There are today significant knowledge gaps on phytovirus populations and on the drivers impacting them but also on the comparative performance-methodological approaches for their study. We used and compared two viral sequence enrichment approaches, double-stranded RNAs (dsRNA) and virion-associated nucleic acids (VANA), for phytovirome description in complex pools representative of the most prevalent plant species in unmanaged and cultivated ecosystems. Viral richness was assessed by determining operational taxonomic units (OTU) following the clustering of conserved viral domains. There is some limited evidence of an impact of cultivation on viral populations. These results provide data allowing us to reason the methodological choices in virome studies. For researchers primarily interested in RNA viruses, the dsRNA approach is recommended because it consistently provided a more comprehensive description of the analyzed phytoviromes, but it understandably underrepresented DNA viruses and bacteriophages.
KEYWORDS: metagenomics, double-stranded RNA (dsRNA), virion-associated nucleic acids (VANA), virome, OTU, viral diversity, phytovirome
ABSTRACT
Metagenomic studies have indicated that the diversity of plant viruses was until recently far underestimated. As important components of ecosystems, there is a need to explore the diversity and richness of the viruses associated with plant populations and to understand the drivers shaping their diversity in space and time. Two viral sequence enrichment approaches, double-stranded RNA (dsRNA) and virion-associated nucleic acids (VANA), have been used and compared here for the description of the virome of complex plant pools representative of the most prevalent plant species in unmanaged and cultivated ecosystems. A novel bioinformatics strategy was used to assess viral richness not only at the family level but also by determining operational taxonomic units (OTU) following the clustering of conserved viral domains. A large viral diversity dominated by novel dsRNA viruses was detected in all sites, while a large between-site variability limited the ability to draw a clear conclusion on the impact of cultivation. A trend for a higher diversity of dsRNA viruses was nevertheless detected in unmanaged sites (118 versus 77 unique OTUs). The dsRNA-based approach consistently revealed a broader and more comprehensive diversity for RNA viruses than the VANA approach, whatever the assessment criterion. In addition, dissimilarity analyses indicated both approaches to be largely reproducible but not necessarily convergent. These findings illustrate features of phytoviromes in various ecosystems and a novel strategy for precise virus richness estimation. These results allow us to reason methodological choices in phytovirome studies and likely in other virome studies where RNA viruses are the focal taxa.
IMPORTANCE There are today significant knowledge gaps on phytovirus populations and on the drivers impacting them but also on the comparative performance-methodological approaches for their study. We used and compared two viral sequence enrichment approaches, double-stranded RNAs (dsRNA) and virion-associated nucleic acids (VANA), for phytovirome description in complex pools representative of the most prevalent plant species in unmanaged and cultivated ecosystems. Viral richness was assessed by determining operational taxonomic units (OTU) following the clustering of conserved viral domains. There is some limited evidence of an impact of cultivation on viral populations. These results provide data allowing us to reason the methodological choices in virome studies. For researchers primarily interested in RNA viruses, the dsRNA approach is recommended because it consistently provided a more comprehensive description of the analyzed phytoviromes, but it understandably underrepresented DNA viruses and bacteriophages.
INTRODUCTION
Until recently, plant virology was largely focused on important crops and on destructive viruses impacting agricultural production, limiting our understanding of plant virus diversity (1). In particular, viruses infecting wild plants have been largely neglected, even if they represent reservoirs both for known viruses and for novel, emerging agents. The few metagenomic studies to date have shown that viruses are common in wild plants, even in the absence of symptoms, with a prevalence as high as 60% and a majority of novel agents (2–5). These studies also showed that in wild plants a majority of the detected agents are persistent viruses which are mostly asymptomatic and transmitted vertically through host cell division and sexual reproduction (6, 7).
Building on these pioneering efforts, metagenomics and virus ecology are now trying to answer some fundamental questions centered on the identity and diversity of plant-associated viruses, the evolutionary drivers influencing the assembly in space and time of these viral communities, and their contribution to the functioning of plant communities (8–10).
High-throughput sequencing (HTS) coupled with bioinformatic analyses is at the core of metagenomics but has also largely superseded all previously used approaches in virus discovery and etiology because of the ability to efficiently detect known and novel viruses without any a priori information (11–16). Moreover, metagenomics studies (17–19) have greatly contributed to a redefinition of the RNA virosphere of invertebrates and to a reshaping of our understanding of the origin and evolution of RNA viruses. HTS has been successfully used for a large range of plants (20–23), demonstrating its wide applicability. These efforts also show that a variety of nucleic acid populations can be used, with consequences for the range of identified viruses (23). So far, the main approaches have targeted double-stranded RNA (dsRNA) (7, 22), virus-derived small interfering RNA (siRNA) (24, 25), virion-associated nucleic acids (VANA) (26–28), total RNA with or without rRNA depletion (29, 30), and polyadenylated RNA (31). Their respective advantages and disadvantages for virus discovery and etiology efforts involving single plant samples or samples of low complexity have been discussed in detail (21, 23, 32).
Compared to these efforts, the study of viromes associated with plant populations requires the analysis of a large number of plants. Two strategies have essentially been used, the “ecogenomics” (23) or “geometagenomics” approach (33), which involves the analysis of single plants or of low-complexity pools in a highly multiplexed format, and the “metagenomics” or “lawnmower” approach, which relies on the direct analysis of complex plant pools (30). While the first strategy retains information on the host(s) of each agent, the second allows a more direct virome characterization for multiple environmental points. However, with the currently used multiplexing strategies, a low-level index-hopping may ultimately have a negative impact on data quality (34, 35). Given budgetary constraints, both approaches have so far relied almost exclusively on the two strategies, providing an enrichment of viral sequences, dsRNA, and VANA (7, 33, 36).
Unfortunately, there is little information on the comparative performance of these two approaches for virome description. Some elements can be gleaned, however, from virus discovery efforts. Candresse et al. (37) used both siRNA and VANA and showed that higher genome coverage and longer contigs were obtained using VANA for a DNA mastrevirus. Another study compared siRNA and VANA to test if the same representation of within-host viral population structure could be obtained (38). Both approaches provided similar viral mutational landscapes, but VANA performed better in complete viral genome reconstruction and allowed recombinants to be more readily detected (38).
In 2016, a comparison of siRNA and rRNA-depleted total RNA for Citrus tristeza virus (CTV) ([+]single-stranded RNA [ssRNA], Closteroviridae) and Citrus dwarfing viroid (Pospiviroidae) characterization on grapefruit indicated that rRNA-depleted total RNA is superior to siRNA in de novo assembly and genome coverage for CTV but not for the viroid (30). The same approaches have also been compared for the detection of various viruses and viroids in different plants (29). The main conclusion was that the performance of these two approaches was virus-dependent but that consistent with reference 34, longer contigs and higher genome coverage were generated using rRNA-depleted total RNA. Additionally, a Cytorhabdovirus was detected only from rRNA-depleted total RNA (29).
In the sole study to date that compared dsRNA and VANA for wide-scale metagenomics, Thapa et al. (36) used the two approaches to describe viral diversity in six native plant species from the Tallgrass Prairie Preserve in Oklahoma and test the effects of host identity, location, and sampling year on the virome composition. More operational viral taxonomic units (OTUs) were discovered using the dsRNA approach (29 compared to 7 for VANA). In addition, 86% of VANA-OTUs were also detected by dsRNA. The two approaches also showed different performances when the effects of sampling site on virome composition were analyzed (36). It should be pointed out that similar to that of Thapa et al., most studies to date have used a quite broad definition for OTUs, considering relatively wide taxonomic categories such as families or genera (33, 36).
Overall, while the available approaches have proven effective in a virus detection context in a range of plant-virus combinations, there is still limited information allowing researchers to make methodological choices in plant virus metagenomics. Here, we directly compared the performance of dsRNA and VANA for virome description using complex metagenomics plant pools from unmanaged and cultivated environments. The results uncovered rich viromes and suggest that the dsRNA approach should be preferred when analyzing such complex plant pools since it consistently provided a more comprehensive description of the analyzed phytoviromes, with the exception of the DNA viruses.
RESULTS
Summary of HTS data sets and sequencing depth normalization.
The phytoviromes of 6 different study sites were analyzed using pools of equal complexity composed of 200 plants assembled using 5 individual plants of each of the 40 most prevalent species. Following double-stranded RNA (dsRNA) or virion-associated nucleic acid (VANA) extractions, target nucleic acid populations were converted to cDNA and submitted to random whole-genome amplification (WGA) before Illumina sequencing. In order to evaluate the reproducibility of the WGA, all samples were amplified in duplicates involving different multiplex identifier (MID) tags. This situation is reflected in the names of the various libraries, which indicate the name of the target nucleic acids (ds or VANA) followed by the study site and PCR1 or PCR2 to indicate the WGA replicate. A total of 20 million reads (paired-end reads and singletons) were generated from the 12 dsRNA libraries, 16 million reads from the 12 VANA libraries, and 1 million reads from 1 negative-control blank reagent-only library (see Table S1 in the supplemental material). Following demultiplexing and quality trimming, each library provided 0.5 to 3 million reads, but in order to limit intersample cross-talk, only pair members for which the expected MID tag was identified in both pair members (122,295 to 997,993 pairs, depending on the library) were used for subsequent steps. To allow meaningful comparisons between approaches and sites, a normalization was performed by randomly subsampling all libraries to a depth of 122,259 pairs, corresponding to the library with the fewest reads, VANA-IT-PCR2 (Table S1). All further analyses were performed on these normalized data sets. The average read length for the dsRNA libraries is 120.9 ± 1.3 nucleotides (nt), which is not significantly different from the 121.2 ± 1.5 nt obtained for VANA libraries (P = 0.6075) (Table 1 and Table S1).
TABLE 1.
Comparisons of high-throughout sequencing (HTS) average output and virus richness per library based on normalized data sets obtained for dsRNA and VANA targets
| Approach | Length of reads (nt) | Percent reads in contigs | No. of contigs | N50 (bp) | Percent viral contigs | Percent reads in viral contigs | No. of viral families identified | Viral RdRp OTUsb |
|---|---|---|---|---|---|---|---|---|
| dsRNA | 120.9 ± 1.3 | 80.4 ± 4.3 | 614 ± 128 | 796 ± 110.0 | 33.3 ± 7.3 | 49.9 ± 14.3 | 13.3 ± 1.7 | 39.8 ± 13.4 |
| VANA | 121.2 ± 1.5 | 63.4 ± 12.4 | 565 ± 121 | 578 ± 114.9 | 20.7 ± 10.3 | 40.5 ± 16.6 | 9.3 ± 2.6 | 13.3 ± 6.2 |
| P valuea | 0.6075 | 0.0009 | 0.4393 | 4.185e-06 | 1.296e-06 | 0.2193 | 0.0001 | 1.273e-05 |
Significance value was calculated using a paired t test; bold text indicates a statistically significant difference at the 5% level.
Operational taxonomy units (OTUs) were defined by clustering, using a 10% distance cutoff for contigs encoding virus-specific conserved RNA-dependent RNA polymerase protein motifs.
Comparisons of contig assembly and annotation.
Overall, a significantly higher proportion of reads from dsRNA libraries was assembled into contigs (average, 80.4% ± 4.3%) than from the VANA libraries (average, 63.4% ± 12.4%) (36% of reads in the blank control) (Table 1 and Table S1). Statistically significant differences between the two approaches were observed in all parameters describing contig length (total length of contigs, mean, median, N50, and N90 [Table 1 and Table S1]), with dsRNA library assemblies providing, on average, longer contigs than VANA library assemblies. Taken together, these results would suggest a higher complexity, leading to a decreased assembly efficiency for the VANA libraries.
Contigs were then annotated using BLASTN and BLASTX analyses against the GenBank database and an E value cutoff of 10−4 (39). For those contigs annotated as viruses, taxonomic assignation was retained at the family level, if available, since in many cases, our own and others’ observations have shown that assignation at a lower taxonomic level (genus or species) is frequently unreliable (36, 40). Those viruses with no family information were either kept as unclassified viruses or, if genome-type information was available, annotated as single-stranded RNA or double-stranded RNA unassigned viruses. The proportions of plants, viruses, or unknown contigs proved highly variable between libraries. In addition, the VANA libraries obtained from two sampling sites showed the significant presence of contigs of bacterial origin (average of 47.3% and 50.3% of contigs for the VO and SP sites, respectively) (Table S1). On average, dsRNA libraries yielded 33.3% of viral contigs (standard deviation, 7.3%; extremes, 23.6% to 44.8%) compared to 20.7% ± 10.3% for VANA libraries (extremes, 8.2% to 34.5%) (Table 1 and Table S1), a statistically significant difference (P = 1.3e-06). When taking into consideration the reads integrated in the different contigs, 49.9% ± 14.3% of dsRNA reads were annotated as viral, compared to 40.5% ± 16.6% for VANA reads. However, this difference was not found to be statistically significant (P = 0.2193) (Table 1). Unsurprisingly, 94.3% of contigs in the blank control library were annotated as being of bacterial origin, and no viral contigs were identified (Table S1).
Family-level viral diversity as reflected by contig annotation.
The results of the BLAST annotation show that at the family level, the dsRNA-based approach consistently detected more viral families per study site (extremes, 11 to 16 families; average, 13.3 ± 1.7 families) than VANA (extremes, 6 to 15 families; average, 9.3 ± 2.6 families) (Table 1 and Table S1). Therefore, on average, dsRNAs allowed the identification of 3.9 ± 2.3 more viral families per study site than VANA. However, when considering all samples together, VANA allowed the identification of a total of 24 viral families, compared to 21 for dsRNA. This difference is largely due to the infrequent detection of DNA viral or phage families not detected by dsRNA (Metaviridae, Siphoviridae, Podoviridae, Genomoviridae, Geminiviridae, and Circoviridae). Conversely, dsRNA allowed the detection of RNA viral families not detected by VANA (Reoviridae, Cystoviridae, Rhabdoviridae, and Narnaviridae) (Table S2). Overall, phages represented only a very minor fraction of the detected viral contigs (16 contigs or 1.2% of viral contigs for VANA and 11 contigs or 0.5% of viral contigs for dsRNA).
While most DNA virus families were only detected by VANA from a few sites, many dsRNA or ssRNA families, such as the Amalgaviridae, Chrysoviridae, Closteroviridae, Benyviridae, Luteoviridae, and Secoviridae, were detected from significantly more study sites using dsRNA than VANA (Table S2). This situation is particularly striking for the Chrysoviridae (6 sites versus 1) and the Closteroviridae (6 sites versus 3). On the other hand, as judged by read number, the ssDNA Nanoviridae family was very poorly detected by the dsRNA approach compared with the VANA approach (Table S2).
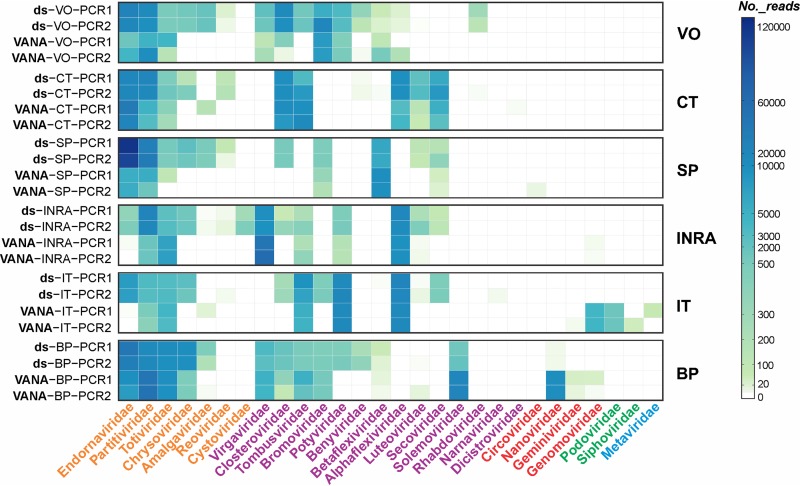
Representation of viral families as estimated by read number.
The numbers of reads for each viral family varied significantly between study sites and, for a given site, between the two virome sequencing approaches (Fig. 1). The most represented viral family in the dsRNA approach is the Endornaviridae, and overall, it accounts for nearly 4 times the reads observed with the VANA approach (410,884 versus 107,904). Intuitively, Endornaviridae reads may have saturated the dsRNA libraries of the SP site, reaching 66% to 70% of the viral reads (Table S2). For other dsRNA viral families, the same general trend of a higher representation in dsRNA libraries is also observed. This is particularly clear for the Chrysoviridae but generally applies to all dsRNA viruses. There are, however, some exceptions, for example, for Partitiviridae at the BP study site or for Totiviridae at the INRA study site (Table S2). Conversely, the expected better representation of DNA viral families in the VANA approach is observed, but these families were only detected in a minority of the study sites (Fig. 1). For ssRNA viral families, the picture is more complex. However, it seems noteworthy that viral families showing the strongest overrepresentation in the VANA libraries, with up to 3 times more reads than in the dsRNA libraries, tend to have particularly stable particles, such as for the Virgaviridae or Solemoviridae (41, 42). On the other hand, the Closteroviridae, which have unstable and hard to purify particles, or the low-titer Luteoviridae showed, with over two times more reads, a tendency to be more represented in the dsRNA libraries (Fig. 1 and Table S2).
FIG 1.
Heatmap showing the number of reads corresponding to 28 viral families in each library, as estimated from the results of BLASTN and BLASTX analyses. The library names and sampling sites are indicated on the left side, and viral families are indicated below. Viral families are color-coded orange (dsRNA viral families), purple (ssRNA viral families), red (ssDNA families), green (dsDNA viral families), and blue (retro-transcribing viral family). Cell color intensity is proportional to the number of reads, following the scale on the right.
OTU-based assessment of viral richness with the dsRNA and VANA approaches.
For a variety of reasons, including the absence of universally conserved genomic elements and the frequently incomplete genome coverages, the in-depth characterization of viromes at a level close to taxonomic species has remained largely elusive. However, a possible strategy to circumvent these difficulties has been proposed, involving the clustering of contigs encoding proteins sharing conserved motifs (40). We have developed a pipeline which sequentially identifies such contigs for a range of conserved viral motifs using reverse-position-specific (RPS)-BLAST against the Pfam database (Table S3), aligns the contigs, and finally, performs a clustering, allowing the definition of operational taxonomic units (OTUs) on the basis of a defined identity cutoff value (43).
The dsRNA and VANA data sets were analyzed using this strategy and a 10% cutoff value, which reasonably approximates in many families the envelope of viral species variability (43). RPS-BLAST of all contigs identified contigs encoding 47 different viral conserved protein motifs, including those corresponding to well-known signature sequences such as RNA-dependent RNA polymerases (RdRp) and viral helicases (Table S3). For example, the matches for the different RdRp signatures (RdRp_1, 2, 3, and 4) collectively consist of sequences covering a very wide range of plant or fungal RNA virus families. Contigs corresponding to motifs with a much more restricted taxonomic signature were also identified, such as pfam01787, a protein family specific to the coat protein of ilarviruses in the family Bromoviridae (Table S3).
In order to avoid counting the same contig multiple times if it contained multiple signature sequences, the analysis was focused on the four RdRp protein families, RdRp_1, 2, 3, and 4, which are specific to RNA viruses and cover the broadest diversity of these agents. This has, however, the side effect of focusing the analysis on RNA viruses, so that a detailed analysis of DNA viruses would require, in addition, also considering some DNA virus-specific motifs. Using a 10% identity clustering cutoff value, a total of 239 RdRp OTUs were identified when taking into consideration all dsRNA and VANA data sets. Annotation of contigs representative of each OTU by BLASTX allowed the identification of 16 RNA virus families (Table S4) to be compared with the 18 RNA virus families detected by the direct annotation of contigs (Fig. 1). This difference might be explained by families for which a low coverage has resulted in incomplete genome assemblies in which the conserved viral RdRp motif is missing. The two families detected by direct BLAST annotation of contigs but not by OTU annotation were Reoviridae and Rhabdoviridae.
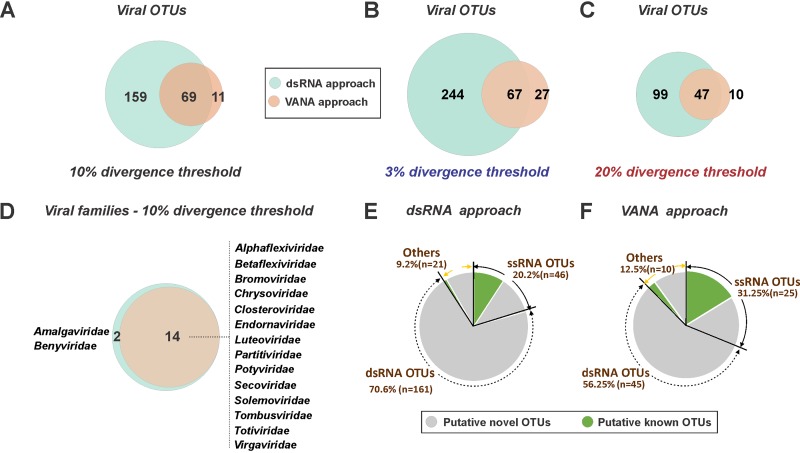
When the VANA and dsRNA approaches are compared, VANA detected 14 of the 16 RNA virus families detected using dsRNA, missing only two families, the Amalgaviridae and Benyviridae (Fig. 2D). As indicated above, this is likely due to the low read numbers for these families in the VANA approach (Fig. 1), resulting in incomplete genome coverage and in an absence of contigs covering the RdRp conserved domain for the viruses in these families. This result confirms that when considering the viral families detected, the performances of the VANA and dsRNA approaches are significantly but not widely different.
FIG 2.
Virus richness and known/novel status assessed at both the family and operational taxonomic unit (OTU) levels using dsRNA or VANA approaches. (A to C) Scaled Venn diagrams showing the numbers of OTUs discovered using the dsRNA or VANA approaches and a 10% divergence criterion for OTU definition (A) or using 3% (B) and 20% (C) divergence criteria. (D) Scaled Venn diagrams showing the number and identity of OTU families discovered using the dsRNA or VANA approaches. (E and F) Pie charts illustrating for the dsRNA (E) and VANA (F) approaches the proportions and known or novel RNA-dependent RNA polymerase (RdRp) OTUs for dsRNA viruses, ssRNA viruses, and others (unclassified viruses and virus-associated RNAs).
The dsRNA strategy detected a total of 228 OTUs, while VANA only detected 80 OTUs, of which 69 were detected by both strategies (Fig. 2A). A large number of dsRNA OTUs (n = 159) were not detected by VANA (Fig. 2A). Of these 159 OTUs, 60% were annotated as corresponding to Totiviridae members, while the remaining 40% came from other families (Table S4). This difference is also observed if different, lower (3%) or higher (20%), cutoff thresholds are used in the clustering phase for the definition of OTUs (Fig. 2B and C).
If the OTUs for which there are no sequences in GenBank that share less than the 10% clustering cutoff criterion are defined as novel, the majority of the VANA (81.2%) and dsRNA (89.5%) OTUs correspond to novel agents (Fig. 2E and F). In both approaches, the putative novel OTU group integrates almost all the dsRNA OTUs, while only around half of the ssRNA OTUs (48% to 54%) appear to correspond to novel agents (Fig. 2E and F).
Comparison of the dsRNA and VANA approaches at the level of individual plant populations.
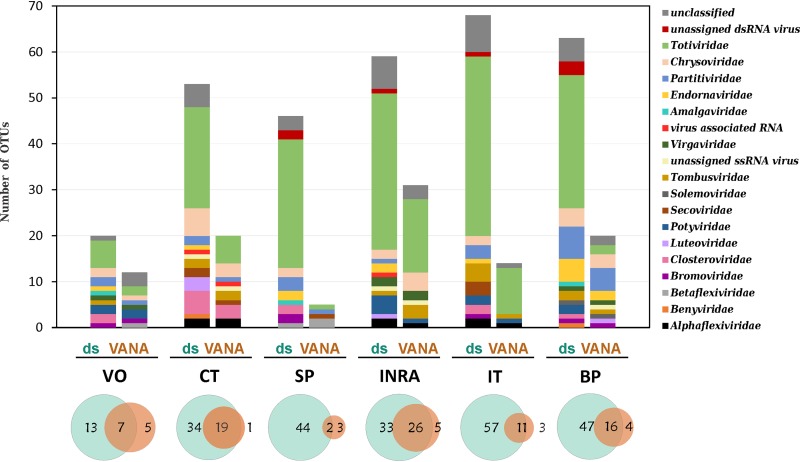
When each sampling site was analyzed independently, the same pattern emerged, and significantly more OTUs were identified using the dsRNA approach than the VANA approach (Fig. 3). The virome compositions at the family level were also more diverse (Fig. 3). On average, 9.8 ± 1.3 families were identified using the dsRNA approach per sampling site compared to only 6.2 ± 1.9 for VANA (P = 0.0007), with the SP site showing the lowest viral richness (5 OTUs and 4 viral families; Fig. 3). In most sites, Totiviridae was the most represented family by OTU, with OTUs making up, on average, 49.2% ± 12.0% of the virome for the dsRNA approach, compared to 33.5% ± 23.6% for VANA (P = 0.2132).
FIG 3.
Comparison of the viral diversity identified at each individual sampling site using the dsRNA and VANA approaches. The bar chart shows the RdRp OTU-based virome composition for the different viral families using the dsRNA and VANA approaches. Scaled Venn diagrams show the number of RdRp OTUs discovered by either the dsRNA (light blue) or the VANA (light orange) approach or by both approaches simultaneously.
At the individual OTU level also, the dsRNA approach revealed a significantly higher diversity, with an average of 51.5 ± 17.0 OTUs per sampling site compared to 17.2 ± 8.9 OTUs for VANA (P = 0.003). In addition, a large proportion of the OTUs identified using VANA (73% ± 20%; extremes, 40% to 95%) were also discovered using the dsRNA approach, while a large majority of dsRNA OTUs were not detected by VANA (73% ± 15%; extremes, 56% to 96%).
Reproducibility of the VANA and dsRNA approaches.
Since two random amplifications and ensuing library sequencing were performed for each complex pool, it is possible to evaluate the reproducibility of the viromes obtained from the two whole-genome amplification (WGA) replicates but also virome composition specificity in the different study sites. For most variables, there were no statistically significant differences between the two libraries obtained from each sample, including for variables such as number of assembled reads, number of contigs, N50, number of viral contigs, number of viral families, and number of OTUs identified (Table S5).
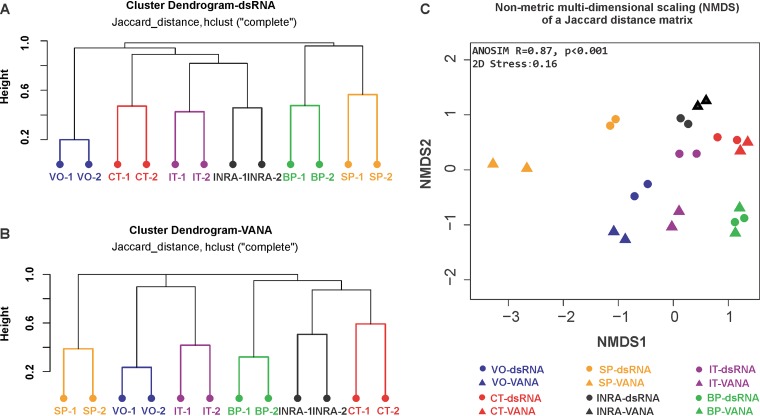
In addition, the reproducibility of the viromes from either different WGA replicates or different enrichment strategies (dsRNA or VANA) was further evaluated based on OTU presence/absence data (Fig. 4). The results of hierarchical clustering analyses based on these data show that even if some variability is observed between replicates, the distance between replicates is systematically much lower than the distances between samples (Fig. 4A and B). In addition, when comparing the results obtained with the dsRNA and VANA approaches, it is clear that the replicates for each site-technique combination end up very close (Fig. 4C). As shown above, there is a very significant clustering of libraries corresponding to a given site (analysis of similarity [ANOSIM], R = 0.87, P < 0.001) (Fig. 4C), also illustrating the fact that each virome showed strong site specificity with 41% to 71% of site-specific OTUs (Table 2). The ecosystem type (cultivated or unmanaged) had only limited impact on virome composition (ANOSIM test, R = 0.2, P = 0.002).
FIG 4.
Dissimilarity analyses of the RdRp OTU virome composition between sites. The dissimilarity (distance) matrix was calculated using a Jaccard method on OTU presence/absence data. Hierarchical clustering dendrograms of the 12 dsRNA libraries (A) and of the 12 VANA libraries (B) corresponding to the 6 sampling sites were prepared using hclust and the “complete” algorithm. (C) Nonmetric multidimensional scaling (NMDS) of a Jaccard distance matrix generated using the presence/absence data of all dsRNA and VANA libraries. Circles represent the dsRNA libraries, and triangles, the VANA libraries. The symbols are color-coded according to the sampling site.
TABLE 2.
Virome characteristics in the six different study sites based on RdRp OTUs
| Parameter | Result for site and ecosystem type |
|||||
|---|---|---|---|---|---|---|
| VO cultivated | CT cultivated | IT cultivated | SP unmanaged | INRA unmanaged | BP unmanaged | |
| No. of viral families | 11 | 10 | 10 | 9 | 9 | 13 |
| No. of OTUs | 25 | 54 | 71 | 49 | 64 | 68 |
| Site specificity | ||||||
| No. of site-specific OTUs | 12 | 28 | 29 | 35 | 31 | 46 |
| Site-specific OTUs (%) | 48.0 | 51.9 | 40.8 | 71.4 | 48.4 | 67.6 |
| Novelty | ||||||
| No. of putative novel OTUs (RdRp identity <90%) | 20 | 41 | 64 | 49 | 54 | 63 |
| Novel OTUs (%) | 80.0 | 75.9 | 90.1 | 100.0 | 84.4 | 92.6 |
Impact of management practices on virome richness and composition.
There were no statistically significant differences in the number of OTUs or of viral families between the cultivated and unmanaged sites (Table 2). Similarly, although a small trend could be seen in the average values (92.3% ± 7.8% novel OTUs for unmanaged sites versus 82.7% ± 7.3% for cultivated ones), the difference in the proportion of novel OTUs was not statistically significant (Table 2). Conversely, OTUs corresponding to already known viruses proved more frequent in cultivated sites than in unmanaged ones (18 versus 12 OTUs).
While the large variability seen at the level of individual sites limited the ability to draw clear conclusions, comparison of aggregated OTU numbers for viral families or viral groups supported the notion of a higher dsRNA virus diversity in unmanaged sites (118 versus 77 unique OTUs). For ssRNA viruses, the trend was reversed, with a marginally higher diversity (31 versus 28 unique OTUs) in managed sites. This trend was particularly clear for Closteroviridae (7 versus 2 OTUs) and Secoviridae (4 versus 1 OTU). Conversely, persistent viruses showed an overall higher richness in unmanaged sites, in particular, Totiviridae (84 versus 56 OTUs), Chrysoviridae (8 versus 3 OTUs), and Endornaviridae (10 versus 6 OTUs).
DISCUSSION
In this study, we compared the effectiveness for phytovirome description of the two most widely used nucleic acid enrichment approaches, double-stranded RNA (dsRNA) and virion-associated nucleic acids (VANA). The richness of the analyzed viromes was assessed with two strategies, direct BLASTN or BLASTX-based taxonomic annotation of assembled contigs, providing a virome richness estimate at the family level and the identification of viral OTUs based on a clustering of contigs encoding viral RdRp conserved motifs (43). The BLAST-based annotation of contigs representative of each OTU also allows a richness estimate at the family level.
The OTU-based analysis is expected to provide a lower-bound richness estimate, because agents for which the RdRp-encoding region is not covered cannot be identified as an OTU. This may explain why direct contig annotation identified, on average, a slightly higher RNA virus family-based richness than the OTU approach (paired t test, P = 0.0001) (Table S1). For example, for dsRNA libraries, an average of 13.3 ± 1.7 RNA virus families were identified using direct BLAST annotation compared to 11.2 ± 1.5 families using OTU clustering (paired t test, P = 0.008). Similarly significant differences were observed using the VANA approach (paired t test, P = 0.01). A possible strategy to increase the completeness of the OTU-based approach would be to also take into account the OTUs defined by other conserved viral motifs, such as viral helicases or viral coat proteins (Table S3). A virus for which the RdRp region has no coverage could then be taken into account if its helicase is among the sequence data. This has the potential advantage of improving the ability to detect viral contigs. Indeed, of the 1,393 contigs identified by RPS-BLAST as containing at least one virus-specific motif, 337 (24.2%) were not annotated as viral by the BLAST initial analysis. However, this strategy would likely provide an overrepresentation of the true viral richness, since a fully sequenced virus would then give rise to as many OTUs as it has conserved motifs. It is interesting to notice that the low frequency of phage sequences identified by the BLAST-based annotation is confirmed by the RPS-BLAST search for encoded protein motifs since, overall, a single VANA contig could be identified as encoding a phage-specific motif.
It should be stressed that the family-level annotation of contigs or OTUs performed here is based on the first BLAST hit and therefore does not guarantee that the agents indeed belong to the identified family. Phylogenetic analyses performed with the contigs representative of OTUs have, however, shown in other experiments a good general fit between the BLAST-assigned family and phylogenetic affinities. Metagenomics studies (17–19) have, for example, greatly contributed to a redefinition of the RNA virosphere of invertebrates. While a wealth of novel OTUs were identified here, our results do not point to the existence of a large number of novel higher-order viral taxa (family and above) associated with the sampled plant populations.
Broadly speaking, when taking into account all data sets, the dsRNA and VANA approaches recovered largely the same viral families, with only a few viral families not recovered by one or the other approach. Interestingly, Endornaviridae members that lack a true capsid or particle but that produce host-derived vesicles containing their nucleic acids were abundantly found from several VANA libraries, confirming similar observations in other studies (33, 44) and indicating that the VANA approach is not limited to virion-producing agents. As expected, for the dsRNA approach, DNA viruses were not efficiently recovered, even if some Nanoviridae were identified. Indeed, the detection of DNA viruses using dsRNA has been reported in the literature (20, 45, 46). For the VANA approach, a low efficiency of detection was observed for viruses or families with low titer and/or less stable particles, although Closteroviridae, which are known to have quite labile particles, have been detected here and elsewhere (33, 47). It should be mentioned that the excess of reads annotated as having a bacterial origin detected in two sites by VANA may in fact represent the detection of phages, since many integrated phages, which can make up to 10% to 20% of bacterial genomes (48, 49), have been sequenced and annotated as part of bacterial genomes. Overall, the results obtained would, however, point to a limited presence of phage in the analyzed plant-associated viromes. One possible explanation could be that the concentration in phage particles could be low in the analyzed samples and that they could have been outcompeted during the sequencing phase by more frequent phyto- or mycoviruses. In any case, the search for phage-specific motifs using the VirAnnot pipeline allows researchers to specifically search for evidence of phage presence, so it will be possible in the future to confirm the results reported here.
Comparable to other studies, the characterized viromes were dominated by novel dsRNA viruses, while a significant fraction of the less abundant ssRNA viruses proved to correspond to already known agents. Although some tentative trends were observed, no statistically supported differences could be identified between cultivated and unmanaged sites, raising the question of the impact of cultivation practices on the virome of wild plants and weeds growing nearby. Among the strongest trends was the finding of a higher diversity of dsRNA viruses which largely have persistent lifestyles in unmanaged environments. This might reflect an indirect impact of the fungicide treatments applied to crops (see below) or have other causes yet to be established.
Whatever the viral richness evaluation strategy and the sample analyzed, the dsRNA approach provided a more complete, richer virome representation. This statistically significant difference was observed at both the family and OTU levels (Table 1, Fig. 2 and 3) and is also observed if different, lower (3%) or higher (20%), cutoff thresholds are used in the clustering phase for the definition of OTUs (Fig. 2B and C). The reasons for this differential performance are unclear. One possibility is that the dsRNA purification protocol used allows for a greater enrichment of viral sequences. This could in turn lead to an ability to assemble longer, more efficiently annotated contigs (Table 1). An alternative hypothesis would involve the possible existence, in the case of VANA, of stronger competition effects between viruses in the complex pools analyzed. In this scenario, highly concentrated and stable viruses could outcompete less stable and/or concentrated viruses during the amplification of VANA targets, resulting in a less complete representation. Under both hypotheses, the use of less complex pools and/or deeper sequencing is likely to improve VANA and dsRNA performances.
Both approaches proved to have a good (but not perfect) reproducibility. Indeed, while the libraries prepared using independent amplifications of the same target pool always showed tight clustering in nonmetric multidimensional scaling (NMDS) (Fig. 4C), the corresponding viromes frequently showed a differential detection of a small fraction of the OTUs (Fig. 4). A careful analysis shows that most of the differential OTUs are represented by low read numbers so that small variations in representation in the data set may strongly affect the ability to assemble contigs for them and, ultimately, their identification. However, a few OTUs with significant coverage were also observed to be differentially detected between duplicate amplification libraries, which might point to other artifactual effects.
A rich diversity for mycovirus-like viruses from the Totiviridae and, to a lower extent, Chrysoviridae families was identified at all study sites. Given that the plant holobionts were used for sampling, this raises the possibility that a proportion of these agents might infect endophytic, epiphytic, or parasitic fungi associated with the sampled plants. Indeed, a lower richness is observed overall for these families from cultivated sites, a possible consequence of fungicide applications on overall fungal diversity (50, 51). At the same time, many typical fungal virus families, such as Hypoviridae, Narnaviridae, Fusariviridae, and Birnaviridae, were not detected here, further complicating the issue.
Overall, unless DNA viruses are of particular interest in metagenomics efforts involving the analysis of complex sample pools using the “lawnmower” strategy (3), the results presented here suggest that a preference should be given to the dsRNA-based approach since it consistently provides a more comprehensive vision of the virome. It should, however, be stressed that this recommendation may not apply when analyzing less complex samples such as individual plants or pools of plants of a single species, such as in ecogenomics or geometagenomics (23, 33), since VANA has been shown to perform efficiently in virus discovery and etiology studies (21, 23, 27).
MATERIALS AND METHODS
Study sites and plant samples.
To analyze plant virus richness in different cultivated or unmanaged environments, six different sites were selected in southwest France (Table S6). The VO site near Bordeaux is a cultivated horticultural agrosystem in which the main crops are vegetables such as tomato and lettuce. The nearby unmanaged site (INRA) corresponds to a prairie and adjoining path borders within the INRA research center. Near the town of Bergerac, two cultivated agrosystems (CT and IT) with, respectively, carrot and tobacco crops were selected, together with two unmanaged areas (SP and BP) corresponding, respectively, to a dry prairie and to a deciduous forest border.
For each site, a total of 200 individual plants were collected in spring 2016 (5 individual plants of each of the 40 locally most abundant species; Table S6). In the agrosystems, the cropped species were not collected. No specific efforts were made to select symptomatic plants, and plants with obvious fungal attack, insect colonization, or necrotized parts were excluded. All collected plants were identified to the species level or, when not possible, to the genus level by a trained researcher.
Sample processing and plant pool preparation.
For each sampling site, 4 different bulked samples (50 plants each, 10 different species) were prepared for dsRNA extraction, while 8 different bulked samples (25 plants each, 5 different species) were used for VANA extraction. In each case, the pools were composed of 0.1 g of fresh tissue of each sampled plant, yielding a total of 5 g of plant material for dsRNA pools and 2.5 g for VANA pools.
Viral nucleic acid enrichment, library preparation, and Illumina HiSeq sequencing.
Double-stranded RNAs were purified from each pool by two rounds of CF11 cellulose chromatography and converted to cDNA according to the protocol described by Marais et al. (22). In parallel, a negative-control blank was similarly prepared using only buffer. In order to evaluate the reproducibility of the whole-genome amplification (WGA) procedure, duplicate WGA PCRs involving different MID tags (19) were performed on each cDNA sample. PCR products were purified using the MinElute PCR purification kit (Qiagen) and their concentration determined spectrophotometrically. Finally, equal DNA amounts of the identically tagged WGA PCR products obtained from the 4 separate plant pools of each study site were pooled, generating a superpool corresponding to the 200 sampled plants.
Virion-associated nucleic acids (VANA) were extracted from each bulked sample following the protocol described by Candresse et al. (37). Synthesis and amplification of cDNAs prepared from nucleic acid extracts were performed by combining reverse transcriptase priming as described in the dsRNA strategy and a Klenow fragment polymerization step so as to allow the detection of both RNA and DNA viruses simultaneously (33). The resulting products were submitted to WGA in duplicates involving different multiplex identifier (MID) tags, purified, quantified, and assembled in superpools as described for the dsRNA strategy. The various libraries were named based on the target nucleic acids (ds or VANA) followed by the study site and PCR1 or PCR2 to indicate the WGA replicates (e.g., ds-VO-PCR1).
In total, 12 libraries were thus prepared for the dsRNA approach (corresponding to duplicate WGA for each of the 6 sampling sites), and there was one blank pool library for all the negative controls (Table S1). WGA was also performed in duplicate for the VANA samples, again yielding a total of 12 libraries. The 25 resulting libraries, each having a different MID tag, were separately used for preparation of independent sequencing libraries and sequenced in multiplexed format (2 × 150 bp) on an Illumina HiSeq 3000 system at the GenoToul platform (INRA Toulouse, France).
Bioinformatics analyses: read cleaning, normalization, and contig assembly.
Following demultiplexing, adapters and MID tags were removed with Cutadapt (52), and reads were quality trimmed (minimum quality score, 20; minimum length, 70 nucleotides). In order to limit intersample cross-talk associated with index-hopping (34), only reads having identical MID tags on both pair members were retained for further analyses. Cleaned virome HTS reads have been deposited in the INRA National Data Portal. To compensate for uneven sequencing depth between libraries, libraries were normalized by random subsampling to the same depth (122,295 pairs) using the Seqtk tool (https://github.com/lh3/seqtk) (Table S1). Contigs were de novo assembled for each library using IDBA-UD (53).
Contig annotation and operational taxonomic unit (OTU) clustering.
All contigs were annotated using BLASTN and BLASTX against the NCBI GenBank nonredundant (nr) nucleotide or protein database with a conservative E value cutoff of 10−4. In this way, contigs were assigned to one of the following categories: virus, eukaryote, bacteria, algae, and unknown. A heatmap illustrating the representation (absolute number of reads) of viral families (Table S2) in each library/site was prepared using the ComplexHeatmap package without clustering in R (54).
A clustering approach (43) was used to define and count operational taxonomy units, as initially highlighted (36, 40). Briefly, a search of all contigs against the Pfam database (55) was performed using reverse-position-specific BLAST (RPS-BLAST) (56). The contigs encoding a virus-specific conserved protein motif (Table S4) were retrieved and aligned with reference sequences and distance matrices computed with the ETE3 toolkit (57). These matrices were used to perform a clustering, allowing regrouping in a single operational taxonomic unit (OTU) of all contigs differing by less than a set cutoff divergence value (58). We used a 10% divergence cutoff value, which has been shown to generate in many viral families OTUs that are a relatively good approximation of taxonomic species (43). OTUs were thus defined and counted for each virus-specific conserved motif, allowing generation of an OTU table indicating for each approach-sampling site combination the presence/absence of each identified OTU. With the exception of the reproducibility analysis, all other analyses were performed by regrouping the data of the duplicate normalized libraries corresponding to the two separate PCR amplifications performed for each approach-sampling site combination.
Dissimilarity analyses between duplicate PCRs and among sampling pools/sites.
The availability of two random amplifications and the ensuing libraries (PCR1 and PCR2) for each approach-sampling site combination allowed us to evaluate the virome description reproducibility. Dissimilarity analyses were performed on OTU presence/absence binary data to generate a Jaccard distance matrix. Based on this distance matrix, hierarchical clusterings and nonmetric multidimensional scaling (NMDS) ordination were performed using hclust with the “complete” algorithm and the R “vegan” package (58, 59). The significance of comparisons among different sites and between different ecosystem types (cultivated and unmanaged) was assessed using the nonparametric statistical test analysis of similarity (ANOSIM) in the R “vegan” package (59–61).
Data availability.
The cleaned virome HTS reads have been deposited in the INRA National Data Portal under the identifier TVWBCQ (https://doi.org/10.15454/TVWBCQ).
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Raoult and F. Villeneuve (Centre Technique Interprofessionnel des Fruits et Légumes, Lanxade) and F. Dorlhac de Borne (Imperial Tobacco, Bergerac) for access to some sampling sites, P. Roumagnac (UMR BGPI, CIRAD) for sharing the VANA protocol, and the Genotoul Platform (INRA, Toulouse, France) for the Illumina sequencing.
Y. Ma was supported by a China Scholarship Council grant.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wren JD, Roossinck MJ, Nelson RS, Scheets K, Palmer MW, Melcher U. 2006. Plant virus biodiversity and ecology. PLoS Biol 4:e80. doi: 10.1371/journal.pbio.0040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roossinck MJ. 2015. Metagenomics of plant and fungal viruses reveals an abundance of persistent lifestyles. Front Microbiol 5:767. doi: 10.3389/fmicb.2014.00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roossinck MJ. 2012. Plant virus metagenomics: biodiversity and ecology. Annu Rev Genet 46:359–369. doi: 10.1146/annurev-genet-110711-155600. [DOI] [PubMed] [Google Scholar]
- 4.Cooper I, Jones RA. 2006. Wild plants and viruses: under-investigated ecosystems. Adv Virus Res 67:1–47. doi: 10.1016/S0065-3527(06)67001-2. [DOI] [PubMed] [Google Scholar]
- 5.Roossinck MJ. 2011. The big unknown: plant virus biodiversity. Curr Opin Virol 1:63–67. doi: 10.1016/j.coviro.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Roossinck MJ. 2012. Persistent plant viruses: molecular hitchhikers or epigenetic elements?, p 177–186. In Witzany G. (ed), Viruses: essential agents of life. Springer, Dordrecht, Netherlands. [Google Scholar]
- 7.Roossinck MJ, Saha P, Wiley GB, Quan J, White JD, Lai H, Chavarria F, Shen GA, Roe BA. 2010. Ecogenomics: using massively parallel pyrosequencing to understand virus ecology. Mol Ecol 19:81–88. doi: 10.1111/j.1365-294X.2009.04470.x. [DOI] [PubMed] [Google Scholar]
- 8.Roossinck MJ. 2013. Plant virus ecology. PLoS Pathog 9:e1003304. doi: 10.1371/journal.ppat.1003304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roossinck MJ. 2015. Plants, viruses and the environment: ecology and mutualism. Virology 479–480:271–277. doi: 10.1016/j.virol.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 10.Malmstrom CM, Melcher U, Bosque-Pérez NA. 2011. The expanding field of plant virus ecology: historical foundations, knowledge gaps, and research directions. Virus Res 159:84–94. doi: 10.1016/j.virusres.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 11.Goodwin S, McPherson JD, McCombie WR. 2016. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams IP, Glover RH, Monger WA, Mumford R, Jackeviciene E, Navalinskiene M, Samuitiene M, Boonham N. 2009. Next-generation sequencing and metagenomic analysis: a universal diagnostic tool in plant virology. Mol Plant Pathol 10:537–545. doi: 10.1111/j.1364-3703.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaire L, Wang Y, Gonzalez-Ibeas D, Mayer KF, Aranda MA, Llave C. 2009. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 392:203–214. doi: 10.1016/j.virol.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Kreuze JF, Perez A, Untiveros M, Quispe D, Fuentes S, Barker I, Simon R. 2009. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 388:1–7. doi: 10.1016/j.virol.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 15.Jones S, Baizan-Edge A, MacFarlane S, Torrance L. 2017. Viral diagnostics in plants using next generation sequencing: computational analysis in practice. Front Plant Sci 8:1770. doi: 10.3389/fpls.2017.01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al Rwahnih M, Daubert S, Golino D, Islas C, Rowhani A. 2015. Comparison of next-generation sequencing versus biological indexing for the optimal detection of viral pathogens in grapevine. Phytopathology 105:758–763. doi: 10.1094/PHYTO-06-14-0165-R. [DOI] [PubMed] [Google Scholar]
- 17.Wolf YI, Kazlauskas D, Iranzo J, Lucia-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. doi: 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang YZ, Chen YM, Wang W, Qin XC, Holmes EC. 2019. Expanding the RNA virosphere by unbiased metagenomics. Annu Rev Virol 6:119–139. doi: 10.1146/annurev-virology-092818-015851. [DOI] [PubMed] [Google Scholar]
- 19.Dolja VV, Koonin EV. 2018. Metagenomics reshapes the concepts of RNA virus evolution by revealing extensive horizontal virus transfer. Virus Res 244:36–52. doi: 10.1016/j.virusres.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rott M, Xiang Y, Boyes I, Belton M, Saeed H, Kesanakurti P, Hayes S, Lawrence T, Birch C, Bhagwat B, Rast H. 2017. Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis 101:1489–1499. doi: 10.1094/PDIS-03-17-0306-RE. [DOI] [PubMed] [Google Scholar]
- 21.Villamor DEV, Ho T, Al Rwahnih M, Martin RR, Tzanetakis IE. 2019. High throughput sequencing for plant virus detection and discovery. Phytopathology 109:716. doi: 10.1094/PHYTO-07-18-0257-RVW. [DOI] [PubMed] [Google Scholar]
- 22.Marais A, Faure C, Bergey B, Candresse T. 2018. Viral double-stranded RNAs (dsRNAs) from plants: alternative nucleic acid substrates for high-throughput sequencing. Methods Mol Biol 1746:45–53. doi: 10.1007/978-1-4939-7683-6_4. [DOI] [PubMed] [Google Scholar]
- 23.Roossinck MJ, Martin DP, Roumagnac P. 2015. Plant virus metagenomics: advances in virus discovery. Phytopathology 105:716–727. doi: 10.1094/PHYTO-12-14-0356-RVW. [DOI] [PubMed] [Google Scholar]
- 24.Susi H, Filloux D, Frilander MJ, Roumagnac P, Laine A-L. 2019. Diverse and variable virus communities in wild plant populations revealed by metagenomic tools. PeerJ 7:e6140. doi: 10.7717/peerj.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pooggin MM. 2018. Small RNA-omics for plant virus identification, virome reconstruction, and antiviral defense characterization. Front Microbiol 9:2779. doi: 10.3389/fmicb.2018.02779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filloux D, Dallot S, Delaunay A, Galzi S, Jacquot E, Roumagnac P. 2015. Metagenomics approaches based on virion-associated nucleic acids (VANA): an innovative tool for assessing without a priori viral diversity of plants. Methods Mol Biol 1302:249–257. doi: 10.1007/978-1-4939-2620-6_18. [DOI] [PubMed] [Google Scholar]
- 27.Filloux D, Fernandez E, Comstock JC, Mollov D, Roumagnac P, Rott P. 2018. Viral metagenomic-based screening of sugarcane from Florida reveals occurrence of six sugarcane-infecting viruses and high prevalence of Sugarcane yellow leaf virus. Plant Dis 102:2317–2323. doi: 10.1094/PDIS-04-18-0581-RE. [DOI] [PubMed] [Google Scholar]
- 28.Palanga E, Filloux D, Martin DP, Fernandez E, Gargani D, Ferdinand R, Zabré J, Bouda Z, Neya JB, Sawadogo M, Traore O, Peterschmitt M, Roumagnac P. 2016. Metagenomic-based screening and molecular characterization of cowpea-infecting viruses in Burkina Faso. PLoS One 11:e0165188. doi: 10.1371/journal.pone.0165188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pecman A, Kutnjak D, Gutiérrez-Aguirre I, Adams I, Fox A, Boonham N, Ravnikar M. 2017. Next generation sequencing for detection and discovery of plant viruses and viroids: comparison of two approaches. Front Microbiol 8:1998. doi: 10.3389/fmicb.2017.01998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visser M, Bester R, Burger JT, Maree HJ. 2016. Next-generation sequencing for virus detection: covering all the bases. Virol J 13:85. doi: 10.1186/s12985-016-0539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wylie SJ, Luo H, Li H, Jones MGK. 2012. Multiple polyadenylated RNA viruses detected in pooled cultivated and wild plant samples. Arch Virol 157:271–284. doi: 10.1007/s00705-011-1166-x. [DOI] [PubMed] [Google Scholar]
- 32.Boone M, De Koker A, Callewaert N. 2018. Capturing the ‘ome’: the expanding molecular toolbox for RNA and DNA library construction. Nucleic Acids Res 46:2701–2721. doi: 10.1093/nar/gky167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernardo P, Charles-Dominique T, Barakat M, Ortet P, Fernandez E, Filloux D, Hartnady P, Rebelo TA, Cousins SR, Mesleard F, Cohez D, Yavercovski N, Varsani A, Harkins GW, Peterschmitt M, Malmstrom CM, Martin DP, Roumagnac P. 2018. Geometagenomics illuminates the impact of agriculture on the distribution and prevalence of plant viruses at the ecosystem scale. ISME J 12:173–184. doi: 10.1038/ismej.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Illumina. 2017. Effects of index misassignment on multiplexing and downstream analysis. https://www.illumina.com/content/dam/illumina-marketing/documents/products/whitepapers/index-hopping-white-paper-770-2017-004.pdf.
- 35.van der Valk T, Vezzi F, Ormestad M, Dalén L, Guschanski K. 8 March 2019. Index hopping on the Illumina HiseqX platform and its consequences for ancient DNA studies. Mol Ecol Resour doi: 10.1111/1755-0998.13009. [DOI] [PubMed] [Google Scholar]
- 36.Thapa V, McGlinn DJ, Melcher U, Palmer MW, Roossinck MJ. 2015. Determinants of taxonomic composition of plant viruses at the Nature Conservancy’s Tallgrass Prairie Preserve, Oklahoma. Virus Evol 1:vev007. doi: 10.1093/ve/vev007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Candresse T, Filloux D, Muhire B, Julian C, Galzi S, Fort G, Bernardo P, Daugrois J-H, Fernandez E, Martin DP, Varsani A, Roumagnac P. 2014. Appearances can be deceptive: revealing a hidden viral infection with deep sequencing in a plant quarantine context. PLoS One 9:e102945. doi: 10.1371/journal.pone.0102945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutnjak D, Rupar M, Gutierrez-Aguirre I, Curk T, Kreuze JF, Ravnikar M. 2015. Deep sequencing of virus-derived small interfering RNAs and RNA from viral particles shows highly similar mutational landscapes of a plant virus population. J Virol 89:4760–4769. doi: 10.1128/JVI.03685-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. 2008. NCBI BLAST: a better Web interface. Nucleic Acids Res 36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmonds P. 2015. Methods for virus classification and the challenge of incorporating metagenomic sequence data. J Gen Virol 96:1193–1206. doi: 10.1099/vir.0.000016. [DOI] [PubMed] [Google Scholar]
- 41.Adams MJ, Adkins S, Bragard C, Gilmer D, Li D, MacFarlane SA, Wong S-M, Melcher U, Ratti C, Ryu KH, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Virgaviridae. J Gen Virol 98:1999–2000. doi: 10.1099/jgv.0.000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Truve E, Fargette D. 2011. ICTV virus taxonomy profile: Sobemovirus. ICTV 9th Report. https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/291/sobemovirus.
- 43.Lefebvre M, Theil S, Ma Y, Candresse T. 21 October 2019. The VirAnnot pipeline: a resource for automated viral diversity estimation and operational taxonomy units (OTU) assignation for virome sequencing data. Phytobiomes J doi: 10.1094/PBIOMES-07-19-0037-A. [DOI] [Google Scholar]
- 44.Maclot F, Candresse T, Filloux D, Roumagnac P, Massart S. 2019. Effect of species composition on virome diversity in various ecosystemic communities of Poaceae, abstr Rencontres de Virologie Végétale (RVV 2019), Aussois, France. [Google Scholar]
- 45.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. 2006. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol 80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kesanakurti P, Belton M, Saeed H, Rast H, Boyes I, Rott M. 2016. Screening for plant viruses by next generation sequencing using a modified double strand RNA extraction protocol with an internal amplification control. J Virol Methods 236:35–40. doi: 10.1016/j.jviromet.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Roumagnac P, Mollov D, Daugrois J, Filloux D. 2018. Viral metagenomics and sugarcane pathogens, p 183–200. In Rott P. (ed), Achieving sustainable cultivation of sugarcane. Burleigh Dodds Science Publishing, Cambridge, United Kingdom. [Google Scholar]
- 48.Casjens S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol 49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- 49.de Sousa AL, Maués D, Lobato A, Franco EF, Pinheiro K, Araújo F, Pantoja Y, da Costa da Silva AL, Morais J, Ramos RTJ. 2018. PhageWeb: Web interface for rapid identification and characterization of prophages in bacterial genomes. Front Genet 9:644. doi: 10.3389/fgene.2018.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Newton A, Gravouil C, Fountaine J. 2010. Managing the ecology of foliar pathogens: ecological tolerance in crops. Ann Appl Biol 157:343–359. doi: 10.1111/j.1744-7348.2010.00437.x. [DOI] [Google Scholar]
- 51.Karlsson I, Friberg H, Steinberg C, Persson P. 2014. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS One 9:e111786. doi: 10.1371/journal.pone.0111786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 53.Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 54.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 55.Bateman A, Smart A, Luciani A, Salazar GA, Mistry J, Richardson LJ, Qureshi M, El-Gebali S, Potter SC, Finn RD, Eddy SR, Sonnhammer ELL, Piovesan D, Paladin L, Tosatto SCE, Hirsh L. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huerta-Cepas J, Bork P, Serra F. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murtagh F, Legendre P. 2014. Ward’s hierarchical agglomerative clustering method: which algorithms implement Ward’s criterion? J Classif 31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 59.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin P, O’Hara R, Simpson G, Solymos P, Stevens M, Wagner H. 2017. Vegan: community ecology package. R package version 2.3–0. 2015. https://github.com/vegandevs/vegan.
- 60.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Austral Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 61.Buttigieg PL, Ramette A. 2014. A guide to statistical analysis in microbial ecology: a community-focused, living review of multivariate data analyses. FEMS Microbiol Ecol 90:543–550. doi: 10.1111/1574-6941.12437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cleaned virome HTS reads have been deposited in the INRA National Data Portal under the identifier TVWBCQ (https://doi.org/10.15454/TVWBCQ).