Abstract
Background
Myocardial infarction (MI) generally leads to heart failure and sudden death. The hearts of people with MI undergo remodeling with the features of expanded myocardial infarct size and dilated left ventricle. Many microRNAs (miRs) have been revealed to be involved in the remodeling process; however, the participation of miR‐101 remains unknown. Therefore, this study aims to find out the regulatory mechanism of miR‐101 in MI‐induced cardiac remodeling.
Methods and Results
Microarray data analysis was conducted to screen differentially expressed genes in MI. The rat model of MI was established by left coronary artery ligation. In addition, the relationship between miR‐101 and runt‐related transcription factor 1 (RUNX1) was identified using dual luciferase reporter assay. After that, the rats injected with lentiviral vector expressing miR‐101 mimic, inhibitor, or small interfering RNA against RUNX1 were used to examine the effects of miR‐101 and RUNX1 on transforming growth factor β signaling pathway, cardiac function, infarct size, myocardial fibrosis, and cardiomyocyte apoptosis. RUNX1 was highly expressed, while miR‐101 was poorly expressed in MI. miR‐101 was identified to target RUNX1. Following that, it was found that overexpression of miR‐101 or silencing of RUNX1 improved the cardiac function and elevated left ventricular end‐diastolic and end‐systolic diameters. Also, miR‐101 elevation or RUNX1 depletion decreased infarct size, myocardial fibrosis, and cardiomyocyte apoptosis. Moreover, miR‐101 could negatively regulate RUNX1 to inactivate the transforming growth factor β1/Smad family member 2 signaling pathway.
Conclusions
Taken together, miR‐101 plays a protective role against cardiac remodeling following MI via inactivation of the RUNX1‐dependent transforming growth factor β1/Smad family member 2 signaling pathway, proposing miR‐101 and RUNX1 as potential therapeutic targets for MI.
Keywords: cardiac remodeling, microRNA‐101, myocardial infarction, runt‐related transcription factor 1, transforming growth factor β1/Smad2 signaling pathway
Subject Categories: Myocardial Infarction
Clinical Perspective
What Is New?
Our study highlights the potential importance of microRNA‐101 in myocardial infarction–induced cardiac remodeling and reveals a novel molecular mechanism that the involvement of runt‐related transcription factor 1–dependent transforming growth factor β1/Smad family member 2 signaling pathway is responsible for the protective role of microRNA‐101.
What Are the Clinical Implications?
The current study offers a possible therapeutic target for future intervention against myocardial infarction and theoretical basis for clinical applications of molecule‐targeted therapies.
Introduction
Myocardial infarction (MI) is induced by temporary or permanent thrombosis in the coronary arteries, leading to insufficient blood supply to the left ventricular (LV) beating heart muscle.1 MI could induce inflammatory and fibrotic responses in both infarcted and noninfarcted areas, leading to harmful cardiac remodeling and dysfunctional ventricle.2 Generally speaking, balloon dilatation and stenting of the coronary artery is found to be effective in controlling tissue necrosis and contributing to positive treatment outcome in MI, especially in acute MI.3 Also, stem cell therapy has been extensively demonstrated to be effective in treating MI and heart failure (HF) through regenerating cardiomyocytes and recruiting permanent stem cells.4 Moreover, gene‐targeted therapies overexpressing or repressing specific genes or translational modifiers, which may be associated with the dysfunction and remodeling of the left ventricle, are likely to become an uninterrupted endogenous source of therapy against oxidation.5 In spite of the reduced death rate attributable to early treatment, survivors still experience cardiac remodeling and HF.6 Therefore, it is important to find more treatment modalities, especially therapeutic targets to protect against cardiac remodeling following MI.
Runt‐related transcription factor (RUNX) 1 (also known as AML1) is a member of the transcription factor family composed of RUNX1, RUNX2, and RUNX3.7 RUNX1 is closely involved in mediating various cellular progression, including cell apoptosis and proliferation.8 As depicted in the published microarray gene expression profile of MI (GSE46395), RUNX1 was an upregulated gene in MI. It was previously found that the interval of App and RUNX1 played an important part in the cardiac defect in the mouse model of Down syndrome.9 RUNX1 deficiency is able to alleviate MI‐induced cardiac remodeling.10 In addition, RUNX1 was reported to be targeted and negatively regulated by microRNA (miR) 101.11 As small noncoding RNAs, miRs could regulate gene expression post‐transcriptionally and extensively participate in the development of various human diseases, including acute MI.12, 13 For instance, miR‐130 could function to ameliorate cardiac remodeling and dysfunction induced by MI.14 miR‐101a was found to be expressed at a lower level in hearts after infarction, the restoration of which could mitigate cardiac interstitial fibrosis.15 Moreover, forced expression of miR‐101 was proven to inactivate the transforming growth factor β (TGF‐β) signaling pathway, thus alleviating hypoxia‐caused cardiac fibrosis.16 TGF‐β is a widely known regulator of fibrogenesis, and drosophila mothers against decapentaplegic proteins (Smads) are key intracellular effectors belonging to the TGF‐β1 class.17, 18 TGF‐β1 acts as an essential part in remodeling and fibrosis and thus is regarded as a promising target for treating MI and cardiac remodeling.19, 20 Strikingly, overexpression of miR‐101 could alleviate liver fibrosis by blocking the activation of the TGF‐β signaling pathway.21 Based on these findings, we hypothesized that miR‐101 is involved in the cardiac remodeling following MI by regulating RUNX1. This study was performed to verify this hypothesis based on an MI model in rats induced by the left coronary artery ligation and the potential regulation associated with TGF‐β1/Smad family member 2 (Smad2) signaling pathway.
Materials and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethical Statements
This study protocol was approved by the Experimental Animal Ethics Committee of Linyi People's Hospital. The animal experiment strictly adhered to principles expressed in the National Institutes of Health's Guide for the Care and Use of Laboratory Animals and the principle of using the least number of animals to complete the experiment and to minimize the pain of experimental animals.
Microarray‐Based Gene Expression Data Analysis
The microarray data related to MI were retrieved from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). Standardized pretreatment of the gene expression data was conducted using the affy package in the R language programming package.22 Based on the microarray data GSE46395, differentially expressed genes (DEGs) were screened using the limma package with |log2FC| >2.0 and adj.P.Val (P value after correction) <0.05 as the threshold.23 The heatmap of the DEGs were constructed. MI‐related genes and protein‐protein interaction were searched in the DisGeNET (http://www.disgenet.org/web/DisGeNET/menu/search?4) and String databases (https://string-db.org/).24, 25 Cytoscape 3.6.0 software was applied to obtain information about gene interaction and construct gene interaction network.26 The putative miRs regulating DEGs were predicted in the TargetScan (http://www.targetscan.org/vert_71/).
Dual Luciferase Reporter Assay
In order to confirm the relationship between miR‐101 and RUNX1, the 3′untranslated region (UTR) sequence of RUNX1 (containing putative miR‐101 binding sites) was synthesized and then introduced into the pMIR‐reporter (Huayueyang Biotechnology Co., Ltd.) using the endonuclease sites SpeI and Hind III. The mutant form in which the potential binding sites of miR‐101 were mutated was also constructed. The pMIR‐RUNX1‐wild type or pMIR‐RUNX1‐mutant (Mut) were cotransfected with miR‐101 into human embryonic kidney (HEK)‐293T cells (Beinuo Life Science). Cells were collected and lysed 48 hours after transfection. The luciferase activity was detected using the Glomax20/20 luminometer (Promega Corporation) according to the instructions of the luciferase assay kits (K801‐200, BioVision).
Surgical Ligation Model of MI
A total of 140 specific pathogen‐free healthy Sprague‐Dawley rats (70 male and 70 female) aged 2 months and weighing 220 to 250 g were purchased from Shanghai Experimental Animal Center (Chinese Academy of Sciences). The rats were fed with normal diet and had free access to water under 12 hours cycle of light/dark. First, 60 male and 60 female rats were randomly selected to construct the rat model of MI using surgical ligation as previously published.27 Briefly, the rats were anesthetized and the left chest was opened at the fourth intercostal space with an incision (1.2 cm). Then the pectoral muscles were bluntly separated to expose the fourth intercostal space, the pericardium was opened using hemostatic forceps, and then the heart was exposed. The left anterior descending branch of the left coronary artery was ligated 5 mm beneath the left atrial appendage using a 6‐0 suture. The model was successfully established if the anterior wall of the left ventricle turned pale and ST elevation of 0.2 mV occurred in >2 limb leads. After that, the heart was put back into the chest, which was then closed with layered sutures. The rats were intraperitoneally injected with 80 000 U of penicillin for 3 consecutive days post‐operation to avoid infection. The rats were lying in the right lateral position and housed in single cages with a table lamp set above to keep them warm. The remaining 10 rats were threaded without left coronary artery ligation as sham control.
Construction of Lentivirus Vector
HEK‐293T cells in the logarithmic phase were employed for lentivirus package. After trypsinization and counting, the cells (6×105 cells/mL) were inoculated into a 15 cm2 culture dish. The complete medium was replaced with serum‐free medium 2 hours before cell infection. When cell confluence reached about 80%, cells were infected using Lipofectamine 2000. The 2.5 mL Solution A (Opti‐MEM supplemented with 20 μg GV320 lentiviral vector expressing miR‐101 mimic, miR‐101 inhibitor or small interfering RNA targeting RUNX1 [si‐RUNX1], 15 μg pHelper1.0, and 10 μg pHelper2.0) and Solution B (10 μL Lipofectamine 2000 and 2.5 mL Opti‐MEM) were mixed at room temperature for 20 minutes. The HEK‐293T cells were cultured with the mixture of Solution A and B with 5% CO2 at 37°C. After 8 hours of culture, the cells were fully washed with PBS with the removal of culture medium and continued to be cultured with medium containing 10% fetal bovine serum for 48 hours. Subsequently, the supernatant of HEK‐239T cells was collected and centrifuged at 4000g at 4°C for 10 minutes to remove the cell debris. The cells filtered using 0.45 μm filter membrane were collected and preserved at −80°C.
Animal Treatment
The 10 male and 10 female sham‐operated rats (only puncture but no ligation of coronary artery) and 10 male and 10 female rats in the MI model were not subjected to any delivery of vectors. Rats from the MI model (n=100, half male and half female) were injected via tail vein with lentiviral vectors carrying the following plasmids, respectively: empty vector, miR‐101 mimic, miR‐101 inhibitor, si‐RUNX1, and combination of miR‐101 inhibitor and si‐RUNX1, with 10 male and 10 female rats in each group. The lentiviral vector injection (1×107 TU per mouse) via tail vein above was conducted within 5 minutes after modeling and 3 days after modeling again. On the seventh day post‐modeling, the cardiac function was evaluated using a Vevo770 ultrasonic imaging system (Visual Sonics), after which the rats were euthanized and the tissues were collected for subsequent experiments.
Echocardiography
On the seventh day post‐modeling, the rats were anesthetized with 1% isoflurane. During echocardiography, isoflurane (1.5–2.0%) and oxygen were supplied to maintain anesthesia. The cardiac function parameters including LV end‐diastolic diameter (LVEDD), LV end‐systolic diameter (LVESD), LV end‐diastolic volume, and LV end‐systolic volume were measured and recorded using Ultrasound Biomicroscopy InviVue (Vevo770; Visual Sonics) with an RMV 704 probe of 40 MHz central frequency. After that, LV ejection fraction (LVEF), and LV fractional shortening (LVFS) were calculated,28 and the average was taken after 3 successive cardiac cycles.
Tissue Collection
The rats were euthanized after echocardiography and the hearts were excised quickly. Myocardial tissues were extracted from the apex of the heart to the ligature site along the short axis of the heart using a blade, and then the tissues were sliced into 2‐mm thick sections, followed by triphenyltetrazolium chloride (TTC) staining. Meanwhile, myocardial tissues were extracted from the infarct areas in rats. Parts of myocardial tissues were preserved for later reverse transcription quantitative polymerase chain reaction (RT‐qPCR) and western blot analysis. The rest of the myocardial tissues were fixed in 10% formaldehyde solution at 4°C for 24 hours, paraffin‐embedded, and sliced into 3‐μm sections. After being conventionally dewaxed and hydrated, the sections were washed with ultrapure water and double distilled water and preserved for hematoxylin‐eosin (H&E) staining, Masson staining, and terminal deoxynucleotidyl transferase‐mediated dUTP‐biotin nick end labeling (TUNEL) assays.
H&E Staining
The tissue sections were dewaxed with xylene twice, and hydrated with gradient ethanol. After that, the sections were stained with hematoxylin, and differentiated with 1% hydrochloric acid ethanol. Afterwards, the sections were dehydrated using gradient ethanol, cleared twice with xylene, and mounted using neutral balsam. The pathological changes of the tissues were observed under microscope (H‐7500, Hitachi Chemical).
TTC Staining
Myocardial tissue sections were immersed in 2% TTC liquor and then water‐bathed at 37°C avoiding exposure to light for 15 minutes with the sections turned over for staining every 5 minutes. After that, the extra TTC liquor was removed and the sections were fixed with 4% polyformaldehyde for 20 minutes. Images were captured using the scanner. The MI areas were stained pale and noninfarct areas were stained brick red. The ratio (percent) of myocardial infarct area to ventricle area was calculated using ImageJ software to analyze the MI size of rats.
Masson Staining
Five dewaxed sections were stained with Ponceau S for 2 minutes and immersed in 0.2% glacial acetic acid solution, 5% phosphomolybdic acid solution, and 0.2% glacial acetic acid solution (2 minutes for each) successively. The sections were subsequently stained using methyl green for 3 minutes, followed by color separation using 95% ethanol and dehydration using ethanol. Next, the sections were cleared with xylene and mounted with neutral balsam. The mounted sections were observed under the optical microscope (×400). The areas with collagen fiber deformation were stained blue and the cellular matrix was stained red. With 10 visual fields randomly selected in each section, the sections were photographed and then analyzed using ImageJ software. The degree of myocardial fibrosis was expressed as the percentage of area of myocardial fibrosis to the area of whole left ventricle, and then the myocardial fibrosis of rats was evaluated.
TUNEL Staining
Five dewaxed sections were incubated with 50 μL 1% protease K at 37°C for 30 minutes, and then washed with PBS. The sections were then incubated with methanol containing 0.3% H2O2 at 37°C for 30 minutes to eliminate the endogenous peroxidase. The sections were then washed with PBS and stained with TUNEL reagents at 37°C for 1 hour, followed by PBS washing again. The sections were incubated with 50 μL converter‐peroxidase and then visualized by 2% diaminobenzidine at room temperature for 15 minutes. The reaction was terminated with the addition of distilled water when the nucleus became tan under microscope. Subsequently, the sections were counterstained using hematoxylin, dehydrated by gradient alcohol, cleared by xylene, and mounted by neutral balsam. Afterwards, the mounted sections were observed under the optical microscope. With 10 visual fields selected randomly from each section, the positive cells and total cardiomyocytes were counted to detect the cell apoptosis in myocardial tissues of rats.
Immunohistochemical Staining
The tissues were dewaxed and sliced into 5 sections. After dewaxing and dehydration, antigen retrieval was proceeded at a high temperature and high pressure for 2 minutes, and naturally cooled at room temperature. The tissue sections were incubated with avidin for 10 minutes at room temperature, rinsed 3 times with PBS, followed by incubation with D‐biotin solution for 10 minutes at room temperature and PBS washing. The endogenous peroxidase activity was blocked for 15 minutes at room temperature, followed by PBS washing. Next, the tissue sections were incubated with 10% goat serum for 10 minutes at room temperature. The sections were incubated with diluted antibody to phosphorylated Smad2/3 (ab63399, 1:100, Abcam) overnight at 4°C. After PBS washing, the sections were incubated with secondary antibody, horseradish peroxidase–labeled goat anti‐rabbit IgG (ab6721, 1:1000, Abcam) at room temperature for 10 minutes. After PBS washing, the sections were incubated with streptomycin avidin‐peroxidase solution at 37°C for 10 minutes. The sections developed with freshly prepared diaminobenzidine for 20 seconds were observed under microscope. After hematoxylin counterstaining, the sections were rinsed with water for 15 minutes, dehydrated by ethanol, followed by xylene clearing and neutral gum sealing. The staining was observed under microscope.
Reverse Transcription Quantitative Polymerase Chain Reaction
RT‐qPCR was performed to determine the mRNA expression of the predicted 8 miRNAs, RUNX1, MI‐related genes, and TGF‐β1/Smad2 pathway‐related genes. Total RNA was extracted from the myocardial tissues in strict accordance to the instructions of TRIzol (Invitrogen Inc.). After determination of RNA purity and concentration, the RNA was reversely transcribed into complementary DNA. The RT‐qPCR was then performed using Fast SYBR Green PCR kit with the ABI PRISM 7300 RT‐PCR system (both from Applied Biosystems). Reaction condition was as follows: predegeneration at 95°C for 10 minutes, 40 cycles of degeneration at 94°C for 30 seconds, annealing at 59°C for 30 seconds, and extension at 72°C for 30 seconds. The primers (Table 1) used were synthesized by the GenScript. U6 was used as the endogenous control for miR‐101 and β‐actin was used as the internal control for other genes. The fold changes of genes between the experimental group and the control group were calculated by means of relative quantification (2−ΔΔCt method).29
Table 1.
Prime Sequence for RT‐qPCR
| Gene | Primer Sequence |
|---|---|
| miR‐9a‐5p | F: 5′‐ACGGCGGTCTTTGGTTATCTA‐3′ |
| R: 5′‐CAGTGCAGGGTCCGAGGTAT‐3′ | |
| miR‐18a‐5p | F: 5′‐ACGTAAGGTGCATCTAGTGCAGATA‐3′ |
| R: 5′‐GTGCAGGGTCCGAGGT‐3′ | |
| miR‐199a‐3p | F: 5′‐GCCCCAGTGTTCAGACTACC‐3′ |
| R: 5′‐CTGGCCCTTGCCCAGTCTAA‐3′ | |
| miR‐144a‐3p | F: 5′‐TACTGCATCAGGAACTGACTGGA‐3′ |
| R: 5′‐GTGCAGG GTCCGAGGT‐3′ | |
| miR‐128‐3p | F: 5′‐CAGCCGTCACAGTGAACCG‐3′ |
| R: 5′‐TCCACACCACCTGAGCCG‐3′ | |
| miR‐27b‐3p | F: 5′‐TCTAGATGCTCCTCCAGAAACCGTGG‐3′ |
| R: 5′‐GAATTCCCTTTCAGACAGATGTCCGA‐3′ | |
| miR‐27a‐3p | F: 5′‐TTCACAGTGGCTAAGTTCCGC‐3′ |
| R: 5′‐AGGGCTTAGCTGCTTGTGAGCA‐3′ | |
| miR‐101 | F: 5′‐CTGTGCAGCAAACAGTCCAC‐3′ |
| R: 5′‐AGAGAGGGTGCTACAGGAG‐3′ | |
| RUNX1 | F: 5′‐CTCCGTGCTACCCACTCACT‐3′ |
| R: 5′‐GTCGTTGAATCTCGCTACCTG‐3′ | |
| RANTES | F: 5′‐CCCCATATGGCTCGGACACCA‐3′ |
| R: 5′‐CTAGCTCATCTCCAATATGTTGAT‐3′ | |
| CCR5 | F: 5′‐AACCTGGCCATCTCTGACCTG‐3′ |
| R: 5′‐GTAGCAGATGACCATGAC‐3′ | |
| TGF‐β1 | F: 5′‐TCGACATGGAGCTGGTGAAA‐3′ |
| R: 5′‐GAGCCTTAGTTTGGACAGGATCTG‐3′ | |
| Smad2 | R: 5′‐GTGTTTGCCGAGTGCCTAAGT‐3′ |
| R: 5′‐TTACAGCCTGGTGGGATTTG‐3′ | |
| U6 | F: 5′‐CTCGCTTCGGCAGCACA‐3′ |
| R: 5′‐AACGCTTCACGAATTTGCGT‐3′ | |
| β‐actin | F: 5′‐GGCATGGGTCAGAAGGATTCC‐3′ |
| R: 5′‐ATGTCACGCACGATTTCCCGC‐3′ |
CCR5 indicates chemokine receptor 5; F, forward; miR, microRNA; R, reverse; RANTES, regulated upon activation normal T‐cell expressed and secreted; RT‐qPCR, reverse transcription quantitative polymerase chain reaction; RUNX1, runt‐related transcription factor 1; Smad2, Smad family member 2; TGF‐β1, transforming growth factor β1.
Western Blot Analysis
Western blot analysis was conducted to examine the expression of the MI‐related proteins and TGF‐β1/Smad2 pathway‐related proteins. Myocardial tissues from rats were lysed in precooled TRIzol lysis buffer on ice and the supernatant was collected after centrifugation. The protein concentration was determined using bicinchoninic acid kits (PD‐BCA‐500, Shanghai Yan Xi Biotechnology Co., Ltd.). A total of 50 μg proteins were separated with 10% SDS‐PAGE and then transferred onto the nitrocellulose membrane (HATF00010, Beijing Solarbio Science & Technology Co. Ltd.). After that, the membrane was blocked with 5% skimmed milk at room temperature for 2 hours, rinsed with Tris‐buffered saline, and incubated overnight at 4°C with the diluted primary polyclonal rabbit antibodies (1:1000) to RUNX1 (ab92336), regulated upon activation normal T‐cell expressed and secreted (RANTES; ab9783), chemokine receptor 5 (CCR5; ab65850), Bcl‐2–associated X protein (Bax; ab32503), B‐cell CLL/lymphoma 2 (Bcl‐2; ab59348), SMAD2 (ab33875), TGF‐β1 (ab179695), and GAPDH (ab8245). Subsequently, the membrane was probed with horseradish peroxidase–labeled secondary antibody and polyclonal goat anti‐rabbit IgG (1:500, ab20272) at 37°C for 1 hour. These antibodies were all from Abcam. The membrane was then reacted with electrochemiluminescence reagents (ECL808‐25, Biomiga) at room temperature for 1 minute. With the removal of the liquid, the membrane was covered with preservative film and observed under an X‐ray machine (36209ES01, Qcbio Science and Technologies Co., Ltd). With GAPDH as the endogenous control, the relative expression of the proteins was expressed as the gray value ratio of the target gene band to that of the GAPDH band.
Statistical Analysis
The data were analyzed using SPSS 21.0 software (IBM). The measurement data were presented as mean±SD. The data among multiple groups were compared using 1‐way ANOVA and the data between 2 groups were compared using independent sample 2‐tailed t test. P<0.05 was considered statistically significant.
Results
RUNX1 is Highly Expressed in MI and a Putative miR Targeting RUNX1, miR‐101, is Poorly Expressed in MI
DEGs were screened out from the GSE46395 microarray data with |log2FC| >2.0 and adj.P.Val<0.05 as the threshold, and the heatmap of the top 15 GEGs was constructed (Figure 1A). The MI‐related genes were retrieved in the DisGeNET database and the top 15 MI genes were selected. The String database was used for gene interaction analysis of the DEGs and MI genes and the gene‐gene interaction network was constructed (Figure 1B), from which it was found that the MI genes were tightly interacted with each other, and B‐cell antigen receptor and RUNX1 had a more close association with the MI genes. B‐cell antigen receptor was rarely studied in MI. Although RUNX1 depletion has been previously reported to alleviate cardiac remodeling following MI,10 the specific molecule mechanism of RUNX1 in MI remains unclear. Therefore, this study focused on the underlying mechanism of differentially expressed RUNX1 in MI. The potential miRs regulating RUNX1 were predicted in the TargetScan database and the information of the top 10 predicted miRs with 8 nonoverlapping miRs are presented in Table 2. After successful establishment of the MI rat model, the expression of the 8 nonoverlapping miRs in myocardial tissues was detected using RT‐qPCR, showing that miR‐101a‐3p downregulated in the myocardial tissues from rats of MI models as compared with the sham‐operated rats, which displayed the largest fold change among the above 8 miRs (Figure 1C). It was previously reported that miR‐101 overexpression could ameliorate interstitial fibrosis and the deterioration of cardiac function in postinfarct rats.15 We assumed that miR‐101 might influence MI by targeting RUNX1.
Figure 1.
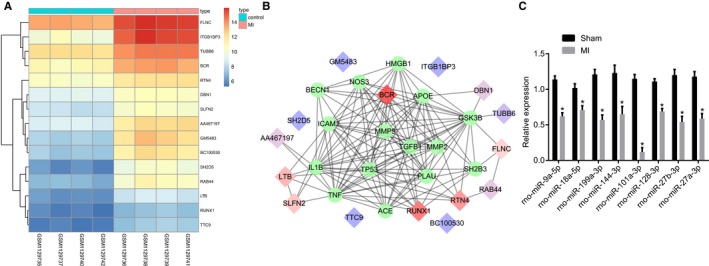
MicroRNA‐101 (miR‐101) could target runt‐related transcription factor 1 (RUNX1), which was an upregulated gene in myocardial infarction (MI). A, The heatmap of the top 15 differentially expressed genes (DEGs) in the microarray data GSE46395. The abscissa refers to the sample number, the ordinate refers to the DEGs, and the upper right histogram represents color grade. Each rectangle in the image represents the expression of a sample. B, Gene interaction network of the DEGs and MI genes, in which the diamonds refer to MI genes and the circles refer to DEGs. C, Expression of 8 nonoverlapping miRs in myocardial tissues in the sham‐operated and MI rats determined by reverse transcription quantitative polymerase chain reaction. BCR indicates B cell receptor; DBN1, Drebrin; FLNC, filamin C; LTB, heat‐lable enterotoxin B unit; RTN4, reticulon 4; SH2D5, SH2 domain‐containing 5; SLFN2, Schlafen; TTC9, Tetratricopeptide repeat domain 9; Tubb6, beta 6 class V β‐tubulin.
Table 2.
Top 10 Putative miRs Regulating RUNX1 Predicted in the TargetScan Database
| miR | Position in the UTR | Seed Match | Context++ Score | Context++ Score Percentile | Weighted Context++ Score | Conserved Branch Length | Pct |
|---|---|---|---|---|---|---|---|
| rno‐miR‐27a‐3p | 68–75 | 8mer | −0.37 | 98 | −0.37 | 6.627 | 0.95 |
| rno‐miR‐27b‐3p | 68–75 | 8mer | −0.37 | 98 | −0.37 | 6.627 | 0.95 |
| rno‐miR‐128‐3p | 68–74 | 7mer‐1A | −0.16 | 86 | −0.16 | 6.627 | 0.76 |
| rno‐miR‐27b‐3p | 95–101 | 7mer‐1A | −0.2 | 91 | −0.2 | 6.768 | 0.73 |
| rno‐miR‐27a‐3p | 95–101 | 7mer‐1A | −0.2 | 91 | −0.2 | 6.768 | 0.73 |
| rno‐miR‐9a‐5p | 154–160 | 7mer‐1A | −0.14 | 84 | −0.14 | 4.505 | 0.74 |
| rno‐miR‐18a‐5p | 229–236 | 8mer | −0.54 | 99 | −0.54 | 5.853 | 0.83 |
| rno‐miR‐199a‐3p | 480–486 | 7mer‐m8 | −0.37 | 97 | −0.23 | 6.643 | 0.86 |
| rno‐miR‐144‐3p | 482–488 | 7mer‐1A | −0.08 | 79 | −0.05 | 6.306 | 0.43 |
| rno‐miR‐101a‐3p | 482–488 | 7mer‐1A | −0.07 | 75 | −0.04 | 6.306 | 0.43 |
miR indicates microRNA; RUNX1, runt‐related transcription factor 1; UTR, untranslated region. Pct, probabilities of conserved targeting.
RUNX1 is a Target Gene of miR‐101
Here, the relationship between miR‐101 and RUNX1 was predicted in the TargetScan website available at http://www.targetscan.org/vert_71/, finding that a specific binding site of miR‐101 existed in the 3′UTR of RUNX1 (Figure 2A). Subsequently, a dual luciferase reporter assay was performed to confirm whether RUNX1 was a target gene of miR‐101, which suggested that the luciferase activity of RUNX1‐3′UTR‐Wt was attenuated by the cotransfection with miR‐101 mimic (P<0.05), while that of RUNX1‐3′UTR‐Mut did not change in response to the cotransfection of miR‐101 mimic (P>0.05) (Figure 2B). It was elucidated that miR‐101 could specifically bind to RUNX1.
Figure 2.
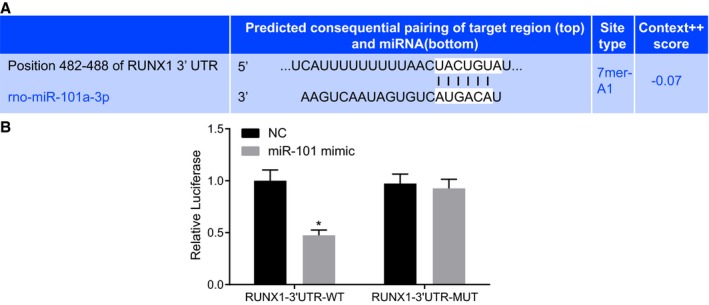
MicroRNA‐101 (miR‐101) could directly bind to runt‐related transcription factor 1 (RUNX1) gene. A, The binding site of miR‐101 in the RUNX1‐3′untranslated region (3′UTR) predicted in the biological website. B, Luciferase activity of RUNX1‐3′UTR‐wild‐type (WT) and RUNX1‐3′UTR‐mutant (MUT) followed by the cotransfection with miR‐101 mimic detected by dual luciferase reporter assay. The data were expressed as mean±SD and tested using independent sample t test. The experiment was repeated 3 times. *P<0.05 compared with the negative control (NC) group.
miR‐101 Improves Cardiac Function Following MI Through Targeting RUNX1
Since RUNX1 was a target gene of miR‐101, its effects on MI were explored in the MI rats injected with lentiviral vectors carrying empty vector, miR‐101 mimic, miR‐101 inhibitor, or si‐RUNX1. The sham‐operated rats were used as controls. Subsequently, the indicators of cardiac function (LVEDD, LVESD, LVFS, and LVEF) were measured with the aim to find out the effects of miR‐101 and RUNX1 on cardiac function. As revealed by echocardiography (Figure 3A and 3B), compared with the sham‐operated rats, the levels of LVEDD and LVESD were significantly elevated, while the levels of LVFS and LVEF were markedly reduced in the MI rats (all P<0.05). The treatment of lentiviral vector expressing miR‐101 mimic or si‐RUNX1 could decrease the levels of LVEDD and LVESD and increase the levels of LVFS and LVEF in MI rats (all P<0.05). On the contrary, the delivery of miR‐101 inhibitor unregulated the levels of LVEDD and LVESD and downregulated the levels of LVFS and LVEF in MI rats (all P<0.05). However, compared with the MI rats injected with both lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1, the MI rats injected with lentiviral vector expressing si‐RUNX1 alone displayed low LVEDD and LVESD levels but high LVFS and LVEF levels (all P<0.05), opposite to the results caused by treatment with lentiviral vector expressing miR‐101 inhibitor alone (P<0.05). All in all, the MI‐induced cardiac dysfunction could be relieved with upregulation of miR‐101 or downregulation of RUNX1.
Figure 3.
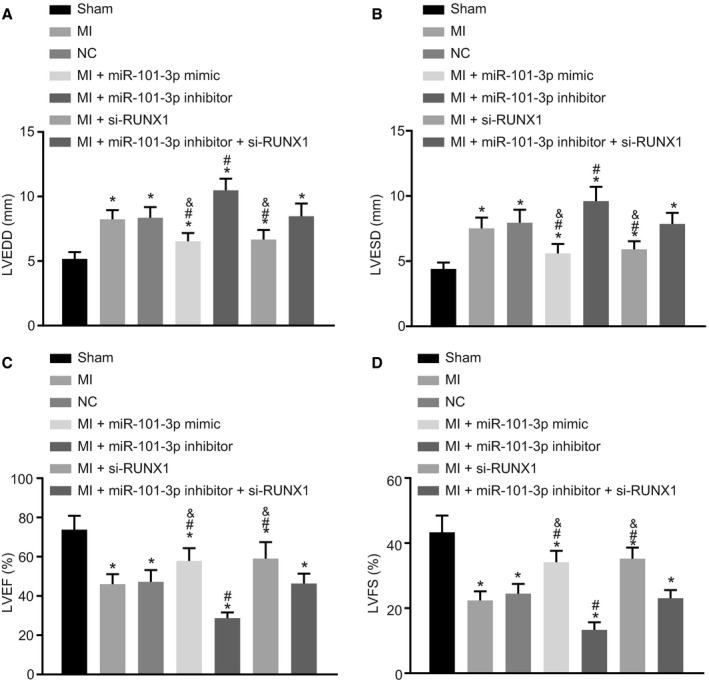
MicroRNA‐101 (miR‐101) overexpression or runt‐related transcription factor 1 (RUNX1) silencing led to improved cardiac function in rats with myocardial infarction (MI). The MI rats used for following measurements were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, The level of left ventricular end‐diastolic diameter (LVEDD) in sham‐operated and MI rats. B, The levels of left ventricular end‐systolic diameter (LVESD) in sham‐operated and MI rats. C, The level of left ventricular ejection fraction (LVEF) in sham‐operated and MI rats. D, The level of left ventricular fractional shortening (LVFS) in sham‐operated and MI rats. The data were expressed as mean±SD and analyzed using ANOVA (n=20 in each group). *P<0.05 compared with the sham‐operated rats; # P<0.05 compared with the MI rats injected with empty vector; & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. NC indicates negative control.
miR‐101 Alleviates the MI‐Induced Damage to Cardiomyocytes in Rats by Targeting RUNX1
Next, H&E staining was performed to observe the pathological changes of the cardiomyocytes. It revealed that the sham‐operated rats exhibited normal ventricular wall, orderly arranged cardiomyocytes with normal size, homogeneous cytoplasm, and uniform nuclear staining. MI rats presented with cardiomyocyte enlargement (P<0.05), accompanied with ventricular wall thickening, obvious inflammatory cell infiltration, swelled myocardial fiber, and myocardial edema. The overexpression of miR‐101 or silencing of RUNX1 in MI rats resulted in a decline in the thickness of ventricular wall, less enlarged cardiomyocytes, and smaller surface area of cardiomyocytes (P<0.05). In MI rats injected with miR‐101 inhibitor, the ventricular wall was further thickened, the surface area of cardiomyocytes was further increased (P<0.05), and the cardiomyocytes were disorderly arranged with deeply stained cytoplasm, expanded intercellular space, and increased unstained nuclei. The thickness of the ventricular wall, cardiomyocyte enlargement, and surface area of cardiomyocytes showed no evident difference in the MI rats following the treatment of lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1 (Figure 4A and 4B). All of these findings provide evidence that the MI rats exhibit reduced damage to cardiomyocytes after overexpressing miR‐101 or silencing RUNX1.
Figure 4.
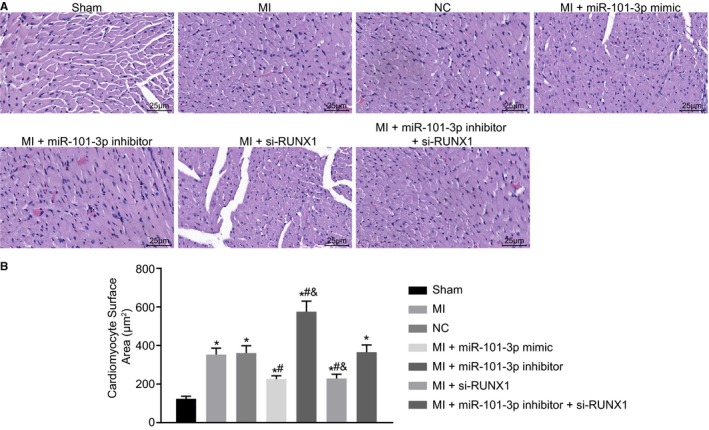
Myocardial infarction (MI)–induced damage to cardiomyocytes was alleviated with the enhancement of microRNA‐101 (miR‐101) or depletion of runt‐related transcription factor 1 (RUNX1). The MI rats used for following assays were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, Representative images of cardiomyocytes in sham‐operated and MI rats observed by hematoxylin‐eosin staining (×400). B, The quantitative surface areas of cardiomyocytes in sham‐operated and MI rats. The data were expressed as mean±SD and analyzed using ANOVA (n=20 in each group). *P<0.05 compared with the sham‐operated rats. # P<0.05 compared with the MI rats injected with empty vector. & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. NC indicates negative control.
miR‐101 Reduces Myocardial Infarct Area in MI Rats Through Targeting RUNX1
Here, TTC staining was conducted in order to explore the roles of miR‐101 and RUNX1 in myocardial infarct size of MI rats. In comparison with the sham‐operated rats, the MI rats all experienced larger heart volume, thinner LV wall, and obvious white myocardial infarct area. The myocardial infarct area of MI rats was decreased following the treatment with lentiviral vector expressing miR‐101 mimic or si‐RUNX1 but increased following the treatment with lentiviral vector expressing miR‐101 inhibitor (all P<0.05). The myocardial infarct area did not differ in the MI rats injected with lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1 relative to that in the MI rats injected with lentivirus expressing empty vector. Compared with the MI rats injected with lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1, the myocardial infarct area was reduced in MI rats injected with lentiviral vector expressing si‐RUNX1 alone but increased in those injected with miR‐101 inhibitor alone (both P<0.05) (Figure 5A and 5B). Based on the above findings, the conclusion could be made that overexpressing miR‐101 or silencing RUNX1 contributed to decreased myocardial infarct size of MI rats.
Figure 5.
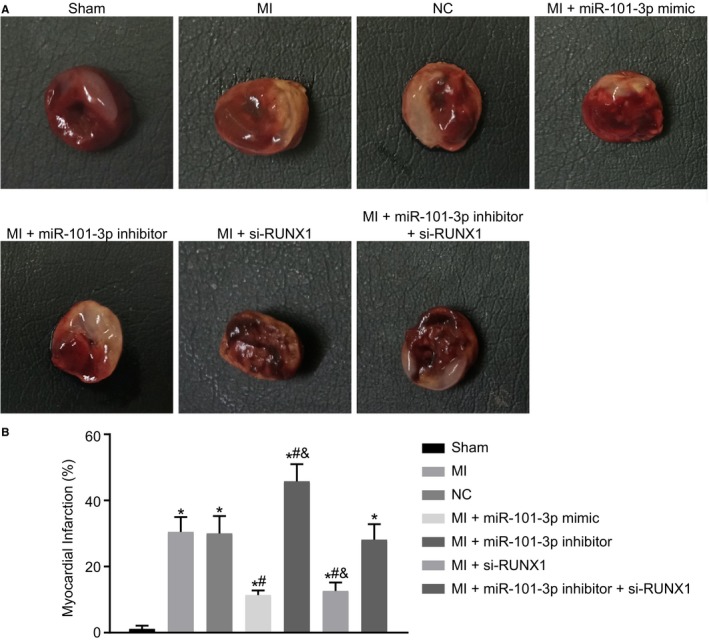
Myocardial infarct size in rats with myocardial infarction (MI) was reduced in response to microRNA‐101 (miR‐101) elevation or runt‐related transcription factor 1 (RUNX1) depletion. The MI rats used for following assays were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, Representative images of myocardial infarct in sham‐operated and MI rats detected by triphenyltetrazolium chloride staining. B, The percentage of myocardial infarct area of sham‐operated and MI rats. The data were expressed as mean±SD and analyzed using ANOVA (n=10 in each group). *P<0.05 compared with the sham‐operated rats. # P<0.05 compared with the MI rats injected with empty vector. & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. NC indicates negative control.
miR‐101 Alleviates Myocardial Fibrosis After MI by Targeting RUNX1
Masson staining was performed to assess myocardial fibrosis of MI rats after overexpression of miR‐101 or silence of RUNX1. The results suggested that the LV cavity was obviously dilated and the ventricular wall was thinner with significant myocardial fibrosis in rats with left coronary artery ligation–induced MI compared with the sham‐operated rats. In addition, it was found that miR‐101 overexpression or RUNX1 silencing led to decreased fibrosis area (P<0.05), while miR‐101 inhibition resulted in increased fibrosis area in MI rats (P<0.05). The fibrosis area showed no great difference in the MI rats injected with lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1 versus the MI rats injected with lentivirus expressing empty vector. In contrast to the combined injection with miR‐101 inhibitor and si‐RUNX1, injection with si‐RUNX1 alone resulted in reduced fibrosis area, and injection with miR‐101 inhibitor resulted in increased fibrosis area (both P<0.05) (Figure 6A and 6B). Taken together, myocardial fibrosis after MI was attenuated by miR‐101 overexpression or RUNX1 silencing.
Figure 6.
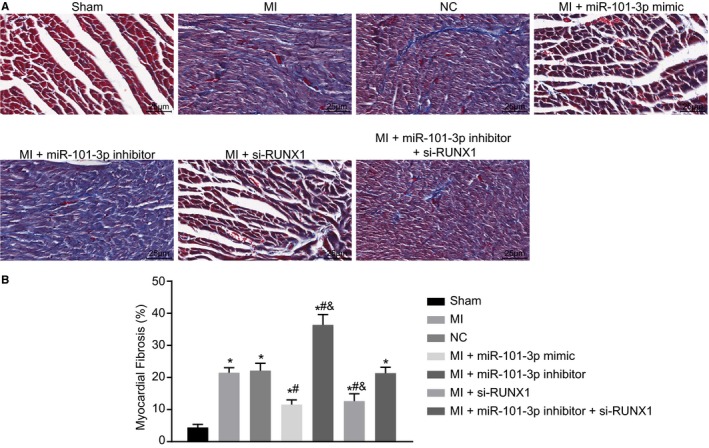
Myocardial fibrosis following myocardial infarction (MI) was ameliorated in response to microRNA‐101 (miR‐101) elevation or runt‐related transcription factor 1 (RUNX1) depletion. The MI rats used for following assays were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, Representative images showing myocardial fibrosis in myocardial tissues of sham‐operated and MI rats assessed by Masson staining (×400). B, The percentage of myocardial fibrosis area in sham‐operated and MI rats. The data were expressed as mean±SD and analyzed using ANOVA (n=20 in each group). *P<0.05 compared with the sham‐operated rats. # P<0.05 compared with the MI rats injected with empty vector. & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. NC indicates negative control.
miR‐101 Suppresses Cardiomyocyte Apoptosis in MI Rats by Targeting RUNX1
As revealed above, miR‐101 elevation or RUNX1 depletion could lead to mitigated myocardial fibrosis. The effects of miR‐101 and RUNX1 on cardiomyocyte apoptosis were then analyzed with alterations in the expression of miR‐101 or RUNX1 in cardiomyocytes. As shown by TUNEL staining (Figure 7A and 7B), the MI rats displayed elevated apoptotic index (AI) of cardiomyocytes (P<0.05) compared with the sham‐operated rats. Compared with the MI rats injected with empty vector, MI rats injected with lentiviral vector expressing miR‐101 mimic or si‐RUNX1 presented a notable reduced AI of cardiomyocytes, while MI rats injected with lentiviral vector expressing miR‐101 inhibitor presented with a markedly increased AI of cardiomyocytes (all P<0.05). The AI of cardiomyocytes did not differ greatly in the cardiomyocytes of MI rats injected with both miR‐101 inhibitor and si‐RUNX1 relative to those injected with empty vector (P>0.05). The silencing of RUNX1 alone could contribute to a reduction in AI, while inhibition of miR‐101 alone could lead to obviously upregulated AI in contrast to the combined treatment of lentiviral vectors expressing miR‐101 inhibitor and si‐RUNX1 (all P<0.05). Collectively, miR‐101 upregulation or RUNX1 downregulation could contribute to inhibited cardiomyocyte apoptosis after MI.
Figure 7.
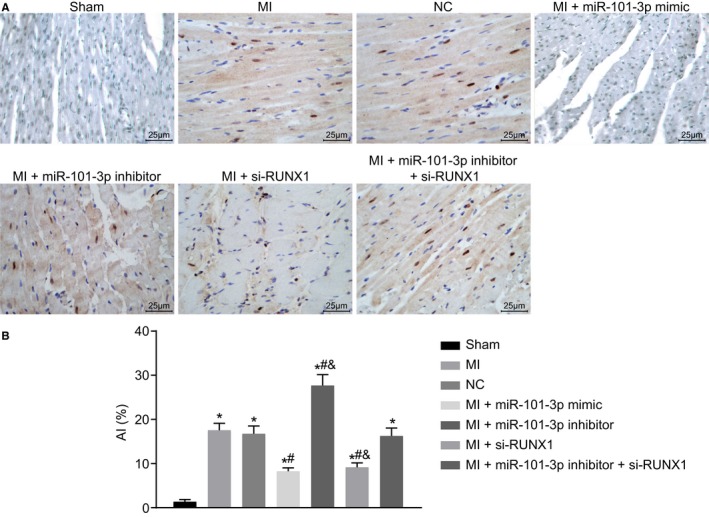
Cardiomyocyte apoptosis after myocardial infarction (MI) was found to be repressed with the elevation of microRNA‐101 (miR‐101) or depletion of runt‐related transcription factor 1 (RUNX1). The MI rats used for following assays were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, Representative images showing cell apoptosis in myocardial tissues of sham‐operated and MI rats detected by terminal deoxynucleotidyl transferase‐mediated dUTP‐biotin nick end labeling staining (×400). B, The cell apoptotic rate in myocardial tissues. The data were expressed as mean±SD and analyzed using ANOVA (n=20 in each group). *P<0.05 compared with the sham‐operated rats. # P<0.05 compared with the MI rats injected with empty vector. & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. AI indicates apoptotic index; NC indicates negative control.
miR‐101 Blocks the TGF‐β1/Smad2 Signaling Pathway and Suppresses Cardiomyocyte Apoptosis After MI by Targeting RUNX1
To understand the underlying mechanism of miR‐101 in MI, the expression of MI‐related factors (RANTES and CCR5), TGF‐β1 and Smad2 and apoptosis‐related factors (Bax and Bcl‐2) were measured using RT‐qPCR and western blot analysis. It revealed that expression of miR‐101 and mRNA and protein expression of Bcl‐2 were significantly decreased (all P<0.05), while the expression of RUNX1, RANTES, CCR5, TGF‐β1, Smad2, and Bax was markedly increased in myocardial tissues of MI rats than the sham‐operated rats (all P<0.05). It was found that the expression of miR‐101 was obviously elevated by injection of lentiviral vector expressing miR‐101 mimic but significantly reduced in response to the delivery of lentiviral vector expressing miR‐101 inhibitor (P<0.05). The expression of miR‐101 was not affected by the silencing of RUNX1 (P>0.05). In addition, the overexpression of miR‐101 or silencing of RUNX1 in MI rats upregulated Bcl‐2, while downregulated RUNX1, RANTES, CCR5, TGF‐β1, Smad2, and Bax at both the mRNA and protein levels (P<0.05), while the depletion of miR‐101 in MI rats resulted in opposite results (P<0.05) (Figure 8A through 8C). In addition, immunohistochemistry was employed to examine the expression of phosphorylated Smad2 (p‐Smad2) in the myocardial tissues of rats. The result displayed that the positive rate of p‐Smad2 significantly increased in MI rats relative to the sham‐operated rats (P<0.05). Overexpression of miR‐101 or silencing of RUNX1 significantly reduced the positive rate of p‐Smad2, but while the depletion of miR‐101 led to an increase in the positive rate of p‐Smad2 (P<0.05). The positive rate of p‐Smad2 inhibited by RUNX1 silencing was obviously rescued upon inhibition of miR‐101 (Figure 8D). Taken together, miR‐101 overexpression could inactivate the TGF‐β1/Smad2 signaling pathway, inhibit cardiomyocyte apoptosis, and attenuate MI through downregulation of RUXN1.
Figure 8.
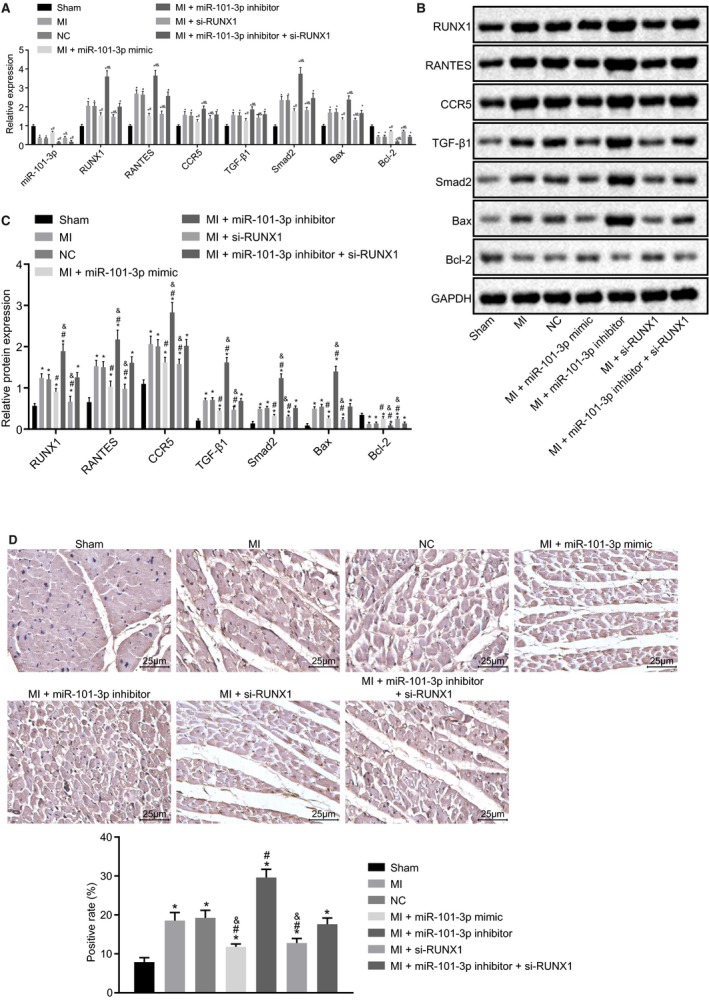
Overexpression of microRNA‐101 (miR‐101) downregulated the expression of runt‐related transcription factor 1 (RUNX1), thus inactivating the transforming growth factor β1 (TGF‐β1)/Smad family member 2 (Smad2) signaling pathway and repressing cardiomyocyte apoptosis after myocardial infarction (MI). The MI rats used for following assays were injected with lentiviral vectors expressing empty vector, miR‐101 mimic, miR‐101 inhibitor, and/or small interfering RNA targeting RUNX1 (si‐RUNX1). A, Expression of miR‐101, RUNX1, regulated upon activation normal T‐cell expressed and secreted (RANTES), chemokine receptor 5 (CCR5), TGF‐β1, Smad2, B‐cell CLL/lymphoma 2 (Bcl‐2), and Bcl‐2–associated X protein (Bax) in myocardial tissues of sham‐operated and MI rats determined by reverse transcription quantitative polymerase chain reaction. B and C, Protein expression of RUNX1, RANTES, CCR5, TGF‐β1, Smad2, Bcl‐2, and Bax relative to GAPDH in myocardial tissues of sham‐operated and MI rats measured by western blot analysis. D, The positive rate of phosphorylated Smad2 in myocardial tissues of sham‐operated and MI rats detected by immunohistochemistry. The data were expressed as mean±SD and analyzed using ANOVA (n=20 in each group); *P<0.05 compared with the sham‐operated rats. # P<0.05 compared with the MI rats injected with empty vector. & P<0.05 compared with the MI rats injected with both miR‐101 inhibitor and si‐RUNX1. NC indicates negative control.
Discussion
Patients with MI at young ages are associated with high reinfarction and death rates and frequent serious HF caused by cardiovascular events.30 In spite of the improvements made in the treatments of MI such as adjunctive pharmacotherapy, procedural techniques, and stent technology, MI still leads to increased morbidity and mortality globally and it remains a challenge to effectively treat.31 A previous study by Devaux and colleagues32 revealed that patients with acute MI presenting with downregulated expression of miR‐101 were likely to experience impaired LV contractility, and thus it was crucial in prognosticating the outcomes postacute MI. Upregulation of miR‐101a could alleviate interstitial fibrosis and prevent the deterioration of cardiac function in rats after infarction, which suggested that miR‐101a displayed a therapeutic role in cardiac disease related to fibrosis.15 In addition, miR‐101 improved osteogenic differentiation by activating the EZH2‐mediated Wnt/β‐Catenin signaling pathway.33 In our study, it was intriguingly suggested that miR‐101 could attenuate cardiac remodeling following MI and block the TGF‐β1/Smad2 signaling pathway by downregulating RUNX1.
Initially, it was found that RUNX1 was highly expressed in MI. Consistently, it was also previously demonstrated that both patients with MI and animals of the MI model exhibited elevated expression of RUNX1 at both mRNA and protein levels, the loss of which may potentially protect against cardiac remodeling following MI.10 Likewise, high expression of AML1 (RUNX1) was also detected in chronic ischemic human HF.34 RUNX1 is involved in the death or MI of patients with cardiovascular disease through directly targeting phosphatidylcholine transfer protein at the transcriptional level.35 RUNX1 depletion in mesenchymal stem cells derived from prostate or bone marrow could inhibit proliferation and induce differentiation of myofibroblasts.36 In line with those previous studies, this study confirms that inhibition of RUNX1 protected cardiac function, reduced myocardial infarct area and fibrosis, and suppressed cardiomyocyte apoptosis following MI.
RANTES, a member belonging to the CC chemokine group, is a mediator that induces inflammation and generally works together with its cognate receptor CCR5.37 In this study, miR‐101 was shown to improve cardiac function, reduce myocardial infarct size and fibrosis after MI, which were further evidenced by increased LVFS and LVEF, and decreased RANTES and CCR5. The miR‐101 family has been proposed as antifibrotic mediators, whose overexpression could mitigate fibrosis in the damaged hearts.38 It was recently reported that upregulation of miR‐101 led to alleviated cardiac hypertrophy, suggesting that miR‐101 functioned as an essential mediator in cardiac hypertrophy.39 Similarly, miR‐101 exerts a cardioprotective effect that could prevent MI‐induced LV dysfunction by alleviating fibrosis.32 Especially, miR‐101 has also been proposed as a promising target in cardiovascular diseases through its suppression in cardiac fibrosis and LV dysfunction, and improvement of LV relaxation.40 Zhao and colleagues41 have proved that miR‐101a could inhibit hypoxia‐induced apoptosis of cardiac fibroblasts via inactivation of the TGF‐β signaling pathway, illustrated by increased Bcl‐2 expression and decreased Bax expression. In the present study, we also found that upregulation of miR‐101 inhibited cardiomyocyte apoptosis following MI, which was indicated by elevated Bcl‐2 and reduced Bax.
Furthermore, the current study demonstrated that the regulatory effects of miR‐101 on MI were achieved by negatively targeting RUNX1. RUNX1 was reported to be targeted by miR‐101 in lung cancer cells.11 Consistently, the current study provided evidence that miR‐101 could specifically bind to RUNX1 and downregulate its expression in MI. Inhibition of RUNX1 reversed the aggravated myocardial fibrosis and cardiac dysfunction induced by depletion of miR‐101. In addition, the present study further suggested that miR‐101 inactivated the TGF‐β1/Smad2 signaling pathway through targeting RUNX1. A previous study demonstrated that RUNX1 interacted with TGF‐β/Smad signaling pathways in the process of neurogenesis.42 Another recent study reported that RUNX1 activated the TGF‐β signaling pathway to enhance renal fibrosis.43 The scar tissues and border zone region following MI exhibited elevated TGF‐β, which was linked to the deposition of fibrous tissues via synthesis of extracellular matrix protein.44 Also, inactivation of the TGF‐β1/Smad2 signaling pathway underlies the induction of myocardial fibrosis, accompanied by pathogenic cardiac remodeling.45 It has been reported that the restoration of miR‐101a induced by controlled intermittent aerobic exercise could inactivate the TGF‐β signaling pathway, thus alleviating cardiac fibrosis and scar formation.46 Moreover, miR‐101a has been previously verified to repress cardiac fibrosis and improve cardiac function after infarction by inhibiting the TGF‐β1 signaling pathway.47 Similarly, miR‐10a is demonstrated to suppress atrial fibrillation–induced cardiac fibrosis via disrupting the TGF‐β1/Smads signaling pathway.48
Conclusions
The findings in the present study demonstrate that restoration of miR‐101 could protect against MI‐induced cardiac remodeling through inhibiting RUNX1 in the rat model of MI. Meanwhile, miR‐101 disrupts the TGF‐β1/Smad2 signaling pathway by downregulating the expression of RUNX1 (Figure 9), which suggests that the miR‐101/RUNX1/TGF‐β1/Smad2 axis could provide new insight for deeper understanding of the underlying mechanism of MI. However, lack of validation with the TGF‐β1 signaling pathway activator or inhibitor was one of the limitations of our study. Although the results in the present study show promising future treatment for MI, further efforts are needed to validate whether miR‐101–targeted therapies would be effective in clinical treatments. Moreover, the effects of overexpressed miR‐101 in HF based on the animal model are expected to be investigated in the future.
Figure 9.
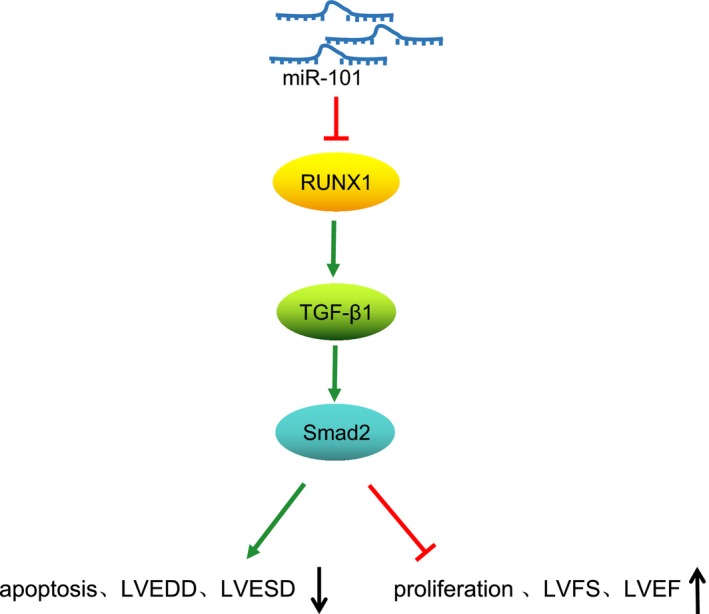
The regulatory mechanism of microRNA‐101 (miR‐101) in cardiac remodeling following myocardial infarction (MI). miR‐101 downregulated the expression of runt‐related transcription factor 1 (RUNX1) to inactivate the transforming growth factor β1 (TGF‐β1)/Smad family member 2 (Smad2) signaling pathway, thus inhibiting cardiomyocyte apoptosis, preventing cardiac dysfunction, and eventually alleviating cardiac remodeling following MI.
Disclosures
None.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this article.
(J Am Heart Assoc. 2019;8:e013112 DOI: 10.1161/JAHA.119.013112.)
References
- 1. Ruvinov E, Cohen S. Alginate biomaterial for the treatment of myocardial infarction: progress, translational strategies, and clinical outlook: from ocean algae to patient bedside. Adv Drug Deliv Rev. 2016;96:54–76. [DOI] [PubMed] [Google Scholar]
- 2. Sanchez‐Mas J, Lax A, Asensio‐Lopez MC, Fernandez‐Del Palacio MJ, Caballero L, Garrido IP, Pastor F, Januzzi JL, Pascual‐Figal DA. Galectin‐3 expression in cardiac remodeling after myocardial infarction. Int J Cardiol. 2014;172:e98–e101. [DOI] [PubMed] [Google Scholar]
- 3. Kempf T, Zarbock A, Vestweber D, Wollert KC. Anti‐inflammatory mechanisms and therapeutic opportunities in myocardial infarct healing. J Mol Med (Berl). 2012;90:361–369. [DOI] [PubMed] [Google Scholar]
- 4. Cai M, Shen R, Song L, Lu M, Wang J, Zhao S, Tang Y, Meng X, Li Z, He ZX. Bone marrow mesenchymal stem cells (BM‐MSCs) improve heart function in swine myocardial infarction model through paracrine effects. Sci Rep. 2016;6:28250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kral BG, Kraitchman DL. From mice to men: gene therapy's future for treatment of myocardial infarction. Circ Cardiovasc Imaging. 2013;6:360–362. [DOI] [PubMed] [Google Scholar]
- 6. Wang X, Meng H, Chen P, Yang N, Lu X, Wang ZM, Gao W, Zhou N, Zhang M, Xu Z, Chen B, Tao Z, Wang L, Yang Z, Zhu T. Beneficial effects of muscone on cardiac remodeling in a mouse model of myocardial infarction. Int J Mol Med. 2014;34:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu D, Ozaki T, Yoshihara Y, Kubo N, Nakagawara A. Runt‐related transcription factor 1 (RUNX1) stimulates tumor suppressor p53 protein in response to DNA damage through complex formation and acetylation. J Biol Chem. 2013;288:1353–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Luo MC, Zhou SY, Feng DY, Xiao J, Li WY, Xu CD, Wang HY, Zhou T. Runt‐related transcription factor 1 (RUNX1) binds to p50 in macrophages and enhances TLR4‐triggered inflammation and septic shock. J Biol Chem. 2016;291:22011–22020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arbogast T, Raveau M, Chevalier C, Nalesso V, Dembele D, Jacobs H, Wendling O, Roux M, Duchon A, Herault Y. Deletion of the App‐Runx1 region in mice models human partial monosomy 21. Dis Model Mech. 2015;8:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCarroll CS, He W, Foote K, Bradley A, McGlynn K, Vidler F, Nixon C, Nather K, Fattah C, Riddell A, Bowman P, Elliott EB, Bell M, Hawksby C, MacKenzie SM, Morrison LJ, Terry A, Blyth K, Smith GL, McBride MW, Kubin T, Braun T, Nicklin SA, Cameron ER, Loughrey CM. Runx1 deficiency protects against adverse cardiac remodeling after myocardial infarction. Circulation. 2018;137:57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang X, Zhao Y, Qian H, Huang J, Cui F, Mao Z. The miR‐101/RUNX1 feedback regulatory loop modulates chemo‐sensitivity and invasion in human lung cancer. Int J Clin Exp Med. 2015;8:15030–15042. [PMC free article] [PubMed] [Google Scholar]
- 12. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43:D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shyu KG, Wang BW, Cheng WP, Lo HM. MicroRNA‐208a increases myocardial endoglin expression and myocardial fibrosis in acute myocardial infarction. Can J Cardiol. 2015;31:679–690. [DOI] [PubMed] [Google Scholar]
- 14. Lu C, Wang X, Ha T, Hu Y, Liu L, Zhang X, Yu H, Miao J, Kao R, Kalbfleisch J, Williams D, Li C. Attenuation of cardiac dysfunction and remodeling of myocardial infarction by microRNA‐130a are mediated by suppression of PTEN and activation of PI3K dependent signaling. J Mol Cell Cardiol. 2015;89:87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pan Z, Sun X, Shan H, Wang N, Wang J, Ren J, Feng S, Xie L, Lu C, Yuan Y, Zhang Y, Wang Y, Lu Y, Yang B. MicroRNA‐101 inhibited postinfarct cardiac fibrosis and improved left ventricular compliance via the FBJ osteosarcoma oncogene/transforming growth factor‐beta1 pathway. Circulation. 2012;126:840–850. [DOI] [PubMed] [Google Scholar]
- 16. Yu Y, Zhao Y, Sun XH, Ge J, Zhang B, Wang X, Cao XC. Down‐regulation of miR‐129‐5p via the Twist1‐Snail feedback loop stimulates the epithelial‐mesenchymal transition and is associated with poor prognosis in breast cancer. Oncotarget. 2015;6:34423–34436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu L, Wang Y, Yan R, Li S, Shi M, Xiao Y, Guo B. Oxymatrine inhibits renal tubular EMT induced by high glucose via upregulation of SnoN and inhibition of TGF‐beta1/Smad signaling pathway. PLoS One. 2016;11:e0151986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pang L, Li Q, Wei C, Zou H, Li S, Cao W, He J, Zhou Y, Ju X, Lan J, Wei Y, Wang C, Zhao W, Hu J, Jia W, Qi Y, Liu F, Jiang J, Li L, Zhao J, Liang W, Xie J, Li F. TGF‐beta1/Smad signaling pathway regulates epithelial‐to‐mesenchymal transition in esophageal squamous cell carcinoma: in vitro and clinical analyses of cell lines and nomadic Kazakh patients from northwest Xinjiang, China. PLoS One. 2014;9:e112300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frangogiannis NG. The role of transforming growth factor (TGF)‐beta in the infarcted myocardium. J Thorac Dis. 2017;9:S52–S63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang M, Pan X, Zou Q, Xia Y, Chen J, Hao Q, Wang H, Sun D. Notch3 ameliorates cardiac fibrosis after myocardial infarction by inhibiting the TGF‐beta1/Smad3 pathway. Cardiovasc Toxicol. 2016;16:316–324. [DOI] [PubMed] [Google Scholar]
- 21. Tu X, Zhang H, Zhang J, Zhao S, Zheng X, Zhang Z, Zhu J, Chen J, Dong L, Zang Y, Zhang J. MicroRNA‐101 suppresses liver fibrosis by targeting the TGFbeta signalling pathway. J Pathol. 2014;234:46–59. [DOI] [PubMed] [Google Scholar]
- 22. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- 23. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 24. Pinero J, Bravo A, Queralt‐Rosinach N, Gutierrez‐Sacristan A, Deu‐Pons J, Centeno E, Garcia‐Garcia J, Sanz F, Furlong LI. DisGeNET: a comprehensive platform integrating information on human disease‐associated genes and variants. Nucleic Acids Res. 2017;45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta‐Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhindi R, Witting PK, McMahon AC, Khachigian LM, Lowe HC. Rat models of myocardial infarction. Pathogenetic insights and clinical relevance. Thromb Haemost. 2006;96:602–610. [PubMed] [Google Scholar]
- 28. Xiang L, Wang M, You T, Jiao Y, Chen J, Xu W. Prognostic value of ventricular wall motion score and global registry of acute coronary events score in patients with acute myocardial infarction. Am J Med Sci. 2017;354:27–32. [DOI] [PubMed] [Google Scholar]
- 29. Morimoto A, Kannari M, Tsuchida Y, Sasaki S, Saito C, Matsuta T, Maeda T, Akiyama M, Nakamura T, Sakaguchi M, Nameki N, Gonzalez FJ, Inoue Y. An HNF4alpha‐microRNA‐194/192 signaling axis maintains hepatic cell function. J Biol Chem. 2017;292:10574–10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shojaie M, Rajpout MY, Abtahian A, Pour AE, Ghobadifar MA, Akbarzadeh A. Dehydroepiandrosterone sulfate as a risk factor for premature myocardial infarction: a comparative study. Korean J Fam Med. 2015;36:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reddy K, Khaliq A, Henning RJ. Recent advances in the diagnosis and treatment of acute myocardial infarction. World J Cardiol. 2015;7:243–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, Wagner DR, Squire IB. A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS One. 2013;8:e70644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen Y, Cheng Y, Shi J, Guo Y. MiR‐101 targets the EZH2/Wnt/beta‐catenin the pathway to promote the osteogenic differentiation of human bone marrow‐derived mesenchymal stem cells. Sci Rep. 2016;6:36988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gattenloner S, Waller C, Ertl G, Bultmann BD, Muller‐Hermelink HK, Marx A. [The overexpression of NCAM (CD56) in human hearts is specific for ischemic damage]. Verh Dtsch Ges Pathol. 2004;88:246–251. [PubMed] [Google Scholar]
- 35. Li MY, Liu DW, Mao YG. [Advances in the research of effects of exosomes derived from stem cells on wound repair]. Zhonghua Shao Shang Za Zhi. 2017;33:180–184. [DOI] [PubMed] [Google Scholar]
- 36. Lee HD, Kim YH, Kim DS. Exosomes derived from human macrophages suppress endothelial cell migration by controlling integrin trafficking. Eur J Immunol. 2014;44:1156–1169. [DOI] [PubMed] [Google Scholar]
- 37. Baturcam E, Abubaker J, Tiss A, Abu‐Farha M, Khadir A, Al‐Ghimlas F, Al‐Khairi I, Cherian P, Elkum N, Hammad M, John J, Kavalakatt S, Lehe C, Warsame S, Behbehani K, Dermime S, Dehbi M. Physical exercise reduces the expression of RANTES and its CCR37 receptor in the adipose tissue of obese humans. Mediators Inflamm. 2014;2014:627150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McVicker BL, Bennett RG. Novel anti‐fibrotic therapies. Front Pharmacol. 2017;8:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei L, Yuan M, Zhou R, Bai Q, Zhang W, Zhang M, Huang Y, Shi L. MicroRNA‐101 inhibits rat cardiac hypertrophy by targeting Rab1a. J Cardiovasc Pharmacol. 2015;65:357–363. [DOI] [PubMed] [Google Scholar]
- 40. Zhou Y, Shiok TC, Richards AM, Wang P. MicroRNA‐101a suppresses fibrotic programming in isolated cardiac fibroblasts and in vivo fibrosis following trans‐aortic constriction. J Mol Cell Cardiol. 2018;121:266–276. [DOI] [PubMed] [Google Scholar]
- 41. Zhao X, Wang K, Hu F, Qian C, Guan H, Feng K, Zhou Y, Chen Z. MicroRNA‐101 protects cardiac fibroblasts from hypoxia‐induced apoptosis via inhibition of the TGF‐beta signaling pathway. Int J Biochem Cell Biol. 2015;65:155–164. [DOI] [PubMed] [Google Scholar]
- 42. Logan TT, Villapol S, Symes AJ. TGF‐beta superfamily gene expression and induction of the Runx1 transcription factor in adult neurogenic regions after brain injury. PLoS One. 2013;8:e59250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhou T, Luo M, Cai W, Zhou S, Feng D, Xu C, Wang H. Runt‐related transcription factor 1 (RUNX1) promotes TGF‐beta‐induced renal tubular epithelial‐to‐mesenchymal transition (EMT) and renal fibrosis through the PI3K subunit p110delta. EBioMedicine. 2018;31:217–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He Y, Zhou X, Zheng X, Jiang X. Exogenous high‐mobility group box 1 protein prevents postinfarction adverse myocardial remodeling through TGF‐beta/Smad signaling pathway. J Cell Biochem. 2013;114:1634–1641. [DOI] [PubMed] [Google Scholar]
- 45. Bhandary B, Meng Q, James J, Osinska H, Gulick J, Valiente‐Alandi I, Sargent MA, Bhuiyan MS, Blaxall BC, Molkentin JD, Robbins J. Cardiac fibrosis in proteotoxic cardiac disease is dependent upon myofibroblast TGF‐beta signaling. J Am Heart Assoc. 2018;7:e010013 DOI: 10.1161/JAHA.118.010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao L, He H, Ma L, Da M, Cheng S, Duan Y, Wang Q, Wu H, Song X, Duan W, Tian Z, Hou Y. Effects of miR‐29a and miR‐101a expression on myocardial interstitial collagen generation after aerobic exercise in myocardial‐infarcted rats. Arch Med Res. 2017;48:27–34. [DOI] [PubMed] [Google Scholar]
- 47. Zhao X, Wang K, Liao Y, Zeng Q, Li Y, Hu F, Liu Y, Meng K, Qian C, Zhang Q, Guan H, Feng K, Zhou Y, Du Y, Chen Z. MicroRNA‐101a inhibits cardiac fibrosis induced by hypoxia via targeting TGFbetaRI on cardiac fibroblasts. Cell Physiol Biochem. 2015;35:213–226. [DOI] [PubMed] [Google Scholar]
- 48. Li PF, He RH, Shi SB, Li R, Wang QT, Rao GT, Yang B. Modulation of miR‐10a‐mediated TGF‐beta1/Smads signaling affects atrial fibrillation‐induced cardiac fibrosis and cardiac fibroblast proliferation. Biosci Rep. 2019;39:BSR20181931. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]


