While APH-2 is the only viral protein consistently expressed in infected carriers, its role in HTLV-2 infection is poorly understood. In this study, we characterized the interaction between the ESCRT-0 component HRS and APH-2 and explored the role of HRS in HTLV-2 replication. HRS is a master regulator of protein sorting for lysosomal degradation, a feature that is manipulated by several viruses to promote replication. Unexpectedly, we found that HRS targets APH-2 and possibly Tax2B for lysosomal degradation and has an overall negative impact on HTLV-2 replication and release. The negative impact of interactions between HTLV-2 regulatory proteins and HRS, and by extension the ESCRT machinery, may represent an important strategy used by HTLV-2 to limit virus production and to promote persistence, features that may contribute to the limited pathogenic potential of this infection.
KEYWORDS: ESCRT, HRS, HTLV-1, HTLV-2, lysosomes
ABSTRACT
The divergent clinical outcomes of human T cell leukemia virus type 1 (HTLV-1) and HTLV-2 infections have been attributed to functional differences in their antisense proteins. In contrast to HTLV-1 bZIP factor (HBZ), the role of the antisense protein of HTLV-2 (APH-2) in HTLV-2 infection is poorly understood. In previous studies, we identified the endosomal sorting complex required for transport 0 (ESCRT-0) subunit HRS as a novel interaction partner of APH-2 but not HBZ. HRS is a master regulator of endosomal protein sorting for lysosomal degradation and is hijacked by many viruses to promote replication. However, no studies to date have shown a link between HTLVs and HRS. In this study, we sought to characterize the interaction between HRS and APH-2 and to investigate the impact of HRS on the life cycle of HTLV-2. We confirmed a direct specific interaction between APH-2 and HRS and showed that the CC2 domain of HRS and the N-terminal domain of APH-2 mediate their interaction. We demonstrated that HRS recruits APH-2 to early endosomes, possibly furnishing an entry route into the endosomal/lysosomal pathway. We demonstrated that inhibition of this pathway using either bafilomycin or HRS overexpression substantially extends the half-life of APH-2 and stabilizes Tax2B expression levels. We found that HRS enhances Tax2B-mediated long terminal repeat (LTR) activation, while depletion of HRS enhances HTLV-2 production and release, indicating that HRS may have a negative impact on HTLV-2 replication. Overall, our study provides important new insights into the role of the ESCRT-0 HRS protein, and by extension the ESCRT machinery and the endosomal/lysosomal pathway, in HTLV-2 infection.
IMPORTANCE While APH-2 is the only viral protein consistently expressed in infected carriers, its role in HTLV-2 infection is poorly understood. In this study, we characterized the interaction between the ESCRT-0 component HRS and APH-2 and explored the role of HRS in HTLV-2 replication. HRS is a master regulator of protein sorting for lysosomal degradation, a feature that is manipulated by several viruses to promote replication. Unexpectedly, we found that HRS targets APH-2 and possibly Tax2B for lysosomal degradation and has an overall negative impact on HTLV-2 replication and release. The negative impact of interactions between HTLV-2 regulatory proteins and HRS, and by extension the ESCRT machinery, may represent an important strategy used by HTLV-2 to limit virus production and to promote persistence, features that may contribute to the limited pathogenic potential of this infection.
INTRODUCTION
Human T cell leukemia virus type 2 (HTLV-2) was the second retrovirus discovered in the late 1970s and is estimated to infect approximately 800,000 people worldwide (1, 2). While HTLV-2 is 70% similar to HTLV-1, the causative agent of adult T cell leukemia (ATL) or HTLV-1-associated myelopathy/tropical spastic paraparesis, HTLV-2 is mainly nonpathogenic, being associated with lymphocytosis, certain inflammatory conditions, and an overall higher mortality risk (3, 4). Despite intensive efforts, mechanisms behind the establishment of such divergent clinical outcomes are still unclear (5).
Most HTLV structural and enzymatic genes (gag, pol, and env) and regulatory genes, such as tax and rex, are expressed from the 5′ long terminal repeat (LTR) using the sense strand of the viral genome. The viral Tax protein promotes viral transactivation by interacting with various host transcriptional factors such as activating transcription factor (ATF)/CREB and p300/CREB-binding protein (CBP), resulting in their recruitment to Tax-responsive elements (TxREs) at the 5′ LTR and transcriptional activation (6). In addition to sense transcription, HTLVs use the antisense strand of their viral genomes to express proteins from the 3′ LTR. This leads to the production of antisense proteins, i.e., HTLV-1 bZIP factor (HBZ) for HTLV-1 and antisense protein of HTLV-2 (APH-2) for HTLV-2 (7, 8).
As opposed to Tax, both antisense proteins are consistently expressed in most infected cells and interact with shared and unique cellular factors. Like HBZ, APH-2 inhibits Tax-mediated viral gene expression by interacting with CREB (9–12). A recent study examined the effects of APH-2 on pathways known to be regulated by HBZ and showed that, while both proteins downregulate the Tax-mediated NF-κB pathway, APH-2 has divergent effects on transforming growth factor β (TGF-β) and interferon regulatory factor 1 (IRF-1) signaling (13). Such opposite effects show that APH-2 and HBZ modulate cellular and viral pathways in a divergent manner, leading to the suggestion that these differences may influence key processes involved in the viral life cycle and disease development in the case of HTLV-1 but not HTLV-2. While both proteins are dispensable for the immortalization of T cells in vitro, studies show that APH-2, unlike HBZ, has no effect on the proliferation of HTLV-2-infected cells and is dispensable for viral replication and persistence in vivo (9, 12, 14). This was shown by the finding that rabbits infected with APH-2-deficient virus displayed higher rates of replication and higher proviral loads (12). This led to the conclusion that APH-2 may have a protective role in HTLV-2 infection and may contribute to the nonpathogenic nature of HTLV-2. To date, however, few studies have examined interactions between APH-2 and cellular factors (12, 15, 16).
To expand our current knowledge on possible cellular interaction partners for APH-2, we performed yeast two-hybrid screening (N. Sheehy and W. W. Hall, unpublished data). This screening showed that APH-2 interacts with several components of the endosomal complex required for transport (ESCRT) machinery. This machinery is involved in membrane remodeling, facilitating membrane budding and vesicle release. This key feature means that the ESCRT machinery regulates many cellular processes, such as trafficking and lysosomal degradation of internalized membrane-bound receptors via the multivesicular body (MVB) pathway, cytokinesis, exosome release, autophagy, neuron pruning, and nuclear envelope reassembly (17).
The ESCRT machinery is composed of multiprotein complexes known as ESCRT-0, I, II, and III and the VPS4 ATPase complex, together with accessory proteins such as Alix. Each ESCRT complex is recruited sequentially to membranes to promote the budding and release of vesicles, which are essential for the trafficking and lysosomal degradation of internalized plasma membrane receptors (18). The role of the ESCRT machinery in the lysosomal degradation of cellular signaling receptors such as epidermal growth factor receptor (EGFR) and TGF-β via the MVB pathway is well characterized (19, 20). The ESCRT-0 protein HRS initiates this process by binding to ubiquitinated cargos and tethering them to the surface of early endosomes (21). HRS subsequently recruits the ESCRT-I complex by binding TSG101 through a conserved PSAP motif (22, 23). ESCRT-I in turn recruits ESCRT-II, which recruits and activates ESCRT-III complexes. Finally, ESCRT-III complexes recruit the VPS4 ATPase, which dissociates the ESCRT machinery from the membrane, completing the release of vesicles to form MVBs (17, 24).
Viruses usurp the ESCRT machinery for replication and release from infected cells. The role of the ESCRT machinery has been extensively studied for retroviruses, but it is now established that most enveloped viruses use this machinery to bud from infected cells (25). The PSAP late domain in the HIV-1 Gag protein mimics the PSAP domain in HRS to interact with the ESCRT-I protein TSG101 and to recruit the ESCRT machinery, ensuring efficient viral budding (26). HTLV-1 also relies on the ESCRT machinery to bud from infected cells. Previous studies have shown that the HTLV-1 Gag protein interacts with TSG101 through a PPPYEPTAP motif, resulting in efficient release of virions (27, 28). In addition to viral budding, the interactions of the HIV-1 accessory proteins Vpu and Nef with the ESCRT-0 protein HRS and the ESCRT accessory protein Alix, respectively, promote viral replication by facilitating the lysosomal degradation of cellular restriction factors such as tetherin and the viral receptor CD4 (29, 30). Overall, it is clear that the ESCRT machinery plays a significant role in the establishment and dissemination of retroviral infections. Interactions between HTLV-2 and the ESCRT machinery have yet to be described.
In this study, we sought to confirm the interaction between APH-2 and HRS highlighted by our yeast two-hybrid screening and to investigate the impact of HRS on the life cycle of HTLV-2. Our study confirms a direct interaction between APH-2 and HRS and reveals that HRS overexpression or bafilomycin treatment, which is known to inhibit the endolysosomal pathway, leads to the recruitment and stabilization of APH-2 into early endosomes. This strongly suggests that this pathway is involved in the degradation of APH-2 and is responsible in part for its instability. We also demonstrate that HRS has an overall negative impact on HTLV-2 replication and dissemination.
RESULTS
APH-2 directly interacts with the ESCRT-0 subunit HRS.
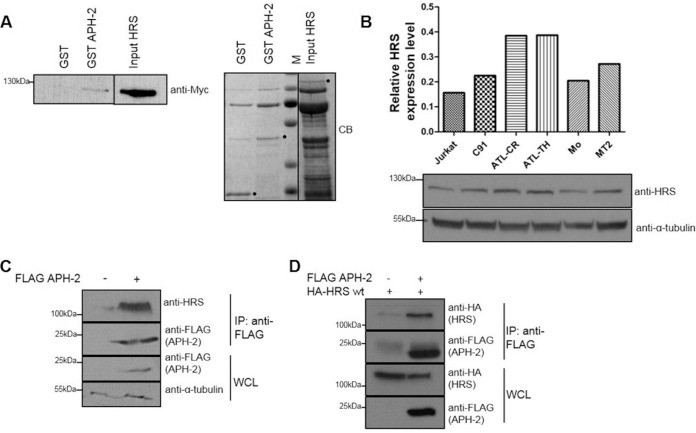
In order to better decipher the function of APH-2 in HTLV-2 infection, a yeast two-hybrid approach was used to screen a number of different T cell cDNA libraries to identify potential APH-2 interactors. From those studies, we discovered that APH-2 interacts with several components of the ESCRT machinery, including the ESCRT-0 subunit HRS, also known as HGS (data not shown). Although retroviral Gag proteins mimic HRS to bind TSG101 and to recruit the ESCRT machinery, which is essential for viral budding, HRS is also known to have a significant impact on viral replication via the endosomal/lysosomal pathway (31). Based on this, we speculated that HRS may also play a role in HTLV-2 replication via its interaction with APH-2 and possibly other viral proteins. To confirm the interaction between APH-2 and HRS, we performed glutathione S-transferase (GST) pulldown assays using GST, GST-APH-2, and HRS-His-Myc proteins purified from Escherichia coli (Fig. 1A). Our results illustrated that HRS-His-Myc bound GST-APH-2 but not GST, thus indicating a direct and specific interaction between APH-2 and HRS.
FIG 1.
APH-2 directly interacts with the ESCRT-0 subunit HRS. (A) GST pulldown assays were performed by incubating purified HRS-His-Myc with GST or GST-APH-2 immobilized on GST resin. Eluates were analyzed by immunoblotting using an anti-Myc antibody (left) and Coomassie blue (CB) staining (right). HRS input corresponds to 10% of the total HRS added to each pulldown. M indicates protein markers. Dots indicate purified GST, GST-APH-2, and HRS. (B) Expression levels of endogenous HRS in Jurkat cells, the HTLV-1-transformed cell lines MT2 and C91, the HTLV-2-transformed cell line Mo, and the ATL cell lines ATL-TH and ATL-CR were determined. Equal amounts of whole-cell lysates were analyzed by Western blotting using antibodies against HRS and α-tubulin. For the densitometric analysis, individual bands for HRS were quantified and normalized to the tubulin signal. (C and D) Coimmunoprecipitation assays were performed on lysates from HEK293T cells cotransfected with 10 μg of FLAG-APH-2 or empty plasmid (C) or 5 μg of FLAG-APH-2 together with either 5 μg of empty plasmid or 5 μg of HA-HRS expression plasmid (D). Proteins from whole-cell lysates (WCL) were immunoprecipitated using anti-FLAG M2 resin and were analyzed by Western blotting using anti-FLAG, anti-HRS, and anti-HA antibodies. IP, immunoprecipitation; wt, wild type.
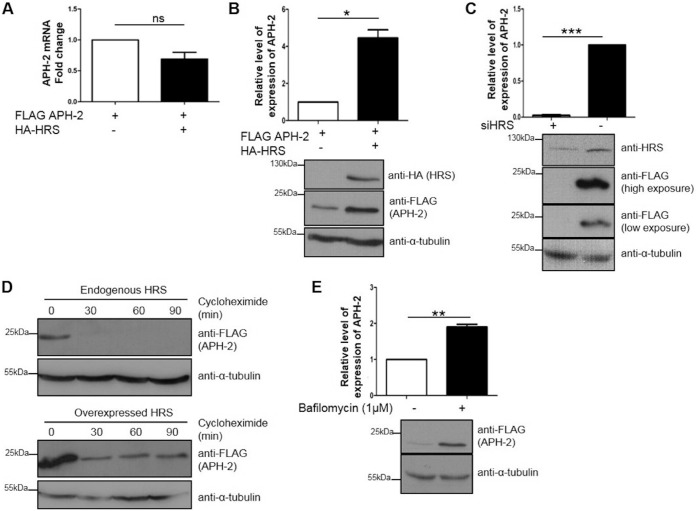
HRS is ubiquitously expressed in a variety of cell types, and HRS overexpression is linked to cancer development (32, 33). Initially, we sought to examine the expression levels of HRS in HTLV-infected cells, compared to HTLV-negative cells, to determine whether HTLV infection affects HRS expression levels. To this end, Western blot analysis was performed on lysates from a HTLV-negative T cell line (Jurkat), two chronically HTLV-1-infected cell lines (MT2 and C91), a chronically HTLV-2-infected cell line (Mo), and two ATL patient cell lines (ATL-CR and ATL-TH) (Fig. 1B). Compared to Jurkat cells, the highest level of HRS expression was observed in the ATL cell lines, followed by MT2, C91, and Mo. In order to confirm the interaction between HRS and APH-2 in mammalian cells, we transfected HEK293T cells with a FLAG-APH-2 expression plasmid or an empty control plasmid. Cellular lysates were subjected to coimmunoprecipitation assays using an anti-FLAG M2 resin, and precipitates were analyzed by Western blotting. We showed that endogenous HRS could be precipitated in the presence of APH-2 but not in its absence (Fig. 1C). Because endogenous levels of HRS in HEK293T cells were not detectable in whole-cell lysates by Western blotting, we decided to perform the same coimmunoprecipitation assays in cells overexpressing HRS (Fig. 1D). We confirmed that hemagglutinin (HA)-HRS was coprecipitated with FLAG-APH-2 when the two proteins were coexpressed but not in the absence of FLAG-APH-2, thus confirming the interaction.
The CC2 domain of HRS interacts with APH-2.
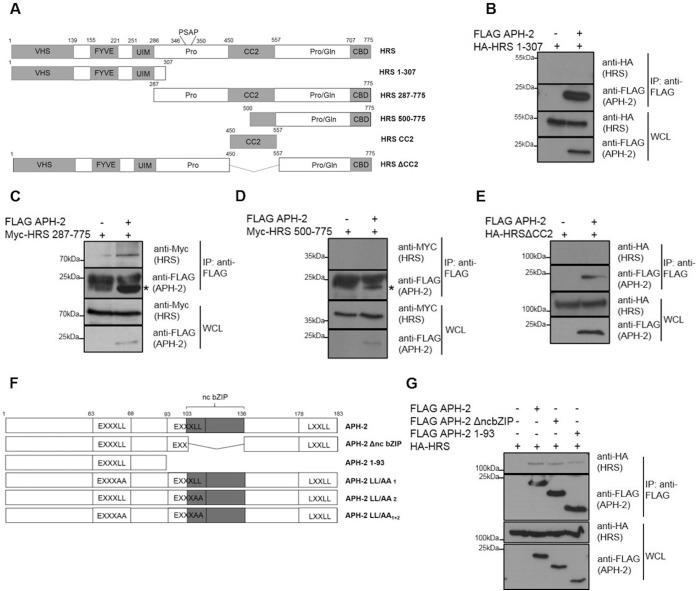
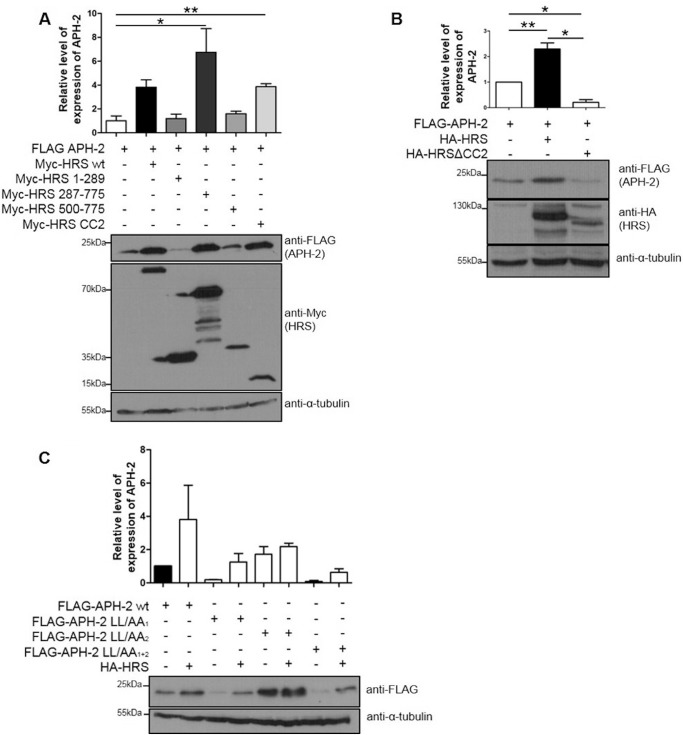
HRS possesses several well-characterized domains, including a VHS domain, an FYVE domain, a ubiquitin interaction motif (UIM), a proline-rich domain that contains a PSAP motif, a coiled-coil domain (CC2), a proline-glutamine-rich (Pro/Gln) domain, and a clathrin-binding domain (CBD) (Fig. 2A) (23). These domains are responsible for the capacity of HRS to bind ubiquitinated proteins, to recruit the subsequent ESCRT components, and to sort cargo proteins into the endosomal/lysosomal pathway (34). To better understand the functional significance of the interaction between APH-2 and HRS, we sought to determine which domain of HRS was involved in the interaction with APH-2. To this end, we cotransfected HEK293T cells with FLAG-APH-2 and the indicated HRS deletion mutants and coimmunoprecipitations were performed on cellular lysates using an anti-FLAG M2 resin (Fig. 2B to E). Possible interactions were analyzed by Western blotting. While the HA-HRS 1–307 and Myc-HRS 500–775 mutants did not coprecipitate with FLAG-APH-2, we observed pulldown of the deletion mutant Myc-HRS 287–775 in the presence of APH-2 (Fig. 2C). This suggests that the region of HRS encompassing amino acids 307 to 500 mediates the interaction with APH-2. This region contains a proline-rich domain and, interestingly, the PSAP motif, which is essential for the interaction of HRS with TSG101 and the recruitment of the other ESCRT components (23). This region also contains the first half of the CC2 domain, a domain that is essential for targeting HRS to early endosomes (35). To assess whether the CC2 domain facilitates the interaction with APH-2, we performed coimmunoprecipitation on lysates from cells cotransfected with expression plasmids encoding FLAG-APH-2 and the deletion mutant HA-HRSΔCC2, lacking the CC2 domain. We showed that HA-HRSΔCC2 was not coprecipitated with FLAG-APH-2 (Fig. 2E). Collectively, these data demonstrate that the CC2 domain is involved in the interaction between HRS and APH-2.
FIG 2.
HRS interacts with the N terminus of APH-2 via its CC2 domain. (A) Schematic representation of the functional domains in HRS and the deletion mutants used in this study. VHS, VPS27-Hrs-STAM; FYVE, FYVE zinc finger domain; UIM, ubiquitin interaction motif; Pro, proline-rich domain containing the PSAP motif; CC2, coiled-coil domain; Pro/Gln, proline- and glutamine-rich domain; CBD, clathrin-binding domain. (B to E) HEK293T cells were cotransfected with FLAG-APH2 or empty plasmid and the indicated HRS deletion plasmids. Cellular lysates were subjected to immunoprecipitation using an anti-FLAG M2 resin, and interactions were analyzed by Western blotting using anti-FLAG, anti-HA, and anti-Myc antibodies. The asterisks indicate the band for FLAG-APH-2. (F) Schematic representation of full-length APH-2 and deletion mutants used in this study. Functional domains are indicated, as follows: EXXXLL, endosomal trafficking motif; ncbZIP, nonconventional basic leucine zipper domain; LXXLL, LXXLL motif. (G) Coimmunoprecipitations with lysates from HEK293T cells cotransfected with HA-HRS and the indicated FLAG-APH-2 deletion mutants were performed using anti FLAG-M2 resin. Antibodies against HA and FLAG epitopes were used to analyze interactions. IP, immunoprecipitation; WCL, whole-cell lysate.
APH-2 binds HRS via the N terminus.
APH-2 contains a nonconventional bZIP domain (ncbZIP), a LXXLL motif in the C terminus, and two putative EXXXLL endosomal trafficking motifs (8, 36). To further characterize the interaction between APH-2 and HRS, we sought to determine the domain in APH-2 involved in the interaction. To this end, we used two different deletion mutants; the APH-2 ΔncbZIP mutant lacks the ncbZIP domain, and APH-2 1–93 contains only the N-terminal portion of APH-2 (Fig. 2F). We performed coimmunoprecipitations using anti-FLAG M2 resin with lysates of HEK293T cells transfected with plasmids encoding HA-HRS and the indicated APH-2 deletion mutants. Western blot analysis of precipitates showed that HA-HRS interacted with APH-2 ΔncbZIP and APH-2 1–93 (Fig. 2G), suggesting that the N-terminal region of APH-2 mediates the interaction with HRS and that the ncbZIP domain plays no role in the interaction.
HRS and APH-2 colocalize in endocytic structures in a CC2-dependent manner.
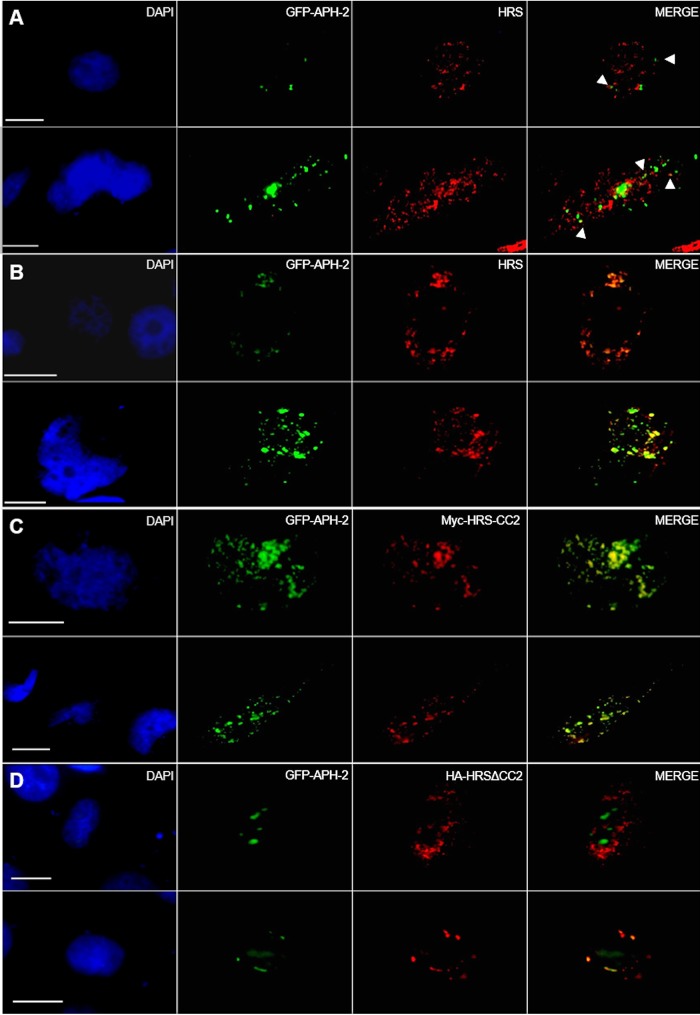
Because HRS plays a crucial role in trafficking proteins through the endocytic pathway to lysosomes for degradation, HRS localizes in the cytoplasm and has been shown to mainly colocalize with EEA1, an early endosome marker. Based on this, we sought to examine the subcellular localization of APH-2 and either exogenous or endogenous HRS in HeLa cells (Fig. 3A and B). HeLa cells were cotransfected with green fluorescent protein (GFP)-APH-2 (green) alone (Fig. 3A) or together with a HA-HRS expression plasmid (Fig. 3B). Both endogenous HRS and exogenous HRS were detected using an anti-HRS antibody followed by the anti-mouse IgG-Alexa Fluor 594 secondary antibody (red). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue), and the subcellular localization of these proteins was examined by fluorescence microscopy. We observed that endogenous HRS localized in the cytoplasm in a granular pattern, which is consistent with previous reports. APH-2, as described previously, displays fluorescence in a speckle-like manner in the cytoplasm and the nucleus (8, 16). In Fig. 3A, we observed some foci that were positive for both APH-2 and HRS, suggesting that the two proteins can colocalize in such structures in the cytoplasm.
FIG 3.
HRS and APH-2 colocalize in endocytic structures in a CC2-dependent manner. HeLa cells were transfected with 750 ng of GFP-APH-2 (green) and 250 ng of control plasmid (A), HA-HRS (B), Myc-HRS-CC2 (C), or HA-HRSΔCC2 (D) expression plasmids; 24 h posttransfection, endogenous HRS and exogenous HRS were detected using anti-HRS (A, B, and D) or anti-Myc antibodies (C), followed by Alexa Fluor 594-conjugated secondary antibody (red). Nuclei were stained with DAPI (blue). Two representative images are shown per condition. The arrowheads indicate colocalization of HRS and GFP-APH-2. Scale bars, 10 μm.
To further investigate the localization of APH-2 and HRS, we performed the previously described staining in cells overexpressing HRS (Fig. 3B). Under these conditions, HRS was detected in large vesicular structures possibly consisting of clustered endocytic structures, as reported previously (37). Interestingly we also observed extensive colocalization of APH-2 and HRS in such structures. This suggests that HRS recruits APH-2 into endocytic structures. Furthermore, we consistently observed that HRS overexpression led to the accumulation of APH-2. Based on our finding that the CC2 domain mediates the interaction between APH-2 and HRS, we sought to determine whether this domain was sufficient to recruit APH-2 into the endocytic structures. To this end, we transfected HeLa cells with GFP-APH-2 and Myc-HRS-CC2 or HA-HRSΔCC2 expression plasmids (Fig. 3C and D). Strikingly, we observed marked colocalization of Myc-HRS-CC2 and GFP-APH-2 in enlarged endosomal clusters, confirming our previous finding that the CC2 domain mediates the interaction between HRS and APH-2 and is sufficient for the accumulation of APH-2 in the endocytic structures (Fig. 3C). This is in stark contrast to the localization of GFP-APH-2 in the presence of HA-HRSΔCC2, where APH-2 predominantly localized in the nucleus and did not accumulate to any great extent in cytoplasmic clusters (Fig. 3D). However, we observed limited colocalization between APH-2 and HA-HRSΔCC2 (Fig. 3D, lower), suggesting that other HRS domains may be involved in the localization of APH-2 to structures distinct from the endocytic structures. Taken together, our results show that the CC2 domain of HRS targets APH-2 to endosomal structures and is responsible for its accumulation and stabilization.
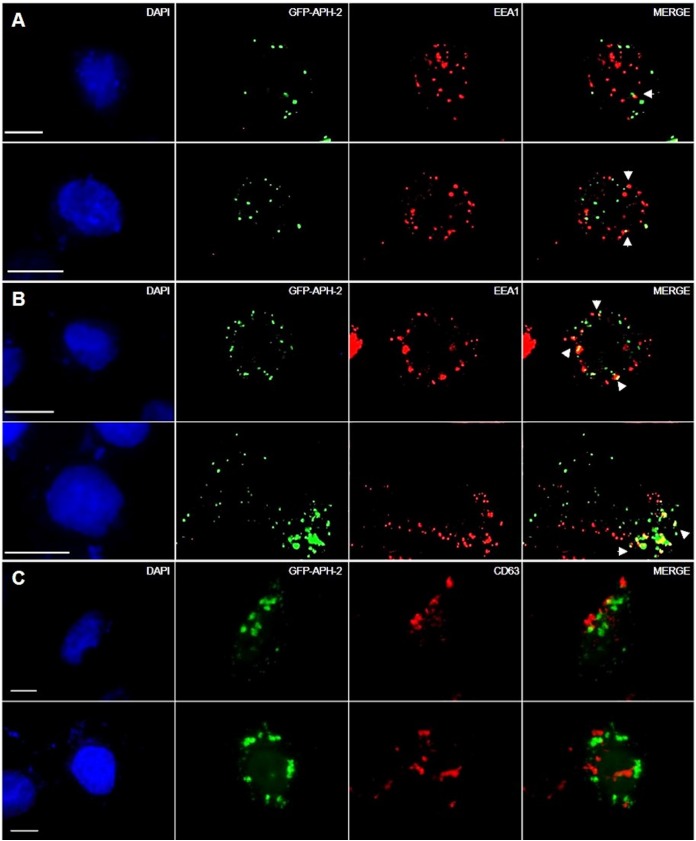
APH-2 and HRS interact in EEA1- but not CD63-positive endosomes.
Cellular proteins destined for lysosomal degradation are initially incorporated into intraluminal vesicles (ILVs) of early endosomes, which mature into late endosomes/MVBs that eventually fuse with lysosomes and are degraded. MVBs can also fuse with the cell membrane and deliver ILVs to the outside of cells or fuse with autophagosomes before lysosomal degradation during autophagy (38). This process is governed by the ESCRT machinery and is initiated by the ESCRT-0 component HRS. Based on this, we postulated that HRS recruits APH-2 to early endosomes for trafficking to lysosomes via the MVB pathway. This was investigated by staining HeLa cells transfected with GFP-APH-2 (Fig. 4A to C) and either an empty control plasmid (Fig. 4A and C) or HA-HRS (Fig. 4B) with an antibody against the early endosome marker EEA1 or the late endosomal marker CD63, followed by the anti-rabbit IgG-Alexa Fluor 594 secondary antibody (red). We observed that GFP-APH-2 colocalized with EEA1 in a limited number of foci (Fig. 4A). Interestingly, HRS overexpression resulted in higher levels of GFP-APH-2 in EEA1-positive structures, compared to levels observed in cells not transfected with exogenous HRS expression plasmids (Fig. 4A and B). Moreover, we observed low levels of colocalization of APH-2 with the late endosomal marker CD63 (Fig. 4C), indicating that HRS overexpression, and hence disruption of the endolysosomal pathway, mainly blocks the trafficking of APH-2 to lysosomes in early endosomes but it can also be found in late endosomes.
FIG 4.
APH-2 and HRS interact in EEA1-positive but not CD63-positive endosomes. HeLa cells were transfected with 750 ng of GFP-APH-2 (green) and 250 ng of control plasmid (A and C) or HA-HRS (B) expression plasmid; 24 h posttransfection, endogenous EEA1 or CD63 was detected using antibodies against EEA1 (A and B) or CD63 (C) epitopes, followed by Alexa Fluor 594-conjugated secondary antibody (red). Nuclei were stained with DAPI (blue). Scale bars, 10 μm.
HRS overexpression extends the half-life of APH-2, and APH-2 is degraded in lysosomes.
Based on our observation from immunofluorescence analysis that HRS enhances APH-2 expression levels, we sought to investigate the mechanisms involved. Initially, we examined the effect of HRS overexpression on APH-2 mRNA levels. We performed reverse transcription-quantitative PCR (RT-qPCR) on mRNA extracted from HEK293T cells cotransfected with FLAG-APH-2 and HA-HRS (Fig. 5A). The APH-2 signal was normalized to the β-actin signal, and the signal for cells transfected with FLAG-APH-2 alone was set as the control, with a value of 1. We observed no statistically significant difference in APH-2 mRNA levels in the presence versus absence of exogenous HRS, suggesting that HRS does not affect the stability of APH-2 mRNA levels. In contrast, Western blot analysis of cellular lysates from HEK293T cells transfected with FLAG-APH-2 and HA-HRS expression plasmids showed a 4-fold increase in APH-2 protein levels in the presence of exogenous HRS, compared to the control condition (Fig. 5B). Unexpectedly, we found that HRS knockdown in HeLa cells expressing APH-2 resulted in a 40-fold reduction in APH-2 levels, compared to those observed with the negative-control small interfering RNA (siRNA), clearly indicating that endogenous HRS stabilizes APH-2 protein levels (Fig. 5C).
FIG 5.
HRS overexpression increases the half-life of APH-2, and APH-2 is degraded in lysosomes. (A and B) Effects of HRS overexpression on APH-2 mRNA and protein levels. (A) HEK293T cells were cotransfected with FLAG-APH-2 and either FLAG-empty (control) or HA-HRS expression plasmids. Twenty-four hours posttransfection, mRNAs were extracted, and APH-2 expression was measured by qRT-PCR. APH-2 mRNA levels were calculated by the ΔCT method. Values were normalized to β-actin expression and compared to the expression of APH-2 when HRS was not transfected, which was set as 1. (B) HEK293T cells were transfected as described for panel A. Twenty-four hours after transfection, cells were lysed and proteins levels were analyzed by Western blotting using anti-HA, anti-FLAG, and anti-α-tubulin antibodies. The APH-2 level in the absence of exogenous HRS was set as 1. (C) Effect of HRS knockdown on APH-2 expression. HeLa cells were transfected twice with a siRNA against HRS or a control siRNA, at times of 0 h and 48 h; cells were transfected with a FLAG-APH-2 expression plasmid 24 h after the second transfection and were lysed after 24 h. Lysates were analyzed by Western blotting using anti-HRS, anti-FLAG, and anti-α-tubulin antibodies. (D) Effect of HRS overexpression on APH-2 half-life. HEK293T cells were transfected with a FLAG-APH-2 plasmid and an empty control plasmid (top) or a HA-HRS expression plasmid (bottom). Twenty-four hours posttransfection, cells were treated with cycloheximide (100 μg/ml) for the indicated times. Cells were lysed with RIPA buffer, and equal amounts of proteins were analyzed by Western blotting using anti-FLAG and anti-α-tubulin antibodies. (E) Effect of the inhibition of lysosomal acidification on APH-2 degradation. HEK293T cells were transfected with a FLAG-APH-2 plasmid. Twenty-four hours posttransfection, cells were treated with bafilomycin (1 μM) for 4 h. Cells were lysed with RIPA buffer, and equal amounts of proteins were analyzed by Western blotting using anti-FLAG and anti-α-tubulin antibodies. Densitometric analyses were performed in three independent experiments. Results are shown as fold changes in comparison with the control condition, which was arbitrarily set as 1. Error bars represent the standard error of the mean. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001, ns, not significant, by two-tailed Student's t test.
Previous studies showed that, in contrast to the HTLV-1 antisense protein HBZ, which has a half-life of approximately 6.4 h, APH-2 has a half-life of ≤30 min (13). To assess the effect of HRS on the turnover of APH-2, HEK293T cells were transfected with a FLAG-APH-2 expression plasmid, with or without a plasmid encoding HRS, and were treated with the translation elongation inhibitor cycloheximide for defined periods. Western blot analysis of lysates showed that APH-2 had a half-life of <30 min in the absence of exogenous HRS (Fig. 5D). In stark contrast, although reduced levels of APH-2 were observed in cells treated with cycloheximide for 30 min, HRS overexpression resulted in steady-state levels of APH-2 and extended its half-life to at least 90 min.
Previous studies reported that HRS overexpression had a dominant negative effect on the endosomal/lysosomal pathway by preventing the recruitment of downstream ESCRT components (22). Hence, our finding that APH-2 expression levels are enhanced under such conditions suggests that APH-2 may be degraded via the lysosomal pathway. Based on this, we sought to determine the role of lysosomes in APH-2 degradation. To this end, we treated cells transfected with an expression plasmid encoding FLAG-APH-2 with bafilomycin (1 μM), a well-characterized lysosomal inhibitor, or dimethyl sulfoxide (DMSO) and examined APH-2 levels by Western blotting (Fig. 5E). We observed a 2-fold increase in APH-2 protein levels in cells treated with bafilomycin, compared to control cells treated with DMSO. This finding firmly establishes that APH-2 is degraded in lysosomes.
HRS enhances the stability of APH-2 in a CC2-dependent manner.
To determine the domain in HRS responsible for APH-2 stabilization, we transfected HEK293T cells with FLAG-APH-2 and Myc-HRS deletion mutant expression plasmids. Protein expression was analyzed by Western blotting, and FLAG-APH-2 signals were normalized to tubulin levels (Fig. 6A). We observed a 6-fold increase in the levels of FLAG-APH-2 in the presence of Myc-HRS 287–775, compared to APH-2 alone. FLAG-APH-2 levels were similarly enhanced by Myc-HRS-CC2 but not by Myc-HRS 1–289 or Myc-HRS 500–775. Surprisingly, loss of the CC2 domain in HRS resulted not only in the loss of the APH-2 stabilization but also in a 4-fold decrease in APH-2 basal expression (Fig. 6B). Overall, this shows that the interaction between HRS and APH-2 via the CC2 domain is required for APH-2 stabilization.
FIG 6.
The CC2 domain of HRS suffices to stabilize APH-2, and an EXXXLL motif in APH-2 targets it for lysosomal degradation. (A and B) Cellular lysates of HEK293T expressing FLAG-APH-2 plasmid and Myc-HRS (A) or HA-HRS (B) deletion mutants were analyzed by Western blotting using anti-FLAG, anti-Myc, anti-HA, and anti-α-tubulin antibodies. Results are shown as fold changes in comparison with the condition in which APH-2 was expressed alone, which was arbitrarily set as 1. Error bars represent the standard error of the mean. *, P ≤ 0.05; **, P ≤ 0.01, by two-tailed Student's t test. Data represent three independent experiments. (C) The effect of the EXXXLL motif on APH-2 stabilization was assessed. HeLa cells were transfected with the indicated FLAG-APH-2 mutants and an empty control plasmid or HA-HRS expression plasmid. Cellular lysates were analyzed by Western blotting using anti-FLAG and anti-α-tubulin antibodies. The value for APH-2 expressed alone was arbitrarily set as 1, and error bars represent the standard error of the mean. Data represent two independent experiments.
Analysis of the protein sequence of APH-2 indicates that it contains two possible endocytic targeting motifs of EXXXLL (residues 6 to 68 and 101 to 106), which resemble the consensus dileucine endocytic targeting motif [D/E]XXXLL/I, where X is any amino acid. These motifs are found in several cellular receptors that traffic through endosomes to lysosomes for degradation (36). Moreover, these domains are also present in viral proteins such as HIV Nef and Vpu, which are involved in the trafficking of cellular proteins for lysosomal degradation. Based on this, we speculated that these domains may play a role in trafficking APH-2 to lysosomes. To examine this possibility, we transfected HEK293T cells with expression plasmids encoding APH-2 proteins with leucine-to-alanine (LL/AA) substitutions in both endocytic motifs, with or without a plasmid encoding HRS (Fig. 6C). We showed that wild-type FLAG-APH-2 and FLAG-APH-2-LL/AA1 were both stabilized by HRS overexpression, whereas there was no difference in the stability of FLAG-APH-2-LL/AA2 in the presence or absence of exogenous HRS. Indeed, the FLAG-APH-2-LL/AA2 expression levels were higher than those of wild-type FLAG-APH-2 in the absence of exogenous HRS, suggesting that this motif is involved in targeting APH-2 for lysosomal degradation. When the two motifs were mutated (FLAG-APH-2-LL/AA1 + 2), however, the stabilization induced by HRS overexpression was restored, suggesting that both motifs are required for APH-2 to be stabilized by HRS.
HRS overexpression enhances Tax2B-mediated LTR activation.
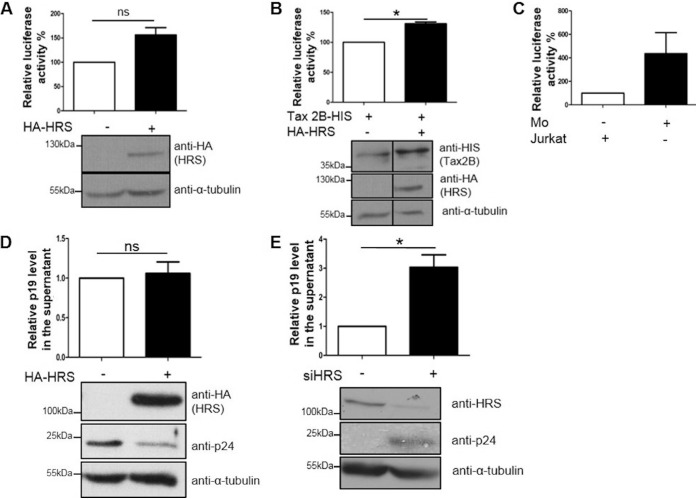
To examine the possible role of HRS in HTLV-2 replication, we investigated the impact of HRS overexpression on basal and Tax2B-mediated LTR activation. We transfected HeLa cells with a plasmid containing the HTLV-2 LTR linked to a luciferase reporter, together with either an empty plasmid or a His-Tax2B expression plasmid and a HA-HRS construct, as indicated (Fig. 7A and B). Twenty-four hours posttransfection, cells were lysed, and luciferase assays were performed. We found that, while HRS overexpression had no effect on basal LTR activation (Fig. 7A), it upregulated Tax2B-mediated activation of the LTR promoter (Fig. 7B). Analysis of cellular lysates by Western blotting showed that, similarly to APH-2, Tax2B levels were increased by HRS overexpression (Fig. 7B). Overall, we conclude that inhibition of the endosomal/lysosomal pathway by HRS overexpression stabilizes Tax2B expression possibly by inhibiting its lysosomal degradation. This may contribute to the elevated levels of Tax2B-mediated LTR activation observed when HRS is overexpressed.
FIG 7.
HRS enhances Tax2B-mediated LTR activation and suppresses HTLV-2 production and release. (A and B) Effects of HRS on basal (A) and Tax2B-mediated (B) LTR activation. HeLa cells were cotransfected with 100 ng of HTLV-2 LTR-luciferase reporter construct, 20 ng of pCAGGS-Tax2B, and 3 μg of HRS or control plasmids as indicated. Luciferase assays were carried out 24 h posttransfection, and results were normalized to the protein concentrations. The mean of three independent experiments is shown. Results are plotted as percentage luciferase activation relative to the control, which was arbitrarily set as 100%. Lysates were subjected to Western blot analysis using anti-HA, anti-His, and anti-α-tubulin antibodies. (C) HTLV-2 infection model. HeLa cells were transfected with 100 ng of HTLV-2 LTR; 24 h posttransfection, similar numbers of Jurkat or Mo cells were added on top of the HeLa cells for 24 h before the cells were washed three times with PBS and incubated for an additional 24 h prior to cell lysis with passive lysis buffer. Luciferase assays were carried out, and values were normalized to protein concentrations. (D and E) Effects of HRS overexpression (D) or knockdown (E) on HTLV-2 Gag levels. HeLa cells were transfected with similar amounts of HA-HRS or control expression plasmids (D) or were double transfected over 24 h with a control siRNA or a siRNA against HRS (E); 24 h posttransfection, similar numbers of Mo cells were added on top of the HeLa cells for 24 h before the cells were washed three times with PBS and incubated for an additional 24 h prior to cell lysis. Protein levels were analyzed by Western blotting using antibodies against HA, HRS, p24, and α-tubulin epitopes. p19gag protein levels in the supernatants were quantified by ELISA. Results are shown as fold changes in comparison with the control condition, which was arbitrarily set as 1. Error bars represent the standard error of the mean. *, P ≤ 0.05; ns, not significant, by two-tailed Student's t test. Data represent three independent experiments.
HRS inhibits HTLV-2 production and release.
We next sought to examine whether HRS affects HTLV-2 production and release. In order to study this, we used an infection model consisting of HeLa cells transfected with our plasmid of interest or siRNA and cocultured with Mo cells for 24 h. To assess the efficiency of the infection, HeLa cells were first transfected with the HTLV-2 LTR-luciferase reporter construct and then cocultured with HTLV-2-infected Mo cells. Twenty-four hours later, Mo cells were removed, and HeLa cells were extensively washed before being cultivated for an additional 24 h. Control cells were cocultured with similar numbers of noninfected Jurkat cells. Luciferase assays showed a 4-fold increase in the luciferase activity for cells cultured in the presence of Mo cells, compared to that detected in the presence of Jurkat cells, thus validating our infection model (Fig. 7C). To examine the effect of HRS on HTLV-2 replication, we performed the same infection in HeLa cells that overexpressed HRS or were knocked down for HRS (Fig. 7D and E). We observed that, while HRS overexpression had no statistically significant effect on the release of p19gag in the supernatant, intracellular p24gag levels were strongly reduced (Fig. 7D). HRS knockdown, however, induced a 3-fold increase in p19gag levels in the supernatant and increased intracellular p24gag levels (Fig. 7E). Overall, our results suggest that inhibition of the endolysosomal pathway by HRS overexpression substantially reduced intracellular levels of p24gag, suggesting that the endolysosomal pathway is involved in the intracellular accumulation of virus. However, HRS knockdown increased p19gag and intracellular p24gag levels, suggesting that HRS has an overall inhibitory effect on HTLV-2 production.
DISCUSSION
It is well established that viruses hijack the ESCRT machinery to promote viral replication. The interaction between the late domain in retroviral Gag proteins and the ESCRT-I subunit TSG101 has been extensively studied and is essential for recruitment of the ESCRT machinery and viral budding and release. Although the PSAP domains in retroviral Gag proteins mimic the PSAP in HRS to bind TSG101, HRS plays an alternative role in viral replication that is mediated mainly by its interactions with viral accessory proteins and the endosomal/lysosomal pathway. For instance, the interaction between the HIV-1 Vpu accessory protein and HRS promotes viral release by facilitating the lysosomal degradation of BST-2/tetherin (39). Our previous studies showed that the HTLV-2 antisense protein APH-2 interacts with several components of the ESCRT machinery, thus implicating this pathway in HTLV-2 infection. In this study, we characterized one of these interactions, namely, the interaction between APH-2 and the ESCRT-0 subunit HRS. We show that HRS recruits APH-2 to early endosomes, possibly furnishing an entry route into the MVB/lysosomal pathway. We demonstrated that inhibition of this pathway using either bafilomycin or HRS overexpression resulted in the stabilization of APH-2 and Tax2B expression levels, clearly indicating that this pathway is involved in the degradation of these viral proteins. Moreover, we found that HRS enhanced Tax2B-mediated LTR activation and may have an overall negative impact on HTLV-2 replication.
The abnormal expression of growth factor receptors is associated with a large number of malignancies (40). Based on its key role in the degradation and hence deactivation of cell surface receptors that regulate cell growth and survival, HRS was speculated to play a protective role against cancer. However, previous studies have shown the opposite, in that HRS is overexpressed in several cancers, compared to corresponding healthy tissue, and disruption of HRS expression in cancer cells diminishes cell proliferation, soft agar colony formation, and in vivo tumorigenesis (33). Our analysis of the expression levels of HRS in HTLV-1- and HTLV-2-infected cells showed that the highest HRS expression levels were detected in ATL-TH and ATL-CR cells, followed by MT2, C91, and Mo cells, compared to Jurkat cells. Given that HRS expression is induced by interleukin 2 (IL-2) stimulation of T cells (32) and can be upregulated by viral infection (41), it would be of great interest to investigate the levels of HRS expression in cells from patients infected with HTLV-1 and HTLV-2.
Ubiquitin is a well-established marker for targeting proteins for ESCRT-dependent lysosomal degradation (42). HRS together with the ESCRT-0 subunit STAM and Eps15b bind to ubiquitinated proteins via UIMs to initiate protein sorting at early endosomes (43). In the present study, using bacterially expressed proteins and GST pulldown assays, we show that APH-2 directly interacts with HRS independently of posttranslational modification. Moreover, we show that the HRS CC2 domain alone and not the UIM in HRS is responsible for its interaction with APH-2. This clearly indicates that this interaction is not ubiquitin dependent. There are several studies showing that HRS interacts with and downregulates cellular proteins in a ubiquitin-independent manner. HRS has been shown to downregulate cytokine receptors such as IL-2 receptor β (IL-2Rβ) and IL-4Rα in a ubiquitin-independent manner (44). Moreover, ubiquitin is not involved in the association of the hepatitis B core (HBc) antigen and HRS (54). The CC2 domain of HRS is responsible for the interaction between HRS and the ESCRT-0 component STAM, and loss of this domain leads to destabilization of the ESCRT-0 complex (32, 35). The CC2 domain is also required for the localization of HRS to early endosomes (21). We initially speculated that the interaction between APH-2 and HRS via the CC2 domain might disrupt the normal function of HRS by preventing the binding of cellular components essential for the stability and function of the ESCRT-0 complex. However, we did not observe that APH-2 altered the normal subcellular localization of HRS; instead, HRS overexpression led to the colocalization of APH-2 in EEA1-positive early endosomes.
A striking observation made in this study is that HRS overexpression significantly enhances APH-2 protein levels, while HRS knockdown promotes its degradation. Previous studies showed that, in contrast to the HTLV-1 antisense protein HBZ, which has a half-life of approximately 6 h, APH-2 has a very short half-life of 20 min in Jurkat cells (13). In the present study, we observed that HRS overexpression resulted in substantially higher levels of APH-2 in EEA1-positive endosomes, in a CC2-dependent manner. Based on our finding that HRS and APH-2 interact via this domain, we propose that HRS recruits APH-2 into such structures. We also demonstrated that, although there was a substantial reduction in APH-2 expression levels after 30 min of cycloheximide treatment, which is consistent with previous studies (13), HRS overexpression extended the half-life of APH-2 to at least 90 min, clearly indicating that APH-2 levels are stabilized under these conditions. HRS overexpression was shown in previous studies to lead to the accumulation of EGFR in early endosomes and to inhibit the trafficking of this receptor from early to late endosomes (22). We also found that HRS overexpression resulted in the accumulation of APH-2 in early endosomes, suggesting that, like EGFR, APH-2 is trafficked through early endosomes on the way to lysosomes. This conclusion was supported by our data showing that bafilomycin, a lysosomal inhibitor, also stabilized APH-2 expression. Although lysosomal degradation of APH-2 has not been reported previously, APH-2 has been shown to be degraded by the proteasome following its SUMOylation (45). Overall, our data support the view that, in addition to the proteasome, APH-2 is degraded by the lysosomal pathway. On the face of it, our data showing that HRS depletion promotes the degradation of APH-2 are counterintuitive, because we would have expected that, if HRS was responsible for targeting APH-2 to early endosomes and lysosomal degradation, we would observe higher levels of APH-2 in the absence of HRS. Although the reasons behind these observations are not clear, they raise the possibility that APH-2 is degraded by alternative means, possibly in the proteasome, in the presence of reduced levels of HRS. Overall our data support the view that the interaction between APH-2 and HRS is essential for the initial recruitment of APH-2 to early endosomes, which is followed by endosomal/lysosomal degradation.
Previous studies showed that the ESCRT machinery plays a role in HTLV-1 replication and release. It was proposed that interactions between the PPPY and PTAP domains in the HTLV-1 Gag protein and the ubiquitin ligase Nedd4 and TSG101, respectively, govern the viral trafficking of Gag proteins through the endocytic pathway to the cell membrane and promote virus release (28). The role of HRS in HTLV replication and release has not been previously addressed. To address this, we initially sought to determine the effects of HRS on basal and Tax2B-mediated LTR activation. We show that, although HRS has no effect on basal LTR activation, HRS overexpression gave rise to a significant increase in Tax2B-mediated activation of the HTLV-2 promoter. Since HRS is not a nuclear protein, the mechanism involved in the stimulation of Tax2B-mediated LTR activation is not clear but, like APH-2, we consistently observed elevated levels of Tax2B expression in the presence of exogenous HRS, which may account in part for the elevated levels of LTR activation by Tax2B detected under such conditions. A previous study examining the host/HTLV protein/protein interactomes showed that Tax1 and Tax2 from HTLV-1 and HTLV-2, respectively, interact with HRS (46). Although not confirmed, the interaction between Tax2B and HRS may influence the capacity of Tax2B to activate the LTR or Tax2B and APH-2 may compete to bind HRS, but this requires further investigation. In terms of HTLV-2 replication and release, we show that inhibition of the endolysosomal pathway by HRS overexpression reduces intracellular p24gag levels while having no effect on viral release. This suggests that this pathway is involved in the control of HTLV-2 production. HRS knockdown, however, enhances intracellular p24gag levels and cell-free p19gag levels. This suggests that HRS may have an overall inhibitory effect on HTLV-2 replication and release. Although the mechanisms involved in the regulation of HTLV-2 replication by HRS are unclear, the ESCRT machinery, and HRS in particular, has been shown to regulate other endosomal degradative processes, such as autophagy (47). Loss of HRS promotes the accumulation of autophagosomes by inhibiting the fusion of these structures with lysosomes. HRS is indispensable for the lysosomal degradation of neurodegeneration-related proteins, and the disruption of HRS is linked to the development of neurological disorders such as Alzheimer’s disease and Parkinson’s disease (48–50). Previous studies showed that Tax1 and Tax2 also promote the accumulation of autophagosomes, which aids viral replication and survival of infected cells (51, 52). The role of APH-2 in autophagy has not been determined, but it may have the opposite effect, compared with Tax2, and promote autophagosome fusion with lysosomes. This may have an overall negative impact on viral replication, which could be related to the observations made in the present study but requires further investigation.
In conclusion, we have identified for the first time the interaction between HTLV-2 and the ESCRT machinery. We demonstrate that HRS leads to the stabilization of APH-2 and Tax2B, two key HTLV-2 regulatory proteins. We show that HRS affects HTLV-2 replication and release, leading us to conclude that the ESCRT-dependent endolysosomal pathway plays a key role in HTLV-2 infection and possibly low pathogenicity.
MATERIALS AND METHODS
Cell culture.
HeLa cells and HEK293T were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS). The HTLV-2-infected cell line Mo was cultured in RPMI 1640 medium containing 20% FBS and 100 μg/ml penicillin-streptomycin (Gibco, Life Technologies). The HTLV-1-infected cell lines MT2 and C91-PL, the ATL cell lines ATL-CR and ATL-TH, and Jurkat cells were maintained in RPMI 1640 medium supplemented with 10% FBS and 100 μg/ml penicillin-streptomycin. Cells were cultured under standard tissue culture conditions.
Plasmid constructs and transfection.
The expression plasmids encoding FLAG-APH-2, FLAG-APH-2Δncbzip, FLAG-APH-2 1–103, and GFP-APH-2 were described previously (15). FLAG-APH-2-LL/AA1 was obtained by site-directed mutagenesis (Phusion site-directed mutagenesis kit; Thermo Scientific), using FLAG-APH-2 as a template for the primers 5′-GATCTTAAAAGCTGCGCAGGAGC-3′ and 5′-TCCCGCTGCTCCTCCG-3′. FLAG-APH-2-LL/AA2 was also obtained by site-directed mutagenesis, using FLAG-APH-2 as a template for the primers 5′-GTGTATAGGAGCCGCGGGTTTTGATG-3′ and 5′-TCCAACTGCTGATGCCTTTCTTCCCTCTC-3′. FLAG-APH-2-LL/AA1 + 2 was generated by site-directed mutagenesis using FLAG-APH-2-LL/AA1 as a template for the primers used for FLAG-APH-2-LL/AA2. To obtain pBAD HRS-His-Myc, we generated an HRS PCR product by using pCIneo3×HA-HRS (29) as a template. The HRS PCR product was then digested with EcoRI and cloned into the pBAD expression vector. HRS-HA Δ307 was constructed by site-directed mutagenesis using pCIneo3×HA-HRS as a template for the primers 5′-TGTACTCTTCACCTGTGAACTCGTC-3′ and 5′-GAATTCAGCGTAATCTGGAACGTC-3′. HA-HRSΔCC2 was generated by site-directed mutagenesis using pCIneo3×HA-HRS as a template for the primers 5′-CTGGAGCAGCAGAAGCAGACGGTC-3′ and 5′-CATGCGTTGATGGACTGGAAGAGTGAG-3′. Myc-HRS, Myc-HRS 1–289, Myc-HRS287-775, Myc-HRS 500–775, and Myc-HRS CC2 were kindly provided by Camilla Raiborg (University of Oslo) (21).
HEK293T cells were transfected using Lipofectamine 2000 (Life Technologies), according to the manufacturer’s guidelines. TurboFect (Thermo Scientific) transfection reagent was used to transfect HeLa cells.
Western blotting.
Cellular lysates were subjected to SDS-PAGE and transferred to nitrocellulose membranes (GE Healthcare). Membranes were blocked with 4% milk in Tris-buffered saline (TBS)-Tween for 1 h at room temperature and incubated overnight with primary antibodies. This study used antibodies against FLAG (product no. F3165; Sigma), α-tubulin (product no. ab7291; Abcam), HA (product no. 26183; Invitrogen), and Myc (product no. 44-0603; Life Technologies), together with antibodies against HRS (product no. sc-271455; Santa Cruz Biotechnology) and HTLV-2 p24 (product no. 0801087; ZeptoMetrix). Anti-mouse IgG and anti-rabbit IgG secondary antibodies were from GE Healthcare. Densitometric analysis of Western blots was performed using LI-COR Image Studio Lite software.
GST pulldown assay.
GST and GST-APH-2 fusion proteins were expressed and purified from E. coli BL21 cells as described previously (53). HRS protein was expressed in E. coli Top10 cells and purified using a nickel resin (Qiagen). Purified GST or GST-APH-2 fusion proteins were immobilized on glutathione-Sepharose Fast Flow beads (GE Healthcare) overnight at 4°C. Following this incubation, resins were incubated with purified HRS for an additional 24 h. Beads were then washed using GST wash buffer (0.5% Triton X-100 in phosphate-buffered saline [PBS]), and bound proteins were eluted using GST elution buffer (50 mM Tris-HCl [pH 8.0] containing 10 mM reduced glutathione). Eluted proteins were analyzed by Western blotting and Coomassie brilliant blue staining (InstantBlue protein stain; Expedeon).
Coimmunoprecipitations.
HEK293T cells were transfected with the relevant amounts of expression constructs. Twenty-four hours posttransfection, cells were lysed in a buffer containing 1× TBS (50 mM Tris-HCl [pH 7.5], 150 mM NaCl), 5 mM EDTA, and 1% Triton X-100, supplemented with protease inhibitors (complete protease inhibitor cocktail, EDTA free; Roche). Cellular lysates were subjected to coimmunoprecipitation with an anti-FLAG M2 resin (Sigma-Aldrich) overnight at 4°C. The beads were washed three times with relevant buffer. Coimmunoprecipitations were analyzed by Western blotting using relevant antibodies, as indicated in the individual figure legends.
Immunofluorescence assays.
HeLa cells were seeded into 2-well chamber slides (Nunc Lab-Tek; Thermo Scientific) and transiently transfected with enhanced GFP or GFP-APH-2 and, where indicated, Myc-HRS, Myc-HRS-CC2, or HA-HRSΔCC2 expression plasmids. After 24 h, cells were fixed with 4% paraformaldehyde for 10 min and permeabilized using 0.5% Triton X-100 in PBS for 10 min at room temperature; 5% bovine serum albumin (BSA) in PBS was used as a blocking buffer for 1 h at room temperature. To stain for HRS, slides were incubated with anti-HRS followed by Alexa Fluor 594-conjugated goat anti-mouse IgG (Life Technologies). EEA1 was stained with anti-EEA1 (product no. ab50313; Abcam), while CD63 was stained with an antibody against CD63 (product no. sc15363; Santa Cruz Biotechnology). Nuclei were stained using DAPI (Sigma-Aldrich). Primary antibodies were incubated overnight at 4°C, and secondary antibodies were incubated at room temperature for 1 h. All antibodies were diluted in the blocking buffer. Slides were mounted in ProLong Gold Anti-Fade (Life Technologies), and images were acquired with a Zeiss Axio Imager microscope.
RT-qPCR.
RT-qPCR was performed on RNA templates extracted from HEK293T cells transfected with the indicated plasmids. RNA extractions were performed using the RNeasy minikit (Qiagen), and equal amounts of RNA were subjected to RT-qPCR using the QuantiNova SYBR green RT-PCR kit (Thermo Fisher Scientific) with specific primers for β-actin (5′-GGTCACCCACACTGTGACTACCTCA-3′ and 5′-GCTTCTCCTTAATGTCACGCACGAT-3′) and for APH-2 (5′-CCCCAAGACTATTTTAGGAGATTGC-3′ and 5′-CCGATCCCGACCCCAGAG-3′). Amplification was carried out on an Applied Biosystems 7300 system, following the manufacturer’s instructions. Relative RNA expression was calculated as PCR efficiency(CT internal control – CT sample), where the efficiency was the mean PCR efficiency for all reactions within the primer set, calculated using the LinRegPCR program.
Knockdown of HRS by siRNA.
The siRNA duplex targeting HRS was 5′-CGACAAGAACCCACACGUC-dTdT-3′, purchased from Dharmacon. Allstars negative-control siRNA from Qiagen was used to monitor any nonspecific effect of the siRNA transfection. Briefly, 160,000 HeLa cells were seeded into individual wells of a 6-well plate and transfected with 50 nM siRNA using Lipofectamine RNAiMAX on day 1. On day 3, cells were replated and retransfected. Cells were transfected with FLAG-APH2 or an empty control plasmid on day 4, using TurboFect. Cell lysis was performed on day 5 using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) supplemented with protease inhibitors. Cellular lysates were analyzed by Western blotting.
Drug treatments.
HEK293T cells were transfected with the indicated expression plasmids. Twenty-four hours after transfection, cells were incubated with bafilomycin (1 μM; Sigma) or cycloheximide (100 μg/ml; Sigma) for the indicated times. Cells were lysed using RIPA buffer supplemented with protease inhibitors. Cellular lysates were subjected to Western blotting.
Luciferase reporter gene assay.
HeLa cells were transfected with HTLV-2 LTR-luciferase reporter construct and different combinations of expression vectors as indicated. Luciferase assays were performed using the dual-luciferase reporter assay system (product no. E1501; Promega), according to the manufacturer’s recommendation. Luciferase activity results were normalized to the protein concentrations quantified using the Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific).
Cell-to-cell infection.
HeLa cells were transfected with the indicated plasmids; 24 h posttransfection, equivalent numbers of Mo cells were added on top of the HeLa cells. Cells were incubated for 24 h, the Mo cells were then removed, and the HeLa cells were washed multiple times with PBS prior to incubation with DMEM with 10% FBS for another 24 h. Cells were then lysed, and luciferase assays and Western blotting were performed.
p19gag ELISAs.
HTLV-2 matrix p19gag protein in the cell culture supernatant was measured using a commercially available enzyme-linked immunosorbent assay (ELISA) kit (RETROtek HTLV-2 p19 antigen ELISA; ZeptoMetrix), following the manufacturer’s recommendations.
Statistics.
Two-tailed Student's t tests were performed on results from three independent experiments using GraphPad Prism 5 software.
ACKNOWLEDGMENTS
This work was supported by the Center for Research in Infectious Diseases, University College Dublin.
We thank Camilla Raiborg for her gift of the HRS deletion mutant constructs and Luis L. P. daSilva for providing the HA-HRS plasmid. We thank Jonathan Dean and Ursula Morley for their assistance with sequencing. We are grateful to Keith Fay for his preliminary work with HRS purification.
REFERENCES
- 1.Kalyanaraman VS, Sarngadharan MG, Robert-Guroff M, Miyoshi I, Golde D, Gallo RC. 1982. A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 218:571–573. doi: 10.1126/science.6981847. [DOI] [PubMed] [Google Scholar]
- 2.Murphy EL, Cassar O, Gessain A. 2015. Estimating the number of HTLV-2 infected persons in the world. Retrovirology 12:O5. doi: 10.1186/1742-4690-12-S1-O5. [DOI] [Google Scholar]
- 3.Bartman MT, Kaidarova Z, Hirschkorn D, Sacher RA, Fridey J, Garratty G, Gibble J, Smith JW, Newman B, Yeo AE, Murphy EL. 2008. Long-term increases in lymphocytes and platelets in human T-lymphotropic virus type II infection. Blood 112:3995–4002. doi: 10.1182/blood-2008-05-155960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orland JR, Wang B, Wright DJ, Nass CC, Garratty G, Smith JW, Newman B, Smith DM, Murphy EL. 2004. Increased mortality associated with HTLV-II infection in blood donors: a prospective cohort study. Retrovirology 1:4. doi: 10.1186/1742-4690-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rende F, Cavallari I, Romanelli MG, Diani E, Bertazzoni U, Ciminale V. 2012. Comparison of the genetic organization, expression strategies and oncogenic potential of HTLV-1 and HTLV-2. Leuk Res Treat 2012:1–14. doi: 10.1155/2012/876153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romanelli MG, Diani E, Bergamo E, Casoli C, Ciminale V, Bex F, Bertazzoni U. 2013. Highlights on distinctive structural and functional properties of HTLV Tax proteins. Front Microbiol 4:271. doi: 10.3389/fmicb.2013.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaudray G, Gachon F, Basbous J, Biard-Piechaczyk M, Devaux C, Mesnard J-M. 2002. The complementary strand of the human T-cell leukemia virus type 1 RNA genome encodes a bZIP transcription factor that down-regulates viral transcription. J Virol 76:12813–12822. doi: 10.1128/jvi.76.24.12813-12822.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halin M, Douceron E, Clerc I, Journo C, Ko NL, Landry S, Murphy EL, Gessain A, Lemasson I, Mesnard J-M, Barbeau B, Mahieux R. 2009. Human T-cell leukemia virus type 2 produces a spliced antisense transcript encoding a protein that lacks a classic bZIP domain but still inhibits Tax2-mediated transcription. Blood 114:2427–2438. doi: 10.1182/blood-2008-09-179879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douceron E, Kaidarova Z, Miyazato P, Matsuoka M, Murphy EL, Mahieux R. 2012. HTLV-2 APH-2 expression is correlated with proviral load but APH-2 does not promote lymphocytosis. J Infect Dis 205:82–86. doi: 10.1093/infdis/jir708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemasson I, Lewis MR, Polakowski N, Hivin P, Cavanagh M-H, Thébault S, Barbeau B, Nyborg JK, Mesnard J-M. 2007. Human T-cell leukemia virus type 1 (HTLV-1) bZIP protein interacts with the cellular transcription factor CREB to inhibit HTLV-1 transcription. J Virol 81:1543–1553. doi: 10.1128/JVI.00480-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Satou Y, Yasunaga J, Yoshida M, Matsuoka M. 2006. HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells. Proc Natl Acad Sci U S A 103:720–725. doi: 10.1073/pnas.0507631103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H, Kannian P, Dissinger N, Haines R, Niewiesk S, Green PL. 2012. Human T-cell leukemia virus type 2 antisense viral protein 2 is dispensable for in vitro immortalization but functions to repress early virus replication in vivo. J Virol 86:8412–8421. doi: 10.1128/JVI.00717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panfil AR, Dissinger NJ, Howard CM, Murphy BM, Landes K, Fernandez SA, Green PL. 2016. Functional comparison of HBZ and the related APH-2 protein provides insight into human T-cell leukemia virus type 1 pathogenesis. J Virol 90:3760–3772. doi: 10.1128/JVI.03113-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnold J, Yamamoto B, Li M, Phipps AJ, Younis I, Lairmore MD, Green PL. 2006. Enhancement of infectivity and persistence in vivo by HBZ, a natural antisense coded protein of HTLV-1. Blood 107:3976–3982. doi: 10.1182/blood-2005-11-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marban C, McCabe Á, Bukong TN, Hall WW, Sheehy N. 2012. Interplay between the HTLV-2 Tax and APH-2 proteins in the regulation of the AP-1 pathway. Retrovirology 9:98. doi: 10.1186/1742-4690-9-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy J, Hall WW, Ratner L, Sheehy N. 2016. Novel interactions between the HTLV antisense proteins HBZ and APH-2 and the NFAR protein family: implications for the HTLV lifecycles. Virology 494:129–142. doi: 10.1016/j.virol.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christ L, Raiborg C, Wenzel EM, Campsteijn C, Stenmark H. 2017. Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem Sci 42:42–56. doi: 10.1016/j.tibs.2016.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Jouvenet N. 2012. Dynamics of ESCRT proteins. Cell Mol Life Sci 69:4121–4133. doi: 10.1007/s00018-012-1035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burke P, Schooler K, Wiley HS. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol Biol Cell 12:1897–1910. doi: 10.1091/mbc.12.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller DSJ, Bloxham RD, Jiang M, Gori I, Saunders RE, Das D, Chakravarty P, Howell M, Hill CS. 2018. The dynamics of TGF-β signaling are dictated by receptor trafficking via the ESCRT machinery. Cell Rep 25:1841–1855.e5. doi: 10.1016/j.celrep.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raiborg C, Bremnes B, Mehlum A, Gillooly DJ, D'Arrigo A, Stang E, Stenmark H. 2001. FYVE and coiled-coil domains determine the specific localisation of Hrs to early endosomes. J Cell Sci 114:2255–2263. [DOI] [PubMed] [Google Scholar]
- 22.Bache KG, Brech A, Mehlum A, Stenmark H. 2003. Hrs regulates multivesicular body formation via ESCRT recruitment to endosomes. J Cell Biol 162:435–442. doi: 10.1083/jcb.200302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Q, Hope LW, Brasch M, Reinhard C, Cohen SN. 2003. TSG101 interaction with HRS mediates endosomal trafficking and receptor down-regulation. Proc Natl Acad Sci U S A 100:7626–7631. doi: 10.1073/pnas.0932599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babst M. 2005. A protein’s final ESCRT. Traffic 6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 25.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 27.Blot V, Perugi F, Gay B, Prévost M-C, Briant L, Tangy F, Abriel H, Staub O, Dokhélar M-C, Pique C. 2004. Nedd4.1-mediated ubiquitination and subsequent recruitment of Tsg101 ensure HTLV-1 Gag trafficking towards the multivesicular body pathway prior to virus budding. J Cell Sci 117:2357–2367. doi: 10.1242/jcs.01095. [DOI] [PubMed] [Google Scholar]
- 28.Bouamr F, Melillo JA, Wang MQ, Nagashima K, de Los Santos M, Rein A, Goff SP. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J Virol 77:11882–11895. doi: 10.1128/jvi.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amorim NA, da Silva EML, de Castro RO, da Silva-Januário ME, Mendonça LM, Bonifacino JS, da Costa LJ, daSilva L. 2014. Interaction of HIV-1 Nef protein with the host protein Alix promotes lysosomal targeting of CD4 receptor. J Biol Chem 289:27744–27756. doi: 10.1074/jbc.M114.560193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kueck T, Neil S. 2012. A cytoplasmic tail determinant in HIV-1 Vpu mediates targeting of tetherin for endosomal degradation and counteracts interferon-induced restriction. PLoS Pathog 8:e1002609. doi: 10.1371/journal.ppat.1002609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clague MJ, Urbé S. 2003. Hrs function: viruses provide the clue. Trends Cell Biol 13:603–606. doi: 10.1016/j.tcb.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, Kasai H, Takeshita T, Endo Y, Fujita T, Sugamura K. 1997. Hrs is associated with STAM, a signal-transducing adaptor molecule: its suppressive effect on cytokine-induced cell growth. J Biol Chem 272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- 33.Toyoshima M, Tanaka N, Aoki J, Tanaka Y, Murata K, Kyuuma M, Kobayashi H, Ishii N, Yaegashi N, Sugamura K. 2007. Inhibition of tumor growth and metastasis by depletion of vesicular sorting protein Hrs: its regulatory role on E-cadherin and β-catenin. Cancer Res 67:5162–5171. doi: 10.1158/0008-5472.CAN-06-2756. [DOI] [PubMed] [Google Scholar]
- 34.Schuh AL, Audhya A. 2014. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol 49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi H, Tanaka N, Asao H, Miura S, Kyuuma M, Semura K, Ishii N, Sugamura K. 2005. Hrs, a mammalian master molecule in vesicular transport and protein sorting, suppresses the degradation of ESCRT proteins signal transducing adaptor molecule 1 and 2. J Biol Chem 280:10468–10477. doi: 10.1074/jbc.M409969200. [DOI] [PubMed] [Google Scholar]
- 36.Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem 72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 37.Komada M, Masaki R, Yamamoto A, Kitamura N. 1997. Hrs, a tyrosine kinase substrate with a conserved double zinc finger domain, is localized to the cytoplasmic surface of early endosomes. J Biol Chem 272:20538–20544. doi: 10.1074/jbc.272.33.20538. [DOI] [PubMed] [Google Scholar]
- 38.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janvier K, Pelchen-Matthews A, Renaud J-B, Caillet M, Marsh M, Berlioz-Torrent C. 2011. The ESCRT-0 component HRS is required for HIV-1 Vpu-mediated BST-2/tetherin down-regulation. PLoS Pathog 7:e1001265. doi: 10.1371/journal.ppat.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaoka T, Kusumoto S, Ando K, Ohba M, Ohmori T. 2018. Receptor tyrosine kinase-targeted cancer therapy. Int J Mol Sci 19:E3491. doi: 10.3390/ijms19113491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo Z, Ge M, Chen J, Geng Q, Tian M, Qiao Z, Bai L, Zhang Q, Zhu C, Xiong Y, Wu K, Liu F, Liu Y, Wu J. 2017. HRS plays an important role for TLR7 signaling to orchestrate inflammation and innate immunity upon EV71 infection. PLoS Pathog 13:e1006585. doi: 10.1371/journal.ppat.1006585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 43.Roxrud I, Raiborg C, Pedersen NM, Stang E, Stenmark H. 2008. An endosomally localized isoform of Eps15 interacts with Hrs to mediate degradation of epidermal growth factor receptor. J Cell Biol 180:1205–1218. doi: 10.1083/jcb.200708115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amano Y, Yamashita Y, Kojima K, Yoshino K, Tanaka N, Sugamura K, Takeshita T. 2011. Hrs recognizes a hydrophobic amino acid cluster in cytokine receptors during ubiquitin-independent endosomal sorting. J Biol Chem 286:15458–15472. doi: 10.1074/jbc.M110.191924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dubuisson L, Lormières F, Fochi S, Turpin J, Pasquier A, Douceron E, Oliva A, Bazarbachi A, Lallemand-Breitenbach V, De Thé H, Journo C, Mahieux R. 2018. Stability of HTLV-2 antisense protein is controlled by PML nuclear bodies in a SUMO-dependent manner. Oncogene 37:2806. doi: 10.1038/s41388-018-0163-x. [DOI] [PubMed] [Google Scholar]
- 46.Simonis N, Rual J-F, Lemmens I, Boxus M, Hirozane-Kishikawa T, Gatot J-S, Dricot A, Hao T, Vertommen D, Legros S, Daakour S, Klitgord N, Martin M, Willaert J-F, Dequiedt F, Navratil V, Cusick ME, Burny A, Van Lint C, Hill DE, Tavernier J, Kettmann R, Vidal M, Twizere J-C. 2012. Host-pathogen interactome mapping for HTLV-1 and 2 retroviruses. Retrovirology 9:26. doi: 10.1186/1742-4690-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rusten TE, Stenmark H. 2009. How do ESCRT proteins control autophagy? J Cell Sci 122:2179–2183. doi: 10.1242/jcs.050021. [DOI] [PubMed] [Google Scholar]
- 48.Tamai K, Tanaka N, Nara A, Yamamoto A, Nakagawa I, Yoshimori T, Ueno Y, Shimosegawa T, Sugamura K. 2007. Role of Hrs in maturation of autophagosomes in mammalian cells. Biochem Biophys Res Commun 360:721–727. doi: 10.1016/j.bbrc.2007.06.105. [DOI] [PubMed] [Google Scholar]
- 49.Edgar JR, Willén K, Gouras GK, Futter CE. 2015. ESCRTs regulate amyloid precursor protein sorting in multivesicular bodies and intracellular amyloid-β accumulation. J Cell Sci 128:2520–2528. doi: 10.1242/jcs.170233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oshima R, Hasegawa T, Tamai K, Sugeno N, Yoshida S, Kobayashi J, Kikuchi A, Baba T, Futatsugi A, Sato I, Satoh K, Takeda A, Aoki M, Tanaka N. 2016. ESCRT-0 dysfunction compromises autophagic degradation of protein aggregates and facilitates ER stress-mediated neurodegeneration via apoptotic and necroptotic pathways. Sci Rep 6:24997. doi: 10.1038/srep24997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang S-W, Chen C-Y, Klase Z, Zane L, Jeang K-T. 2013. The cellular autophagy pathway modulates human T-cell leukemia virus type 1 replication. J Virol 87:1699–1707. doi: 10.1128/JVI.02147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren T, Dong W, Takahashi Y, Xiang D, Yuan Y, Liu X, Loughran TP, Sun S-C, Wang H-G, Cheng H. 2012. HTLV-2 Tax immortalizes human CD4+ memory T lymphocytes by oncogenic activation and dysregulation of autophagy. J Biol Chem 287:34683–34693. doi: 10.1074/jbc.M112.377143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuji T, Sheehy N, Gautier VW, Hayakawa H, Sawa H, Hall WW. 2007. The nuclear import of the human T lymphotropic virus type I (HTLV-1) Tax protein is carrier- and energy-independent. J Biol Chem 282:13875–13883. doi: 10.1074/jbc.M611629200. [DOI] [PubMed] [Google Scholar]
- 54.Chou S-F, Tsai M-L, Huang J-Y, Chang Y-S, Shih C. 2015. The dual role of an ESCRT-0 component HGS in HBV transcription and naked capsid secretion. PLoS Pathog 11:e1005123. doi: 10.1371/journal.ppat.1005123. [DOI] [PMC free article] [PubMed] [Google Scholar]