Abstract
Rationale: Analyzing the molecular heterogeneity of different forms of organ fibrosis may reveal common and specific factors and thus identify potential future therapeutic targets.
Objectives: We sought to use proteome-wide profiling of human tissue fibrosis to (1) identify common and specific signatures across end-stage interstitial lung disease (ILD) cases, (2) characterize ILD subgroups in an unbiased fashion, and (3) identify common and specific features of lung and skin fibrosis.
Methods: We collected samples of ILD tissue (n = 45) and healthy donor control samples (n = 10), as well as fibrotic skin lesions from localized scleroderma and uninvolved skin (n = 6). Samples were profiled by quantitative label-free mass spectrometry, Western blotting, or confocal imaging.
Measurements and Main Results: We determined the abundance of more than 7,900 proteins and stratified these proteins according to their detergent solubility profiles. Common protein regulations across all ILD cases, as well as distinct ILD subsets, were observed. Proteomic comparison of lung and skin fibrosis identified a common upregulation of marginal zone B- and B1-cell–specific protein (MZB1), the expression of which identified MZB1+/CD38+/CD138+/CD27+/CD45−/CD20− plasma B cells in fibrotic lung and skin tissue. MZB1 levels correlated positively with tissue IgG and negatively with diffusing capacity of the lung for carbon monoxide.
Conclusions: Despite the presumably high molecular and cellular heterogeneity of ILD, common protein regulations are observed, even across organ boundaries. The surprisingly high prevalence of MZB1-positive plasma B cells in tissue fibrosis warrants future investigations regarding the causative role of antibody-mediated autoimmunity in idiopathic cases of organ fibrosis, such as idiopathic pulmonary fibrosis.
Keywords: fibrosis, proteomics, interstitial lung disease, localized scleroderma (morphea), autoimmunity
At a Glance Commentary
Scientific Knowledge on the Subject
Organ fibrosis is a major clinical problem with limited to no therapeutic options, depending on organ manifestation. Fibrosis can occur as a result of persistent tissue injury and inflammation, impaired regeneration or repair pathways, distorted proteostasis (e.g., during aging), or autoimmunity. It is unclear whether organ-specific fibrotic diseases such as idiopathic pulmonary fibrosis have a common underlying pathophysiology compared with other fibrotic syndromes or whether tissue-specific mechanisms of fibrosis exist that allow targeted therapeutic intervention.
What This Study Adds to the Field
The analysis of tissue fibrosis has relied mainly on gene expression data to date, but full-scale quantitative proteome approaches to fibrosis are limited. It is well documented that changes in protein abundance are not necessarily reflected at the mRNA level, and novel therapeutic compounds act largely on proteins. We provide the most comprehensive proteomic resource of human tissue fibrosis, containing information about the abundance, stoichiometry, and detergent solubility of proteins. We identified common and distinct features of lung fibrosis in comparison with skin fibrosis of patients with localized scleroderma. The most significant commonality of different interstitial lung diseases and skin fibrosis was the prevalent occurrence of marginal zone B- and B1-cell–specific protein–positive plasma B cells, which points to a common involvement of antibody-mediated autoimmunity in at least two forms of tissue fibrosis.
Tissue fibrosis is a major health burden, accounting for about 45% of deaths in the developed world, both directly and indirectly (1). Replacement of normal tissue architecture by extracellular matrix (ECM)-rich scar tissue in fibrosis impedes organ functionality and regeneration after injury. More than 200 different chronic lung disorders are characterized by lung fibrosis. Many of these interstitial lung diseases (ILDs) exhibit poor prognosis, such as idiopathic pulmonary fibrosis (IPF), with a median survival time of 3–5 years after diagnosis (2). In localized scleroderma (morphea), autoimmune-mediated chronic inflammation leads to severe fibrotic plaques restricted to the skin (3). This disease represents a particularly good model system for study of fibrotic reactions because involved and uninvolved areas can be directly identified and compared in the same patient.
In many cases, the true origin and cause of fibrosis remain unknown. Possible causes of idiopathic fibrosis discussed today include persistent tissue injury or inflammation, impaired tissue regeneration or repair, distorted proteostasis (e.g., during aging), or autoimmunity (4). ILD caused by autoimmunity is well known in connective tissue disease (CTD) (5), but the involvement of autoimmunity in IPF has also been discussed, owing to the presence of circulating immune complexes (6). However, definitive evidence remains limited, owing to a lack of specific diagnostic tests. Recent experimental evidence shows that autoimmunity to a lung-specific autoantigen can drive pulmonary fibrosis (7), suggesting that the presence of other unidentified autoantigens may drive IPF. Furthermore, impaired regeneration and subsequent fibrosis upon injury are associated with dysregulated developmental pathways, such as the Wnt, bone morphogenetic protein/transforming growth factor-β, or sonic hedgehog (Shh) signaling pathways (8–11). The interactions of secreted morphogens of these pathways with the ECM affect its function as an “instructing niche” (12), which has motivated the recently growing interest in ECM structure and function (13).
The tissue- and disease-specific composition of the ECM proteome (matrisome) in vivo, as well as its specific architecture and dynamic association with secreted proteins, is still largely unexplored because of challenging technical limitations. We recently developed a quantitative detergent solubility profiling (QDSP) method that greatly improved the in-depth analysis of tissue proteomes and matrisomes (14). In the present study, we used the QDSP method to characterize human tissue proteomes from lung and skin fibrosis to identify common and distinct molecular alterations in cases of ILD and morphea. We provide a comprehensive resource of protein regulation in human tissue fibrosis and describe a surprisingly high prevalence of marginal zone B- and B1-cell–specific protein (MZB1)-positive plasma B cells in IPF.
Methods
Human Patient Material
Resected human lung tissue and explant material was obtained from the bioarchive at the Comprehensive Pneumology Center in Munich. Biopsies were obtained from 10 healthy donors and 45 patients with end-stage ILD (see Tables E3 and E7 in the online supplement for clinical baseline characteristics). Segments of the resected fresh frozen lung tissue that were histologically characterized with fibrosis were used for the proteomic analysis. All participants gave written informed consent, and the study was approved by the local ethics committee of Ludwig-Maximilians University of Munich, Germany (333-10). Skin biopsies were taken from six patients (three females, three males; mean age, 66 yr) with localized scleroderma (morphea). From each patient, one biopsy was obtained from an involved area that was clinically characterized by sclerosis and inflammation, and another one was taken from a distant, clinically uninvolved site. Samples were immediately snap frozen in liquid nitrogen. The study was approved by the local ethics committee at University Hospital of Cologne, Germany (08144). Human lung tissue–derived proteins for the University of Colorado at Denver cohort were obtained from the National Jewish Health Interstitial Lung Disease Program, including IPF (n = 4) and nonfibrotic control (n = 5) samples. Control tissue was obtained from transplant specimens that failed regional lung selection (at National Jewish Health). The diagnosis of IPF was determined by a pathology core consisting of two pulmonary pathologists, a radiology core consisting of three pulmonary radiologists, and a clinical core consisting of five pulmonary physicians. All diagnoses were made in accordance with established criteria. The institutional review board at National Jewish Health approved the collection and use of tissue.
Results
QDSP of Human Fibrotic Lung and Skin
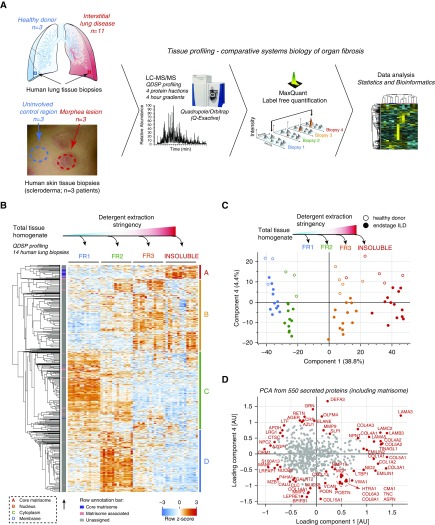
We used mass spectrometry to analyze human tissue fibrosis biopsies. Segments of the resected lung and skin tissue were histologically analyzed to confirm fibrosis in this region and then used for the proteomic analysis. From each sample, the proteins were extracted with increasing stringency into four fractions by changing the detergent and buffer conditions as described in the QDSP protocol (14). We then subjected each protein fraction individually to our shotgun proteomic analysis pipeline using a 4-hour gradient measurement on a Quadrupole-Orbitrap mass spectrometer (Q Exactive; Thermo Scientific, Waltham, MA) and subsequently to label-free protein quantification and data analysis with the MaxQuant (15) and Perseus (16) software packages, as well as custom-built analysis scripts (Figure 1A). We quantified 7,907 proteins in the ILD analysis (Table E1) and 5,826 proteins in the analysis of the morphea biopsies (Table E2). The QDSP method adds an additional dimension to the tissue proteome by separating proteins by their detergent solubility. As expected, we observed a significant separation of cytoplasmic, membrane, nuclear, and ECM proteins, with ECM proteins being most insoluble (Figure 1B). This analysis is particularly interesting for secreted proteins, which might stay soluble upon secretion, or they may become incorporated into the ECM, which renders them insoluble. We used UniProt keywords and the Matrisome annotations (17) to identify 550 proteins in our dataset that were previously annotated to be secreted by cells, and we performed a principal component analysis (PCA) with this subset of the data. This analysis efficiently separated the four protein fractions in component 1, which accounted for 38.8% of the data variability, and separated healthy donor controls from end-stage ILD in component 4, which accounted for 4.4% of the data variability (Figure 1C). Principal component 4 was significantly enriched for the gene categories “antimicrobial” and “innate immunity,” which were higher in the healthy controls, and “proteoglycans” and “extracellular matrix,” which were higher in the ILD proteomes. A scatterplot of the loadings of the PCA revealed the position of individual proteins in the data space (Figure 1D).
Figure 1.
Quantitative detergent solubility profiling (QDSP) of human lung and skin fibrosis. (A) Experimental design (graphs adapted from Reference 14). (B) Proteins in the four indicated QDSP detergent solubility fractions were quantified individually, and the z-score of relative protein mass spectrometric (MS) intensities across 14 interstitial lung disease (ILD) proteomes and the four protein fractions was used for unsupervised hierarchical clustering (using Pearson correlation of rows). Clusters A–D were significantly enriched for the indicated gene categories. Core matrisome and matrisome-associated proteins are assigned to the cluster by the indicated color code. (C) Principal component analysis (PCA) of the relative MS intensities of 550 secreted proteins was used to separate the four QDSP protein fractions (indicated by the color code) in component 1 and end-stage ILD tissues (closed circles) from healthy donor lungs (open circles) in component 4. (D) Scatterplot depicting the protein loadings used for the PCA in (C). AU = arbitrary units; FR = fraction; LC-MS/MS = liquid chromatography–tandem mass spectrometry.
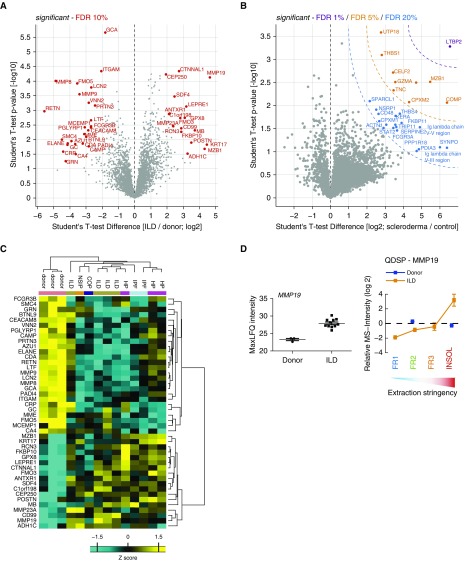
We next determined the total abundance of proteins in the tissue biopsies by summing up the mass spectrometric intensities of the four individual protein fractions. We performed a t test to compare ILD and donor lung tissue proteomes (Figure 2A), as well as the skin lesions from patients with localized scleroderma with the respective healthy skin from the same patient (Figure 2B). To identify common factors in different forms of ILD, we began our proteomic investigation with a heterogeneous group of patients (see Table E3 for clinical baseline characteristics). Regardless of the expected heterogeneity of the patient biopsies, we observed significant alterations in both ILD and localized scleroderma compared with the respective controls. At a false discovery rate (FDR) of 10%, 44 proteins were regulated in the ILD cohort (Figure 2A). Hierarchical clustering analysis (Pearson correlation) of these 44 proteins sorted patients by diagnostic classes (Figure 2C). The most significant common factor in all forms of ILD analyzed was matrix metalloproteinase 19 (MMP19), which was previously shown to be upregulated in pulmonary fibrosis in both mice and humans (18). MMP19 was enriched mostly in the detergent-insoluble fraction, indicating its association with the ECM upon secretion (Figure 2D). We also found common upregulation of the collagen chaperone FKBP10 that we previously identified to be upregulated in the bleomycin model of lung fibrosis and IPF (19). Furthermore, we also observed increased expression of the prolyl 3-hydroxylase 1 protein (LEPRE1), which is involved in collagen hydroxylation (20, 21) and thus may serve the increased production of collagen in fibrotic tissue. We confirmed the upregulation of KRT17 and SDF4 using Western blot analysis of IPF samples from an independent U.S. cohort (Figures E1A and E1B).
Figure 2.
Total tissue protein abundance changes. The depicted volcano plots show significantly altered proteins relative to controls at the indicated false discovery rates (FDRs) in (A) lung biopsy samples from patients with interstitial lung disease (ILD) and (B) fibrotic skin lesions from patients with localized scleroderma. (C) Hierarchical clustering (Pearson correlation of z-score) of the 44 significant proteins in (A). Gene names and clinical classification of patients are shown. (D) In the left panel, the summed mass spectrometric (MS) intensities of matrix metalloproteinase 19 (MMP19) (total tissue protein abundance) are shown on log2 scale for donor and ILD samples. In the right panel, the relative MS intensities across detergent solubility fractions of MMP19 are shown. The mean and standard error of the mean is shown. COP = cryptogenic organizing pneumonia; FR = fraction; HP = hypersensitivity pneumonitis; INSOL = insoluble; IPF = idiopathic pulmonary fibrosis; LFQ = label-free protein quantification; NSIP = nonspecific interstitial pneumonia; QDSP = quantitative detergent solubility profiling.
Finally, we compared our ILD proteomic dataset with the currently largest (to our knowledge) available transcriptomic dataset of human ILD (n = 194) and control tissue (n = 91) (Gene Expression Omnibus dataset GSE47460), published by the Lung Tissue Research Consortium, and we identified many proteins that are regulated on both the RNA and proteome levels, including the proteins KRT17 and MZB1 (Figure E1C). In this comparison, the Pearson correlation of protein and mRNA copy numbers was weak (Figures E1D and E1E), confirming the known fact that protein and mRNA abundances do not always correlate well, even in matched samples. Of note, some of the proteins we identified as upregulated in ILD by mass spectrometry, such as LEPRE1 and MMP19, were not found to be increased in total ILD tissue mRNA abundance by microarray analysis (Figure E1C).
In localized scleroderma, 1 protein (LTBP2) was detected at less than 1% FDR, 10 proteins were detected at less than 5% FDR, and a total of 27 proteins were detected at less than 20% FDR (Figure 2B). One of the most upregulated proteins in the fibrotic skin lesions was the cartilage oligomeric matrix protein (Figure 2B), which we previously showed to be increased in skin fibrosis to regulate dermal collagen ultrastructure (22, 23) and collagen secretion (24). The analysis identified several interesting proteins that are not well studied in the context of fibrosis, including LTBP2 and CPXM2. Interestingly, LTBP2 was recently shown to bind basic fibroblast growth factor FGF-2 in hypertrophic scars, thereby blocking cell proliferation (25, 26). In summary, we provide a comprehensive biochemical characterization of the ECM proteome in human lung and skin fibrosis, and we identify previously known as well as novel alterations in protein abundance.
Molecular Heterogeneity of ILD Tissue Proteomes
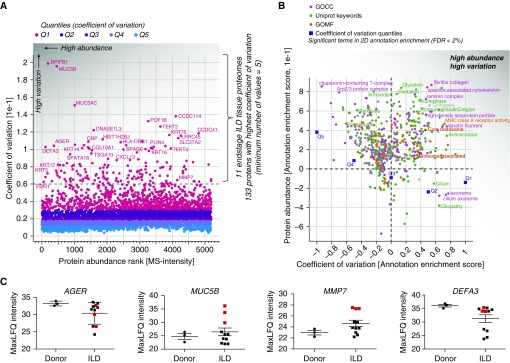
ILD pathophysiology can be highly heterogeneous. Thus, it was conceivable that we would encounter wide variability between patients, even though all 11 lung biopsies were taken from diseased areas that underwent fibrotic remodeling and showed uniform upregulation of fibrosis markers such as MMP19 (Figure 2). To identify the proteins with highest differences between ILD samples, we calculated the coefficient of variation (CV) for each protein and plotted it against the protein abundance rank (Figure 3A). We identified 133 proteins with a high CV between patient samples, which were quantified in at least 5 of 11 ILD biopsies (Figure 3A). To reveal gene categories that show high variation between patients, we performed two-dimensional annotation enrichment analyses (27) for protein abundance ranks versus CV ranks. We also calculated the enrichment score of the CV quantiles, showing that there is a mild increase in data dispersion with decreasing abundance (Figure 3B and Table E4). Interestingly, the upper 20% quantile (Q1) with highest CV breaks the trend and shows a slightly higher abundance rank than Q2. This indicates that many highly abundant proteins also showed high data dispersion. In the upper right quadrant of the two-dimensional annotation enrichment plot, the gene categories enriched within highly abundant proteins with high CV (including ECM proteins, antibodies, and antimicrobial peptides) are depicted (Figure 3B).
Figure 3.
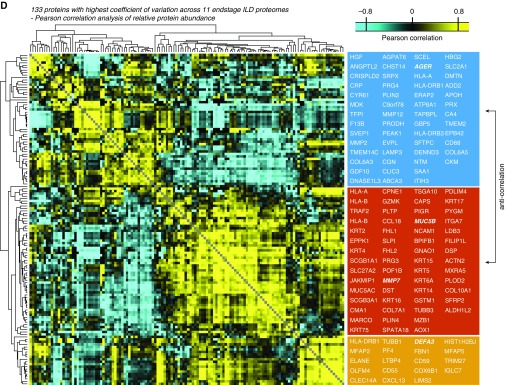
Identification of proteins and gene categories with high variance in interstitial lung disease (ILD) proteomes. (A) Scatterplot depicting proteins sorted from highest to lowest abundance (mass spectrometric [MS] intensity normalized by number of theoretical tryptic peptides by intensity-based absolute quantification) and their coefficients of variation (CVs) across the ILD tissue proteomes. The indicated color code shows the five quantiles of CVs. The dashed line shows the chosen cutoff resulting in 133 proteins with high CV used for analysis in D. (B) Scatterplot depicting significantly enriched gene categories along two dimensions, with annotation enrichment scores (−1 to +1) for both CV and protein abundance. The indicated gene categories in the upper right quadrant are highly abundant while nevertheless having high CVs. (C) Normalized MS intensities of the indicated proteins are shown on log2 scale. (D) Correlogram visualizing the color-coded Pearson correlations of 133 high-CV proteins across the ILD proteomes. Gene names within the three main clusters of proteins are indicated in the boxes. FDR = false discovery rate; GOCC = Gene Ontology Cellular Compartment; GOMF = Gene Ontology Molecular Function; LFQ = label-free protein quantification; Q = quantile.
Interestingly, many of the high CV proteins are cell type–specific genes, such as BPIFB1, mucin 5B (MUC5B; goblet cells), AGER (type 1 pneumocytes), KRT5, or KRT14 (basal cells), indicating that we do observe differences in cellular composition between samples. The average levels of AGER were only mildly reduced in ILD samples compared with donor lungs; however, a few patient samples showed drastic changes with at least 60-fold reduced protein levels, explaining its high CV. Interestingly, a subset of three ILD patient biopsies showed at least 60-fold increased levels of MUC5B, which is normally expressed by goblet cells in the bronchi. The same three patient samples were strongly enriched for matrilysin (MMP7), which was shown to be a key regulator of pulmonary fibrosis in mice and humans (28) and one of the most upregulated genes in microarray studies of IPF (29). Interestingly, these samples did not display significantly different levels of neutrophil defensin 3 (DEFA3), which was shown to be a marker of acute exacerbations of IPF (29), compared with donor lungs (Figure 3C).
We next determined the Pearson correlation coefficients between the mass spectrometric intensity profiles of the 133 proteins (Table E5) and group proteins by similarity. Unsupervised hierarchical clustering (Pearson correlation) of these correlation coefficients revealed three main groups of proteins, which were anticorrelated (Figure 3C). In group 1, we detected markers for type 1 (AGER) and type 2 (SFTPC) pneumocytes, whereas in group 2, we found markers of lung fibrosis, such as MMP7 (28), as well as the basal stem cell markers KRT5 and KRT14 (30). The third distinct group showed higher correlation with group 2 than with group 1 and contained proteins with functions in innate immune defense (DEFA3, ELANE), as well as immunoregulatory proteins (CXCL13) (Figure 3D).
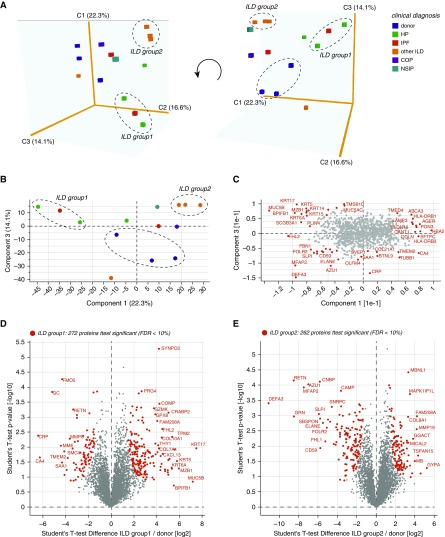
To visualize patient heterogeneity, we used PCA of 1,037 proteins representing the upper 20% quantile (Q1) of the CV (Figure 4A), and we selected two ILD subgroups that were characterized by a distinct protein profile compared with healthy donor controls (Figures 4B and 4C). Clinically, ILD group 1 (one IPF and two hypersensitivity pneumonitis) had a lower DlCO than group 2 (three unclassifiable ILDs) (see Table E3). To determine which proteins in these patient subsets were significantly different from healthy donor controls, we used a two-sided t test, which produced 272 significantly regulated proteins (FDR, <10%) in group 1 and 262 significantly regulated proteins (FDR, <10%) in group 2 (Figures 4D and 4E and Table E1). In summary, the application of unsupervised exploratory statistics on the proteomic data uncovered correlated groups of proteins and enabled the stratification of patients with ILD, revealing patient groups with distinct protein composition.
Figure 4.
Distinct molecular signatures of subsets of patients with interstitial lung disease (ILD). (A) Principal component analysis (PCA) of 5,707 quantified proteins was used to separate the 14 human lung tissue proteomes. A three-dimensional visualization of components 1–3 enabled separation of patient subsets and healthy donor controls as indicated. Clinical diagnosis is color coded as indicated. Two distinct ILD subsets and the donor controls are labeled with dashed ovals. (B) Two-dimensional PCA plot of components 1 and 3. Two distinct ILD subsets and the donor controls are labeled with dashed ovals. (C) The loadings of the PCA in (B) are shown. Proteins with the highest loadings are labeled with their gene names. (D) Volcano plot depicting 272 significantly regulated proteins (false discovery rate [FDR], <10%) in ILD subset 1 (indicated in A and B) compared with healthy donor controls. (E) Volcano plot depicting 262 significantly regulated proteins (FDR, <10%) in ILD subset 2 (indicated in A and B) compared with healthy donor controls. COP = cryptogenic organizing pneumonia; HP = hypersenstivity pneumonitis; IPF = idiopathic pulmonary fibrosis; NSIP = nonspecific interstitial pneumonia.
Enrichment of MZB1-Positive Tissue-Resident Plasma B Cells Is a Highly Prevalent Feature of Lung and Skin Fibrosis
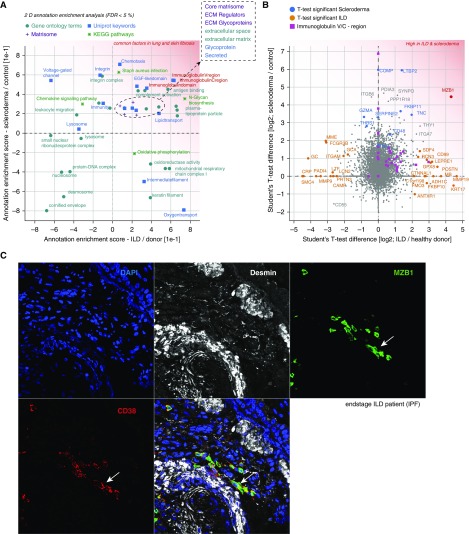
We matched the two proteomic datasets and compared fold changes in ILD and localized scleroderma biopsies. To reveal common and distinct gene categories in lung and skin fibrosis, we first performed two-dimensional annotation enrichment analysis (27) (Table E6). We observed common upregulation of ECM genes, complement activation, N-glycan biosynthesis, and plasma lipoprotein particles, and most significantly, we found a common increase in the abundance of antibodies (Figures 5A and 5B). Comparing the protein outliers with the highest fold changes, we identified a number of interesting differences as well as similarities between both datasets. For instance, the ECM protein tenascin C, which is known to be increased in IPF (31), was upregulated in both datasets. Surprisingly, the most significant similarity with highest fold changes in both lung and skin fibrosis was an upregulation of MZB1 (Figure 5B), which are known to be expressed in certain B-cell subsets to diversify peripheral B-cell functions by regulating Ca2+ stores, antibody secretion, and integrin activation (32). We validated this finding by staining MZB1 in tissue sections from patients with ILD and patients with scleroderma (Figures 5C and E3–E7). MZB1 localized to cells with a considerable volume of cytoplasm that were found in higher numbers in fibrotic tissue than in controls and were localized typically in perivascular regions.
Figure 5.
High prevalence of marginal zone B- and B1-cell–specific protein (MZB1)-positive tissue-resident plasma B cells is a common feature in human lung and skin fibrosis. (A) Scatterplot depicting significantly enriched gene categories along two dimensions, with annotation enrichment scores (−1 to +1) for both lung fibrosis (interstitial lung disease [ILD]/donor) and skin fibrosis (scleroderma/control). The indicated gene categories in the upper right quadrant are common factors in ILD and localized scleroderma. (B) Scatterplot showing the t test difference in human skin and lung fibrosis, respectively. Proteins found to be significant (by t test) in the ILD comparison (orange) and the scleroderma comparison (blue) are color coded. MZB1 protein was significant in both comparisons. Text in red in A and B shows proteins that are up-regulated in both ILD and scleroderma. (C) Representative confocal image of four-color immunostainings with antibodies to the indicated proteins in a formalin-fixed, paraffin-embedded tissue section of a patient with ILD. Arrows indicate MZB1/CD38 double-positive cells. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), and desmin stained vascular smooth muscle cells and some mesenchymal cells in fibrotic tissues. ECM = extracellular matrix; FDR = false discovery rate; IPF = idiopathic pulmonary fibrosis; KEGG = Kyoto Encyclopedia of Genes and Genomes.
MZB1 is an endoplasmic reticulum (ER)-resident protein that is important for antibody secretion and is thus upregulated in cells with high antibody secretory activity (33). We confirmed this finding by treating human peripheral blood mononuclear cells with IL-2 and the Toll-like receptor 7/8 ligand R848, which induces differentiation of memory B cells to immunoglobulin-secreting cells (34), and analyzing MZB1 expression. IL-2/R848 treatment induced IgG production and expression of BLIMP1, a transcription factor essential for plasma cell function (35). Indeed, expression of MZB1 on both transcript and protein levels was drastically increased under these conditions (Figure E2), confirming its specific expression in antibody-secreting cells.
To establish MZB1 as a marker for antibody-secreting plasma B cells in human ILD tissue, we performed coimmunostaining with several lineage markers. The MZB1-positive cells were negative for the T-lymphocyte lineage marker CD3 (Figures E3A and E6A), and they were also completely negative for the B-cell lineage marker CD20 (Figures E3B and E6B) and the leukocyte lineage marker CD45 (Figures E4A and E7A). Coexpression of MZB1 with CD38 (Figure 5C), CD138 (Figure E4B), and CD27 (Figure E5A) clearly identified the MZB1-positive cells as terminally differentiated plasma B cells in ILD tissues (36). Similarly, we also found MZB1+/CD38+ double-positive cells that were CD20−/CD45− double negative (Figures E6 and E7) in the skin. Furthermore, consistent with the notion that we identified tissue-resident plasma B cells, we also found positive staining of MZB1-positive cells with an antibody against human IgG (Figure E5B).
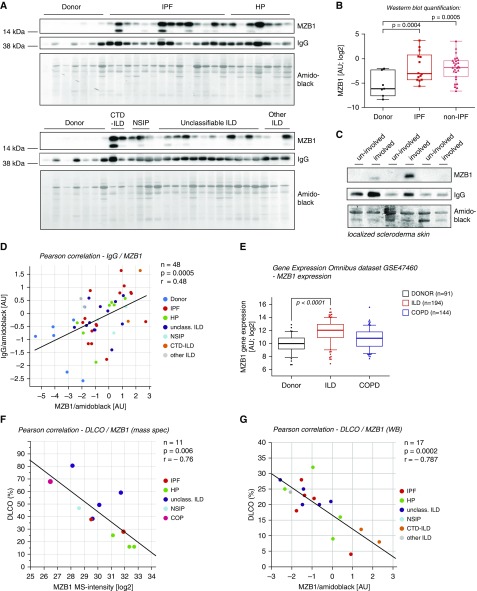
Finally, to increase the overall number of samples and validate our findings in an independent cohort, we performed Western blot analysis of 34 additional ILD tissues (IPF, n = 14; HP, n = 7; CTD-ILD, n = 2; non-specific interstitial pneumonia, n = 3; unclassifiable ILD, n = 12; other ILD, n = 3) and 7 healthy donor controls (see Table E7 for clinical baseline characteristics). We found that MZB1 was significantly increased in both IPF and non-IPF ILD tissue compared with healthy donor tissue (Figures 6A and 6B). We also reconfirmed our findings by Western blotting of MZB1 protein in three additional samples from patients with localized scleroderma (Figure 6C). Importantly, the quantification of total tissue IgG and MZB1 levels showed a significant positive correlation, again indicating that MZB1 amounts are predictive for local antibody secretion (Figure 6D). Increased abundance of MZB1 transcripts in ILD compared with healthy donor controls and chronic obstructive pulmonary disease cases were also found in an independent large U.S. cohort microarray study (Gene Expression Omnibus dataset GSE47460) (Figure 6E). Of note, the same dataset shows increased abundance of CD38 in ILD tissues (Figure E1), thus providing an independent confirmation for the prevalence of plasma B cells. MZB1 levels were independent of age, vital capacity, sex, and treatment with steroids or antifibrotics (Figure E8), but they showed a significant negative correlation with DlCO (%) in both cohorts analyzed (Figures 6F and 6G). In summary, the unbiased proteomic analysis of human lung and skin fibrosis uncovered a surprising prevalence of MZB1+/CD38+/CD138+/CD27+/CD20−/CD45− plasma B cells, which is an indication of a common involvement of antibody-mediated autoimmunity in idiopathic organ fibrosis.
Figure 6.
The number of marginal zone B- and B1-cell–specific protein (MZB1)-positive cells correlates with tissue IgG and predicts lower diffusing capacity of the lung for carbon monoxide (DlCO). (A) Tissue homogenates of the indicated groups were subjected to Western blot analysis with antibodies against MZB1 and human IgG. Blots were stained with amido black for quantification of total protein loading. (B) Box-and-whisker plot showing densitometric quantification of the MZB1 bands in the Western blot shown in A, which were normalized to amido black staining. The box-and-whisker plot shows the median, lower and upper quartiles, and the maximum range of values. (C) Tissue homogenates of the indicated groups were subjected to Western blot analysis with antibodies against MZB1 and human IgG. Blots were stained with amido black for quantification of total protein loading. (D, F, G) Linear regression analyses. Clinical classifications are color coded as indicated, and the P values of the linear regressions and the Pearson correlation coefficients (r) are shown. (D) Positive correlation of IgG and MZB1 levels (normalized to total protein analyzed using amido black quantification). (E) Quantification of MZB1 tissue abundance by microarray in a large U.S. cohort (Gene Expression Omnibus dataset GSE47460 published by the Lung Tissue Research Consortium). The box-and-whisker plot shows the median, lower and upper quartiles, and the maximum range of values. The outliers show the 2nd and the 98th percentiles. (F) Negative correlation of DlCO (%) and MZB1 levels (quantified by mass spectrometry [mass spec]). (G) Negative correlation of DlCO (%) and MZB1 levels (normalized to total protein analyzed using amido black quantification). AU = arbitrary units; COPD = chronic obstructive pulmonary disease; CTD = connective tissue disease; HP = hypersensitivity pneumonitis; ILD = interstitial lung disease; IPF = idiopathic pulmonary fibrosis; NSIP = nonspecific interstitial pneumonia; unclass. ILD = unclassifiable ILD; WB = Western blot.
Discussion
Mass spectrometry–driven proteomics has evolved into a highly sensitive and accurate technology that enables the precise quantification of thousands of proteins at once (37). In this study, we used the recently developed QDSP method to analyze tissue biopsies of human lung and skin fibrosis at a depth of more than 7,900 proteins quantified. We provide the most comprehensive proteomic resource of human tissue fibrosis, containing information about the abundance, stoichiometry, and detergent solubility of proteins, as well as the first cross-organ comparison of tissue fibrosis. Profiling lung biopsies from a heterogeneous cohort of human ILD together with skin biopsies from patients with localized scleroderma (morphea) enabled the identification of common and distinct protein regulation in various forms and stages of fibrotic remodeling.
Proteomic analysis of tissue composition is particularly powerful for secreted proteins, whose protein abundance very often does not correlate with total tissue mRNA quantification in RNA sequencing assays (14). Thus, our data represents an essential addition to existing transcriptomic studies of human lung and skin fibrosis. Furthermore, the QDSP method captures the interactions of morphogens and other secreted proteins with the ECM in an unbiased way, revealing those that are bound to the matrix by their decreased detergent solubility. Thus, we added an additional dimension to the human lung and skin proteome, which for the first time revealed the association of secreted proteins with the ECM.
The use of unsupervised statistical tests such as PCA clearly showed the high degree of molecular heterogeneity between samples. This was no surprise, because we intentionally selected a diverse patient cohort to screen for molecular events that are commonly present in all forms of fibrosis. We made use of patient heterogeneity by grouping proteins with high abundance variation across samples by their Pearson correlation. The correlation of proteins in this analysis can be explained by, for instance, their cell type–specific expression and thus their capacity to report differences in the relative amount of cell types in the respective biopsies. Along these lines, the recent development of high-throughput technologies in the field of single-cell mRNA sequencing (38) will likely enable future attempts to study cellular heterogeneity in chronic lung disease in great detail. Given the high amount of biological variation and heterogeneity of cellular composition observed in our ILD cohort, it is remarkable that we identified a substantial number of common factors that were increased in all samples.
The most significant common factor across ILD and scleroderma samples was the protein MZB1, which we localized to CD38+/CD138+/CD27+/CD20−/CD45− B cells. In respiratory immunity, B cells can be recruited to tertiary lymphoid organs around the bronchi, where they are organized in B-cell follicles (39, 40). In our analysis, MZB1-positive cells were found to be quite dispersed in the tissue and not necessarily associated with tertiary lymphoid structures, but with predominant perivascular abundance. MZB1 has important functions in the ER of B cells that undergo ER stress upon high antibody secretory activity (32, 33). In our immunostaining experiments, we observed MZB1-high and MZB1-low cells, indicating that the expression level is tightly regulated in B cells. MZB1-high cells had a large cytoplasm and were positive for a comprehensive panel of known mature plasma B-cell markers, which clearly identifies them as terminally differentiated antibody-producing tissue-resident plasma cells. Because most of the samples in our study were biopsies from idiopathic forms of ILD, we believe that this observation warrants future investigation regarding the causative role of antibody-mediated autoimmunity in idiopathic cases of organ fibrosis. Of note, it has been recognized that many patients with idiopathic interstitial pneumonia have clinical features that suggest an underlying autoimmune process but do not meet established criteria for a CTD. To address this problem, a European Respiratory Society/American Thoracic Society task force recently proposed the term “interstitial pneumonia with autoimmune features” and offered several classification criteria (41).
Circulating autoantibodies in IPF were first described long time ago (6), and a causative role for B-cell–mediated autoimmunity for idiopathic ILD has been discussed (42, 43). In localized scleroderma, the role of autoantibodies is unclear, but the histology of the fibrotic reaction, involving a strong inflammatory infiltrate around the blood vessels, is identical to the lesions found in patients with systemic scleroderma, who all have circulating autoantibodies (44). A recent study demonstrated that autoantibodies against the lung-specific protein BPIFB1 occur in 12% of patients with idiopathic ILD (7). Importantly, the authors of this landmark study also demonstrated that T cells specific for a single autoantigen (Bpifb1) are sufficient to induce full-blown and irreversible lung fibrosis in mice (7). It is thus conceivable that (1) the presence of autoantibodies and autoreactive T cells against unknown antigens may cause or at least perpetuate many if not most idiopathic interstitial pneumonias and (2) the identification of these unknown autoantigens in patient plasma may serve as a powerful tool for both patient stratification and future immunotherapy-based approaches to treatment of ILD. The recent use of chimeric antigen receptor T cells specific for autoantigen-producing B cells for targeted therapy of autoimmune disease (45) introduces an exciting new avenue for eliminating autoreactive B-cell clones while maintaining protective adaptive immunity. Such future therapeutic approaches will depend on the identification of disease-specific autoantigens and appropriate preclinical models to test if indeed certain antigens have causative roles in idiopathic forms of organ fibrosis.
Acknowledgments
Acknowledgment
The authors thank Silvia Weidner for technical assistance and Christoph Schaab for providing data analysis scripts. The authors also thank Gabi Sowa, Igor Paron, and Korbinian Mayr for expert support of the proteomic pipeline. The authors also thank Britta Peschel and Marion Frankenberger for support with bioarchive material.
Footnotes
Supported by the German Center for Lung Research (DZL), the Max Planck Society for the Advancement of Science, German Research Foundation (DFG) grant KR558 (T.K.), and the Helmholtz Association.
Author Contributions: H.B.S. and O.E.: initiated, conceptualized, and designed the study; H.B.S.: acquired and analyzed the proteomic data and wrote the paper; C.H.M., G.L., M.S., and C.S.-W.: performed immunofluorescence and immunoblot analysis of tissue samples; S.P.: performed MZB1 analysis on plasma cell differentiation; G.L.: evaluated clinical patient data; and B.E., P.M., T.K., R.A.H., J.B., D.A.S., and M.M.: provided patient material and/or important analytical tools. All authors read and approved the final version of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201611-2263OC on June 27, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech. 2011;4:165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez IE, Eickelberg O. New cellular and molecular mechanisms of lung injury and fibrosis in idiopathic pulmonary fibrosis. Lancet. 2012;380:680–688. doi: 10.1016/S0140-6736(12)61144-1. [DOI] [PubMed] [Google Scholar]
- 3.Kreuter A, Krieg T, Worm M, Wenzel J, Moinzadeh P, Kuhn A, Aberer E, Scharffetter-Kochanek K, Horneff G, Reil E, et al. German guidelines for the diagnosis and therapy of localized scleroderma. J Dtsch Dermatol Ges. 2016;14:199–216. doi: 10.1111/ddg.12724. [DOI] [PubMed] [Google Scholar]
- 4.Thannickal VJ, Zhou Y, Gaggar A, Duncan SR. Fibrosis: ultimate and proximate causes. J Clin Invest. 2014;124:4673–4677. doi: 10.1172/JCI74368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vij R, Strek ME. Diagnosis and treatment of connective tissue disease-associated interstitial lung disease. Chest. 2013;143:814–824. doi: 10.1378/chest.12-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dreisin RB, Schwarz MI, Theofilopoulos AN, Stanford RE. Circulating immune complexes in the idiopathic interstitial pneumonias. N Engl J Med. 1978;298:353–357. doi: 10.1056/NEJM197802162980701. [DOI] [PubMed] [Google Scholar]
- 7.Shum AK, Alimohammadi M, Tan CL, Cheng MH, Metzger TC, Law CS, Lwin W, Perheentupa J, Bour-Jordan H, Carel JC, et al. BPIFB1 is a lung-specific autoantigen associated with interstitial lung disease. Sci Transl Med. 2013;5:206ra139. doi: 10.1126/scitranslmed.3006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng T, Frank DB, Kadzik RS, Morley MP, Rathi KS, Wang T, Zhou S, Cheng L, Lu MM, Morrisey EE. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Königshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, et al. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. J Clin Invest. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JH, Bhang DH, Beede A, Huang TL, Stripp BR, Bloch KD, Wagers AJ, Tseng YH, Ryeom S, Kim CF. Lung stem cell differentiation in mice directed by endothelial cells via a BMP4-NFATc1-thrombospondin-1 axis. Cell. 2014;156:440–455. doi: 10.1016/j.cell.2013.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martino MM, Briquez PS, Güç E, Tortelli F, Kilarski WW, Metzger S, Rice JJ, Kuhn GA, Müller R, Swartz MA, et al. Growth factors engineered for super-affinity to the extracellular matrix enhance tissue healing. Science. 2014;343:885–888. doi: 10.1126/science.1247663. [DOI] [PubMed] [Google Scholar]
- 13.Hynes RO. Stretching the boundaries of extracellular matrix research. Nat Rev Mol Cell Biol. 2014;15:761–763. doi: 10.1038/nrm3908. [DOI] [PubMed] [Google Scholar]
- 14.Schiller HB, Fernandez IE, Burgstaller G, Schaab C, Scheltema RA, Schwarzmayr T, Strom TM, Eickelberg O, Mann M. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol Syst Biol. 2015;11:819. doi: 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 16.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 17.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, Hynes RO. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11:M111.014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu G, Kovkarova-Naumovski E, Jara P, Parwani A, Kass D, Ruiz V, Lopez-Otín C, Rosas IO, Gibson KF, Cabrera S, et al. Matrix metalloproteinase-19 is a key regulator of lung fibrosis in mice and humans. Am J Respir Crit Care Med. 2012;186:752–762. doi: 10.1164/rccm.201202-0302OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staab-Weijnitz CA, Fernandez IE, Knüppel L, Maul J, Heinzelmann K, Juan-Guardela BM, Hennen E, Preissler G, Winter H, Neurohr C, et al. FK506-binding protein 10, a potential novel drug target for idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2015;192:455–467. doi: 10.1164/rccm.201412-2233OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willaert A, Malfait F, Symoens S, Gevaert K, Kayserili H, Megarbane A, Mortier G, Leroy JG, Coucke PJ, De Paepe A. Recessive osteogenesis imperfecta caused by LEPRE1 mutations: clinical documentation and identification of the splice form responsible for prolyl 3-hydroxylation. J Med Genet. 2009;46:233–241. doi: 10.1136/jmg.2008.062729. [DOI] [PubMed] [Google Scholar]
- 21.Vranka JA, Sakai LY, Bächinger HP. Prolyl 3-hydroxylase 1, enzyme characterization and identification of a novel family of enzymes. J Biol Chem. 2004;279:23615–23621. doi: 10.1074/jbc.M312807200. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal P, Zwolanek D, Keene DR, Schulz JN, Blumbach K, Heinegård D, Zaucke F, Paulsson M, Krieg T, Koch M, et al. Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J Biol Chem. 2012;287:22549–22559. doi: 10.1074/jbc.M111.335935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal P, Schulz JN, Blumbach K, Andreasson K, Heinegård D, Paulsson M, Mauch C, Eming SA, Eckes B, Krieg T. Enhanced deposition of cartilage oligomeric matrix protein is a common feature in fibrotic skin pathologies. Matrix Biol. 2013;32:325–331. doi: 10.1016/j.matbio.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Schulz JN, Nüchel J, Niehoff A, Bloch W, Schönborn K, Hayashi S, Kamper M, Brinckmann J, Plomann M, Paulsson M, et al. COMP-assisted collagen secretion – a novel intracellular function required for fibrosis. J Cell Sci. 2016;129:706–716. doi: 10.1242/jcs.180216. [DOI] [PubMed] [Google Scholar]
- 25.Menz C, Parsi MK, Adams JR, Sideek MA, Kopecki Z, Cowin AJ, Gibson MA. LTBP-2 has a single high-affinity binding site for FGF-2 and blocks FGF-2-induced cell proliferation. PLoS One. 2015;10:e0135577. doi: 10.1371/journal.pone.0135577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sideek MA, Teia A, Kopecki Z, Cowin AJ, Gibson MA. Co-localization of LTBP-2 with FGF-2 in fibrotic human keloid and hypertrophic scar. J Mol Histol. 2016;47:35–45. doi: 10.1007/s10735-015-9645-0. [DOI] [PubMed] [Google Scholar]
- 27.Cox J, Mann M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics. 2012;13(Suppl 16):S12. doi: 10.1186/1471-2105-13-S16-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, et al. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci USA. 2002;99:6292–6297. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konishi K, Gibson KF, Lindell KO, Richards TJ, Zhang Y, Dhir R, Bisceglia M, Gilbert S, Yousem SA, Song JW, et al. Gene expression profiles of acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2009;180:167–175. doi: 10.1164/rccm.200810-1596OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smirnova NF, Schamberger AC, Nayakanti S, Hatz R, Behr J, Eickelberg O. Detection and quantification of epithelial progenitor cell populations in human healthy and IPF lungs. Respir Res. 2016;17:83. doi: 10.1186/s12931-016-0404-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estany S, Vicens-Zygmunt V, Llatjós R, Montes A, Penín R, Escobar I, Xaubet A, Santos S, Manresa F, Dorca J, et al. Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFβ1. BMC Pulm Med. 2014;14:120. doi: 10.1186/1471-2466-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flach H, Rosenbaum M, Duchniewicz M, Kim S, Zhang SL, Cahalan MD, Mittler G, Grosschedl R. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity. 2010;33:723–735. doi: 10.1016/j.immuni.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbaum M, Andreani V, Kapoor T, Herp S, Flach H, Duchniewicz M, Grosschedl R. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014;28:1165–1178. doi: 10.1101/gad.240762.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tellier J, Shi W, Minnich M, Liao Y, Crawford S, Smyth GK, Kallies A, Busslinger M, Nutt SL. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat Immunol. 2016;17:323–330. doi: 10.1038/ni.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- 37.Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- 38.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.John-Schuster G, Hager K, Conlon TM, Irmler M, Beckers J, Eickelberg O, Yildirim AÖ. Cigarette smoke-induced iBALT mediates macrophage activation in a B cell-dependent manner in COPD. Am J Physiol Lung Cell Mol Physiol. 2014;307:L692–L706. doi: 10.1152/ajplung.00092.2014. [DOI] [PubMed] [Google Scholar]
- 40.Moyron-Quiroz JE, Rangel-Moreno J, Kusser K, Hartson L, Sprague F, Goodrich S, Woodland DL, Lund FE, Randall TD. Role of inducible bronchus associated lymphoid tissue (iBALT) in respiratory immunity. Nat Med. 2004;10:927–934. doi: 10.1038/nm1091. [DOI] [PubMed] [Google Scholar]
- 41.Fischer A, Antoniou KM, Brown KK, Cadranel J, Corte TJ, du Bois RM, Lee JS, Leslie KO, Lynch DA, Matteson EL, et al. ERS/ATS Task Force on Undifferentiated Forms of CTD-ILD. An official European Respiratory Society/American Thoracic Society research statement: interstitial pneumonia with autoimmune features. Eur Respir J. 2015;46:976–987. doi: 10.1183/13993003.00150-2015. [DOI] [PubMed] [Google Scholar]
- 42.Xue J, Kass DJ, Bon J, Vuga L, Tan J, Csizmadia E, Otterbein L, Soejima M, Levesque MC, Gibson KF, et al. Plasma B lymphocyte stimulator and B cell differentiation in idiopathic pulmonary fibrosis patients. J Immunol. 2013;191:2089–2095. doi: 10.4049/jimmunol.1203476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donahoe M, Valentine VG, Chien N, Gibson KF, Raval JS, Saul M, Xue J, Zhang Y, Duncan SR. Autoantibody-targeted treatments for acute exacerbations of idiopathic pulmonary fibrosis. PLoS One. 2015;10:e0127771. doi: 10.1371/journal.pone.0127771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleischmajer R, Nedwich A. Generalized morphea. I. Histology of the dermis and subcutaneous tissue. Arch Dermatol. 1972;106:509–514. doi: 10.1001/archderm.106.4.509. [DOI] [PubMed] [Google Scholar]
- 45.Ellebrecht CT, Bhoj VG, Nace A, Choi EJ, Mao X, Cho MJ, Di Zenzo G, Lanzavecchia A, Seykora JT, Cotsarelis G, et al. Reengineering chimeric antigen receptor T cells for targeted therapy of autoimmune disease. Science. 2016;353:179–184. doi: 10.1126/science.aaf6756. [DOI] [PMC free article] [PubMed] [Google Scholar]