Abstract
Liquid biopsy to identify epidermal growth factor receptor (EGFR) gene mutations from circulating tumor DNA (ctDNA) for lung adenocarcinoma is less invasive than traditional tissue biopsy. Most patients have concordant results in liquid/tissue biopsy, while the clinical significance of concordant results remains unclear. Our study aimed to evaluate the predicting factors and clinical outcomes associated with concordant results in liquid/tissue biopsy in newly diagnosed lung adenocarcinoma patients with EGFR mutations. In the 80 patients of stage III or IV lung adenocarcinoma, 51 patients had EGFR mutations detected in tissue samples, while 33 (65%) of them had concordant results shown in liquid biopsy. Multivariable regression analysis showed that lymph node involvement (adjusted odds ratio (95% CI): 8.71 (1.88–40.35), p = 0.0057) and bone metastasis (adjusted odds ratio (95% CI): 9.65 (1.72–54.05), p = 0.0099) were the independent predicting factors for concordant results. Forty of these 51 patients were stage IV and were treated with EGFR tyrosine kinase inhibitors (TKIs). The concordant results in liquid/tissue samples were associated with significantly poorer progression-free survival (PFS) in univariate analysis. However, multivariable analysis showed that lymph node involvement was the only independent predicting factor for poorer PFS, while concordant results in liquid/tissue samples were excluded during variable selection. The concordant results in liquid/tissue samples might indicate a larger tumor burden, which actually contributes to poorer PFS.
Keywords: lung cancer, adenocarcinoma, EGFR, liquid biopsy
1. Introduction
Lung cancer is the most common cause of cancer-related mortality worldwide [1]. About 80% of all cases are non-small cell lung cancer (NSCLC), among which the most common cell type is adenocarcinoma. More than half of these NSCLC were diagnosed in advanced stage [2]. In the past, the only treatment for advanced NSCLC was platinum-based doublet chemotherapy, resulting in a median overall survival (OS) period of around 8 months [3]. Clinical practice has changed since the development of epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), with the discovery of EGFR driver gene mutations in NSCLC. Patients with NSCLC harboring such mutations, such as exon 21 L858R point mutation and exon 19 deletion, have better progression-free survival (PFS) when treated with EGFR TKIs [4,5,6]. It is therefore very important to determine the presence of EGFR mutation in NSCLC.
Initially, tissue samples, biopsied from either primary tumor or metastatic lesions, had been used for the EGFR mutation testing. The procedures of tissue biopsy, including bronchoscopic biopsy, computed tomography-guided biopsy, and surgical biopsy, are all invasive, and bring risks of some complications, such as hemoptysis, pneumothorax, and pneumonitis [7,8,9]. Another limitation of tissue biopsy is the tumor heterogeneity, especially in patients with advanced stages. The results of EGFR gene testing might be different in various parts of the cancer, especially in the metastatic sites, so tissue biopsy from one part of a solitary tumor might miss the intra-tumoral and inter-metastatic molecular heterogeneity [10,11]. Furthermore, malignant cells could be found in 72.9% of specimens under pathological examination [12]. Repeated biopsies may sometimes be required to obtain sufficient cancer tissue for gene testing [13].
Liquid biopsy, identifying the genotype of tumor cells from circulating tumor DNA (ctDNA) of the patients’ blood, is a less invasive method. Since the report by Mandel and Metais in 1948, fragmented DNA in the cell-free component of serum has been a field of active research [11]. Investigation of cell-free DNA has been conducted in many disciplines, such as exercise, end-stage renal failure, stroke, myocardial infarction, surgery, and trauma [14,15,16,17]. In the oncological field, studies of cell-free DNA derived from tumors, also known as ctDNA, has dramatically increased recently, mainly because of the development of the genomic technologies that allow detection of rare gene mutant variants of DNA [11]. In recent studies of NSCLC, liquid biopsy was shown to detect driver gene mutations, such as EGFR and anaplastic lymphoma kinase (ALK) mutation, and tumor mutational burden [18,19]. Compared to tissue biopsy, liquid biopsy done at disease progression can reduce the necessity of invasive biopsy procedures, the risk of biopsy related complications, and the cost of the complication-related hospitalization [20].
Most patients have concordant results in liquid/tissue biopsy, while a few patients have discordant results in the gene testing of tissue biopsy and liquid biopsy. The factors related to concordant results and the clinical significance of concordant results remain unclear. Our study aimed to evaluate the predicting factors and clinical outcomes associated with concordant results in liquid/tissue biopsy in newly diagnosed lung adenocarcinoma patients with EGFR mutations.
2. Materials and Methods
2.1. Study Population
This retrospective study was conducted in Kaohsiung Medical University Hospital between June 2016 and August 2018. Treatment naïve stage III or IV lung adenocarcinoma patients with EGFR mutation tested from both their biopsied tumor tissue and liquid biopsy at their diagnosis were enrolled. All patients received imaging studies, including computed tomography of the chest, brain magnetic resonance imaging (MRI), and whole-body bone scan, to determine the extent of cancer invasion, lymph node involvement, and distant metastasis. The clinical stage was determined according to the American Joint Committee on Cancer (AJCC) Cancer Staging Manual, 7th Edition.
The Institutional Review Board (IRB) of Kaohsiung Medical University Hospital (KMUH) approved this study (KMUHIRB-G(II)-20190024) and waived the need for written informed consent from all patients.
2.2. DNA Extraction, Amplification, and Detection
The genomic DNA was extracted from formalin-fixed and paraffin-embedded (FFPE) tumor tissue samples, and EGFR mutation was tested by using Qiagen® EGFR RGQ PCR kit (QIAGEN, Hilden, Germany) as in our previous studies [21,22,23,24,25,26] or Cobas® EGFR Mutation Test v2 (Roche Diagnostics, Rotkreuz, Switzerland) (for the latest 6 samples). The previous validation exam in the Department of Laboratory Medicine, KMUH showed excellent correlation between the results obtained from these two kits. For liquid biopsy, the plasma ctDNA was extracted and then analyzed by Cobas® EGFR Mutation Test v2, which was a real-time PCR test for the quantitative detection and identification of mutations in exons 18, 19, 20, and 21 of the EGFR gene.
2.3. Definitions of Variables
The EGFR mutations detected in tissue samples were taken as the reference, to which the mutations detected in plasma ctDNA were compared. The patients with the same EGFR mutation patterns detected in both biopsied tissue samples and plasma ctDNA were classified as “concordant” group, whereas those having different mutation patterns in their plasma ctDNA versus their tissue samples were classified as “discordant” group.
Patients with stage IV disease who received first-line EGFR TKIs were further extracted for outcome analyses, while those who had discontinued the EGFR TKI for personal reasons or side effects were excluded. The objective treatment response was assessed according to the Response Evaluation Criteria in Solid Tumors (RECIST) version 1.1. A computed tomography of the chest was obtained three months after the initiation of a EGFR TKI to determine the 3-month treatment response. To determine the progression-free survival, these patients were followed till either disease progression or 16 January 2019.
2.4. Statistical Analysis
The baseline characteristics, including sex, age, performance status, tumor stage, primary tumor size, lymph node involvement, distant metastases, and EGFR mutation in tissue sample, were compared between the concordant and discordant groups. Categorical and continuous variables were analyzed using Chi-square test and Student’s t-test, respectively. The effects of factors in predicting concordant EGFR mutation test results in liquid/tissue biopsy were assessed using logistic regression analysis. The odds ratio (OR) with 95% confidence interval (CI) was reported. Following univariate analyses, all factors were included to build a maximal model of multivariable analysis. The reduced multivariable model was then developed with backward variable selection method, keeping only variables with p value less than 0.1, from the maximal model.
In the outcome analyses, which included only stage IV patients receiving first-line EGFR TKIs, the initial objective response rate (ORR) and disease control rate (DCR) were calculated. The PFS of patients in concordant and discordant groups were assessed with the Kaplan–Meier method and compared with log-rank test. The effects of factors in predicting PFS were assessed using Cox regression analysis. The hazard ratio (HR) with 95% CI was reported. Following univariate analyses, all factors were included to build maximal models of multivariable analyses. The reduced multivariable models were then developed with backward variable selection method, keeping only variables with p value less than 0.1, from the maximal models.
The statistical analyses were performed using SAS system (version 9.4 for Windows, SAS Institute Inc., Cary, NC, USA). A two-sided p value of <0.05 was taken as the statistical significance level.
3. Results
3.1. Predicting Factors for Concordant EGFR Mutation Detected in Liquid/Tissue Biopsy
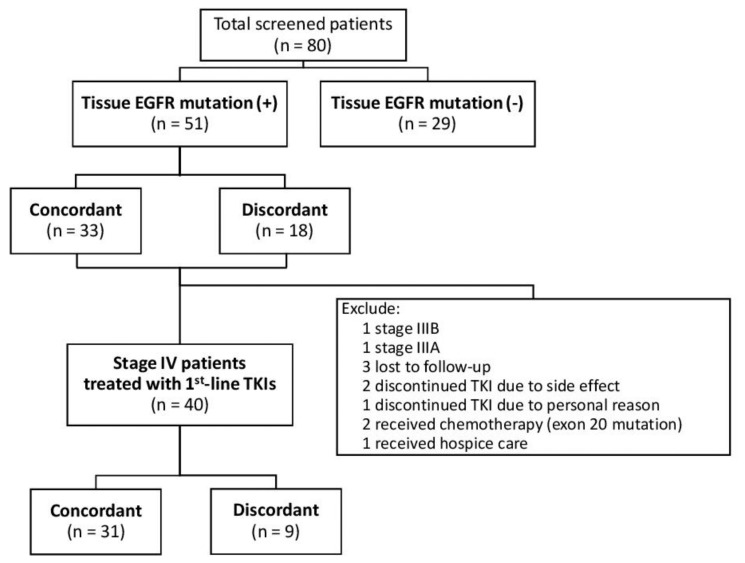
We identified 80 treatment-naïve stage III or IV lung adenocarcinoma patients during the study period, and 51 (63.75%) patients had EGFR mutation detected in their tumor tissue samples (Figure 1). These patients had a mean (±standard deviation) age of 64.2 (±10.5) years, and 17 (33.3%) patients were male. Two patients had stage IIIA cancer, one patient had stage IIIB cancer, and the remaining 48 (94%) patients had stage IV disease.
Figure 1.
Flow diagram of eligible study population. From the 80 screened patients, 51 patients had detectable EGFR mutation in their tissue samples. From these 51 patients, 40 patients who received treatment with a first-line EGFR TKI were enrolled in the outcome analysis. Abbreviations: EGFR = epidermal growth factor receptor; TKI = tyrosine kinase inhibitor.
In the 51 patients with EGFR mutation detected in the tumor tissue, 33 (65%) patients had concordant EGFR mutation test results in liquid biopsy, while 18 (35%) patients had discordant results (Figure 1, Table 1). The patients with concordant results had similar sex distribution, age, and performance status as those with discordant results. The patients with concordant results had significantly higher rate of lymph node involvement (N1–3 disease) than those with discordant results (82% vs. 39%, p = 0.0019). The patients with concordant results were all stage IV, whereas only 83% of those with discordant results were stage IV (p = 0.0156). The patients with concordant results, compared with those having discordant results, had significantly higher rates of metastases to brain (36% vs. 6%, p = 0.0158) and bone (61% vs. 17%, p = 0.0026). No patient with concordant results had mutation in exon 20, while 5 (28%) patients in the discordant group had exon 20 mutation (p = 0.0014).
Table 1.
Baseline characteristics of the study population, stage III or IV lung adenocarcinoma patients having EGFR mutations detected in the tissue samples.
| Characteristics | EGFR Mutation Test Results in Liquid/Tissue Biopsy | p Value | |
|---|---|---|---|
| Concordant (n = 33) | Discordant (n = 18) | ||
| Sex | 0.5342 | ||
| Female | 21 (64%) | 13 (72%) | |
| Male | 12 (36%) | 5 (28%) | |
| Age (year) | 63.2 ± 10.2 | 66.2 ± 11.0 | 0.3358 |
| Age: | 0.9448 | ||
| <65 years | 15 (45%) | 8 (44%) | |
| ≥65 years | 18 (55%) | 10 (56%) | |
| Performance status: | 0.2412 | ||
| ECOG 0–1 | 32 (97%) | 16 (89%) | |
| ECOG 2–4 | 1 (3%) | 2 (11%) | |
| Advanced primary tumor (T3–4) | 27 (82%) | 12 (67%) | 0.2228 |
| Lymph node involvement (N1–3) | 27 (82%) | 7 (39%) | 0.0019 |
| Metastasis (M1) | 33 (100%) | 15 (83%) | 0.0156 |
| Metastasis to: | |||
| Brain | 12 (36%) | 1 (6%) | 0.0158 |
| Lung | 12 (36%) | 8 (44%) | 0.5722 |
| Bone | 20 (61%) | 3 (17%) | 0.0026 |
| Pleural space | 18 (55%) | 9 (50%) | 0.7560 |
| Liver | 5 (15%) | 1 (6%) | 0.3094 |
| Pericardial space | 7 (21%) | 1 (6%) | 0.1418 |
| Adrenal gland | 5 (15%) | 1 (6%) | 0.3094 |
| Other site | 4 (12%) | 0 (0%) | 0.1239 |
| Mutation site: | |||
| Exon 18 | 1 (3%) | 1 (6%) | 0.6571 |
| Exon 19 | 17 (52%) | 7 (39%) | 0.3880 |
| Exon 20 | 0 (0%) | 5 (28%) | 0.0014 |
| Exon 21 | 15 (45%) | 6 (33%) | 0.4006 |
Data were presented as mean ± standard deviation or number (percentage). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group.
Using univariate logistic regression analysis, we found a few predicting factors for concordant EGFR mutation test results in liquid/tissue biopsy, including lymph node involvement (N1–3), brain metastasis, and bone metastasis (Table 2). In the reduced model of multivariable analysis, which was developed with backward variable selection method, only lymph node involvement (adjusted OR (95% CI): 8.71 (1.88–40.35), p = 0.0057) and bone metastasis (adjusted OR (95% CI): 9.65 (1.72–54.05), p = 0.0099) remained the independent predicting factors for concordant EGFR mutation test results in liquid/tissue biopsy.
Table 2.
Predicting factors for concordant EGFR mutation detected in liquid/tissue biopsy.
| Variables | Univariate Analysis | Multivariable Analysis—Maximal Model | Multivariable Analysis—Reduced Model † | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| Male (vs. female) | 1.49 (0.42–5.19) | 0.5354 | 1.83 (0.28–12.11) | 0.5296 | ||
| Age ≥65 (vs. <65) | 0.96 (0.30–3.05) | 0.9448 | 1.46 (0.19–11.27) | 0.7161 | ||
| ECOG ≥2 (vs. ≤1) | 0.25 (0.02–2.97) | 0.2722 | 0.06 (0.00–21.15) | 0.3423 | ||
| T3–4 (vs. T1–2) | 2.25 (0.60–8.42) | 0.2286 | 10.48 (0.87–126.59) | 0.0645 | 5.45 (0.98–30.24) | 0.0527 |
| N1–3 (vs. N0) | 7.07 (1.93–25.85) | 0.0031 | 10.06 (1.28–78.79) | 0.0280 | 8.71 (1.88–40.35) | 0.0057 |
| Brain metastasis (with vs. without) | 9.71 (1.15–82.39) | 0.0371 | 5.87 (0.36–96.65) | 0.2153 | ||
| Lung metastasis (with vs. without) | 0.71 (0.22–2.30) | 0.5728 | 0.31 (0.04–2.47) | 0.2666 | ||
| Bone metastasis (with vs. without) | 7.69 (1.85–31.91) | 0.0049 | 8.15 (0.99–66.95) | 0.0510 | 9.65 (1.72–54.05) | 0.0099 |
| Pleural metastasis (with vs. without) | 1.20 (0.38–3.79) | 0.7561 | 0.49 (0.06–3.80) | 0.4954 | ||
| Liver metastasis (with vs. without) | 3.04 (0.33–28.23) | 0.3291 | 5.26 (0.29–96.02) | 0.2630 | ||
| Pericardial metastasis (with vs. without) | 4.57 (0.52–40.56) | 0.1721 | 0.99 (0.04–22.00) | 0.9943 | ||
| Adrenal metastasis (with vs. without) | 3.04 (0.33–28.23) | 0.3291 | 3.85 (0.19–78.24) | 0.3808 | ||
| Metastasis to other sites (with vs. without) | ‡ | ‡ | ||||
Abbreviations: EGFR = epidermal growth factor receptor; OR = odds ratio; CI = confidence interval; ECOG = Eastern Cooperative Oncology Group. † Reduced multivariable models were developed with backward variable selection method, keeping only variables with p value less than 0.1, from the maximal model. ‡ Because all cases with metastasis to other sites were in the concordant group, odds ratio could not be estimated. This variable was therefore not included in the models.
3.2. Predicting Factors for PFS in Stage IV Patients Receiving EGFR TKI
From the 51 patients (Figure 1), 40 patients with stage IV lung adenocarcinoma treated with an EGFR TKI as their first-line therapy were enrolled in the following outcome analysis (Table 3). These patients included 31 (78%) patients with concordant EGFR mutation detected in liquid/tissue biopsy and nine (23%) patients with discordant results (Table 3). The patients with concordant results had similar sex, age, and performance status as those with discordant results. The patients with concordant results had significantly higher rate of lymph node involvement (N1–3 disease) than those with discordant results (84% vs. 33%, p = 0.0028). The EGFR mutation sites did not differ significantly between two groups and no patient in either group had exon 20 mutation. In terms of the first-line EGFR TKI used, 18 (58%) patients in the concordant group took erlotinib, while 6 (67%) of patients in the discordant group took afatinib (p = 0.0243).
Table 3.
Characteristics of the stage IV patients receiving first-line EGFR TKIs.
| Characteristics | EGFR Mutation Test Results in Liquid/Tissue Biopsy | p Value | |
|---|---|---|---|
| Concordant (n = 31) | Discordant (n = 9) | ||
| Sex | 0.4546 | ||
| Female | 20 (65%) | 7 (78%) | |
| Male | 11 (35%) | 2 (22%) | |
| Age (year) | 63.4 ± 10 | 63.0 ± 9.9 | 0.9192 |
| Age: | 0.5825 | ||
| <65 years | 14 (45%) | 5 (56%) | |
| ≥65 years | 17 (55%) | 4 (44%) | |
| Performance status: | 0.3393 | ||
| ECOG 0–1 | 30 (97%) | 8 (89%) | |
| ECOG 2–4 | 1 (3%) | 1 (11%) | |
| Advanced primary tumor (T3–4) | 25 (81%) | 5 (56%) | 0.1260 |
| Lymph node involvement (N1–3) | 26 (84%) | 3 (33%) | 0.0028 |
| Metastasis (M1) | 31 (100%) | 9 (100%) | 0.0401 |
| Metastasis to: | |||
| Brain | 11 (35%) | 0 (0%) | 0.0358 |
| Lung | 11 (35%) | 5 (56%) | 0.2792 |
| Bone | 19 (61%) | 2 (22%) | 0.0388 |
| Pleural space | 17 (55%) | 6 (67%) | 0.5274 |
| Liver | 5 (16%) | 1 (11%) | 0.7105 |
| Pericardial space | 7 (23%) | 0 (0%) | 0.1165 |
| Adrenal gland | 5 (16%) | 1 (11%) | 0.7105 |
| Other site | 4 (13%) | 0 (0%) | 0.2560 |
| Mutation site: | |||
| Exon 18 | 1 (3%) | 0 (0%) | 0.5853 |
| Exon 19 | 16 (52%) | 6 (67%) | 0.4242 |
| Exon 21 | 14 (45%) | 3 (33%) | 0.5274 |
| TKI used: | 0.0243 | ||
| Gefitinib | 6 (19%) | 2 (22%) | |
| Erlotinib | 18 (58%) | 1 (11%) | |
| Afatinib | 7 (23%) | 6 (67%) | |
| Initial treatment response: | 0.4320 | ||
| Partial response (PR) | 18 (58%) | 5 (56%) | |
| Stable disease (SD) | 9 (29%) | 4 (44%) | |
| Progressive disease (PD) | 4 (13%) | 0 (0%) | |
| Objective response rate | 18 (58%) | 5 (56%) | 0.8934 |
| Disease control rate | 27 (87%) | 9 (100%) | 0.2560 |
Data were presented as mean ± standard deviation or number (percentage). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group; TKI = tyrosine kinase inhibitors.
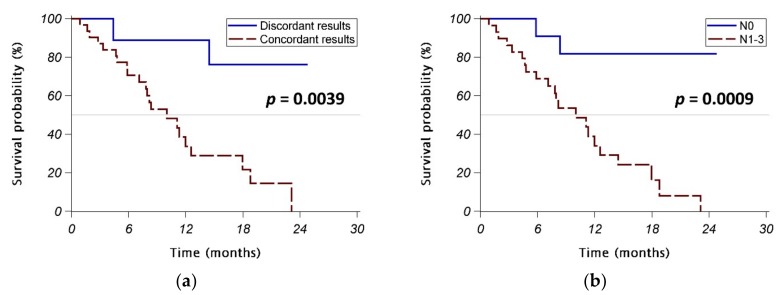
The initial objective response rate was similar in the concordant group and the discordant group (58% vs. 56%, p = 0.8934), as was the disease control rate (87% vs. 100%, p = 0.2560). However, the patients with concordant EGFR mutation test results in liquid/tissue biopsy had a significantly poorer PFS than those with discordant results (p = 0.0039) (Figure 2a). Interestingly, patients with lymph node involvement (N1–3 disease) also had significantly poorer PFS than those without lymph node involvement (N0 disease) (p = 0.0009) (Figure 2b).
Figure 2.
Progression-free survival (PFS) in stage IV patients receiving first-line EGFR TKIs. (a) Patients with concordant EGFR mutation test results in liquid/tissue biopsy vs. those with discordant results. (b) Patients with lymph node involvement (N1–3 disease) vs. those without lymph node involvement (N0 disease). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group; TKI = tyrosine kinase inhibitors.
Univariate Cox regression analyses identified factors significantly associated with poorer PFS included concordant EGFR mutation detected in liquid/tissue biopsy, lymph node involvement, brain metastasis, and bone metastasis (Table 4). Three EGFR TKIs showed similar effects in these patients. Multivariable analyses revealed that lymph node involvement was the only independent prognostic factor for poorer PFS (adjusted HR (95% CI): 7.53 (1.70–33.26), p = 0.0078 in reduced model 1 and 8.34 (1.92–36.22), p = 0.0047 in reduced model 2).
Table 4.
Predicting factors for progression-free survival (PFS) in stage IV patients receiving first-line EGFR TKIs.
| Variable | Univariate Analysis | Multivariable Analysis—Maximal Model 1 | Multivariable Analysis—Reduced Model 1 † | Multivariable Analysis—Maximal Model 2 | Multivariable Analysis—Reduced Model 2 † | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | HR (95% CI) | p Value | |
| Concordant (vs. discordant) ‡ | 6.71 (1.54–29.23) | 0.0112 | 4.70 (0.32–69.53) | 0.2604 | 5.11 (0.69–37.59) | 0.1092 | ||||
| Male (vs. female) | 1.34 (0.59–3.04) | 0.4849 | 0.85 (0.26–2.76) | 0.7890 | 1.55 (0.56–4.28) | 0.3969 | ||||
| Age ≥65 (vs. <65) | 1.22 (0.54–2.73) | 0.6337 | 2.69 (0.53–13.72) | 0.2349 | 1.81 (0.62–5.25) | 0.2752 | ||||
| ECOG ≥ 2 (vs. ≤1) | 0.75 (0.10–5.59) | 0.7783 | 1.02 (0.08–13.35) | 0.9858 | 0.98 (0.11–8.65) | 0.9826 | ||||
| T3–4 (vs. T1–2) | 2.11 (0.71–6.22) | 0.1780 | 1.11 (0.18–6.90) | 0.9086 | 0.91 (0.24–3.43) | 0.8950 | ||||
| N1–3 (vs. N0) | 8.34 (1.92–36.22) | 0.0047 | 6.42 (1.09–37.92) | 0.0401 | 7.53 (1.70–33.26) | 0.0078 | 4.35 (0.80–23.49) | 0.0879 | 8.34 (1.92–36.22) | 0.0047 |
| Number of metastatic site(s) ≥2 (vs. ≤1) | 2.46 (0.97–6.27) | 0.0585 | 3.23 (1.03–10.14) | 0.0447 | ||||||
| Metastasis to: (with vs. without) | ||||||||||
| Brain | 3.12 (1.32–7.35) | 0.0094 | 1.75 (0.43–7.08) | 0.4351 | ||||||
| Lung | 0.96 (0.42–2.19) | 0.9314 | 0.57 (0.10–3.07) | 0.5098 | ||||||
| Bone | 2.76 (1.15–6.65) | 0.0234 | 2.57 (0.84–7.92) | 0.0994 | 2.26 (0.92–5.58) | 0.0766 | ||||
| Pleural space | 1.59 (0.67–3.75) | 0.2911 | 3.09 (0.83–11.6) | 0.0938 | ||||||
| Liver | 0.77 (0.26–2.26) | 0.6308 | 0.35 (0.06–2.02) | 0.2414 | ||||||
| Pericardial space | 1.55 (0.57–4.18) | 0.3913 | 0.72 (0.14–3.63) | 0.6877 | ||||||
| Adrenal gland | 1.42 (0.42–4.82) | 0.5712 | 2.20 (0.29–16.54) | 0.4436 | ||||||
| Other site | 1.59 (0.46–5.55) | 0.4637 | 0.48 (0.06–3.68) | 0.4776 | ||||||
| TKI used: | ||||||||||
| Gefitinib | ref | ref | ref | |||||||
| Erlotinib | 1.07 (0.38–3.04) | 0.8927 | 1.54 (0.25–9.62) | 0.6413 | 0.60 (0.17–2.12) | 0.4254 | ||||
| Afatinib | 0.75 (0.23–2.48) | 0.6429 | 1.79 (0.27–11.96) | 0.5473 | 1.28 (0.30–5.43) | 0.7344 | ||||
Abbreviation: HR = hazard ratio; CI = confidence interval; EGFR = epithelial growth factor receptor; TKI = tyrosine kinase inhibitors. † Reduced multivariable models were developed with backward variable selection method, keeping only variables with p value less than 0.1, from the maximal model. ‡ Concordant vs. discordant EGFR mutation test results in tissue and liquid biopsies.
3.3. Sensitivity Analyses
To eliminate the potential bias introduced by different EGFR mutation test kits used for tissue specimens, we performed another set of analyses (sensitivity analyses). The six patients using Cobas® EGFR Mutation Test v2 for their tissue samples were excluded (Table A1). In consistence with our previous findings, univariate logistic regression analysis showed the same predicting factors for concordant EGFR mutation test results in liquid/tissue biopsy, including lymph node involvement (N1–3), brain metastasis, and bone metastasis (Table A2). In the reduced model of multivariable analysis, developed with the backward variable selection method, only lymph node involvement (adjusted OR (95% CI): 9.90 (1.91–51.37), p = 0.0064) and bone metastasis (adjusted OR (95% CI): 9.91 (1.62–60.61), p = 0.0131) remained the independent predicting factors for concordant EGFR mutation test results in liquid/tissue biopsy (Table A3).
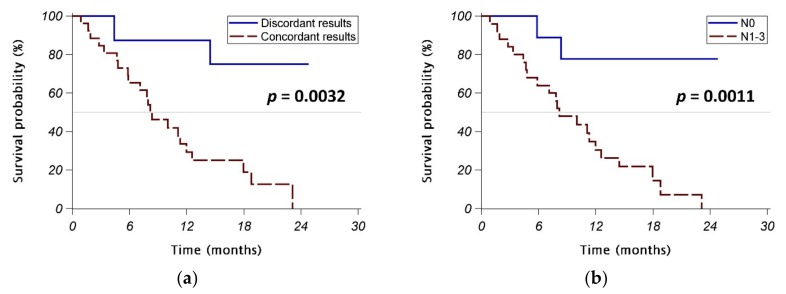
In the 34 patients with stage IV lung adenocarcinoma treated with a first-line EGFR TKI (Table A3), those with concordant EGFR mutation test results in liquid/tissue biopsy had significantly poorer PFS than those with discordant results (p = 0.0032) (Figure A1a). Similarly, patients with lymph node involvement (N1–3 disease) also had significantly poorer PFS than those without lymph node involvement (N0 disease) (p = 0.0011) (Figure A1b).
4. Discussion
Determining EGFR mutation status is important in guiding the treatment for advanced lung adenocarcinoma. Liquid biopsy, which uses plasma ctDNA as surrogates for tissue samples, has been increasingly used in clinical practice. However, it remains unclear whether plasma ctDNA provides the same information about EGFR mutation status as do tissue samples. In the current study, we found that 65% of patients with newly diagnosed advanced lung adenocarcinoma harboring EGFR mutation in their tissue samples had concordant EGFR mutation testing results in the testing using liquid biopsy. The factors independently associated with the concordant results in liquid/tissue biopsy included lymph node involvement and bone metastasis. We further showed that the concordant results in liquid/tissue biopsy was associated with significantly poorer PFS in stage IV patients treated with EGFR TKIs. However, multivariable analysis showed that only lymph node involvement was the independent predicting factor for poorer PFS, while the concordant results in liquid/tissue biopsy was not an independently predicting factor.
There are several advantages of using liquid biopsy to determine the tumor genotypes. It is less invasive than traditional tissue biopsy, so the risk of biopsy-related complications can be eliminated. Liquid biopsy may provide similar results of EGFR testing as tissue biopsy. In a recent study analyzing the association between plasma genotyping and treatment outcomes of osimertinib in advanced NSCLC patients who failed to the first-line EGFR TKIs therapy, patients with T790M mutation detected by either liquid biopsy or tissue biopsy had similar outcomes [27]. In addition, liquid biopsy provides the results of tumor genotypes more rapidly than traditional tissue biopsy. A recent study revealed that the median turnaround time for EGFR gene analysis in newly diagnosed lung cancer patients was three business days while using liquid biopsy and was twelve business days while using tissue biopsy [13]. Finally, investigating plasma ctDNA from cancer patients can account for molecular heterogeneity, because ctDNA fragments from all parts of cancer tissues throughout the patient’s body are collected [28,29,30,31]. In our study, two patients had different EGFR genotypes shown in tumor tissue and ctDNA, which might be related to molecular heterogeneity.
The association between clinical features and detectable ctDNA has been investigated in some previous studies. Although a few studies found no correlation between ctDNA levels and tumor burden [32,33], further studies still suggested that the presence of ctDNA was significantly associated with a larger tumor burden [34,35,36,37]. A recent study of late-staged NSCLC patients even showed that patients with bone metastasis had significantly higher ctDNA quantities than those without bone metastases [38]. A recent Korean study of 57 patients with adenocarcinoma harboring activating EGFR mutations found that bone metastasis was the only independent factor predicting ctDNA detection [39]. Similar to their findings, our current study found that the concordant EGFR testing results in liquid/tissue biopsy was associated with lymph node involvement, brain metastasis, and bone metastasis. Multivariable analysis showed that lymph node involvement and bone metastasis were independent predicting factors for the concordant results in liquid/tissue biopsy, while a trend of association between larger original tumor burden (T3–4) and the concordant results was also noted. Based on our findings, liquid biopsy, rather than repeated tissue biopsies, might be considered first for patients with lung adenocarcinoma with lymph node involvement and/or bone metastasis, especially for patients with high risk of biopsy-related complications. Our findings also suggested that the concordant results in liquid/tissue biopsy might be related to a more extensive tumor burden.
The detectable ctDNA might suggest poorer clinical outcomes because circulating mutant DNA has been found quite useful in assessing tumor dynamics [40]. In the BENEFIT study, a multicenter, single-arm, phase 2 clinical trial in 15 centers in China, patients with clearance of EGFR mutations in ctDNA at week 8 had longer PFS than those whose EGFR mutations persisted at week 8 [41]. The recent Korean study found that ctDNA detection was associated with poorer PFS in patients treated with EGFR TKIs, and also identified ctDNA detection and extrathoracic lymph node metastasis as independent factors predicting poorer PFS [39]. Similar to their findings, our current study showed that concordant EGFR testing results in liquid/tissue biopsy was significantly associated with a poor PFS. In contrast to their findings, the predicting effect of concordant EGFR testing results in liquid/tissue biopsy became insignificant after adjusting with other variables, especially lymph node involvement. Our finding suggested that lymph node involvement might be a confounding factor intervening between concordant results in liquid/tissue biopsy and poorer PFS. Our study was different from the Korean study in several aspects: Firstly, we included only stage IV patients actually receiving EGFR TKIs in the outcome analysis, whereas the Korean study included some patients with earlier stage (M0), which might bias the analysis. Secondly, the TKI used was included in the analysis of our study, whereas the Korean study did not include TKI used in their analysis because almost all of their patients used gefitinib (55 patients used gefitinib, one patient used erlotinib, and one patient used afatinib). Thirdly, we included lymph node involvement in the analysis and found it was independently associated with poorer PFS.
Our study still has some limitations. Firstly, the number of enrolled patients was relatively small. Nevertheless, our study was one of the largest studies discussing this topic. Currently, liquid biopsy to detect EGFR mutation is usually performed on the failure of first-line EGFR TKI in clinical practice, so not many patients received liquid biopsy before starting their first-line treatment. Secondly, the follow-up time was relatively short, so overall survival could not be assessed. Further follow-up study is needed to investigate the association between concordant EGFR testing results in liquid/tissue biopsy and overall survival. Finally, our study adopted different approaches to tissue analysis (Qiagen® EGFR RGQ PCR kit or Cobas® EGFR Mutation Test v2) and plasma ctDNA analysis (Cobas® EGFR Mutation Test v2). The two approaches might have different detection limits. Due to low concentrations of ctDNA in plasma, the detection rate of gene mutation by liquid biopsy might be relatively lower. This might affect the results of our study. However, the tests have been adopted in many previous studies [21,22,23,24,25,26,27,42], and previous validation examination in our hospital showed excellent correlation between the results obtained from these two kits. We also performed sensitivity analysis, i.e., another set of analysis excluding those using Cobas® EGFR Mutation Test v2 for their tissue samples and found consistent results. Furthermore, the aim of this study is to identify the predicting factors for concordant results from both tests. All patients in the sensitivity analyses had their tumor tissue examined with the Qiagen® kit and their plasma examined with the Cobas® kit. We therefore believe that using different analyzing methods for different sample types might minimally affect the results of this study.
5. Conclusions
Based on our findings, in advanced lung adenocarcinoma patients having insufficient tissue samples for EGFR mutation testing, liquid biopsy to determine EGFR mutation status might be particularly useful if they have lymph node involvement and/or bone metastasis. We also demonstrated that the concordant results in liquid/tissue samples might indicate a larger tumor burden, as evidenced by the presence of lymph involvement, which actually contributes to poorer PFS. Physicians should be cautious in interpreting results of cell-free DNA assay.
Acknowledgments
The authors thank the personnel from the Statistical Analysis Laboratory, Department of Internal Medicine and the Statistical Analysis Laboratory, Department of Medical Research, Kaohsiung Medical University Hospital for their help.
Appendix A
Table A1.
Baseline characteristics of the study population, stage III or IV lung adenocarcinoma patients having EGFR mutations detected in the tissue samples (only patients using Qiagen® EGFR RGQ PCR kit for their tissue samples).
| Characteristics | EGFR Mutation Test Results in Liquid/Tissue Biopsy | p Value | |
|---|---|---|---|
| Concordant (n = 28) | Discordant (n = 17) | ||
| Sex | 0.3671 | ||
| Female | 16 (57%) | 12 (71%) | |
| Male | 12 (43%) | 5 (29%) | |
| Age (year) | 64 ± 10.4 | 66.6 ± 11.2 | 0.4348 |
| Age: | 0.9119 | ||
| <65 years | 12 (43%) | 7 (41%) | |
| ≥65 years | 16 (57%) | 10 (59%) | |
| Performance status: | 0.2854 | ||
| ECOG 0–1 | 27 (96%) | 15 (88%) | |
| ECOG 2–4 | 1 (4%) | 2 (12%) | |
| Advanced primary tumor (T3–4) | 22 (79%) | 11 (65%) | 0.3078 |
| Lymph node involvement (N1–3) | 23 (82%) | 7 (41%) | 0.0047 |
| Metastasis (M1) | 28 (100%) | 14 (82%) | 0.0214 |
| Metastasis to: | |||
| Brain | 10 (36%) | 1 (6%) | 0.0240 |
| Lung | 10 (36%) | 7 (41%) | 0.7141 |
| Bone | 16 (57%) | 3 (18%) | 0.0093 |
| Pleural space | 16 (57%) | 8 (47%) | 0.5109 |
| Liver | 5 (18%) | 1 (6%) | 0.2519 |
| Pericardial space | 5 (18%) | 1 (6%) | 0.2519 |
| Adrenal gland | 4 (14%) | 1 (6%) | 0.3845 |
| Other site | 3 (11%) | 0 (0%) | 0.1624 |
| Mutation site: | |||
| Exon 18 | 1 (4%) | 1 (6%) | 0.7153 |
| Exon 19 | 14 (50%) | 7 (41%) | 0.5651 |
| Exon 20 | 0 (0%) | 5 (29%) | 0.0023 |
| Exon 21 | 13 (46%) | 5 (29%) | 0.2586 |
Data were presented as mean ± standard deviation or number (percentage). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group.
Table A2.
Predicting factors for concordant EGFR mutation detected in liquid/tissue biopsy (only patients using Qiagen® EGFR RGQ PCR kit for their tissue samples).
| Variables | Univariate Analysis | Multivariable Analysis—Maximal Model | Multivariable Analysis—Reduced Model † | |||
|---|---|---|---|---|---|---|
| OR (95% CI) | p Value | OR (95% CI) | p Value | OR (95% CI) | p Value | |
| Male (vs. female) | 1.80 (0.50–6.50) | 0.3697 | 1.99 (0.29–13.58) | 0.4828 | ||
| Age ≥65 (vs. <65) | 0.93 (0.28–3.17) | 0.9120 | 1.41 (0.17–11.93) | 0.7537 | ||
| ECOG ≥2 (vs. ≤1) | 0.28 (0.02–3.32) | 0.3118 | 0.06 (0.00–26.27) | 0.3675 | ||
| T3–4 (vs. T1–2) | 2.00 (0.52–7.66) | 0.3118 | 10.10 (0.75–136.82) | 0.0820 | 4.95 (0.87–28.27) | 0.0720 |
| N1–3 (vs. N0) | 6.57 (1.68–25.78) | 0.0069 | 12.91 (1.28–130.23) | 0.0301 | 9.90 (1.91–51.37) | 0.0064 |
| Brain metastasis (with vs. without) | 8.89 (1.02–77.32) | 0.0477 | 2.43 (0.15–38.28) | 0.5288 | ||
| Lung metastasis (with vs. without) | 0.79 (0.23–2.73) | 0.7143 | 0.27 (0.03–2.63) | 0.2583 | ||
| Bone metastasis (with vs. without) | 6.22 (1.45–26.64) | 0.0138 | 8.43 (0.92–76.96) | 0.0589 | 9.91 (1.62–60.61) | 0.0131 |
| Pleural metastasis (with vs. without) | 1.50 (0.45–5.04) | 0.5118 | 0.62 (0.08–4.86) | 0.6525 | ||
| Liver metastasis (with vs. without) | 3.48 (0.37–32.66) | 0.2754 | 5.49 (0.30–101.12) | 0.2520 | ||
| Pericardial metastasis (with vs. without) | 3.48 (0.37–32.66) | 0.2754 | 1.01 (0.03–29.49) | 0.9965 | ||
| Adrenal metastasis (with vs. without) | 2.67 (0.27–26.09) | 0.3993 | 4.61 (0.19–114.06) | 0.3507 | ||
| Metastasis to other site (with vs. without) | ‡ | ‡ | ||||
Abbreviations: EGFR = epidermal growth factor receptor; OR = odds ratio; CI = confidence interval; ECOG = Eastern Cooperative Oncology Group. † Reduced multivariable models were developed with the backward variable selection method, keeping only variables with p value less than 0.1, from the maximal model. ‡ Because all cases with metastasis to other sites were in the concordant group, odds ratio could not be estimated. This variable was therefore not included in the models.
Table A3.
Characteristics of the stage IV patients receiving first-line EGFR TKIs (only patients using Qiagen® EGFR RGQ PCR kit for their tissue samples).
| Characteristics | EGFR Mutation Test Results in Liquid/Tissue Biopsy | p Value | |
|---|---|---|---|
| Concordant (n = 26) | Discordant (n = 8) | ||
| Sex | 0.3784 | ||
| Female | 15 (58%) | 6 (75%) | |
| Male | 11 (42%) | 2 (25%) | |
| Age (year) | 64.3 ± 10.2 | 63.5 ± 10.5 | 0.8466 |
| Age: | 0.7016 | ||
| <65 years | 11 (42%) | 4 (50%) | |
| ≥65 years | 15 (58%) | 4 (50%) | |
| Performance status: | 0.3630 | ||
| ECOG 0–1 | 25 (96%) | 7 (88%) | |
| ECOG 2–4 | 1 (4%) | 1 (13%) | |
| Advanced primary tumor (T3–4) | 20 (77%) | 4 (50%) | 0.1439 |
| Lymph node involvement (N1–3) | 22 (85%) | 3 (38%) | 0.0083 |
| Metastasis (M1) | 26 (100%) | 8 (100%) | 0.0656 |
| Metastasis to: | |||
| Brain | 9 (35%) | 0 (0%) | 0.0523 |
| Lung | 9 (35%) | 4 (50%) | 0.4336 |
| Bone | 15 (58%) | 2 (25%) | 0.1058 |
| Pleural space | 15 (58%) | 5 (63%) | 0.8091 |
| Liver | 5 (19%) | 1 (13%) | 0.6623 |
| Pericardial space | 5 (19%) | 0 (0%) | 0.1793 |
| Adrenal gland | 4 (15%) | 1 (13%) | 0.8403 |
| Other site | 3 (12%) | 0 (0%) | 0.3143 |
| Mutation site: | |||
| Exon 18 | 1 (4%) | 0 (0%) | 0.5734 |
| Exon 19 | 13 (50%) | 6 (75%) | 0.2130 |
| Exon 21 | 12 (46%) | 2 (25%) | 0.2877 |
| TKI used: | 0.0312 | ||
| Gefitinib | 5 (19%) | 2 (25%) | |
| Erlotinib | 16 (62%) | 1 (13%) | |
| Afatinib | 5 (19%) | 5 (63%) | |
| Initial treatment response: | 0.3889 | ||
| Partial response (PR) | 14 (54%) | 4 (50%) | |
| Stable disease (SD) | 8 (31%) | 4 (50%) | |
| Progressive disease (PD) | 4 (15%) | 0 (0%) | |
| Objective response rate | 14 (54%) | 4 (50%) | 0.8488 |
| Disease control rate | 22 (85%) | 8 (100%) | 0.2376 |
Data were presented as mean ± standard deviation or number (percentage). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group; TKI = tyrosine kinase inhibitors.
Figure A1.
Progression-free survival (PFS) in stage IV patients receiving first-line EGFR TKIs (only patients using Qiagen® EGFR RGQ PCR kit for their tissue samples). (a) Patients with concordant EGFR mutation test results in liquid/tissue biopsy vs. those with discordant results. (b) Patients with lymph node involvement (N1–3 disease) vs. those without lymph node involvement (N0 disease). Abbreviations: EGFR = epidermal growth factor receptor; ECOG = Eastern Cooperative Oncology Group; TKI = tyrosine kinase inhibitors.
Author Contributions
Conceptualization, C.-Y.K., M.-J.T. and J.-Y.H.; Methodology, M.-J.T. and J.-Y.H.; Formal analysis, M.-J.T.; Investigation, C.-Y.K., M.-H.L., M.-J.T., C.-J.Y., J.-Y.H. and I.-W.C.; Data curation, C.-Y.K. and J.-Y.H.; Writing—original draft preparation, C.-Y.K., M.-H.L., M.-J.T. and J.-Y.H.; Writing—review and editing, C.-Y.K., M.-H.L., M.-J.T., C.-J.Y., J.-Y.H. and I.-W.C.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bai H., Mao L., Wang H.S., Zhao J., Yang L., An T.T., Wang X., Duan C.J., Wu N.M., Guo Z.Q., et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J. Clin. Oncol. 2009;27:2653–2659. doi: 10.1200/JCO.2008.17.3930. [DOI] [PubMed] [Google Scholar]
- 2.Dai L.J., Wang C., Ding Z.Y. A Case-control Study Supporting the Use of Liquid Biopsy in the Targeted Therapy for Lung Cancer. Asian Pac. J. Cancer Prev. 2018;19:1761–1766. doi: 10.22034/APJCP.2018.19.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiller J.H., Harrington D., Belani C.P., Langer C., Sandler A., Krook J., Zhu J., Johnson D.H., Eastern Cooperative Oncology G. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N. Engl. J. Med. 2002;346:92–98. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 4.Mok T.S., Wu Y.L., Thongprasert S., Yang C.H., Chu D.T., Saijo N., Sunpaweravong P., Han B., Margono B., Ichinose Y., et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N. Engl. J. Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Mitsudomi T., Morita S., Yatabe Y., Negoro S., Okamoto I., Tsurutani J., Seto T., Satouchi M., Tada H., Hirashima T., et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 6.Zhou C., Wu Y.L., Chen G., Feng J., Liu X.Q., Wang C., Zhang S., Wang J., Zhou S., Ren S., et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 7.Nosaki K., Satouchi M., Kurata T., Yoshida T., Okamoto I., Katakami N., Imamura F., Tanaka K., Yamane Y., Yamamoto N., et al. Re-biopsy status among non-small cell lung cancer patients in Japan: A retrospective study. Lung Cancer. 2016;101:1–8. doi: 10.1016/j.lungcan.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Lee J.M., Hays J.L., Noonan A.M., Squires J., Minasian L., Annunziata C., Wood B.J., Yu M., Calvo K.R., Houston N., et al. Feasibility and safety of sequential research-related tumor core biopsies in clinical trials. Cancer. 2013;119:1357–1364. doi: 10.1002/cncr.27916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overman M.J., Modak J., Kopetz S., Murthy R., Yao J.C., Hicks M.E., Abbruzzese J.L., Tam A.L. Use of research biopsies in clinical trials: Are risks and benefits adequately discussed? J. Clin. Oncol. 2013;31:17–22. doi: 10.1200/JCO.2012.43.1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlinger M., Rowan A.J., Horswell S., Math M., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz L.A., Jr., Bardelli A. Liquid biopsies: Genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao B.C., Bai Y.Y., Lee J.H., Lin C.C., Lin S.Y., Lee Y.F., Ho C.C., Shih J.Y., Chang Y.C., Yu C.J., et al. Outcomes of research biopsies in clinical trials of EGFR mutation-positive non-small cell lung cancer patients pretreated with EGFR-tyrosine kinase inhibitors. J. Formos. Med. Assoc. 2018;117:326–331. doi: 10.1016/j.jfma.2017.04.018. [DOI] [PubMed] [Google Scholar]
- 13.Sacher A.G., Paweletz C., Dahlberg S.E., Alden R.S., O’Connell A., Feeney N., Mach S.L., Janne P.A., Oxnard G.R. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016;2:1014–1022. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atamaniuk J., Kopecky C., Skoupy S., Saemann M.D., Weichhart T. Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol. Dial. Transplant. 2012;27:902–905. doi: 10.1093/ndt/gfr695. [DOI] [PubMed] [Google Scholar]
- 15.Jing R.R., Wang H.M., Cui M., Fang M.K., Qiu X.J., Wu X.H., Qi J., Wang Y.G., Zhang L.R., Zhu J.H., et al. A sensitive method to quantify human cell-free circulating DNA in blood: Relevance to myocardial infarction screening. Clin. Biochem. 2011;44:1074–1079. doi: 10.1016/j.clinbiochem.2011.06.083. [DOI] [PubMed] [Google Scholar]
- 16.Macher H., Egea-Guerrero J.J., Revuelto-Rey J., Gordillo-Escobar E., Enamorado-Enamorado J., Boza A., Rodriguez A., Molinero P., Guerrero J.M., Dominguez-Roldan J.M., et al. Role of early cell-free DNA levels decrease as a predictive marker of fatal outcome after severe traumatic brain injury. Clin. Chim. Acta. 2012;414:12–17. doi: 10.1016/j.cca.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Tsai N.W., Lin T.K., Chen S.D., Chang W.N., Wang H.C., Yang T.M., Lin Y.J., Jan C.R., Huang C.R., Liou C.W., et al. The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin. Chim. Acta. 2011;412:476–479. doi: 10.1016/j.cca.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 18.Hofman P. ALK Status Assessment with Liquid Biopsies of Lung Cancer Patients. Cancers (Basel) 2017;9:106. doi: 10.3390/cancers9080106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chae Y.K., Davis A.A., Agte S., Pan A., Simon N.I., Iams W.T., Cruz M.R., Tamragouri K., Rhee K., Mohindra N., et al. Clinical Implications of Circulating Tumor DNA Tumor Mutational Burden (ctDNA TMB) in Non-Small Cell Lung Cancer. Oncologist. 2019;24:820–828. doi: 10.1634/theoncologist.2018-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ilie M., Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl. Lung Cancer Res. 2016;5:420–423. doi: 10.21037/tlcr.2016.08.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu K.L., Tsai M.J., Yang C.J., Chang W.A., Hung J.Y., Yen C.J., Shen C.H., Kuo T.Y., Lee J.Y., Chou S.H., et al. Liver metastasis predicts poorer prognosis in stage IV lung adenocarcinoma patients receiving first-line gefitinib. Lung Cancer. 2015;88:187–194. doi: 10.1016/j.lungcan.2015.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Tsai M.J., Hung J.Y., Lee M.H., Kuo C.Y., Tsai Y.C., Tsai Y.M., Liu T.C., Yang C.J., Huang M.S., Chong I.W. Better Progression-Free Survival in Elderly Patients with Stage IV Lung Adenocarcinoma Harboring Uncommon Epidermal Growth Factor Receptor Mutations Treated with the First-line Tyrosine Kinase Inhibitors. Cancers (Basel) 2018;10:434. doi: 10.3390/cancers10110434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang C.J., Hung J.Y., Tsai M.J., Wu K.L., Liu T.C., Chou S.H., Lee J.Y., Hsu J.S., Huang M.S., Chong I.W. The salvage therapy in lung adenocarcinoma initially harbored susceptible EGFR mutation and acquired resistance occurred to the first-line gefitinib and second-line cytotoxic chemotherapy. BMC Pharmacol. Toxicol. 2017;18:21. doi: 10.1186/s40360-017-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang C.J., Tsai M.J., Hung J.Y., Lee M.H., Tsai Y.M., Tsai Y.C., Hsu J.F., Liu T.C., Huang M.S., Chong I.W. The clinical efficacy of Afatinib 30 mg daily as starting dose may not be inferior to Afatinib 40 mg daily in patients with stage IV lung Adenocarcinoma harboring exon 19 or exon 21 mutations. BMC Pharmacol. Toxicol. 2017;18:82. doi: 10.1186/s40360-017-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang C.J., Tsai M.J., Hung J.Y., Liu T.C., Chou S.H., Lee J.Y., Hsu J.S., Tsai Y.M., Huang M.S., Chong I.W. Pemetrexed had significantly better clinical efficacy in patients with stage IV lung adenocarcinoma with susceptible EGFR mutations receiving platinum-based chemotherapy after developing resistance to the first-line gefitinib treatment. Onco Targets Ther. 2016;9:1579–1587. doi: 10.2147/OTT.S100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang C.J., Tsai M.J., Hung J.Y., Tsai Y.M., Lee J.Y., Chou S.H., Liu T.C., Shen M.C., Huang M.S., Chong I.W. Poorer prognosis in Taiwanese female ever smokers with stage IV lung adenocarcinoma who were readministered a tyrosine kinase inhibitor. Onco Targets Ther. 2016;9:1511–1518. doi: 10.2147/OTT.S100169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oxnard G.R., Thress K.S., Alden R.S., Lawrance R., Paweletz C.P., Cantarini M., Yang J.C., Barrett J.C., Janne P.A. Association Between Plasma Genotyping and Outcomes of Treatment with Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2016;34:3375–3382. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murtaza M., Dawson S.J., Tsui D.W., Gale D., Forshew T., Piskorz A.M., Parkinson C., Chin S.F., Kingsbury Z., Wong A.S., et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 29.Yi X., Ma J., Guan Y., Chen R., Yang L., Xia X. The feasibility of using mutation detection in ctDNA to assess tumor dynamics. Int. J. Cancer. 2017;140:2642–2647. doi: 10.1002/ijc.30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan K.C., Jiang P., Zheng Y.W., Liao G.J., Sun H., Wong J., Siu S.S., Chan W.C., Chan S.L., Chan A.T., et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 31.De Mattos-Arruda L., Weigelt B., Cortes J., Won H.H., Ng C.K., Nuciforo P., Bidard F.C., Aura C., Saura C., Peg V., et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: A proof-of-principle. Ann. Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nygaard A.D., Holdgaard P.C., Spindler K.L., Pallisgaard N., Jakobsen A. The correlation between cell-free DNA and tumour burden was estimated by PET/CT in patients with advanced NSCLC. Br. J. Cancer. 2014;110:363–368. doi: 10.1038/bjc.2013.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Saenz J.A., Ayllon P., Laig M., Acosta-Eyzaguirre D., Garcia-Esquinas M., Montes M., Sanz J., Barquin M., Moreno F., Garcia-Barberan V., et al. Tumor burden monitoring using cell-free tumor DNA could be limited by tumor heterogeneity in advanced breast cancer and should be evaluated together with radiographic imaging. BMC Cancer. 2017;17:210. doi: 10.1186/s12885-017-3185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oxnard G.R., Paweletz C.P., Kuang Y., Mach S.L., O’Connell A., Messineo M.M., Luke J.J., Butaney M., Kirschmeier P., Jackman D.M., et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin. Cancer Res. 2014;20:1698–1705. doi: 10.1158/1078-0432.CCR-13-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman A.M., Bratman S.V., To J., Wynne J.F., Eclov N.C., Modlin L.A., Liu C.L., Neal J.W., Wakelee H.A., Merritt R.E., et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014;20:548–554. doi: 10.1038/nm.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanagita M., Redig A.J., Paweletz C.P., Dahlberg S.E., O’Connell A., Feeney N., Taibi M., Boucher D., Oxnard G.R., Johnson B.E., et al. A Prospective Evaluation of Circulating Tumor Cells and Cell-Free DNA in EGFR-Mutant Non-Small Cell Lung Cancer Patients Treated with Erlotinib on a Phase II Trial. Clin. Cancer Res. 2016;22:6010–6020. doi: 10.1158/1078-0432.CCR-16-0909. [DOI] [PubMed] [Google Scholar]
- 37.Perez-Ramirez C., Canadas-Garre M., Robles A.I., Molina M.A., Faus-Dader M.J., Calleja-Hernandez M.A. Liquid biopsy in early stage lung cancer. Transl. Lung Cancer Res. 2016;5:517–524. doi: 10.21037/tlcr.2016.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia J., Huang B., Zhuang Z., Chen S. Circulating tumor DNA as prognostic markers for late stage NSCLC with bone metastasis. Int. J. Biol. Markers. 2018;33:222–230. doi: 10.1177/1724600817753576. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y., Park S., Kim W.S., Lee J.C., Jang S.J., Choi J., Choi C.M. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac. Cancer. 2018;9:1104–1110. doi: 10.1111/1759-7714.12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diehl F., Schmidt K., Choti M.A., Romans K., Goodman S., Li M., Thornton K., Agrawal N., Sokoll L., Szabo S.A., et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008;14:985–990. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z., Cheng Y., An T., Gao H., Wang K., Zhou Q., Hu Y., Song Y., Ding C., Peng F., et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): A phase 2, single-arm, multicentre clinical trial. Lancet Respir. Med. 2018;6:681–690. doi: 10.1016/S2213-2600(18)30264-9. [DOI] [PubMed] [Google Scholar]
- 42.Mok T.S., Wu Y.L., Ahn M.J., Garassino M.C., Kim H.R., Ramalingam S.S., Shepherd F.A., He Y., Akamatsu H., Theelen W.S., et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]