Influenza epidemics are caused by influenza A and influenza B viruses (IAV and IBV, respectively). IBV causes substantial disease; however, it is far less studied than IAV. While IAV originates from animal reservoirs, IBV circulates in humans only. Virus spread requires that the viral hemagglutinin (HA) is active and sufficiently stable in human airways. We resolve here how these mechanisms differ between IBV and IAV. Whereas human IAVs rely on one particular protease for HA activation, this is not the case for IBV. Superior activation of IBV by several proteases should enhance shedding of infectious particles. IBV HA exhibits acid stability and a preference for 33°C, indicating pronounced adaptation to the human upper airways, where the pH is mildly acidic and a cooler temperature exists. These adaptive features are rationalized by the long existence of IBV in humans and may have broader relevance for understanding the biology and evolution of respiratory viruses.
KEYWORDS: acid stability, airway proteases, hemagglutinin, host adaptation, influenza A virus, influenza B virus, membrane fusion, temperature
ABSTRACT
Influenza A virus (IAV) and influenza B virus (IBV) cause yearly epidemics with significant morbidity and mortality. When zoonotic IAVs enter the human population, the viral hemagglutinin (HA) requires adaptation to achieve sustained virus transmission. In contrast, IBV has been circulating in humans, its only host, for a long period of time. Whether this entailed adaptation of IBV HA to the human airways is unknown. To address this question, we compared two seasonal IAVs (A/H1N1 and A/H3N2) and two IBVs (B/Victoria and B/Yamagata lineages) with regard to host-dependent activity of HA as the mediator of membrane fusion during viral entry. We first investigated proteolytic activation of HA by covering all type II transmembrane serine protease (TTSP) and kallikrein enzymes, many of which proved to be present in human respiratory epithelium. The IBV HA0 precursor is cleaved by a broader panel of TTSPs and activated with much higher efficiency than IAV HA0. Accordingly, knockdown of a single protease, TMPRSS2, abrogated spread of IAV but not IBV in human respiratory epithelial cells. Second, the HA fusion pH values proved similar for IBV and human-adapted IAVs (with one exception being the HA of 1918 IAV). Third, IBV HA exhibited higher expression at 33°C, a temperature required for membrane fusion by B/Victoria HA. This indicates pronounced adaptation of IBV HA to the mildly acidic pH and cooler temperature of human upper airways. These distinct and intrinsic features of IBV HA are compatible with extensive host adaptation during prolonged circulation of this respiratory virus in the human population.
IMPORTANCE Influenza epidemics are caused by influenza A and influenza B viruses (IAV and IBV, respectively). IBV causes substantial disease; however, it is far less studied than IAV. While IAV originates from animal reservoirs, IBV circulates in humans only. Virus spread requires that the viral hemagglutinin (HA) is active and sufficiently stable in human airways. We resolve here how these mechanisms differ between IBV and IAV. Whereas human IAVs rely on one particular protease for HA activation, this is not the case for IBV. Superior activation of IBV by several proteases should enhance shedding of infectious particles. IBV HA exhibits acid stability and a preference for 33°C, indicating pronounced adaptation to the human upper airways, where the pH is mildly acidic and a cooler temperature exists. These adaptive features are rationalized by the long existence of IBV in humans and may have broader relevance for understanding the biology and evolution of respiratory viruses.
INTRODUCTION
The global burden of seasonal influenza is estimated at 3 to 5 million severe cases (1) and 290,000 to 650,000 fatalities per year (2). In addition, influenza A virus (IAV) causes sporadic pandemics with an even more severe impact on public health. Influenza B virus (IBV) does not cause pandemics but is responsible for a significant proportion of seasonal influenza (3). IBV may account for 22 to 44% of influenza deaths in children (3), and elderly persons are also at high risk (4). Between 1997 and 2009, IBV was the predominant cause of influenza-associated death in 4 out of 12 seasons (5). It was estimated that IAV and IBV diverged approximately 4,000 years ago (6). Current IBV strains fall into two lineages, B/Victoria and B/Yamagata (B/Vic and B/Yam, respectively), which diverged about 40 years ago and are now cocirculating globally (7). Gene reassortment between the two lineages is common yet not seen for the hemagglutinin (HA), PB1, and PB2 gene segments (7). Whereas IAV is widespread in birds and mammals (8), IBV is restricted to humans, with no sustained animal reservoir (9) and only rare spillover events (10). When a zoonotic IAV enters the human population, adaptive changes in multiple viral proteins are required to achieve sustained human-to-human transmissibility (8). Which adaptive features were acquired by IBV as a result of its long existence in humans has hardly been investigated (9).
One major protein involved in this host adaptation is the viral HA, the mediator of viral entry (11, 12). HA binds to sialylated glycans on the cell surface to enable virus uptake by endocytosis. Next, HA mediates fusion of the viral envelope and endosomal membrane, allowing release of the viral genome into the cytoplasm. Membrane fusion occurs when virus-containing endosomes mature from early (pH ∼6) to late (pH ∼5) endosomes, providing the low-pH trigger for drastic refolding of HA and expulsion of its fusion peptide (13). To gain membrane fusion competence, HA relies on host cell proteases for posttranslational cleavage of the HA0 precursor into two polypeptides, HA1 and HA2, a process termed priming or activation (14). In human IAV and IBV strains, HA0 has a monobasic (single arginine) cleavage site that is recognized by trypsin-like serine proteases. Understanding the protease dependency of HA might reveal biological differences between IAV and IBV and direct drug concepts to target these proteases for influenza therapy (15, 16). A series of proteases have been proposed for IAV (reviewed by Garten et al. [17] and Galloway et al. [18]), but for IBV hardly any information is available. The type II transmembrane serine protease (TTSP) TMPRSS2 (transmembrane protease serine 2) is an activator of IAV HA0 in cell culture and essential for IAV replication in mice, at least for some IAV subtypes (19–22). In humans, a gene polymorphism leading to higher TMPRSS2 expression was correlated with the risk for developing severe influenza (23). In cell culture, IAV can also be activated by some other TTSPs and kallikreins, but which of these support(s) spread of IAV in human airways remains to be demonstrated (reviewed in references 15 and 24). Regarding IBV, TMPRSS2 was shown to cleave IBV HA0 (16) and mediate virus spread in some human airway cell culture models (25); however, this protease appeared dispensable for IBV pathogenesis in mice (26). Hence, the protease recognition profile of IBV HA is largely unknown.
Two other adaptive features of HA are related to its pH and temperature dependence. Again, data for IBV are very limited. Successful virus replication and transmission require a balance between the low pH that triggers HA refolding and membrane fusion during viral entry, and acid stability of progeny virus in the respiratory tract and environment (27, 28). Extracellular virions are sensitive to inactivation in mildly acidic parts of the upper respiratory tract (URT) like the nasal cavity (27, 29, 30). The acid stability of HA appears to increase when a zoonotic IAV enters the human population and evolves into a strain with human-to-human transmissibility (31, 32). Studies in ferrets showed that increased IAV HA acid stability contributes to airborne transmissibility (31, 33, 34). In addition, human airways exhibit a temperature gradient, from ∼30 to 32°C in the nasal mucosa (35), ∼32°C in the upper trachea, and ∼36°C in the bronchi (36). Avian IAVs, adapted to the temperature of the avian enteric tract (40°C), show restricted growth at cooler temperatures (∼32°C). Whereas the temperature dependency is fairly understood for the PB2 subunit of the viral polymerase (37), this is not the case for the viral surface glycoproteins, HA and neuraminidase (NA) (38). It is conceivable that HA proteins of IBV or human IAV might show intrinsic adaptation to the temperature of human airways, including the cooler temperature of the URT.
To understand how these human airway-specific factors may influence the membrane fusion activity of HA, we here compared the HA proteins of two seasonal human IAVs and two IBVs. Their proteolytic activation was analyzed by covering all members of the human TTSP (39) and kallikrein (KLK) families (40). We demonstrate that IBV HA exhibits much more efficient activation by a broad range of airway proteases, explaining why TMPRSS2 alone was required for spread of IAV but not IBV in human respiratory epithelial cells. Whereas the human IAV and two IBV HA proteins showed similar low-pH dependence to trigger membrane fusion, a distinction was seen in terms of temperature dependence, with IBV HA having a clear preference for 33°C. For the IBV HA of the B/Victoria lineage, this cooler temperature was required to achieve membrane fusion. We propose that these distinct and intrinsic properties of IBV HA might reflect extensive adaptation of IBV to the human respiratory tract.
(This article was submitted to an online preprint archive [41].)
RESULTS
Human lung tissue and airway epithelial cell lines are rich in TTSP and KLK enzymes.
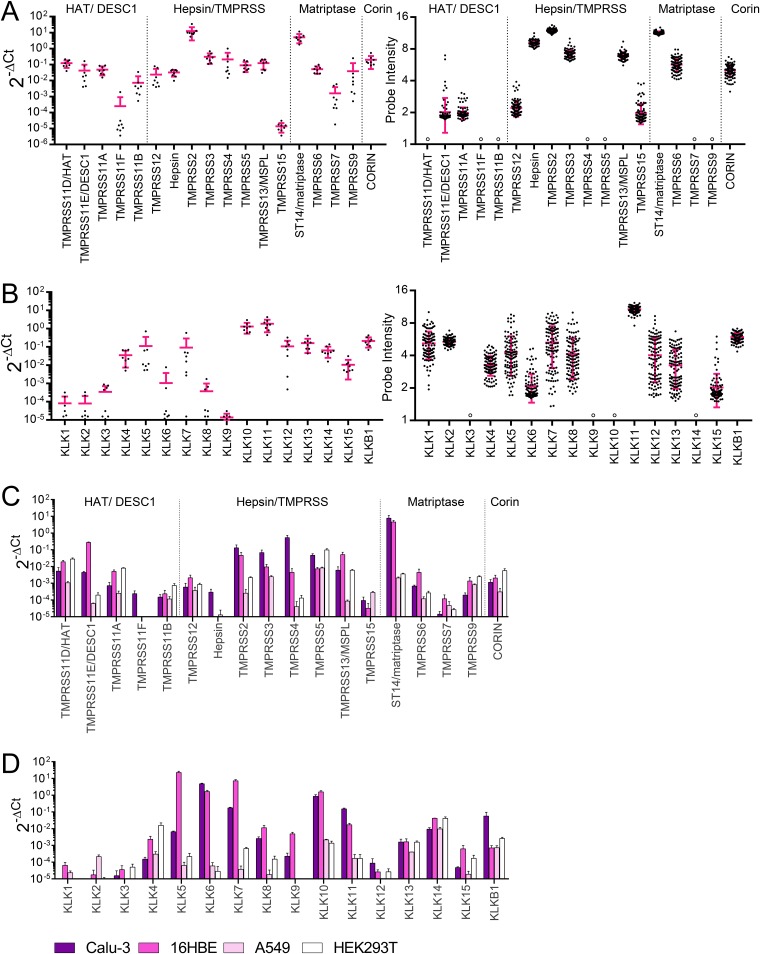
Before investigating which TTSPs or KLKs are involved in HA activation of IAV or IBV, we assessed expression of these proteases in human airways. Their mRNA levels were quantified in tissue samples of healthy human lungs (from eight different donors) (Fig. 1A and B) and three continuous cell lines (Calu-3, 16HBE, and A549) derived from human airways (Fig. 1C and D). Lung tissue was shown to express abundant levels of TMPRSS2 and ST14 (matriptase). All other TTSPs showed high and comparable transcript levels, except for TMPRSS11F and TMPRSS15 which were present at lower levels (Fig. 1A, left). These results agreed with microarray data for 108 healthy human lungs, retrieved from the Gene Expression Omnibus (GEO) database (Fig. 1A, right). Human lung tissue contained high mRNA levels for several KLKs, but these showed more variable expression than the TTSPs (compare right panels of Fig. 1A and B). Calu-3 and 16HBE cells (Fig. 1C and D) showed abundant expression of ST14 (matriptase), and Calu-3 cells also contained high levels of TMPRSS2, TMPRSS3, and TMPRSS4. Both cell lines expressed several KLK transcripts. In A549 and HEK293T cells, expression of these proteases was generally lower. As expected, human lung tissue and the four cell lines abundantly expressed FURIN, which recognizes multibasic HA0 cleavage sites of highly pathogenic avian IAVs (42) (data not shown).
FIG 1.
Human lung tissue and airway epithelial cell lines are rich in TTSP and KLK enzymes. TTSP (A) and KLK (B) transcript levels in healthy human lung, measured by RT-qPCR on samples from eight different donors (left) or retrieved from the NCBI GEO database (GEO accession number GSE47460) (right) are shown. Black, individual data points; pink, SEM; open circle, no data available. (C and D) Transcript levels in human airway epithelial Calu-3, 16HBE, and A549 cells and human embryonic kidney HEK293T cells. The four TTSP subfamilies are indicated at the top of the graphs.
IBV HA0 is efficiently cleaved by a broad range of TTSPs.
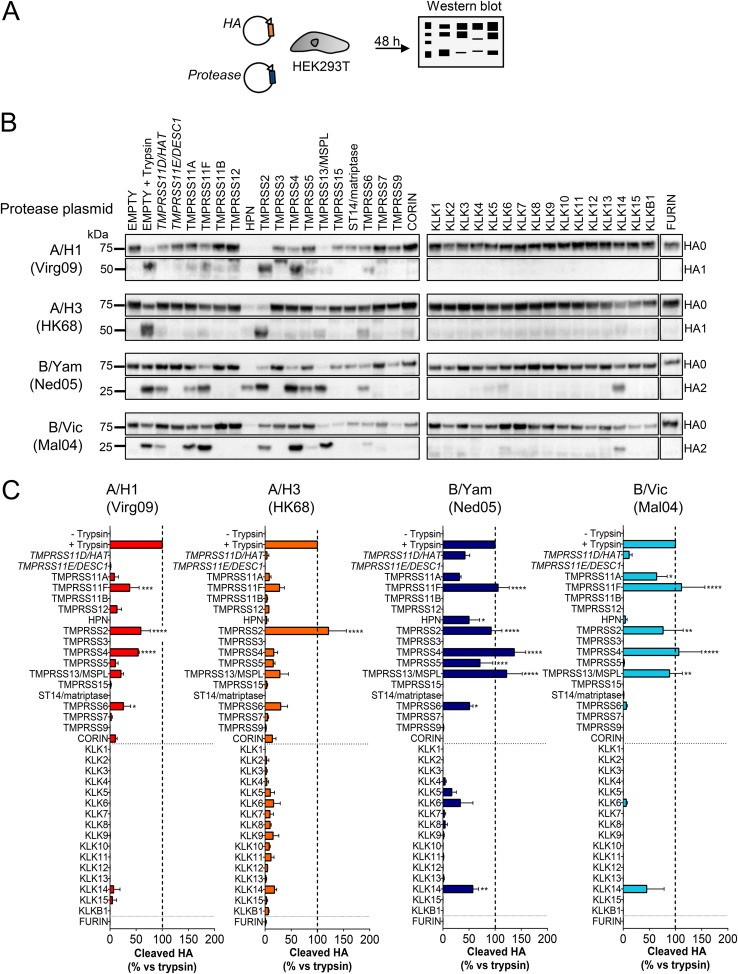
To investigate which of these proteases can activate the HA proteins of IAV or IBV, expression plasmids were generated for all 18 human TTSPs and 16 KLKs bearing a C-terminal FLAG tag (for details, see Table S1 in the supplemental material). When we analyzed expression in transfected cells, most proteases showed a protein level similar to that of TMPRSS2, regarded as the reference (Fig. S1C). For TMPRSS11B, TMPRSS12, TMPRSS7, and KLK13, expression was 3-fold lower. For TMPRSS11D/HAT and TMPRSS11E/DESC1 expression was very weak, meaning that the results from the HA cleavage assay are an underestimate for these two proteases. In this assay, the proteases were coexpressed with the HAs from four seasonal IAVs (A/H1 and A/H3) and IBVs (B/Yam and B/Vic) (Fig. 2A). HA0 cleavage was assessed by Western blotting for the HA1 or HA2 cleavage product, depending on which epitope was recognized by the anti-HA antibody. The percent cleaved HA was calculated relative to that of the trypsin control, consisting of cells transfected with HA plus an empty plasmid and briefly exposed to exogenous trypsin.
FIG 2.
IBV HA0 is efficiently cleaved by a broad range of TTSPs. (A) Experiment setup. HEK293T cells were transfected with two plasmids, one encoding IAV or IBV HA and one encoding the indicated TTSP, KLK, or furin. The HA cleavage state was determined at 48 h posttransfection. (B) Representative Western blots showing the bands of uncleaved HA0 and cleaved HA1 or HA2. The trypsin controls (second lanes on each row) consisted of cells transfected with HA and a protease-lacking empty plasmid and exposed to trypsin for 15 min just before cell extraction. (C) Quantitative data for TTSP- and KLK-mediated HA0 cleavage. For each HA, the intensity of the HA1 or HA2 band was normalized to that of clathrin, and the percent cleaved HA (mean ± SEM; N = 3) was expressed relative to that of the trypsin control. P values for results versus the no-trypsin sample are indicated as follows: *, ≤0.05; **, ≤0.01; ***, ≤0.001; ****, ≤0.0001 (ordinary one-way ANOVA, followed by Dunnett’s test).
TMPRSS2 was the only protease that efficiently cleaved all four IAV/IBV HA0 proteins tested. It was the only protease with high activity on A/H3 HA0 (Fig. 2B and C), while A/H1 HA0 was equally well cleaved by TMPRSS4. On the other hand, the two IBV HA0 proteins were efficiently processed by four TTSPs (Fig. 2B and C), namely, TMPRSS11F, TMPRSS2, TMPRSS4, and TMPRSS13/MSPL. In addition, IBV HA0 was cleaved by TMPRSS11A and TMPRSS11D/HAT, yet low expression of the latter protease (see the paragraph above) precluded conclusions on cleavage efficiency. B/Yam HA0 was also recognized by TMPRSS5 and TMPRSS6. One TTSP, namely, hepsin, generated weak bands for both HA0 and HA1/HA2 (Fig. 2B), indicating HA degradation as observed previously (43). Finally, none of the kallikreins was found to be a broad HA activator. KLK14 cleaved both IBV proteins, and KLK5 and KLK6 cleaved B/Yam HA0 with low efficacy. In summary, compared to the IAV counterparts, the two IBV HA0 proteins that we studied were demonstrated to use a broad panel of TTSPs for efficient proteolytic cleavage.
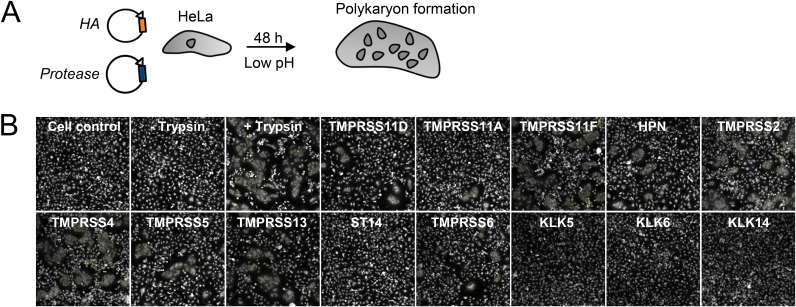
All HA0-cleaving TTSPs generate membrane fusion-competent IBV HA.
We next examined whether HA0 cleavage translates into HA activation. We first employed a polykaryon assay (Fig. 3A) in which HeLa cells that coexpress HA and protease are exposed to acidic pH to trigger HA-mediated cell-cell fusion. IBV HA generated abundant polykaryons (Fig. 3B) when coexpressed with TMPRSS11F, TMPRSS2, TMPRSS4, and TMPRSS13/MSPL, consistent with efficient cleavage by these four TTSPs in the Western blot assay (see above). A lower number of polykaryons was seen for TMPRSS11D/HAT, TMPRSS11A, hepsin, TMPRSS5, and TMPRSS6. All of these TTSPs thus generate a fusion-competent HA1-HA2 protein, meaning that they cleave IBV HA0 at the correct position. No polykaryons were formed in cells expressing KLK5, KLK6, or KLK14 (Fig. 3B). For KLK5 and KLK6, the levels of cleaved HA (Fig. 2) were probably too low to induce membrane fusion. KLK14 likely cleaves the IBV HA0 protein at an incorrect position since the HA2 product generated by KLK14 migrated slightly more slowly (Fig. 2B) than that produced by trypsin or an activating TTSP.
FIG 3.
TTSP cleavage generates fusion-competent IBV HA. (A) Experiment setup. HeLa cells expressing IBV (B/Yam) HA plus a TTSP or KLK protease were exposed to an acidic buffer 0.1 units below the fusion pH. (B) Representative photographs showing polykaryon formation in cells expressing IBV (B/Yam) HA. Polykaryon formation was similar in cells expressing B/Vic HA (data not shown). The trypsin control (third photograph) received an empty (i.e., protease-lacking) plasmid and underwent HA activation by a 15- min exposure to trypsin just before the acidic pulse.
IBV shows superior TTSP activation for viral entry.
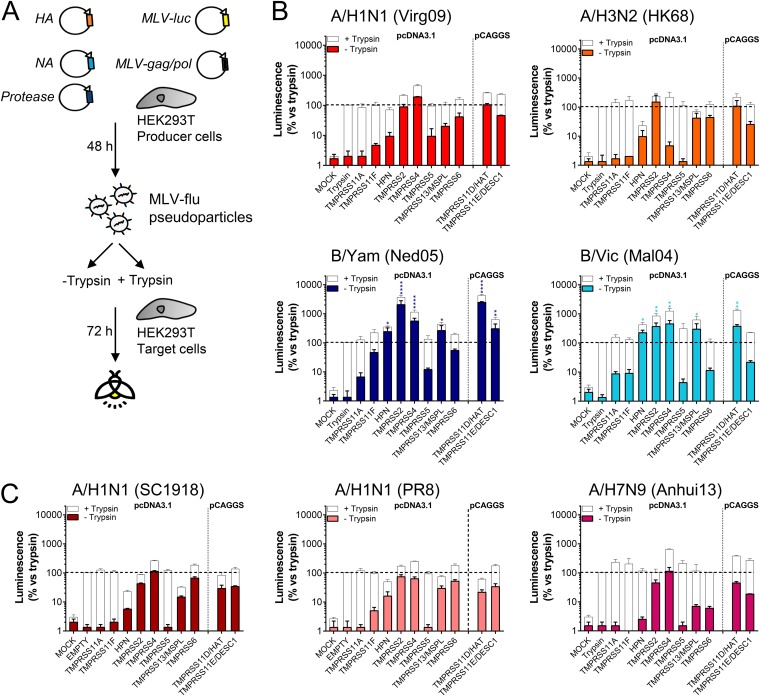
To verify that the TTSPs activate HA for membrane fusion during viral entry, we employed a retroviral pseudotyping system (Fig. 4A). Each HA was combined with its cognate NA to ensure efficient particle release (44–46). Pseudoparticles were produced in the presence or absence of TTSPs, treated with PBS or trypsin, and then examined for their ability to transduce target cells (Fig. 4A). In Fig. 4B and C, the colored bars show the transduction efficiencies of TTSP-activated particles, whereas the white bars show total particle infectivity levels upon additional treatment with trypsin.
FIG 4.
Pseudoparticles carrying IBV HA are efficiently activated by different TTSPs. (A) Experiment setup. HEK293T producer cells were transfected with plasmids encoding the HA and NA from IAV or IBV, MLV-backbone, MLV-luciferase reporter, and a TTSP enzyme. The produced pseudoparticles were left untreated (to assess TTSP-induced infectivity) or were secondarily treated with trypsin (to measure total particle infectivity). After HEK293T target cells were transduced, luminescence was measured at day 3 p.i. (B and C) Transduction efficiency (relative to that of the trypsin control) of TTSP-activated pseudoparticles tested as such (in color) or after additional trypsin treatment (in white). Data are the means ± SEM (n = 3 with triplicate readouts). P values for results versus those with the trypsin control are indicated as follows: *, ≤0.05; ***, ≤0.001; ****, ≤0.0001 (Kruskal-Wallis followed by Dunnett’s test).
For the two seasonal IAVs, pseudoparticles produced in the presence of TMPRSS2 and TMPRSS11D/HAT (now generated from a pCAGGS-based plasmid to bypass low expression [see above]) transduced cells with the same efficiency as trypsin-treated control particles (indicated with dashed lines in Fig. 4B and C). Particles carrying A/H1 HA, but not A/H3 HA, were also fully activated by TMPRSS4. Accordingly, transduction efficiency by these particles was only slightly increased when they were treated with trypsin. In sharp contrast, IBV pseudotypes produced in the presence of hepsin, TMPRSS2, TMPRSS4, TMPRSS13/MSPL, TMPRSS11D/HAT, and TMPRSS11E/DESC1 (for B/Yam) transduced cells with markedly higher efficiency than control particles activated by trypsin (Fig. 4B). For instance, B/Yam particles activated by TMPRSS2 or TMPRSS11D/HAT generated approximately 2,000-fold-higher signals than the trypsin control. This superior activation of the IBV particles was also evident from the observation that subsequent trypsin treatment gave a lower signal increase for the IBV than for the IAV particles (Fig. 4B, white bars).
To investigate the possibility that the efficiency of HA activation might be linked to IAV virulence or pandemic potential, we included pseudotypes derived from the 1918 A/H1N1 IAV and avian A/H7N9 IAV, both of which possess a monobasic HA0 cleavage site. The former virus caused the devastating 1918 pandemic, and its HA protein was identified as a virulence determinant (47, 48) although the underlying mechanism is not understood (49, 50). A high case fatality rate in humans is also seen with avian A/H7N9 IAV, which is feared for its pandemic potential (51). The three A/H1N1 pseudotypes tested, i.e., derived from the 1918 IAV, the 2009 pandemic virus, and the laboratory strain A/Puerto Rico/8/1934 (PR8), as well as the A/H7N9 subtype, shared TMPRSS2 and TMPRSS4 as the most effective HA activators (Fig. 4C). Hence, whereas TTSP activation was clearly more efficient for the two IBVs than for the IAV pseudotypes, such a difference was not seen between highly virulent and seasonal IAVs. On the other hand, the 1918 IAV pseudoparticles showed 10- and 100-fold-higher transduction efficiencies than the PR8 and A/Virginia/ATCC3/2009 (Virg09) pseudotypes, respectively (Fig. S2). Since the three A/H1N1 pseudotypes showed comparable incorporation of HA and MLV Gag and a similar level of HA0 cleavage (Fig. S2), the difference might be related to the HA fusion pH (see below) (28). An alternative explanation may be that the 1918 IAV NA, present on these pseudoparticles, may have enhanced their entry process (52).
To summarize, these polykaryon and pseudoparticle experiments established 10 TTSPs as moderate to strong HA activators. The two IBV HAs tested exhibited superior activation by a broader range of TTSPs than the IAV proteins.
IBV but not IAV employs several proteases for spread in human airway-derived Calu-3 cells.
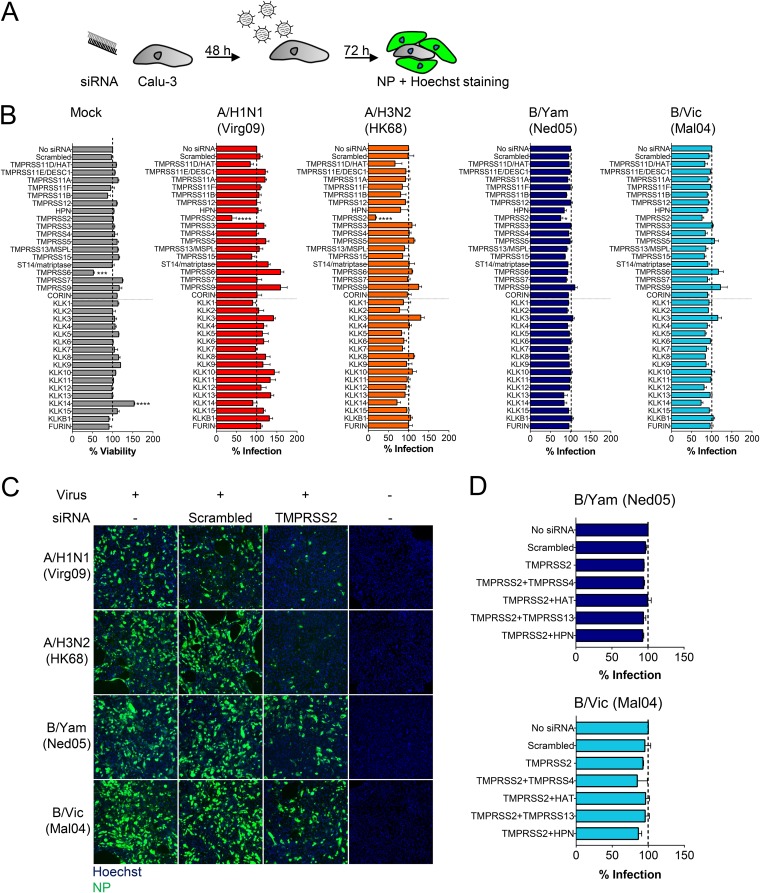
Since Calu-3 cells show a TTSP and KLK expression profile similar to that of healthy human lungs (see above), they are a good model to study the role of these proteases in virus activation in the human respiratory tract. For the four IAV/IBV strains, i.e., Virg09 (A/H1N1), A/HK/2/68 (HK68; A/H3N2), B/Ned/537/05 (Ned05; B/Yam), and B/Malaysia/2506/04 (Mal04; B/Vic), multicycle (i.e., zanamivir-sensitive) (Table S5) replication in Calu-3 cells was the same whether trypsin was added or not (P > 0.1) (Fig. S3), meaning that these cells express one or more HA0-activating proteases. The four viruses were similarly inhibited by three serine protease inhibitors (see antiviral 50% effective concentration [EC50] values in Table S5, which includes a brief description of the assay) but were not blocked by a furin inhibitor, consistent with furin’s inability to process monobasic HA0 cleavage sites (42). We also observed no effect with inhibitors of lysosomal cathepsins, which activate some unrelated viruses that enter by endocytosis (53). Thus, HA activation in Calu-3 cells relies on one or more serine proteases.
After optimizing the procedure for robust small interfering RNA (siRNA)-mediated gene knockdown in Calu-3 cells (Fig. S4), we performed broad knockdown for the 18 TTSPs and 16 KLKs. To assess the impact of protease knockdown on IAV or IBV replication, virus was added 48 h after siRNA transfection, and virus infection was monitored at day 3 postinfection (p.i.) by high-content imaging of nucleoprotein (NP) immunostaining (Fig. 5A). None of the siRNAs produced unwanted cytotoxic effects, with the exception of TMPRSS6 knockdown, which reduced cell viability by 40% (Fig. 5B). TMPRSS2 was the only protease for which knockdown caused a significant (P = 0.0001) reduction in replication of A/H1N1 and A/H3N2 virus (Fig. 5B and C). In sharp contrast, IBV infection was only marginally (Fig. 5B) (B/Yam, P = 0.047) reduced or not reduced (Fig. 5B) (B/Vic, P = 0.48) by TMPRSS2 knockdown. Finally, IBV was not affected by single knockdown of any TTSP or KLK, nor by combined knockdown of TMPRSS2 and the most efficacious IBV HA activators identified above (Fig. 5D). Hence, the two IBV strains appear to rely on redundant proteases for their spread, supporting the above findings that, in addition to TMPRSS2, several proteases effectively activate the IBV HA protein.
FIG 5.
TMPRSS2 is a crucial protease for replication of IAV but not IBV. (A) Experiment setup. Calu-3 cells were transfected with a TTSP- or KLK-targeting siRNA and infected with IAV or IBV. The percentage of infected cells was quantified by high-content imaging of NP. (B) Impact of protease knockdown on cell viability (gray bars) or virus growth (colored bars), expressed relative to the condition receiving no siRNA. Data are the means ± SEM (n = 3 with four replicates). P values for results versus those with a scrambled siRNA are indicated as follows: *, ≤0.05; ***, ≤0.001; ****, ≤0.0001 (ordinary one-way ANOVA, followed by Dunnett’s test). (C) TMPRSS2 knockdown markedly reduces IAV but not IBV replication, as shown by NP staining. (D) IBV infection in Calu-3 cells receiving combinations of siRNAs.
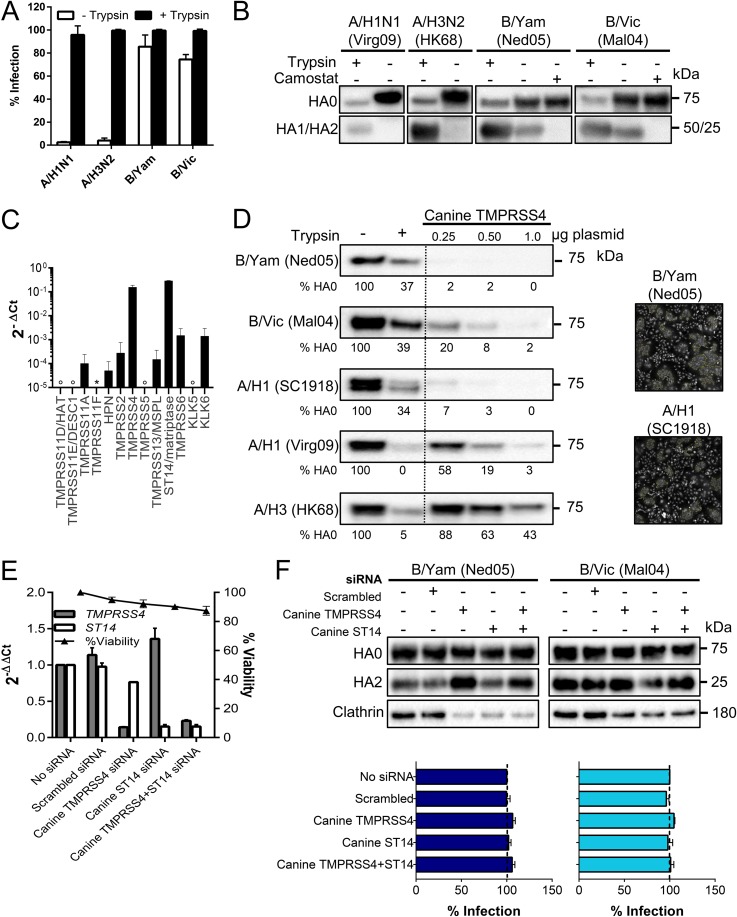
TMPRSS4 is abundant in MDCK cells yet dispensable for spread of IBV in these cells.
To assess whether some TTSPs might stand out as activators of IBV, we included Madin-Darby canine kidney (MDCK) cells which, like Calu-3 cells, support replication of IBV in the absence of trypsin (54). This observation was also made for the 1918 IAV (52, 55) which, in this regard, is unique among human IAV strains. Indeed, the two IBVs but not seasonal IAVs did not require exogenous trypsin to replicate in MDCK cells (Fig. 6A). HA0 cleavage during IBV replication was inhibited by the broad serine protease inhibitor camostat (Fig. 6B), indicating that it is executed by one or more proteases of this family. mRNA quantification in MDCK cells (see Table S3 for canine protease-specific primers) revealed abundant expression of ST14 (matriptase), as observed previously (56, 57), and TMPRSS4 (Fig. 6C). Another study (52) found no TMPRSS4 mRNA in MDCK cells, an observation which is potentially related to the use of different MDCK cell lines (58). Canine TMPRSS4 was cloned into an expression plasmid to investigate its HA0-cleaving capacity. Canine TMPRSS4 proved a strong activator for IBV HA and more effective on A/H1 HA from the 1918 IAV than from the 2009 pandemic (Virg09) IAV (Fig. 6D). The lowest efficiency was seen for A/H3 HA. The polykaryon assay confirmed that fusion-competent HA was formed (Fig. 6D, photographs).
FIG 6.
MDCK cells contain high levels of HA-activating TMPRSS4. (A and B) IAV or IBV replication in MDCK cells in the presence or absence of trypsin. (A) Percentage of infected cells at day 3 p.i., quantified by high-content imaging of NP. Values are means ± SEM of two experiments performed in quadruple. (B) HA0 cleavage state at day 3 p.i. in cells receiving 0 or 10 μM camostat. (C) Expression of TTSP and KLK proteases in MDCK cells (normalized to canine GAPDH and HMBS). Open circle, undetermined; *, no data (no canine mRNA sequence was available to design primers). (D) Canine TMPRSS4 activates the HAs of IAV and IBV. Western blot lanes, from left to right, are as follows: HA0 band in HEK293T cells receiving no protease, exogenous trypsin, or the canine TMPRSS4 plasmid at three different concentrations. Under each lane, the HA0 band intensity is given (normalized to that of clathrin and expressed relative to the no-trypsin condition). Photographs show polykaryon formation in HeLa cells undergoing coexpression of HA and canine TMPRSS4. (E and F) Effect of TMPRSS4 and/or ST14/matriptase knockdown in MDCK cells. (E) mRNA levels at 24 h after siRNA transfection, normalized to the levels of canine HMBS and GAPDH and shown as the fold change relative to the level in the untransfected control. (F) siRNA-transfected cells were infected with B/Yam or B/Vic virus; at day 3 p.i., the HA0 cleavage state was determined by Western blotting, and virus infection was assessed by high-content imaging of NP.
The mRNA data suggested that canine TMPRSS4 or ST14/matriptase might be responsible for spread of IBV in MDCK cells. For IAV HA, published data on the role of matriptase are not consistent (57, 59, 60). Although our data thus far argued against a direct role for matriptase, an indirect role (e.g., as an activator of TMPRSS4) could not be excluded. With this is mind, we performed knockdown for canine TMPRSS4 and matriptase individually or in combination (Fig. 6E and F). Knockdown efficiency was >85% based on reverse transcription-quantitative PCR (RT-qPCR) for mRNA. Virus replication and HA0 cleavage in IBV-infected MDCK cells proved unaffected by knockdown of TMPRSS4 or matriptase and by a combination of the two (Fig. 6F). Thus, in both Calu-3 and MDCK cells, the two IBV strains do not seem to rely on one single protease for their replication.
IBV HA exhibits a similar fusion pH as human-adapted IAV HAs.
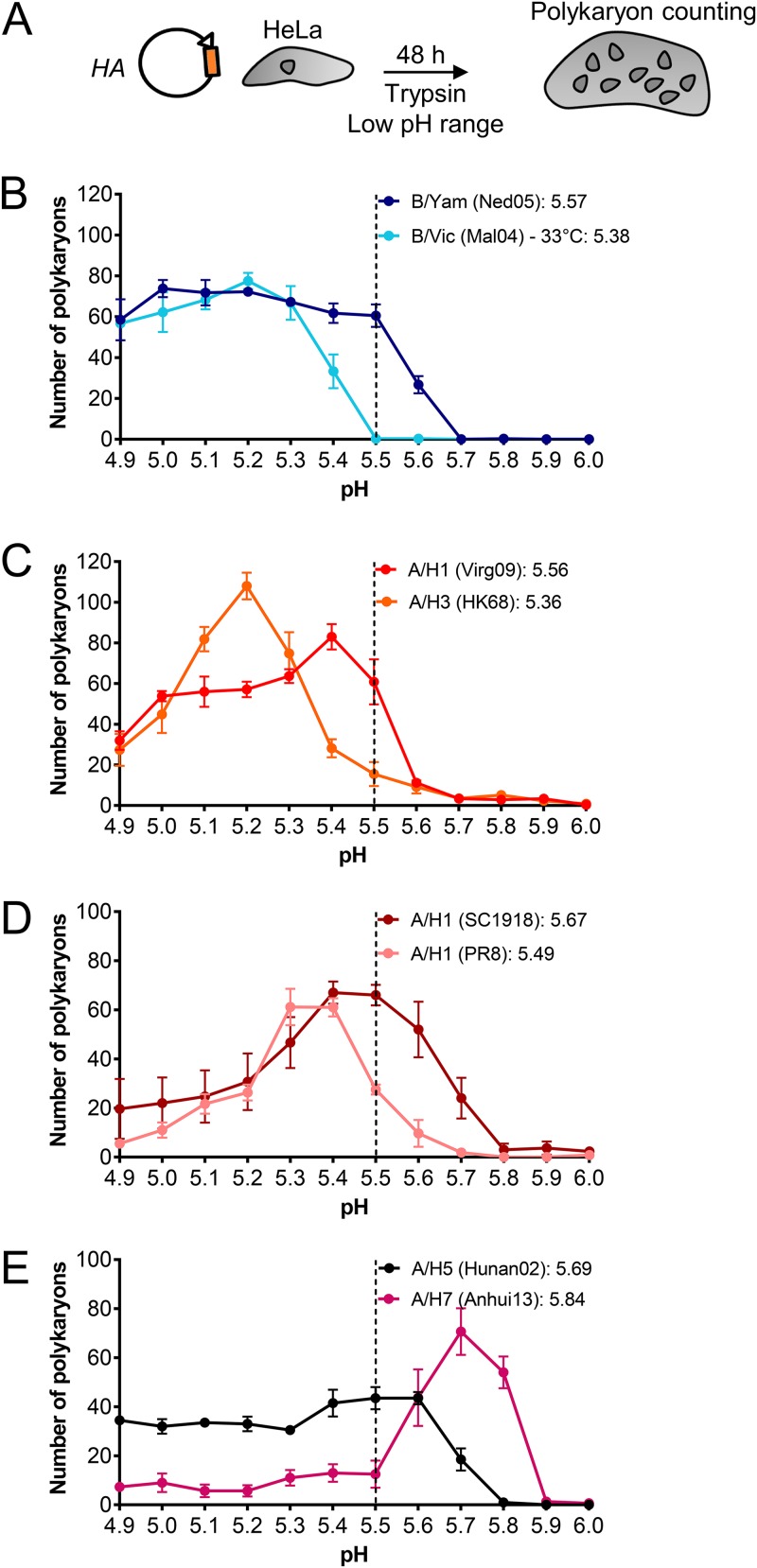
Cleavage of HA0 into HA1-HA2 activates the protein for membrane fusion but also renders HA susceptible to inactivation at acidic pH. The human nasal cavity is mildly acidic (average pH of 6.3 in healthy adults and 5.9 in children) (30, 61). For pandemic IAV, acquirement of a more acid-stable HA is considered a prerequisite to achieve human-to-human transmissibility (27, 31, 44, 62). Since IBV is restricted to humans, we asked whether IBV possesses an acid-stable HA. Indeed, the polykaryon assay showed that the two IBV HA (i.e., B/Yam and B/Vic) proteins have a fusion pH value in the same range (5.4 to 5.6) as their human IAV counterparts (Fig. 7B to D), while avian A/H5 and A/H7 HAs had higher values (5.7 and 5.8, respectively) (Fig. 7E), in line with previous reports (34, 44, 63, 64). On the other hand, the shape of the pH curves was distinct for IBV HA. For human-adapted IAV HAs, the curves showed a discrete pH value at which the number of polykaryons was maximal and a steep decline at more acidic pH due to HA inactivation (65). In contrast, both IBV HAs produced high numbers of polykaryons over the entire low-pH range. Hence, at least for the two strains in our study, IBV HA seems to be triggered for fusion at an acidic pH similar to that of IAV HA; however, the IBV proteins may be more resistant to acidic conditions. In addition, we noticed that the 1918 A/H1 HA (Fig. 7D) had the highest fusion pH among the tested human IAV HAs, with a value (5.7) as high as that of avian A/H5 HA (Fig. 7E). This might explain the superior transduction efficiency of 1918 IAV pseudoparticles (see above) since an increased fusion pH enables earlier endosomal escape and more efficient infection (28).
FIG 7.
IBV HA exhibits a similar fusion pH as human-adapted IAV HAs. (A) Experiment setup. HA-expressing HeLa cells were activated with exogenous trypsin and then exposed to a range of acidic buffers to induce cell-cell fusion. (B to E) Curves showing the number of polykaryons as a function of the pH applied to trigger HA. Data are the means ± SEM of two independent experiments, each performed in triplicate. The insets show the fusion pH values, defined as the pH at which the number of polykaryons was 50% of the maximum number.
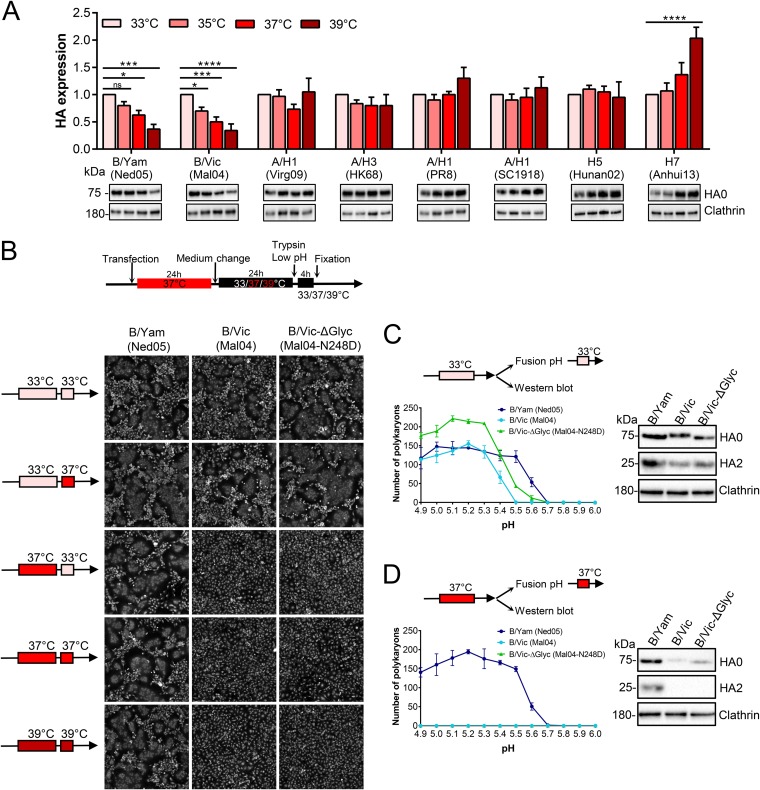
A temperature of 33°C is preferred by IBV HA and required for fusion by the B/Victoria lineage.
The human URT has a temperature (∼30 to 32°C) below body temperature (35, 36) and well below that of the avian intestinal tract (∼40°C) (66, 67). Since a temperature of 33°C is preferred for propagating IBV in cell culture, we investigated whether HA has an intrinsic role in this temperature preference. Protein expression of IBV HA proved to be temperature dependent, with expression highest at 33°C and gradually decreasing at higher temperatures (Fig. 8A). The 33°C preference was seen for the two strains representing both IBV lineages but was particularly significant for the B/Vic HA strain (P value of 0.0006 for a comparison of protein levels at 33°C and 37°C). For human IAV and avian A/H5 HAs, expression levels were similar within the range of 33 to 39°C although for A/H1 HAs expression tended to be highest at 39°C. Avian A/H7 HA manifested a clear preference for 39°C (Fig. 8A).
FIG 8.
IBV HA prefers a temperature of 33°C for protein expression, explaining temperature-restricted fusion of B/Vic HA. (A) HeLa cells were transfected with the indicated HA plasmids and incubated during 48 h at four different temperatures. The graphs show quantitative Western blot data for HA0 band intensity, normalized to that of clathrin and expressed relative to the HA0 band seen at 33°C. Data are the means ± SEM (n = 3). *, P < 0.05; **, P < 0.01; ****, P ≤ 0.0001; ns, not significant (ordinary one-way ANOVA, followed by Dunnett’s test). (B to D) Polykaryon formation in IBV HA-transfected HeLa cells exposed to different temperatures during the stages of HA expression (starting 24 h posttransfection) or cell-cell fusion. Panel B shows fusion induced by a pH 5.0 buffer. Panels C and D show the number of polykaryons as a function of pH, after HA expression at 33°C or 37°C, as indicated. The Western blots show the corresponding HA levels after trypsin activation.
When the polykaryon assay was conducted at 37°C, B/Mal04 HA (B/Vic lineage) proved unable to induce membrane fusion at any pH tested (Fig. 8D); however, abundant polykaryons were formed at 33°C (Fig. 8B and C). Varying the temperature during protein expression or membrane fusion (see arrow schemes in Fig. 8B) revealed that the 33°C requirement was due to inefficient HA expression at 37°C. Whether cell-cell fusion took place at 33°C or 37°C made no difference. Due to low expression at 37°C, the levels of activated HA, generated by trypsin, were undetectable (Fig. 8D). The strict 33°C requirement was not seen with B/Ned05 HA (B/Yam lineage) since this protein generated polykaryons at all temperatures, including at 39°C (Fig. 8B). The HA of the B/Vic lineage carries a globular head glycan at residue N248 that is lacking in the B/Yam lineage (7, 68, 69). When we replaced N248 in B/Mal04 HA by D248, the corresponding residue in B/Ned05 HA, the pH threshold to induce polykaryons shifted from 5.4 to 5.5 (Fig. 8C). Polykaryons induced by the N248D mutant were larger and more numerous than those of wild-type B/Mal04 HA (Fig. 8B and C). However, the mutation did not change the 33°C preference for protein expression and membrane fusion (Fig. 8B to D), indicating that this N248 glycan is not responsible for the temperature-sensitive phenotype of B/Mal04 HA.
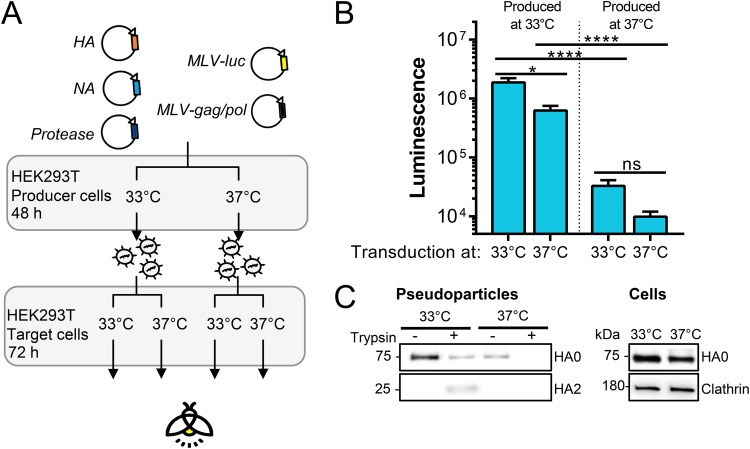
Accordingly, pseudoparticles carrying B/Mal04 HA produced at 33°C had much higher infectivity than those generated at 37°C (P < 0.0001) (Fig. 9B). Transduction efficiency was similar at both temperatures. At the cooler temperature, higher HA protein levels were visible in the producer cells and particularly in released pseudoparticles (Fig. 9C), suggesting that this reduced temperature may be required for efficient posttranslational transport and membrane incorporation of B/Mal04 HA and possibly also other strains of the B/Vic lineage.
FIG 9.
Pseudoparticles carrying B/Victoria HA require 33°C for infectivity. (A) Experiment setup. Pseudoparticles carrying B/Vic (Mal04) HA and NA were produced at 33°C or 37°C, activated with trypsin, and transduced into HEK293T cells at 33°C or 37°C. (B) Infectivity of produced particles as measured by luminescence readout 3 days p.i. *, P < 0.05; ****, P ≤ 0.0001; ns, not significant. (C) HA levels in released pseudoparticles collected by ultracentrifugation and in HEK293T producer cells.
In combination, these results indicate a possible correlation between HA expression and host temperature, with IBV HA preferring 33°C, the temperature of the human URT, and avian A/H7 HA preferring the body temperature of birds. The 33°C preference was stricter for the B/Vic HA than for the B/Yam HA strain, a difference unrelated to the lineage-specific N248 head glycan.
DISCUSSION
Although there is a large body of literature on the HA proteins from human and zoonotic IAVs and on how some of their properties reflect viral host adaptation (8, 27), the HA of IBV is far less characterized (9). In this study, we compared the HA proteins of human IAV and IBV in terms of proteolytic activation, pH stability, and temperature preference as markers for host adaptation. Our results provide evidence for more pronounced adaptation of IBV HA to the human airways, in keeping with the long and exclusive circulation of IBV in humans.
In order to be membrane fusion competent, HA requires cleavage by a host cell protease. Several TTSPs and KLKs have been linked to IAV HA activation (reviewed in references 17 and 18), yet a comprehensive analysis was, thus far, missing for IAV and especially for IBV. To close this gap, we compared all 18 human TTSP and 16 KLK enzymes for their capacities to cleave and activate the IAV and IBV HA proteins. Our results confirm the leading role of TMPRSS2 in activation of monobasic IAV HAs, consistent with data obtained in cell culture, knockout mice, and humans (19, 20, 23). The report that TMPRSS2 proved dispensable for spread of IBV in mice (26) suggested either that TMPRSS2 is not involved at all or that several redundant proteases may activate IBV HA. Our findings support the latter assumption since the cleavability of IBV HA proved clearly superior to that of IAV. We demonstrated that two IBV HAs, representing both lineages, are cleaved and activated by a broad range (a total of 10) of TTSP enzymes, with the strongest activators being TMPRSS2, TMPRSS4, TMPRSS11F, TMPRSS13/MSPL, hepsin, and TMPRSS11D/HAT, followed by four other proteases (TMPRSS5, TMPRSS6, TMPRSS11A, and TMPRSS11E/DESC1). All of these TTSPs are expressed in human lung tissue and generate a functional, i.e., membrane fusion-competent, IBV HA protein. In contrast, activation of A/H1N1 and A/H3N2 HA was largely limited to TMPRSS2 and TMPRSS11D/HAT and to TMPRSS4 in the case of A/H1N1. We saw no evidence for a role of TMPRSS15/enterokinase (70) or KLK5, KLK12 (71, 72) or of any other kallikrein. It is possible that the KLK levels attained in our cell systems were below those applied during incubation with recombinant KLKs (71). Finally, our data do not support involvement of ST14/matriptase in cleavage of A/H1 HA (59, 60), in line with another report (57).
Limburg et al. recently showed that TMPRSS2 was crucial for IBV activation in human primary type II alveolar epithelial cells but dispensable for spread in primary human bronchial epithelial cells and Calu-3 cells, pointing to as yet unspecified protease(s) (25). We show that single knockdown of none of the 18 TTSPs had an impact on IBV replication in Calu-3 cells and also that dual knockdown of TMPRSS2 plus another HA-activating TTSP had no effect, supporting the hypothesis that IBV can rely on redundant proteases for HA activation. This is also evident from our observation in MDCK cells, in which knockdown of the neither abundantly expressed TMPRSS4 nor matriptase could halt IBV replication. In contrast, replication of A/H1N1 and A/H3N2 viruses in Calu-3 cells proved strongly dependent on TMPRSS2. This is noteworthy considering that TMPRSS4 efficiently activated A/H1 HA in transfected cells and proved abundantly expressed in Calu-3 cells. How can this central role of TMPRSS2 in activation of IAV HA be explained? Unlike other TTSPs, TMPRSS2 may be present at high levels in all compartments of the constitutive secretory pathway (17), which is also followed by HA. Only TMPRSS2 appears to extensively colocalize with HA (73). Also, TMPRSS2 might activate not only HA but also the viral M2 ion channel (74). Why is IBV HA more efficiently activated than IAV HA? It is plausible that IBV HA may exhibit higher TTSP accessibility, perhaps governed by the dynamics of HA glycoprotein folding, maturation, or transport toward the cell membrane. This might be influenced by N-glycans in the HA head or stem (at sites very distant from the cleavage loop) having an effect on HA folding rate (75). An N-glycan (N8) located at the bottom of the A/H3 HA stem was shown to retard HA folding (75) and be lost when an A/H3N2 virus was passaged in TMPRSS2 knockout mice (76). Another factor is the HA0 cleavage site itself, for which two differences between IBV and IAV may be relevant: (i) at position P3 (P1 being the scissile arginine), IBV HA0 contains an extra basic lysine residue while A/H1 and A/H3 HAs carry a glutamine; (ii) the P2′ residue is phenylalanine in IBV but leucine, a less hydrophobic residue, in human IAV HAs. In addition, the IBV HA0 cleavage loop might be structurally distinct from the A/H1 and A/H3 HA0 loops. While the latter two have been resolved (77, 78), an X-ray structure of IBV HA0 is still missing.
Since the clinical picture of IBV is similar to that of IAV (9), the high cleavability of IBV HA appears not to be linked to virulence. However, it ensures shedding of infectious virus. Human-to-human transmissibility also requires optimal HA acid stability since this renders the virus more stable under mildly acidic conditions in the human URT or environment (27). For zoonotic IAVs, adaptation to the human host is associated with HA stabilization, reducing the HA fusion pH to 5.2 to 5.5 (31). This fits with the fusion pH values that we measured for IBV HA, i.e., 5.4 and 5.6 for the two strains representing the B/Victoria and B/Yamagata lineages, respectively. Literature data for IBV are scarce, but our values are in agreement with those of other reports (79, 80). The difference in fusion pH values between the two lineages appears partially related to the B/Victoria lineage-specific N248 HA head glycan, positioned adjacent to the receptor binding site (7). Removing this N-glycan caused a small increase in the fusion pH threshold, pointing to an allosteric effect of the HA head domain on the fusion process, which is consistent with a role for receptor binding in fusion peptide dynamics (13).
For both IBV lineages, HA exhibits an intrinsic preference for 33°C which is particularly pronounced for the B/Mal04 strain of the B/Victoria lineage. Analysis of additional IBV strains should verify our hypothesis that the B/Victoria HA might be even better adapted than the B/Yamagata lineage to the proximal airways, given its stricter 33°C dependence and lower fusion pH. Its more stable HA could perhaps explain higher prevalence of this lineage in children (9) which typically have a more acidic nasal pH (27). A preference for cooler temperature was also reported for the hemagglutinin-esterase-fusion (HEF) protein of influenza C virus, which mainly infects humans (81). We did not observe this temperature effect for human IAV HAs and saw the opposite pattern for avian A/H7 HA. This leads us to hypothesize that fine-tuning of HA protein expression to fit the temperature of the host target organs could be another viral adaptation strategy, in addition to well-known mechanisms related to receptor use, polymerase activity, or immune evasion (8). The temperature-sensitive expression of IBV HA is an intrinsic feature seen in the absence of any other IBV proteins; hence, it is unrelated to the reduced RNA synthesis seen with some temperature-sensitive IBV strains (82). As for the biochemical basis, for influenza C virus HEF, trimer formation and surface expression proved more efficient at 33°C than at 37°C (81). A similar temperature effect probably applies to IBV HA since pseudoparticles carrying B/Mal04 (B/Victoria lineage) HA contained much higher HA levels when produced at 33°C than at 37°C. For IAV HA, a link exists between N-glycosylation and temperature sensitivity of the processes of HA folding or transport toward the cell membrane (75, 83–86). We therefore examined the role of the B/Victoria lineage-specific N248 glycan; however, removal of this HA head N-glycan did not change the dependence on 33°C.
Although our study was focused on the HA proteins of seasonal human IAVs and IBVs, we also made an interesting observation for the HA of 1918 IAV. Its HA fusion pH (5.7) proved to be exceptionally high for a human IAV but identical to that of highly pathogenic avian A/H5N1 virus. This may help to explain the exceptional virulence of the 1918 IAV since IAVs with high HA fusion pH evade interferon control (87) and are more virulent in mice (88). It could also rationalize a mouse study in which the 1918 IAV HA protein by itself generated a highly pathogenic phenotype when introduced in a contemporary backbone virus (48). In addition, we showed that TTSP activation is comparable for the 1918 IAV HA and other A/H1 HAs. Hence, the unique capacity of the 1918 IAV to replicate in MDCK cells in the absence of trypsin (52, 55) seems not, or at least not entirely, explained by HA activation by TMPRSS4 or another TTSP expressed in MDCK cells. As proposed (55, 89), a role for the 1918 IAV NA is plausible.
To conclude, the findings for the two IBV strains investigated in our study suggest that the IBV HA protein combines broad and efficient activation capacity, favorable acid stability, and a preference for the cooler temperature of the human URT. These distinct properties likely reflect host adaptation resulting from sustained presence of this respiratory pathogen in the human population.
MATERIALS AND METHODS
Ethics statement.
Lung tissue samples from eight different healthy donors were obtained under the approval of the ethical committee from the University Hospital Leuven (UZ Leuven Biobanking S51577). All patients were adult and provided written informed consent.
Cells, media, and compounds.
Calu-3 (ATCC HTB-55), A549 (ATCC CCL-185), 16HBE (a gift from P. Hoet, Leuven, Belgium), and HeLa (ATCC CCL-2) cells were grown in minimum essential medium (MEM) supplemented with 10% fetal calf serum (FCS), 0.1 mM nonessential amino acids, 2 mM l-glutamine, and 10 mM HEPES. HEK293T cells (HCL4517; Thermo Fisher Scientific) and Madin-Darby canine kidney (MDCK) cells, a gift from M. Matrosovich (Marburg, Germany), were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS, 1 mM sodium pyruvate, and 0.075% sodium bicarbonate. Medium with reduced (i.e., 0.2%) FCS content was used during protease expression and virus infection experiments. Except as stated otherwise, all cell incubations were done at 37°C. The following compounds were purchased from Sigma-Aldrich: N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin, camostat, nafamostat, aprotinin, and leupeptin. Chloromethylketone, E64-d, and CA-074Me were from Enzo.
Viruses.
The four human influenza virus strains and their abbreviations used in the text and figures are as follows: A/Virginia/ATCC3/2009 (Virg09; A/H1N1 subtype; ATCC VR-1737); A/HK/2/68 (HK68; A/H3N2 subtype) and B/Ned/537/05 (Ned05; B/Yamagata [B/Yam] lineage), generously donated by R. Fouchier (Rotterdam, The Netherlands); and B/Malaysia/2506/04 (Mal04; B/Victoria [B/Vic] lineage) (NR-12280; BEI resources)). Viruses were expanded in 10-day-old embryonated chicken eggs. For virus titration, a virus dilution series was added in quadruple to Calu-3 cells. At day 3 postinfection (p.i.), virus positivity was assessed by immunostaining for viral nucleoprotein (NP) (see below). Virus titers were expressed as the 50% cell culture infective dose (CCID50), calculated by the method of Reed and Muench (90).
Plasmids.
The panel of 35 pcDNA3.1+/C-(K)DYK plasmids containing the coding sequences for the 18 TTSPs, 16 KLKs, and furin (for accession numbers and clone identification numbers, see Table S1 in the supplemental material), was purchased from GenScript. The lengths of the open reading frames (ORFs) were checked by PCR (Fig. S1). Expression of the different proteases was verified at 48 h posttransfection of HEK293T cells using a dot blot assay. Cell lysates, prepared in radioimmunoprecipitation assay (RIPA) buffer, were spotted onto a nitrocellulose membrane. The membrane was dried, blocked with 5% low-fat milk powder in phosphate-buffered saline (PBS), and incubated for 1 h with horseradish peroxidase (HRP)-conjugated anti-FLAG antibody (see the full list of antibodies in Table S2). The dots were detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and a ChemiDoc XRS+ system (Bio-Rad).
The expression plasmid for canine TMPRSS4 was constructed by extracting total RNA from MDCK cells, followed by reverse transcription and high-fidelity PCR using primers extended with EcoRV and XbaI sites to allow subcloning into the pCAGEN vector provided by C. Cepko (Boston, MA) via Addgene (plasmid 11160). A similar cloning procedure was used to prepare pCAGEN vectors with the HA and NA coding sequences from Virg09, HK68, Ned05, and Mal04, starting from allantoic virus stocks. These HA proteins were >99% identical to the following GenBank sequences published in NCBI (www.ncbi.nlm.nih.gov): AGI54750.1 (for Virg09), AFG71887.1 (for HK68), AGX16237.1 (for Ned05), and CY040449.1 (for Mal04).
The coding sequences for the A/H1 HA from A/South Carolina/1/1918 (SC1918) (GenBank accession number AF117241.1) and the N248D mutant form of Mal04 HA were ordered from IDT. The cDNAs for A/H5 HA (GenBank accession number ACA47835.1) from A/duck/Hunan/795/2002 (Hunan02), and the A/H7 HA and A/N9 NA (EPI439507 and EPI439509, respectively; GISAID EpiFlu database) from A/Anhui/1/2013 (Anhui13) were purchased from Sino Biological. cDNAs were amplified by Platinum SuperFi PCR (Invitrogen) and subcloned in the pCAGEN expression vector as described above. A/H1 HA from A/Puerto Rico/8/1934 (PR8; GenBank accession number AYA81842.1) was subcloned into pCAGEN starting from a pVP-HA reverse genetics plasmid, kindly donated by M. Kim (Daejeon, South Korea).
Protease gene expression analysis in cells and human lung tissue.
Lung tissue samples from eight different healthy donors were used. Calu-3, 16HBE, A549, and HEK293T lysates were made from three different cell passages. Total RNA was extracted using a ReliaPrep RNA Cell Miniprep System (Promega), and 0.5 μg of RNA was converted to cDNA with a high-capacity cDNA reverse transcription kit (Thermo Fisher Scientific). BRYT Green dye-based quantitative PCR (qPCR) was performed with GoTaq qPCR Master Mix (Promega) and intron-spanning primer pairs (see Table S3 for a list of all primers) in an ABI 7500 Fast real-time PCR system (Applied Biosciences). Expression data were normalized to the geometric mean of three housekeeping genes (GAPDH, HMBS, and ACTB) and analyzed using the 2−ΔCT (where CT is threshold cycle) method. All primers were checked for PCR efficiency and specificity by melt curve analysis. Microarray data for 108 healthy lung samples were obtained from GEO data set GSE47460 (www.ncbi.nlm.nih.gov/geo) (91).
Western blot assay to monitor HA0 cleavage or HA protein expression.
To assess HA0 cleavage in protease-expressing cells, HEK293T cells were seeded in growth medium at 300,000 cells per well in 12-well plates and transfected with 0.5 μg of pCAGEN-HA plasmid and 0.5 μg of pcDNA3.1+/C-(K)DYK-protease plasmid using Lipofectamine 2000 (Life Technologies). Four hours later, the growth medium was replaced by medium with 0.2% FCS, and cells were further incubated at 37°C or 33°C (for B/Vic HA). At 48 h posttransfection, the control well was exposed to 5 μg/ml TPCK-treated trypsin for 15 min at 37°C. Cells were then lysed in RIPA buffer supplemented with protease inhibitor cocktail (both from Thermo Fisher Scientific). The lysates were boiled for 5 min in 1× XT sample buffer containing 1× XT reducing agent (both from Bio-Rad) and resolved on 4 to 12% Bis-Tris XT precast 26-well gels (Bio-Rad). Proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad), blocked with 5% low-fat milk powder, and probed for 1 h with primary antibody followed by 45 min with HRP-conjugated secondary antibody. Clathrin served as the loading control (see Table S2 for a list of all antibodies). The bands were detected and visualized as explained above for the dot blot assay. To quantify HA protein expression at different temperatures, HEK293T or HeLa cells seeded in 12-well plates were transfected with pCAGEN-HA plasmid, incubated during 48 h at 33, 35, 37, or 39°C, and then exposed to exogenous trypsin when indicated. Cell extraction and Western blot analysis were carried out as described above.
Polykaryon assay to measure protease-mediated activation of HA or its fusion pH.
The method was adapted from Vanderlinden et al. (92) to enable a high-throughput format with high-content imaging. HeLa cells in black 96-well plates (20,000 cells per well) were reverse transfected with 50 ng of pCAGEN-HA plasmid and 12.5 ng of pcDNA3.1+/C-(K)DYK protease-plasmid using Fugene 6 (Promega). After 24 h, the growth medium was replaced by medium with 0.2% FCS, and cells were further incubated at 37°C or 33°C (for B/Vic HA). Another 24 h later, the control conditions (which received HA plasmid only) were exposed for 15 min to MEM containing 5 μg/ml TPCK-treated trypsin. After gentle washing with PBS plus Ca2+ and Mg2+ (PBS-CM), the cells were exposed for 15 min to PBS-CM that had been pH adjusted with citric acid to the required pH (i.e., a pH 0.1 unit lower than the fusion pH of that HA). After this pulse, the acidic PBS-CM was removed, and growth medium with 10% FCS was added to stop the reaction. The cells were allowed to fuse during 4 h at 37°C (33°C for B/Vic HA or another temperature if specifically mentioned), then fixed with 4% paraformaldehyde in PBS (15 min), permeabilized with 0.1% Triton X-100 (15 min), and stained for 30 min with 2 μg/ml HCS Cell Mask stain (Life Technologies). The plates were imaged using a CellInsight CX5 high-content imaging platform (Thermo Scientific). Nine images were taken per well, and polykaryons were identified and counted using the SpotDetector protocol of the HCI software.
To determine the fusion pH of the different HAs, HeLa cells were transfected with the pCAGEN-HA plasmids, and HA0 was cleaved with exogenous trypsin as described above. During the acidic pulse, a range of acidic buffers was used having a pH between 4.9 to 6.0 with 0.1 increments. High-content imaging-based quantification of polykaryons was done as described above. The fusion pH was defined as the pH at which the number of polykaryons was 50% of the maximum number (91).
Production of protease-activated HA pseudoparticles and transduction experiments.
The method to produce firefly luciferase (fLuc)-expressing retroviral vectors pseudotyped with a viral glycoprotein was previously described (93). In brief, HEK293T cells were transfected in six-well plates, using calcium phosphate precipitation, with a mixture of plasmids encoding murine leukemia virus (MLV) Gag-Pol (1.5 μg), an MLV vector coding for fLuc (3 μg), HA and NA expression plasmids (both 0.75 μg), and the protease expression plasmids specified above (0.125 μg). The two additional expression vectors pCAGGS-HAT and pCAGGS-DESC1 were described previously (73). At 16 h posttransfection, the medium was replaced by medium with 2% FCS. At 48 h, the pseudoparticle-containing supernatants were harvested and clarified by centrifugation. To verify that all TTSP conditions contained a similar total number of pseudoparticles, they were subsequently exposed to trypsin. Namely, one half of the supernatant was left untreated, and the other half was treated with 20 μg/ml trypsin for 15 min at 37°C, after which 20 μg/ml soybean trypsin inhibitor was added. To measure particle infectivity, HEK293T target cells were seeded in 96-well plates at a density of 20,000 cells per well. One day later, the cells were exposed to 100 μl of virus stock, and 6 h later, fresh medium was added. To measure fLuc activity at day 3 posttransduction, the cells were lysed for 10 min in 50 μl of cold PBS with 0.5% Triton-X. The lysates were transferred to a white, opaque-walled 96-well plate and, after the addition of fLuc substrate (Beetle-Juice kit; PJK Biotech), the signal was read in a microplate reader (Plate Chameleon V; Hidex) using MicroWin2000 software (version 4.44; Mikrotek Laborsysteme GmbH).
Virus replication following siRNA-mediated protease knockdown or exposure to trypsin.
On-TARGETplus siRNA SMARTpools targeting the 35 human proteases and a nontargeting (scrambled) control were ordered from Dharmacon (for catalogue numbers, see Table S4). siRNAs targeting canine TMPRSS4 and canine ST14/matriptase were custom synthesized by IDT (see sequences provided in Table S4). Cells suspended in a black 96-well plate (for Calu-3 cells, 35,000 cells per well; for MDCK cells, 7,500 cells per well) were reverse transfected with 10 nM siRNA (each condition in quadruple) using Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific). One day later, the transfection medium with 10% FCS was replaced by medium containing 0.2% FCS. Another 24 h later, the cells were infected with Virg09, HK68, Ned05, or Mal04 virus at a multiplicity of infection (MOI) of 100 CCID50s. At 72 h p.i., immunostaining for viral NP was performed. The cells were fixed for 5 min in 2% paraformaldehyde, permeabilized for 10 min with 0.1% Triton X-100, and blocked for 4 h in 1% bovine serum albumin (BSA). Next, the plates were stained overnight at 4°C with anti-NP antibody diluted in 1% BSA (for IAV, 3IN5 at 1/2,000 [HyTest]; for IBV, RIF17 at 1/2,000 [HyTest]). After plates were washed in PBS containing 0.01% Tween, Alexa Fluor 488 secondary antibody (A21424 at 1/500; Invitrogen) was applied for 1 h at room temperature. Cell nuclei were stained with Hoechst (Thermo Fisher Scientific). The plates were imaged using the high-content platform specified above. Nine images per well were analyzed to determine the total (Hoechst) and infected (NP) cell numbers.
To measure their potential cytotoxic effects, the siRNAs were added to a parallel plate containing mock-infected cells. After 5 days of incubation, cell viability was measured by the colorimetric MTS [3,4-(5-dimethylthiazol-2-yl)-5-(3-carboxymethoxy phenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt] assay (CellTiter 96 AQueous MTS reagent; Promega) (94).
To quantify virus replication in the absence or presence of trypsin, the Calu-3 or MDCK cells were infected with virus as described above, using infection medium with or without 2 μg/ml TPCK-treated trypsin and 0.2% FCS. After 3 days of incubation, the infection rate was determined by NP staining and high-content imaging.
Data analysis.
Unless stated otherwise, data shown are the means ± standard errors of the means (SEM) of three independent experiments. GraphPad Prism software (version 7.0) was used to analyze the data and construct the graphs. One-way analysis of variance (ANOVA) or Kruskal-Wallis with a post hoc test for multiple comparisons was performed to compare groups, as indicated in the figure legends. To compare two groups, an unpaired Student's t test was used.
Supplementary Material
ACKNOWLEDGMENTS
Part of this research work was performed using the Caps-It research infrastructure (project ZW13-02) that was financially supported by the Hercules Foundation (FWO) and Rega Foundation, KU Leuven.
We thank John McDonough for assisting the GEO expression analysis, Dirk Daelemans for providing the high-content imaging infrastructure, and Wim van Dam for technical assistance.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2018. Influenza (seasonal)—fact sheet no. 211. http://www.who.int/mediacentre/factsheets/fs211/en/.
- 2.Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson MA, Bresee JS. 2018. Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet 391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glezen PW, Schmier JK, Kuehn CM, Ryan KJ, Oxford J. 2013. The burden of influenza B: a structured literature review. Am J Public Health 103:e43–e51. doi: 10.2105/AJPH.2012.301137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen J, Vestergaard LS, Richter L, Schmid D, Bustos N, Asikainen T, Trebbien R, Denissov G, Innos K, Virtanen MJ, Fouillet A, Lytras T, Gkolfinopoulou K, Heiden MA, Grabenhenrich L, Uphoff H, Paldy A, Bobvos J, Domegan L, O’Donnell J, Scortichini M, de Martino A, Mossong J, England K, Melillo J, van Asten L, de Lange MM, Tonnessen R, White RA, da Silva SP, Rodrigues AP, Larrauri A, Mazagatos C, Farah A, Carnahan AD, Junker C, Sinnathamby M, Pebody RG, Andrews N, Reynolds A, McMenamin J, Brown CS, Adlhoch C, Penttinen P, Molbak K, Krause TG. 2019. European all-cause excess and influenza-attributable mortality in the 2017/18 season: should the burden of influenza B be reconsidered? Clin Microbiol Infect 25:1266–1276. doi: 10.1016/j.cmi.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 5.Matias G, Taylor R, Haguinet F, Schuck-Paim C, Lustig R, Shinde V. 2014. Estimates of mortality attributable to influenza and RSV in the United States during 1997–2009 by influenza type or subtype, age, cause of death, and risk status. Influenza Other Respir Viruses 8:507–515. doi: 10.1111/irv.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki Y, Nei M. 2002. Origin and evolution of influenza virus hemagglutinin genes. Mol Biol Evol 19:501–509. doi: 10.1093/oxfordjournals.molbev.a004105. [DOI] [PubMed] [Google Scholar]
- 7.Vijaykrishna D, Holmes EC, Joseph U, Fourment M, Su YC, Halpin R, Lee RT, Deng YM, Gunalan V, Lin X, Stockwell TB, Fedorova NB, Zhou B, Spirason N, Kuhnert D, Boskova V, Stadler T, Costa AM, Dwyer DE, Huang QS, Jennings LC, Rawlinson W, Sullivan SG, Hurt AC, Maurer-Stroh S, Wentworth DE, Smith GJ, Barr IG. 2015. The contrasting phylodynamics of human influenza B viruses. Elife 4:e05055. doi: 10.7554/eLife.05055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17:67–81. doi: 10.1038/s41579-018-0115-z. [DOI] [PubMed] [Google Scholar]
- 9.Koutsakos M, Nguyen TH, Barclay WS, Kedzierska K. 2016. Knowns and unknowns of influenza B viruses. Future Microbiol 11:119–135. doi: 10.2217/fmb.15.120. [DOI] [PubMed] [Google Scholar]
- 10.Osterhaus AD, Rimmelzwaan GF, Martina BE, Bestebroer TM, Fouchier RA. 2000. Influenza B virus in seals. Science 288:1051–1053. doi: 10.1126/science.288.5468.1051. [DOI] [PubMed] [Google Scholar]
- 11.Byrd-Leotis L, Cummings RD, Steinhauer DA. 2017. The interplay between the host receptor and influenza virus hemagglutinin and neuraminidase. Int J Mol Sci 18:1541. doi: 10.3390/ijms18071541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross KJ, Burleigh LM, Steinhauer DA. 2001. Mechanisms of cell entry by influenza virus. Expert Rev Mol Med 3:1–18. doi: 10.1017/S1462399401003453. [DOI] [PubMed] [Google Scholar]
- 13.Das DK, Govindan R, Nikic-Spiegel I, Krammer F, Lemke EA, Munro JB. 2018. Direct visualization of the conformational dynamics of single influenza hemagglutinin trimers. Cell 174:926–912. doi: 10.1016/j.cell.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klenk HD, Rott R, Orlich M, Blödorn J. 1975. Activation of influenza A viruses by trypsin treatment. Virology 68:426–439. doi: 10.1016/0042-6822(75)90284-6. [DOI] [PubMed] [Google Scholar]
- 15.Laporte M, Naesens L. 2017. Airway proteases: an emerging drug target for influenza and other respiratory virus infections. Curr Opin Virol 24:16–24. doi: 10.1016/j.coviro.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Böttcher-Friebertshäuser E, Lu Y, Meyer D, Sielaff F, Steinmetzer T, Klenk HD, Garten W. 2012. Hemagglutinin activating host cell proteases provide promising drug targets for the treatment of influenza A and B virus infections. Vaccine 30:7374–7380. doi: 10.1016/j.vaccine.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Garten W, Braden C, Arendt A, Peitsch C, Baron J, Lu Y, Pawletko K, Hardes K, Steinmetzer T, Böttcher-Friebertshäuser E. 2015. Influenza virus activating host proteases: identification, localization and inhibitors as potential therapeutics. Eur J Cell Biol 94:375–383. doi: 10.1016/j.ejcb.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 18.Galloway SE, Liang B, Steinhauer DA. 2018. Activation of the hemagglutinin of influenza viruses, p 3–26. In Böttcher-Friebertshäuser E, Garten W, Klenk HD (ed), Activation of viruses by host proteases. Springer International Publishing AG, Cham, Switzerland. [Google Scholar]
- 19.Hatesuer B, Bertram S, Mehnert N, Bahgat MM, Nelson PS, Pöhlmann S, Pöhlman S, Schughart K. 2013. Tmprss2 is essential for influenza H1N1 virus pathogenesis in mice. PLoS Pathog 9:e1003774. doi: 10.1371/journal.ppat.1003774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tarnow C, Engels G, Arendt A, Schwalm F, Sediri H, Preuss A, Nelson PS, Garten W, Klenk HD, Gabriel G, Böttcher-Friebertshäuser E. 2014. TMPRSS2 is a host factor that is essential for pneumotropism and pathogenicity of H7N9 influenza A virus in mice. J Virol 88:4744–4751. doi: 10.1128/JVI.03799-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakai K, Ami Y, Tahara M, Kubota T, Anraku M, Abe M, Nakajima N, Sekizuka T, Shirato K, Suzaki Y, Ainai A, Nakatsu Y, Kanou K, Nakamura K, Suzuki T, Komase K, Nobusawa E, Maenaka K, Kuroda M, Hasegawa H, Kawaoka Y, Tashiro M, Takeda M. 2014. The host protease TMPRSS2 plays a major role in in vivo replication of emerging H7N9 and seasonal influenza viruses. J Virol 88:5608–5616. doi: 10.1128/JVI.03677-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambertz RLO, Gerhauser I, Nehlmeier I, Leist SR, Kollmus H, Pohlmann S, Schughart K. 2019. Tmprss2 knock-out mice are resistant to H10 influenza A virus pathogenesis. J Gen Virol 100:1073–1078. doi: 10.1099/jgv.0.001274. [DOI] [PubMed] [Google Scholar]
- 23.Cheng Z, Zhou J, To KK, Chu H, Li C, Wang D, Yang D, Zheng S, Hao K, Bosse Y, Obeidat M, Brandsma CA, Song YQ, Chen Y, Zheng BJ, Li L, Yuen KY. 2015. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A(H1N1) influenza and A(H7N9) influenza. J Infect Dis 212:1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton BS, Whittaker GR, Daniel S. 2012. Influenza virus-mediated membrane fusion: determinants of hemagglutinin fusogenic activity and experimental approaches for assessing virus fusion. Viruses 4:1144–1168. doi: 10.3390/v4071144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limburg H, Harbig A, Bestle D, Stein DA, Moulton HM, Jaeger J, Janga H, Hardes K, Koepke J, Schulte L, Koczulla AR, Schmeck B, Klenk HD, Böttcher-Friebertshäuser E. 7 August 2019. TMPRSS2 is the major activating protease of influenza A virus in primary human airway cells and influenza B virus in human type II pneumocytes. J Virol doi: 10.1128/jvi.00649-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakai K, Ami Y, Nakajima N, Nakajima K, Kitazawa M, Anraku M, Takayama I, Sangsriratanakul N, Komura M, Sato Y, Asanuma H, Takashita E, Komase K, Takehara K, Tashiro M, Hasegawa H, Odagiri T, Takeda M. 2016. TMPRSS2 independency for haemagglutinin cleavage in vivo differentiates influenza B virus from influenza A virus. Sci Rep 6:29430. doi: 10.1038/srep29430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell CJ, Hu M, Okda FA. 2018. Influenza hemagglutinin protein stability, activation, and pandemic risk. Trends Microbiol 26:841–853. doi: 10.1016/j.tim.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singanayagam A, Zambon M, Barclay W. 2019. Influenza virus with increased pH of HA activation has improved replication in cell culture but at the cost of infectivity in human airway epithelium. J Virol 93:e00058-19. doi: 10.1128/JVI.00058-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer H, Widdicombe JH. 2006. Mechanisms of acid and base secretion by the airway epithelium. J Membr Biol 211:139–150. doi: 10.1007/s00232-006-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.England RJ, Homer JJ, Knight LC, Ell SR. 1999. Nasal pH measurement: a reliable and repeatable parameter. Clin Otolaryngol Allied Sci 24:67–68. doi: 10.1046/j.1365-2273.1999.00223.x. [DOI] [PubMed] [Google Scholar]
- 31.Russier M, Yang G, Rehg JE, Wong SS, Mostafa HH, Fabrizio TP, Barman S, Krauss S, Webster RG, Webby RJ, Russell CJ. 2016. Molecular requirements for a pandemic influenza virus: an acid-stable hemagglutinin protein. Proc Natl Acad Sci U S A 113:1636–1641. doi: 10.1073/pnas.1524384113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumann J, Kouassi NM, Foni E, Klenk HD, Matrosovich M. 2016. H1N1 swine influenza viruses differ from avian precursors by a higher pH optimum of membrane fusion. J Virol 90:1569–1577. doi: 10.1128/JVI.02332-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linster M, van Boheemen S, de Graaf M, Schrauwen EJA, Lexmond P, Manz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, Osterhaus A, Matrosovich M, Fouchier RAM, Herfst S. 2014. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157:329–339. doi: 10.1016/j.cell.2014.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindemann J, Leiacker R, Rettinger G, Keck T. 2002. Nasal mucosal temperature during respiration. Clin Otolaryngol Allied Sci 27:135–139. doi: 10.1046/j.1365-2273.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 36.McFadden ER Jr, Pichurko BM, Bowman HF, Ingenito E, Burns S, Dowling N, Solway J. 1985. Thermal mapping of the airways in humans. J Appl Physiol 58:564–570. doi: 10.1152/jappl.1985.58.2.564. [DOI] [PubMed] [Google Scholar]
- 37.Aggarwal S, Dewhurst S, Takimoto T, Kim B. 2011. Biochemical impact of the host adaptation-associated PB2 E627K mutation on the temperature-dependent RNA synthesis kinetics of influenza A virus polymerase complex. J Biol Chem 286:34504–34513. doi: 10.1074/jbc.M111.262048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scull MA, Gillim-Ross L, Santos C, Roberts KL, Bordonali E, Subbarao K, Barclay WS, Pickles RJ. 2009. Avian Influenza virus glycoproteins restrict virus replication and spread through human airway epithelium at temperatures of the proximal airways. PLoS Pathog 5:e1000424. doi: 10.1371/journal.ppat.1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohler A, Becker-Pauly C. 2012. TMPRSS4 is a type II transmembrane serine protease involved in cancer and viral infections. Biol Chem 393:907–914. doi: 10.1515/hsz-2012-0155. [DOI] [PubMed] [Google Scholar]
- 40.Kalinska M, Meyer-Hoffert U, Kantyka T, Potempa J. 2016. Kallikreins—the melting pot of activity and function. Biochimie 122:270–282. doi: 10.1016/j.biochi.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laporte M, Stevaert A, Raeymaekers V, Boogaerts T, Nehlmeier I, Chiu W, Benkheil M, Vanaudenaerde B, Pöhlmann S, Naesens L. 2019. Evidence for influenza B virus hemagglutinin adaptation to the human host: high cleavability, acid-stability and preference for cool temperature. bioRxiv 10.1101/736678. [DOI] [PMC free article] [PubMed]
- 42.Klenk H-D, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39. doi: 10.1016/0966-842X(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 43.Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pöhlmann S. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol 84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 9:e1003151. doi: 10.1371/journal.ppat.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gamblin SJ, Skehel JJ. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch V, Kramer B, Pfeiffer T, Starck L, Steinhauer DA. 2001. Inhibition of release of lentivirus particles with incorporated human influenza virus haemagglutinin by binding to sialic acid-containing cellular receptors. J Gen Virol 82:2485–2494. doi: 10.1099/0022-1317-82-10-2485. [DOI] [PubMed] [Google Scholar]
- 47.Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N, Usui T, Murata T, Maeda Y, Watanabe S, Suresh M, Suzuki T, Suzuki Y, Feldmann H, Kawaoka Y. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- 48.Watanabe T, Tisoncik-Go J, Tchitchek N, Watanabe S, Benecke AG, Katze MG, Kawaoka Y. 2013. 1918 Influenza virus hemagglutinin (HA) and the viral RNA polymerase complex enhance viral pathogenicity, but only HA induces aberrant host responses in mice. J Virol 87:5239–5254. doi: 10.1128/JVI.02753-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Short KR, Kedzierska K, van de Sandt CE. 2018. Back to the future: lessons learned from the 1918 influenza pandemic. Front Cell Infect Microbiol 8:343. doi: 10.3389/fcimb.2018.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quan C, Shi W, Yang Y, Yang Y, Liu X, Xu W, Li H, Li J, Wang Q, Tong Z, Wong G, Zhang C, Ma S, Ma Z, Fu G, Zhang Z, Huang Y, Song H, Yang L, Liu WJ, Liu Y, Liu W, Gao GF, Bi Y. 2018. New threats from H7N9 influenza virus: spread and evolution of high- and low-pathogenicity variants with high genomic diversity in wave five. J Virol 92:e00301-18. doi: 10.1128/JVI.00301-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaipan C, Kobasa D, Bertram S, Glowacka I, Steffen I, Tsegaye TS, Takeda M, Bugge TH, Kim S, Park Y, Marzi A, Pöhlmann S. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol 83:3200–3211. doi: 10.1128/JVI.02205-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Y, Vedantham P, Lu K, Agudelo J, Carrion R Jr, Nunneley JW, Barnard D, Pöhlmann S, McKerrow JH, Renslo AR, Simmons G. 2015. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res 116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noma K, Kiyotani K, Kouchi H, Fujii Y, Egi Y, Tanaka K, Yoshida T. 1998. Endogenous protease-dependent replication of human influenza viruses in two MDCK cell lines. Arch Virol 143:1893–1909. doi: 10.1007/s007050050428. [DOI] [PubMed] [Google Scholar]
- 55.Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77–80. doi: 10.1126/science.1119392. [DOI] [PubMed] [Google Scholar]
- 56.Godiksen S, Selzer-Plon J, Pedersen ED, Abell K, Rasmussen HB, Szabo R, Bugge TH, Vogel LK. 2008. Hepatocyte growth factor activator inhibitor-1 has a complex subcellular itinerary. Biochem J 413:251–259. doi: 10.1042/BJ20071496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Baron J, Tarnow C, Mayoli-Nussle D, Schilling E, Meyer D, Hammami M, Schwalm F, Steinmetzer T, Guan Y, Garten W, Klenk HD, Böttcher-Friebertshäuser E. 2013. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol 87:1811–1820. doi: 10.1128/JVI.02320-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lugovtsev VY, Melnyk D, Weir JP. 2013. Heterogeneity of the MDCK cell line and its applicability for influenza virus research. PLoS One 8:e75014. doi: 10.1371/journal.pone.0075014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamilton BS, Gludish DW, Whittaker GR. 2012. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J Virol 86:10579–10586. doi: 10.1128/JVI.00306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beaulieu A, Gravel E, Cloutier A, Marois I, Colombo E, Desilets A, Verreault C, Leduc R, Marsault E, Richter MV. 2013. Matriptase proteolytically activates influenza virus and promotes multicycle replication in the human airway epithelium. J Virol 87:4237–4251. doi: 10.1128/JVI.03005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McShane D, Davies JC, Davies MG, Bush A, Geddes DM, Alton EW. 2003. Airway surface pH in subjects with cystic fibrosis. Eur Respir J 21:37–42. doi: 10.1183/09031936.03.00027603. [DOI] [PubMed] [Google Scholar]
- 62.Russell CJ. 2014. Acid-induced membrane fusion by the hemagglutinin protein and its role in influenza virus biology. Curr Top Microbiol Immunol 385:93–116. doi: 10.1007/82_2014_393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zaraket H, Bridges OA, Russell CJ. 2013. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus replication and pathogenesis in mice. J Virol 87:4826–4834. doi: 10.1128/JVI.03110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaraket H, Baranovich T, Kaplan BS, Carter R, Song MS, Paulson JC, Rehg JE, Bahl J, Crumpton JC, Seiler J, Edmonson M, Wu G, Karlsson E, Fabrizio T, Zhu H, Guan Y, Husain M, Schultz-Cherry S, Krauss S, McBride R, Webster RG, Govorkova EA, Zhang J, Russell CJ, Webby RJ. 2015. Mammalian adaptation of influenza A(H7N9) virus is limited by a narrow genetic bottleneck. Nat Commun 6:6553. doi: 10.1038/ncomms7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mittal A, Shangguan T, Bentz J. 2002. Measuring pKa of activation and pKi of inactivation for influenza hemagglutinin from kinetics of membrane fusion of virions and of HA expressing cells. Biophys J 83:2652–2666. doi: 10.1016/S0006-3495(02)75275-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prinzinger R, Preßmar A, Schleucher E. 1991. Body temperature in birds. Comp Biochem Phys 99:499–506. doi: 10.1016/0300-9629(91)90122-S. [DOI] [Google Scholar]
- 67.Massin P, van der Werf S, Naffakh N. 2001. Residue 627 of PB2 is a determinant of cold sensitivity in RNA replication of avian influenza viruses. J Virol 75:5398–5404. doi: 10.1128/JVI.75.11.5398-5404.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.An Y, Cipollo JF. 2011. An unbiased approach for analysis of protein glycosylation and application to influenza vaccine hemagglutinin. Anal Biochem 415:67–80. doi: 10.1016/j.ab.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 69.Ni F, Kondrashkina E, Wang Q. 2013. Structural basis for the divergent evolution of influenza B virus hemagglutinin. Virology 446:112–122. doi: 10.1016/j.virol.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hayashi H, Kubo Y, Izumida M, Takahashi E, Kido H, Sato K, Yamaya M, Nishimura H, Nakayama K, Matsuyama T. 2018. Enterokinase enhances influenza A virus infection by activating trypsinogen in human cell lines. Front Cell Infect Microbiol 8:91. doi: 10.3389/fcimb.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hamilton BS, Whittaker GR. 2013. Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J Biol Chem 288:17399–17407. doi: 10.1074/jbc.M112.440362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Magnen M, Elsasser BM, Zbodakova O, Kasparek P, Gueugnon F, Petit-Courty A, Sedlacek R, Goettig P, Courty Y. 2018. Kallikrein-related peptidase 5 and seasonal influenza viruses, limitations of the experimental models for activating proteases. Biol Chem 399:1053–1064. doi: 10.1515/hsz-2017-0340. [DOI] [PubMed] [Google Scholar]
- 73.Zmora P, Blazejewska P, Moldenhauer AS, Welsch K, Nehlmeier I, Wu Q, Schneider H, Pöhlmann S, Bertram S. 2014. DESC1 and MSPL activate influenza A viruses and emerging coronaviruses for host cell entry. J Virol 88:12087–12097. doi: 10.1128/JVI.01427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhirnov OP, Manykin AA, Rossman JS, Klenk HD. 2016. Intravirion cohesion of matrix protein M1 with ribonucleocapsid is a prerequisite of influenza virus infectivity. Virology 492:187–196. doi: 10.1016/j.virol.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 75.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. 1997. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol 139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakai K, Sekizuka T, Ami Y, Nakajima N, Kitazawa M, Sato Y, Nakajima K, Anraku M, Kubota T, Komase K, Takehara K, Hasegawa H, Odagiri T, Tashiro M, Kuroda M, Takeda M. 2015. A mutant H3N2 influenza virus uses an alternative activation mechanism in TMPRSS2 knockout mice by loss of an oligosaccharide in the hemagglutinin stalk region. J Virol 89:5154–5158. doi: 10.1128/JVI.00124-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409–417. doi: 10.1016/s0092-8674(00)81771-7. [DOI] [PubMed] [Google Scholar]
- 78.Stevens J, Corper AL, Basler CF, Taubenberger JK, Palese P, Wilson IA. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- 79.Brassard DL, Lamb RA. 1997. Expression of influenza B virus hemagglutinin containing multibasic residue cleavage sites. Virology 236:234–248. doi: 10.1006/viro.1997.8749. [DOI] [PubMed] [Google Scholar]
- 80.Nakowitsch S, Waltenberger AM, Wressnigg N, Ferstl N, Triendl A, Kiefmann B, Montomoli E, Lapini G, Sergeeva M, Muster T, Romanova JR. 2014. Egg- or cell culture-derived hemagglutinin mutations impair virus stability and antigen content of inactivated influenza vaccines. Biotechnol J 9:405–414. doi: 10.1002/biot.201300225. [DOI] [PubMed] [Google Scholar]
- 81.Takashita E, Muraki Y, Sugawara K, Asao H, Nishimura H, Suzuki K, Tsuji T, Hongo S, Ohara Y, Ohara Y, Kawaoka Y, Ozawa M, Matsuzaki Y. 2012. Intrinsic temperature sensitivity of influenza C virus hemagglutinin-esterase-fusion protein. J Virol 86:13108–13111. doi: 10.1128/JVI.01925-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen Z, Aspelund A, Kemble G, Jin H. 2008. Molecular studies of temperature-sensitive replication of the cold-adapted B/Ann Arbor/1/66, the master donor virus for live attenuated influenza FluMist vaccines. Virology 380:354–362. doi: 10.1016/j.virol.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 83.Ueda M, Sugiura A. 1984. Physiological characterization of influenza virus temperature-sensitive mutants defective in the haemagglutinin gene. J Gen Virol 65:1889–1897. doi: 10.1099/0022-1317-65-11-1889. [DOI] [PubMed] [Google Scholar]
- 84.Schuy W, Will C, Kuroda K, Scholtissek C, Garten W, Klenk HD. 1986. Mutations blocking the transport of the influenza virus hemagglutinin between the rough endoplasmic reticulum and the Golgi apparatus. EMBO J 5:2831–2836. doi: 10.1002/j.1460-2075.1986.tb04576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gallagher PJ, Henneberry JM, Sambrook JF, Gething MJ. 1992. Glycosylation requirements for intracellular transport and function of the hemagglutinin of influenza virus. J Virol 66:7136–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roberts PC, Garten W, Klenk HD. 1993. Role of conserved glycosylation sites in maturation and transport of influenza A virus hemagglutinin. J Virol 67:3048–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerlach T, Hensen L, Matrosovich T, Bergmann J, Winkler M, Peteranderl C, Klenk HD, Weber F, Herold S, Pohlmann S, Matrosovich M. 2017. pH optimum of hemagglutinin-mediated membrane fusion determines sensitivity of influenza A viruses to the interferon-induced antiviral state and IFITMs. J Virol 91:e00246-17. doi: 10.1128/JVI.00246-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koerner I, Matrosovich MN, Haller O, Staeheli P, Kochs G. 2012. Altered receptor specificity and fusion activity of the haemagglutinin contribute to high virulence of a mouse-adapted influenza A virus. J Gen Virol 93:970–979. doi: 10.1099/vir.0.035782-0. [DOI] [PubMed] [Google Scholar]
- 89.Takahashi T, Kurebayashi Y, Ikeya K, Mizuno T, Fukushima K, Kawamoto H, Kawaoka Y, Suzuki Y, Suzuki T. 2010. The low-pH stability discovered in neuraminidase of 1918 pandemic influenza A virus enhances virus replication. PLoS One 5:e15556. doi: 10.1371/journal.pone.0015556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 91.McDonough JE, Kaminski N, Thienpont B, Hogg JC, Vanaudenaerde BM, Wuyts WA. 2019. Gene correlation network analysis to identify regulatory factors in idiopathic pulmonary fibrosis. Thorax 74:132–140. doi: 10.1136/thoraxjnl-2018-211929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vanderlinden E, Göktas F, Cesur Z, Froeyen M, Reed ML, Russell CJ, Cesur N, Naesens L. 2010. Novel inhibitors of influenza virus fusion: structure-activity relationship and interaction with the viral hemagglutinin. J Virol 84:4277–4288. doi: 10.1128/JVI.02325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Simmons G, Reeves JD, Grogan CC, Vandenberghe LH, Baribaud F, Whitbeck JC, Burke E, Buchmeier MJ, Soilleux EJ, Riley JL, Doms RW, Bates P, Pöhlmann S. 2003. DC-SIGN and DC-SIGNR bind Ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 305:115–123. doi: 10.1006/viro.2002.1730. [DOI] [PubMed] [Google Scholar]
- 94.Vrijens P, Noppen S, Boogaerts T, Vanstreels E, Ronca R, Chiodelli P, Laporte M, Vanderlinden E, Liekens S, Stevaert A, Naesens L. 2019. Influenza virus entry via the GM3 ganglioside-mediated platelet-derived growth factor receptor beta signalling pathway. J Gen Virol 100:583–601. doi: 10.1099/jgv.0.001235. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.