 | ND |
| ZLR003 | NZ_CP014786.1 | Pig | Complete | 2,234,097 | 1 | 2 | 1 | 4 | ND |
| 20-2 | 2599185332 (IMG) | Pig | Scaffold | 2,232,947 | 0 | 0 | 0 | 0 | ND |
| lp167-67 | 2599185361 (IMG) | Pig | Scaffold | 2,015,596 | 1 | 0 | 0 | 1 | No |
| 3c6 | 2599185333 (IMG) | Pig | Scaffold | 1,934,800 | 0 | 0 | 1 | 1 | No |
| P43 | GCA_001705505.1 | Pig | Contig | 2,151,063 | 2 | 0 | 4 | 6 | ND |
| JCM 1081 | GCA_002112225.1 | Chicken | Scaffold | 2,313,528 | 2 | 1 | 3 | 6 | Yes |
| 1366 | GCA_002112185.1 | Chicken | Scaffold | 2,062,915 | 0 | 0 | 3 | 3 | No |
| CSF8 | GCA_002112245.1 | Chicken | Scaffold | 1,952,049 | 0 | 0 | 1 | 1 | Yes |
| An71 | GCA_002159305.1 | Chicken | Contig | 2,280,851 | 1 | 0 | 2 | 3 | ND |
IMG, Integrated Microbial Genomes database by Joint Genome Institute (JGI).
ND, not determined.
FIG 1.
Distribution of active prophages in L. reuteri. (A) The distribution of active (green) and nonactive (gray) prophages as determined by mitomycin C induction in L. reuteri strains with different host origins. “n” represents the number of strains tested in total (n = 106) or per origin. (B) Schematic of the experimental design to identify prophage excision showing oligonucleotides (P1 and P2) flanking the prophage (Φ) and the attB sites. The table shows select L. reuteri strains with their predicted prophage(s) (see the text for details). The agarose gel shows PCR amplicons using oligonucleotides flanking the predicted prophage(s). Strains and their prophages are listed above the gel. “N” indicates the negative (water) control.
Since mitomycin C may be toxic to certain strains at certain concentrations, we performed PCR with oligonucleotides flanking the prophage genome to predict excision upon mitomycin C treatment. From the pool of 106 strains, we selected 16 strains (indicated with an asterisk in Fig. S2) that lysed upon mitomycin C induction and for which a genome sequence is available. Using the online tool PHASTER we identified 21 presumptive intact prophage genomes distributed over these 16 strains. Eleven putative prophage genomes were located in a single contig, which did not allow us to pinpoint the integration site in the bacterial genome. For the remaining 10 prophage genomes, distributed over six strains, we designed oligonucleotides flanking the predicted prophage integration site. Each strain was exposed to mitomycin C, and PCR analysis yielded amplicons in all reactions (Fig. 1B), suggesting prophage excision. Collectively, our analyses demonstrate that biologically active prophages are widely distributed in L. reuteri.
Construction of an experimental system to study the effect of L. reuteri 6475 prophages on their host.
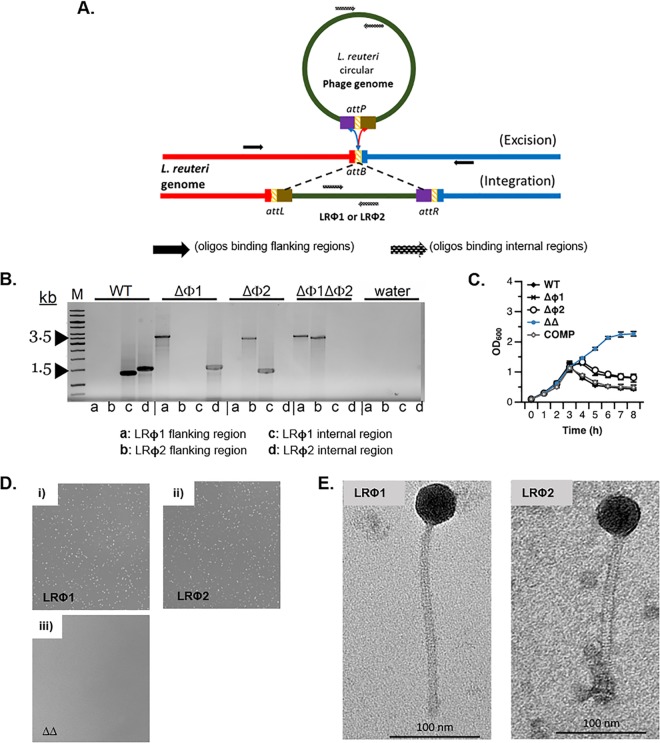
Recently, we developed a derivative of L. reuteri 6475, a human breast milk isolate (32), lacking both intact prophage genomes to yield the L. reuteri ΔΦ1 ΔΦ2 double mutant strain, and we developed a sensitive host to enumerate phage production by L. reuteri 6475 (15). The sensitive host is a derivative of the L. reuteri ΔΦ1 ΔΦ2 strain, which lacks the corresponding attB sites in the genome. Here, we expanded the platform by construction of single prophage deletions to yield the L. reuteri ΔΦ1 and L. reuteri ΔΦ2 single mutant strains (see Materials and Methods for details). The corresponding genotypes were confirmed by PCR (Fig. 2A and B). Whole-genome sequencing of the phage deletion derivatives revealed that the L. reuteri ΔΦ1 ΔΦ2 strain acquired four single nucleotide polymorphisms (SNPs), one insertion, and one deletion mutation (Table 2). To link potential phenotypical differences with deletion of prophage(s) rather than the acquired SNPs, we generated a derivative of the L. reuteri ΔΦ1 ΔΦ2 mutant in which we restored each prophage in the chromosome, which here we refer to as the prophage-complemented strain (COMP). We confirmed by Sanger sequencing that the complemented version is a derivative of the L. reuteri ΔΦ1 ΔΦ2 strain as we identified the same SNPs in both the L. reuteri ΔΦ1 ΔΦ2 mutant and the complemented strain (data not shown).
FIG 2.
Development of a model system to study prophages in L. reuteri 6475. (A) Location of oligonucleotides on the prophage and bacterial chromosome to screen for integrated or deleted prophages. (B) PCR amplification to confirm prophage deletion(s). Lane M is the molecular size marker lane. (C) Growth curves following mitomycin C induction of L. reuteri wild-type (WT), the isogenic mutants lacking prophage 1 (ΔΦ1), prophage 2 (ΔΦ2), or both prophages (ΔΔ), and a derivative of the ΔΔ mutant in which both prophages were restored (COMP). (D) Plaque assay with supernatants derived from mitomycin C-treated cultures of the L. reuteri ΔΦ2 (i), ΔΦ1 (ii), and ΔΦ1 ΔΦ2 (iii) mutants which contain LRΦ1, LRΦ2, or no phage, respectively. (E) Transmission electron micrographs of the LRΦ1 (left) and LRΦ2 (right) particles.
TABLE 2.
Mutations identified following whole-genome sequencing in L. reuteri-derivatives used in this study
| Gene ID (JCM1112) | Annotation | DNA sequence change (amino acid sequence change)a
|
|||
|---|---|---|---|---|---|
| ΔФ1 | ΔФ2 | SH | ΔФ1 ΔФ2 | ||
| LAR_0044 | FmtB (Mrp) protein | - | - | - | 1268–1270delATA; 3-bp deletion |
| LAR_0099 | ParB, chromosome partitioning | - | - | 268G→T (A90S) | 268G→T (A90S) |
| LAR_0011 | DNA-binding response regulator | - | - | - | 371C→T (T124I) |
| LAR_1262 | GentR family transcription regulator | 298G→A (V100I) | - | 298G→A (V100I) | 298G→A (V100I) |
| LAR_1266 | IS30 family transposon | 257G→A (S86N) | - | 257G→A (S86N) | 257G→A (S86N) |
| LAR_1268 | Dextransucrase protein | - | - | - | 107_108insC; frameshift mutation |
-, identical to the wild-type sequence; ΔΦ1, L. reuteri 6475 mutant in which prophage 1 is deleted; ΔΦ2, L. reuteri 6475 mutant in which prophage 2 is deleted; SH, sensitive host lacking both prophages and their attB sites; ΔΦ1 ΔΦ2, derivative of SH in which attB sites were restored.
Next, we tested the dynamics of lysis of the L. reuteri wild-type (WT) strain and its derivatives following induction with MMC. We observed that the L. reuteri wild type lysed most efficiently upon MMC treatment; lysis was delayed in each of the single-prophage-deletion strains (Fig. 2C). The strain in which we restored both prophages showed a lysis pattern very similar to that of the wild type, while the L. reuteri ΔΦ1 ΔΦ2 double mutant continued to grow following exposure to MMC. The latter indicates that LRΦ1 and LRΦ2 are the only MMC-inducible prophages in the L. reuteri 6475 genome. Although the dynamics of lysis are different between the single-prophage-deletion strains and the strains harboring both prophages, the growth rates were comparable between strains (see Fig. S3A in the supplemental material).
The supernatants derived from MMC-induced cultures of the L. reuteri ΔΦ1 and ΔΦ2 single mutant strains, containing LRΦ2 and LRΦ1, respectively, were exposed to the sensitive host strain. Each phage yielded plaques (Fig. 2D), which shows that each phage can be detected and quantified. Through these single-prophage-deletion variants, the individual phage particles can be produced following MMC treatment, allowing their characterization. Transmission electron microscopy (TEM) analysis revealed that phage particles derived from Φ1 and Φ2 have a structure typical for Siphoviridae (Fig. 2E). The phage particles share a similar morphology, with the only noticeable difference that LRΦ1 has a slightly longer tail compared to LRΦ2. In contrast to previous findings (37), where a linear correlation was found between the amino acid length of the tail tape measure protein and the tail length of the corresponding virus particle, we found that the annotated tail tape measure proteins of both L. reuteri 6475 phages were similar in size (data not shown). Collectively, our analysis showed that L. reuteri 6475 has two biologically active phages, and we developed a model system to study and quantify each of the prophages.
Prophages reduce fitness of L. reuteri 6475 during gastrointestinal transit.
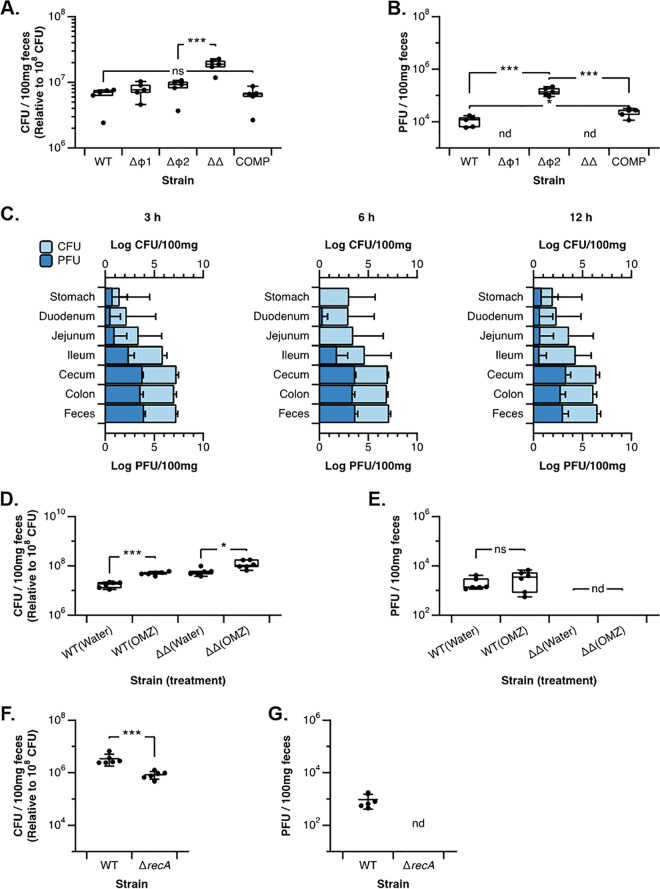
We previously demonstrated that both prophages reduce the fitness of L. reuteri following gastrointestinal (GI) transit (15); however, the impact of the individual prophages on gastrointestinal survival of L. reuteri was unknown. Therefore, we compared the fitness of the L. reuteri wild type to the single-prophage-deletion strains and the prophage-complemented strain. We administered conventional mice with 109 bacteria for two consecutive days, followed by plate count analysis 24 h following the last gavage. L. reuteri 6475 does not colonize conventional mice but the strain can be detected up to 5 days in fecal samples (data not shown). In our experimental setup, we are thus measuring the impact that prophages have on the fitness of L. reuteri 6475 following gastrointestinal transit. As shown in Fig. 3A, we recovered a similar bacterial load of wild-type and COMP strains (6.19 × 106 versus 6.18 × 106 CFU/100 mg for WT versus COMP, respectively; P = 0.99). Inactivation of single prophages only led to minor and nonsignificant 1.2- and 1.4-fold increases for the L. reuteri ΔΦ1 and ΔΦ2 single mutants, respectively (P > 0.05), while deletion of both prophages increased bacterial counts 3-fold (P = 0.002). Thus, carriage of prophages reduces the fitness of L. reuteri 6475 during gastrointestinal transit.
FIG 3.
Characterization of L. reuteri 6475 prophages during gastrointestinal transit. (A) Cell numbers of the L. reuteri wild-type strain (WT), the L. reuteri ΔΦ1, ΔΦ2, and ΔΦ1 ΔΦ2 (ΔΔ) mutant strains, and the L. reuteriΔΦ1 ΔΦ2∷Φ1∷Φ2 complemented strain (COMP) as determined by bacterial culture from feces. ns, not significant; ***, P < 0.001. (B) Phage numbers expressed as PFU derived from mice administered the WT, ΔΦ1, ΔΦ2, ΔΔ, and COMP strains. n = 5 animals per group. nd, not detected; ***, P < 0.001. (C) Temporal and spatial analyses of the cell numbers of L. reuteri 6475 wild-type (light blue bars, top axis) and PFU (dark blue bars, bottom axis) normalized to 100 mg tissue/feces following administration of 108 CFU at t = 0 h. Per time point, five animals were sacrificed. (D and E) Bacterial (D) and phage (E) counts following administration of L. reuteri strain 6475 (WT) or the ΔΦ1 ΔΦ2 mutant (ΔΔ) to mice that received regular drinking water (Water) or drinking water supplemented with omeprazole (OMZ). n = 6 animals per treatment group. (F and G) Bacterial (F) and phage (G) counts following administration of L. reuteri 6475 (WT) or 6475 ΔrecA (ΔrecA). For all panels, CFU data are expressed per 100 mg feces and normalized to 108 administered CFU and PFU data are expressed per 100 mg feces. ns, not significant; nd, not detected. *, P < 0.05; ***, P < 0.001 (t test analyses).
Temporal and spatial scale of phage production of L. reuteri in digestive tract.
In an attempt to determine the impact of each prophage on reducing the fitness of L. reuteri during gastrointestinal transit, we first determined how many phages were produced. Although there was a statistically significant difference, between the L. reuteri COMP and wild-type strains, the two strains still produced very similar amounts of PFU as detected in fecal samples (2.33 × 104 versus 1.11 × 104 PFU/100 mg feces for COMP versus WT, respectively; P = 0.02) (Fig. 3B). Administration of the L. reuteri ΔΦ2 strain yielded 10-fold more PFU than the L. reuteri wild type (1.48 × 105 versus 1.11 × 104 PFU/100 mg feces for ΔΦ2 mutant versus WT, respectively; P < 0.001), while no PFU were identified in the feces of animals gavaged with the L. reuteri ΔΦ1 or L. reuteri ΔΦ1 ΔΦ2 mutant.
Understanding where phages are induced during gastrointestinal transit may lead to identification of the trigger(s) of phage production in the intestinal tract. To get a sense of the temporal and spatial scale of phage production in the intestinal tract, we performed a time course experiment whereby the L. reuteri wild type was administered with a single dose of 109 CFU. We analyzed the stomach, three distinct regions in the small intestine (duodenum, jejunum, and ileum), the cecum, the colon, and the feces at 3, 6, and 12 h postgavage. At all three time points, phages were mainly recovered from the ileum and the large intestine, with the highest densities detected in the cecum, colon, and feces (Fig. 3C). These data suggest that the distal intestinal tract (cecum and colon) yields more phage production compared to the small intestinal regions.
Stomach acid does not induce phage production in the GI tract.
The experiments above established that during GI transit prophages are induced in L. reuteri, which impacts host survival. We aimed next to determine what environmental cues contribute to the findings presented above. During GI transit, bacteria encounter several conditions, including acid stress in the stomach, which could activate prophages. To determine the effect of stomach acids on phage production by L. reuteri, we performed an experiment in which animals were treated with omeprazole, a proton pump inhibitor that increases the pH in the stomach (38), which has been used previously to determine the role of stomach acidity in gut colonization of L. reuteri (39). Animals (n = 4/group) were administered with vehicle or with omeprazole for 2 days, followed by administration of 109 bacteria (L. reuteri wild type or L. reuteri ΔΦ1 ΔΦ2 mutant). Supplementation of omeprazole increased gastrointestinal survival of both the L. reuteri wild type (1.73 × 107 versus 4.93 × 107 CFU/100 mg feces for water versus omeprazole, respectively; P < 0.0001 [Fig. 3D]) and L. reuteri ΔΦ1 ΔΦ2 mutant (5.87 × 107 versus 1.19 × 108 CFU/100 mg feces for water versus omeprazole, respectively; P = 0.007 [Fig. 3D]). We recovered similar levels of phage following GI transit of the L. reuteri wild type in the omeprazole and water groups (2.05 × 103 versus 3.38 × 103 PFU/100 mg feces for water versus omeprazole, respectively; P = 0.557 [Fig. 3E]). These data demonstrate that prophages do not influence the survival of L. reuteri in stomach acid and stomach acid is not a major contributor of phage induction.
In vivo phage production is dependent on activation of the SOS response.
Next, we addressed the question of whether L. reuteri phage production during gastrointestinal transit is induced by stress on the bacterial cells. The SOS response is among the best-studied stress-response systems, which induces DNA repair and recombination to promote survival when exposed to stressful conditions. A key player in activation of the SOS response is RecA, which—at least in E. coli—together with the repressor LexA regulates the expression of genes involved in DNA repair (40). Prophages are induced following activation of the SOS response, including L. reuteri (15). However, knowledge is limited to what extent the stress response triggers phage production during gastrointestinal transit. To test this, we used a mutant in which recA is inactivated (L. reuteri ΔrecA). We administered 109 CFU of the L. reuteri wild type or L. reuteri ΔrecA to mice, and 2 days after administration we determined the bacterial and viral loads in the feces. Inactivation of recA significantly reduced the ability of L. reuteri to survive GI transit (Fig. 3F; 3.42 × 106 versus 8.38 × 105 CFU/100 mg feces for WT versus ΔrecA mutant, respectively; P < 0.001), while phage levels following transit of the L. reuteri ΔrecA mutant were below the detection limit (Fig. 3G). Thus, although our data show that the SOS response likely induces phages during gastrointestinal transit, there are other factors driven by RecA—or the SOS response—that influence fitness independent of phages.
Production of phages provides L. reuteri with a competitive advantage in the gastrointestinal tract when in direct competition.
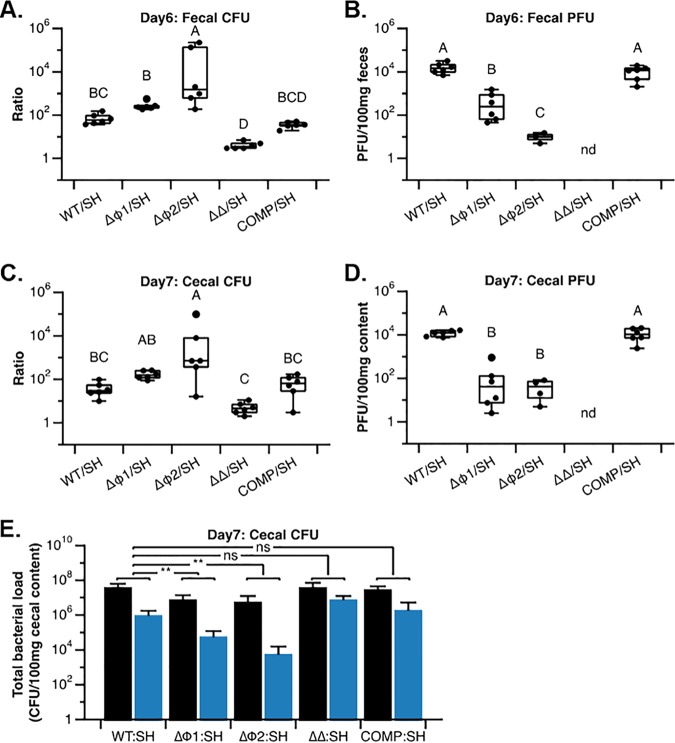
So far, our findings show that active prophages are widely distributed in the mutualistic host-associated species Lactobacillus reuteri. Using L. reuteri 6475 as our model organism, we found that phages are produced during gastrointestinal transit in a SOS-dependent manner; however, phages reduce the fitness in the main habitat of L. reuteri. This raises the question: why do so many strains—from different lineages—maintain prophages in their genome over evolutionary times despite the fact prophages could be inactivated by mutations or deleted by homologous recombination? We hypothesized that production of phages by L. reuteri provides a competitive advantage by reducing the colonization of a strain that is susceptible to these phages. To test this, we coadministered 1:1 mixtures of the sensitive host with either the L. reuteri wild type, or the three prophage deletion variants (with single or double deletions), or the complemented strain. Each mixture was administered to a group of germfree Swiss-Webster mice (n = 6/group). To distinguish between the strains by standard plate counts, we tagged the strains to yield different antibiotic resistance profiles (Table 3). We confirmed that all tagged strains had similar growth rates (Fig. S3B and C). Six days postadministration, fecal material was analyzed for bacterial and phage counts, and animals were sacrificed at day 7, after which we analyzed the cecal contents. Based on the fecal counts, the L. reuteri wild type outcompeted the sensitive host by 74-fold, which was comparable to the competition ratio of the complemented strain (37-fold; Fig. 4A). The L. reuteri ΔΦ1 and ΔΦ2 single mutants outcompeted the sensitive host by 288- and 6.0 × 104-fold, respectively. This fitness advantage was not detectable for the L. reuteri ΔΦ1 ΔΦ2 double mutant, which was recovered in similar proportions to the sensitive host. Thus, in our model system, the prophage-containing strains contribute to the in vivo competitive advantage of L. reuteri. While the ratio of the L. reuteri wild-type and complemented strains to the sensitive host was lower than those of the single-prophage-deletion strains, we recovered the highest viral load from the wild-type and the complemented strains (1.64 × 104 versus 1.06 × 104 PFU/100 mg feces for WT versus COMP, respectively [Fig. 4B]). Competition experiments with the L. reuteri ΔΦ1 or ΔΦ2 mutant yielded 502 and 10 PFU/100 mg feces for the ΔΦ1 and ΔΦ2 mutants, respectively. Similar results were obtained when we examined the competition ratios and the viral load in the ceca (Fig. 4C and D).
TABLE 3.
Bacterial strains used in this studya
| Species | Strain | Description | Reference or source |
|---|---|---|---|
| E. coli | EC1000 | In trans RepA provider, Kanr (cloning host) | 41 |
| VPL3002 | EC1000 harboring pVPL3002, Emr | 35 | |
| VPL3590 | EC1000 harboring pVPL3590, Emr | 15 | |
| VPL3593 | EC1000 harboring pVPL3593, Emr | 15 | |
| VPL3746 | EC1000 harboring pVPL3746, Emr | 15 | |
| VPL3749 | EC1000 harboring pVPL3749, Emr | 15 | |
| VPL3810 | EC1000 harboring pVPL3810, Emr | 15 | |
| VPL3886 | EC1000 harboring pVPL3886, Cmr | This study | |
| L. lactis | VPL2042 | L. lactis NZ9000 harboring pVPL2042, Emr | 35 |
| L. reuteri | ATCC PTA 6475 | Wild type, human breast milk isolate | BioGaia AB. (Fig. S2) |
| VPL4079 | VPL1014 ΔLRФ1 ΔattB1 | 15 | |
| VPL4104 | VPL1014 ΔLRФ2 ΔattB2 | 15 | |
| VPL4090 (LH) | VPL4079 ΔLRФ2 ΔattB2 | 15 | |
| VPL4119 (ΔФ1) | VPL4079::attB1 | This study | |
| VPL4120 (ΔФ2) | VPL4104::attB2 | This study | |
| VPL4121 (ΔФ1 ΔФ2) | VPL4150::attB2 | 15 | |
| VPL4126 (WT Rifr) | VPL1014 rpoB::oVPL236 (T487S H488R) Rifr | 15 | |
| VPL4132 (ΔФ1 Rifr) | VPL4119 rpoB::oVPL236 (T487S H488R) Rifr | This study | |
| VPL4135 (ΔФ2 Rifr) | VPL4120 rpoB::oVPL236 (T487S H488R) Rifr | This study | |
| VPL4129 (ΔФ1 ΔФ2 Rifr) | VPL4121 rpoB::oVPL236 (T487S H488R) Rifr | 15 | |
| VPL4150 | VPL4121::LRФ1 | This study | |
| VPL4152 | VPL4121::LRФ2 | This study | |
| VPL4159 (COMP) | VPL4152::LRФ1 | This study | |
| VPL4154 | VPL4129::LRФ1 Rifr | This study | |
| VPL4156 | VPL4129::LRФ2 Rifr | This study | |
| VPL4161 (COMP Rifr) | VPL4156::LRФ1 Rifr | This study | |
| VPL4178 (LH Cmr) | VPL4090::Cmr | This study | |
| VPL4167 (ΔФ1 Cmr) | VPL4119::Cmr | This study | |
| VPL4181 (ΔФ2 Emr) | VPL4120::Emr | This study | |
| DSM 20016 | Human isolate | ATCC (Fig. S2) | |
| SD2112 | Human isolate | BioGaia AB (Fig. S2) | |
| ATCC PTA 4659 | Human isolate | BioGaia AB (Fig. S2) | |
| PNG008B_M | Human isolate | Jens Walter (Fig. S2) | |
| PNG008-2c_8_1 | Human isolate | Jens Walter (Fig. S2) | |
| PNG008_48h | Human isolate | Jens Walter (Fig. S2) | |
| PNG008_24h | Human isolate | Jens Walter (Fig. S2) | |
| PNG008_ANA | Human isolate | Jens Walter (Fig. S2) | |
| PNG008A_M | Human isolate | Jens Walter (Fig. S2) | |
| PNG008C_M | Human isolate | Jens Walter (Fig. S2) | |
| DSM 20056 | Human isolate | JGI 642555135 (Fig. S2) | |
| mlc3 | Mouse isolate | JGI 2506381016 (Fig. S2) | |
| Lpuph-1 | Mouse isolate | JGI 2506381017 (Fig. S2) | |
| Lr4020 | Mouse isolate | 31 (Fig. S2) | |
| 100-93 | Mouse isolate | 31 (Fig. S2) | |
| 6799jm-1 | Mouse isolate | 31 (Fig. S2) | |
| 6798jm-1 | Mouse isolate | 31 (Fig. S2) | |
| ML1 | Mouse isolate | 31 (Fig. S2) | |
| one-one | Mouse isolate | 31 (Fig. S2) | |
| L1600-1 | Mouse isolate | 31 (Fig. S2) | |
| lpupjm1 | Mouse isolate | 31 (Fig. S2) | |
| L1604-1 | Mouse isolate | 31 (Fig. S2) | |
| Mouse 2 | Mouse isolate | 31 (Fig. S2) | |
| Lr4000 | Mouse isolate | BioGaia AB (Fig. S2) | |
| 100-23 | Rat isolate | JGI 2500069000 (Fig. S2) | |
| FUA3043 | Rat isolate | 31 (Fig. S2) | |
| FUA3048 | Rat isolate | 31 (Fig. S2) | |
| N2D | Rat isolate | Siv Ahrné (Fig. S2) | |
| N4I | Rat isolate | 31 (Fig. S2) | |
| Rat 19 | Rat isolate | 31 (Fig. S2) | |
| R2LC | Rat isolate | Siv Ahrné (Fig. S2) | |
| CR | Rat isolate | 31 (Fig. S2) | |
| 2010 | Rat isolate | BioGaia AB (Fig. S2) | |
| N2J | Rat isolate | Siv Ahrné (Fig. S2) | |
| AD 23 | Rat isolate | 31 (Fig. S2) | |
| bmc2 | Rat isolate | Stefan Roos (Fig. S2) | |
| LK139 | Chicken isolate | Jens Walter (Fig. S2) | |
| 11284 | Chicken isolate | Jens Walter (Fig. S2) | |
| KS6 | Chicken isolate | Jens Walter (Fig. S2) | |
| KE1 | Chicken isolate | Jens Walter (Fig. S2) | |
| LB54 | Chicken isolate | Jens Walter (Fig. S2) | |
| 1204 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK146 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK20 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK94 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK159 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK75 | Chicken isolate | Jens Walter (Fig. S2) | |
| KYE26 | Chicken isolate | Jens Walter (Fig. S2) | |
| KY21 | Chicken isolate | Jens Walter (Fig. S2) | |
| HWB7 | Chicken isolate | Jens Walter (Fig. S2) | |
| HWH3 | Chicken isolate | Jens Walter (Fig. S2) | |
| HW8 | Chicken isolate | Jens Walter (Fig. S2) | |
| CSA9 | Chicken isolate | Jens Walter (Fig. S2) | |
| CSB7 | Chicken isolate | Jens Walter (Fig. S2) | |
| L1 | Chicken isolate | Jens Walter (Fig. S2) | |
| L2 | Chicken isolate | Jens Walter (Fig. S2) | |
| KL3B | Chicken isolate | Jens Walter (Fig. S2) | |
| L3S | Chicken isolate | Jens Walter (Fig. S2) | |
| L4 | Chicken isolate | Jens Walter (Fig. S2) | |
| L5 | Chicken isolate | Jens Walter (Fig. S2) | |
| JCM1081 | Chicken isolate | JGI 2684623011 (Fig. S2) | |
| 1366 | Chicken isolate | JGI 2684623010 (Fig. S2) | |
| CSF8 | Chicken isolate | JGI 2684623009 (Fig. S2) | |
| 11283 | Chicken isolate | Jens Walter (Fig. S2) | |
| LK150 | Chicken isolate | Jens Walter (Fig. S2) | |
| NCK983 | Chicken isolate | Jens Walter (Fig. S2) | |
| ATCC 53608 | Pig isolate | BioGaia AB (Fig. S2) | |
| I5007 | Pig isolate | JGI 2554235423 (Fig. S2) | |
| LP167-67 | Pig isolate | BioGaia AB (Fig. S2) | |
| 3C6 | Pig isolate | JGI 2599185333 (Fig. S2) | |
| I5007 | Pig isolate | JGI 2554235423 (Fig. S2) | |
| 3c6 | Pig isolate | JGI 2599185333 (Fig. S2) | |
| 53608 | Pig isolate | EMBL LN906634 (Fig. S2) | |
| LP167-67 | Pig isolate | JGI 2599185361 (Fig. S2) | |
| 13S14 | Pig isolate | Jens Walter (Fig. S2) | |
| 10C2 | Pig isolate | Jens Walter (Fig. S2) | |
| 393 | Pig isolate | Jens Walter (Fig. S2) | |
| 6S15 | Pig isolate | Jens Walter (Fig. S2) | |
| 4S17 | Pig isolate | Jens Walter (Fig. S2) | |
| 104R | Pig isolate | Jens Walter (Fig. S2) | |
| LEM83 | Pig isolate | Jens Walter (Fig. S2) | |
| 23012 | Pig isolate | Jens Walter (Fig. S2) | |
| JW2015 | Pig isolate | Jens Walter (Fig. S2) | |
| JW2016 | Pig isolate | Jens Walter (Fig. S2) | |
| JW2017 | Pig isolate | Jens Walter (Fig. S2) | |
| JW2019 | Pig isolate | Jens Walter (Fig. S2) | |
| 20/2 | Pig isolate | JGI 2599185332 (Fig. S2) | |
| 27/4 | Pig isolate | Jens Walter (Fig. S2) | |
| 69/3 | Pig isolate | Jens Walter (Fig. S2) | |
| 146/2 | Pig isolate | Jens Walter (Fig. S2) | |
| 173/3 | Pig isolate | Jens Walter (Fig. S2) | |
| 173/4 | Pig isolate | Jens Walter (Fig. S2) | |
| 173/5 | Pig isolate | Jens Walter (Fig. S2) | |
| 32 | Pig isolate | Jens Walter (Fig. S2) | |
| 676 | Pig isolate | Jens Walter (Fig. S2) | |
| 1704 | Pig isolate | Jens Walter (Fig. S2) | |
| 1013 | Pig isolate | Jens Walter (Fig. S2) | |
| 1048 | Pig isolate | Jens Walter (Fig. S2) | |
| 1068 | Pig isolate | Jens Walter (Fig. S2) | |
| 1063 | Pig isolate | Jens Walter (Fig. S2) | |
| LPA1 | Pig isolate | Jens Walter (Fig. S2) | |
| Cp415 | Pig isolate | Jens Walter (Fig. S2) | |
| Cp447 | Pig isolate | Jens Walter (Fig. S2) | |
| P26 | Pig isolate | Jens Walter (Fig. S2) | |
| P97 | Pig isolate | Jens Walter (Fig. S2) | |
Rifr, rifampin resistant; RpoB, DNA-directed RNA polymerase (HMPREF0536_0828 for L. reuteri); Cmr, chloramphenicol resistant; Emr, erythromycin resistant; VPLxxxx, van Pijkeren Lab culture collection identification number. JGI and EMBL numbers are found at the Joint Genome Institute (JGI) genome portal (http://genome.jgi.doe.gov) and the European Molecular Biology Laboratory (https://www.embl.de/), respectively.
FIG 4.
Phages provide L. reuteri with a competitive advantage in the gastrointestinal tract. (A) Competition ratios at day 6 between the L. reuteri wild type and the sensitive host (WT/SH), the ΔΦ1 mutant and the sensitive host (ΔΦ1/SH), the ΔΦ2 mutant and the sensitive host (ΔΦ2/SH), the ΔΦ1 ΔΦ2 mutant and the sensitive host (ΔΔ/SH), and the L. reuteri ΔΦ1 ΔΦ2∷Φ1∷Φ2 complemented strain and the sensitive host (COMP/SH). Bacterial counts of the competing strains (rifampin resistant) and the sensitive host strain (chloramphenicol resistant) were normalized to 100 mg fecal material followed by determination of the ratio. (B) Phage numbers at day 6 following competition between the different competing strains and the sensitive host (see panel A for details). Data are expressed as PFU normalized to 100 mg feces. (C) Competition ratios at day 7 of the competing strains (see panel A for details). Bacterial counts of the competing strains (rifampin resistant) and the sensitive host strain (chloramphenicol resistant) were normalized to 100 mg intestinal content followed by determining the ratio. (D) Phage numbers in the ceca at day 7 following competition between the different competing strains and the sensitive host (see panel A for details). Data are expressed as PFU normalized to 100 mg intestinal content. (A to D) Means with different capital letters indicate statistical significance (ANOVA; P < 0.05; Tukey’s HSD test). nd, not detected. (E) Total bacterial counts of the competing strains (black bars) and the sensitive host (SH [blue bars]) at day 7 in the ceca normalized to 100 mg intestinal content. **, P < 0.01 as determined by t test; ns, not significant.
In an effort to determine why the L. reuteri wild type versus sensitive host and L. reuteri ΔΦ2 mutant versus sensitive host yielded the lowest and highest competition ratio yet yielded the most and fewest phage, respectively, we compared the total cecal bacterial load in each of these competition experiments. The total bacterial load of competitions between the wild-type and the sensitive host was 8-fold higher than competitions between the L. reuteri ΔΦ2 mutant and the sensitive host (Fig. 4E), which leads us to suggest that differences in the size of the bacterial community could explain differences in the phage population. Collectively, our data demonstrate that phage production by the gut symbiont Lactobacillus reuteri provides the organism with a competitive advantage in the gut when in competition with a strain that competes for the same nutrients.
DISCUSSION
In this work, we demonstrated that active prophages are present in the majority of Lactobacillus reuteri strains originating from different vertebrate hosts. Phages are produced throughout the gastrointestinal tract, which appears to be dependent on activation of the SOS response. In addition, we established that prophages have a double-edged sword effect on the fitness of this mutualistic microbe in the gut ecosystem.
Nearly all cellular life forms harbor parasites, which includes bacteria harboring prophages (42). Metagenome analysis of human feces revealed that the bacteriophage community is remarkably stable, which—combined with a low virus-to-microbe ratio in the gut—collectively reflects a lysogenic lifestyle (3, 43, 44). A study by Kim and Bae investigating the mouse microbiota also revealed that lysogeny is prevalent and widely distributed, where a large fraction of commensal lysogens were identified as Firmicutes (45). Indeed, nearly all human- and rodent-derived L. reuteri isolates we tested (n = 37) contained active prophages, but a lower distribution of active prophages was observed in chicken (63%) and pig (44%) lineages. In hog and poultry farming, antibiotics are broadly used to treat and prevent disease and to improve the animal’s growth rate (46). Several antibiotics, including carbadox and ASP250, are known to induce prophages in the gastrointestinal tract of swine (47, 48). Thus, extensive antibiotic use may have selected for L. reuteri strains that did not harbor, have lost, or contain a cryptic prophage (a defective prophage that cannot enter the lytic cycle). For example, the pig-derived L. reuteri strains I5007 and LP167-67 do contain a presumptive intact prophage (Table 1), yet mitomycin C treatment did not lead to cell lysis, which indeed suggests these prophages are cryptic. While we cannot exclude that prophages are induced via a mechanism other than the SOS pathway, overall, we provided experimental evidence that active prophages are broadly distributed in the gut symbiont species Lactobacillus reuteri (69% prevalence). This suggests that prophages are important for this mutualistic microbe because they are evolutionary conserved.
Despite their assumed importance, we established that in conventional mice, prophage carriage comes at a fitness cost. Similar observations were made in a study by De Paepe et al., who found that phage λ of E. coli was induced in the intestine of monoxenic mice, which consequently reduced the fitness of the lysogen (14). Their finding was elegantly validated by a strain with a mutation in the phage repressor—abolishing phage induction—that was fitter than the inducible strain (14). Another example where prophage carriage can be a burden has been established in Staphylococcus aureus. In the nasopharynx—the upper part of the throat behind the nose—S. aureus is replaced the hydrogen-peroxide producing microbe Streptococcus pneumoniae (49). Exposure of S. aureus to H2O2 activates its prophages, which consequently reduces the fitness of S. aureus and which allows the H2O2-resistant S. pneumoniae to invade the niche. Thus, both host-derived as well as microbe-derived triggers can activate prophages in a lysogen thereby reducing fitness.
In an attempt to understand the mechanism by which L. reuteri prophages are induced in the intestine, we found that prophage production in the gut is strictly dependent on activation of the SOS response. While exposure to a low pH can activate prophages (50, 51), this does not seem to be a conserved mechanism. We found that the acidity in the stomach does not contribute to L. reuteri phage production, which is in-line with previous observations in Lactococcus lactis where in vitro growth in media with a lower pH did not boost prophage induction (52). Thus, while the stomach environment reduced L. reuteri survival, this does not seem to trigger activation of the SOS response leading to phage production. This means that other mechanisms are at play, potentially driven by exposure to short-chain fatty acids (SCFAs). Throughout the intestine, the dominant SCFAs are acetic acid, butyric acid, and propionic acid (53). We previously demonstrated that exposure to biologically relevant levels of SCFAs activates the Ack pathway, which accordingly activates the SOS response, leading to a 3-log increase in phage production (15). These, and potentially other, stressors could drive phage production in L. reuteri, thereby reducing the fitness in the gut ecosystem.
Now we have established that the gut environment activates the SOS response leading to prophage production and reduced fitness, why are lysogens dominant in the gut ecosystem (3, 8, 43–45) while evidence exists that prophages can be lost (54, 55)? Carriage of prophages can alter the biology of their host and potentially provides the host with an advantage (reviewed in references 56 and 57). Briefly, prophages can alter cellular transcription (58), introduce new functions such as immunity to infection by other phages (59), promote DNA transfer that allows the cell to acquire, for example, antibiotic resistance (60), and induced prophages can provide the host with a competitive advantage by killing competitor strains (27). In our study, we tested the hypothesis that phage production provides the gut symbiont Lactobacillus reuteri with a competitive advantage by killing a competitor strain. We found that L. reuteri wild type—harboring two prophages—outcompeted the sensitive host less efficiently than the single-prophage-deletion strains did. A study in E. coli investigated within-host competition of prophages and identified that the presence of multiple prophages can reduce lytic productivity of the lysogen (61), which could explain our observed differences in competition ratios. Interestingly, in each of our competition experiments, we observed that the sensitive strain is outcompeted, yet, not eradicated by the phage-producing strain. This could indicate there is frequency-dependent selection (62), meaning that the sensitive strain seems to determine the amount of phage present. If the cell density of the sensitive host is reduced, then phage levels are reduced. Perhaps, this explains in our competition experiments why most phages are recovered from communities that contain the densest population of the sensitive host and why the sensitive host does not go extinct.
In this work, we used carefully controlled experiments with genetically modified strains to elucidate the nature of the interrelationship of prophages with a bacterial gut symbiont. Although prophages are induced and produced in the gut and thereby display a fitness burden to their host, the relationship between L. reuteri and the prophage cannot primarily be described as parasitism. Instead, prophages can provide L. reuteri with a competitive advantage against direct competitors (e.g., strains of the same species), suggesting an element to prophages that supports a mutualistic relationship with its host. Overall, these findings provide potential explanations for why bacterial gut symbionts maintain prophages in their genomes and the large amount of prophage-derived virus particles in gut microbiomes. The findings are relevant as they provide basic information on how host-phage interplay influences bacterial interactions in gut ecosystems. Future work should determine if fitness trade-offs associated with bearing prophages can stabilize the coexistence of sensitive and resistant strains and thus contribute to strain diversity in gut ecosystems. Lastly, increased knowledge of prophage-mediated lysis in the intestinal tract will be important to continue the development of recombinant L. reuteri to deliver therapeutics in the gut following prophage-mediated lysis (63, 64).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The strains and plasmids used in this study are listed in Table 3 and Table 4. All L. reuteri strains were cultured at 37°C in deMan-Rogosa-Sharpe medium (MRS; BD BioSciences). Escherichia coli and Lactococcus lactis strains were cultured in lysogeny broth (LB [Teknova]) and M17 (BD BioSciences), respectively. M17 broth was supplemented with 0.5% (wt/vol) glucose. If applicable, antibiotics were added as follows: erythromycin and chloramphenicol were supplemented at 5 μg/ml for L. reuteri and L. lactis strains, and rifampin or vancomycin was supplemented at 25 μg/ml or 500 μg/ml, respectively, for L. reuteri. E. coli was supplemented with 300 μg/ml or 20 μg/ml erythromycin or chloramphenicol, respectively.
TABLE 4.
Plasmids and bacteriophages used in this studya
| Plasmid or bacteriophage | Genotype | Description | Source |
|---|---|---|---|
| Plasmids | |||
| pVPL2042 | pNZ8048::Emr | This study | |
| pVPL3002 | pORI19::ddlA F258Yreuteri, Emr | Suicide shuttle vector with vancomycin counterselection marker | 35 |
| pVPL3048 | pVPL3002::gene insertion cassette (PCR with oVPL265-266), Emr | This study | |
| pVPL3590 | pVPL3002::LRФ1 deletion cassette, Emr | Deletion cassette targets entire LRФ1 and attB1 | 15 |
| pVPL3593 | pVPL3002::LRФ2 deletion cassette, Emr | Deletion cassette targets entire LRФ2 and attB2 | 15 |
| pVPL3746 | pVPL3002::attB1 recovery cassette, Emr | For attB1 recovery on ΔLRФ1 and ΔattB1 background | 15 |
| pVPL3749 | pVPL3002::attB2 recovery cassette, Emr | For attB2 recovery on ΔLRФ2 and ΔattB2 background | 15 |
| pVPL3810 | pVPL3002::Cmr gene insertion cassette, Emr | For Cmr gene insertion in L. reuteri VPL1014 genome | This study |
| pVPL3886 | pVPL3002::Emr gene insertion cassette, Cmr | For Emr gene insertion in L. reuteri VPL1014 genome | This study |
| pJP028 | pNZ8048::Phelp::Cmr Emr | Lab stock | |
| pNZ8048 | Nisin-inducible promoter, Cmr | 65 | |
| Bacteriophages | |||
| VPL1014 Ф1 (LRФ1) | LAR0766∼LAR0809 in L. reuteri JCM1112 reference genome | LRФ1 was isolated from VPL4120 (GenBank accession no. MH837542 [https://www.ncbi.nlm.nih.gov/genome/?term=MH837542]) | 15 |
| VPL1014 Ф2 (LRФ2) | LAR1081∼LAR1041 in L. reuteri JCM1112 reference genome | LRФ2 was isolated from VPL4119 (GenBank accession no. MH837543 [https://www.ncbi.nlm.nih.gov/genome/?term=MH837543]) | 15 |
Cmr, chloramphenicol resistant; Emr, erythromycin resistant; pVPLxxxx, van Pijkeren Lab plasmid collection identification number; DdlA, d-alanine-d-alanine ligase (HMPREF0536_1572).
Reagents and enzymes.
DNA fragments were cloned by ligation cycle reaction (LCR) (66). Prior to electroporation, LCRs were precipitated with Pellet Paint (Novagen). For cloning purposes or screen purposes, we used Phusion Hot Start polymerase II (Fermentas) or Taq polymerase (Denville Scientific), respectively. For standard ligations, we used T4 DNA ligase (Thermo Scientific). Oligonucleotides and synthetic double-stranded DNA fragments were synthesized by Integrated DNA Technologies (IDT), and are listed in Table 5.
TABLE 5.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′→3′)a | Descriptionb |
|---|---|---|
| oVPL49 | ACAATTTCACACAGGAAACAGC | Oligo paired with oVPL97 used for screening pVPL3002 constructs |
| oVPL97 | CCCCCATTAAGTGCCGAGTGC | Oligo paired with oVPL49 used for screening pVPL3002 constructs |
| oVPL187 | TACCGAGCTCGAATTCACTGG | Rev, internal oligo for pVPL3002 backbone amplification |
| oVPL188 | ATCCTCTAGAGTCGACCTGC | Fwd, internal oligo for pVPL3002 backbone amplification |
| oVPL202 | ATGAACTTTAATAAAATTGATTTAGAC | Fwd, Cmr gene from pNZ8048 |
| oVPL203 | TTATAAAAGCCAGTCATTAGGCC | Rev, Cmr gene from pNZ8048 |
| oVPL236 | TCAAACCACCAGGACCAAGCGCTGAAAGACGACGCTTtctgcTTAATTCACCTAATGGGTTGGTTTGATCCATGAACTGG | Recombineering oligos targeting L. reuteri VPL1014 RpoB gene |
| oVPL265 | TCTGTGGGGATACACTGCGATTACATG | Fwd, paired with oVPL266 used for Emr and Cmr gene insertion cassette (u/s and d/s) |
| oVPL266 | ACTATGCTGATGGAATTGATACTAGCTGG | Rev, paired with oVPL265 used for Emr and Cmr gene insertion cassette (u/s and d/s) |
| oVPL271 | TTAAAAATTAATCTTTCCAGTAATAATCAACATC | Fwd, internal oligo for pVPL3048 backbone for cloning Emr and Cmr genes |
| oVPL272 | TTAAAATGTAGGTTTAATTTTTAGGGC | Rev, internal oligo for pVPL3048 backbone for cloning Emr and Cmr genes |
| oVPL279 | TGCGCTGATGTTGATTATTACTGGAAAGATTAATTTTTAACATTATGCTTTGGCAGTTTATTCTTGACATG | Fwd, paired with oVPL280 for Phelp::Cmr gene amplification from pJP028 |
| oVPL280 | AAGCAGTCAAAAAGCCCTAAAAATTAAACCTACATTTTAATTTGATTGATAGCCAAAAAGCAGCAG | Rev, paired with oVPL279 for Phelp::Cmr gene amplification from pJP028 |
| oVPL304 | AACAGCTTGCCGTTGCATGTTAGC | Oligo paired with oVPL305, oVPL306 for MAMA PCR screening of L. reuteri VPL1014 rpoB recombinants |
| oVPL305 | AAAAGGGTGATACGGTAACCAAGG | Oligo paired with oVPL304, oVPL306 for MAMA PCR screening of L. reuteri VPL1014 rpoB recombinants |
| oVPL306 | AAGCGCTGAAAGACGACGCTTTCTG | Oligo paired with oVPL304, oVPL305 for MAMA PCR screening of L. reuteri VPL1014 rpoB recombinants |
| oVPL334 | AACTTTCGCCATTAATGTGTTTTATCGG | Fwd, for SCO and DCO screening of Cmr and Emr gene insertion |
| oVPL335 | AGACAGATGACAAGCCCTTTAGC | Rev, for SCO and DCO screening of Cmr and Emr gene insertion |
| oVPL1377 | TGCCCGTAATTTGCGAGTTC | Fwd, internal of recT1 in LRФ1 |
| oVPL1378 | ACAGGTTGCCAATCCTCTTTG | Rev, internal of recT1 in LRФ1 |
| oVPL1379 | GCTAGAAACGGGTTGCCAAT | Fwd, internal of recT2 in LRФ2 |
| oVPL1380 | TCTACCTGCTGATGTTATGGGA | Rev, internal of recT2 in LRФ2 |
| oVPL1390 | TAAGTTAAGGGATGCATAAACTGCATCC | Fwd, internal oligo for pVPL3048 backbone for cloning Cmr gene |
| oVPL1391 | TATAACCCTCTTTAATTTGGTTATATG | Rev, internal oligo for pVPL3048 backbone for cloning Cmr gene |
| oVPL1436 | AGGATTCCGACAACGTGACT | Fwd, screening oligo paired with oVPL1438 used for screening LRФ1 excision after MitC induction, Fwd, for SCO and DCO screening of LRФ1deletion |
| oVPL1437 | AGTAGCGACGGCGATTAAGA | Fwd, screening oligo paired with oVPL1439 used for screening LRФ1 excision after MitC induction |
| oVPL1438 | TATGCTGCGCTCAGTAATGG | Rev, screening oligo paired with oVPL1436 used for screening LRФ1 excision after MitC induction |
| oVPL1439 | ATCTGCCATTGTTGCTTTCC | Rev, screening oligo paired with oVPL1437 used for screening LRФ1 excision after MitC induction, Rev, oligo SCO and DCO screening of LRФ1 deletion |
| oVPL1440 | AGATGTTATTTCAGCGGTGGCG | Fwd, screening oligo paired with oVPL1441 used for screening LRФ2 excision after MitC induction |
| oVPL1441 | AATGCCACGAGGATTGATCGGG | Rev, screening oligo paired with oVPL1440 used for screening LRФ2 excision after MitC induction |
| oVPL1442 | ACTTAAAAACTGAGCAGCAATTGC | Sequencing oligo starting 49 bases d/s of oVPL1440 |
| oVPL1443 | ATTATAACTCCAATATAATTTTCGCGC | Sequencing oligo starting 398 bases d/s of oVPL1440 |
| oVPL1444 | TTGGAACAAAGTGAAGGTCTTTAATTGC | Sequencing oligo starting 53 bases d/s of oVPL1436 |
| oVPL1445 | ATAATTGAGTTACTACCAAATTGGTGAGC | Sequencing oligo starting 522 bases d/s of oVPL1436 |
| oVPL1446 | AAACGGAGATACCGAATTTAGGC | Sequencing oligo starting 41 bases d/s of oVPL1437 |
| oVPL1516 | AATTGGTGAGCCATTGGAAC | Fwd, 978 bp 5' upstream flanking sequence of LRФ1 omitting attB1L sequence |
| oVPL1517 | TGGGATCGGGGGCCAGCTTGGAC | Rev, 978 bp 5' upstream flanking sequence of LRФ1 omitting attB1L sequence |
| oVPL1518 | TGTAAGTGGTAAGCCGAGTAAC | Fwd, 1,257 bp 5' downstream flanking sequence of LRФ1 omitting attB1R sequence |
| oVPL1519 | TGGCTCAACAAGACACAAGC | Rev, 1,257 bp 5' downstream flanking sequence of LRФ1 omitting attB1R sequence |
| oVPL1520 | AAACGACGGCCAGTGAATTCGAGCTCGGTAAATTGGTGAGCCATTGGAACAACAAGCCCG | Bridging oligo for LCR assembly to construct pVPL3590 |
| oVPL1521 | GCTAAGTGTCCAAGCTGGCCCCCGATCCCATGTAAGTGGTAAGCCGAGTAACAAACCAAT | Bridging oligo for LCR assembly to construct pVPL3590 |
| oVPL1522 | AACCTTATCTGCTTGTGTCTTGTTGAGCCAATCCTCTAGAGTCGACCTGCAGGCATGCAA | Bridging oligo for LCR assembly to construct pVPL3590 |
| oVPL1528 | ATCGCAGCCTTAAGGAAATG | Fwd, 1,217 bp 5' upstream flanking sequence of LRФ2 omitting attB2L sequence |
| oVPL1529 | AGAAGTACCGGCATGCAAAG | Rev, 1,217 bp 5' upstream flanking sequence of LRФ2 omitting attB2L sequence |
| oVPL1530 | ACCAGTACGTTCACGTAAGTAG | Fwd, 1,139 bp 5' downstream flanking sequence of LRФ2 omitting attB2R sequence |
| oVPL1531 | GATGCGATTGCGGCTAATAC | Rev, 1,139 bp 5' downstream flanking sequence of LRФ2 omitting attB2R sequence |
| oVPL1532 | AAACGACGGCCAGTGAATTCGAGCTCGGTAATCGCAGCCTTAAGGAAATGGGAGACTTTT | Bridging oligo for LCR assembly to construct pVPL3593 |
| oVPL1533 | ACTGAATCCACTTTGCATGCCGGTACTTCTACCAGTACGTTCACGTAAGTAGTAGAGCTT | Bridging oligo for LCR assembly to construct pVPL3593 |
| oVPL1534 | CGTTTCAGCGGTATTAGCCGCAATCGCATCATCCTCTAGAGTCGACCTGCAGGCATGCAA | Bridging oligo for LCR assembly to construct pVPL3593 |
| oVPL1585 | CGAATCGACCCTGCTAAGTT | Fwd, oligo SCO and DCO screening of LRФ2 deletion |
| oVPL1586 | GCCTTAACTGGTGGGTTTGA | Rev, oligo SCO and DCO screening of LRФ2 deletion |
| oVPL1716 | TGCAAAGGTTCTTGATGCTG | Fwd, Emr gene from pNZ8048_Emr |
| oVPL1717 | CCGTTTATTATGCTCGCGTTA | Rev, Emr gene from pNZ8048_Emr |
| oVPL2414 | TTGATTATTACTGGAAAGATTAATTTTTAATGCAAAGGTTCTTGATGCTGAAACGGGGGA | Bridging oligo for LCR assembly to construct pVPL3886 |
| oVPL2415 | TATTGTCGATAACGCGAGCATAATAAACGGTTAAAATGTAGGTTTAATTTTTAGGGCTTT | Bridging oligo for LCR assembly to construct pVPL3886 |
| oVPL2529 | ATTCATATAACCAAATTAAAGAGGGTTATAATGAACTTTAATAAAATTGATTTAGACAAT | Bridging oligo for LCR assembly to construct pVPL3886 |
| oVPL2530 | TCAGATAGGCCTAATGACTGGCTTTTATAATAAGTTAAGGGATGCATAAACTGCATCCCT | Bridging oligo for LCR assembly to construct pVPL3886 |
| oVPL3148 | CAACGCCGACCAAACTTATT | Fwd, screening oligo paired with oVPL1439 used for screening L. reuteri 6475 and DSM 20016T LRФ1 excision |
| oVPL3150 | TGCTTCTGATATTGCCAACG | Fwd, screening oligo paired with oVPL1586 used for screening L. reuteri 6475 and DSM 20016T LRФ2 excision |
| oVPL3597 | GCTGCAGCTCACTTTGGATT | Fwd, screening oligo paired with oVPL3599 used for screening L. reuteri R2lc Ф1 excision |
| oVPL3599 | TGCTGACCATTGGGATAGTG | Rev, screening oligo paired with oVPL3597 used for screening L. reuteri R2lc Ф1 excision |
| oVPL3600 | TCATCGGTGCAGTACTTTGC | Fwd, screening oligo paired with oVPL3601 used for screening L. reuteri JCM 1081 Ф1 excision |
| oVPL3601 | ATTCCGTGCCGGTGATACTA | Rev, screening oligo paired with oVPL3600 used for screening L. reuteri JCM 1081 Ф1 excision |
| oVPL3605 | GTAAGCGACTAGGCCATCCA | Fwd, screening oligo paired with oVPL3601 used for screening L. reuteri JCM 1081 Ф2 excision |
| oVPL3607 | AACACGCCGAAAGTAGTTGG | Rev, screening oligo paired with oVPL3600 used for screening L. reuteri JCM 1081 Ф2 excision |
| oVPL3608 | GGTGAACCAACGCTCTCAAT | Fwd, screening oligo paired with oVPL3601 used for screening L. reuteri JCM 1081 Ф3 excision |
| oVPL3609 | GTTGGATCATCTCAGCGTCA | Rev, screening oligo paired with oVPL3600 used for screening L. reuteri JCM 1081 Ф3 excision |
| oVPL3626 | GGTACGGAGGATGTCGTTGT | Fwd, screening oligo paired with oVPL3601 used for screening L. reuteri 2010 Ф1 excision |
| oVPL3627 | CTGTTCCGCATATCCAATCA | Rev, screening oligo paired with oVPL3600 used for screening L. reuteri 2010 Ф1 excision |
| oVPL3628 | CAGGGCTTGGATATTGGAGA | Fwd, screening oligo paired with oVPL3601 used for screening L. reuteri Lr4020 Ф1 excision |
| oVPL3629 | TTAGCAAGCGGCTAGGTCAT | Rev, screening oligo paired with oVPL3600 used for screening L. reuteri Lr4020 Ф1 excision |
Sequence letters in lowercase are the sequence changes for the recombineering oligonucleotide that are identical to the lagging strand of DNA.
pVPLxxxx, van Pijkeren Lab oligonucleotide collection identification number; oligo, oligonucleotide; d/s, downstream; u/s, upstream; SCO, single crossover; DCO, double crossover; Fwd, forward; Rev, reverse; MAMA-PCR, mismatch amplification mutation assay PCR.
Bioinformatic analyses.
Prophages in L. reuteri genomes were identified using the PHAge Search Tool (PHASTER) (67). To accomplish this, L. reuteri genome sequences, retrieved from the databases Integrated Microbial Genomes (IMG) at Joint Genome Institute (JGI) and National Center for Biotechnology Information (NCBI), were uploaded to PHASTER followed by standard analysis. Prophages identified in L. reuteri ATCC PTA 6475 were manually annotated following a nonredundant search against the DNA database at NCBI. To compare both prophage genomes in L. reuteri ATCC PTA 6475, we used the Mauve alignment tool (at the default progressive alignment setting) (68).
Prophage induction.
Overnight cultures of L. reuteri were transferred to an optical density at 600 nm (OD600) of 0.1 in 40 ml prewarmed MRS. At an OD600 of 0.2 to 0.3, mitomycin C (0.5 μg/ml) was added, and we determined the OD600 every hour for up to 8 h. Subsequently, we harvested the bacterial supernatants containing presumptive phages by centrifugation (1 min at 5,000 rpm), followed by filtration (0.22-μm-pore polyvinylidene difluoride [PVDF] filter [Millipore]).
Identification of prophage attB sites in L. reuteri 6475.
Prophage excision in L. reuteri 6475 yielded PCR amplicons with oligonucleotide pairs oVPL1436-1438 and oVPL1437-1439, which flank prophage 1 (LRΦ1), and oVPL1440-1441, which flank prophage 2 (LRΦ2). Following Sanger sequencing (GeneWiz), we determined the attB sites based on the presence of palindrome repeats.
Determination of prophage excision following mitomycin C induction.
Presumptive prophage genomes in select L. reuteri genome sequences were identified with PHASTER. Oligonucleotides were designed that flanked the predicted attB sites. L. reuteri strains were subjected to mitomycin C (0.5 μg/ml) at an OD600 of 0.3. Two hours following induction, cells were subjected to PCR using oligonucleotide pairs oVPL3148-1439 and oVPL3150-1586 for L. reuteri 6475 or DSM 20016T, oVPL3597-3599 for L. reuteri R2lc, oVPL3600-3601, oVPL3605-3607, and oVPL3608-3609 for L. reuteri JCM 1081, oVPL3626-3627 for L. reuteri 2010, and oVPL3628-3629 for L. reuteri Lr4020 (Table 5).
Construction of LRΔΦ1, LRΔΦ2, and LRΔΦ1Φ2.
To generate markerless deletions in L. reuteri, we used the recently in-house-developed counterselection plasmid pVPL3002, which is broadly applicable in lactic acid bacteria (35). pVPL3002 is a derivative of pORI19 (69) and encodes the dipeptide ligase enzyme d-Ala-d-Ala (Ddl), which modifies the peptidoglycan cell wall, which consequently increases the binding affinity of vancomycin in bacteria that are intrinsically resistant to vancomycin. Thus, the presence of Ddl (e.g., following single-crossover recombination) reduces the MIC to vancomycin of bacteria that are intrinsically resistant to vancomycin. Cells that have undergone a second homologous recombination event lose the plasmid harboring the ddl gene, regain their vancomycin resistance, and have either the wild-type or recombinant genotype. First, we delete each prophage, including the attB sites. Briefly, by LCR we cloned the upstream and downstream flank of each prophage in pVPL3002 to yield pVPL3590 and pVPL3593 to delete LRΦ1 and LRΦ2, respectively (see Table 5 for oligonucleotides). Five micrograms of each plasmid was electroporated in L. reuteri 6475 as described previously (35), followed by recovery in MRS and plating on MRS agar containing 5 μg/ml erythromycin. Erythromycin-resistant (Emr) colonies were screened by colony PCR to confirm single-crossover homologous recombination using oligonucleotides oVPL49-1436-1439/97-1436-1439 and oVPL49-1585-1586/97-1585-1586 (upstream/downstream) for pVPL3590 and pVPL3593, respectively. Upon confirmation of single-crossover homologous recombination, a single colony was cultured in MRS for 20 generations and plated on MRS agar containing 500 μg/ml vancomycin, which yields colonies only after a second homologous recombination event. Deletion of LRΔΦ1 ΔattB and LRΔΦ2 ΔattB was confirmed with oligonucleotides oVPL1436-oVPL1439 and oVPL1585-1586, respectively, and the strains were named VPL4079 and VPL4101, respectively. To generate a double-prophage-deletion variant (LRΔΦ1 ΔattB ΔΦ2 ΔattB), we used LRΔΦ1 ΔattB as our genetic background and deleted LRΦ2 and its attB site in a manner identical to that described above to yield strain VPL4090. Each deletion variant was subjected to colony purification (3 times), and by PCR, we verified that each strain was cured from the corresponding phage. Next, we restored the attB sites of each phage to yield a genotype that is identical to when the phage would naturally excise from the genome. Cell pellets derived from mitomycin C-induced wild-type cells were used as a template for PCR to amplify the attBΦ1 recovery cassette (oVPL1516-1519) and attBΦ2 recovery cassettes (oVPL1528-1531). Each recovery cassette was cloned into the pVPL3002 by LCR with bridging oligonucleotides oVPL1520 and oVPL1522 (attBΦ1) and oVPL1532 and oVPL1534 (attBΦ2) to yield pVPL3746 and pVPL3749. In a manner identical to that described above, we restored by two-step homologous recombination the attB sites in VPL4079, VPL4101, and VPL4090. To identify single-crossover and double-crossover recombination, we used the same oligonucleotides as for the construction of VPL4079 and VPL4101. The resultant double-crossover recombinants were named VPL4119 (LRΔΦ1), VPL4120 (LRΔΦ2), and VPL4121 (LRΔΦ1 ΔΦ2). A schematic of all strain constructions is displayed in Fig. S1 in the supplemental material.
Plaque assay for L. reuteri 6475 phages.
Unlike the LRΔΦ1 ΔΦ2 strain, the LRΔΦ1 ΔattB ΔΦ2 ΔattB strain forms plaques when exposed to LRΦ1 or LRΦ2. Therefore, we used this strain as a sensitive host to determine the PFU/ml using an MRS double-layer agar method. For plaque assays, we followed a method established in a previous study (15).
Restoring the prophages in LRΔΦ1 ΔΦ2.
Mitomycin C-induced supernatants derived from LRΔΦ2 or LRΔΦ1 contain phages LRФ1 or LRФ2, respectively. LRΔФ1 ΔФ2 (VPL4121) was harvested at an OD600 of 4.0, washed once with phage diluent, and resuspended to approximately 2 × 109 CFU/ml. LRΔФ1 ΔФ2 cells were mixed with LRΦ1 supernatant at a 1:1,000 CFU/PFU ratio and incubated for 1 h at 37°C. After infection, cells were harvested by centrifugation (5,000 rpm, 1 min) and resuspended in 1 volume of MRS followed by 3 h of incubation at 37°C. Cells were serially diluted and plated on an MRS agar plate. To assess for the presence of prophage LRΦ1 in LRΔФ1 ΔФ2 following infection, we performed colony PCR (oVPL1377-1378) to amplify part of the LRΦ1 recT gene. Next, double-purified colonies that tested positive for the presence of LRΦ1 were subjected to colony PCR (oVPL1436-1439 and oVPL1377-1378) to confirm integration of LRΦ1 in the attB site. Mitomycin C induction was performed to assess prophage induction. The strain in which we recovered LRΦ1 in LRΔФ1 ΔФ2 was named VPL4150. Next, we infected VPL4150 with LRΦ2 in a manner identical to that described above. We used oligonucleotide pair oVPL1379-1380 to target the internal region of LRΦ2 recT2. Insertion of LRΦ2 in the corresponding attB site was confirmed by PCR using oligonucleotides oVPL1440-1441 and oVPL1379-1380. The derivative of LRΔФ1 ΔФ2 in which we restored both LRΦ1 and LRΦ2 was named VPL4159, and is also referred to as the complemented (COMP) strain (Fig. S1).
Whole-genome sequencing of L. reuteri strains.
Bacterial genome sequencing was performed by the University of Wisconsin—Madison Biotechnology Center. Genomic DNAs were prepared with the Wizard genomic DNA purification kit (Promega) and quantified by Qubit (Life Technologies). DNA libraries were run on the Illumina MiSeq system with 2 × 250-bp reads at the University of Wisconsin—Madison Biotechnology Center. Samples were prepared per the TruSeq Nano DNA LT Library Prep kit (Illumina Inc.) with minor modifications. Samples were sheared using a Covaris M220 ultrasonicator (Covaris, Inc.) and were size selected for an average insert size of 550 bp using SPRI bead-based size exclusion. The quality and quantity of the finished libraries were analyzed using an Agilent DNA1000 chip and Qubit double-stranded DNA (dsDNA) HS assay kit. Libraries were standardized to 2 nM. Paired end, 250-bp sequencing was performed using the Illumina MiSeq Sequencer and a MiSeq 500 bp (v2) sequencing cartridge. Images were analyzed using the standard Illumina Pipeline, version 1.8.2. To identify single nucleotide polymorphisms (SNPs) and indels, comparative genome analysis was performed in SegMan Pro (DNASTAR). All SNPs identified were subjected to Sanger sequencing analysis, and the oligonucleotides for this analysis are listed in Table 5.
Construction of rifampin-resistant derivatives.
To render rifampin-resistant derivatives of the L. reuteri wild-type strain, the LRΔΦ1, LRΔΦ2, and LRΔΦ1 ΔΦ2 mutant strains, and the complemented strain, we performed single-stranded DNA recombineering (SSDR). Bacteria were transformed with 100 μg oVPL236 to modify rpoB. SSDR was performed as described before (34), with the exception that we did not make use of heterologous expression of RecT. Recombinant cells were screened on MRS containing 25 μg/ml rifampin. The genotype of rifampin-resistant colonies was confirmed by mismatch mutation assay PCR (MAMA-PCR) (oVPL304-305-306).
Cmr gene insertion in L. reuteri sensitive host and ΔФ1.
To insert the gene encoding chloramphenicol resistance (Cmr), we first amplified the flanking sequence of a noncoding region from the L. reuteri wild type with oVPL265-266. By standard blunt-end ligation, the amplicon was fused to the pVPL3002 backbone, which was generated with oVPL187-188 to yield pVPL3048. Subsequently, the pVPL3048 backbone was amplified with oVPL271-272 and the Phelp::Cmr cassette amplified from pJP028 (unpublished) with oVPL279-280 followed by Gibson assembly (70) to yield pVPL3810. After transforming the L. reuteri sensitive host and LRΔФ1 with 5 μg pVPL3810, we confirmed single-crossover recombinants with oligonucleotide combinations oVPL203-334-335 (upstream single crossover) and oVPL202-334-335 (downstream single crossover). Double-crossover recombinants were confirmed by PCR (oVPL334-335) and confirmation of erythromycin-sensitive and chloramphenicol-resistant phenotypes. The Cmr mutants of the sensitive host and LRΔФ1 were named VPL4178 (sensitive host Cmr) and VPL4167 (LRΔФ1 Cmr), respectively (Fig. S1).
Emr gene insertion in LRΔФ2.
To insert the gene encoding erythromycin resistance (Emr), we amplified two fragments of pVPL3048 plasmid backbone with oVPL271-1391 and oVPL272-1390 to omit the Emr gene and Cmr gene expression cassette. The Cmr gene was amplified from pNZ8048 with oVPL202-203, and the Emr gene expression cassette was amplified from pVPL2042 with oVPL1716-1717. pVPL3048 backbones, the Cmr gene, and the Emr gene expression cassette were assembled by LCR with bridging oligonucleotides (oVPL2414, -2415, -2529, and -2530) to yield pVPL3886. After transformation of LRΔФ2 with 5 μg pVPL3886, we confirmed single crossover with oligonucleotide combinations oVPL49-334-335 (upstream single crossover) and oVPL97-334-335 (downstream single crossover). Double-crossover recombinants were confirmed by PCR (oVPL334-335) and confirmation of Cms and Emr phenotypes. The Emr mutant of LRΔФ2 was named VPL4181 (LRΔФ2 Emr) (Fig. S1).
Transmission electron microscopy.
Filtered cell-free phage suspensions (0.22-μm-pore PVDF filter) were resuspended in phage diluent (about 105 PFU/ml). Five microliters of phage suspension was adsorbed onto a carbon-coated 200 mesh copper TEM grid (3.05 mm diameter [Gilder]). The phage particles were negatively stained with 2% (vol/vol) uranyl acetate staining solution and examined with a Tecnai T-12 electron microscope at an acceleration voltage of 120 kV and a magnification range of 20,000 to 200,000×.
Phage production and survival of L. reuteri strains during murine GI transit.
Twenty 6-week-old male B6 mice (C57BL/6J) were purchased from Jackson Laboratory (Bar Harbor, ME). After arrival, animals were adjusted for 1 week to the new environment prior to the start of the experiment. Animals were housed at an environmentally controlled facility with a 12-h light and dark cycle. Food (standard chow [LabDiet, St. Louis, MO]) and water were provided ad libitum. Five groups (n = 4 per group) were gavaged for two consecutive days with a 100-μl phosphate-buffered saline (PBS) suspension containing 109 CFU of rifampin-resistant derivatives of (i) the L. reuteri wild-type, (ii) the LRΔФ1 mutant, (iii) the LRΔФ2 mutant, (iv) the LRΔФ1 ΔФ2 mutant, and (v) the COMP strain. Fecal samples were collected from the bedding 16 h after the last oral administration and collected every following day after up to 6 days. Fecal content was resuspended in PBS to 100 mg/ml and plated on MRS agar plates containing 25 μg/ml rifampin. The fecal suspension was also used to determine the PFU/ml by a plaque assay as described above.
In vivo colonization and competition in germfree mice.
Thirty-six germfree Swiss-Webster mice (12 to 16 weeks old) were maintained in sterile biocontainment cages in the gnotobiotic animal facility at the University of Alberta. Six treatment groups (n = 6 per group) were colonized following a single oral gavage of 100 μl L. reuteri cocktail in PBS (1:1 ratio, ∼109 CFU). Each group was gavaged with a mixture of (i) WT Rifr strain and sensitive host with Cmr, (ii) LRΔΦ1 Rifr mutant and sensitive host with Cmr, (iii) LRΔΦ2 Rifr and sensitive host with Cmr, (iv) LRΔΦ1 ΔΦ2 Rifr mutant and sensitive host with Cmr, (v) complemented strain with Rifr and sensitive host with Cmr, and (vi) LRΔΦ1 mutant with Cmr and LRΔΦ2 mutant with Emr. Six days following colonization, fecal samples were collected from individual mice to determine the fecal CFU and PFU. At day 7, mice were sacrificed by CO2 asphyxiation and cecal contents and jejunal digesta were collected accordingly to determine the CFU and PFU per 100 mg content. To enumerate L. reuteri phages from intestinal contents, we performed the fecal plaque assay as described above. Mouse experiments were performed with approval of the Institutional Animal Care and Use Committee of the University of Alberta (Project ID 2099).
Statistics.
Data representation was performed using DataGraph 4.3 (Visual Data Tools, Inc., Chapel Hill, NC). Statistical comparisons were performed using paired t test, one-way analysis of variance (ANOVA), and Tukey’s honestly significant difference (HSD) test (JMP Pro, version 11.0.0) for the multiple comparison between treatment groups in the animal experiment. Three biological replicates were performed for all in vitro studies. All samples were included in the analyses, and experiments were performed without blinding.
Supplementary Material
ACKNOWLEDGMENTS
We thank the College of Agricultural Life Sciences Statistical Consulting Lab for assistance with the statistical analyses. We thank Stefan Roos (BioGaia, Stockholm, Sweden) for providing Lactobacillus reuteri ATCC PTA 6475, the University of Wisconsin—Madison Biotechnology Center for microbial genome sequencing support, and Alan Attie for sharing the animal facility.
J.-P.V.P. thanks the University of Wisconsin—Madison Food Research Institute (233-PRJ75PW) and BioGaia (Stockholm, Sweden; 233-AAB1492) for financial support. M.Ö. is the recipient of the Robert H. and Carol L. Deibel Distinguished Graduate Fellowship in Probiotic Research and C.D. was sponsored by the Summer Undergraduate Research Program, both awarded by the Food Research Institute (University of Wisconsin—Madison). S.Z. was in part supported by the by the United States Department of Agriculture, National Institute of Food and Agriculture (USDA NIFA) Hatch Award MSN185615.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Breitbart M, Haynes M, Kelley S, Angly F, Edwards RA, Felts B, Mahaffy JM, Mueller J, Nulton J, Rayhawk S, Rodriguez-Brito B, Salamon P, Rohwer F. 2008. Viral diversity and dynamics in an infant gut. Res Microbiol 159:367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 2.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. 2003. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 185:6220–6223. doi: 10.1128/jb.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckling A, Rainey PB. 2002. Antagonistic coevolution between a bacterium and a bacteriophage. Proc Biol Sci 269:931–936. doi: 10.1098/rspb.2001.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weitz JS, Hartman H, Levin SA. 2005. Coevolutionary arms races between bacteria and bacteriophage. Proc Natl Acad Sci U S A 102:9535–9540. doi: 10.1073/pnas.0504062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohannan BJM, Lenski RE. 2000. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett 3:362–377. doi: 10.1046/j.1461-0248.2000.00161.x. [DOI] [Google Scholar]
- 7.Carding SR, Davis N, Hoyles L. 2017. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther 46:800–815. doi: 10.1111/apt.14280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shkoporov AN, Hill C. 2019. Bacteriophages of the human gut: the “known unknown” of the microbiome. Cell Host Microbe 25:195–209. doi: 10.1016/j.chom.2019.01.017. [DOI] [PubMed] [Google Scholar]
- 9.Canchaya C, Proux C, Fournous G, Bruttin A, Brüssow H. 2003. Prophage genomics. Microbiol Mol Biol Rev 67:238–276. doi: 10.1128/mmbr.67.2.238-276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glick BR. 1995. Metabolic load and heterologous gene expression. Biotechnol Adv 13:247–261. doi: 10.1016/0734-9750(95)00004-A. [DOI] [PubMed] [Google Scholar]
- 11.Ramisetty BCM, Sudhakari PA. 2019. Bacterial ‘grounded’ prophages: hotspots for genetic renovation and innovation. Front Genet 10:65–65. doi: 10.3389/fgene.2019.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozkov A, Avignone-Rossa CA, Ertl PF, Jones P, O'Kennedy RD, Smith JJ, Dale JW, Bushell ME. 2004. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol Bioeng 88:909–915. doi: 10.1002/bit.20327. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira J, Mahony J, Hanemaaijer L, Kouwen T, Neve H, MacSharry J, van Sinderen D. 2017. Detecting Lactococcus lactis prophages by mitomycin C-mediated induction coupled to flow cytometry analysis. Front Microbiol 8:1343. doi: 10.3389/fmicb.2017.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Paepe M, Tournier L, Moncaut E, Son O, Langella P, Petit M-A. 2016. Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet 12:e1005861. doi: 10.1371/journal.pgen.1005861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh JH, Alexander LM, Pan M, Schueler KL, Keller MP, Attie AD, Walter J, van Pijkeren JP. 2019. Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe 25:273–284. doi: 10.1016/j.chom.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Breitbart M, Miyake JH, Rohwer F. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol Lett 236:249–256. doi: 10.1016/j.femsle.2004.05.042. [DOI] [PubMed] [Google Scholar]
- 17.Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. 2013. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci U S A 110:20236–20241. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howe A, Ringus DL, Williams RJ, Choo ZN, Greenwald SM, Owens SM, Coleman ML, Meyer F, Chang EB. 2016. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J 10:1217–1227. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennon JT, Khatana SA, Marston MF, Martiny JB. 2007. Is there a cost of virus resistance in marine cyanobacteria? ISME J 1:300–312. doi: 10.1038/ismej.2007.37. [DOI] [PubMed] [Google Scholar]
- 20.Lopez-Pascua L, Buckling A. 2008. Increasing productivity accelerates host-parasite coevolution. J Evol Biol 21:853–860. doi: 10.1111/j.1420-9101.2008.01501.x. [DOI] [PubMed] [Google Scholar]
- 21.McGrath S, Fitzgerald GF, Sinderen D. 2002. Identification and characterization of phage‐resistance genes in temperate lactococcal bacteriophages. Mol Microbiol 43:509–520. doi: 10.1046/j.1365-2958.2002.02763.x. [DOI] [PubMed] [Google Scholar]
- 22.Bondy-Denomy J, Davidson AR. 2014. When a virus is not a parasite: the beneficial effects of prophages on bacterial fitness. J Microbiol 52:235–242. doi: 10.1007/s12275-014-4083-3. [DOI] [PubMed] [Google Scholar]
- 23.Baugher JL, Durmaz E, Klaenhammer TR. 2014. Spontaneously induced prophages in Lactobacillus gasseri contribute to horizontal gene transfer. Appl Environ Microbiol 80:3508–3517. doi: 10.1128/AEM.04092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Denou E, Pridmore RD, Ventura M, Pittet AC, Zwahlen MC, Berger B, Barretto C, Panoff JM, Brussow H. 2008. The role of prophage for genome diversification within a clonal lineage of Lactobacillus johnsonii: characterization of the defective prophage LJ771. J Bacteriol 190:5806–5813. doi: 10.1128/JB.01802-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bossi L, Fuentes JA, Mora G, Figueroa-Bossi N. 2003. Prophage contribution to bacterial population dynamics. J Bacteriol 185:6467–6471. doi: 10.1128/jb.185.21.6467-6471.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams H. 2013. Phage-induced diversification improves host evolvability. BMC Evol Biol 13:17. doi: 10.1186/1471-2148-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duerkop BA, Clements CV, Rollins D, Rodrigues JLM, Hooper LV. 2012. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci U S A 109:17621–17626. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41(Suppl 1):S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 29.Walter J, Britton RA, Roos S. 2011. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc Natl Acad Sci U S A 108(Suppl 1):4645–4652. doi: 10.1073/pnas.1000099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, Peterson DA, Blom J, Wenzel CQ, Szymanski CM, Walter J. 2017. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol 83:e00132-17. doi: 10.1128/AEM.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frese SA, Benson AK, Tannock GW, Loach DM, Kim J, Zhang M, Oh PL, Heng NCK, Patil PB, Juge N, Mackenzie DA, Pearson BM, Lapidus A, Dalin E, Tice H, Goltsman E, Land M, Hauser L, Ivanova N, Kyrpides NC, Walter J. 2011. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet 7:e1001314. doi: 10.1371/journal.pgen.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh PL, Benson AK, Peterson DA, Patil PB, Moriyama EN, Roos S, Walter J. 2010. Diversification of the gut symbiont Lactobacillus reuteri as a result of host-driven evolution. ISME J 4:377–387. doi: 10.1038/ismej.2009.123. [DOI] [PubMed] [Google Scholar]
- 33.van Pijkeren JP, Britton RA. 2012. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res 40:e76. doi: 10.1093/nar/gks147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh JH, van Pijkeren JP. 2014. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res 42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Oh JH, Alexander LM, Ozcam M, van Pijkeren JP. 2018. d-Alanyl-d-alanine ligase as a broad-host-range counterselection marker in vancomycin-resistant lactic acid bacteria. J Bacteriol 200:e00607-17. doi: 10.1128/JB.00607-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spinler JK, Sontakke A, Hollister EB, Venable SF, Oh PL, Balderas MA, Saulnier DM, Mistretta TA, Devaraj S, Walter J, Versalovic J, Highlander SK. 2014. From prediction to function using evolutionary genomics: human-specific ecotypes of Lactobacillus reuteri have diverse probiotic functions. Genome Biol Evol 6:1772–1789. doi: 10.1093/gbe/evu137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahony J, Alqarni M, Stockdale S, Spinelli S, Feyereisen M, Cambillau C, Sinderen DV. 2016. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci Rep 6:36667. doi: 10.1038/srep36667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Betton GR, Dormer CS, Wells T, Pert P, Price CA, Buckley P. 1988. Gastric ECL-cell hyperplasia and carcinoids in rodents following chronic administration of H2-antagonists SK&F 93479 and oxmetidine and omeprazole. Toxicol Pathol 16:288–298. doi: 10.1177/019262338801600222. [DOI] [PubMed] [Google Scholar]
- 39.Krumbeck JA, Marsteller NL, Frese SA, Peterson DA, Ramer-Tait AE, Hutkins RW, Walter J. 2016. Characterization of the ecological role of genes mediating acid resistance in Lactobacillus reuteri during colonization of the gastrointestinal tract. Environ Microbiol 18:2172–2184. doi: 10.1111/1462-2920.13108. [DOI] [PubMed] [Google Scholar]
- 40.Butala M, Zgur-Bertok D, Busby SJ. 2009. The bacterial LexA transcriptional repressor. Cell Mol Life Sci 66:82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leenhouts K, Bolhuis A, Venema G, Kok J. 1998. Construction of a food-grade multiple-copy integration system for Lactococcus lactis. Appl Microbiol Biotechnol 49:417–423. doi: 10.1007/s002530051192. [DOI] [PubMed] [Google Scholar]
- 42.Koonin EV. 2016. Viruses and mobile elements as drivers of evolutionary transitions. Philos Trans R Soc Lond B Biol Sci 371:20150442. doi: 10.1098/rstb.2015.0442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. 2012. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M-S, Bae J-W. 2018. Lysogeny is prevalent and widely distributed in the murine gut microbiota. ISME J 12:1127–1141. doi: 10.1038/s41396-018-0061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson TA, Looft T, Severin AJ, Bayles DO, Nasko DJ, Wommack KE, Howe A, Allen HK. 2017. The in-feed antibiotic carbadox induces phage gene transcription in the swine gut microbiome. mBio 8:e00709-17. doi: 10.1128/mBio.00709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen HK, Looft T, Bayles DO, Humphrey S, Levine UY, Alt D, Stanton TB. 2011. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2:e00260-11. doi: 10.1128/mBio.00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selva L, Viana D, Regev-Yochay G, Trzcinski K, Corpa JM, Lasa Í, Novick RP, Penadés JR. 2009. Killing niche competitors by remote-control bacteriophage induction. Proc Natl Acad Sci U S A 106:1234–1238. doi: 10.1073/pnas.0809600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi J, Kotay SM, Goel R. 2010. Various physico-chemical stress factors cause prophage induction in Nitrosospira multiformis 25196—an ammonia oxidizing bacteria. Water Res 44:4550–4558. doi: 10.1016/j.watres.2010.04.040. [DOI] [PubMed] [Google Scholar]
- 51.de Matos AA, Lehours P, Timóteo A, Roxo-Rosa M, Vale F. 2013. Comparison of induction of B45 Helicobacter pylori prophage by acid and UV radiation. Microsc Microanal 19:27–28. doi: 10.1017/S1431927613000755. [DOI] [Google Scholar]
- 52.Lunde M, Aastveit AH, Blatny JM, Nes IF. 2005. Effects of diverse environmental conditions on ΦLC3 prophage stability in Lactococcus lactis. Appl Environ Microbiol 71:721–727. doi: 10.1128/AEM.71.2.721-727.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 54.Bobay L-M, Touchon M, Rocha E. 2014. Pervasive domestication of defective prophages by bacteria. Proc Natl Acad Sci U S A 111:12127–12132. doi: 10.1073/pnas.1405336111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Kim Y, Ma Q, Hong SH, Pokusaeva K, Sturino JM, Wood TK. 2010. Cryptic prophages help bacteria cope with adverse environments. Nat Commun 1:147. doi: 10.1038/ncomms1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. 2017. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J 1:97. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harrison E, Brockhurst MA. 2017. Ecological and evolutionary benefits of temperate phage: what does or doesn’t kill you makes you stronger. Bioessays 39. doi: 10.1002/bies.201700112. [DOI] [PubMed] [Google Scholar]
- 58.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. 2015. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol 13:641–650. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 59.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL. 2016. Prophages mediate defense against phage infection through diverse mechanisms. ISME J 10:2854–2866. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haaber J, Leisner JJ, Cohn MT, Catalan-Moreno A, Nielsen JB, Westh H, Penadés JR, Ingmer H. 2016. Bacterial viruses enable their host to acquire antibiotic resistance genes from neighbouring cells. Nat Commun 7:13333–13333. doi: 10.1038/ncomms13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Refardt D. 2011. Within-host competition determines reproductive success of temperate bacteriophages. ISME J 5:1451–1460. doi: 10.1038/ismej.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Levin B. 1988. Frequency-dependent selection in bacterial populations. Philos Trans R Soc Lond B Biol Sci 319:459–472. doi: 10.1098/rstb.1988.0059. [DOI] [PubMed] [Google Scholar]
- 63.Alexander LM, Oh JH, Stapleton DS, Schueler KL, Keller MP, Attie AD, van Pijkeren JP. 2019. Exploiting prophage-mediated lysis for biotherapeutic release by Lactobacillus reuteri. Appl Environ Microbiol 85:e02335-18. doi: 10.1128/AEM.02335-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hendrikx T, Duan Y, Wang Y, Oh JH, Alexander LM, Huang W, Starkel P, Ho SB, Gao B, Fiehn O, Emond P, Sokol H, van Pijkeren JP, Schnabl B. 2018. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut 68:1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Ruyter PG, Kuipers OP, de Vos WM. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, Newman JD, Chandran SS. 2014. Rapid and reliable DNA assembly via ligase cycling reaction. ACS Synth Biol 3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- 67.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J Bacteriol 177:7011–7018. doi: 10.1128/jb.177.24.7011-7018.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.