Abstract
Armadillo repeat and Armadillo-like helical domain containing proteins form a large family with diverse and fundamental functions in many eukaryotes. Herein we investigated the spatiotemporal expression pattern of Armadillo-like helical domain containing 4 (or Armh4) as an uncharacterized protein coding mouse gene, within the mouse embryo during the initial stages of heart morphogenesis. We found Armh4 is initially expressed in both first heart field as well as the second heart field progenitors and subsequently within predominantly their cardiomyocyte derivatives. Armh4 expression is initially cardiac-restricted in the developing embryo and is expressed in second heart field subpharyngeal mesoderm prior to cardiomyocyte differentiation, but Armh4 diminishes as the embryonic heart matures into the fetal heart. Armh4 is subsequently expressed in craniofacial structures and neural crest-derived dorsal root and trigeminal ganglia. Whereas lithium chloride-induced stimulation of Wnt/β-catenin signaling elevated Armh4 expression in both second heart field subpharyngeal mesodermal progenitors and outflow tract, right ventricle and atrial cardiomyocytes, neither a systemic loss of Islet-1 nor an absence of cardiac neural crest cells had any effect upon Armh4 expression. These results confirm that Wnt/β-catenin-responsive Armh4 is a useful specific biomarker of the FHF and SHF cardiomyocyte derivatives only.
Keywords: Armh4, Armadillo-like helical domain, gene expression, first and second heart field, beta-catenin/Wnt signaling
1. Introduction
Although the heart is the first organ to develop in the mammalian embryo and can even function before it is fully formed (Conway et al., 2003), astonishingly the majority of the mature heart is not even part of the initial beating heart primordium (Koushik et al., 2001). Indeed several transgenic indicator, lineage tracing and gene reporter studies have all clearly established that most of the definitive heart is actually derived from at least two spatially and temporally separate progenitor cell regions. Initially, the first heart field (FHF) progenitors differentiate into cardiomyocytes in the pre-crescent medial cardiac plate mesoderm to give rise to the linear heart tube and eventually contribute to the mature left ventricle, half of the ventricular septum and the inflow tract myocardium (Meilhac et al., 2004; Watanabe and Buckingham, 2010; Cohen et al., 2012; Später et al., 2013). However, multipotent second heart field (SHF) progenitors in more lateral regions of the cardiac crescent migrate and remain proliferative longer, but subsequently undergo delayed differentiation within the subpharyngeal mesoderm (outside the early beating heart) and eventually contribute to the mature right ventricle, the right half of the septum, the outflow tract and atria (Kelly et al., 2001; Cai et al., 2003; Meilhac et al., 2004; Abu‐Issa and Kirby, 2007; Black, 2007; Watanabe and Buckingham, 2010).
The FHF predominantly gives rise to cardiomyocyte (cTnt+, Mlc2a/v+) derivatives and FHF progenitors is known to express the nucleotide-gated channel protein Hcn4 (Später et al., 2013), the chromatin remodeling factor Smarcd3 (Devine et al., 2014) and established cardiac progenitor markers including Nkx2.5 and Tbx5 transcription factors (Cai et al., 2003; Black, 2007; Prall et al., 2007; Meilhac and Buckingham, 2018). In contrast, although Nkx2.5 is similarly required for SHF progenitor specification, undifferentiated SHF cells also differentially express a diverse set of other molecular markers including Fgf8/10, Foxh1, Gata4, Mef2c, Tbx1, Wnt5/11 transcription factors, growth factors and intracellular glycoproteins. Moreover, the SHF progenitors can give rise to heterogeneous derivatives including cardiomyocytes (cTnt+, Mef2c+, Mlc2a/v+), vascular smooth muscle (αSMA+) and endothelial/endocardial (Flk-1+, Ets+) cells (Moretti et al., 2006; Kattman et al., 2006; Black, 2007; Watanabe and Buckingham, 2010; Cohen et al., 2012; Später et al., 2013; Zhang et al., 2014). Consequently, the coordinated temporal control of cardiac SHF-derived progenitor cell proliferation and subsequent spatial differentiation into diverse sublineages is emerging as a central feature of delayed SHF verses early FHF morphogenesis. Combinatorial signals, including BMPs, FGFs and WNT/β-catenin signals are thought to temporally regulate key upstream FHF and SHF transcriptional regulators (Lin et al., 2007; Ueno et al., 2007; Cohen et al., 2012). Indeed, WNT/β-catenin has been shown to have a biphasic temporal effect on cardiogenesis, initially promoting mesoderm induction of cardiac progenitors and later inhibiting their differentiation (Buikema et al., 2013). Moreover, the WNT/β-catenin pathway can directly regulate Islet-1 and Nkx2.5 expression (Cohen et al., 2007; Lin et al., 2007; Nathan et al., 2008) and WNT/β-catenin signaling drives SHF progenitor cell proliferation (Klaus and Birchmeier, 2009).
Within the developing embryo, canonical WNT signaling (or WNT/β-catenin pathway) results in an accumulation of stable β-catenin in the cytoplasm and its eventual translocation into the nucleus to act as a transcriptional coactivator of transcription factors, including key FHF and SHF effectors (Cohen et al., 2007, 2012; Lin et al., 2007; Nathan et al., 2008). WNT signals are primarily transduced β-catenin and loss of β-catenin leads to decreased numbers of SHF progenitors, whereas stabilization of β-catenin expands the number of SHF progenitors (Lin et al., 2007; Tian et al., 2010). Β-catenin contains a repetitive ~42 amino acid sequence, called an Armadillo repeat, that fold together to form a single, rigid Armadillo domain (ARM). ARM containing proteins can often combine structural roles, such as cell-contact and cytoskeleton-associated proteins, as well as signaling functions by generating and transducing signals affecting gene expression (Tewari et al., 2010). ARM proteins form a large and diverse family, as ~204 known and novel proteins are predicted via InterPro Predicted Protein Domain Annotations dataset (http://www.ebi.ac.uk/interpro/; Mitchell et al., 2015) to contain an Armadillo repeat or an Armadillo-like helical domain. During our searches for genes highly enriched in the mouse embryo outflow tract, we identified Armadillo-like helical domain containing 4 (or Armh4) as an uncharacterized protein coding mouse gene. Although the developmental role nor the postnatal functional requirement of Armh4 is not yet known, it has been demonstrated that C14orf37 (the human ortholog of mouse Armh4) can interact with and inhibit the function of mTORC2 kinase activity via negative regulation of AKT (Lee et al., 2014) and STAT3 (Lee et al., 2016) signaling within adult hematopoietic malignancies. Significantly, AKT signaling regulates SHF progenitor cell proliferation via β-catenin activity (Luo et al., 2015) and STAT3 is necessary for the in vitro differentiation of mouse embryonic stem cells into cardiomyocytes (Foshay et al., 2005). Establishing when and where uncharacterized genes are expressed is of crucial importance to understanding or predicting their physiological role and how they may interact to form the complex expression networks that underlie embryonic organ development and function. Therefore, given the presence of Armadillo-like repeats and the important biphasic role that WNT/β-catenin is known to play during embryonic heart formation, we carried out detailed spatiotemporal expression profiling of all four uncharacterized Armadillo-like helical domain containing genes (Armh1–4) during cardiovascular morphogenesis in mice.
2. Materials and methods
2.1. Quantitative PCR analysis
Total RNA was isolated using RNAEasy (QIAGEN) kit from pooled C57BL/6J isolated embryo, fetal, newborn and adult hearts and organs (n=3 samples/stage). mRNA was reverse transcribed using SuperScript II Reverse Transcriptase and cDNAs amplified within the linear range. PCR primers were designed to amplify nucleotides 150–316bp of Mus musculus Armadillo-like helical domain containing-1 (also known as 1700012C08Rik or C14orf37); 648–813bp of Mus musculus Armadillo-like helical domain containing-2 (also known as 1700016G14Rik); 67–230bp of Mus musculus Armadillo-like helical domain containing-3 (also known as 9130011E15Rik); and 196–306bp of Mus musculus Armadillo-like helical domain containing-4 (also known as 3632451O06Rik) mRNAs. The following primers were used: Armh1: 5’ GCACCCTCCTGTGAAGGTT and 3’ CGTGGGTGACTCCACTACTC; Armh2: 5’ TCCACGGCCGACTACATTTT and 3’ TCATGTACTCGCTGGCGAAC; Armh3: 5’ GGGCTCGGTTCTCTAGCCAT and 3’ AGTTCCTCCCAAAACCGTGG; Armh4: 5’ ACCAGGGTTTACTCTGCCTG and 3’ TCCCACTCGATGGTCTCTGG, and PCR products were cloned into pCRII-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced to confirm identity. qPCR was performed in technical triplicate for each sample and qPCR reactions carried out using SyberGreenER (Invitrogen). Loading control and normalization was via GAPDH as described (Snider et al., 2014, 2019). The relative quantification of gene expression between developmental stages, hearts and/or organs was calculated by the 2−ΔΔCt approximation method. All data are presented as means ± SEM.
2.2. Isolation of mouse Armh4 cDNA probe
A mouse Armh4 cDNA probe was generated via PCR amplification of C57BL/6J E11.5 whole embryo cDNA using established methods (Kruzynska-Frejtag et al., 2001; Lindsley et al., 2005). PCR primers were designed to amplify a fragment from 1,874–2,243bps and across the 2,231/2,232bp intron-exon junction. The following primers were used: 5’ TCCATCTATCAGCCGCCCAA, 3’ CATGTAGCCTGCCTTGTCTTTT that generated a 370bp product that was cloned into pCRII-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced to confirm identity.
2.3. In situ hybridization and molecular marker immunohistochemistry
Mouse embryos and microdissected embryonic and fetal hearts; as well as Islet-1 null mutant (Pfaff et al., 1996), Pax3 null mutant (Olaopa et al., 2011) and lithium chloride-treated embryos (see below), were fixed overnight in 4% cold paraformaldehyde, washed in RNA-free phosphate buffered saline and then either processed for sectioning or whole embryo analysis, as described (Simmons et al., 2014). Both sense and antisense non-radioactive RNA probes were synthesized from the cloned Armh4 cDNA and labeled with digoxigenin using the DIG RNA Labeling kit (Roche). Specific signal was only observed when sections or whole embryos were hybridized with the anti-sense probe, and at least 3–5 mouse embryo serial sections were examined for comparable spatiotemporal patterns in a minimum of 3 consecutive sections and in at least three individual embryos or organs at each stage of development. Immunostaining of cardiomyocyte lineage marker MF20 (1:100, Developmental Studies Hybridoma Bank) and monoclonal anti-βCatenin (1:200; Abcam) using the ABC kit (Vector) following manufacturer’s directions, was performed on adjacent sections to verify Armh4 co-localization and lithium chloride-mediated induction of Wnt/βCatenin signaling.
2.4. Lithium chloride stimulation, Islet-1 null and Pax3 null embryos
In vivo stimulation of the Wnt/β-catenin signaling pathway by lithium chloride was carried out as described (Cohen et al., 2007; Tian et al., 2010). Pregnant C57BL/6J dams were injected daily intraperitoneally with 30μl of a 600mM LiCl (Sigma) or NaCl (Sigma) control solution at E7.5, 8.5 and E9.5. Treated embryos (n=13 LiCl-treated and 8 NaCl-treated) were collected at E10 for in situ hybridization or qPCR analysis. To verify that this lithium chloride regime resulted in stimulation of the Wnt/β-catenin signaling pathway and upregulation of known SHF targets (as identified by Xie et al., 2012), qPCR analysis of microdissected E10 hearts (following either LiCl or NaCl control treatment) was performed. The following primers were used: T-box transcription factor-5 (Tbx5): 5’ ATCCCCAGCACAAACTCCAG and 3’ GCGAGGTTCTATTCTCGCTC; Odd-skipped related-1 zinc finger (Osr1): 5’ CGGGACCACAGATATATTCACTCC and 3’ GTCTTGTGGACAGCGAGAGT, and PCR products were cloned into pCRII-TOPO vector (Invitrogen, Carlsbad, CA) and sequenced to confirm identity. qPCR was performed in technical triplicate and normalization was via GAPDH as described (Snider et al., 2014). For in situ analysis, E9 Islet-1 knockout (n=3) and wildtype (n=5) age-matched littermate control embryos were genotyped, 4% PFA fixed and processed as described (Cai et al., 2003, generously provided by Prof. Chen-Leng Cai, Indiana University School of Medicine). Similarly, E9.5 and E10.5 Pax3 knockout (n=4/stage) and wildtype (n=4/stage) age-matched littermate control embryos were genotyped, 4% PFA fixed and processed as described (Olaopa et al., 2011). Noon on the day of finding a copulation plug is designated 0.5 embryonic (E) days of gestation. Animal procedures and experimental conditions were refined to minimize harm to animals and performed with the approval of the Institutional Animal Care and Use Committee of Indiana University School of Medicine.
2.5. Statistical analysis
The relative quantification of qPCR gene expression between the developmental ages and microdissected organs was calculated by the 2−ΔΔCt approximation method. Three samples per gene from three separate cDNA samples (n=3 developmental stage or organ cDNAs) were obtained to calculate the mean +/− sd. Statistical analysis was conducted using the two-tailed unpaired Student’s t-test. Differences were considered statistically significant for those with P<0.05. Statistical analysis was performed with Prism software version 5.02 (Graphpad).
3. Results and discussion
3.1. Armh4 is robustly expressed in embryonic hearts
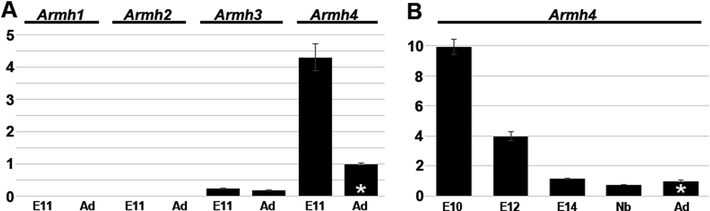
To investigate when and where the four related Armadillo-like helical domain containing protein coding genes are expressed in the heart, we used qPCR to examine mRNA expression levels within isolated E11 mouse embryo hearts. Expression profiling revealed that only Armh4 (MGI:1914669 on mouse chromosome 14) was robustly expressed in E11 hearts and that Armh3 (MGI:1918867 on chromosome 19) was expressed at very low levels (~17x fold less, Fig. 1A). To verify Armh1 (MGI:2686507 on chromosome 4) and Armh2 (MGI:1916676 on chromosome 13) PCR primers, adult testis and brain and E14 placenta were used positive controls and the expected amplification detected (based on Mouse ENCODE transcriptome data, not shown). To further characterize the functional significance of Armh4 in embryonic heart, developmental qPCR revealed that Armh4 mRNA expression levels decrease as heart morphogenesis advances, with lower level expression in isolated fetal E14 (~8.6x fold less), newborn and postnatal adult hearts (Fig. 1B). Armh4 is an uncharacterized gene that is predicted to be an intracellular, membrane-associated protein. Mouse Armh4 contains 9 putative exons, whilst its human ortholog ARMH4 (HGNC:19846) is also on chromosome 14 but contains 12 exons. NCBI data reveals Armh4 is broadly expressed, with elevated levels in adult cerebellum and embryonic central nervous system (from E11.5 to E18). Via HomoloGene (https://www.ncbi.nlm.nih.gov/homologene/12127) ARMH4 is highly conserved and orthologs are present in chimpanzee (99.01%), mouse (72.38%), lizard (88%) and chicken (54.11%). Combined, these expression profiling results indicate Armh4 is the major armadillo-like helical domain containing gene expressed in utero and within mouse embryo hearts.
Figure 1. Comparative qPCR analysis of uncharacterized Armh1–4 genes in isolated hearts.
(A) Comparative qPCR of Armh1, Armh2, Armh3 and Armh4 mRNA relative expression levels in isolated E11 embryo and adult hearts. Neither Armh1 nor Armh2 were detectible in embryo or adults hearts, whilst some Armh3 expression was present it was at considerably lower levels than Armh4 (32.1 Armh3 vs. 27.9 Armh4 median cycles in E11). (B) Quantitative PCR of Armh4 mRNA levels during heart morphogenesis, in microdissected isolated pooled (n=3) hearts (E10, E12 and E14) and isolated newborn and adult (8 week old) hearts. Note Armh4 levels decrease as the heart matures. qPCR analysis was performed in technical triplicate and data are presented as a logarithmic plot of relative expression, where a value of 1 indicates no difference between adult heart in A and B (indicated via *); and values <1 indicate reduced and >1 indicate increased expression. Error bars represent sd (fold change).
3.2. Spatiotemporal expression of Armh4 during initial heart morphogenesis
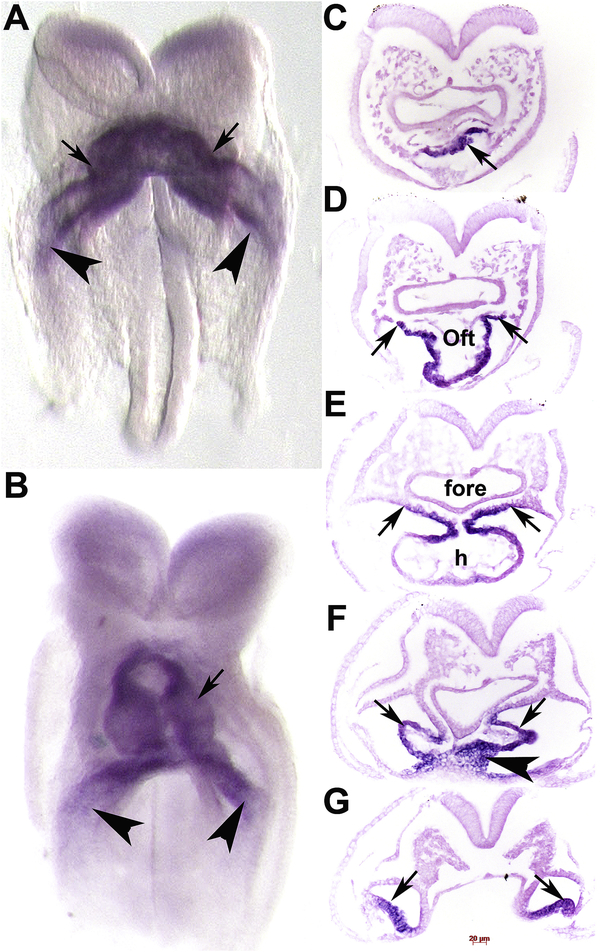
To confirm cardiac expression and determine where within embryonic hearts Armh4 was expressed, we used non-radioactive in situ hybridization. Both control sense and anti-sense digoxigenin-labeled Armh4 cDNA probes were transcribed and used for both wholemount in situ hybridization (Simmons et al., 2014). Specific signal was only observed when embryos were hybridized with the anti-sense Armh4 probe and not the control sense probe, and positive expression was only reported when a comparable expression pattern was observed in three consecutive serial sections and in at least 3 age-matched embryo samples. The earliest detectable Armh4 is within the midline FHF cardiac crescent and the more lateral sinus venosus (Fig. 2A), but is absent from the rest of the embryo. The FHF contains predominantly myocardial progenitors and differentiating cardiomyocytes that are precursors of distinct myocyte populations within the developing heart, whereas the sinus venosus is mainly mesodermal. Similarly, Armh4 is restricted to the 6 somite-containing embryonic heart and is expressed within the entire heart tube (including the arterial and venous poles) as the single primitive heart tube relocates to the midline, and in the adjacent lateral sinus venosus (Fig. 2B). Serial histology confirmed Armh4 is expressed within the arterial mesodermal SHF progenitors (Fig. 2C), the outflow tract cardiomyocyte layer (Fig. 2D), common atrial chamber wall cardiomyocytes (Fig. 2E), as well as the inflow tract mesodermal sinus venosus and primitive septum transversum (Fig. 2F,G). However, unlike the key FHF expressed Nkx2.5 molecular regulator expression, Armh4 is initially confined to cardiac structures and is not expressed in the adjacent ventral foregut endoderm, endocardium nor the 1st pharyngeal arch (Cai et al., 2003; Prall et al., 2007). Thus, Armh4 initially exhibits cardiac-restriction and during early heart formation and prior to heart looping, Armh4 is localized within FHF cardiomyocytes and both SHF early cardiomyocyte and mesodermal progenitor lineages.
Figure 2. Armh4 mRNA is expressed in the early heart.
(A,B) Wholemount in situ hybridization detection using DIG-labelled Armh4 antisense cDNA probe on 3 somite (A) and 6 somite (B) embryos. Note ~E7.75–8.25, Armh4 mRNA (purple stain) is cardiac-restricted and is strongly expressed in the FHF cardiac crescent and early heart tube (arrows) and sinus venosus (arrowheads), and in the single primitive heart tube (B). (C-G) Serial sections through heart region in panel B, counterstained with eosin (pink). Armh4 mRNA is localized to the cardiogenic plate (C, arrow), OFT outer myocardial layer and SHF (D,E, arrows), but is absent in the foregut endoderm. More posteriorly, Armh4 is expressed in the cardiogenic left and right horns of the sinus venosus (F,G, arrows) and early septum transversum (F, arrowhead). Scale bars: C-G=20μm. Abbreviations: h, primitive heart tube.
3.3. Armh4 expression in the SHF and its derivatives
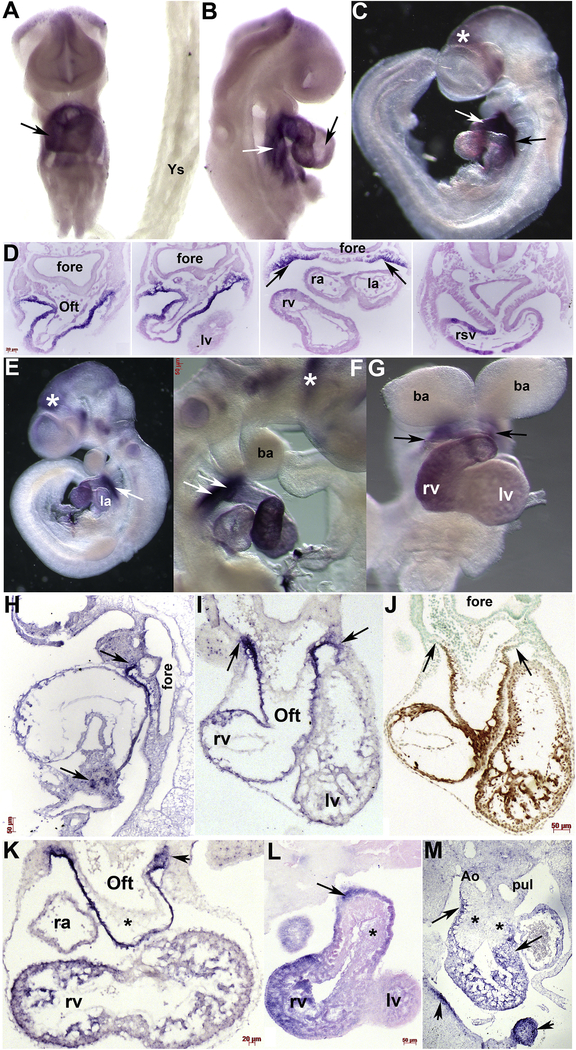
During SHF morphogenesis, robust Armh4 mRNA continues throughout the heart and sinus venosus but is absent from the embryonically-derived yolk sac (Fig. 3A). Beginning around E9 (containing ~15–18 somites), Armh4 expression becomes less prevalent in the left ventricle and left atria compared to the rest of the heart (Fig. 3A–G). In E10.5 embryos, Armh4 expression in the left atria is almost completely undetectable (Fig. 3E). Histology and serial section in situ hybridization analysis confirms Armh4 is predominately FHF and SHF cardiomyocyte and subpharyngeal mesodermally restricted (Fig. 3D), as well as a subpopulation of septum transversum cells (Fig. 3H). Moreover, when adjacent serial E10.5 sections were probed with either Armh4 or the cardiomyocyte marker MF20 (Han et al., 1992), undifferentiated subpharyngeal mesodermal SHF progenitors robustly express Armh4 but only co-express Armh4 and MF20 once the SHF progenitors have migrated in the arterial outflow tract outer myocardial layer and undergo differentiation (Fig. 3I,J). Both wholemount and section in situ hybridization analysis (both methods used to enable efficient 3D analysis and ensure efficient probe access to target mRNA) revealed Armh4 is not expressed in either the endothelium nor the endocardial cushion lineages, either the neural crest- or endocardial-derived regions (Fig. 3I,K–M). Similar to the reduced E14 qPCR levels detected (see Fig. 1B), in situ hybridization is unable to efficiently detect Armh4 expression in fetal mouse hearts onwards (negative data not shown). Collectively, these expression data indicate that Armh4 is transiently expressed throughout mouse embryo hearts, within both the FHF and SHF progenitors and their cardiomyocyte derivatives.
Figure 3. Armh4 is expressed in second heart field and its derivatives.
(A,B) Wholemount in situ hybridization of E9 embryo (A, frontal view; B, right side view) reveals Armh4 is expressed throughout the heart and SHF (white arrow, B) but is expressed at lower levels in the left ventricle (black arrow, B). However, Armh4 expression is absent from the yolk sac (Ys). (C) Armh4 is robustly expressed in E9.5 SHF (black arrow) and OFT (white arrow) but only weakly expressed in the left ventricle and atria. Armh4 is also expressed in the head, posterior to the forebrain (*). (D) Serial sections through heart region in panel C, counterstained with eosin. Armh4 is localized to the OFT outer myocardial layer, the right ventricle and atria, the SHF (arrows) and the right sinuous venous (rsv), but is absent in the foregut endoderm. (E-G) At E10.5 (E, left; F, right; G, frontal views) Armh4 is only robustly expressed in the SHF (white arrows in E,F; black arrows in G) and right ventricle. Armh4 in the cranial region persists and is also present in trigeminal ganglia (* in F). (H) Serial section in situ hybridization analysis confirmed Armh4 mRNA in the SHF extends from the branchial arches all the way down along the OFT and arterial chamber wall to the septum transversum below the heart (arrows). (I,J) Armh4 section in situ hybridization (I) and MF20 expression (J) in adjacent transverse E10.5 embryo sections. Note Armh4 mRNA and MF20 protein overlap in the OFT cardiomyocyte outer layer and right ventricle (rv) cardiomyocytes, but Armh4 extends further into the branchial arch/SHF region (arrows, I) that is negative for MF20 (arrows, J). (K) Armh4 section in situ hybridization in transverse E11 embryos confirms Armh4 is expressed throughout the proximal pharyngeal arch/SHF region (arrowhead), SHF, and OFT and predominantly right ventricular cardiomyocytes. Note Armh4 mRNA is absent from the early OFT endocardial cushions (*). (L) A single representative example of serial sections through E12.5 wholemount Armh4 stained embryo, counterstained with eosin. Armh4 is localized to the OFT outer myocardial layer, the right ventricle and right atria, but is absent from endocardial cushions (*). (M) Section in situ hybridization of transverse E13 embryo reveals Armh4 is expressed in the OFT and right ventricular cardiomyocytes (arrows) but is absent from both the conal and truncal endocardial cushions (*). Armh4 is also present in the maxillar and lateral nasal process (arrowheads). Scale bars: D,K=20μm; F, H-J,L=50μm. Abbreviations: Ao, ascending aorta; ba, branchial arch; Oft, outflow tract; fore, foregut; la, left atria; lv, left ventricle; pul, pulmonary trunk; ra, right atria.
3.4. Neuronal Armh4 expression
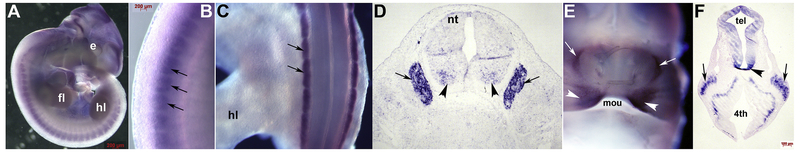
Although Armh4 mRNA is initially heart-restricted, Armh4 is expressed in the E9.5–10.5 forebrain/midbrain boundary (Fig. 3C,E), along with E10.5–12.5 cranial structures and trigeminal ganglia (Figs. 3F, 4A,F). Similarly, Armh4 is expressed in E12–13.5 dorsal root ganglia (Fig. 4A–D), within select craniofacial frontonasal (Figs. 3M, 4E) and CNS structures (Fig. 4E,F). These in situ localization data are similar to the Mouse ENCODE transcriptome database which shows significant Armh4 expression levels in the E11 CNS and adult brain. Thus, these results demonstrate that Armh4 is expressed more broadly that just the heart, as the embryo transitions into a fetus.
Figure 4. Armh4 expression in neural lineages.
(A,B) Wholemount in situ hybridization detection of Armh4 mRNA in E12 embryo illustrating expression in the developing segmental dorsal root ganglia (DRGs) and in select cranial structures, with enlarged view of DRGs (arrows, B). (C,D) Wholemount Armh4 expression in E13.5 DRGs (arrows, C) and section in situ hybridization in transverse sections confirms Armh4 is present through the DRGs (arrows, D) and punctate expression is present within ventral regions of the neural tube (arrowheads, D). (E) Armh4 is also expressed in the E12.5 frontonasal region, within the maxillary prominences (arrowheads) above the primordial mouth and either side (arrows) of the nasal septum. (F) E12.5 transverse view of head reveals Armh4 is expressed in the trigeminal ganglia (arrows), as well as telencephalon (tel) walls, median sulcus (arrowhead) and marginal layer around the forth (4th) ventricle. Scale bars: A,B =200μm; F=100μm. Abbreviations: e, eye; fl, forelimb; hl, hindlimb; mou, mouth; nt, neural tube.
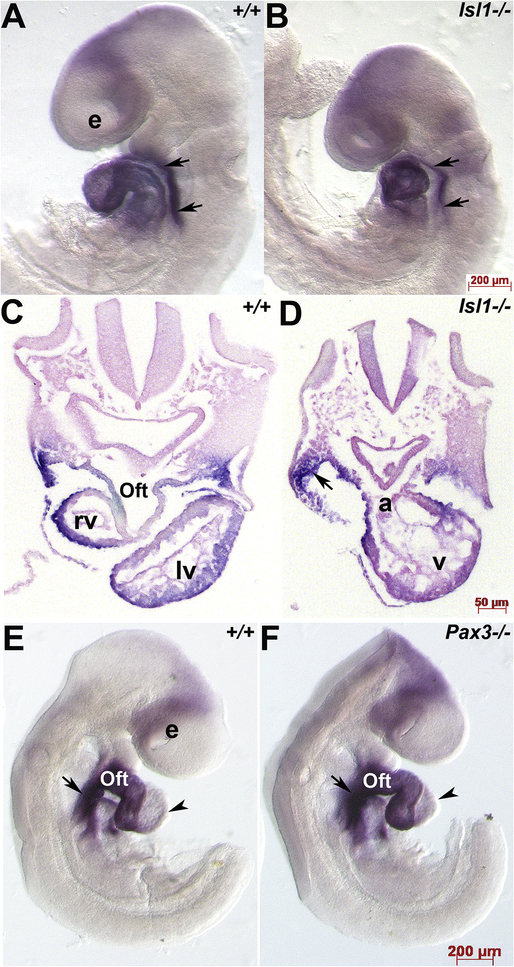
3.5. Armh4 expression persists despite an absence of Islet-1 or the cardiac neural crest lineage
As Islet-1 is a key marker of cardiac progenitor cells derived from the SHF and that Islet-1 function is required for these progenitors to contribute to the heart (Cai et al., 2007), we used Islet-1 null mutants (Islet-1−/− which exhibit 100% lethality ~E9.5) as a model to examine whether Armh4 expression is dependent upon Islet-1 presence and whether the absence of an outflow tract and right ventricle in Islet-1−/− embryos would alter Armh4 expression. Surprisingly, despite obvious SHF-derivative structural defects, Armh4 mRNA appears unaltered in E9 Islet-1−/− when compared with age-matched littermate controls (Fig. 5A–D). Given this lack of difference and that Islet-1 has also been shown to be expressed in some Wnt1-Cre lineage mapped cardiac neural crest cells that contributes to normal outflow septation (Engleka et al., 2012), used a mouse model that completely lacks the cardiac neural crest lineage. We have previously shown that transcription factor Pax3 null (Pax3−/− which exhibit 100% lethality ~E14) embryos exhibit decreased neural crest stem cell expansion and lack any migratory Wnt1-Cre mapped cardiac neural crest resulting in conotruncal and interventricular septal heart defects (Conway et al., 2000; Oloapa et al., 2011). Once again, we did not observe any Armh4 expression differences, and the normal Armh4 downregulation observed in wildtype left ventricle was similarly observed in Pax3−/− left ventricle (Fig. 5E,F). Hence, these data revealed that Armh4 expression within the SHF and heart itself, occurs independently of Islet-1 (and Pax3) expression and is not affected by a reduction of SHF progenitors present in Islet-1−/− embryos nor an absence of cardiac neural crest cells migrating through the pharyngeal arches and into the outflow tract.
Figure 5. Armh4 expression is unaffected by loss of either SHF or cardiac neural crest cell lineages.
(A,B) Wholemount mRNA in situ hybridization in E9 wildtype (+/+, A) and Islet-1 null (−/−, B) littermates illustrating Armh4 expression. Left view reveals Armh4 is expressed throughout both hearts as well as in the SHF (arrows). (C,D) Transverse section histology confirmed that Armh4 continues to be expressed in Islet-1−/− mutant SHF and dysmorphic heart, despite the lack of outflow tract (Oft) and right ventricular (RV) segments in the Islet-1−/− (D). (E,F) Wholemount mRNA in situ hybridization in E9.5 wildtype (+/+, E) and Pax3 null (−/−, F) littermates illustrating Armh4 expression. Right view reveals Armh4 is unperturbed in Pax3−/− cardiac-neural crest deficient mutant embryo outflow tract and SHF (arrows), and Armh4 expression is similarly diminished in both age-matched wildtype (E, arrowhead) and Pax3−/− mutant (F, arrowhead) left ventricles. Scale bars: A,B,E,F=200μm; C,D=50μm; E-G=50μm. Abbreviations: a, atria; e, optic vesicle; v, ventricle.
3.6. Armh4 is Wnt/β-catenin responsive
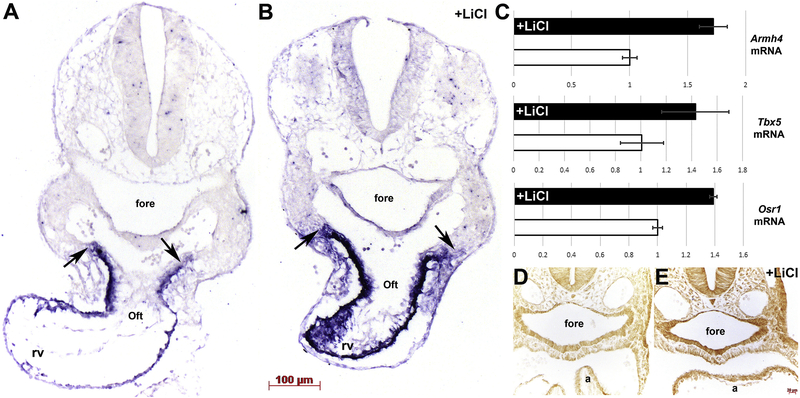
Given the presence of the Armadillo-like helical domain in Armh4, its co-expression in the FHF and SHF, the fact that Wnt/β-catenin signaling has a biphasic effect on cardiogenesis (Cohen et al., 2007; 2017), that β-catenin is upstream of Islet-1 and a number of SHF genes (Lin et al., 2007), and that MesP1-Cre-driven conditional ablation of β–catenin affects only the SHF (Klaus et al., 2007); we tested whether Armh4 expression was Wnt/β-catenin responsive. We employed a pharmacological approach using lithium chloride (LiCl), an inhibitor of Gsk-3β and strong activator of Wnt/β-catenin signaling (Tian et al., 2010), to induce Wnt/β-catenin signaling. Either LiCl or saline (NaCl) was injected daily for 3 days into pregnant females, and resultant E10 harvested and examined via section in situ hybridization. We established that Armh4 mRNA is upregulated in E10 SHF progenitors, outflow tract and right ventricle cardiomyocytes (Fig. 6A–C). Significantly, LiCl-treatment resulted in a dramatic increase in the number of Armh4 expressing cells within the subpharyngeal mesoderm SHF progenitors (Fig. 6B). Moreover, qPCR confirmed Armh4 upregulation in only LiCl-treated embryos hearts and both SHF transcriptional effectors T-box5 and Odd-skipped related-1 (Osr1) known to be Wnt/β-catenin responsive (Xie et al., 2012) were similarly upregulated via LiCl-treatment (Fig. 6C). Finally, we used immunohistochemistry to verify that our LiCl pharmacological approach resulted in β-catenin stabilization, elevated levels of nuclear β-catenin accumulation (Tian et al., 2010) and henceforth increased Wnt/β-catenin signaling levels (Fig. 6D,E).
Figure 6. Armh4 is induced following stimulation of the Wnt/β-catenin signaling.
(A,B) Section in situ hybridization in saline (NaCl)-treated control (A) and lithium chloride (LiCl)-treated age-matched E10 embryos illustrating Armh4 expression. Note, Armh4 is significantly upregulated within the LiCl-treated embryo SHF progenitors and OFT (arrows) and right ventricle cardiomyocytes (B), when compared to NaCl control (A). (C) Quantitative qPCR analysis of microdissected E10 hearts verified Armh4 mRNA levels are induced (1.73x fold) via LiCl treatment, when compared to NaCl treatment (open bar). Moreover, Tbx5 (FHF and SHF effector) and Osr1 (SHF effector and direct target of Tbx5) mRNA levels are also similarly upregulated in LiCl-treated but not in NaCl-treated control samples. Relatively, at this stage, Armh4 (25 cycles) is expressed at higher overall levels than Osr1 (27 cycles) and Tbx5 (31 cycles). Error bars represent sd (fold change). (D,E) Immunohistochemistry using anti-βCatenin antibody verified that LiCl-treatment (E) but not NaCl-treatment (D) results in β–catenin elevation. Scale bars: A,B=100μm; D,E=20μm. Abbreviations: a, atria; fore, foregut.
4. Summary
Wnt/β-catenin signaling and the canonical Wnt effector β-catenin play a pivotal biphasic role in development of cardiac progenitors of the SHF, initially promoting their induction but later inhibiting their differentiation (Cohen et al., 2007; 2012; Lin et al., 2007; Ueno et al., 2007; Tian et al., 2010; Buikema et al., 2013; Luo et al., 2015). The Wnt/β-catenin signaling pathway is also essential for development of heart progenitors in human embryonic stem cells (Paige et al., 2010), and is robustly re-activated after a variety of cardiac injuries (Zhao et al., 2018). Thus, understanding factors the genes that respond to Wnt/β-catenin signaling is critical to understanding factors that drive cardiac progenitor proliferation, survival, and migration, both in the context of normal development and congenital heart defect pathogenic, but also for potential application to cell therapies using cardiac progenitors.
Our results demonstrate Armh4 is initially cardiac-restricted and transiently expressed in both the FHF and SHF progenitors, and that Armh4 mRNA is limited to only the subpharyngeal mesoderm SHF progenitors and cardiomyocytes within the outflow tract, right ventricle and right atria. Although SHF progenitors can also give rise to vascular smooth muscle and endothelial/endocardial cell lineages in vivo and in vitro (Black, 2007; Später et al., 2013; Meilhac and Buckingham, 2018), our data suggests that Armh4 is specific marker of the FHF and SHF cardiomyocyte derivatives only. Thus, future lineage mapping of the transient Armh4-expressing progeny may be informative in understanding how segregation into distinct lineages may occur within FHF and SHF progenitor subpopulations. Furthermore, it will be important to examine whether Armh4 can drive and/or merely biomark differentiation of cardiac progenitor cells into cardiomyocytes for future regenerative therapies.
The finding that Armh4 mRNA is unperturbed via the absence of Islet-1 transcriptional regulator or a lack of any cardiac neural crest cells, suggests Armh4 may be upstream of Islet-1 or act independently of Islet-1. This also suggests that SHF cardiomyocyte derivatives and the cardiac neural crest lineage do not intersect nor do they appear dependent upon each other. Similarly, the locations of neuronal Armh4 expression are unlikely to reflect a common lineage, as both the neuroepithelial-derived CNS and neural crest-derived dorsal root ganglia/trigeminal ganglia are distinct from the FHF and SHF lineages. However, it is intriguing that Islet-1 (Sun et al., 2008) is also actively expressed in dorsal root ganglia and select neuronal cells and Wnt/β–catenin signaling is implicated in every step of neuronal development (Brafman and Willert, 2017).
Although the embryonic requirement of Armh4 is unknown, as there is presently no Armh4 knockout mouse, the fact that Armh4 encodes a putative type-I transmembrane protein with an intracytoplasmic tail that has no clear functional homologies (Lee et al., 2014), suggests a possible cell-surface localization and the ability to detect growth-regulated signals from the FHF and SHF microenvironment. Indeed, the LiCl-treatment results demonstrate that Armh4 is responsive to β-catenin (also contains armadillo repeats) accumulation and increased Wnt/β-catenin signaling, suggesting that the Armadillo-like helical domain in Armh4 can directly respond in Wnt/β-catenin stimulation. Significantly, LiCl-treatment resulted in a dramatic increase in the number of Armh4-expressing SHF progenitors, suggesting Wnt activation of SHF development may include increased expansion of Armh4–positive progenitors. However, although LiCl is a well-known inhibitor of Gsk-3β, it can also affect other signaling pathways including Akt signaling (Nemoto et al., 2008), thus as Armh4 may inhibit the function of mTORC2 kinase activity via negative regulation of Akt (Lee et al., 2014) caution should be used when extrapolating from systemic pharmacological data. Finally, although we do not yet know if Armh4 is required for normal FHF and SHF morphogenesis, there are at least two examples of proteins that contain armadillo repeats include β-catenin and Plakoglobin, whose gene knockouts result in SHF defects and early cardiovascular-related lethality (Lin et al., 2007; Bierkamp et al., 1996). These spatiotemporal expression analysis will help to further the understanding of this uncharacterized gene in normal heart development and to ultimately pave the way for examination of any future potential role in congenital heart defect pathogenesis and to purify FHF and SHF cardiomyocyte lineages from human pluripotent stem cell lines for tissue engineering purposes.
Highlights.
Uncharacterized Armh4 is initially expressed in both first and second heart fields
Armh4 is expressed in subpharyngeal mesoderm prior to cardiomyocyte differentiation
Armh4 is restricted to SHF-derived cardiomyocytes and diminishes as the heart matures
Lithium chloride-induced activation of WNT/β-catenin signaling elevates Armh4 mRNA
Armh4 is unperturbed in Islet-1 null and Pax3 null embryos with dysmorphic hearts
Acknowledgements
We are very grateful to Prof. Chen-Leng Cai (IUSM) for providing Islet-1 null and control littermate embryos. The MF20 antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Funding sources
These studies were supported, in part, by the Riley Children’s Foundation, Indiana University Department of Pediatrics and National Institutes of Health grant HL148165 (SJC).
Footnotes
Competing interest statement
The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu‐Issa R, Kirby ML, 2007. Heart field: from mesoderm to heart tube. Annu Rev Cell Dev Biol 23, 45–68. 10.1146/annurev.cellbio.23.090506.123331 [DOI] [PubMed] [Google Scholar]
- Black BL, 2007. Transcriptional pathways in second heart field development. Semin Cell Dev Biol. 18, 67–76. 10.1016/j.semcdb.2007.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierkamp C, Mclaughlin KJ, Schwarz H, Huber O, Kemler R, 1996. Embryonic heart and skin defects in mice lacking plakoglobin. Dev Biol. 180, 780–5. 10.1006/dbio.1996.0346 [DOI] [PubMed] [Google Scholar]
- Brafman D,, Willert K, 2017. Wnt/β-catenin signaling during early vertebrate neural development. Dev Neurobiol. 77, 1239–1259. 10.1002/dneu.22517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buikema JW, Mady AS, Mittal NV, Atmanli A, Caron L, Doevendans PA, Sluijter JP, Domian IJ, 2013. Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development. 140, 4165–76. https://dev.biologists.org/content/140/20/4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S, 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 5, 877–89. 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE, 2007. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest. 117, 1794–804. 10.1172/JCI31731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen ED, Miller MF, Wang Z, Moon RT, Morrisey EE, 2012. Wnt5a and Wnt11 are essential for second heart field progenitor development. Development. 139, 1931–40. 10.1242/dev.069377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway SJ, Bundy J, Chen J, Dickman E, Rogers R, Will BM, 2000. Decreased neural crest stem cell expansion is responsible for the conotruncal heart defects within the splotch (Sp(2H))/Pax3 mouse mutant. Cardiovasc Res. 47, 314–28. 10.1016/S0008-6363(00)00098-5 [DOI] [PubMed] [Google Scholar]
- Conway SJ, Kruzynska-Frejtag A, Kneer PL, Machnicki M, Koushik SV, 2003. What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis. 35, 1–21. 10.1002/gene.10152 [DOI] [PubMed] [Google Scholar]
- Devine WP, Wythe JD, George M, Koshiba-Takeuchi K, Bruneau BG, 2014. Early patterning and specification of cardiac progenitors in gastrulating mesoderm. Elife. 3. doi: 10.7554/eLife.03848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM, Epstein JA, 2012. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res. 110, 922–6. 10.1161/CIRCRESAHA.112.266510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foshay K, Rodriguez G, Hoel B, Narayan J, Gallicano GI, 2005. JAK2/STAT3 directs cardiomyogenesis within murine embryonic stem cells in vitro. Stem Cells. 23, 530–43. https://10.1634/stemcells.2004-0293 [DOI] [PubMed] [Google Scholar]
- Han Y, Dennis JE, Cohen-Gould L, Bader DM, Fischman DA, 1992. Expression of sarcomeric myosin in the presumptive myocardium of chicken embryos occurs within six hours of myocyte commitment. Dev Dyn. 193, 257–65. 10.1002/aja.1001930306 [DOI] [PubMed] [Google Scholar]
- Lee D, Sykes SM, Kalaitzidis D, Lane AA, Kfoury Y, Raaijmakers MH, Wang YH, Armstrong SA, Scadden DT, 2014. Transmembrane Inhibitor of RICTOR/mTORC2 in Hematopoietic Progenitors. Stem Cell Reports. 3, 832–40. 10.1016/j.stemcr.2014.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Wang YH, Kalaitzidis D, Ramachandran J, Eda H, Sykes DB, Raje N, Scadden DT, 2016. Endogenous transmembrane protein UT2 inhibits pSTAT3 and suppresses hematological malignancy. J Clin Invest. 126, 1300–10. 10.1172/JCI84620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Cui L, Zhou W, Dufort D, Zhang X, Cai CL, Bu L, Yang L, Martin J, Kemler R, Rosenfeld MG, Chen J, Evans SM, 2007. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A. 104, 9313–8. 10.1073/pnas.0700923104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley A, Li W, Wang J, Maeda N, Rogers R, Conway SJ, 2005. Comparison of the four mouse fasciclin-containing genes expression patterns during valvuloseptal morphogenesis. Gene Expr Patterns. 5, 593–600. 10.1016/j.modgep.2005.03.005 [DOI] [PubMed] [Google Scholar]
- Luo W, Zhao X, Jin H, Tao L, Zhu J, Wang H, Hemmings BA, Yang Z, 2015. Akt1 signaling coordinates BMP signaling and β-catenin activity to regulate second heart field progenitor development. Development. 142, 732–42. 10.1242/dev.119016 [DOI] [PubMed] [Google Scholar]
- Kattman SJ, Huber TL, Keller GM, 2006. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 11, 723–32. 10.1016/j.devcel.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Kelly RG, Brown NA, Buckingham ME, 2001. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev Cell. 1, 435–40. 10.1016/S1534-5807(01)00040-5 [DOI] [PubMed] [Google Scholar]
- Klaus A, Birchmeier W, 2009. Developmental signaling in myocardial progenitor cells: a comprehensive view of Bmp- and Wnt/beta-catenin signaling. Pediatr Cardiol. 30, 609–16. https://link.springer.com/article/10.1007%2Fs00246-008-9352-7 [DOI] [PubMed] [Google Scholar]
- Koushik SV, Wang J, Rogers R, Moskophidis D, Lambert N, Creazzo TL, Conway SJ, 2001. Targeted inactivation of the sodium-calcium exchanger (Ncx1) results in the lack of a heartbeat and abnormal myofibrillar organization. FASEB J. 15, 1209–11. 10.1096/fj.00-0696fje [DOI] [PubMed] [Google Scholar]
- Kruzynska-Frejtag A, Machnicki M, Rogers R, Markwald R, Conway SJ, 2001. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 103, 183–8. 10.1016/S0925-4773(01)00356-2 [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME, 2004. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev. Cell 6, 685–698. 10.1016/S1534-5807(04)00133-9 [DOI] [PubMed] [Google Scholar]
- Meilhac SM, Buckingham ME, 2018. The deployment of cell lineages that form the mammalian heart. Nat Rev Cardiol. 15, 705–724. https://www.nature.com/articles/s41569-018-0086-9 [DOI] [PubMed] [Google Scholar]
- Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, Sangrador-Vegas A, Scheremetjew M, Rato C, Yong SY, Bateman A, Punta M, Attwood TK, Sigrist CJ, Redaschi N, Rivoire C, Xenarios I, Kahn D, Guyot D, Bork P, Letunic I, Gough J, Oates M, Haft D, Huang H, Natale DA, Wu CH, Orengo C, Sillitoe I, Mi H, Thomas PD, Finn RD, 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43, D213–21. 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, Sun Y, Evans SM, Laugwitz KL, Chien KR, 2006. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 127, 1151–65. 10.1016/j.cell.2006.10.029 [DOI] [PubMed] [Google Scholar]
- Nathan E, Monovich A, Tirosh-Finkel L, Harrelson Z, Rousso T, Rinon A, Harel I, Evans SM, Tzahor E, 2008. The contribution of Islet1-expressing splanchnic mesoderm cells to distinct branchiomeric muscles reveals significant heterogeneity in head muscle development. Development. 135, 647–57. https://dev.biologists.org/content/135/4/647.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto T, Kanai T, Yanagita T, Satoh S, Maruta T, Yoshikawa N, Kobayashi H, Wada A, 2008. Regulation of Akt mRNA and protein levels by glycogen synthase kinase-3beta in adrenal chromaffin cells: effects of LiCl and SB216763. Eur J Pharmacol. 586, 82–9. 10.1016/j.ejphar.2008.02.075 [DOI] [PubMed] [Google Scholar]
- Olaopa M, Zhou HM, Snider P, Wang J, Schwartz RJ, Moon AM, Conway SJ, 2011. Pax3 is essential for normal cardiac neural crest morphogenesis but is not required during migration nor outflow tract septation. Dev Biol. 356, 308–22. 10.1016/j.ydbio.2011.05.583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Osugi T, Afanasiev OK, Pabon L, Reinecke H, Murry CE, 2010. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 5, e11134 10.1371/journal.pone.0011134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM, 1996. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell. 84, 309–20. 10.1016/S0092-8674(00)80985-X [DOI] [PubMed] [Google Scholar]
- Prall OW, Menon MK, Solloway MJ, Watanabe Y, Zaffran S, Bajolle F, Biben C, McBride JJ, Robertson BR, Chaulet H, Stennard FA, Wise N, Schaft D, Wolstein O, Furtado MB, Shiratori H, Chien KR, Hamada H, Black B, Saga Y, Robertson EJ, Buckingham ME, Harvey RP, 2007. An Nkx2–5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 128, 947–59. 10.1016/j.cell.2007.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Später D, Abramczuk MK, Buac K, Zangi L, Stachel MW, Clarke J, Sahara M, Ludwig A, Chien KR, 2013. A HCN4+ cardiomyogenic progenitor derived from the first heart field and human pluripotent stem cells. Nat Cell Biol. 15, 1098–106. 10.1038/ncb2824 [DOI] [PubMed] [Google Scholar]
- Snider P, Simmons O, Wang J, Hoang CQ, Conway SJ, 2014. Ectopic Noggin in a Population of Nfatc1 Lineage Endocardial Progenitors Induces Embryonic Lethality. J Cardiovasc Dev Dis. 1, 214–236. 10.3390/jcdd1030214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider P, Snider E, Simmons O, Lilly B, Conway SJ, 2019. Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis. J Cardiovasc Dev Dis. 6, pii: E24 10.3390/jcdd6020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons O, Bolanis E, Wang J, Conway SJ, 2014. In situ hybridization (both radioactive and nonradioactive) and spatiotemporal gene expression analysis. Methods Mol Biol. 1194, 225–44. 10.1007/978-1-4939-1215-5_12 [DOI] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE, 2008. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 11, 1283–93. 10.1038/nn.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R, Bailes E, Bunting KA, Coates JC, 2010. Armadillo-repeat protein functions: questions for little creatures. Trends Cell Biol. 20, 470–81. 10.1016/j.tcb.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE, 2010. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Dev Cell. 18, 275–87. 10.1016/j.devcel.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S, Weidinger G, Osugi T, Kohn AD, Golob JL, Pabon L, Reinecke H, Moon RT, Murry CE, 2007. Biphasic role for Wnt/beta-catenin signaling in cardiac specification in zebrafish and embryonic stem cells. Proc Natl Acad Sci U S A. 104, 9685–90. 10.1073/pnas.0702859104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Buckingham M, 2010. The formation of the embryonic mouse heart: heart fields and myocardial cell lineages. Ann N Y Acad Sci. 1188, 15–24. 10.1111/j.1749-6632.2009.05078.x [DOI] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP, 2012. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 23, 280–91. 10.1016/j.devcel.2012.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang C, Wang C, Hong X, Miao J, Liao Y, Zhou L, Liu Y, 2018. An essential role for Wnt/β-catenin signaling in mediating hypertensive heart disease. Sci Rep. 8, 8996 10.1038/s41598-018-27064-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Nomura-Kitabayashi A, Sultana N, Cai W, Cai X, Moon AM, Cai CL, 2014. Mesodermal Nkx2.5 is necessary and sufficient for early second heart field development. Dev Biol. 390, 68–79. 10.1016/j.ydbio.2014.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]