Rapid mutation rates and correspondingly high levels of intra- and interhost diversity are often cited as key features of viruses with the capacity for emergence and sustained transmission in a new host species. However, most of this information comes from studies of RNA viruses, with relatively little known about evolutionary processes in viruses with single-stranded DNA (ssDNA) genomes. Here, we provide a unique model of virus evolution, integrating both long-term global-scale and short-term intrahost evolutionary processes of an ssDNA virus that emerged to cause a pandemic in a new host animal. Our analysis reveals that successful host jumping and sustained transmission does not necessarily depend on a high level of intrahost diversity nor result in the continued accumulation of high levels of long-term evolution change. These findings indicate that all aspects of the biology and ecology of a virus are relevant when considering their adaptability.
KEYWORDS: canine parvovirus, deep sequencing, viral cross-species transmission, viral evolution, viral host adaption, viral intrahost diversity
ABSTRACT
Canine parvovirus (CPV) is a highly successful pathogen that has sustained pandemic circulation in dogs for more than 40 years. Here, integrating full-genome and deep-sequencing analyses, structural information, and in vitro experimentation, we describe the macro- and microscale features that accompany CPV’s evolutionary success. Despite 40 years of viral evolution, all CPV variants are more than ∼99% identical in nucleotide sequence, with only a limited number (<40) of substitutions becoming fixed or widespread during this time. Notably, most substitutions in the major capsid protein (VP2) gene are nonsynonymous, altering amino acid residues that fall within, or adjacent to, the overlapping receptor footprint or antigenic regions, suggesting that natural selection has channeled much of CPV evolution. Among the limited number of variable sites, CPV genomes exhibit complex patterns of variation that include parallel evolution, reversion, and recombination, compromising phylogenetic inference. At the intrahost level, deep sequencing of viral DNA in original clinical samples from dogs and other host species sampled between 1978 and 2018 revealed few subconsensus single nucleotide variants (SNVs) above ∼0.5%, and experimental passages demonstrate that substantial preexisting genetic variation is not necessarily required for rapid host receptor-driven adaptation. Together, these findings suggest that although CPV is capable of rapid host adaptation, a relatively low mutation rate, pleiotropy, and/or a lack of selective challenges since its initial emergence have inhibited the long-term accumulation of genetic diversity. Hence, continuously high levels of inter- and intrahost diversity are not necessarily required for virus host adaptation.
IMPORTANCE Rapid mutation rates and correspondingly high levels of intra- and interhost diversity are often cited as key features of viruses with the capacity for emergence and sustained transmission in a new host species. However, most of this information comes from studies of RNA viruses, with relatively little known about evolutionary processes in viruses with single-stranded DNA (ssDNA) genomes. Here, we provide a unique model of virus evolution, integrating both long-term global-scale and short-term intrahost evolutionary processes of an ssDNA virus that emerged to cause a pandemic in a new host animal. Our analysis reveals that successful host jumping and sustained transmission does not necessarily depend on a high level of intrahost diversity nor result in the continued accumulation of high levels of long-term evolution change. These findings indicate that all aspects of the biology and ecology of a virus are relevant when considering their adaptability.
INTRODUCTION
The emergence of viruses to cause epidemics in new hosts poses a constant threat to humans and other animals, as their appearance in immunologically naive populations may result in widespread disease. In many instances, newly emerging viruses acquire adaptive mutations that increase replication and facilitate transmission in the new host species (1, 2). However, it is unclear at what rate phenotypically relevant mutations arise and become selected under different circumstances, how many such changes are needed for host adaptation and sustained transmission, and whether there is a complex series of mutational changes that must occur in a specific order. Likewise, the relationship between intrahost genetic diversity and long-term interhost evolution and adaptation remain poorly defined. Clarifying the sources, extent, and dynamics of viral variation during known emergence events could inform our understanding of the risks of similar events occurring in the future.
Canine parvovirus (CPV), a nonenveloped single-stranded DNA (ssDNA) virus (family Parvoviridae, genus Protoparvovirus), is an extremely successful viral pathogen that overcame the evolutionary hurdles associated with cross-species transmission to emerge and spread in multiple host species. The original CPV strain (designated CPV type 2 [CPV-2] to distinguish it from the distantly related canine bocavirus [minute virus of canines]) arose as a variant from a group of closely related parvoviruses circulating among other carnivores. The virus was first recognized in dogs early in 1978 as the causative agent of a disease characterized by vomiting and diarrhea, primarily among young dogs, and myocarditis in neonatal puppies (3–5). By mid-1978, CPV-2 had reached pandemic proportions, but it was rapidly replaced by a new genetic variant by the end of 1980 (6, 7). This new variant, designated CPV-2a, contained a group of five nonsynonymous substitutions in the virus capsid protein (VP) gene (7) that enabled it to expand its host range among carnivores, most notably gaining the ability to infect and likely transmit between domestic cats (8). All CPV variants circulating today appear to be descendants of the CPV-2a lineage, and they remain significant pathogens of dogs despite the widespread use of effective vaccines (9, 10). The literature on these viruses has identified a number of more recently derived CPV-2a genetic and antigenic variants, sometimes designated as “CPV-2b,” “CPV-2c,”or “New CPV-2a”, among others. However, these variants are based on the presence of one or two specific amino acid substitutions and do not reflect the complexity and multitude of changes across entire viral genomes. Therefore, for the purposes of this study, our reference to “CPV-2a” encompasses all CPV variants and strains descendent from the initial CPV-2a global sweep that occurred in the late 1970s and early 1980s.
Central to understanding the cross-species transmission and continued evolution of CPV and the closely related carnivore parvoviruses is their interaction with the host cell receptor, transferrin receptor type 1 (TfR) (11). Indeed, acquiring the ability to properly bind the canine TfR to allow cell infection was a critical evolutionary step for CPV (12, 13). Comparisons of CPV and closely related feline panleukopenia virus (FPV) sequences revealed that the canine host range was determined by three or four amino acid changes within a small region of the VP2 capsid surface, which involved at least three surface loops contributed by two symmetrically arranged VP2 molecules (14–16). The capsid changes in this region overcame a biochemical canine host range barrier—the presence of an N-linked glycosylation site associated with residue 384 of the canine TfR—that blocked receptor binding by the capsids of the ancestor-like viruses from cats, mink, and raccoons (12, 17). Mutations that arose later in the evolution of CPV, particularly those that gave rise to the CPV-2a variant, may have evolved in other hosts besides dogs, as changes of the CPV-2a-specific codons to other residues have been found in viruses from raccoons, foxes, and mink (18, 19). Such mutations likely reflect adaptation of the viruses in overcoming host barriers to infection or optimizing their interactions with TfR binding sites (19). Recently, the TfR-CPV interaction has been further defined using cryo-electron microscopy (cryoEM) (30), providing atomic resolution structural information of the viral capsid-host cell receptor interface.
Other important interactions at the virus-host interface include antibody binding, and selection for antigenic variation has likely played a key role in determining the evolutionary success of different CPV variants in nature (7, 20). Studies defining the binding sites of eight different mouse or rat monoclonal antibody (MAb) antigen binding fragments (Fabs) revealed that more than 65% of the accessible viral capsid surface was covered by antibody footprints, although the mutations that controlled natural or experimentally selected antigenic variation fell within two relatively small regions (21, 22). Importantly, the TfR binding site overlaps with some key MAb binding sites, and many capsid mutations may alter both antibody and TfR binding. Thus, there are likely complex relationships between selection pressures on the CPV capsid associated with host TfR and antibody binding (12, 23).
While many studies have identified specific consensus-level host-adaptive mutations within the major viral capsid protein gene, none have sought to survey the full suite and distribution of mutations that have become fixed or widespread during CPV’s emergence in dogs and evolution on the epidemiological scale. Moreover, the link between intrahost evolution and that seen at the epidemiological scale remains unclear. For example, high levels of standing genetic variation within individual hosts could enable viruses to rapidly adapt to selective pressures associated with host immunity or infection within new host species. The ability to perform genome-wide deep sequencing on natural and experimental viral samples now allows us to more completely define the variability and evolution within the viral population. Other studies on emerging viruses have primarily focused on RNA viruses, revealing that individual hosts can harbor viruses with multiple single nucleotide variants (SNVs) (reviewed in reference 24), the consequence of highly error-prone replication (25, 26). Far less is known about the extent and properties of sequence variation within populations of DNA viruses, including ssDNA viruses such as the parvoviruses. However, despite being replicated by DNA polymerases that have very high fidelity when replicating the host cell genome, CPV evolved relatively rapidly in nature during its initial emergence, with estimates for the evolutionary rate in the range of ∼10−4 substitutions/site/year (27, 28). In addition, the virus is able to rapidly adapt to both variant host receptors and to antibody selection in tissue culture (19, 22).
Herein, we describe the emergence and sustained transmission of CPV over 40 years across multiple evolutionary scales. We combine genome-wide and deep-sequence analyses of a collection of CPV and CPV-related viruses in original clinical samples from dogs and alternative host species collected in 1978 and during the subsequent 40 years of virus evolution, as well as experimental studies of host adaptation to determine (i) the full suite of genetic changes that have become fixed or widespread during emergence and pandemic circulation of CPV and (ii) the extent and structure of intrahost genetic diversity that underpins this long-term evolution. These data, informed by recently resolved high-resolution molecular structures, provide a model encompassing both the macro- and microscale features enabling a virus to emerge and sustain pandemic spread in a new host.
(This article was submitted to an online preprint archive [29].)
RESULTS
The viruses sequenced as part of this study represent a record spanning the entire 41 years of evolution of CPV in dogs, as well as that occurring in related viruses recovered from raccoons, arctic foxes, and raccoon dogs. Most of these samples originated from the United States, but samples from Australia, Nigeria, France, Germany, and Finland were also included. A total of viral 46 genomes from original fecal or intestinal material were sequenced, of which 35 were deep sequenced (Table 1). Viral genomes from various tissues from an acutely infected puppy, as well as from experimentally passaged viruses in different host cells in culture, were also included in our analyses.
TABLE 1.
Naturally circulating viruses sequenced in this study
| Sample | Collection
yr |
Collection
country |
Host | Virus
type |
Consensus
accession no. |
SRA accession no.a |
|---|---|---|---|---|---|---|
| CPV12 | 1978 | USA | Dog | CPV-2 | MN451655 | SAMN12739316 |
| CPV6 | 1979 | USA | Dog | CPV-2 | MN451653 | SAMN12739317 |
| CPV9 | 1979 | USA | Dog | CPV-2 | MN451654 | SAMN12739318 |
| CPV5b | 1980 | USA | Dog | CPV-2 | EU659116.1 | SAMN12739319 |
| CPV63 | 1980 | USA | Dog | CPV-2 | MN451668 | SAMN12739320 |
| RDPV122 | 1980 | Finland | Raccoon dog | CPV-2 | MN451693 | SAMN12739321 |
| RDPV123 | 1980 | Finland | Raccoon dog | CPV-2 | MN451694 | SAMN12739322 |
| CPV13b | 1981 | USA | Dog | CPV-2a | EU659118 | SAMN12739323 |
| CPV81 | 1982 | Australia | Dog | CPV-2a | MN451669 | SAMN12739324 |
| BFPV | 1983 | Finland | Blue fox | FPV | MN451652 | SAMN12739325 |
| CPV27 | 1983 | USA | Dog | CPV-2a | MN451656 | SAMN12739326 |
| CPV29 | 1983 | USA | Dog | CPV-2a | MN451657 | SAMN12739327 |
| CPV30 | 1983 | USA | Dog | CPV-2a | MN451658 | SAMN12739328 |
| CPV31 | 1983 | USA | Dog | CPV-2a | MN451659 | SAMN12739329 |
| CPV33 | 1983 | USA | Dog | CPV-2a | MN451660 | SAMN12739330 |
| CPV58 | 1983 | France | Dog | CPV-2a | MN451667 | SAMN12739331 |
| CPV35 | 1984 | USA | Dog | CPV-2a | MN451661 | SAMN12739332 |
| CPV36 | 1984 | USA | Dog | CPV-2a | MN451662 | SAMN12739333 |
| CPV39 | 1984 | USA | Dog | CPV-2a | MN451663 | SAMN12739334 |
| CPV54 | 1984 | France | Dog | CPV-2a | MN451666 | SAMN12739335 |
| CPV48 | 1985 | USA | Dog | CPV-2 | MN451664 | SAMN12739336 |
| CPV87 | 1985 | Australia | Dog | CPV-2a | MN451670 | SAMN12739337 |
| CPV50 | 1986 | USA | Dog | CPV-2 | MN451665 | SAMN12739338 |
| RDPV124 | 1986 | Finland | Raccoon dog | CPV-2 | MN451695 | SAMN12739339 |
| CPV220 | 1993 | USA | Dog | CPV-2a | MN451671 | NA |
| CPV305 | 1993 | USA | Dog | CPV-2a | MN451672 | SAMN12739340 |
| CPV307 | 1993 | USA | Red wolf | CPV-2a | MN451673 | SAMN12739341 |
| CPV353 | 1996 | USA | Dog | CPV-2a | MN451674 | SAMN12739342 |
| CPV604 | 2007 | USA | Dog | CPV-2a | MN451677 | SAMN12739343 |
| RACCPV1 | 2009 | USA | Raccoon | CPV-2a | MN451691 | SAMN12739344 |
| CPV617c | 2011 | USA | Dog | CPV-2a | MN451690 | SAMN12739350 to SAMN12739357 |
| RACFPV1 | 2011 | USA | Raccoon | FPV | MN451692 | SAMN12739345 |
| CPV606 | 2014 | USA | Dog | CPV-2a | MN451679 | SAMN12739346 |
| CPV601 | 2016 | USA | Dog | CPV-2a | MN451675 | SAMN12739347 |
| CPV603 | 2017 | USA | Dog | CPV-2a | MN451676 | SAMN12739348 |
| CPV605 | 2018 | Nigeria | Dog | CPV-2a | MN451678 | SAMN12739349 |
| CPV607 | 2018 | Nigeria | Dog | CPV-2a | MN451680 | NA |
| CPV608 | 2018 | Nigeria | Dog | CPV-2a | MN451681 | NA |
| CPV609 | 2018 | Nigeria | Dog | CPV-2a | MN451682 | NA |
| CPV610 | 2018 | Nigeria | Dog | CPV-2a | MN451683 | NA |
| CPV614 | 2018 | Nigeria | Dog | CPV-2a | MN451687 | NA |
| CPV615 | 2018 | Nigeria | Dog | CPV-2a | MN451688 | NA |
| CPV616 | 2018 | Nigeria | Dog | CPV-2a | MN451689 | NA |
| CPV611 | 2019 | USA | Dog | CPV-2a | MN451684 | NA |
| CPV612 | 2019 | USA | Dog | CPV-2a | MN451685 | NA |
| CPV613 | 2019 | USA | Dog | CPV-2a | MN451686 | NA |
SRA accession numbers are listed for deep-sequenced samples. NA, not available.
Sample previously sequenced (consensus level) in another study and deep sequenced in this study.
Multiple tissue specimens sequenced from single individual (see Results and Fig. 5).
Sequencing provided nearly complete genome coverage, spanning all major reading frames and lacking only the distal portions of the 5′- and 3′-terminal untranslated regions (UTRs) (see Materials and Methods and see Fig. S1 in the supplemental material). For samples that were deep sequenced to identify subconsensus single nucleotide variants (SNVs), ∼5,000-fold coverage per site was achieved after read processing and coverage normalization. A short region (∼150 nucleotides [nt] in length) flanked by a poly(A)- and G-rich sequence region near the start of the VP2 gene did not sequence or assemble efficiently in any of the natural or experimental samples or control plasmids. While the low coverage in this region (the “low-coverage region” [LCR] in Fig. 4 to 6) did not affect consensus calls, it did affect our ability to accurately call SNV frequencies and was therefore excluded when measuring intrahost diversity. To confirm and define the sensitivity of our subconsensus variant calling in the rest of the CPV genome, artificially created heterogenous virus populations were created by serially diluting two different infectious clones that differed at 39 nucleotide positions (i.e., 39 SNVs). These control samples, which were processed alongside the natural samples, provided evidence that our methods could consistently detect SNVs at very low proportions (<1%) across the genome (Fig. S2). We therefore set a cutoff of 0.5% for SNV detection. For all analyses in this study, genome nucleotide numbering starts at the first position in the NS1/2 gene coding region, while amino acid numbering begins at the first methionine for each protein.
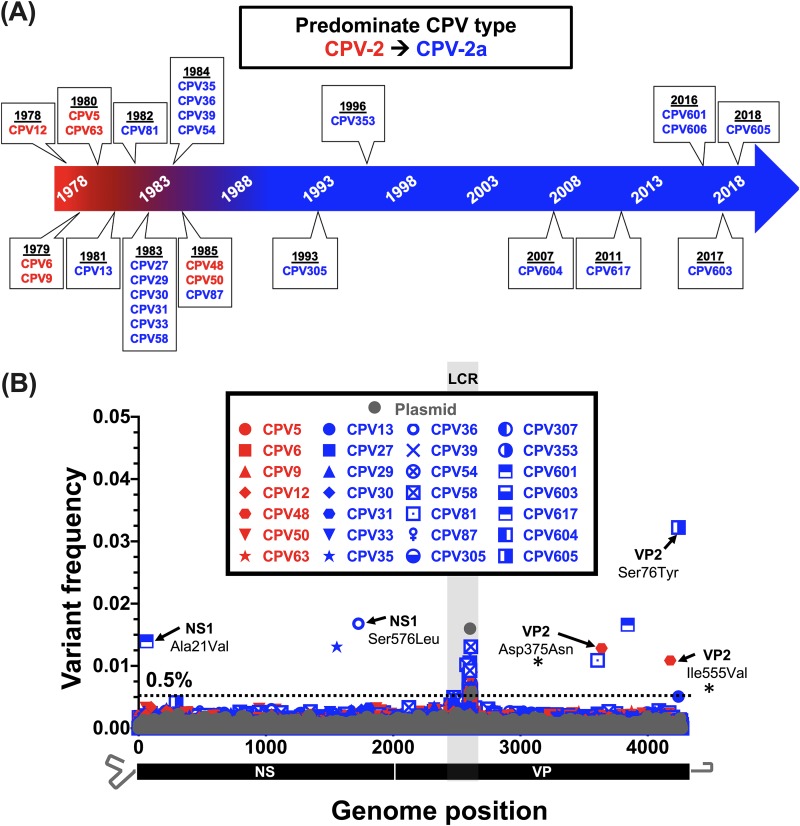
FIG 4.
Intrahost viral diversity among dogs infected with CPV-2 or CPV-2a sampled during 40 years of circulation. (A) Timeline displaying dates of CPV from original fecal material samples that were deep sequenced as part of this study. The dominant circulating strain and isolates are indicated by red and blue timeline shading and text, respectively. (B) Position and frequency of all dominant subconsensus SNVs with red and blue symbol coloring as in panel A. SNVs resulting in nonsynonymous changes are indicated. The nonsynonymous changes also found in a survey of natural samples (Fig. 1 and Table 2) are indicated with a black asterisk. Deep-sequenced plasmid (in gray) provides a baseline sequencing error rate. The low-coverage region (LCR) is indicated by gray shading, and positions in this region were omitted from variant calling analyses.
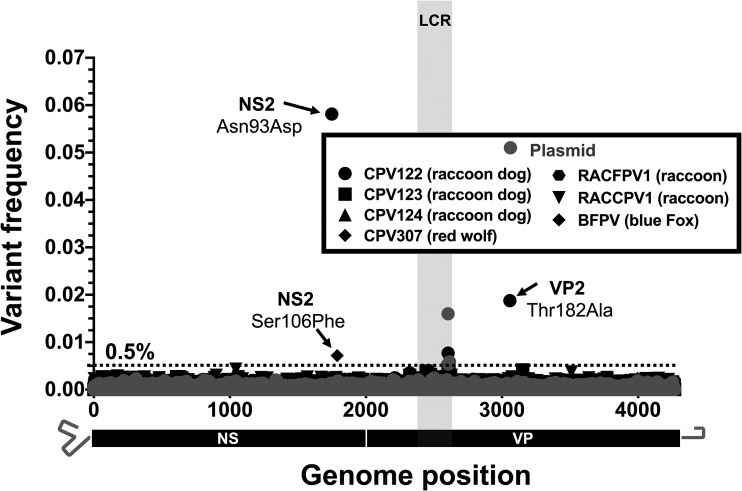
FIG 6.
Intrahost viral diversity among alternative host species infected with CPV-2, CPV-2a, or FPV. The positions and frequencies of all dominant SNVs are displayed. Sequenced viruses include CPV-2 isolates from raccoon dogs (CPV122-124), CPV-2a isolates from a red wolf (CPV307) and a raccoon (RACCPV1), and FPV isolates from an arctic blue fox (BFPV) and raccoon (RACFPV1). Deep-sequenced plasmid (in gray) provides a baseline sequencing error rate. The low-coverage region (LCR) is indicated by gray shading, and positions in this region were omitted from variant calling analyses.
Limited background evolution of CPV during 40 years of circulation.
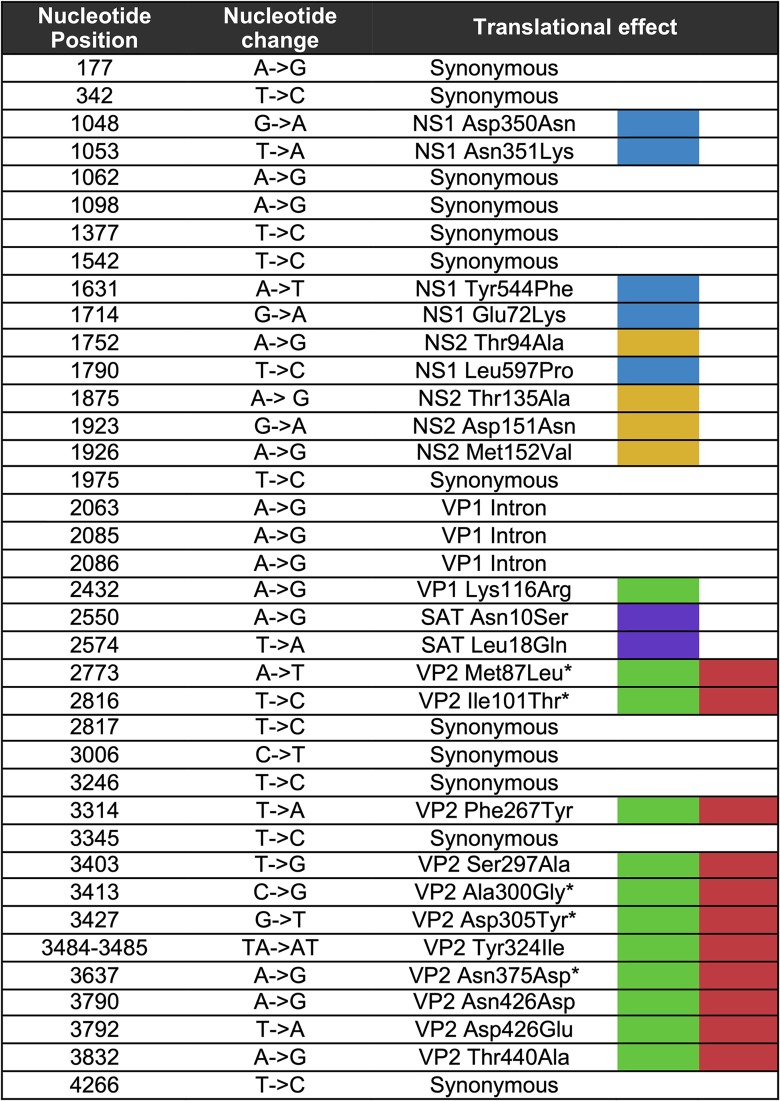
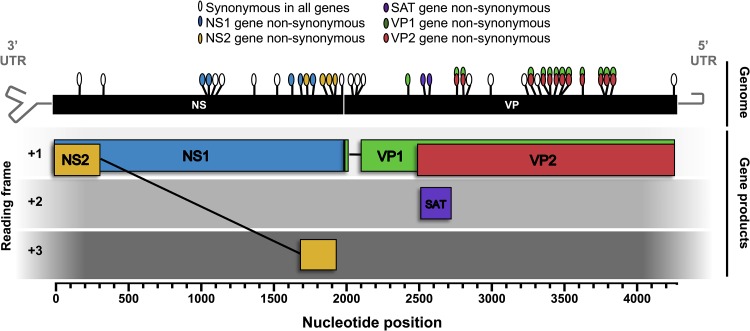
New CPV consensus sequences determined for the viruses analyzed here were combined with all other GenBank CPV sequences isolated directly from dogs and raccoons spanning both the NS and VP coding regions (hereafter referred to as the “full genome”) and for which isolation location and date were available. CPV sequences derived from raccoons were included, as these hosts may play a major role in the long-term evolution of these viruses (18). This data set (222 genomes) was used to survey the distribution and nature of mutations that have arisen and become fixed or widespread in all open reading frames and introns of the CPV genome during its circulation (Table 2 and Fig. 1). We included all those mutations that differed from the nucleotides in the earliest CPV full-genome sequence (CPV12 sequenced in this study) and present in >10% (arbitrarily chosen) of the sequences analyzed. Overall, 15 synonymous and 23 nonsynonymous mutations met these criteria. Of the 15 synonymous mutations, 7 fell within the region of the genome encoding the nonstructural genes, 3 in the VP1 gene intron, and 5 within the VP coding regions. Of the 23 nonsynonymous changes, 5 were found in the NS1 gene, 4 were in the NS2 gene, 1 was exclusive to the VP1 gene, 11 were in the VP1/2 gene overlapping region, and 2 were in the small alternatively translated protein (SAT) open reading frame, contained within the VP2 N-terminus-encoding genomic region (Fig. 1).
TABLE 2.
Translational effects are colored as in Fig. 1. Nucleotide numbering begins at the first coding site of the NS1/2 gene. Amino acid numbering is based on each respective gene translational start site. VP2 position is given for substitutions within the VP1/VP2 overlapping open reading frames. An asterisk indicates that the CPV substitution arose during the CPV-2 to CPV-2a global sweep.
FIG 1.
Survey of mutations in the CPV genome that have arisen and become widespread or fixed since the virus first emerged in dogs in 1978. Schematic diagram of the CPV genome and gene products with locations and coding effects of substitutions (colored ovals). The analysis was based on 222 full-genome CPV sequences from virus isolated in dogs or raccoons. The genome region of analysis (“NS” and “VP” black boxes) includes all coding regions as well as the NS2 and VP1 introns. Nucleotide position numbering begins at the NS1/2 translational start site. Gray hairpins in the genome represent terminal untranslated regions (UTRs) that were not included in this analysis.
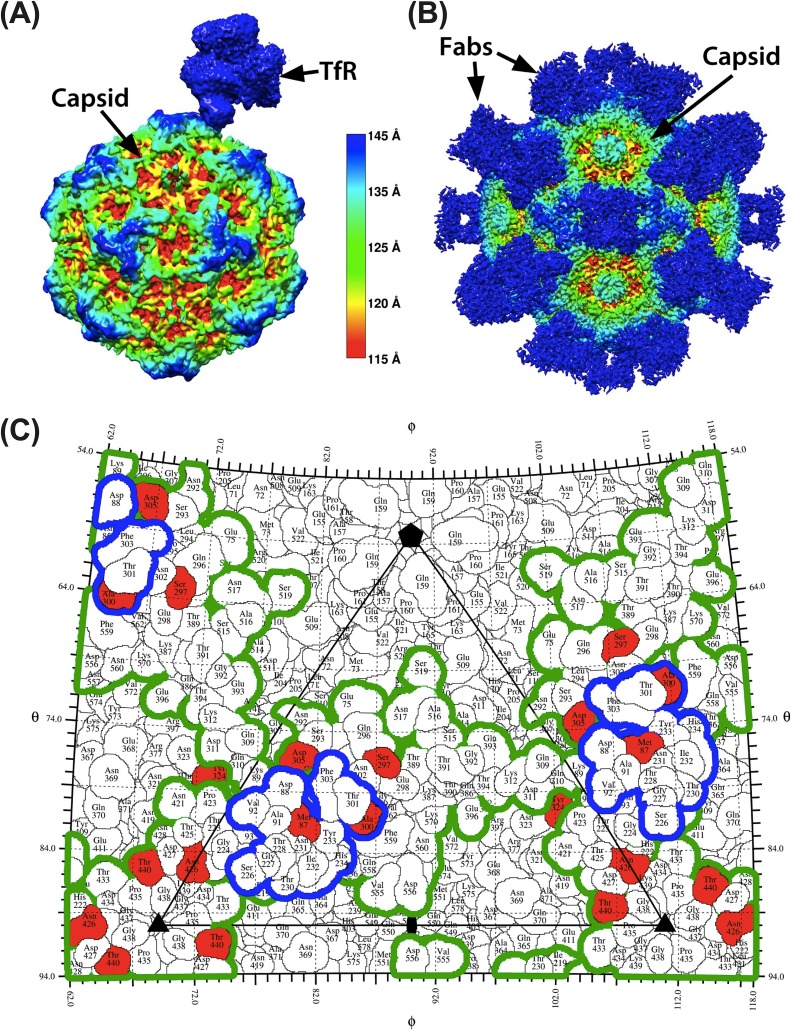
Recently determined structures showing the complex of the black-backed jackal transferrin receptor (TfR) and the CPV-2 capsid (30), or of antibody Fab-capsid costructures, highlight their binding sites (footprints) on the capsid at atomic resolution (21, 22, 31). This information was used to characterize the structural context of the capsid nonsynonymous substitutions identified in our survey (Fig. 2). Of the nonsynonymous substitutions within the VP2 gene, three altered amino acid residues that fell within the footprint of the TfR or immediately adjacent to that footprint (VP2 positions 87, 305, and 300). These residues also fell within or immediately adjacent to defined monoclonal antibody (MAb) antigen binding fragment (Fab) footprint regions (Fig. 2). All remaining naturally variable capsid surface residues fell within or immediately adjacent to the Mab Fab footprints, but they did not overlap the TfR footprint (Fig. 2). Additionally, three naturally occurring nonsynonymous mutations in VP2 altered residues located beneath the capsid surface (VP2 101, 267, and 375), perhaps resulting in indirect and relatively subtle capsid changes. The overlap of receptor and MAb Fab footprints and limited number of mutations on the surface suggests competing selective pressures of TfR and antibody interactions may have channeled and constrained the evolutionary trajectory of capsid.
FIG 2.
Structural context of natural CPV variants and their relation to TfR and MAb Fab footprints. (A and B) High-resolution structure of the transferrin receptor (A) and monoclonal antibody Fabs (B) bound to the CPV capsid colored by capsid radial distance identifies the sites of contact. (C) One asymmetric unit of CPV capsid, showing the footprint of the TfR (blue outline) and the outline of the combined footprints of eight different antibodies (green outline). Red residues varied during evolution of CPV in dogs. All structures were determined by cryoEM. See main text for references.
Less is known about the possible phenotypic effects of the mutations that occurred within the viral nonstructural genes encoding NS1, NS2, and SAT. However, all these nonstructural proteins interact with cellular proteins as demonstrated in CPV or other closely related parvoviruses (32–39), and different selective pressures may therefore exist within different host species. In NS1, most nonsynonymous mutations fell within the C-terminal region, outside the nickase and helicase regions as defined in closely related parvoviruses (40, 41) that are likely conserved in structure, amino acid sequence, and function. Further experimental mutagenesis is needed to understand the phenotypic effects of naturally occurring changes in the nonstructural genes of these viruses.
Phylogenetic analyses reveal complex patterns of interhost evolution and recombination.
To determine the evolutionary patterns of CPV at the epidemiological scale, we performed full-genome phylogenetic analyses. All full-genome CPV sequences isolated directly from dogs, raccoons, or raccoon dogs and for which isolation location and date were available were included in this analysis.
Screening for recombination in these data using the Recombination Detection Program (RDP4; default settings) (42) revealed a common topological break point within the exclusive VP1-encoding region as previously reported (43). However, as most of the putative recombination events received limited bootstrap support in the phylogenetic analysis, it is difficult to conclusively determine whether this breakpoint results from recombination, differential selective pressures on the NS and VP genes, or a combination of both factors. Whereas six potential distinct recombination events were identified that account for 26 of the 225 CPV genomes analyzed, there were discrepancies between the various detection methods, with the RDP (44), GENECONV (45), and BOOTSCAN (46) methods all failing to detect a single recombination event. Only four potential recombination events, accounting for six genomes in total, were supported by two or more of the remaining detection methods (i.e., CHIMERA [47], MAXCHI [48], SISCAN [49], and 3Seq [50]). Of the six candidate recombinant genomes, three were recent isolates from Nigeria sequenced here (CPV610, CPV614, and CPV615), with the NS region of these sequences resembling that of a 2014 Chinese isolate (GenBank accession number KT382542) but with an unknown VP region parent. The remaining three genomes were database sequences (accession numbers KM457139.1, KX774252, and KR002800), one of which (KM457139.1) has been described previously as a recombinant (43). To simplify the subsequent evolutionary analysis, the six putatively recombinant genomes were removed from the phylogenetic analysis.
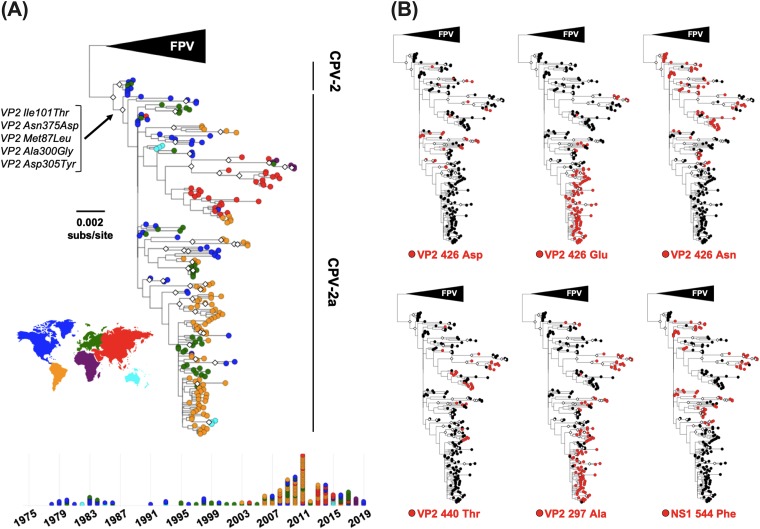
A maximum likelihood tree was then inferred from the remaining 219 nonrecombinant full-genome sequences and annotated by country of sample origin and by mutations of interest (Fig. 3). This analysis identified the five nonsynonymous changes associated with the rise and pandemic spread of CPV-2a viruses in the early 1980s (Fig. 3A) (6, 51). However, after this bifurcation the tree topology was marked by a lack of phylogenetic resolution. Poor phylogenetic support is an important issue, although one that is often overlooked in studies of CPV natural evolution or avoided by focusing on select sequences. While some small clades of high bootstrap support were apparent in the phylogeny, such as a clade of recent Chinese viruses, a single Italian isolate of Chinese origin, and several Nigerian viruses, most sequences sampled appear to be minor variants of a common “pan genome” template. In addition, many of the mutations observed are short-lived, in that they fall on tip branches, and both parallel evolution and reversion appear to be commonplace. For example, VP2 residue 426 is an Asn in FPV-like viruses, CPV-2 and CPV-2a genotypes, but antigenically distinct CPVs resulting from Asp or Glu mutations at this position have arisen and been named CPV-2b and CPV-2c, respectively. Importantly, however, these amino acid changes do not correspond to strongly supported monophyletic groups of viruses and hence likely arose multiple times after the emergence of CPV-2a (Fig. 3B). Other mutations in the capsid that likely have multiple evolutionary origins and have potential phenotypic impacts based on their proximity to the TfR and MAb Fab footprints include Ala at VP2 residue 297 and Glu at VP2 residue 440 (Fig. 3B). Additionally, a Phe at NS1 residue 544, which is often cited as a characteristic mutation among various CPV lineages (52–54), but which phenotypic relevancy has not yet been demonstrated, appears to have multiple evolutionary origins (Fig. 3B). Overall, it is difficult to estimate the relative contributions of parallel evolution, reversion, and recombination on the observed patterns of genetic homoplasy.
FIG 3.
Evolutionary relationships among CPV full-genome sequences. Sequences analyzed include all complete CPV genomes available that were isolated from dogs, raccoons, and raccoon dogs. An FPV-like virus clade was used as an outgroup, and bootstrap values of >75% are indicated with white diamonds at the nodes. (A) Phylogeny with tip shapes colored by geographic origin (continent). The branch and mutations leading to the emergence and pandemic replacement CPV-2a viruses are indicated. A timeline of samples (below the tree; colored by country as in the tree) shows the temporal and geographical origins of sequences analyzed. (B) Phylogenies annotated by mutations of interest that appear to have multiple evolutionary origins.
Extremely low levels of intrahost diversity among CPVs circulating in dogs during 40 years of virus evolution.
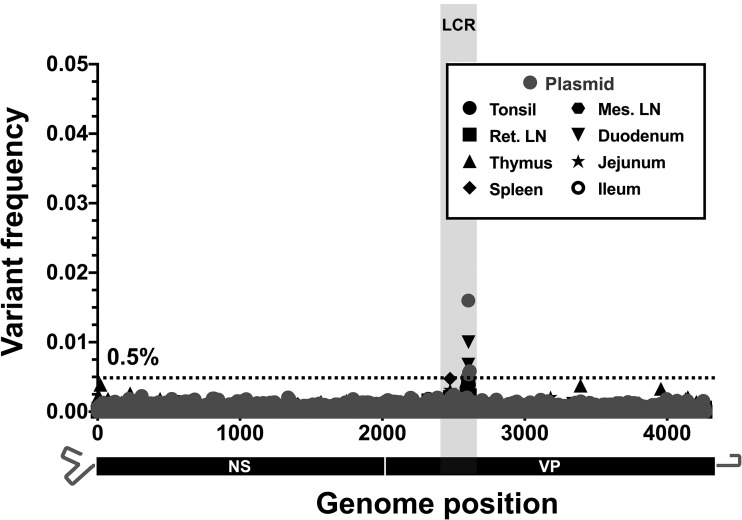
The natural virus samples collected from dogs sampled at any time during the CPV pandemic exhibited few subconsensus SNVs within the deep sequences of any viral DNA (Fig. 4). Importantly, several deep-sequence samples were collected in the late 1970s and early 1980s, around the time of the global spread of CPV-2 and its replacement by CPV-2a. Sequences from this time have been underrepresented in databases and could shed light on this important evolutionary transition. Among the 28 natural clinical samples deep sequenced from dogs (>150,000 bases covered), we observed a total of only eight subconsensus SNVs above 0.5%. Of these, only two (resulting in nonsynonymous changes of VP2 Asp375Asn and VP2 Ile555Val) were also present as mutations in the consensus sequence analysis (Fig. 1 and Table 2), and both were found at frequencies below 2% in a single viral sample (CPV48) collected in 1985 in the United States. In addition, no variant sequences or subconsensus SNVs above 0.5% were observed in viral DNA from different tissues from a naturally infected dog (Fig. 5), suggesting that tissue-specific adaptation of the virus is not a feature in the case analyzed here. Individual coverage and variant call plots for all samples collected from dogs are provided in Fig. S3A to C.
FIG 5.
Intrahost viral diversity in different tissues of a single dog during an acute CPV-2a infection. The positions and frequencies of all dominant subconsensus SNVs are displayed. Deep-sequenced plasmid (in gray) provides a baseline sequencing error rate. The low-coverage region (LCR) is indicated by gray shading, and positions in this region were omitted from variant calling analyses. Ret. LN, retropharyngeal lymph nodes; Mes. LN, mesenteric lymph nodes.
Extremely low levels of intrahost diversity among CPVs from other host species.
While CPV has spread and been maintained in the large domestic dog population of the world, CPV-related viruses infecting other hosts often show specific capsid changes that are likely associated with adaptation to the specific TfRs in those animals (15, 19). Notably, our deep sequencing of five CPV and two FPV intrahost populations from raccoons (Procyon lotor) in the United States, raccoon dogs (Nyctereutes procyonoides) in Finland, a red wolf (Canis lupus rufus) in the United States, and an Arctic (blue) fox (Vulpes lagopus) in Finland similarly revealed low levels of intrahost diversity, with a total of only three subconsensus SNVs detected above ∼0.5% (Fig. 6). All three of these subconsensus SNVs were nonsynonymous in at least one gene product, but none were at positions known to be involved in host adaptation. Individual coverage and variant call plots for all samples collected from nondog hosts are provided in Fig. S3D.
Substantial intrahost diversity is not a prerequisite for rapid host receptor-driven adaptation in cell culture.
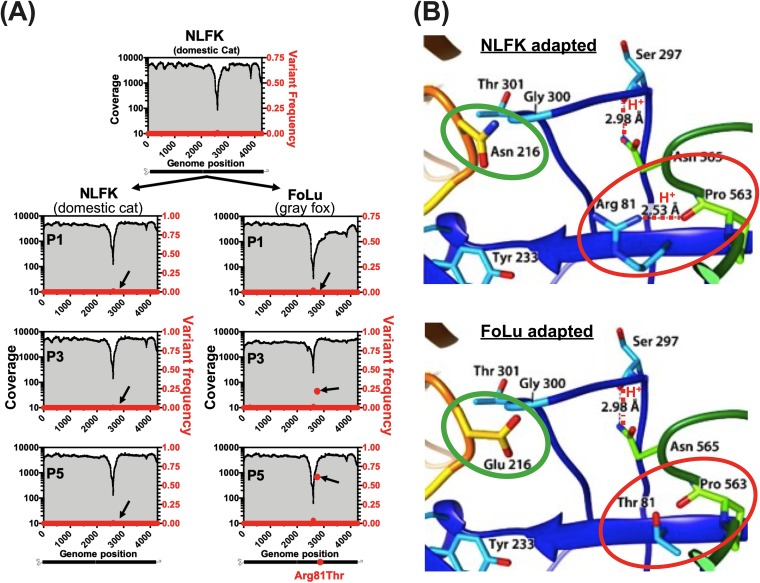
To examine the selective dynamics of the host-specific mutations in these viruses, we passaged a CPV-2a-derived virus in cells from a domestic cat (Felis catus) and from a gray fox (Urocyon cinereoargenteus). These host cells have previously been shown to select for mutations in CPV-related viruses from different sources (15, 19). The virus used was a 1984 isolate (sequence “CPV39” in this study) (6, 20), and an infectious stock was prepared from an infectious plasmid clone by three passages in feline cells in culture (55). Importantly, no subconsensus SNVs were present at frequencies above 0.5% within the virus stock (Fig. 7A). Hence, the starting populations for these experimental viruses resembled those observed in natural samples.
FIG 7.
Receptor-driven adaptation of CPV-2a during a simulated cross-species transmission event. (A) CPV-2a stock virus originating from transfection of NLFK cells with infectious plasmid (p440, sequence CPV39 in this study) was passaged simultaneously on NLFK and FoLu cell lines and deep sequenced at passage 1, 3, and 5 (P1, P3, and P5). Coverage (black lines and gray shading) and dominant SNV frequencies (red circles) are displayed. A single nonsynonymous substitution (G→C) at nucleotide position 2756 (black arrows) resulting in a VP2 R81T residue change reached majority consensus by P5 in FoLu cells only. (B) Comparison of cat (top) and fox (bottom) transferrin receptor (TfR) apical domain (orange loop) amino acid sequences and interacting viral capsid regions (blue and green loops). Residues that differentiate cat and fox TfRs are circled in green. The fox-selected capsid substitution (VP2 R81T), circled in red, results in the loss of a stabilizing hydrogen bond (red dotted line).
After five additional passages in feline cells, all SNVs remained below 0.5%. However, after three passages in fox cells, a single nonsynonymous mutation, changing VP2 residue 81 from Arg to Thr, rose to 20% of the reads. After two additional passages in fox cells, that mutation represented 60% of the sequence reads (and hence in the majority consensus sequence) (Fig. 7A). Similar results were observed in two additional experimental replicates of both NLFK and FoLu-passaged virus (data not shown). The host-specific VP2 Arg81Thr mutation has been described previously (15). It falls within a region of the capsid structure that stabilizes a loop containing VP2 residues 300 and 301, which interact directly with the TfR (Fig. 7B) (30). By removing a hydrogen bond interacting with this loop, the VP2 Arg81Thr substitution likely permits greater flexibility of the capsid. This would allow improved binding or infection through the fox TfR, which differs from the cat in a number of key residues in the two loops of the receptor apical domain that interact with the viral capsid, most notably at residue 216 (Fig. 7B, green circles).
DISCUSSION
CPV first appeared in dogs in the mid-1970s, rapidly emerged to reach pandemic proportions within a few years, and has now maintained global circulation for more than 40 years, despite the widespread use of effective vaccines and other control measures. While there have been many studies published on the sequence variation in CPV (56, 57), largely considering the capsid protein genes, we focused on the extent and pattern of intrahost genomic variation and its connections to long-term virus evolution. We show that the success of the virus in its new host was accomplished with only a limited number of long-term sustained amino acid changes and with individual hosts containing little intrahost genetic diversity.
After the initial emergence of CPV-2, and then CPV-2a, there is no evidence of global sweeps of additional CPV genotypes, and the evolutionary (i.e., phylogenetic) history of these viruses becomes much more obscure. This feature is reflected in low bootstrap support for most nodes and the limited accumulation of mutation variation despite 40 years of continuous circulation in dogs and other animals. A lack of accumulated interhost diversity may mean that most of the host adaptation in CPV was achieved early in its evolutionary history in dogs. While some later lineages have seemingly accumulated fixed mutational changes, there are several positions in the CPV genome that appear to have experienced recurrent parallel evolution or reversion, and thus are variable both within and among different clades. Many of the variable sites in the capsid protein gene are likely to be associated with antibody escape or receptor interaction or both, and it is possible that some of these mutations facilitate the fine-tuning of the virus to the specific host in question, although this will require careful experimental verification.
There are a number of possible explanations for the relatively limited accumulation of CPV interhost diversity over 40 years. The simplest is that mutations accumulate at a rate dependent on the background mutation rate of the virus, which is itself lower than the rates seen in RNA viruses (58). This effect may be compounded by the compact nature of the CPV genome (Fig. 1) and the conservation of secondary structures and/or capsid-interacting sequences within the ssDNA viral genome (59). In this model, most synonymous mutations within the CPV genome would impact multiple functions (i.e., show pleiotropy) and hence reduce viral fitness. Similarly, the extensive overlap of the TfR and Fab footprints means that mutations changing the structures of these capsid regions would likely affect both functions (21, 22, 30). It is therefore possible that only a few sites in the genome can accommodate neutral mutations.
Consistent with the observed low levels of accumulated long-term background evolution over 40 years, our deep sequencing of natural and experimental viral populations show that high intrahost diversity is not a common feature of these viruses, nor is it a specific barrier to rapid host adaptation in experimental systems (15, 19). While replication of the parvoviral DNA and other similar ssDNA viruses by the host cell DNA polymerases appears to be of lower fidelity than that seen for the replication of the host DNA, it is still of higher fidelity than that of RNA viruses (28). This difference between RNA and ssDNA virus error rates is supported by the lack of intrahost diversity observed in our samples. Similarly, our results clearly show that coinfection is not a common feature among the CPV samples collected from either dogs or other hosts over the 40 years of sampling, which differs from the results of other studies reporting coinfections between CPV strains or with FPV reported in dogs or cats (43, 60–62). However, the finding of recombinant CPV genomes indicates that coinfections occur on occasion in nature (43, 63–65). Thus, while mixed infections are not a dominant feature of CPV infections, they may still have important roles in long-term virus evolution.
Despite the evolutionary constraints associated with relatively low mutation rates and limited intrahost diversity, CPV can still rapidly adapt to acquire new host ranges and evade host humoral immunity, likely by relying on the strong and direct positive selection of relatively small numbers of mutations. For example, during adaptation to alternative host TfRs, single amino acid changes in the CPV capsid structure may directly alter the interaction of the capsid with the TfR apical domain, or the flexibility of the receptor-interacting surface of the capsid, as reported for CPV binding to the canine TfR (66, 67) and in the adaptation of CPV to the fox TfR (15). Hence, the acquisition of new host ranges may occur in a step-wise fashion, in which one or a small number of mutations enables the initial establishment in the new host, as seen here after growth on gray fox cells, and perhaps followed by a small number of additional mutations that optimize virus-host interactions. Similar dynamics may occur with antibody epitopes and CPV’s evasion of humoral immunity, and single mutations have been shown to affect the binding of monoclonal antibodies (14, 22, 68).
Taken together, our comparison of both multidecade-long global-scale and short-term intrahost evolutionary processes provides a uniquely detailed view of the evolution of an emerging virus in the context of rapid host adaptation, for which high levels of intra- and interhost diversity are not defining features. This is in contrast to the models that are typically presented for RNA viruses of similar or greater evolutionary capacity due to the low fidelity of their RNA polymerases. We therefore propose that while the waiting time for beneficial mutations in CPV is likely longer than that seen in many RNA viruses, once such mutations appear, natural selection can rapidly fix them in the population, facilitated by relatively rapid virus replication and spread. During the early emergence of CPV in dogs, there were many opportunities for fitness enhancement such that multiple advantageous mutations were successfully fixed in the population. Following this initial period of host adaptation, the relative proportion of beneficial mutations declined and, combined with the pleiotropy inherent in CPV, reduced the overall fixation rate. In sum, our findings emphasize a need to consider multiple facets of a virus’s biology, pathogenesis, and ecology, and not simply mutation rate, when considering its adaptive capabilities.
MATERIALS AND METHODS
Viral genome amplification, library preparation, and consensus sequencing.
Naturally circulating virus specimens were obtained as diagnostic fecal, intestinal material or intestinal tissue samples collected directly from infected animals and stored at −80°C. Other tissues were also used to investigate any tissue-specific mutation in an acutely infected dog (“CPV617”). The origins and collection dates of all natural samples are listed in Table 1. In addition, experimental studies were conducted with virus derived from infectious clones (55). DNA was extracted from all samples regardless of material type using the E.Z.N.A. tissue DNA kit. For experimentally passaged viruses, prior to DNA extraction, culture supernatant was treated with DNase I to remove any residual plasmid DNA not protected by a viral capsid that may have been carried over from transfection. For all samples sequenced, input CPV genome copy numbers were determined via quantitative PCR (qPCR) performed as in reference 69, using primers 5′-AAATGAAACCAGAAACCGTTGAA-3′ and 5′-TCCCGCGCTTTGTTTCC-3′ and probe (5′-ACAGTGACGACAGCAC-3′) targeting a conserved region of the NS1 gene. In most cases, 106 to 107 copies of the viral DNA were used to initiate the PCRs and sample inputs below 104 were not used, ensuring proper amplification with minimal PCR cycling and reduced any sampling effects on the detection of SNVs among samples that were deep sequenced (70). In addition, all PCRs for deep-sequenced samples were run in triplicate and pooled before library preparation to minimize individual PCR errors and sampling effects. A region of the genome spanning all major reading frames and portions of the 5′ and 3′ UTRs was amplified (see Fig. S1 for primer sequences and locations) and Q5 high-fidelity DNA polymerase (New England BioLabs) under the recommended conditions and 25 rounds of amplification. Amplification products were purified with 0.45× volume AMPure XP beads (Beckman Coulter) and 1 ng input DNA used to construct barcoded sequencing libraries with the Nextera XT kit (Illumina). Libraries were multiplexed, and sequences were determined using Miseq 2x250 Illumina sequencing. Raw sequencing reads were trimmed using BBDuk (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/) to remove all adaptors and low-quality regions from reads. The reads were then merged and mapped to a CPV-2 reference (EU659116) with two iterations using Geneious Prime v. 2019.0.4 (71), and the majority consensus sequence for each sample was determined.
Minor variant calling.
For samples that were deep sequenced, additional read processing was performed. Using BBNorm (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbnorm-guide/), reads were error corrected and normalized to target 5,000-fold coverage per site. Reads for each sample were remapped to their consensus sequence, and an additional filtering step (BAMUtil:Filt [72]) was performed to clip base miscalls at the termini of reads which often result in poor assemblies and SNV call errors. Coverage and site frequencies across the region of interest were determined using the PileupParam and ScanBamParam features in Rsamtools (73), and the most abundant minor variant frequency was called for each position.
To define the accuracy and sensitivity of our ability to detect rare subconsensus SNVs in viral samples, we performed control sequencing studies of artificially created heterogeneous virus populations. These virus populations were made from a CPV-2 plasmid (74) spiked with 25% FPV plasmid and serially diluted four times at a 1:4 ratio (Fig. S2A). An input amount of 0.2 ng of mixed template DNA (equivalent to ∼2.5 × 107 copies of the viral genome) was then prepared by PCR as for all of the natural viral samples. The spiked genomes were easily detected in a dose-dependent manner at approximately the proportions that were prepared (Fig. S2B and C).
Phylogenetic analysis.
For the full-genome CPV phylogenies, we excluded all recombinant viruses (n = 6 in total) as determined by more than two different methods in the RDP4 program and employing default settings (42). Evolutionary relationships among the remaining sequences (n = 219) were then determined using the maximum likelihood (ML) method available in the PhyML program (75), employing a general time-reversible (GTR) substitution model, gamma-distributed (Γ) rate variation among sites, and bootstrap resampling (1,000 replications). Mutations of interest were catalogued manually and annotated on tree tips using the MicroReact web-server (76).
Experimental passages.
To experimentally simulate, in vitro, a cross-species transmission event, a plasmid-derived A CPV-2b virus (p440, sequence CPV39 in this study) (55) was passaged in domestic cat (Felis catus) kidney (NLFK) and gray fox (Urocyon cinereoargenteus) lung (FoLu) cells. NLFK cells were maintained in McCoy’s 5A and Liebovitz L15 media with 5% fetal calf serum (FCS), while FoLu cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% FCS, and both cell lines were grown at 37°C and 5% CO2 atmosphere. A common virus stock was generated by transfection of plasmid DNA into NLFK cell culture using Lipofectamine 2000 (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. CPV genome copy number per microliter of virus stock was quantified to ensure adequate input populations (>1 × 108 viral genomes/μl). The stock was then used to infect individual flasks of FoLu and NLFK cells run in triplicate. For each virus passage, cells were seeded 8 to 16 h prior to infection at ∼1 × 105 cells/ml in 12.5-cm2 flasks. Prior to inoculation, growth media were removed and cells were washed twice with sterile phosphate-buffered saline (PBS). Virus (from stock or previous passage supernatant) (250 μl) was added directly to cells. The inoculated cells were then incubated for 1 h to allow infection, after which time conditioned medium was added back to the flask for a final volume of 2.5 ml. Infected cells were grown until confluent (4 to 6 days), supernatant was harvested, and viral DNA amplification, deep sequencing, and variant calling were performed on stock and passaged viruses as previously described.
Data availability.
All newly generated CPV genome consensus sequences derived from natural clinical samples have been deposited in GenBank under accession numbers MN451652 to MN451695. Raw sequence reads from deep-sequenced samples have been deposited to the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA565251. See Table 1 for details.
Supplementary Material
ACKNOWLEDGMENTS
We thank Wendy Weichert and Renee Anderson for their expert technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Antia R, Regoes RR, Koella JC, Bergstrom CT. 2003. The role of evolution in the emergence of infectious diseases. Nature 426:658–661. doi: 10.1038/nature02104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parrish CR, Holmes EC, Morens DM, Park E-C, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. 2008. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev 72:457–470. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nelson DT, Eustis SL, McAdaragh JP, Stotz I. 1979. Lesions of spontaneous canine viral enteritis. Vet Pathol 16:680–686. doi: 10.1177/030098587901600606. [DOI] [PubMed] [Google Scholar]
- 4.Robinson WF, Wilcox GE, Flower RL. 1980. Canine parvoviral disease: experimental reproduction of the enteric form with a parvovirus isolated from a case of myocarditis. Vet Pathol 17:589–599. doi: 10.1177/030098588001700508. [DOI] [PubMed] [Google Scholar]
- 5.Parrish CR. 1999. Host range relationships and the evolution of canine parvovirus. Vet Microbiol 69:29–40. doi: 10.1016/s0378-1135(99)00084-x. [DOI] [PubMed] [Google Scholar]
- 6.Parrish CR, O’Connell PH, Evermann JF, Carmichael LE. 1985. Natural variation of canine parvovirus. Science 230:1046–1048. doi: 10.1126/science.4059921. [DOI] [PubMed] [Google Scholar]
- 7.Parrish CR, Have P, Foreyt WJ, Evermann JF, Senda M, Carmichael LE. 1988. The global spread and replacement of canine parvovirus strains. J Gen Virol 69:1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- 8.Truyen U, Evermann JF, Vieler E, Parrish CR. 1996. Evolution of canine parvovirus involved loss and gain of feline host range. Virology 215:186–189. doi: 10.1006/viro.1996.0021. [DOI] [PubMed] [Google Scholar]
- 9.Pratelli A, Cavalli A, Martella V, Tempesta M, Decaro N, Carmichael LE, Buonavoglia C. 2001. Canine parvovirus (CPV) vaccination: comparison of neutralizing antibody responses in pups after inoculation with CPV2 or CPV2b modified live virus vaccine. Clin Diagn Lab Immunol 8:612–615. doi: 10.1128/CDLI.8.3.612-615.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spibey N, Greenwood NM, Sutton D, Chalmers WSK, Tarpey I. 2008. Canine parvovirus type 2 vaccine protects against virulent challenge with type 2c virus. Vet Microbiol 128:48–55. doi: 10.1016/j.vetmic.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 11.Parker JSL, Murphy WJ, Wang D, O’Brien SJ, Parrish CR. 2001. Canine and feline parvoviruses can use human or feline transferrin receptors to bind, enter, and infect cells. J Virol 75:3896–3902. doi: 10.1128/JVI.75.8.3896-3902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaway HM, Welsch K, Weichert W, Allison AB, Hafenstein SL, Huang K, Iketani S, Parrish CR. 2018. Complex and dynamic interactions between parvovirus capsids, transferrin receptors, and antibodies control cell infection and host range. J Virol 92:e00460-18. doi: 10.1128/JVI.00460-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hueffer K, Parker JSL, Weichert WS, Geisel RE, Sgro J-Y, Parrish CR. 2003. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol 77:1718–1726. doi: 10.1128/jvi.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang SF, Sgro JY, Parrish CR. 1992. Multiple amino acids in the capsid structure of canine parvovirus coordinately determine the canine host range and specific antigenic and hemagglutination properties. J Virol 66:6858–6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allison AB, Organtini LJ, Zhang S, Hafenstein SL, Holmes EC, Parrish CR. 2015. Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J Virol 90:753–767. doi: 10.1128/JVI.02636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker JS, Parrish CR. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J Virol 71:9214–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palermo LM, Hueffer K, Parrish CR. 2003. Residues in the apical domain of the feline and canine transferrin receptors control host-specific binding and cell infection of canine and feline parvoviruses. J Virol 77:8915–8923. doi: 10.1128/jvi.77.16.8915-8923.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison AB, Harbison CE, Pagan I, Stucker KM, Kaelber JT, Brown JD, Ruder MG, Keel MK, Dubovi EJ, Holmes EC, Parrish CR. 2012. Role of multiple hosts in the cross-species transmission and emergence of a pandemic parvovirus. J Virol 86:865–872. doi: 10.1128/JVI.06187-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allison AB, Kohler DJ, Ortega A, Hoover EA, Grove DM, Holmes EC, Parrish CR. 2014. Host-specific parvovirus evolution in nature is recapitulated by in vitro adaptation to different carnivore species. PLoS Pathog 10:e1004475. doi: 10.1371/journal.ppat.1004475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol 65:6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafenstein S, Bowman VD, Sun T, Nelson CDS, Palermo LM, Chipman PR, Battisti AJ, Parrish CR, Rossmann MG. 2009. Structural comparison of different antibodies interacting with parvovirus capsids. J Virol 83:5556–5566. doi: 10.1128/JVI.02532-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strassheim ML, Gruenberg A, Veijalainen P, Sgro J-Y, Parrish CR. 1994. Two dominant neutralizing antigenic determinants of canine parvovirus are found on the threefold spike of the virus capsid. Virology 198:175–184. doi: 10.1006/viro.1994.1020. [DOI] [PubMed] [Google Scholar]
- 23.Callaway HM, Feng KH, Lee DW, Allison AB, Pinard M, McKenna R, Agbandje-McKenna M, Hafenstein S, Parrish CR. 2017. Parvovirus capsid structures required for infection: mutations controlling receptor recognition and protease cleavages. J Virol 91:e01871-16. doi: 10.1128/JVI.01871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirier EZ, Vignuzzi M. 2017. Virus population dynamics during infection. Curr Opin Virol 23:82–87. doi: 10.1016/j.coviro.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11:327–336. doi: 10.1038/nrmicro3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holmes EC. 2009. The evolution and emergence of RNA viruses, 1st ed Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 27.Shackelton LA, Parrish CR, Truyen U, Holmes EC. 2005. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc Natl Acad Sci U S A 102:379–384. doi: 10.1073/pnas.0406765102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duffy S, Shackelton LA, Holmes EC. 2008. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet 9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- 29.Voorhees IEH, Lee H, Allison AB, Lopez-Astacio R, Goodman LB, Oyesola OO, Omobowale O, Fagbohun O, Dubovi EJ, Hafenstein SL, Holmes EC, Parrish CR. 2019. Limited intra-host diversity and background evolution accompany 40 years of canine parvovirus host adaptation and spread. bioRxiv 10.1101/714683. [DOI] [PMC free article] [PubMed]
- 30.Lee H, Callaway HM, Cifuente JO, Bator CM, Parrish CR, Hafenstein SL. 2019. Transferrin receptor binds virus capsid with dynamic motion. Proc Natl Acad Sci U S A 91:20462–20471. doi: 10.1073/pnas.1904918116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Organtini LJ, Lee H, Iketani S, Huang K, Ashley RE, Makhov AM, Conway JF, Parrish CR, Hafenstein S. 2016. Near-atomic resolution structure of a highly neutralizing Fab bound to canine parvovirus. J Virol 90:9733–9742. doi: 10.1128/JVI.01112-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta SK, Sahoo AP, Rosh N, Gandham RK, Saxena L, Singh AK, Harish D, Tiwari AK. 2016. Canine parvovirus NS1 induced apoptosis involves mitochondria, accumulation of reactive oxygen species and activation of caspases. Virus Res 213:46–61. doi: 10.1016/j.virusres.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Wang D, Yuan W, Davis I, Parrish CR. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240:273–281. doi: 10.1006/viro.1997.8946. [DOI] [PubMed] [Google Scholar]
- 34.Zádori Z, Szelei J, Tijssen P. 2005. SAT: a late NS protein of porcine parvovirus. J Virol 79:13129–13138. doi: 10.1128/JVI.79.20.13129-13138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christensen J, Cotmore SF, Tattersall P. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J Virol 69:5422–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christensen J, Cotmore SF, Tattersall P. 1997. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol 71:1405–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young PJ, Jensen KT, Burger LR, Pintel DJ, Lorson CL. 2002. Minute virus of mice small nonstructural protein NS2 interacts and colocalizes with the Smn protein. J Virol 76:6364–6369. doi: 10.1128/jvi.76.12.6364-6369.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller CL, Pintel DJ. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J Virol 76:3257–3266. doi: 10.1128/jvi.76.7.3257-3266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brockhaus K, Plaza S, Pintel DJ, Rommelaere J, Salomé N. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J Virol 70:7527–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tewary SK, Liang L, Lin Z, Lynn A, Cotmore SF, Tattersall P, Zhao H, Tang L. 2015. Structures of minute virus of mice replication initiator protein N-terminal domain: insights into DNA nicking and origin binding. Virology 476:61–71. doi: 10.1016/j.virol.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nüesch JP, Cotmore SF, Tattersall P. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122–135. doi: 10.1006/viro.1995.1236. [DOI] [PubMed] [Google Scholar]
- 42.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pérez R, Calleros L, Marandino A, Sarute N, Iraola G, Grecco S, Blanc H, Vignuzzi M, Isakov O, Shomron N, Carrau L, Hernández M, Francia L, Sosa K, Tomás G, Panzera Y. 2014. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS One 9:e111779. doi: 10.1371/journal.pone.0111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin D, Rybicki E. 2000. RDP: detection of recombination amongst aligned sequences. Bioinformatics 16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- 45.Padidam M, Sawyer S, Fauquet CM. 1999. Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- 46.Martin DP, Posada D, Crandall KA, Williamson C. 2005. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- 47.Posada D, Crandall KA. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA 98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JM. 1992. Analyzing the mosaic structure of genes. J Mol Evol 34:126–129. doi: 10.1007/bf00182389. [DOI] [PubMed] [Google Scholar]
- 49.Gibbs MJ, Armstrong JS, Gibbs AJ. 2000. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- 50.Boni MF, Posada D, Feldman MW. 2007. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parrish CR, Aquadro CF, Carmichael LE. 1988. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink, and raccoon parvoviruses. Virology 166:293–307. doi: 10.1016/0042-6822(88)90500-4. [DOI] [PubMed] [Google Scholar]
- 52.Grecco S, Iraola G, Decaro N, Alfieri A, Alfieri A, Gallo Calderón M, da Silva AP, Name D, Aldaz J, Calleros L, Marandino A, Tomás G, Maya L, Francia L, Panzera Y, Pérez R. 2018. Inter- and intracontinental migrations and local differentiation have shaped the contemporary epidemiological landscape of canine parvovirus in South America. Virus Evol 4:vey011. doi: 10.1093/ve/vey011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mira F, Purpari G, Di Bella S, Colaianni ML, Schirò G, Chiaramonte G, Gucciardi F, Pisano P, Lastra A, Decaro N, Guercio A. 29 June 2019. Spreading of canine parvovirus type 2c mutants of Asian origin in southern Italy. Transbound Emerg Dis doi: 10.1111/tbed.13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stucker KM, Pagan I, Cifuente JO, Kaelber JT, Lillie TD, Hafenstein S, Holmes EC, Parrish CR. 2012. The role of evolutionary intermediates in the host adaptation of canine parvovirus. J Virol 86:1514–1521. doi: 10.1128/JVI.06222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Organtini LJ, Allison AB, Lukk T, Parrish CR, Hafenstein S. 2015. Global displacement of canine parvovirus by a host-adapted variant: structural comparison between pandemic viruses with distinct host ranges. J Virol 89:1909–1912. doi: 10.1128/JVI.02611-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li C, Tang J, Chen Z, Niu G, Liu G. 2019. A divergent canine parvovirus type 2c (CPV-2c) isolate circulating in China. Infect Genet Evol 73:242–247. doi: 10.1016/j.meegid.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 57.de Oliveira PSB, Cargnelutti JF, Masuda EK, Weiblen R, Flores EF. 2019. New variants of canine parvovirus in dogs in southern Brazil. Arch Virol 164:1361–1369. doi: 10.1007/s00705-019-04198-w. [DOI] [PubMed] [Google Scholar]
- 58.Holmes EC, Dudas G, Rambaut A, Andersen KG. 2016. The evolution of Ebola virus: insights from the 2013–2016 epidemic. Nature 538:193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chapman MS, Rossmann MG. 1995. Single-stranded DNA-protein interactions in canine parvovirus. Structure 3:151–162. doi: 10.1016/s0969-2126(01)00146-0. [DOI] [PubMed] [Google Scholar]
- 60.Battilani M, Gallina L, Vaccari F, Morganti L. 2007. Co-infection with multiple variants of canine parvovirus type 2 (CPV-2). Vet Res Commun 31(Suppl 1):209–212. doi: 10.1007/s11259-007-0007-6. [DOI] [PubMed] [Google Scholar]
- 61.Battilani M, Balboni A, Ustulin M, Giunti M, Scagliarini A, Prosperi S. 2011. Genetic complexity and multiple infections with more parvovirus species in naturally infected cats. Vet Res 42:43. doi: 10.1186/1297-9716-42-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Balboni A, Bassi F, De Arcangeli S, Zobba R, Dedola C, Alberti A, Battilani M. 2018. Molecular analysis of carnivore protoparvovirus detected in white blood cells of naturally infected cats. BMC Vet Res 14:41. doi: 10.1186/s12917-018-1356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mochizuki M, Ohshima T, Une Y, Yachi A. 2008. Recombination between vaccine and field strains of canine parvovirus is revealed by isolation of virus in canine and feline cell cultures. J Vet Med Sci 70:1305–1314. doi: 10.1292/jvms.70.1305. [DOI] [PubMed] [Google Scholar]
- 64.Wang J, Cheng S, Yi L, Cheng Y, Yang S, Xu H, Zhao H, Yan X, Wu H. 2012. Evidence for natural recombination between mink enteritis virus and canine parvovirus. Virol J 9:252. doi: 10.1186/1743-422X-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohshima T, Mochizuki M. 2009. Evidence for recombination between feline panleukopenia virus and canine parvovirus type 2. J Vet Med Sci 71:403–408. doi: 10.1292/jvms.71.403. [DOI] [PubMed] [Google Scholar]
- 66.Govindasamy L, Hueffer K, Parrish CR, Agbandje-McKenna M. 2003. Structures of host range-controlling regions of the capsids of canine and feline parvoviruses and mutants. J Virol 77:12211–12221. doi: 10.1128/jvi.77.22.12211-12221.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llamas-Saiz AL, Agbandje-McKenna M, Parker JS, Wahid AT, Parrish CR, Rossmann MG. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65–71. doi: 10.1006/viro.1996.0575. [DOI] [PubMed] [Google Scholar]
- 68.Parrish CR, Carmichael LE. 1983. Antigenic structure and variation of canine parvovirus type-2, feline panleukopenia virus, and mink enteritis virus. Virology 129:401–414. doi: 10.1016/0042-6822(83)90179-4. [DOI] [PubMed] [Google Scholar]
- 69.Hoelzer K, Shackelton LA, Holmes EC, Parrish CR. 2008. Within-host genetic diversity of endemic and emerging parvoviruses of dogs and cats. J Virol 82:11096–11105. doi: 10.1128/JVI.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McCrone JT, Lauring AS. 2016. Measurements of intrahost viral diversity are extremely sensitive to systematic errors in variant calling. J Virol 90:6884–6895. doi: 10.1128/JVI.00667-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jun G, Wing MK, Abecasis GR, Kang HM. 2015. An efficient and scalable analysis framework for variant extraction and refinement from population scale DNA sequence data. Genome Res 25:918–925. doi: 10.1101/gr.176552.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morgan M, Pagès H, Obenchain V, Hayden N. 2019. .Rsamtools: binary alignment (BAM), FASTA, variant call (BCF), and tabix file import.
- 74.Parrish CR. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195–205. doi: 10.1016/0042-6822(91)90132-u. [DOI] [PubMed] [Google Scholar]
- 75.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 76.Argimón S, Abudahab K, Goater RJE, Fedosejev A, Bhai J, Glasner C, Feil EJ, Holden MTG, Yeats CA, Grundmann H, Spratt BG, Aanensen DM. 2016. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genom 2:e000093. doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly generated CPV genome consensus sequences derived from natural clinical samples have been deposited in GenBank under accession numbers MN451652 to MN451695. Raw sequence reads from deep-sequenced samples have been deposited to the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA565251. See Table 1 for details.