Summary
The ocean is home to myriad small planktonic organisms that underpin the functioning of marine ecosystems. However, their spatial patterns of diversity and the underlying drivers remain poorly known, precluding projections of their responses to global changes. Here we investigate the latitudinal gradients and global predictors of plankton diversity across archaea, bacteria, eukaryotes, and major virus clades using both molecular and imaging data from Tara Oceans. We show a decline of diversity for most planktonic groups toward the poles, mainly driven by decreasing ocean temperatures. Projections into the future suggest that severe warming of the surface ocean by the end of the 21st century could lead to tropicalization of the diversity of most planktonic groups in temperate and polar regions. These changes may have multiple consequences for marine ecosystem functioning and services and are expected to be particularly significant in key areas for carbon sequestration, fisheries, and marine conservation.
Video Abstract
Keywords: plankton functional groups, macroecology, latitudinal diversity gradient, temperature, climate warming, Tara Oceans, trans-kingdom diversity, high-throughput sequencing, high-throughput imaging
Graphical Abstract
Highlights
-
•
Most epipelagic planktonic groups exhibit a poleward decline of diversity
-
•
No latitudinal diversity gradient was observed below the photic zone
-
•
Temperature emerges as the best predictor of epipelagic plankton diversity
-
•
Global warming may increase plankton diversity, particularly at high latitudes
The drivers of ocean plankton diversity across archaea, bacteria, eukaryotes, and major virus clades are inferred from both molecular and imaging data acquired by the Tara Oceans project and used to predict the effects of severe warming of the surface ocean on this critical ecosystem by the end of the 21st century.
Introduction
Our planet is dominated by interconnected oceans that harbor a tremendous diversity of microscopic planktonic organisms. They form complex ecological networks (Lima-Mendez et al., 2015) that sustain major biogeochemical cycles (Falkowski et al., 2008, Field et al., 1998) and provide a wide range of ecosystem services (Guidi et al., 2016, Ptacnik et al., 2008, Worm et al., 2006). The ongoing increase in atmospheric carbon dioxide concentrations is having knock-on effects on the ocean by altering, among others, temperature, salinity, circulation, oxygenation, and pH levels (Pachauri et al., 2014, Rhein et al., 2013). These changes have already left visible imprints on marine plankton, fish, mammals, and birds, with shifts in species phenology and distribution (Beaugrand et al., 2009, Boyce et al., 2010, Poloczanska et al., 2013, Richardson and Schoeman, 2004). Future increases in ocean temperatures are expected to modify phytoplankton diversity and distribution directly by altering metabolic rates and growth (Thomas et al., 2012, Toseland et al., 2013) or indirectly through changes in ocean circulation and, consequently, the supply of nutrients to surface waters (Bopp et al., 2013). Given that such modifications will most likely impair the functions, goods, and services provided by the ocean (Brun et al., 2019, Hutchins and Fu, 2017, Worm et al., 2006), predicting how plankton diversity will respond to climate change has become a pressing challenge (Cavicchioli et al., 2019).
Unraveling patterns of diversity across macroclimatic gradients, such as the latitudinal diversity gradient (LDG), is a way to anticipate the effects of climate change (Algar et al., 2009, Frenne et al., 2013). The LDG, historically studied principally in terrestrial macroorganisms, usually consists of a monotonic poleward decline of local diversity (known as alpha diversity; Whittaker, 1972) for both terrestrial and aquatic organisms (Hillebrand, 2004). The LDG is hypothesized to result from a range of non-exclusive ecological and evolutionary mechanisms that operate at multiple spatial and temporal scales (Clarke and Gaston, 2006, Pontarp et al., 2019, Willig et al., 2003). Among the mechanisms classically invoked, temperature is often thought to be one of the major drivers through two effects. The “physiological tolerance hypothesis” posits that temperature structures the LDG by imposing abiotic constraints on species distribution range (Currie et al., 2004), with fewer species tolerating cold conditions and tropical temperatures being generally below the upper thermal tolerance limit of most organisms. The “kinetic energy hypothesis” relates to the metabolic theory (reviewed in Brown, 2014), which posits that higher temperatures increase the rate of metabolic reactions, resulting in shorter generation times, faster ecological or physiological processes, and, ultimately, higher mutation and speciation rates, leading to higher local diversity. Beyond temperature, the “productivity/resources hypothesis” posits that greater resource availability and/or primary production in tropical terrestrial areas can support larger population sizes and limit local extinction, promoting species coexistence (reviewed in Clarke and Gaston [2006]). The “environmental stability hypothesis” asserts that short- to long-term environmental instability in extratropical latitudes should cause greater local extinction rates because life in such unstable environments requires particular and costly physiological adaptations, which would ultimately preclude speciation (Clarke and Gaston 2006). The LDG has also been explained by stronger biotic interactions in the tropics because of higher energy availability, which would increase diversity through complexification and specialization of trophic, mutualistic, or parasitic interactions (reviewed in Willig et al. [2003]). However, this hypothesis has found little support in the literature (Hillebrand, 2004) and further relies on the mechanisms exposed above.
In contrast, current knowledge regarding the global trends and drivers of oceanic plankton diversity, ranging from viruses to microbes and zooplankton, remains highly fragmentary. It is mainly based on meta-analyses, which are sensitive to heterogeneous datasets (Brown et al., 2016) and do not systematically capture the diversity of dominant planktonic groups. Therefore, the form of the LDG remains equivocal for marine bacteria, copepods, and diatoms, whose diversity has been reported to either decline linearly poleward (Fuhrman et al., 2008, Righetti et al., 2019, Sul et al., 2013, Woodd-Walker et al., 2002), peak in extratropical regions (Ladau et al., 2013, Raes et al., 2018, Rombouts et al., 2009), or adopt weak or inverted latitudinal trends (e.g., Chust et al., 2013, Ghiglione et al., 2012). Virus LDGs have been described only recently and seem to exhibit an increase in diversity in the Arctic Ocean (Gregory et al., 2019). Consequently, the extent to which the abovementioned hypotheses apply to the world of marine plankton remains unclear. For example, marine plankton are expected to have huge population sizes, high dispersal abilities, short life cycles, and dormancy stages that would prevent local extinctions and reduce speciation rates. The peak of diversity in temperate to high latitudes has also been suggested to support the productivity/resource hypothesis (Ladau et al., 2013, Raes et al., 2018), which is in agreement with the oligotrophic status of most tropical waters (Field et al., 1998). On the other hand, the environmental stability hypothesis is expected to highly constrain marine plankton at high latitudes, which experience strong seasonality in temperature, nutrients, and light, as suggested for phytoplankton (Behrenfeld et al., 2015, Righetti et al., 2019). These constraints may also cascade across trophic levels, as suggested for copepods (e.g., Rombouts et al., 2009). All of these uncertainties seriously hamper our ability to understand the drivers of these essential components of marine ecosystems and estimate their potential responses in a changing ocean.
Here we provide a unified view of plankton LDGs using systematically collected data from the Tara Oceans global expedition. We combine DNA sequencing of filtered seawater and imaging of net catches to study the diversity in molecular operational taxonomic units (MOTUs) and morphotype diversity of major groups from all domains of life as well as both small and large double-stranded DNA viruses (Karsenti et al., 2011). We then separately examine their respective LDGs while determining their best environmental correlates because they may be influenced by different drivers. Finally, to identify the regions that may experience the most drastic changes in plankton diversity in the future, we model the trends of plankton diversity at the global scale for the beginning (years 1996–2005) and end of the century (years 2090–2099, representative concentration pathway [RCP] 8.5).
Results and Discussion
LDG across Marine Plankton Groups and Water Layers
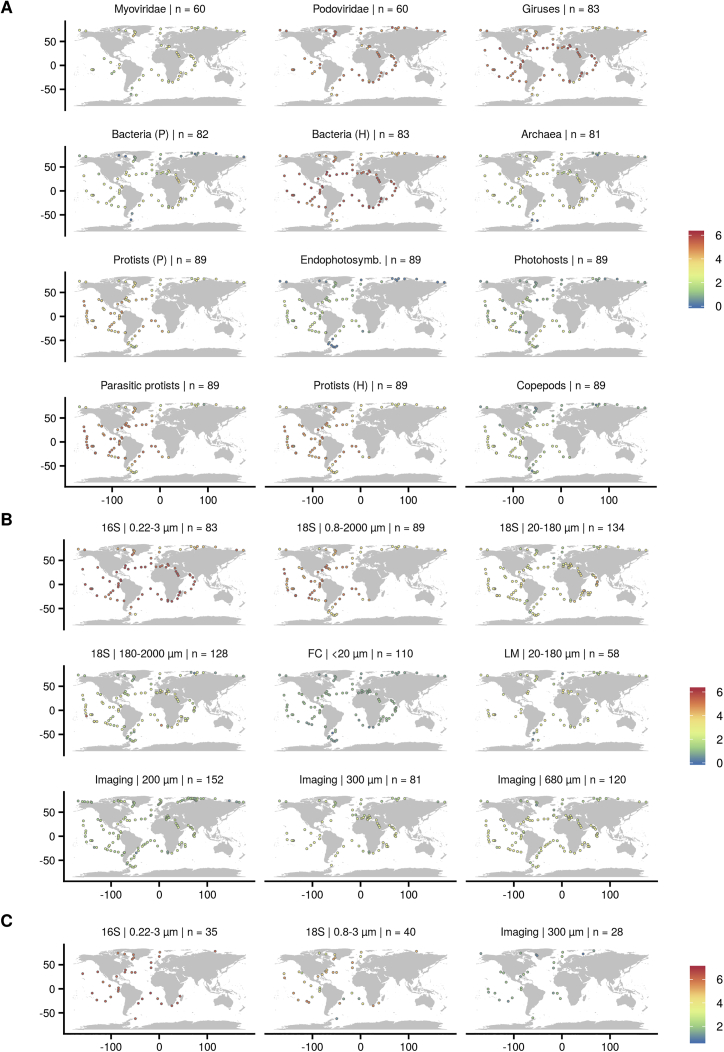
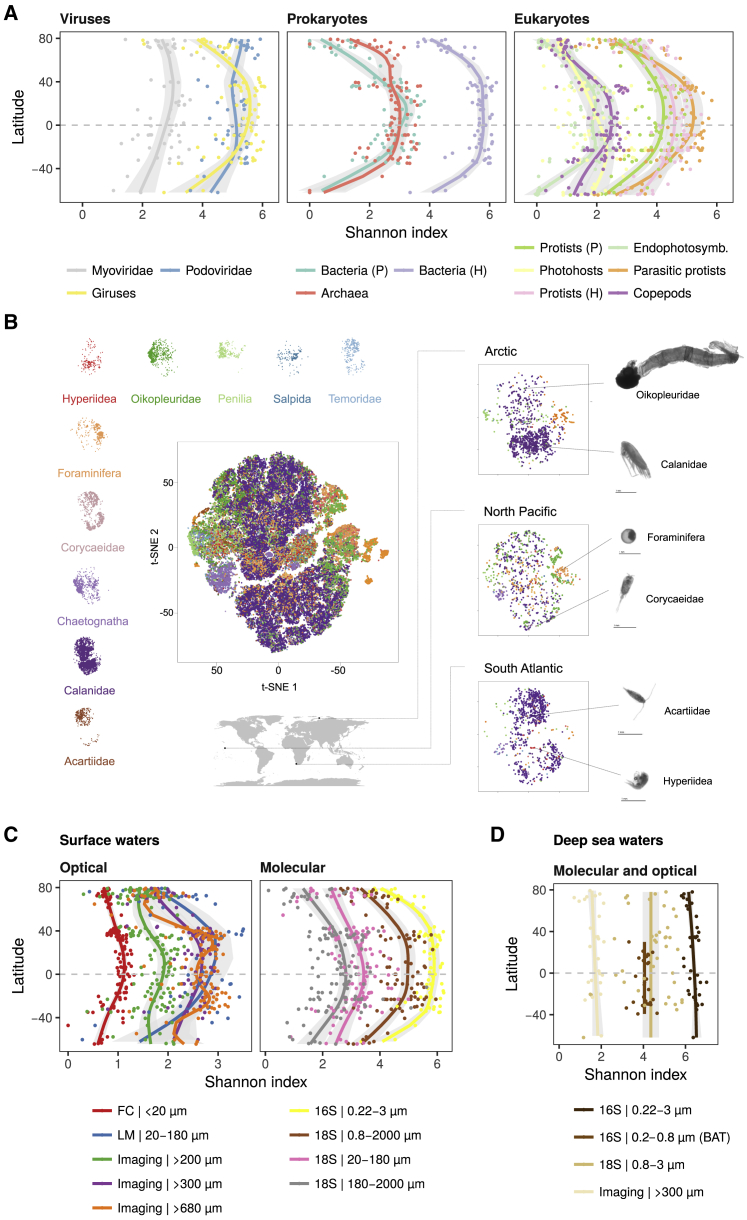
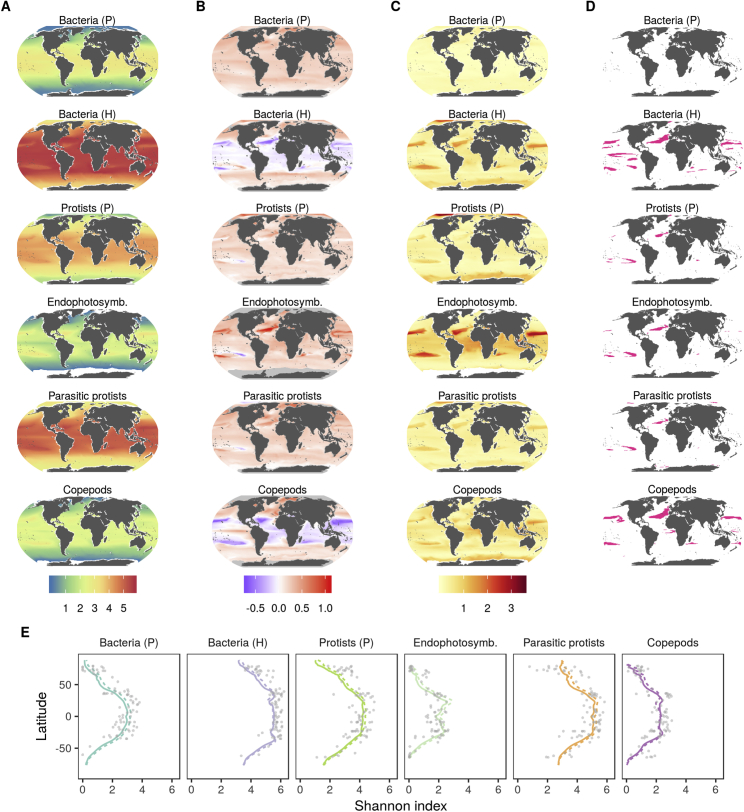
In this study, we used a wide collection of uniformly collected datasets with broad latitudinal coverage to explore the local diversity trends of all organismal groups that make up plankton communities (Table S1A). Besides already publicly available resources from Tara Oceans, we included newly released metabarcoding data of the V9 region of the 18S rRNA gene and flow cytometry (FC) abundances from the Arctic Ocean. We further complemented these observations with new global-scale datasets obtained with amplicon sequencing, microscopy, and imaging techniques. Our datasets were derived from 189 sampling stations distributed worldwide (Figure S1; STAR Methods), where multiple water depths were sampled (surface, 5 m average depth; deep chlorophyll maximum, 17–188 m; mesopelagic, 200–1000 m). This extensive and standardized sampling of plankton diversity encompasses large gradients in temperature, resource/primary production, and environmental stability (Figure 1A). Using the taxonomic information retrieved from genomics and imaging data (Table S1A), we distinguished 12 marine plankton groups (MPGs; Figures 1B, S1A, and S2; Table S1B; STAR Methods) with different trophic modes (e.g., photosynthetic/mixotrophs versus non-photosynthetic/heterotrophs) and different life history strategies (e.g., parasitic protists, endophytosymbionts) or corresponding to highly dominant taxa having a significant contribution to the marine food web (e.g., copepods).
Figure S1.
Tara Oceans Stations and Shannon Diversity Patterns, Related to Figures 1 and 2
(A) MPGs at the sea surface (< 5 m depth), (B) whole planktonic community using different sampling protocols at the sea surface (except for “Imaging,” integrative depth from 500 m depth to the surface) and (C) whole planktonic community of the mesopelagic realm (200-1000 m depth). Number of stations are specified in the inset titles. Color represents the Shannon index. For more details on the different size fractions and sampling protocols, please refer to the caption in Figure 1 and STAR Methods.
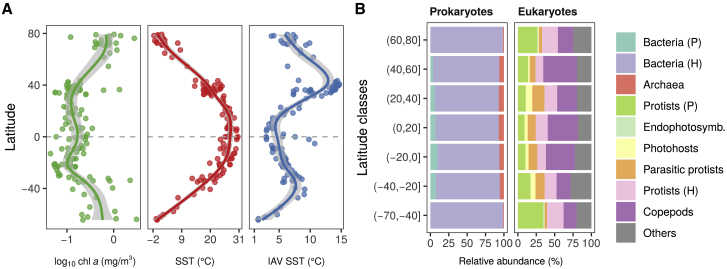
Figure 1.
Latitudinal Trends of Oceanic Conditions and Marine Plankton Composition in Surface Waters
(A) In situ chl a concentrations and sea surface temperatures (SST) across latitude (Tara Oceans expedition), plus IAV of SST (STAR Methods). Solid lines represent the GAM smooth trends and gray ribbons the corresponding 95% confidence intervals of parameter latitudinal trends predicted by the GAMs.
(B) Average relative abundances of MPGs as inferred from molecular datasets across latitude. Prokaryotes: 16S rRNA gene, 0.22–3 μm; eukaryotes: 18S rRNA gene, 0.8–2000 μm (STAR Methods). Dark gray represents other eukaryotic groups. P, photosynthetic/mixotrophic; H, non-photosynthetic/heterotrophic. The three viral groups are not represented here because of the absence of comparable abundance data.
See also Figures S1 and S2 and Tables S1A and S1B.
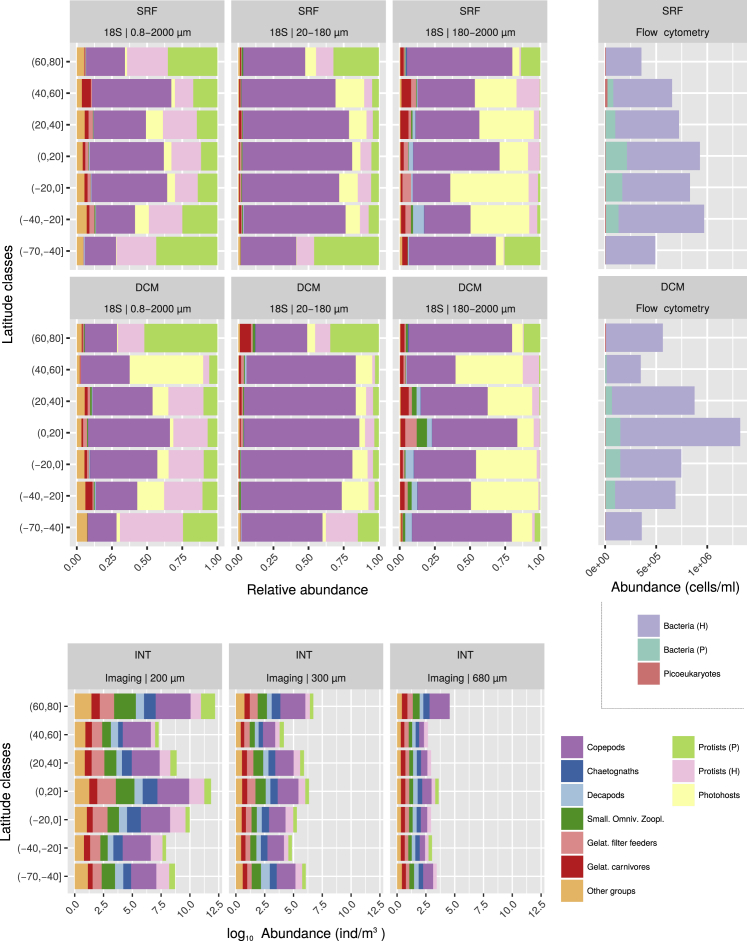
Figure S2.
Average Abundances of a Broader List of Plankton Groups across Latitude, Related to Figures 1 and 2
For 18S rDNA metabarcodes (relative abundances), imaging from net catches and flow cytometry (absolute abundances). Numbers refer to the filter mesh size (μm). H: non-photosynthetic/heterotrophic, P: photosynthetic/mixotrophic. See Figures 1 and S1 for further sampling details. The three viral groups are not represented here due to absence of comparable abundance data. Note that the differences between protocols relate, among other things, to resolution (e.g., potential photohosts from the nets are classified as Protists (H)), marker gene copy number (e.g., high in photohosts), lack of detection (many small copepods are lost when sampling with nets), or water column sampling differences (surface [SRF] or deep chlorophyll maximum [DCM] versus integrative [INT] for molecular/cytometry versus net catches, respectively).
The LDG of each MPG was studied using the Shannon index, a diversity index that relates monotonically to species richness but differs in that it downweighs rare species, whose numbers are highly sensitive to undersampling and molecular artifacts (Figure S3; STAR Methods). Focusing on surface waters first, we found that phyto-, bacterio-, and zooplankton MPGs all exhibited maximal MOTU diversity in tropical to subtropical regions that then decreased poleward (Figure 2A; see also Figure S4 for individualized curves for each MPG as well as specific taxonomic groups). Similar trends were found for parasites of eukaryotes (giruses and parasitic protists mainly composed of marine alveolates [MALVs]) and for eukaryotic photosynthetic intracellular symbionts (endophotosymbionts) as well as their eukaryotic hosts (photohosts). Different patterns emerged for two abundant families of prokaryotic viruses (Myoviridae and Podoviridae) which, unlike their hosts, did not exhibit a clear poleward decline in diversity. Because the diversity of hosts and their symbionts or parasites is often assumed to be linked through eco-evolutionary interactions (Morand, 2015), an explanation for this could be that these virus families have a broader spectrum of host species, which could potentially decouple certain eco-evolutionary constraints (de Jonge et al., 2019). However, other factors may be responsible for this trend as well, such as nutrient availability or bacterial cell density (Gregory et al., 2019). Further data and analyses will be necessary to elucidate the underpinnings of this result.
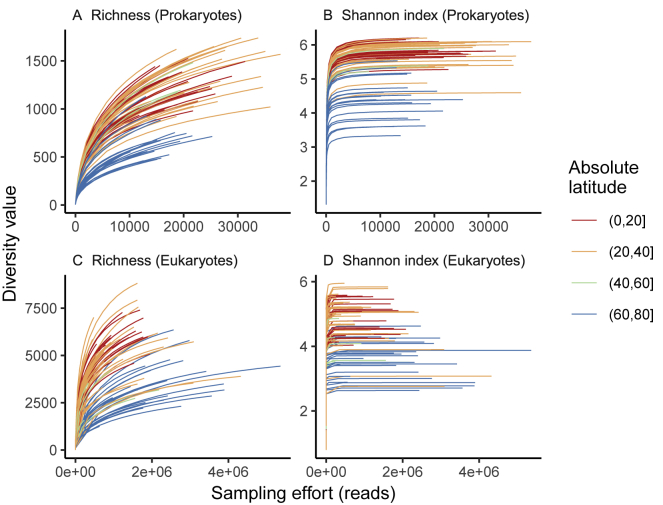
Figure S3.
Rarefaction Curves for the Plankton Community, Related to Figure 2 and STAR Methods
Based on richness (A, C) and Shannon (B, D), for prokaryotes (16S rRNA gene miTags, A, B), and eukaryotes (18S rRNA gene V9 metabarcoding, C, D). Each line corresponds to a surface water sample. Colors correspond to different latitudinal bands (absolute values).
Figure 2.
Latitudinal Patterns of Marine Plankton Diversity
(A) LDGs at the sea surface for all MPGs (STAR Methods).
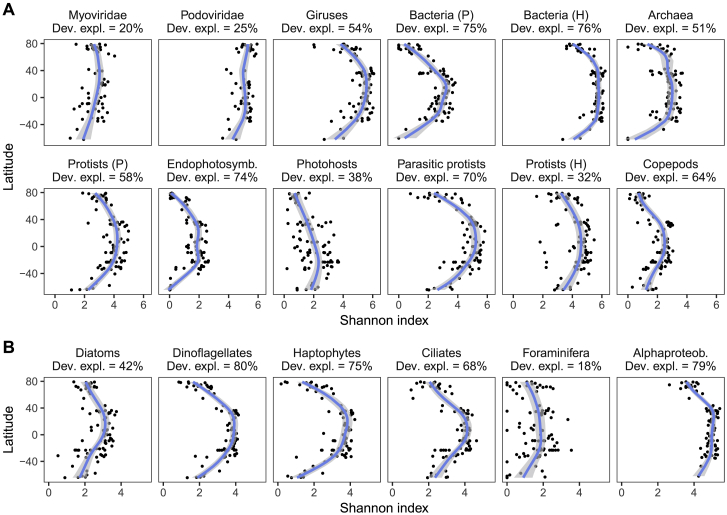
(B) Morphological diversity as analyzed from more than 77,000 organisms collected with the bongo net (imaging | 300 μm). Morphological measurements were normalized and subjected to a t-distributed stochastic neighbor embedding (t-SNE) ordination analysis using all samples (STAR Methods). In the central 2D t-SNE ordination, each dot corresponds to an organism and its color to its taxonomic assignation (>100 taxa). For ease of interpretation, the points corresponding to a subset of abundant groups are displayed separately. The three t-SNE ordinations displayed on the right show dots from three stations distantly located and from different latitudes, as shown in the map. Six images are also presented as examples of the underlying data (STAR Methods); 1-mm scale bars are shown below each picture.
(C and D) Patterns of the whole plankton community using different sampling protocols at (C) the sea surface (16S/18S/FC/LM) or a larger integrative depth of 500 m (imaging) and (D) in mesopelagic (average depth, 540 m) or bathypelagic layers (BAT; average depth, 4000 m, Malaspina expedition). In all cases, solid lines correspond to GAM smooth trends and gray ribbons to the 95% confidence intervals of the Shannon latitudinal trend predicted by the GAMs (see also Figures S4 and S7 for individual curves and explained deviance). These trends are drawn for illustrative purposes and were not used in downstream analyses. 16S and 18S refer to the different rRNA subunit genes used as marker genes for metagenomics and metabarcoding, respectively. Imaging refers to the identification method for large eukaryotes captured with nets. FC refers to flow cytometry for the picoplankton and LM to the light microscopy-based survey of microphytoplankton (STAR Methods). Numbers refer to the filter mesh size.
Figure S4.
Sea Surface LDG, Related to Figure 2A
(A) For viral, prokaryotic and eukaryotic MPGs, and (B) for well-known protist phyla and the dominant class of bacteria, Alphaproteobacteria, for which ∼50% corresponds to the SAR11 clade. Solid lines represent the GAM smooth trends and gray ribbons the corresponding 95% confidence intervals of the Shannon latitudinal trend predicted by the GAMs. The percentages provided below inset titles correspond to the deviance explained by GAMs when significant. Viruses and bacterial diversities are inferred from samples filtered at 0.22-3 μm and analyzed through marker genes derived from metagenomics. Eukaryote diversity shown here is inferred from 18S rDNA metabarcoding of samples filtered at 0.8-2000 μm. H: non-photosynthetic/heterotrophic, P: photosynthetic/mixotrophic.
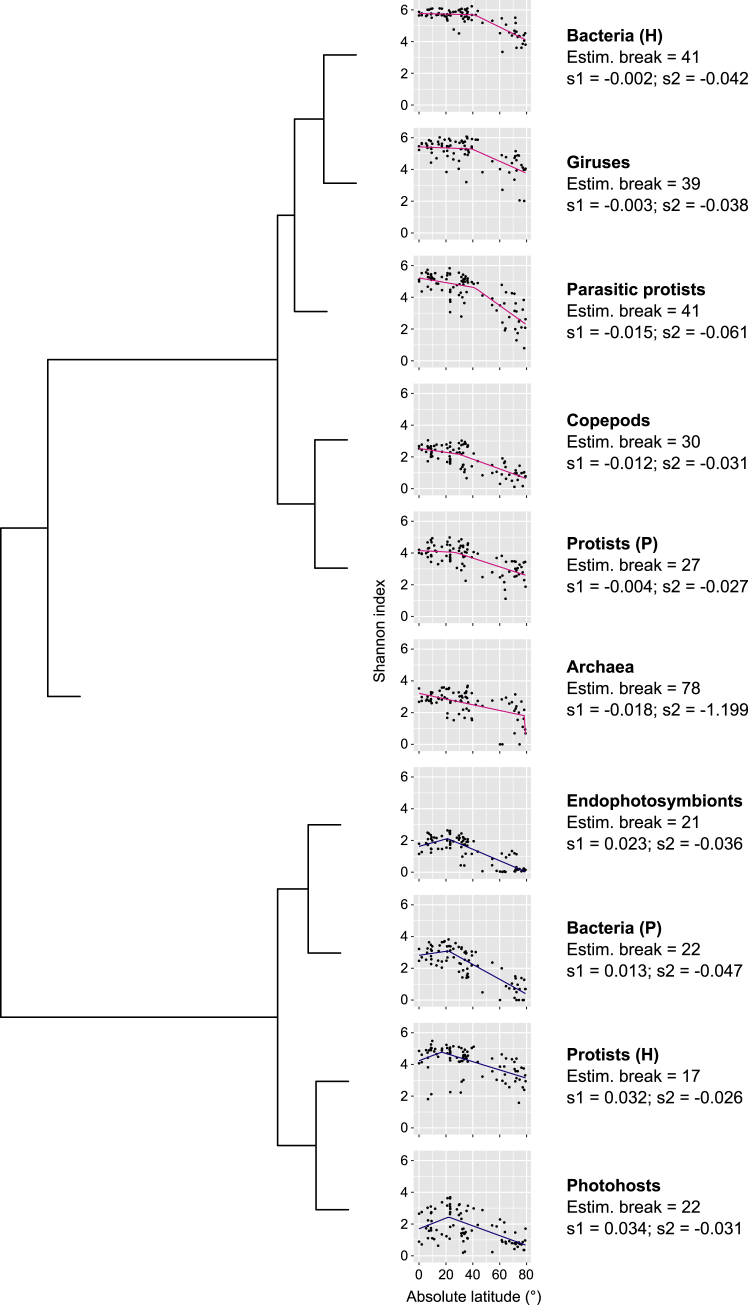
Differences in the form of LDGs have been proposed previously to result from contrasting strategies in energy acquisition and processing (Hillebrand, 2004). To test this hypothesis, we compared LDG forms across MPGs (except for prokaryotic viruses) by conducting a segmented linear regression analysis and using the inferred parameters in a clustering analysis (absolute latitudinal breakpoints and slopes of the segment regressions; STAR Methods; Figure S5). Confirming our above assumption, parasites and endophotosymbionts did not cluster directly with their hosts. Endophotosymbionts have extensive free-living populations (Decelle et al., 2012), and parasitic protists might experience relatively long-lasting free-living stages under the form of resistant cysts waiting for host availability (Siano et al., 2010), which could explain this result. Rather, we found that MPGs with similar broad trophic modes (phototrophic versus heterotrophic/chemotrophic) tended to exhibit similar LDG forms. However, we noticed two particular exceptions: photosynthetic protists clustered with heterotrophs (both prokaryotes and eukaryotes), whereas heterotrophic protists clustered with phototrophs. Whether these features result from the presence of still unknown mixotrophs in heterotrophic protists (i.e., photosymbioses) or a preferential heterotrophy of mixotrophic photosynthetic protists remains to be determined. Top-down or bottom-up controls by other trophic levels as well as interspecific competition could also contribute to these patterns. For example, most copepods preferentially feed on phototrophic protists (e.g., diatoms or dinoflagellates; Saiz and Calbet, 2011), which could explain why both groups exhibited similar LDGs (Figure S5).
Figure S5.
Classification of MPG Sea Surface LDG Analyzed by Segmented Regression, Related to Figure 2A
The estimated break is the absolute latitude between the two segments of slope s1 and s2, respectively. We used pink lines for s1 ≤ 0 (plateau or peak around the equator) and blue lines for s1 > 0 (extra-equatorial peak). The dendrogram is the result of a hierarchical clustering based on the differences in break and slope values across MPGs (Euclidean distance on standardized values).
We then examined the symmetry of the LDGs by performing separate linear regressions for each hemisphere. LDGs are commonly observed to be steeper in the northern hemisphere, supposedly because of stronger climate instability in this area of the globe (Chown et al., 2004). Our results contradict this expectation because we found that LDGs only tended to be asymmetric and steeper in the southern hemisphere for archaea and photosynthetic protists (Figure 2A; Table S1C). Although in agreement with another report on marine bacterioplankton (Sul et al., 2013), we suspect the absence or opposite trend observed here to arise from a significant undersampling at mid- to high latitudes in the southern hemisphere in our dataset (Figure S1). Another explanation could lie in differences in the timing of sampling between the two polar regions; i.e., from the end of spring to the beginning of autumn in the Arctic Ocean but only in a summer month in the Southern Ocean, when diversity is expected to be lowest because of intense blooms (Arrigo et al., 2008). In spite of some variations in the form of LDGs across MPGs or between hemispheres, our results nonetheless show that the poleward decline of diversity is a pervasive feature among marine plankton.
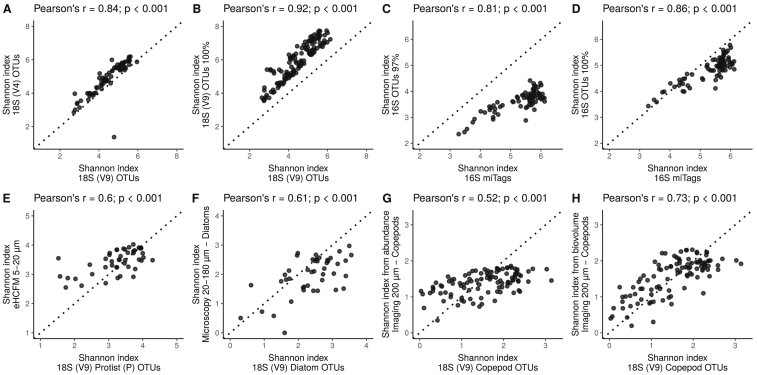
To ensure that the MOTU diversity trends observed with our molecular data (Figure 2A) were not biased by the DNA region studied or molecular approach used (i.e., DNA metabarcoding versus metagenomics for prokaryotes; Salazar et al., 2019), which may vary in taxonomic resolution or overrepresent certain taxa, we compared them against those obtained with other DNA markers (i.e., the V4 region of the 18S rRNA gene for eukaryotes and the V4–V5 regions of the 16S rRNA gene for prokaryotes) or with finer clustering thresholds (STAR Methods). All of these comparisons exhibited high correlation coefficient values, regardless of the clustering similarity thresholds used (Figures S6A–S6D). DNA-based measures of diversity can also be affected by organism size (multicellular organisms) or gene copy number (unicellular organisms), with organisms of smaller size or lower gene copy numbers likely to be more difficult to detect. To examine this potential issue, we compared the diversity trends of different planktonic groups observed with molecular versus optical data (Figures 2B and 2C; STAR Methods). Both zooplankton imaging data, which consist of morphological features (see Figure 2B for examples), as well as photosynthetic protist data obtained through confocal (Colin et al., 2017) or light microscopy (LM) were highly congruent with their corresponding molecularly based diversity trends (Figures 2C and S6E–S6H). Although of much lower taxonomic resolution, FC-based diversity values, comprising mainly prokaryotes, also correlated well with molecularly based prokaryotic diversity (Figure 2C). These results, together with the high correlation of diversity trends between our data and those based on single-copy genes (Milanese et al., 2019) and the fact that relative abundances in DNA-based data correlate well with organism size/biovolume and even abundance at lower taxonomic resolution (de Vargas et al., 2015), suggest that the global diversity trends observed here are unlikely to be biased by differences in body size/gene copy number across taxa.
Figure S6.
Correlation between the Shannon Values Derived from Multiple Datasets of Tara Oceans, Related to STAR Methods
(A) OTUs (as defined with “swarm”; Mahé et al., 2014) obtained with the V9 (x axis) and V4 (y axis) regions of the 18S rRNA gene using surface water samples (SRF); size fraction 0.8-2000 μm; (B) OTUs either as defined with swarm (x axis) or defined at 100% sequence identity from the V9 region of the 18S rRNA gene for SRF samples, size fraction 0.8-2000 μm; (C-D) 16S rRNA gene miTags (x axis) versus OTUs defined at 97% [C] and 100% sequence similarity [D] (y axis) obtained from the V4-V5 regions of the 16S rRNA gene for SRF samples, size fraction 0.22-3 μm. (E) OTUs of photosynthetic protists obtained with the V9 region of the 18S rRNA gene (x axis) versus protists (mostly photosynthetic) as identified with environmental High Content Fluorescence Microscopy (eHCFM; data from Colin et al., 2017) in SRF-DCM samples, size fraction 5-20 μm; (F) Diatom OTUs obtained with V9 region of the 18S rRNA gene (x axis) versus diatom species counted by light microscopy (y axis) in SRF samples; size fraction 20-180 μm; (G/H) Copepod OTUs obtained with the V9 region of the 18S rRNA gene, SRF samples, size fraction 180-2000 μm (x axis) versus abundances [G] and biovolumes [H] of copepods collected by the WP2 net, > 200 μm. Inset titles show the Pearson’s correlation coefficient and its associated p value. Note the differences in axes scales. Dashed line represents 1:1 relation. Refer to STAR Methods for details on each method.
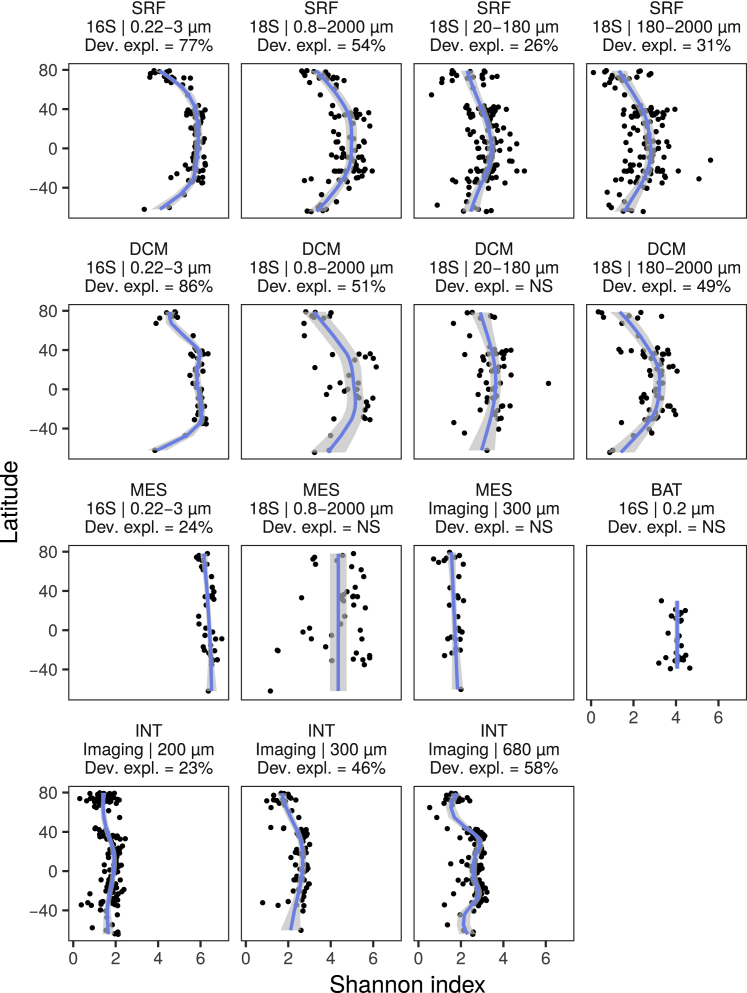
In contrast, we did not observe any LDGs, either in terms of MOTUs or morphological diversity, below the photic zone (>200 m depth, corresponding here to both mesopelagic [Tara Oceans] and bathypelagic waters; Salazar et al., 2016; Figures 2D and S7). These environments are isolated from sunlight and climatic gradients. Accordingly, although we did observe a weak latitudinal trend in temperature in our deep sea samples (linear regression on absolute latitudes: slope = −0.11, R2 = 0.273, p < 0.001), the range of this parameter represented roughly half of the temperature range present in surface waters (0°C to 18°C versus −2°C to 31°C, respectively). Hence, reduced temperature variations could be one of the reasons for an absence of LDGs in the deep sea. In addition, there is overall more carbon export at high latitudes (Henson et al., 2012). This could compensate for the reduction of diversity potentially induced by low temperatures by increasing resource availability in polar deep waters (Pomeroy and Wiebe, 2001). Finally, migration of surface species to deep waters through passive or active vertical flux may also contribute to cancel out temperature effects and perhaps underpin the overall higher diversity values we observe in deep sea waters compared with the surface (Figures 2C and 2D; Mestre et al., 2018). Although the current sampling effort in these aphotic environments is insufficient to firmly support these hypotheses, our results are consistent with previous observations for brittle stars (Woolley et al., 2016) and bacteria (Ghiglione et al., 2012) in the deep sea, whose diversity did not follow LDG trends. Both sediments and water layers below the photic zone are populated by heterotrophic and chemolithoautotrophic organisms, whose diversity and abundance are strongly influenced by organic matter availability (Bergauer et al., 2018, Danovaro et al., 2016, Woolley et al., 2016). This supports the idea that life is sustained by different types of energy supply across water layers, from systems driven by solar energy or kinetic effects of temperature in epipelagic waters to chemically driven environments (i.e., carbon- or mineral-based) in the deep sea.
Figure S7.
LDG across Size and Depth, Related to Figures 2B–2D
For the whole prokaryotic (16S miTags, 0.22-3 μm, and 16S OTUs, 0.2 μm for bathypelagic (BAT)) and eukaryotic communities (18S OTUs, 0.8-2000, 20-180 and 180-2000 μm; imaging, > 300 and > 680 μm) at different depths (SRF: surface, < 5 m; DCM: deep chlorophyll maximum, 17-188 m, and MES: mesopelagic, > 200 m, BAT: bathypelagic, > 4000 m, INT: integrative, depth from 500 m depth to the surface). Non-significant GAMs are denoted with “NS.” See Figure S4 legend for more information on the plot. Note that the particular trend for the regent net, i.e., Imaging | 680 μm might be due to undersampling of small zooplankton.
Global Drivers of MPG Diversity in the Surface Ocean
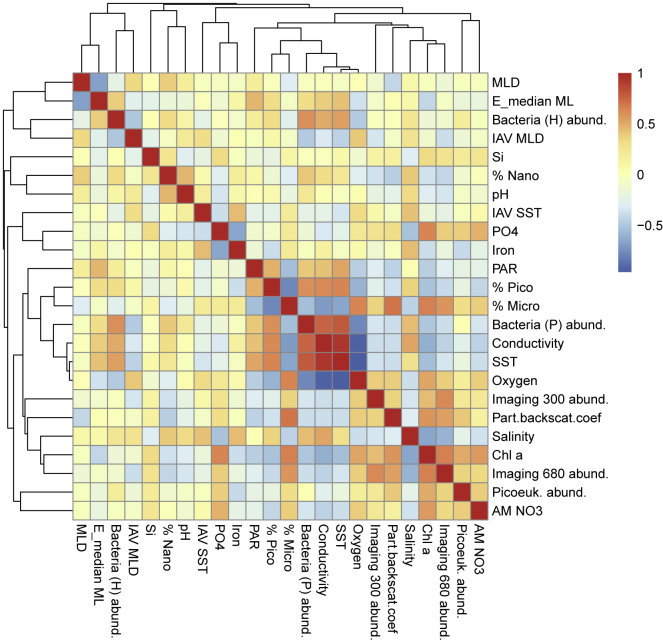
To further understand the mechanisms underlying the observed LDGs in the surface ocean, we considered contextual variables related to the most common LDG hypotheses. First, we used sea surface temperature (SST, in situ measurements) to assess the physiological tolerance and kinetic energy hypotheses. Second, we used chlorophyll a (chl a) concentrations (in situ measurements) and annual maximum of nitrate concentration (AM NO3) to test the productivity/resources hypothesis. Chl a was considered a proxy for phytoplankton biomass. We acknowledge that the latter may be affected by the poleward increase of intracellular pigmentation in phytoplankton to compensate for limitations in light (Behrenfeld et al., 2015). Phytoplankton carbon estimated via particulate backscattering has been proposed as a better proxy (Graff et al., 2015), but we lacked this parameter for many stations. Nevertheless, the available backscattering data exhibited a high correlation with chl a (Spearman’s ρ = 0.6, p < 0.001; Figure S8). Regarding AM NO3, the annual availability of this macronutrient is fundamental for primary production (Moore et al., 2013). We therefore considered this parameter to capture longer-term effects of primary production on the observed plankton diversity. Third, we considered intra-annual variation (IAV) of SST to test the environmental stability hypothesis. In addition, we also included a set of other contextual parameters in our analysis to identify potential drivers of diversity patterns for MPGs that had not been resolved previously (i.e., for viruses and protists; see STAR Methods for more details; Figure S8). Among them, sunlight, which is the fundamental source of energy for photosynthetic groups, was accounted as satellite-derived estimates of photosynthetically active radiation (PAR) and the median light in the mixed layer (STAR Methods). Their more scattered availability in our dataset indicated high correlations with SST and mixed layer depth, respectively (Figure S8), which, together with chl a concentration, reflects well the light conditions at the different sampling stations.
Figure S8.
Multiple Pairwise Spearman Correlation Analysis of the Full Matrix of Contextual Parameters for the Surface Ocean, Related to Figure 3A
Rows and columns were clustered based on the absolute pairwise Spearman correlation turned into distance (1 - |⍴|). MLD: mixed layer depth. E_median ML: median light in the mixed layer. IAV: intra-annual variability. Part.backscat.coef: particle backscattering coefficient. For more information on parameters, see Figure 3 and STAR Methods.
We conducted a combination of correlation analyses and generalized additive models (GAMs; Hastie, 2017), which allows us to deal with non-linear and/or non-monotonic relationships that could be found between diversity and environmental gradients (see below; STAR Methods). We restricted our analysis to surface planktonic communities because of their major contribution to oceanic biogeochemical cycles (Falkowski et al., 2008, Field et al., 1998) and their greater sensitivity to climate change (Bopp et al., 2013) and because of greater Tara Oceans data availability compared with the deep waters, allowing us to make more robust inferences and projections.
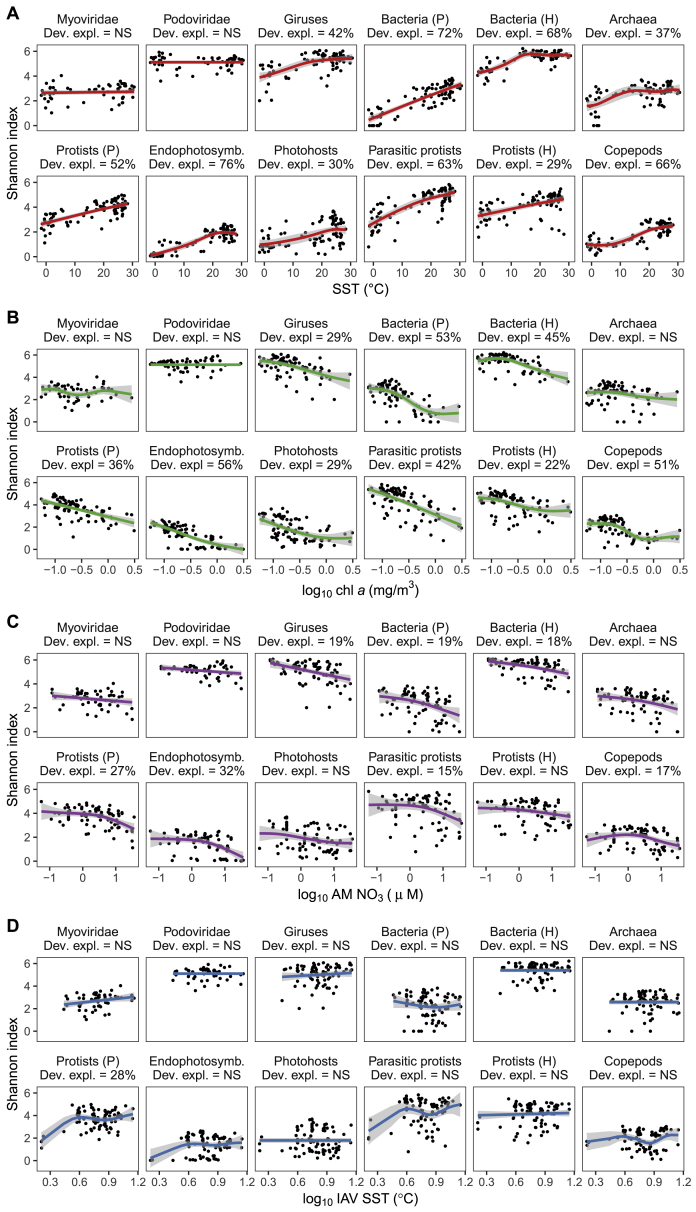
We found that SST was strongly and positively associated with the MOTU diversity patterns of most MPGs (Figures 3A, 3B, and S9A) and, therefore, was the best predictor of MPG diversity among the tested parameters (Figure 3C). Although new for most protist MPGs, these findings are consistent with previous observations for bacterioplankton (Fuhrman et al., 2008), copepods (Rombouts et al., 2009), and larger marine organisms (Tittensor et al., 2010, Woolley et al., 2016). SST and species thermal tolerance limits have been suggested to impose strong constraints on the distribution/abundance of marine ectotherms, including copepods (Beaugrand et al., 2009, Sunday et al., 2012). Our results extend this explanation to unicellular organisms as well; we observed a decline of phototrophic bacteria (mainly cyanobacteria) relative abundance at cooler higher latitudes (Figure 1B), whereas the relative abundance of phototrophic eukaryotes (mainly diatoms) increased. Such differences may result from contrasting thermal niches because diatoms generally have larger thermal breadths and lower minimal thermal growth than cyanobacteria (Chen, 2015). Similarly, the temperature-diversity relationship of several MPGs increased until it reached a plateau, in particular for heterotrophic bacteria and archaea (Figure S9A). This may suggest that these groups have larger ranges of temperature optima, corresponding roughly to those encountered in tropical/subtropical waters, and should be less affected by climate change (Hutchins and Fu, 2017). Greater SST should also increase both speciation and extinction rates, according to the metabolic theory (reviewed in Brown, 2014). This assumption has been proposed for marine foraminifera (Allen et al., 2006) and diatoms (Lewitus et al., 2018), suggesting that temperature-dependent evolutionary processes are likely important in generating patterns of diversity across MPGs. However, our current approach remains correlative, and future phylogenetic studies will be critical to estimate speciation and diversification rates in relation to temperature.
Figure 3.
Drivers of Plankton Diversity in the Surface Ocean
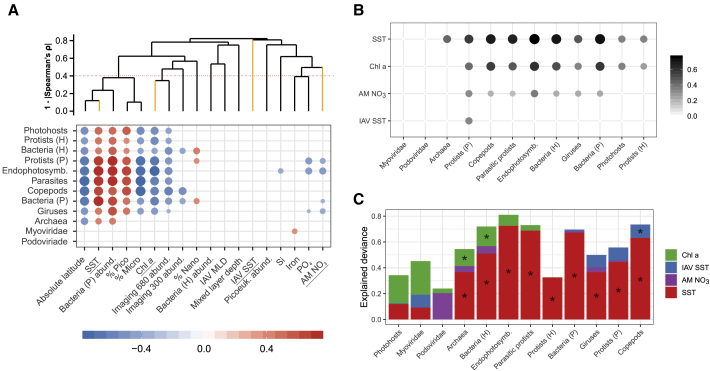
(A) Correlation of contextual variables (abiotic and population densities, x axis) with the Shannon index of each MPG (y axis). The color gradient corresponds to the values of the Spearman ρ correlation coefficient and the dot size to their absolute value. The labels of the x axis are ordered according to a hierarchical clustering analysis of absolute Spearman ρ correlation coefficient values between each pair of contextual variables, whose corresponding dendrogram is shown in the top part of the plot. Yellow leaves correspond to the four variables analyzed in (B) and (C), also underlined below. Variables that do not cluster above the dotted line (|Spearman’s ρ| < 0.6) are considered as non-collinear. Percentages of pico, nano, and micro refer to the relative abundances of fractions of phytoplankton based on pigment analysis. Bacteria and picoeukaryote abundances were determined by FC, whereas imaging abundances refer to counts of individuals caught by nets (STAR Methods). MLD: mixed-layer depth. See also Figure S8.
(B and C) Individual explained deviance (color gradient and dot size) of four variables (B; Figure S9) and additive contribution of the same four variables to the total explained deviance in GAMs, with the Shannon index as a response variable (C; STAR Methods; Tables S1D and S1E).
In (A) and (B), non-significant coefficients or effects are not shown. In (C), significant effects are indicated by asterisks. MPG labels are always ordered according to a hierarchical clustering analysis after a Spearman correlation analysis based on the displayed values in each case (A–C).
Figure S9.
Relationships between Diversity and 4 Contextual Variables for Viral, Prokaryotic, and Eukaryotic MPGs, Related to Figure 3B
(A) SST, (B) chl a, (C) AM NO3, and (D) IAV SST. Solid lines represent the GAM smooth trends and gray ribbons the corresponding 95% confidence intervals of the x-y relationship predicted by the GAMs. The percentages provided below inset titles correspond to the deviance explained by GAMs when significant (p value corrected for multiple comparisons). Non-significant GAMs are denoted with “NS.”
MPG diversity also decreased noticeably and monotonically with increasing standing stocks of chl a and to a lesser extent, of AM NO3 (Figures 3A, 3B, S9B, and S9C). These negative relationships are counterintuitive to the productivity/resources hypothesis, which asserts that greater resource availability should promote species coexistence through niche partitioning. They also contrast with the unimodal biomass-diversity relationship often reported for phytoplankton (Irigoien et al., 2004, Li, 2002, Vallina et al., 2014). As explained above, it is very unlikely that this difference arises from the diversity indices used (i.e., richness versus Shannon index). Rather, we explain this difference by our broader sampling of plankton size classes and the increased detection and taxonomic resolution of our DNA-based identification methods. Accordingly, our results are in agreement with taxon-focused or DNA-based surveys, which have reported a higher diversity of copepods (Rombouts et al., 2009), bacteria (Smith, 2007), and microplankton (Raes et al., 2018) at sites of low primary production. Although our data preclude us to infer the exact mechanisms behind this negative relationship, we propose several potential explanations. First, this observation may be related to the “paradox of the plankton” (Hutchinson, 1961); i.e., the observation that a limited number of resources support unexpectedly highly diverse communities. Non-equilibrium and chaotic environmental and/or population dynamics in aquatic systems can occur at very small temporal and spatial scales, and this, together with the existence of dormant stages in plankton organisms, is usually thought to underlie this feature by preventing local extinction (Roy and Chattopadhyay, 2007, Scheffer et al., 2003, Ser-Giacomi et al., 2018). Also, recent genomic studies in prokaryotes suggest that adaptive gene loss and subsequent microbial feeding interdependencies are selectively favored in aquatic, nutrient-poor environments. These dependencies would constitute additional but currently unmeasurable niche axes, supporting more species (Giovannoni et al., 2014; the “black queen hypothesis”; Morris et al., 2012). More generally, such trophic interdependencies probably do exist in the plankton trophic network without necessarily involving genome streamlining. On the other hand, high-nutrient or -chl a environments can correspond to areas with punctual/mid-term strong physical forcing, such as winds (Demarcq, 2009) or changes in light availability, which we did not measure on site. These environments are usually found to promote the growth of a few species at the expense of others through competitive or trophic interactions (Behrenfeld and Boss, 2014, Huisman et al., 1999, Irigoien et al., 2004, Li, 2002).
IAV of SST (Figure S9D) as well as other abiotic parameters, such as mixed layer depth together with its IAV or silicate or phosphate concentrations, exhibited comparatively weak or no correlation with MPG diversity (Figures 3A–3C; Table S1D). Finally, the GAMs, including the four focus parameters (i.e., SST, chl a, IAV SST, and AM NO3), did not exhibit latitudinal trends in their residuals (Table S1E), suggesting that the LDG was fully explained by these models. SST, followed by chl a concentration, thus appears to be the prominent driver of plankton diversity. This conclusion is further supported by additional GAMs where only SSTs, chl a concentrations, and their interaction were used as explanatory variables (see STAR Methods and their further use below). These latter models had exceptionally high explanatory power for most MPGs (42% to 81% of deviance explained) and also successfully explained the LDGs (Tables S1F and S1G). In addition, SSTs strongly correlated with microbial and photosynthetic abundances, and chl a strongly correlated with abundances of larger metazoans (Figure 3A; Figures S10A and S10B). This supports the idea that these two parameters regulate MPG diversity by controlling their population size and, therefore, also their extinction rates (Clarke and Gaston, 2006). Thus, our results lend support to the interplay of physiological tolerance, kinetic energy, and, to a lesser extent, productivity/resources effects in regulating MPG diversity in planktonic communities and causing latitudinal gradients of diversity in epipelagic waters.
Figure S10.
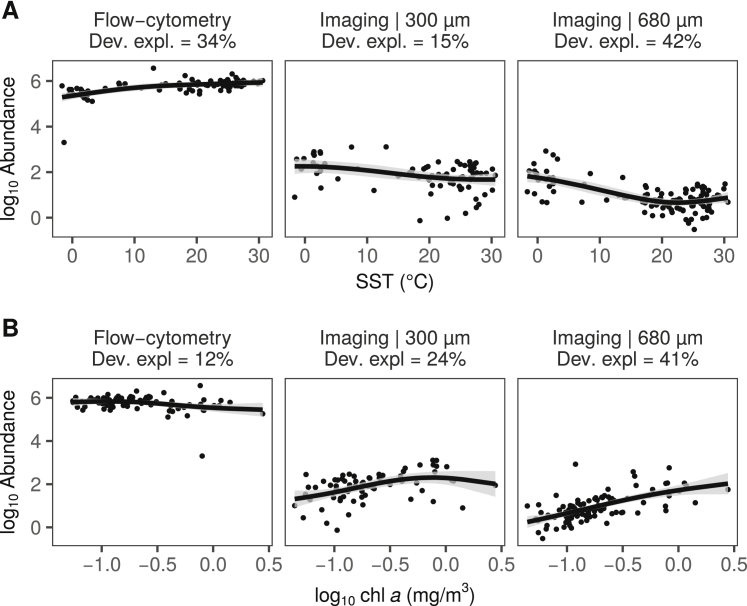
Relationship between Plankton Abundance and Two Contextual Variables, Related to Figure 3A
(A) SST, (B) chl a. Abundance values were obtained with flow cytometry (reported in [cells/ml]) or from counts of individuals captured with nets of different mesh size and identified by imaging (reported in [individuals/m3]), respectively. Solid lines represent the GAM smooth trends and gray ribbons the corresponding 95% confidence intervals of the X-Y relationship predicted by the GAMs. The percentages provided below inset titles correspond to the deviance explained by GAMs when significant.
Although our overall conclusions concur with those reported for marine macroorganisms (Hillebrand, 2004, Tittensor et al., 2010, Woolley et al., 2016), they partially contrast with recent findings regarding planktonic communities in the South Pacific Ocean (Raes et al., 2018), where primary productivity has been found to override temperature effects. This difference could be a consequence of the sampling extent of our study, which covers both the northern and southern hemispheres as well as multiple oceanic provinces that may differ in their diversity gradient (Chown et al., 2004, Sul et al., 2013). In contrast, Raes et al. (2018) characterized the latitudinal trends along a transect exhibiting marked environmental transitions caused by subtropical and subpolar waterfronts. This should be confirmed by analyzing a larger number of sampling points in each basin than those available here. Another possible explanation lies in the different diversity measures used in the two studies. The Shannon index used here, although co-varying with species richness, downweighs the influence of rarest species. The carrying capacity of a given environment strongly relies on resource availability and primary productivity, which control local extinction, in particular of rare species (Vallina et al., 2014). Such processes can solely be detected with species richness, which we did not assess here because of its strong sensitivity to sampling and technical biases (STAR Methods). More generally, several factors are also more confounding at the global scale, and their effects are more difficult to tease apart. For example, SST partially correlates with chl a concentration, PAR, and annual averages and IAVs in solar radiation as well as with the length of the productive season at the global scale (Figure S8; Clarke and Gaston, 2006). Hence, it is possible that both resource/sunlight-energy availability and stability effects partially contribute to the observed temperature effects. This feature may also explain why we could not find clear clustering of MPGs based on the drivers of their diversity according to their broad trophic modes, as found for their LDG patterns (Figure S5). Finally, we acknowledge that we considered environmental stability over short timescales. Past glaciation cycles and associated sea level changes most likely contribute to current MPG diversity, as suggested for marine diatoms (Lewitus et al., 2018) and foraminifera (Yasuhara et al., 2012). Notwithstanding, fossil records do suggest that the poleward decline of zooplankton diversity and its temperature dependence are remarkably stable features through geological times (Yasuhara et al., 2012), albeit with variations in the overall levels of diversity. We therefore believe that paleoclimate effects are unlikely to alter our conclusions.
Future Global Trends of MPG Diversity
Climate change scenarios predict a general increase of SST, with major changes in the Arctic Ocean (Figure S11; Pachauri et al., 2014). Future ocean primary production is expected to decrease in the northern hemisphere and increase in the Southern Ocean, although these projections are more uncertain (Figure S11; Bopp et al., 2013). To search for trends in diversity variation in response to these changes, we mapped the MOTU diversity of several MPGs at the beginning (years 1996–2005; Figure S12) and end of the 21st century (2090–2099) under a scenario of severe climate warming (RCP 8.5; STAR Methods). We used SST and chl a concentration values simulated by Earth System Models of the Coupled Model Intercomparison Project Phase 5 (CMIP5; Bopp et al., 2013; Table S1H) and GAM models from epipelagic plankton that explained 60% or more of deviance (6 MPGs of 12; Table S1F). After ensuring that SST and chl a simulated values were within the range of values used to train our models, we projected current and future MPG diversity at the global scale and calculated diversity anomalies (i.e., percentage of diversity change) between contemporary and future climates to identify areas where plankton diversity is most likely to be affected by the environmental changes in the ocean (Figures 4 and S12). To ensure reliability of our predictions, we generated 13,000 models for each MPG and each projection time to account for the uncertainties in the parameters of the GAMs and the output from different CMIP5 models used in this study (STAR Methods). Here we report averaged predictions from these models (see Figure S12 for prediction uncertainties). We also cross-validated our GAM models with independent datasets from other studies (STAR Methods; Figure S13) and obtained predictions congruent with the observed diversity.
Figure S11.
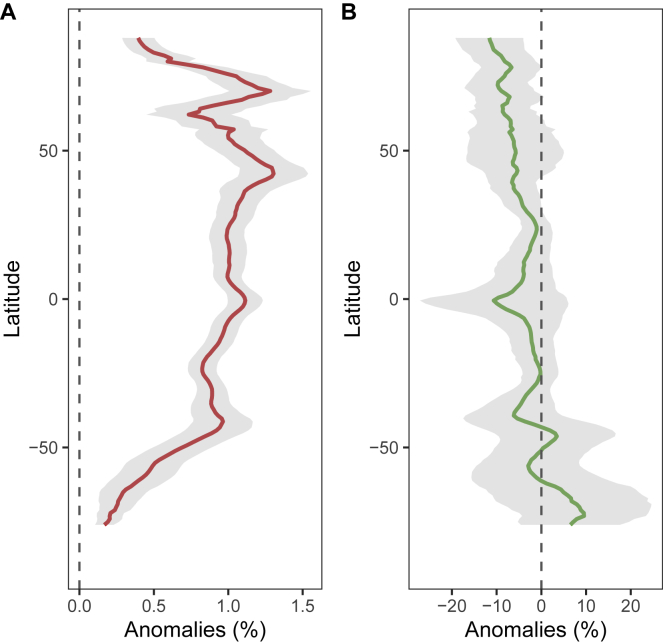
Projected Latitudinal Changes in SST and chl a, Related to Figures 4 and S12
Anomalies (%) in (A) SST and (B) chl a at the end of the 21st century (2090-2099, RCP 8.5) relative to the beginning of the century (1996-2006). Data was obtained from 13 CMIP5 models (Table S1H). Grey ribbons represent the standard error.
Figure S12.
Modeled Patterns of Diversity of MPGs in the Global Ocean, Related to Figure 4
(A) Shannon index modeled at the global scale for oceanic conditions at the beginning of the 21st century (1996-2006). Predicted Shannon values ≤ 0 obtained at high latitudes, particularly for copepods and endophotosymbionts, were excluded. (B) Anomalies were calculated as the difference of their Shannon index at the end (2090-2099, RCP 8.5) and the beginning of the century (1996-2006). A positive value means that diversity will increase by the end of the century. Note that the scale is not symmetric and that white means zero change. (C) Uncertainty maps (standard deviation) for (B). (D) Areas where the effect of chl a on plankton diversity is likely to be higher than the one of SST. To determine this, either chl a or SST were held constant in the projections by the end of the century. Then, if the anomaly caused only by the change of chl a was different than zero and higher (absolute terms) than the one caused only by the change in SST, the pixel was colored. (E) Latitudinal diversity gradient at the beginning (solid line) and the end (dashed line) of the 21st century. Values represent averages over longitude for each latitudinal degree. Dots are observed values (Figure 2). 13 Earth system models from CMIP5 were used (Table S1H).
Figure 4.
Projected Changes in Shannon Diversities by the End of the 21st Century
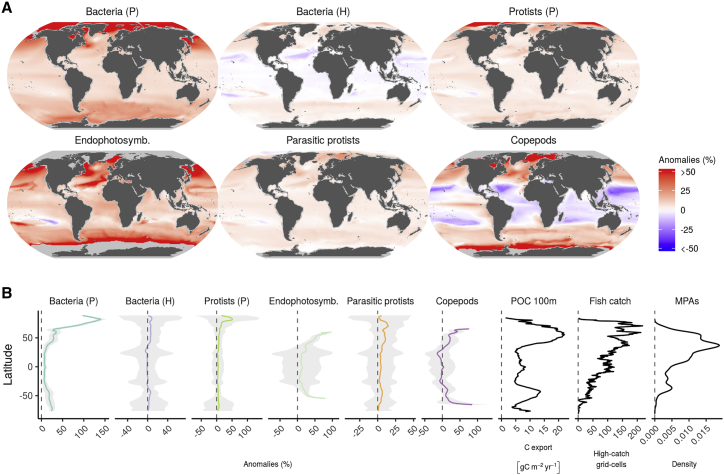
(A) Projected changes by the end of the 21st century relative to the beginning of the century (percent) for MPGs accounting for GAM models with high explained deviance (>60%). Projections were based on SST and chl a data simulated by the CMIP5 models and the GAMs (n = 13,000 for each combination of MPG and time frame; STAR Methods; Tables S1F–S1H; see Figure S12 for SD by grid cell). Copepods, photosynthetic protists, parasitic protists, and endophotosymbiont diversity (Shannon index) was modeled based on 18S rRNA gene metabarcoding data, size fraction 0.8–2000 μm, and diversity of heterotrophic and photosynthetic bacteria on 16S rRNA gene metagenomics data (size fraction 0.22–3 μm), all from the surface layer. Predicted Shannon values of 0 or less obtained at high latitudes, particularly for copepods and endophotosymbionts, were excluded.
(B) Latitudinal averages of values in (A) and their uncertainties. For visualization purposes, average anomalies for endophotosymbionts and copepods were drawn up to latitudes where values remain below 100%, and all plots show the averaged SD reduced by half. The x axis is not fixed. The last three panels refer to latitudinal averages of particulate organic carbon (POC) export at 100 m (Henson et al., 2012), the number of grid cells with a high marine fisheries catch (>200 kg km−2 year−1) (Watson, 2017), and marine protected area (MPA) latitude kernel density plots (Bruno et al., 2018; STAR Methods; Table S1I).
Figure S13.
Correlation between the Shannon Values Observed in Independent Datasets and Those for the Same Locations that Are Predicted by GAM Models Built in This Work, Related to STAR Methods
(A) Heterotrophic bacteria from the surface, open ocean water sites of the International Census of Marine Microbes (ICoMM; Zinger et al., 2011). (B) Eukaryotic community retrieved from Raes et al. (2018). Note that for the latter the mesh size of the filters used were not exactly the same (Tara Oceans > 0.8 μm; Raes et al., 2018 > 0.22 μm). Note also that both datasets have different sampling dates and locations in relation to Tara Oceans. In both cases we used as predictors the temperature and chl a of each site predicted by 13 CMIP5 models at the month of sampling averaged over 1996-2006. Inset titles show the Pearson’s correlation coefficient and its associated p value.
Our projections suggest a general increase in MPG diversity, in particular in the northern hemisphere and at latitudes that encompass the limits of the subtropical gyres (25°–50°) (Figures 4A and S12). These results support tropicalization of temperate planktonic diversity or biomass, as suggested previously for bacterioplankton (Morán et al., 2010, Morán et al., 2017), zooplankton (Beaugrand et al., 2015), and also fish (Cheung et al., 2013, Vergés et al., 2016). Following SST trends, the most dramatic changes in diversity across most MPGs are expected to occur in the Arctic Ocean (more than 100% average increase over latitude; Figure 4B). In this biome, copepods and photosynthetic bacteria should experience the most dramatic increases in diversity, mostly because these communities are currently poorly diverse. The low values for endophotosymbionts resulted in them exhibiting a large relative increase in diversity as well, especially at high latitudes (for absolute anomalies values, see Figure S12). All of these observations are in line with the poleward range expansion predicted for phytoplankton (Barton et al., 2016), in particular the cyanobacterial group Synechococcus (Flombaum et al., 2013) as well as for boreal fish species (Frainer et al., 2017), as a short-term response to poleward shifts in thermal niches (Thomas et al., 2012).
Hence, epipelagic planktonic communities are predicted to be strongly affected in the future, primarily by rising temperatures. Changes in chl a (higher uncertainty), either bound to primary production or photo-acclimation (Behrenfeld et al., 2015), should have more secondary effects, except in restricted areas for heterotrophic bacteria and marine copepods, where their effects seem to override those of SST (Figure S12D). In any case, the changes in MPG diversity predicted to occur by the end of the century will most probably induce cascading changes over the entire marine food web; e.g., by causing trophic mismatches or altering host-parasite/symbiont interactions (Doney et al., 2012, Edwards and Richardson, 2004, Gilg et al., 2012). For example, the increase in diversity and abundance of phototrophic bacteria suggested by our results and others (Hutchins and Fu, 2017) would reduce upward energy flow in marine food webs because these taxa are usually less palatable for higher trophic levels (Ullah et al., 2018). Likewise, increased temperature and diversity may also lead to reduced organism body size (Sommer et al., 2017). If the diversity and abundance of small-bodied organisms are to increase, then this may again reduce the energy transfer to higher trophic levels (Beaugrand et al., 2008).
Finally, we assessed the current ocean socio-economic and conservation status of the most affected latitudes in terms of MPG diversity (i.e., the 25% of latitudes with the highest mean absolute diversity anomalies). We did so by quantifying the current particulate carbon export, the maximum marine fisheries catch, and the number of marine protected areas at each latitude relative to global average expectations. We found that the most affected latitudes in the future currently exhibit a higher fisheries catch (32%–70% above average), carbon export (23%–70% above average), and fraction of marine protected areas (up to 100% above average; Figure 4B; Table S1I). This raises the question of how changes in diversity under the most severe climate warming scenario will affect global biogeochemical processes such as carbon export and sequestration, which are believed to have already been affected by climate change (Brun et al., 2019), and what would be the consequences for marine life in general, from already vulnerable marine animals and fish landings to life in the deep sea.
Concluding Remarks
The present findings and projections need to be interpreted carefully. Although being the largest systematic sampling effort of oceanic plankton to date, our sampling is limited by its punctual nature in space and time. Our models are also correlative and do not directly account for the effects of other abiotic parameters, such as in situ solar irradiance and their seasonal variations as well as biotic interactions and their dynamics, which should all influence plankton diversity. Regarding our projections, there are strong uncertainties about the potential lag between environmental changes and the response of plankton diversity as well as the adaptation potential of planktonic species to climate change, which can be relatively rapid, as proposed recently for zooplankton (Peijnenburg and Goetze, 2013) and diatoms (Schaum et al., 2018). Further studies better accounting for these different and intertwined mechanisms that operate at multiple spatial and temporal scales will be instrumental to improve our understanding of the drivers underlying ocean plankton ecosystems and their feedback with global change. Nevertheless, our approach is a first attempt to embrace this biological complexity at the global scale, and our results broadly agree with other statistical or theoretical projections (Barton et al., 2010, Barton et al., 2016, Righetti et al., 2019, Rombouts et al., 2009, Thomas et al., 2012, Tittensor et al., 2010). Our results should therefore be seen as a baseline and a framework for testing new hypotheses about changes in diversity within the whole plankton community across the global ocean, identifying the most vulnerable areas, and to better appreciate and anticipate functional and socio-economic consequences (Cavicchioli et al., 2019). These results will be helpful for guiding future broad and macroscale strategies to mitigate the effects of climate change on marine diversity and ecosystem services.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Data Deposited and Resource data | ||
| 18S rRNA gene metabarcoding (Tara Oceans) | de Vargas et al., 2015; this paper | European Nucleotide Archive (ENA): PRJEB6610 and PRJEB9737 |
| Metagenomics, 0.22-1.6/3 μm (OM-RGCv2, Tara Oceans) | Salazar et al., 2019 | ENA - See https://zenodo.org/record/3473199 for details |
| Metagenomics, < 0.22 μm (GOV 2.0, Tara Oceans) | Gregory et al., 2019 | ENA - See their Table S3 for details |
| ZooScan imaging - regent net, 680 μm (Tara Oceans) | This paper | EcoTaxa, https://ecotaxa.obs-vlfr.fr/prj/412 |
| ZooScan imaging - bongo net, 300 μm (Tara Oceans) | This paper | EcoTaxa, https://ecotaxa.obs-vlfr.fr/prj/397, https://ecotaxa.obs-vlfr.fr/prj/398, https://ecotaxa.obs-vlfr.fr/prj/395 |
| ZooScan imaging - WP2 net, 200 μm (Tara Oceans) | This paper | EcoTaxa, https://ecotaxa.obs-vlfr.fr/prj/377, https://ecotaxa.obs-vlfr.fr/prj/378 |
| Contextual data (Tara Oceans) | Sunagawa et al., 2015; this paper | https://doi.org/10.1594/PANGAEA.875582 |
| CMIP5 Earth system models | Bopp et al., 2013 | See our Table S1H for details |
| Sample identifiers & Shannon values; flow cytometry abundances | This paper; Sunagawa et al., 2015 | http://dx.doi.org/10.17632/p9r9wttjkm.1 |
| Software and Algorithms | ||
| R package mgcv 1.8-24 | Wood, 2010 | https://cran.r-project.org/web/packages/mgcv/index.html |
| R package segmented 0.5-3.0 | Muggeo, 2008 | https://cran.r-project.org/web/packages/segmented/index.html |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Chris Bowler (cbowler@biologie.ens.fr).
Experimental Model and Subject Details
Samples were derived from 189 stations over the 210 surveyed during the Tara Oceans expedition (2009-2013; Figure S1). They were collected across all major oceanic provinces using the sampling strategy and methodology described in Pesant et al. (2015). Briefly, the sampling was conducted at different water depths, i.e., at the sea surface (< 5 m), the deep chlorophyll maximum (17-188 m) and the mesopelagic realm (200-1000 m). Sampling of the full, trans-kingdom planktonic diversity was performed with different protocols depending on their post processing, i.e., either for DNA-based analyses or for imaging analyses.
For samples dedicated to DNA analyses, we maximized the taxonomic breadth of our diversity assessment by fractionating planktonic communities from pumped seawater with filters of different mesh size. We considered samples collected with filters of 0.22-1.6/3 μm (hereafter 0.22-3 μm) for viruses and bacteria (prokaryotic viruses from the filtrate, giruses and bacteria from the 0.22 μm filter), and 0.8-2000 μm (0.8-3 μm for non-Arctic mesopelagic samples), 5-20 μm, 20–180 μm and 180–2,000 μm for eukaryotes. Prokaryotic viruses were flocculated using iron chloride (John et al., 2011). Preliminary analyses showed that the samples obtained at 0.8-2000 μm mesh size were representative of the whole structure and diversity of protists and even of copepods, whose high dominance allowed a straightforward detection by this protocol too, probably due to the presence of small life-stages or individuals, pieces of large ones, extracellular DNA from cell turnover, or fecal pellets. We hence restricted our analysis of the main eukaryotic planktonic groups on this particular subset of samples, but also provide diversity estimates at the scale of the whole eukaryotic planktonic community for each filter size to support the robustness of the results.
Imaging data acquisition followed different protocols depending on the organisms targeted. First, microphytoplankton were sampled at the sea surface with nets of 20-180 μm mesh size for microscopy analyses (see below), as described in Malviya et al. (2016). Large protists and metazoans were collected with four different nets: WP2, bongo and regent, with mesh sizes of 200, 300 and 680 μm, respectively, which were towed vertically from 500 m to the surface. We also used a multinet, with mesh size of 300 μm, from which only the deepest level matching the mesopelagic realm was analyzed (Pesant et al., 2015). Picoplankton samples were prepared for flow cytometry from three aliquots of 1 ml of seawater (pre-filtered through 200-μm mesh), as described in Hingamp et al. (2013) and Sunagawa et al. (2015). Finally, we made use of data derived from the Malaspina expedition (Salazar et al., 2016) to account for diversity patterns of free-living prokaryotes in the bathypelagic realm (Figures 2D and S7). It should be noted that the numbers of stations and samples examined varied according to the combination of protocol and size fraction being analyzed (Figure S1; Table S1A).
Method Details
Physical and environmental measurements
Measurements of temperature, conductivity, salinity, depth, pressure, and oxygen were carried out at each station with a vertical profile sampling system (CTD-rosette) and Niskin bottles following the sampling package described in Picheral et al. (2014). Chlorophyll a (chl a) concentrations were measured using high-performance liquid chromatography (Ras et al., 2008, Van Heukelem and Thomas, 2001). Phosphate and silicate concentrations were determined using segmented flow analysis (Aminot et al., 2009). The contribution of three pigment size classes (micro-, nano-, and picoplankton) to the total phytoplankton biomass was estimated based on high pressure liquid chromatography (HPLC) analyses (Uitz et al., 2006). A full description of the performed measurements is described in (Pesant et al., 2015). Finally, we complemented these in situ measurements with (i) the average intra-annual variation of sea surface temperature (IAV SST) between years 1997-2017, which we obtained from the Extended Reconstructed Sea Surface Temperature v5 (Huang et al., 2017), (ii) the annual maximum of nitrate concentration (AM NO3) retrieved from the World Ocean Atlas 2009 (Garcia et al., 2010), (iii) the intra-annual variation of the mixed layer depth (IAV MLD), which was derived from a monthly climatology (Holte et al., 2017), (iv) iron levels, which were derived from a global circulation model (Menemenlis et al., 2008), and (v) median sunlight in the mixed layer, which was estimated as in Behrenfeld et al. (2015).
Plankton classification, diversity, and abundance estimates
A combination of molecular and optical methods were used to describe the planktonic diversity of the ocean. A full description of the molecular data production is available in Alberti et al. (2017). Viral and prokaryotic metagenomes were obtained by shotgun metagenomics, for which sequencing, assembly and/or annotation are described in Gregory et al. (2019) for bacterial and archaeal DNA viruses, Hingamp et al. (2013) for nucleo-cytoplasmic large DNA viruses (i.e., giruses, also referred to as NCLDVs or giant viruses in the literature), and Sunagawa et al. (2015) for bacteria and archaea.
Two families of prokaryotic viruses, Myo- and Podoviridae, were studied on the basis of a capsid protein gene (gp23) and a DNA polymerase (polA), respectively (Adriaenssens and Cowan, 2014). We kept populations with a gp23/polA match either via annotation (Pfam, InterProScan and KEGG) or a set of in silico primers to increase the sensitivity (Adriaenssens and Cowan, 2014), and their abundance corresponded to the normalized number of reads that mapped against these genes. Analogously, the diversity of giruses was based on another DNA polymerase gene, polB, specifically recruited with pplacer (Matsen et al., 2010) from a non-redundant gene catalog (OM-RGCv2; Salazar et al., 2019). The corresponding frequency data was obtained by mapping the raw reads to the gene catalog. Note here that ssDNA and RNA viruses were not analyzed.
Prokaryotic taxa were defined on the basis of metagenomic reads that contained signatures of the 16S rRNA genes (referred to as miTags; Logares et al., 2014, Salazar et al., 2019; data accessible at https://www.ocean-microbiome.org/). Briefly, miTags were mapped to cluster centroids of taxonomically annotated 16S/18S reference sequences from the SILVA database (Pruesse et al., 2007) (release 128: SSU Ref NR 99) that had been clustered at 97% sequence identity beforehand using USEARCH v9.2.64 (Edgar, 2010). Mapping of miTags to a unique reference sequence were used to compute the abundances of MOTUs. A MOTU abundance table was built by counting the number of miTags assigned to each reference sequence in each sample. The abundance table was normalized by the total sum for each sample after excluding MOTUs that corresponded to eukaryotes and chloroplasts.
Additionally, we analyzed data obtained by amplicon sequencing of the V4-V5 region of the 16S rRNA gene (primers 515F-Y and 926R; Parada et al., 2016), following the pipeline described in https://github.com/SushiLab/Amplicon_Recipes. Briefly, paired-end reads were merged at a minimum 90% of identity, and those with ≤ 1 mismatches were selected. Primer matching was performed with CUTADAPT v.1.9.1. Dereplication, MOTU clustering at 97% (UPARSE algorithm) and zOTUs denoting 100% similarity (UNOISE algorithm) were performed with USEARCH v.10.0.240 (Edgar 2010). OTUs and zOTUs were taxonomically annotated against the SILVA database v132 (Quast et al., 2013) with the Last Common Ancestor approach. Non-prokaryotic MOTUs (eukaryotes, chloroplast, and mitochondria) were removed, whereas singletons were maintained. This dataset was used to ensure that the diversity estimates obtained with metagenomics and amplicon sequencing approaches were consistent (see below; see Figure S6). For flow cytometry data we defined six different groups: low and high nucleic acid-content heterotrophic bacteria, Prochlorococcus, Synechococcus and two groups of picoeukaryotes (see Data and Code Availability). This latter set of samples was mainly used to determine the cell density of each bacterial/picoeukaryote group, which we considered as contextual data in downstream analyses. Nevertheless, we also retrieved diversity values from these data in order to assess their congruence at broad scale with those obtained through molecular approaches. They were not used to standardize DNA-based taxa abundances.
The taxonomic composition of protists and small metazoan communities was characterized through DNA metabarcoding using mainly the V9 region of the 18S rRNA gene, and the V4 region was also used to assess the congruence of the MOTU diversity estimates between the two DNA markers (Figure S6). For both sequencing reads datasets, we obtained a list of MOTUs as defined by the “swarm” algorithm (Mahé et al., 2014). Each MOTU was represented by a number of sequencing reads, which we used as a proxy for abundance. A full description of the sequencing reads processing (i.e., data curation, clustering into MOTUs, taxonomic classification, etc.) is available at http://taraoceans.sb-roscoff.fr/EukDiv/. Microphytoplankton were also identified and quantified manually using an inverted light microscope, as described in Malviya et al. (2016). The identification was performed by experts and reached the genus level for most of the 440 morphotypes identified (Table S1A). About half of these taxa corresponded to diatoms. Smaller protists in 5-20 μm size fractions from surface and deep chlorophyll maximum layers were retrieved from Colin et al. (2017) and obtained by environmental high content fluorescence microscopy (eHCFM).
The taxonomic classification of mesozooplankton collected with nets was performed on formaldehyde fixed samples scanned with the ZooScan imaging system (Gorsky et al., 2010) and identified with the help of an automatic recognition algorithm to the deepest possible taxonomic level using Ecotaxa (Picheral et al., 2017). The resulting identifications were validated by specialists, and reached different taxonomic levels, mostly the family level (or genus in some cases, e.g., copepods from the WP2 net). All images are accessible within Ecotaxa (https://ecotaxa.obs-vlfr.fr/). Mesozooplankton absolute abundances were calculated by taking into account the volume of water filtered by the nets. Together with images, various morphological measurements were obtained (also accessible within Ecotaxa). Major and minor best ellipsoidal axis were used to calculate the ellipsoidal biovolume of each organism that was used as a proxy of biomass. All other morphological measurements (such as length, elongation, gray level values and distribution; except those related to position of organisms within the initial scan) were recovered, normalized and used in a t-SNE analysis (van der Maaten and Hinton, 2008) using MATLAB software using the default settings (Euclidean distances; Barnes-Hut algorithm; perplexity of 30; exaggeration of 4; learning rate of 500). Different combinations of parameters were tested without clear improvements to the final result shown in Figure 2B for the bongo net. t-SNE results were used to overlay taxonomic information on the morphological overview of the imaging datasets.
Summary statistics of our datasets and their taxonomic resolution are provided in Table S1A. Based on their taxonomic affiliation, we classified all taxa into marine plankton groups (MPGs) following the criteria indicated in Table S1B. We did so not only to separate organisms of different broad functions, but also to minimize biases that could arise when comparing organisms with contrasting body size or marker gene copy number per organism (see below). For viruses we considered the three families mentioned above separately as they are the most abundant groups and have different ecologies (Brum et al., 2015, Hingamp et al., 2013, Roux et al., 2016). For prokaryotes, we distinguished photosynthetic bacteria (i.e., cyanobacteria) from heterotrophic/chemotrophic bacteria and archaea. For protists, we used an extended version of the functional database used in de Vargas et al. (2015), which encompasses a wide variety of protist taxa that are assigned to major functional groups: photosynthetic/mixotrophic protists, endophotosymbionts, hosts with endophotosymbionts (hereafter photohosts), parasitic protists, and free-living heterotrophs or phagotrophs (hereafter heterotrophic protists). Note that the endophotosymbiont group is probably the most incomplete due to the difficulties in currently being able to define comprehensively these organisms. For the mesozooplankton, the categories used corresponded to the most abundant taxonomic groups (such as copepods and chaetognaths) or feeding strategies (Figure S2). We here only considered MPGs for which the total relative abundance in the molecular dataset was > 1%, a threshold under which we considered that the detection level was too low to obtain reliable detection and diversity estimates. In total, we thus studied the diversity of 12 MPGs, of which a full list is provided in Table S1B.
Quantification and Statistical Analysis
Diversity estimates calculation and validation
Plankton diversity was estimated at each station with the Shannon diversity index, a robust measure of entropy. We chose this index rather than richness because, unlike richness, the Shannon index is insensitive to sampling effort, provided that the sampling is not too shallow (Jost, 2006). As such, the sampling effort in our datasets – albeit very deep – varied across samples but the rarefaction curves drawn with the Shannon index were largely saturating contrary to those based on richness (Figure S3). The Shannon index has also been shown to provide more reliable diversity estimates when using DNA-based data (Bálint et al., 2018, Haegeman et al., 2013). Finally, by construction, it also relates monotonically with species richness, and should therefore exhibit similar patterns (Jost, 2006). The Shannon index was calculated separately for each MPG using the samples filtered at < 0.22 μm, 0.22-3 μm and 0.8-2000 μm for prokaryotic viruses, bacteria/giruses (metagenomics) and eukaryotic plankton (DNA metabarcoding), respectively. We also calculated the Shannon index for the full local planktonic communities (i.e., not parsed into MPGs) for each sampling protocol (i.e., metagenomics, metabarcoding, and imaging for each sampling mesh size). To ensure that our Shannon values were robust, we computed their variation from 100 Monte Carlo simulated communities (function EntropyCI, R-package ‘entropart’ v1.6-1, https://cran.r-project.org/web/packages/entropart/; evaluated only for the eukaryotic plankton). Each variation range was very narrow and seldom overlap with the rest (difference between Shannon values from simulated communities, 0.003 ± 0.003; difference between Shannon values from samples; 0.969 ± 0.700), making this uncertainty negligible as compared to that generated by our downstream analyses. Shannon diversity for each MPG retrieved from microscopy/imaging data was in general not assessed due to low taxonomic resolutions, with the exception of microphytoplankton/diatoms identified with light microscopy, and the copepods collected with the WP2 net due to their relatively high taxonomic resolution (Figure S6), as explained above.
Because every method to assess biodiversity has limitations, either due to technical issues (e.g., sampling difficulties, taxonomic resolution, lack of morphological/genetic differences) or more fundamentally due to the difficulties of classifying the diversity of life, we also assessed the congruence of diversity trends across methodologies to ensure the reliability of our conclusions. We therefore provide in Figure S6 several correlation analyses of the diversity patterns observed with a wide array of methodologies used in Tara Oceans, using previously published or newly released datasets. These comparisons include DNA-based diversity trends obtained with different markers, taxonomic resolution, and size fractions, as well as diversity trends obtained with different optical methods. More specifically, we compared the diversity trends for (a) different DNA markers (V9 versus V4 region of the 18S rRNA gene), (b) different taxonomic resolutions using different clustering similarity thresholds, and (c) molecular versus optical approaches (the latter based both on abundance or biovolume). More details about the datasets are available in the legend of Figure S6. Additionally, we evaluated the potential effect of marker gene copy number in prokaryotes by correcting for gene copy numbers of the 16S rRNA gene. The correction was performed using copy number estimates of references in the SILVA database (v136; Louca et al., 2018), from which we could assign a 16S rRNA gene copy number value for almost all miTags in our dataset (99%). After the correction, Shannon values remained essentially unchanged (Pearson’s r correlation between corrected and uncorrected Shannon values = 0.99, p < < 0.001). Furthermore, we relied on the strong correlation we showed in a previous study between Shannon values for bacterial OTUs defined either by 16S rRNA gene or by single-copy genes (Milanese et al., 2019).
Latitudinal diversity gradient
Our first objective was to explore visually the LDG trend across all domains of life using both MOTU and morphotype diversity at each station. To this end, we used generalized additive models (GAMs; Hastie 2017) due to their ability to fit non-linear and/or non-monotonic functions which we expected between diversity and latitude. GAMs are further highly suitable for modeling large scale trends (Guisan et al., 2002). Additionally, we preferred the GAM smoothing approach over simple moving averages because priors are directly learned from the data and the sensitivity to extreme values is relatively low. For this particular analysis, GAMs were used only for visualizing the diversity trendline, and were not used in downstream analyses (see “Diversity modeling of MPGs”). Next, we analyzed the shape of the LDG of each marine planktonic group in two ways. First, a segmented regression analysis was conducted to describe the form of the latitudinal gradient on absolute latitude. More specifically, we aimed at detecting latitudinal breakpoints and changes in slopes across latitude. For this we used the R package ‘segmented’ 0.5-3.0 (https://cran.r-project.org/web/packages/segmented). In order to determine which MPG displayed similar LDG forms, we computed pairwise Euclidean distances between the obtained set of latitudinal breakpoints and slope values for each MPG, and subjected the resulting distance matrix to a hierarchical clustering analysis (Figure S5). In an additional analysis, we further determined whether the LDG of each MPG exhibited a North-South asymmetry. To this end, we performed separate linear regressions for each hemisphere (Table S1C).
Diversity modeling of MPGs
Our second objective was to find predictors for local diversity of marine planktonic groups. To this end, we also used GAMs (see “Latitudinal diversity gradient” for description and references). All GAMs were built using the R library ‘mgcv’ 1.8-24 (https://cran.r-project.org/web/packages/mgcv/), using only MOTU diversity of each MPG as response variable. Smooth terms (‘s’) were based on a thin plate regression splines and estimated by a Laplace approximation marginal likelihood criterion (Wood, 2010). The rest of the parameters were set on default mode.
In order to test the different hypotheses explaining plankton latitudinal diversity gradients and patterns of diversity in general, we first made a selection of variables from the Tara Oceans contextual data and public databases (see “Physical and environmental measurements”) that relate to these hypotheses and/or that were sufficiently available across our global sampling. We then evaluated their redundancy and their link with MPG diversity by conducting multiple pairwise correlation analyses with the Spearman rank correlation test (Figures 3A and S8). We considered the contextual variables associations having |Spearman’s ρ| > 0.6 to be highly correlated and kept only the most representative and biologically meaningful variable among correlated ones to avoid collinearity in downstream analyses. We excluded null hypotheses for the LDG, such as the area (Willig et al., 2003) or mid-domain effect (Colwell and Lees, 2000) due to the high interconnectivity of the global ocean, which should limit the geometric constraints imposed on species distribution on lands.
The associations of plankton diversity with the four selected contextual variables (i.e., SST, chlorophyll a, annual maximum of nitrate concentration, and intra-annual variation of SST) were further analyzed using GAMs, as we expected them to be non-linear or non-monotonic functions based on previous studies (e.g., Tittensor et al., 2010, Vallina et al., 2014). Except for SST, the contextual variables were log10-transformed. For chl a, three low-concentration outlier values were excluded. “Individual” GAMs were built for each MPG and each contextual parameter, from which the explained deviance was used as an association measure and the approximate p value of the smooth term to account for effect significance (Figures 3B and S9). All p values obtained were corrected for multiple comparisons (Holm, 1979). To further test the different LDG hypotheses, we then built “full” GAMs for each MPG that included all four contextual variables, with the settings explained above. Their additive contribution was calculated by a sequential removal of the different parameters and a normalization with a null model (Figure 3C).
From these analyses, we identified temperature and chlorophyll a to be the best correlates with most MPGs diversity. These two variables also capture relatively well other environmental gradients, such as cyanobacteria and mesozooplankton abundance (Figure 3A). Given their strong explanatory power and because their current and future state at the sea surface can be simulated with global ocean circulation models (Figure S11; Bopp et al., 2013), we used them to predict the current global-scale distribution of MPGs diversity, as well as its response to a severe scenario of climate and oceanic change. To this end, we built a set of “reduced” GAMs with surface diversity for each MPG as response variable, and SST, chlorophyll a, as well as an interaction-like term (included as a tensor product, ‘ti’), as explanatory variables. SST and chlorophyll a (in situ measurements) were only partially anti-correlated, probably due to a decoupling in upwelling systems (Demarcq, 2009). Accordingly, the explained deviance of some of our GAMs was increased by 10% or more when considering these two parameters without affecting their parsimony.
Both “full” and “reduced” GAMs were built with the same approaches as described above and were further validated with two additional analyses. First, we quantified the congruence between observed and GAM-modeled Shannon diversity values through a Pearson’s correlation analysis. Second, we ensured that the GAM residuals did not exhibit latitudinal or longitudinal trends, a way to control for spatial autocorrelation (Tables S1E–S1G). Checking the absence of latitudinal trends in the model residuals further indicates if our models successfully explained the latitudinal gradients of diversity. To further assess the performance of the “reduced” models used downstream for predicting current and future trends of diversity, we cross-validated them with other independently collected molecular datasets from open-ocean studies targeting either heterotrophic bacteria (Zinger et al., 2011) or the whole planktonic eukaryotic community (Raes et al., 2018) from different sampling dates and locations. Splitting this latter dataset into MPGs was not possible due to the unavailability of functional databases for the DNA marker used (V4 region of the 18S rRNA gene), so we conducted this cross-validation with a reduced GAM model built for the whole eukaryotic community. Figure S13 shows that the GAM models built with our datasets are able to predict correctly the diversity trends of plankton communities observed in these independent datasets (see complementary details in the caption of Figure S13 and below for CMIP5 models). Additionally, as both datasets include sampling points in the Southern Ocean, which was undersampled in ours, this agreement confirms the decrease in plankton diversity we observed toward the south.
Next, the MOTU diversity of the six MPGs for which “reduced” GAMs yielded a deviance explained ≥ 60% was modeled for the beginning and the end of the 21st century. To do so, we first defined a coarse-grained arrange of 1° grid-cells. SST and chlorophyll a content across space and time were obtained from the Coupled Model Intercomparison Project Phase 5 (CMIP5), a multi-model simulation of the ocean (Bopp et al., 2013). Each model within CMIP5 is an Earth system simulation generated by different research groups (Table S1H), which allows us to account for different mechanistic weights. We extracted the two variables from each CMIP5 model for each grid cell for the beginning of the 21st century (averaged values for years 1996-2005) and the end of the 21st century (averaged values for years 2090-2099), the latter considering RCP 8.5 scenario, the most pessimistic IPCC trajectory for greenhouse gases concentration (radiative forcing level reaches 8.5 W/m2). To obtain an average and assess the uncertainty in our predictions, we generated a combined calculation of the uncertainty of the GAM parameters and the multiple CMIP5 models. We did so by first obtaining posterior distributions of the fitted GAM parameters for the different plankton groups. We then sampled values from these distributions randomly to generate 1,000 models. For each of these models, we used each of the CMIP5 models (n = 13) as current and future temperatures and chl a. Shannon diversity was then predicted with each of the 13,000 models we generated for each MPG and each time of projection (beginning and the end of the century), from which we assessed the uncertainty of our predictions.
Anomalies between future ocean projections and estimates for the beginning of the 21st century were calculated as the difference between the average diversity of each planktonic group projected for the time interval 2090-2099 and the one for 1996-2005 (Figures 4 and S12). In other words, a positive anomaly means that the predicted diversity will increase toward the end of the century. Confidence intervals of anomalies were based on the standard deviations of the average Shannon diversity estimates across the different CMIP5 models. Finally, to assess potential areas in the future ocean where the effect of primary production change on diversity could override the effect of temperature, we performed diversity projections holding either SST or chlorophyll a constant, and then comparing their output. That is, we assessed per grid-cell whether the effect of chlorophyll a on diversity was significantly larger than zero and larger than the one of SST (in absolute values; see Figure S12D). Manipulation and visualization of the CMIP5 spatial data and the corresponding diversity projections were performed combining R packages ‘ncdf4’ 1.16 (https://cran.r-project.org/web/packages/ncdf4), ‘raster’ 2.5-8 (https://cran.r-project.org/web/packages/raster) in R v.3.5.1 (https://www.r-project.org).
Comparison of future trends with current areas of high socioeconomic and conservation value
We identified the latitudes that are expected to experience the most dramatic changes in diversity (defined as the 25% of latitudes with the highest mean absolute diversity anomalies) and analyzed whether these areas overlap with current ecosystem services and reserves. To this end, we compared their corresponding current status in terms of (i) carbon export, using satellite-derived estimates at 100 m depth (Henson et al., 2012), (ii) maximum marine fisheries catch between years 2010-2014 (Watson, 2017), and (iii) number of marine protected areas (Bruno et al., 2018) relative to the global average. In all cases, the difference was expressed as the relative (%) increase or decrease in relation to the global average (Table S1I).
Data and Code Availability
Raw reads of Tara Oceans are deposited at the European Nucleotide Archive (ENA). In particular, newly released 18S rRNA gene metabarcoding reads are available under the number ENA: PRJEB9737. ENA references for the metagenomics reads corresponding to the size fraction < 0.22 μm (for prokaryotic viruses) analyzed in this study are included in Gregory et al. (2019); see their Table S3. ENA references for the metagenomics reads corresponding to the size fraction 0.22-1.6/3 μm (for prokaryotes and giruses) correspond to Salazar et al. (2019) (see https://zenodo.org/record/3473199). Imaging datasets from the nets are available through the collaborative web application and repository EcoTaxa (Picheral et al., 2017) under the address https://ecotaxa.obs-vlfr.fr/prj/412 for regent data, within the 3 projects https://ecotaxa.obs-vlfr.fr/prj/397, https://ecotaxa.obs-vlfr.fr/prj/398, https://ecotaxa.obs-vlfr.fr/prj/395 for bongo data, and within the 2 projects https://ecotaxa.obs-vlfr.fr/prj/377 and https://ecotaxa.obs-vlfr.fr/prj/378 for WP2 data. A table with Shannon values and multiple samples identifiers, plus a table with flow cytometry data split in six groups are available (https://doi.org/10.17632/p9r9wttjkm.1). Contextual data from the Tara Oceans expedition, including those that are newly released from the Arctic Ocean, are available at https://doi.org/10.1594/PANGAEA.875582.
Consortia
The Tara Oceans coordinators are Silvia G. Acinas, Marcel Babin, Peer Bork, Emmanuel Boss, Chris Bowler, Guy Cochrane, Colomban de Vargas, Mick Follows, Gabriel Gorsky, Nigel Grimsley, Lionel Guidi, Pascal Hingamp, Daniele Iudicone, Olivier Jaillon, Stefanie Kandels, Lee Karp-Boss, Eric Karsenti, Fabrice Not, Hiroyuki Ogata, Stéphane Pesant, Nicole Poulton, Jeroen Raes, Christian Sardet, Sabrina Speich, Lars Stemmann, Matthew B. Sullivan, Shinichi Sunagawa, and Patrick Wincker. Affiliations can be found in Document S1.
Acknowledgments
Tara Oceans (which includes both the Tara Oceans and Tara Oceans Polar Circle expeditions) would not exist without the leadership of the Tara Ocean Foundation and the continuous support of 23 institutes (https://oceans.taraexpeditions.org/). We further thank the commitment of the following sponsors: CNRS (in particular Groupement de Recherche GDR3280 and the Research Federation for the Study of Global Ocean Systems Ecology and Evolution FR2022/Tara Oceans-GOSEE), the European Molecular Biology Laboratory (EMBL), Genoscope/CEA, the French Ministry of Research, and the French Government “Investissements d’Avenir” programs OCEANOMICS (ANR-11-BTBR-0008), FRANCE GENOMIQUE (ANR-10-INBS-09-08), MEMO LIFE (ANR-10-LABX-54), the PSL∗ Research University (ANR-11-IDEX-0001-02), as well as EMBRC-France (ANR-10-INBS-02). Funding for the collection and processing of the Tara Oceans data set was provided by NASA Ocean Biology and Biogeochemistry Program under grants NNX11AQ14G, NNX09AU43G, NNX13AE58G, and NNX15AC08G (to the University of Maine); the Canada Excellence research chair on remote sensing of Canada’s new Arctic frontier; and the Canada Foundation for Innovation. We also thank agnès b. and Etienne Bourgois, the Prince Albert II de Monaco Foundation, the Veolia Foundation, Region Bretagne, Lorient Agglomeration, Serge Ferrari, Worldcourier, and KAUST for support and commitment. The global sampling effort was enabled by countless scientists and crew who sampled aboard the Tara from 2009–2013, and we thank MERCATOR-CORIOLIS and ACRI-ST for providing daily satellite data during the expeditions. We are also grateful to the countries who graciously granted sampling permission. We thank Stephanie Henson for providing ocean carbon export data and are also grateful to the other researchers who kindly made their data available. We thank Juan J. Pierella-Karlusich for advice regarding single-copy genes. C.d.V. and N.H. thank the Roscoff Bioinformatics platform ABiMS (http://abims.sb-roscoff.fr) for providing computational resources. C.B. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation Program (grant agreement 835067) as well as the Radcliffe Institute of Advanced Study at Harvard University for a scholar’s fellowship during the 2016-2017 academic year. M.B.S. thanks the Gordon and Betty Moore Foundation (award 3790) and the National Science Foundation (awards OCE#1536989 and OCE#1829831) as well as the Ohio Supercomputer for computational support. S.G.A. thanks the Spanish Ministry of Economy and Competitiveness (CTM2017-87736-R), and J.M.G. is grateful for project RT2018-101025-B-100. F.L. thanks the Institut Universitaire de France (IUF) as well as the EMBRC platform PIQv for image analysis. M.C.B., D.S., and J.R. received financial support from the French Facility for Global Environment (FFEM) as part of the “Ocean Plankton, Climate and Development” project. M.C.B. also received financial support from the Coordination for the Improvement of Higher Education Personnel of Brazil (CAPES 99999.000487/2016-03). The authors declare that all data reported herein are fully and freely available from the date of publication, with no restrictions, and that all of the analyses, publications, and ownership of data are free from legal entanglement or restriction by the various nations whose waters the Tara Oceans expeditions sampled in. This article is contribution number 93 of Tara Oceans.
Author Contributions
Conceptualization, F.M.I., N.H., L.S., L.B., C.d.V., L.K.-B., F.L., C.B., and L.Z.; Data Collection and Production, J.M.G., M.R.-L., K.L., J.F., C.M., S.K., M.P., C.D., J.P., S. Pisarev, M.C., S. Pesant, P.W., E.B., S.G.A., L.K.-B., and F.L.; Data Curation: C.d.V., N.H., F.M., F.L., and S. Pesant; Resources, C.d.V., P.W., F.L., A.A.Z., and J.M.G.; Data Pre-processing, F.M.I., N.H., M.C.B., S.M., G.B., H.B., L.P.C., H.E., A.C.G., F.M., J.R., G.S., I.S.-S., E.S., D.S., A.A.Z., A.Z., E.P., L.B., and F.L.; Formal Analysis and Visualization, F.M.I., N.H., M.C.B., S.M., H.B., F.L., and L.Z.; Writing – Original Draft, F.M.I., C.B., and L.Z; Writing – Review & Editing, M.C.B., M.B., D.I., S.G.A., L.S., M.B.S., S.S., L.B., E.B., C.d.V., L.K.-B., and F.L.; Supervision, C.B. and L.Z.; Funding Acquisition, Tara Oceans Coordinators, M.B., D.I., O.J., P.W., H.O., M.B.S., S.S., E.B., C.d.V., and C.B. M.B., C.B., and L.K.-B. directed the Tara Oceans Polar Circle Expedition.
Declaration of Interests
The authors declare no competing interests.
Published: November 14, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.10.008.
A video abstract is available at https://doi.org/10.1016/j.cell.2019.10.008#mmc3.
Contributor Information
Chris Bowler, Email: cbowler@biologie.ens.fr.
Lucie Zinger, Email: lucie@zinger.fr.
Tara Oceans Coordinators:
Silvia G. Acinas, Marcel Babin, Peer Bork, Emmanuel Boss, Chris Bowler, Guy Cochrane, Colomban de Vargas, Mick Follows, Gabriel Gorsky, Nigel Grimsley, Lionel Guidi, Pascal Hingamp, Daniele Iudicone, Olivier Jaillon, Stefanie Kandels, Lee Karp-Boss, Eric Karsenti, Fabrice Not, Hiroyuki Ogata, Stéphane Pesant, Nicole Poulton, Jeroen Raes, Christian Sardet, Sabrina Speich, Lars Stemmann, Matthew B. Sullivan, Shinichi Sunagawa, and Patrick Wincker
Supplemental Information
[S1A] Summary statistics for the main datasets utilized in this work. Related to Figures 1B, 2A–2C. [S1B] Summary statistics for the 12 marine plankton groups (MPGs). Related to Figures 1B, 2A. [S1C] Analysis of the LDG symmetry for each of the MPGs in surface waters. Related to Figure 2A. [S1D] GAM parameters for the “full” models of diversity of MPGs. Related to Figure 3C and STAR Methods. [S1E] Evaluation of GAM “full” models. Related to Figure 3C and (D). [S1F] GAM parameters for the “reduced” models of diversity of MPGs. Related to Figure 4 and STAR Methods. [S1G] Evaluation of GAM “reduced” models. Related to Figure 4 and (F). [S1H] CMIP5 Earth system models. Related to Figures 4, S12, and S13. [S1I] Latitudinal overlap with areas of socio-economic and conservation value. Related to Figure 4.
References
- Adriaenssens E.M., Cowan D.A. Using signature genes as tools to assess environmental viral ecology and diversity. Appl. Environ. Microbiol. 2014;80:4470–4480. doi: 10.1128/AEM.00878-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti A., Poulain J., Engelen S., Labadie K., Romac S., Ferrera I., Albini G., Aury J.-M., Belser C., Bertrand A., Genoscope Technical Team. Tara Oceans Consortium Coordinators Viral to metazoan marine plankton nucleotide sequences from the Tara Oceans expedition. Sci. Data. 2017;4:170093. doi: 10.1038/sdata.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algar A.C., Kharouba H.M., Young E.R., Kerr J.T. Predicting the future of species diversity: macroecological theory, climate change, and direct tests of alternative forecasting methods. Ecography. 2009;32:22–33. [Google Scholar]
- Allen A.P., Gillooly J.F., Savage V.M., Brown J.H. Kinetic effects of temperature on rates of genetic divergence and speciation. Proc. Natl. Acad. Sci. USA. 2006;103:9130–9135. doi: 10.1073/pnas.0603587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aminot A., Kérouel R., Coverly S. Nutrients in Seawater Using Segmented Flow Analysis. In: Wurl O., editor. Practical Guidelines for the Analysis of Seawater. Taylor & Francis; 2009. [Google Scholar]
- Arrigo K.R., van Dijken G.L., Bushinsky S. Primary production in the Southern Ocean, 1997–2006. J. Geophys. Res. 2008;113:609. [Google Scholar]
- Bálint M., Márton O., Schatz M., Düring R.-A., Grossart H.-P. Proper experimental design requires randomization/balancing of molecular ecology experiments. Ecol. Evol. 2018;8:1786–1793. doi: 10.1002/ece3.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton A.D., Dutkiewicz S., Flierl G., Bragg J., Follows M.J. Patterns of diversity in marine phytoplankton. Science. 2010;327:1509–1511. doi: 10.1126/science.1184961. [DOI] [PubMed] [Google Scholar]
- Barton A.D., Irwin A.J., Finkel Z.V., Stock C.A. Anthropogenic climate change drives shift and shuffle in North Atlantic phytoplankton communities. Proc. Natl. Acad. Sci. USA. 2016;113:2964–2969. doi: 10.1073/pnas.1519080113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaugrand G., Edwards M., Brander K., Luczak C., Ibanez F. Causes and projections of abrupt climate-driven ecosystem shifts in the North Atlantic. Ecol. Lett. 2008;11:1157–1168. doi: 10.1111/j.1461-0248.2008.01218.x. [DOI] [PubMed] [Google Scholar]
- Beaugrand G., Luczak C., Edwards M. Rapid biogeographical plankton shifts in the North Atlantic Ocean. Glob. Change Biol. 2009;15:1790–1803. [Google Scholar]
- Beaugrand G., Edwards M., Raybaud V., Goberville E., Kirby R.R. Future vulnerability of marine biodiversity compared with contemporary and past changes. Nat. Clim. Chang. 2015;5:695. [Google Scholar]
- Behrenfeld M.J., Boss E.S. Resurrecting the ecological underpinnings of ocean plankton blooms. Annu. Rev. Mar. Sci. 2014;6:167–194. doi: 10.1146/annurev-marine-052913-021325. [DOI] [PubMed] [Google Scholar]
- Behrenfeld M.J., O’Malley R.T., Boss E.S., Westberry T.K., Graff J.R., Halsey K.H., Milligan A.J., Siegel D.A., Brown M.B. Revaluating ocean warming impacts on global phytoplankton. Nat. Clim. Chang. 2015;6:323. [Google Scholar]
- Bergauer K., Fernandez-Guerra A., Garcia J.A.L., Sprenger R.R., Stepanauskas R., Pachiadaki M.G., Jensen O.N., Herndl G.J. Organic matter processing by microbial communities throughout the Atlantic water column as revealed by metaproteomics. Proc. Natl. Acad. Sci. USA. 2018;115:E400–E408. doi: 10.1073/pnas.1708779115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp L., Resplandy L., Orr J.C., Doney S.C., Dunne J.P., Gehlen M., Halloran P., Heinze C., Ilyina T., Seferian R. Multiple stressors of ocean ecosystems in the 21st century: projections with CMIP5 models. Biogeosciences. 2013;10:6225–6245. [Google Scholar]
- Boyce D.G., Lewis M.R., Worm B. Global phytoplankton decline over the past century. Nature. 2010;466:591–596. doi: 10.1038/nature09268. [DOI] [PubMed] [Google Scholar]
- Brown J.H. Why are there so many species in the tropics? J. Biogeogr. 2014;41:8–22. doi: 10.1111/jbi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.J., O’Connor M.I., Poloczanska E.S., Schoeman D.S., Buckley L.B., Burrows M.T., Duarte C.M., Halpern B.S., Pandolfi J.M., Parmesan C., Richardson A.J. Ecological and methodological drivers of species’ distribution and phenology responses to climate change. Glob. Change Biol. 2016;22:1548–1560. doi: 10.1111/gcb.13184. [DOI] [PubMed] [Google Scholar]
- Brum J.R., Ignacio-Espinoza J.C., Roux S., Doulcier G., Acinas S.G., Alberti A., Chaffron S., Cruaud C., de Vargas C., Gasol J.M., Tara Oceans Coordinators Ocean plankton. Patterns and ecological drivers of ocean viral communities. Science. 2015;348:1261498. doi: 10.1126/science.1261498. [DOI] [PubMed] [Google Scholar]
- Brun P., Stamieszkin K., Visser A.W., Licandro P., Payne M.R., Kiørboe T. Climate change has altered zooplankton-fuelled carbon export in the North Atlantic. Nat. Ecol. Evol. 2019;3:416–423. doi: 10.1038/s41559-018-0780-3. [DOI] [PubMed] [Google Scholar]
- Bruno J.F., Bates A.E., Cacciapaglia C., Pike E.P., Amstrup S.C., van Hooidonk R., Henson S.A., Aronson R.B. Climate change threatens the world’s marine protected areas. Nat. Clim. Chang. 2018;8:499–503. [Google Scholar]
- Cavicchioli R., Ripple W.J., Timmis K.N., Azam F., Bakken L.R., Baylis M., Behrenfeld M.J., Boetius A., Boyd P.W., Classen A.T. Scientists’ warning to humanity: microorganisms and climate change. Nat. Rev. Microbiol. 2019;17:569–586. doi: 10.1038/s41579-019-0222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. Patterns of thermal limits of phytoplankton. J. Plankton Res. 2015;37:285–292. [Google Scholar]
- Cheung W.W.L., Watson R., Pauly D. Signature of ocean warming in global fisheries catch. Nature. 2013;497:365–368. doi: 10.1038/nature12156. [DOI] [PubMed] [Google Scholar]
- Chown S.L., Sinclair B.J., Leinaas H.P., Gaston K.J. Hemispheric asymmetries in biodiversity--a serious matter for ecology. PLoS Biol. 2004;2:e406. doi: 10.1371/journal.pbio.0020406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chust G., Irigoien X., Chave J., Harris R.P. Latitudinal phytoplankton distribution and the neutral theory of biodiversity. Glob. Ecol. Biogeogr. 2013;22:531–543. [Google Scholar]
- Clarke A., Gaston K.J. Climate, energy and diversity. Proc. Biol. Sci. 2006;273:2257–2266. doi: 10.1098/rspb.2006.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colin S., Coelho L.P., Sunagawa S., Bowler C., Karsenti E., Bork P., Pepperkok R., de Vargas C. Quantitative 3D-imaging for cell biology and ecology of environmental microbial eukaryotes. eLife. 2017;6:e26066. doi: 10.7554/eLife.26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwell R.K., Lees D.C. The mid-domain effect: geometric constraints on the geography of species richness. Trends Ecol. Evol. 2000;15:70–76. doi: 10.1016/s0169-5347(99)01767-x. [DOI] [PubMed] [Google Scholar]
- Currie D.J., Mittelbach G.G., Cornell H.V., Field R., Guegan J.-F., Hawkins B.A., Kaufman D.M., Kerr J.T., Oberdorff T., O’Brien E. Predictions and tests of climate-based hypotheses of broad-scale variation in taxonomic richness. Ecol. Lett. 2004;7:1121–1134. [Google Scholar]
- Danovaro R., Molari M., Corinaldesi C., Dell’Anno A. Macroecological drivers of archaea and bacteria in benthic deep-sea ecosystems. Sci. Adv. 2016;2:e1500961. doi: 10.1126/sciadv.1500961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge P.A., Nobrega F.L., Brouns S.J.J., Dutilh B.E. Molecular and Evolutionary Determinants of Bacteriophage Host Range. Trends Microbiol. 2019;27:51–63. doi: 10.1016/j.tim.2018.08.006. [DOI] [PubMed] [Google Scholar]
- de Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., Tara Oceans Coordinators Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- Decelle J., Probert I., Bittner L., Desdevises Y., Colin S., de Vargas C., Galí M., Simó R., Not F. An original mode of symbiosis in open ocean plankton. Proc. Natl. Acad. Sci. USA. 2012;109:18000–18005. doi: 10.1073/pnas.1212303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarcq H. Trends in primary production, sea surface temperature and wind in upwelling systems (1998–2007) Prog. Oceanogr. 2009;83:376–385. [Google Scholar]
- Doney S.C., Ruckelshaus M., Duffy J.E., Barry J.P., Chan F., English C.A., Galindo H.M., Grebmeier J.M., Hollowed A.B., Knowlton N. Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 2012;4:11–37. doi: 10.1146/annurev-marine-041911-111611. [DOI] [PubMed] [Google Scholar]
- Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edwards M., Richardson A.J. Impact of climate change on marine pelagic phenology and trophic mismatch. Nature. 2004;430:881–884. doi: 10.1038/nature02808. [DOI] [PubMed] [Google Scholar]
- Falkowski P.G., Fenchel T., Delong E.F. The microbial engines that drive Earth’s biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Field C.B., Behrenfeld M.J., Randerson J.T., Falkowski P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science. 1998;281:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- Flombaum P., Gallegos J.L., Gordillo R.A., Rincón J., Zabala L.L., Jiao N., Karl D.M., Li W.K.W., Lomas M.W., Veneziano D. Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. USA. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frainer A., Primicerio R., Kortsch S., Aune M., Dolgov A.V., Fossheim M., Aschan M.M. Climate-driven changes in functional biogeography of Arctic marine fish communities. Proc. Natl. Acad. Sci. USA. 2017;114:12202–12207. doi: 10.1073/pnas.1706080114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenne P., Graae B.J., Rodríguez-Sánchez F., Kolb A., Chabrerie O., Decocq G., Kort H., Schrijver A., Diekmann M., Eriksson O. Latitudinal gradients as natural laboratories to infer species’ responses to temperature. J. Ecol. 2013;101:784–795. [Google Scholar]
- Fuhrman J.A., Steele J.A., Hewson I., Schwalbach M.S., Brown M.V., Green J.L., Brown J.H. A latitudinal diversity gradient in planktonic marine bacteria. Proc. Natl. Acad. Sci. USA. 2008;105:7774–7778. doi: 10.1073/pnas.0803070105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia H.E., Locarnini R.A., Boyer T.P., Antonov J.I., Zweng M.M., Baranova O.K., Johnson D.R. Nutrients (phosphate, nitrate, silicate) In: Levitus S., editor. World Ocean Atlas. U.S. Government Printing Office; 2010. p. 398. [Google Scholar]
- Ghiglione J.-F., Galand P.E., Pommier T., Pedrós-Alió C., Maas E.W., Bakker K., Bertilson S., Kirchmanj D.L., Lovejoy C., Yager P.L., Murray A.E. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc. Natl. Acad. Sci. USA. 2012;109:17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilg O., Kovacs K.M., Aars J., Fort J., Gauthier G., Grémillet D., Ims R.A., Meltofte H., Moreau J., Post E. Climate change and the ecology and evolution of Arctic vertebrates. Ann. N Y Acad. Sci. 2012;1249:166–190. doi: 10.1111/j.1749-6632.2011.06412.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni S.J., Cameron Thrash J., Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorsky G., Ohman M.D., Picheral M., Gasparini S., Stemmann L., Romagnan J.-B., Cawood A., Pesant S., García-Comas C., Prejger F. Digital zooplankton image analysis using the ZooScan integrated system. J. Plankton Res. 2010;32:285–303. [Google Scholar]
- Graff J.R., Westberry T.K., Milligan A.J., Brown M.B., Dall’Olmo G., van Dongen-Vogels V., Reifel K.M., Behrenfeld M.J. Analytical phytoplankton carbon measurements spanning diverse ecosystems. Deep Sea Res. Part I Oceanogr. Res. Pap. 2015;102:16–25. [Google Scholar]
- Gregory A., Zayed A., Conceição-Neto N., Temperton B., Bolduc B., Alberti A., Ardyna M., Arkhipova K., Carmicheal M., Cruaud C. Marine DNA viral macro-and micro-diversity from pole to pole. Cell. 2019;177:1109–1123.e14. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi L., Chaffron S., Bittner L., Eveillard D., Larhlimi A., Roux S., Darzi Y., Audic S., Berline L., Brum J., Tara Oceans coordinators Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–470. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisan A., Edwards T.C., Jr., Hastie T. Generalized linear and generalized additive models in studies of species distributions: setting the scene. Ecological Modelling. 2002;157:89–100. [Google Scholar]
- Haegeman B., Hamelin J., Moriarty J., Neal P., Dushoff J., Weitz J.S. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013;7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T.J. Generalized additive models. In: Chambers J.M., Hastie T.J., editors. Statistical Models in S. Routledge; 2017. pp. 249–307. [Google Scholar]
- Henson S.A., Sanders R., Madsen E. Global patterns in efficiency of particulate organic carbon export and transfer to the deep ocean. Global Biogeochem. Cycles. 2012;26 [Google Scholar]
- Hillebrand H. On the generality of the latitudinal diversity gradient. Am. Nat. 2004;163:192–211. doi: 10.1086/381004. [DOI] [PubMed] [Google Scholar]
- Hingamp P., Grimsley N., Acinas S.G., Clerissi C., Subirana L., Poulain J., Ferrera I., Sarmento H., Villar E., Lima-Mendez G. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013;7:1678–1695. doi: 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics. 1979;6:65–70. [Google Scholar]
- Holte J., Talley L.D., Gilson J., Roemmich D. An Argo mixed layer climatology and database. Geophys. Res. Lett. 2017;44:5618–5626. [Google Scholar]
- Huang B., Thorne P.W., Banzon V.F., Boyer T., Chepurin G., Lawrimore J.H., Menne M.J., Smith T.M., Vose R.S., Zhang H.-M. Extended reconstructed sea surface temperature, version 5 (ERSSTv5): upgrades, validations, and intercomparisons. J. Clim. 2017;30:8179–8205. [Google Scholar]
- Huisman J., Jonker R.R., Zonneveld C., Weissing F.J. Competition for light between phytoplankton species: experimental tests of mechanistic theory. Ecology. 1999;80:211. [Google Scholar]
- Hutchins D.A., Fu F. Microorganisms and ocean global change. Nat. Microbiol. 2017;2:17058. doi: 10.1038/nmicrobiol.2017.58. [DOI] [PubMed] [Google Scholar]
- Hutchinson G.E. The paradox of the plankton. Am. Nat. 1961;95:137–145. [Google Scholar]
- Irigoien X., Huisman J., Harris R.P. Global biodiversity patterns of marine phytoplankton and zooplankton. Nature. 2004;429:863–867. doi: 10.1038/nature02593. [DOI] [PubMed] [Google Scholar]
- John S.G., Mendez C.B., Deng L., Poulos B., Kauffman A.K.M., Kern S., Brum J., Polz M.F., Boyle E.A., Sullivan M.B. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 2011;3:195–202. doi: 10.1111/j.1758-2229.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- Karsenti E., Acinas S.G., Bork P., Bowler C., De Vargas C., Raes J., Sullivan M., Arendt D., Benzoni F., Claverie J.-M., Tara Oceans Consortium A holistic approach to marine eco-systems biology. PLoS Biol. 2011;9:e1001177. doi: 10.1371/journal.pbio.1001177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladau J., Sharpton T.J., Finucane M.M., Jospin G., Kembel S.W., O’Dwyer J., Koeppel A.F., Green J.L., Pollard K.S. Global marine bacterial diversity peaks at high latitudes in winter. ISME J. 2013;7:1669–1677. doi: 10.1038/ismej.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewitus E., Bittner L., Malviya S., Bowler C., Morlon H. Clade-specific diversification dynamics of marine diatoms since the Jurassic. Nat. Ecol. Evol. 2018;2:1715–1723. doi: 10.1038/s41559-018-0691-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.K.W. Macroecological patterns of phytoplankton in the northwestern North Atlantic Ocean. Nature. 2002;419:154–157. doi: 10.1038/nature00994. [DOI] [PubMed] [Google Scholar]
- Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., Chaffron S., Ignacio-Espinosa J.C., Roux S., Vincent F., Tara Oceans coordinators Ocean plankton. Determinants of community structure in the global plankton interactome. Science. 2015;348:1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- Logares R., Sunagawa S., Salazar G., Cornejo-Castillo F.M., Ferrera I., Sarmento H., Hingamp P., Ogata H., de Vargas C., Lima-Mendez G. Metagenomic 16S rDNA Illumina tags are a powerful alternative to amplicon sequencing to explore diversity and structure of microbial communities. Environ. Microbiol. 2014;16:2659–2671. doi: 10.1111/1462-2920.12250. [DOI] [PubMed] [Google Scholar]
- Louca S., Doebeli M., Parfrey L.W. Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem. Microbiome. 2018;6:41. doi: 10.1186/s40168-018-0420-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahé F., Rognes T., Quince C., de Vargas C., Dunthorn M. Swarm: robust and fast clustering method for amplicon-based studies. PeerJ. 2014;2:e593. doi: 10.7717/peerj.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malviya S., Scalco E., Audic S., Vincent F., Veluchamy A., Poulain J., Wincker P., Iudicone D., de Vargas C., Bittner L. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA. 2016;113:E1516–E1525. doi: 10.1073/pnas.1509523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen F.A., Kodner R.B., Armbrust E.V. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menemenlis D., Campin J.M., Heimbach P., Hill C., Lee T., Nguyen A., Schodlok M., Zhang H. ECCO2: High resolution global ocean and sea ice data synthesis. Mercator Ocean Quarterly Newsletter. 2008;31:13–21. [Google Scholar]
- Mestre M., Ruiz-González C., Logares R., Duarte C.M., Gasol J.M., Sala M.M. Sinking particles promote vertical connectivity in the ocean microbiome. Proc. Natl. Acad. Sci. USA. 2018;115:E6799–E6807. doi: 10.1073/pnas.1802470115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milanese A., Mende D.R., Paoli L., Salazar G., Ruscheweyh H.-J., Cuenca M., Hingamp P., Alves R., Costea P.I., Coelho L.P. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat. Commun. 2019;10:1014. doi: 10.1038/s41467-019-08844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C.M., Mills M.M., Arrigo K.R., Berman-Frank I., Bopp L., Boyd P.W., Galbraith E.D., Geider R.J., Guieu C., Jaccard S.L. Processes and patterns of oceanic nutrient limitation. Nat. Geosci. 2013;6:701–710. [Google Scholar]
- Morán X.A.G., López-Urrutia Á., Calvo-Díaz A., Li W.K.W. Increasing importance of small phytoplankton in a warmer ocean. Glob. Change Biol. 2010;16:1137–1144. [Google Scholar]
- Morán X.A.G., Gasol J.M., Pernice M.C., Mangot J.-F., Massana R., Lara E., Vaqué D., Duarte C.M. Temperature regulation of marine heterotrophic prokaryotes increases latitudinally as a breach between bottom-up and top-down controls. Glob. Change Biol. 2017;23:3956–3964. doi: 10.1111/gcb.13730. [DOI] [PubMed] [Google Scholar]
- Morand S. (macro-) Evolutionary ecology of parasite diversity: From determinants of parasite species richness to host diversification. Int. J. Parasitol. Parasites Wildl. 2015;4:80–87. doi: 10.1016/j.ijppaw.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.J., Lenski R.E., Zinser E.R. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. MBio. 2012;3 doi: 10.1128/mBio.00036-12. e00036-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeo V.M. Segmented: an R package to fit regression models with broken-line relationships. R News. 2008;8:20–25. [Google Scholar]
- Pachauri R.K., Allen M.R., Barros V.R., Broome J., Cramer W., Christ R., Church J.A., Clarke L., Dahe Q., Dasgupta P. Climate Change 2014: Synthesis Report. In: Pachauri R.K., Meyer L., editors. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC; 2014. p. 151. [Google Scholar]
- Parada A.E., Needham D.M., Fuhrman J.A. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- Peijnenburg K.T.C.A., Goetze E. High evolutionary potential of marine zooplankton. Ecol. Evol. 2013;3:2765–2781. doi: 10.1002/ece3.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S., Not F., Picheral M., Kandels-Lewis S., Le Bescot N., Gorsky G., Iudicone D., Karsenti E., Speich S., Troublé R., Tara Oceans Consortium Coordinators Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data. 2015;2:150023. doi: 10.1038/sdata.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picheral M., Searson S., Taillandier V., Bricaud A., Boss E., Ras J., Claustre H., Ouhssain M., Morin P., Coppola L. 2014. Vertical profiles of environmental parameters measured on discrete water samples collected with Niskin bottles during the Tara Oceans expedition 2009-2013. PANGAEA. [Google Scholar]
- Picheral M., Colin S., Irisson J.O. 2017. EcoTaxa, a tool for the taxonomic classification of images.http://ecotaxa.obs-vlfr.fr [Google Scholar]
- Poloczanska E.S., Brown C.J., Sydeman W.J., Kiessling W., Schoeman D.S., Moore P.J., Brander K., Bruno J.F., Buckley L.B., Burrows M.T. Global imprint of climate change on marine life. Nat. Clim. Chang. 2013;3:919. [Google Scholar]
- Pomeroy L.R., Wiebe W.J. Temperature and substrates as interactive limiting factors for marine heterotrophic bacteria. Aquat. Microb. Ecol. 2001;23:187–204. [Google Scholar]
- Pontarp M., Bunnefeld L., Cabral J.S., Etienne R.S., Fritz S.A., Gillespie R., Graham C.H., Hagen O., Hartig F., Huang S. The latitudinal diversity gradient: novel understanding through mechanistic eco-evolutionary models. Trends Ecol. Evol. 2019;34:211–223. doi: 10.1016/j.tree.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacnik R., Solimini A.G., Andersen T., Tamminen T., Brettum P., Lepistö L., Willén E., Rekolainen S. Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc. Natl. Acad. Sci. USA. 2008;105:5134–5138. doi: 10.1073/pnas.0708328105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes E.J., Bodrossy L., van de Kamp J., Bissett A., Ostrowski M., Brown M.V., Sow S.L.S., Sloyan B., Waite A.M. Oceanographic boundaries constrain microbial diversity gradients in the South Pacific Ocean. Proc. Natl. Acad. Sci. USA. 2018;115:E8266–E8275. doi: 10.1073/pnas.1719335115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ras J., Claustre H., Uitz J. Spatial variability of phytoplankton pigment distributions in the Subtropical South Pacific Ocean: comparison between in situ and predicted data. Biogeosciences. 2008;5:353–369. [Google Scholar]
- Rhein M., Rintoul S.R., Aoki S., Campos E., Chambers D., Feely R.A., Gulev S., Johnson G.C., Josey S.A., Kostianoy A. Observations: Ocean. In: Stocker T.F., Qin D., Plattner G.-K., Tignor M., Allen S.K., Boschung J., Nauels A., Xia Y., Bex V., Midgley P.M., editors. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; 2013. [Google Scholar]
- Richardson A.J., Schoeman D.S. Climate impact on plankton ecosystems in the Northeast Atlantic. Science. 2004;305:1609–1612. doi: 10.1126/science.1100958. [DOI] [PubMed] [Google Scholar]
- Righetti D., Vogt M., Gruber N., Psomas A., Zimmermann N.E. Global pattern of phytoplankton diversity driven by temperature and environmental variability. Sci. Adv. 2019;5 doi: 10.1126/sciadv.aau6253. eaau6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombouts I., Beaugrand G., Ibanez F., Gasparini S., Chiba S., Legendre L. Global latitudinal variations in marine copepod diversity and environmental factors. Proc. Biol. Sci. 2009;276:3053–3062. doi: 10.1098/rspb.2009.0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Brum J.R., Dutilh B.E., Sunagawa S., Duhaime M.B., Loy A., Poulos B.T., Solonenko N., Lara E., Poulain J., Tara Oceans Coordinators Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature. 2016;537:689–693. doi: 10.1038/nature19366. [DOI] [PubMed] [Google Scholar]
- Roy S., Chattopadhyay J. Towards a resolution of “the paradox of the plankton”: A brief overview of the proposed mechanisms. Ecol. Complex. 2007;4:26–33. [Google Scholar]
- Saiz E., Calbet A. Copepod feeding in the ocean: scaling patterns, composition of their diet and the bias of estimates due to microzooplankton grazing during incubations. Hydrobiologia. 2011;666:181–196. [Google Scholar]
- Salazar G., Cornejo-Castillo F.M., Benítez-Barrios V., Fraile-Nuez E., Álvarez-Salgado X.A., Duarte C.M., Gasol J.M., Acinas S.G. Global diversity and biogeography of deep-sea pelagic prokaryotes. ISME J. 2016;10:596–608. doi: 10.1038/ismej.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G., Paoli L., Alberti A., Huerta-Cepas J., Ruscheweyh H.J., Cuenca M., Field C.M., Coelho L.P., Cruaud C., Engelen S. Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome. Cell. 2019;179:1068–1083. doi: 10.1016/j.cell.2019.10.014. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum C.-E., Buckling A., Smirnoff N., Studholme D.J., Yvon-Durocher G. Environmental fluctuations accelerate molecular evolution of thermal tolerance in a marine diatom. Nat. Commun. 2018;9:1719. doi: 10.1038/s41467-018-03906-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer M., Rinaldi S., Huisman J., Weissing F.J. Why plankton communities have no equilibrium: solutions to the paradox. Hydrobiologia. 2003;491:9–18. [Google Scholar]
- Ser-Giacomi E., Zinger L., Malviya S., De Vargas C., Karsenti E., Bowler C., De Monte S. Ubiquitous abundance distribution of non-dominant plankton across the global ocean. Nat. Ecol. Evol. 2018;2:1243–1249. doi: 10.1038/s41559-018-0587-2. [DOI] [PubMed] [Google Scholar]
- Siano R., Alves-de-Souza C., Foulon E., Bendif E.M., Simon N., Guillou L., Not F. Distribution and host diversity of Amoebophryidae parasites across oligotrophic waters of the Mediterranean Sea. Biogeosciences Discuss. 2010;7:7391–7419. [Google Scholar]
- Smith V.H. Microbial diversity-productivity relationships in aquatic ecosystems. FEMS Microbiol. Ecol. 2007;62:181–186. doi: 10.1111/j.1574-6941.2007.00381.x. [DOI] [PubMed] [Google Scholar]
- Sommer U., Peter K.H., Genitsaris S., Moustaka-Gouni M. Do marine phytoplankton follow Bergmann’s rule sensu lato? Biol. Rev. Camb. Philos. Soc. 2017;92:1011–1026. doi: 10.1111/brv.12266. [DOI] [PubMed] [Google Scholar]
- Sul W.J., Oliver T.A., Ducklow H.W., Amaral-Zettler L.A., Sogin M.L. Marine bacteria exhibit a bipolar distribution. Proc. Natl. Acad. Sci. USA. 2013;110:2342–2347. doi: 10.1073/pnas.1212424110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., Djahanschiri B., Zeller G., Mende D.R., Alberti A., Tara Oceans coordinators Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- Sunday J.M., Bates A.E., Dulvy N.K. Thermal tolerance and the global redistribution of animals. Nat. Clim. Chang. 2012;2:686. [Google Scholar]
- Thomas M.K., Kremer C.T., Klausmeier C.A., Litchman E. A global pattern of thermal adaptation in marine phytoplankton. Science. 2012;338:1085–1088. doi: 10.1126/science.1224836. [DOI] [PubMed] [Google Scholar]
- Tittensor D.P., Mora C., Jetz W., Lotze H.K., Ricard D., Berghe E.V., Worm B. Global patterns and predictors of marine biodiversity across taxa. Nature. 2010;466:1098–1101. doi: 10.1038/nature09329. [DOI] [PubMed] [Google Scholar]
- Toseland A., Daines S.J., Clark J.R., Kirkham A., Strauss J., Uhlig C., Lenton T.M., Valentin K., Pearson G.A., Moulton V. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat. Clim. Chang. 2013;3:979. [Google Scholar]
- Uitz J., Claustre H., Morel A., Hooker S.B. Vertical distribution of phytoplankton communities in open ocean: An assessment based on surface chlorophyll. J. Geophys. Res. 2006;111:57. [Google Scholar]
- Ullah H., Nagelkerken I., Goldenberg S.U., Fordham D.A. Climate change could drive marine food web collapse through altered trophic flows and cyanobacterial proliferation. PLoS Biol. 2018;16:e2003446. doi: 10.1371/journal.pbio.2003446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallina S.M., Follows M.J., Dutkiewicz S., Montoya J.M., Cermeno P., Loreau M. Global relationship between phytoplankton diversity and productivity in the ocean. Nat. Commun. 2014;5:4299. doi: 10.1038/ncomms5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten L., Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Van Heukelem L., Thomas C.S. Computer-assisted high-performance liquid chromatography method development with applications to the isolation and analysis of phytoplankton pigments. J. Chromatogr. A. 2001;910:31–49. doi: 10.1016/s0378-4347(00)00603-4. [DOI] [PubMed] [Google Scholar]
- Vergés A., Doropoulos C., Malcolm H.A., Skye M., Garcia-Pizá M., Marzinelli E.M., Campbell A.H., Ballesteros E., Hoey A.S., Vila-Concejo A. Long-term empirical evidence of ocean warming leading to tropicalization of fish communities, increased herbivory, and loss of kelp. Proc. Natl. Acad. Sci. USA. 2016;113:13791–13796. doi: 10.1073/pnas.1610725113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson R.A. A database of global marine commercial, small-scale, illegal and unreported fisheries catch 1950-2014. Sci. Data. 2017;4:170039. doi: 10.1038/sdata.2017.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker R.H. Evolution and measurement of species diversity. Taxon. 1972;21:213–251. [Google Scholar]
- Willig M.R., Kaufman D.M., Stevens R.D. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 2003;34:273–309. [Google Scholar]
- Wood S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. Series B Stat. Methodol. 2010;73:3–36. [Google Scholar]
- Woodd-Walker R.S., Ward P., Clarke A. Large-scale patterns in diversity and community structure of surface water copepods from the Atlantic Ocean. Mar. Ecol. Prog. Ser. 2002;236:189–203. [Google Scholar]
- Woolley S.N.C., Tittensor D.P., Dunstan P.K., Guillera-Arroita G., Lahoz-Monfort J.J., Wintle B.A., Worm B., O’Hara T.D. Deep-sea diversity patterns are shaped by energy availability. Nature. 2016;533:393–396. doi: 10.1038/nature17937. [DOI] [PubMed] [Google Scholar]
- Worm B., Barbier E.B., Beaumont N., Duffy J.E., Folke C., Halpern B.S., Jackson J.B.C., Lotze H.K., Micheli F., Palumbi S.R. Impacts of biodiversity loss on ocean ecosystem services. Science. 2006;314:787–790. doi: 10.1126/science.1132294. [DOI] [PubMed] [Google Scholar]
- Yasuhara M., Hunt G., Dowsett H.J., Robinson M.M., Stoll D.K. Latitudinal species diversity gradient of marine zooplankton for the last three million years. Ecol. Lett. 2012;15:1174–1179. doi: 10.1111/j.1461-0248.2012.01828.x. [DOI] [PubMed] [Google Scholar]
- Zinger L., Amaral-Zettler L.A., Fuhrman J.A., Horner-Devine M.C., Huse S.M., Welch D.B.M., Martiny J.B.H., Sogin M., Boetius A., Ramette A. Global patterns of bacterial beta-diversity in seafloor and seawater ecosystems. PLoS ONE. 2011;6:e24570. doi: 10.1371/journal.pone.0024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[S1A] Summary statistics for the main datasets utilized in this work. Related to Figures 1B, 2A–2C. [S1B] Summary statistics for the 12 marine plankton groups (MPGs). Related to Figures 1B, 2A. [S1C] Analysis of the LDG symmetry for each of the MPGs in surface waters. Related to Figure 2A. [S1D] GAM parameters for the “full” models of diversity of MPGs. Related to Figure 3C and STAR Methods. [S1E] Evaluation of GAM “full” models. Related to Figure 3C and (D). [S1F] GAM parameters for the “reduced” models of diversity of MPGs. Related to Figure 4 and STAR Methods. [S1G] Evaluation of GAM “reduced” models. Related to Figure 4 and (F). [S1H] CMIP5 Earth system models. Related to Figures 4, S12, and S13. [S1I] Latitudinal overlap with areas of socio-economic and conservation value. Related to Figure 4.
Data Availability Statement
Raw reads of Tara Oceans are deposited at the European Nucleotide Archive (ENA). In particular, newly released 18S rRNA gene metabarcoding reads are available under the number ENA: PRJEB9737. ENA references for the metagenomics reads corresponding to the size fraction < 0.22 μm (for prokaryotic viruses) analyzed in this study are included in Gregory et al. (2019); see their Table S3. ENA references for the metagenomics reads corresponding to the size fraction 0.22-1.6/3 μm (for prokaryotes and giruses) correspond to Salazar et al. (2019) (see https://zenodo.org/record/3473199). Imaging datasets from the nets are available through the collaborative web application and repository EcoTaxa (Picheral et al., 2017) under the address https://ecotaxa.obs-vlfr.fr/prj/412 for regent data, within the 3 projects https://ecotaxa.obs-vlfr.fr/prj/397, https://ecotaxa.obs-vlfr.fr/prj/398, https://ecotaxa.obs-vlfr.fr/prj/395 for bongo data, and within the 2 projects https://ecotaxa.obs-vlfr.fr/prj/377 and https://ecotaxa.obs-vlfr.fr/prj/378 for WP2 data. A table with Shannon values and multiple samples identifiers, plus a table with flow cytometry data split in six groups are available (https://doi.org/10.17632/p9r9wttjkm.1). Contextual data from the Tara Oceans expedition, including those that are newly released from the Arctic Ocean, are available at https://doi.org/10.1594/PANGAEA.875582.