Abstract
The defining feature of the Gram-negative cell envelope is the presence of two cellular membranes with the specialized glycolipid lipopolysaccharide (LPS) exclusively found on the surface of the outer membrane. The surface layer of LPS contributes to the stringent permeability properties of the outer membrane which is particularly resistant to permeation of many toxic compounds, including antibiotics. As a common surface antigen, host immune cells recognize LPS and mount defenses to clear pathogenic organisms. To alter properties of the outer membrane or evade the host immune response, Gram-negative bacteria employ a wide variety of chemical modifications to alter LPS. Here we review key features and physiological consequences of LPS biogenesis and modifications.
Introduction
Molecules at the surface of bacterial cells perform critical roles of interacting with and reacting to the surrounding environment. For Gram-negative bacteria, the surface is composed of an asymmetric outer membrane (OM) with the glycolipid lipopolysaccharide (LPS) exclusively localized to the outer leaflet of the OM1. The asymmetric distribution of LPS on the surface produces a potent barrier that is impermeable to many toxic compounds, including antibiotics2. To maintain the asymmetry and permeability properties of this unique barrier, most Gram-negative bacteria have dedicated pathways for removing glycerophospholipids that are mislocalized to the outer leaflet. In addition, as a major constituent of the OM, LPS is essential in most Gram-negative bacteria (see box 1) and is critical for virulence3.
Box 1: Abandoning Lipid A to become resistant.
Neisseria meningitidis, Moraxella catarrhalis, and Acinetobacter baumannii, have been demonstrated to be viable when lipid A synthesis is disrupted129-131. However, only A. baumannii strains were found to inactivate lipid A synthesis in a selectable manner to provide resistance to cationic AMPs129. Loss of LOS in A. baumannii comes with fitness costs: reduced growth rate, reduced virulence, and sensitivity to many antibiotics132. However, clinical isolates are capable of inactivating lipid A synthesis supporting that this extreme mechanism of resistance could occur in hospitals or patients129. Further, screening for this type of colistin resistance during treatment is needed to determine the clinical relevance. Work on LOS-deficient A. baumannii, have started to unravel how a bacterium can survive when lipid A synthesis is inactivated. Comparing the ability of multiple strains of A. baumannii to inactivate LOS synthesis, it was found that peptidoglycan synthesis is a critical factor; LOS-deficient mutants could be isolated from strains with low protein levels of penicillin binding protein 1A (PBP1A) or with disruption of the encoding gene for PBP1A, ponA, but not from strains expressing higher levels of PBP1A133. Further, LOS-deficient A. baumannii respond by increasing transcription of genes encoding for lipoprotein transport and surface-exposed lipoproteins133,134. These results indicate peptidoglycan and lipoprotein synthesis affect the ability for A. baumannii to survive with an OM consisting of a symmetric glycerophospholipid bilayer. Furthermore, a short-term evolution experiment demonstrated that LOS-deficient bacteria can increase overall fitness through inactivation of genes required for the removal of glycerophospholipids from the outer leaflet of the OM135. Since LOS-deficient A. baumannii need to fill the outer leaflet of the OM with glycerophospholipids, it is logical that enzymes that remove or degrade these lipids in the outer leaflet would be disadvantageous. Specifically, mutations that result in disruption of the Mla transporter, transports mislocalized OM glycerophospholipids back to the IM; disruption of PldA, phospholipase that degrades mislocalized glycerophospholipids; or disruption of both increase growth rate of LOS-deficient A. baumannii strains135. Disruption of Mla and PldA are expected to allow for higher accumulation of glycerophospholipids in the outer leaflet of the OM replacing LOS. Work in A. baumannii has demonstrated that LOS biogenesis is interconnected with biogenesis of other cell envelope components (peptidoglycan, OM lipoproteins, and OM glycerophospholipids) and rewiring these connections is necessary to grow in the absence of LOS. It remains unclear why lipid A synthesis is essential in some organisms and not in others; however, LOS-deficiency in Acinetobacter has provided unique insights into cell envelope biology.
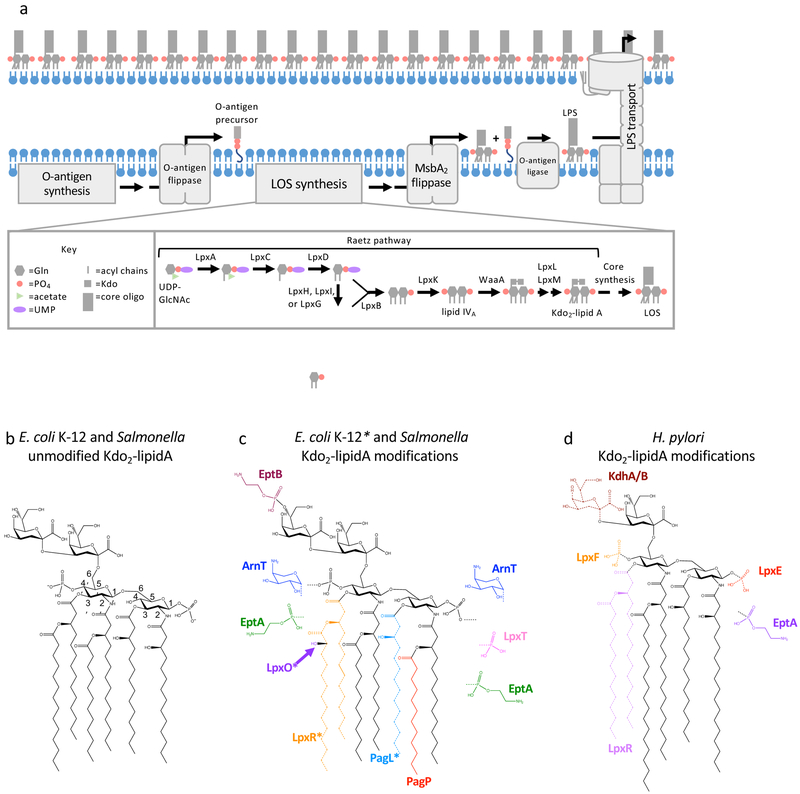
LPS biogenesis begins with synthesis occurring at the cytoplasmic interface of the inner membrane (IM) and then LPS is transported across the IM and to the OM4,5 (Figure 1a). LPS can be divided into three regions: the conserved lipid A anchor, core oligosaccharide, and O antigen. The conserved lipid A unit is a bis-phosphorylated disaccharide of GlcN (glucosamine) typically with 4-7 acyl chains (Figure 1b). LPS synthesis begins with the Raetz pathway (Figure 1a), a series of conserved enzymatic steps that produce lipid A with Kdo (3-deoxy-d-manno-oct-2-ulosonic acid) sugars, the first units of the core oligosaccharide. Briefly, using the model organism Escherichia coli as an example, the Raetz pathway starts with the precursors UDP-GlcNAc (UDP-N-acetylglucosamine) and ACP (acyl carrier protein)-bound fatty acids. The successive actions of LpxA (acyltransferase), LpxC (deacetylase), and LpxD (acyltransferase) produce a UDP-2,3-diacylGlcN3. The first reaction by LpxA is unfavorable and deacetylation by LpxC is the first committed step of synthesis3. A UMP is then cleaved from some of the produced UDP-2,3-diacylGlcN to produce 2,3-diacylGlcN-1-phosphate, also named lipid X3. Lipid X production is performed by one of three non-homologous enzymes: E. coli and most Gram-negative bacteria use LpxH, α-proteobacteria use LpxI3 and Chlamydiae use LpxG6. LpxB then catalyzes formation of a β-1′-6 glycosidic bond between one molecule of lipid X and one molecule of UDP-2,3-diacylGlcN which releases the UDP nucleotide carrier3. The tetra-acylated 1-phosphorylated reaction product is then phosphorylated by LpxK at the 4′-position to produce lipid IVA3. This is followed by the addition of the Kdo sugar(s), the number of which varies in different organisms, by the enzyme WaaA (also known as KdtA)3. Finally, two acyl transferases, LpxL and LpxM, catalyze the transfer of “secondary” acyl chains completing the Kdo-lipid A domain of LPS3. Most bacteria encode homologs of LpxL; however, many bacteria, such as Francisella, do not encode an LpxM homolog and others encode distinct acyl-transferases instead of LpxM: LpxJ in many ε-proteobacteria7 and LpxN in Vibrio cholerae8.
Figure 1. Lipopolysaccharide biogenesis, structure, and modifications.
(a) Overview of LPS biogenesis in E. coli and Salmonella. Briefly, synthesis of the lipid A and core domains of LPS occurs in the cytoplasm and at the cytoplasmic interface of the IM. O antigen, if present, is synthesized separately attached to the carrier lipid, undecaprenyl-pyrophosphate. LOS and O-antigen precursors are flipped across the IM separately by MsbA2 and O-antigen flippases, respectively. O antigen is attached to LOS on the periplasmic side of the IM. Finally, LPS is transported from the IM to the surface of the OM by the Lpt complex. (b) Unmodified Kdo2-lipid A synthesized by E. coli K-12 and Salmonella enterica spp. strains (c and d) Summary of chemical modifications of Kdo2-lipid A that can occur after synthesis in E. coli K12 and Salmonella enterica spp. (c) and Helicobacter pylori (d). Enzymes that catalyze the modification are color coded along with the chemical group. Chemical groups drawn with dotted lines indicate the enzyme catalyzes hydrolysis to remove the group. Asterisks indicate that LpxO, LpxR, and PagL are present in Salmonella enterica spp. strains but not present in E. coli K12 strains.
Next, the core oligosaccharide is extended at the cytoplasmic side of the IM, (not covered here) producing lipooligosaccharide (LOS). Some bacteria, including a number of mucosal pathogens, only produce LOS3. O-antigen precursors, if produced by the bacteria, are synthesized separately at the cytoplasmic interface of the IM and attached to the lipid carrier undecaprenyl-pyrophosphate3. LOS and O-antigen precursors are flipped across the IM by MsbA2 and an O-antigen flippase (vary between organisms), respectively3. Depending on the O-antigen biosynthesis pathway utilized, O antigen is either polymerized before flipping or at the periplasmic leaflet of the IM3. In both scenarios, O antigen is appended to LOS in the periplasm3. LPS or LOS molecules are then transported from the IM to the surface of the OM by the Lpt (LPS transport) machinery9 (Figure 1a).
Properties of LPS impact OM permeability, resistance to antibiotics, virulence, and recognition by the mammalian host’s immune system. Thus, bacteria have evolved many enzymes that modify LPS to alter its properties and allow them to adapt to the dynamic environments they inhabit. Here we review the current state of the field of LPS biogenesis and its modification. We highlight recent insights into how LPS modifications affect stimulation of host immune response and OM vesiculation. Finally, we discuss how a mechanistic understanding of LPS biogenesis and remodeling has led to development of antibacterial therapeutics.
Altering LPS
Variations to the structure of LPS can be introduced at each step of its biogenesis. For the purpose of discussion, variation in the lipid A structure will be compared to the lipid A anchor produced by E. coli K-12 under standard laboratory conditions (Luria-Bertani broth at 37°C, Figure 1b). Under these conditions, the lipid A disaccharide of GlcN has two attached Kdo sugars, is bis-phosphorylated and is hexa-acylated. Two primary N-linked β-hydroxy-acyl chains are attached at the 2 and 2′ positions and two primary O-linked β-hydroxy-acyl chains are attached at the 3 and 3′ positions of the GlcN disaccharide. E. coli lipid A then has two secondary acyl chains attached to the β-hydroxyl groups of the 2′ and 3′ acyl chains.
Variations to LPS introduced during the Raetz pathway alter either the sugars or acyl-chains of the Kdo-lipid A domain (depicted in supplementary information S1). Alterations to sugars, GlcN and Kdo, can occur before incorporation into lipid A. Organisms like Campylobacter jejuni and Leptospira interrogans, utilize the enzymes GnnA and GnnB to oxidize and transaminate UDP-GlcNAc, respectively, converting it into the 3-amino derivative UDP-GlcNAc-3N (2,3-diamino-2,3-dideoxy-d-glucopyranose)10,11. These organisms then encode LpxA homologs that selectively utilize the modified UDP-GlcNAc3N sugar10. The resulting lipid A in these organisms has 4 amide-linked acyl chains and result in reduced recognition by host immune cells and reduced sensitivity to antimicrobials11. Similarly, Kdo is modified by additional enzymes KdnA and KdnB in Shewanella species to produce an 8-amino derivative, Kdo8N (8-amino-3,8-dideoxy-d-manno-octulosonic acid)12. Kdo8N is then activated to its nucleotide-linked form and feeds into lipid A synthesis as a substrate of WaaA. Kdo8N-containing LPS was also found to play a role in protecting cells from antimicrobials12. In addition to the possibility of modifications of sugar precursors, variations to Kdo incorporated into lipid A can also occur during or after the Raetz pathway. WaaA of various organisms have variability in the number of Kdo sugars (one, two, three or four) they attach.13-16 Many organisms will also modify Kdo by hydroxylation (via KdoO)17 or phosphorylation (via KdkA)14,18 during lipid A synthesis. Alterations of GlcN and Kdo introduced during the Raetz pathway are constitutive in their respective organism.
Additional diversity arises from the substrate selectivity of the Lpx acyltransferases of different bacteria altering fatty acyl chain characteristics (length, saturation, branching, etc.). Further, the secondary acyltransferases, LpxL and LpxM, can vary in the position of acyl chain attachment and in the number of acyl chains transferred (supplementary information S1). LpxM from Acinetobacter baumannii, for example, is bifunctional and transfers a laurate to the 2 and 3′ acyl chains19. In addition, many organisms will alter the acyl chains incorporated into lipid A in response to environmental conditions by encoding more than one enzyme that act at the same position (supplementary information S1). This type of variation is well known for organisms like Salmonella enterica spp. and E. coli that express LpxL, a lauroyl (C12:0) transferase, at higher temperatures and LpxP, a palmitoleoyl (C16:1) transferase, at lower temperatures3. In addition, Klebsiella pneumoniae encodes two LpxL homologs that are co-expressed under standard growth conditions and compete to add either a lauroyl (C12:0) group, by LpxL1, or myristoyl (C14:0) group, by LpxL2, at the same position. Finally, organisms like Leptospira interrogans and Francisella tularensis encode two homologs of LpxD (Figure 1a) that incorporate different length acyl chains20,21. This variation in acyl-chain length mediated by Francisella21 LpxD1 and LpxD2 was demonstrated to be in response to temperature, but conditions for which Leptospira LpxD2 are expressed have remained elusive20.
After the Raetz pathway, LPS can be further modified by a diversity of enzymes that generally alter the acyl chains, phosphate groups, the sugar backbone of lipid A, and the core oligosaccharide (Figure 1c-d and supplementary information S2). These modifications have a critical impact on lipid packing, membrane permeability, host recognition, and sensitivity to antimicrobials (discussed later). Alterations to acyl chains of LPS after synthesis typically involve either addition or removal of fatty acids by enzymes in the OM like LpxR, PagL, and PagP22-24. These modifications allow organisms to control the membrane characteristics of LPS already transported to the surface. Other alterations to acyl chains of lipid A can occur at the IM including hydroxylation by the enzyme LpxO25, or, in the unique case by Vibrio cholerae, addition of glycine moieties via AlmG26.
Organisms have evolved a plethora of strategies to alter the charge of lipid A. Briefly, many organisms like Helicobacter pylori cleave the phosphates from lipid A to produce a neutral or less negatively-charged version (Figure 1d). The negative charge of lipid A can also be reduced by masking phosphates with positively-charged, phosphoethanolamine (by EptA27) and 4-aminoarabinose (by ArnT27), or neutral constituents, glucose sugars (by FlmK28). In contrast, many bacteria will append additional phosphates to lipid A with the enzyme LpxT29,30 increasing the negative charge. In addition, Kdo and the core oligosaccharide can be modified by addition of charged moieties, like phosphates or phosphoethanolamine, or addition or removal of sugars27 (supplementary information S2). Finally, Gram-negative bacteria encode a diversity of biosynthetic and modification enzymes that can alter the structure of O antigen that have been reviewed thoroughly elsewhere31,32.
Regulation of LPS modifications
LPS modification enzymes can be either constitutively expressed or controlled by regulatory networks. The human-adapted gastric pathogen H. pylori, for example, constitutively expresses a highly ordered repertoire of lipid A-modifying enzymes resulting in a tetra-acylated lipid A anchor with reduced phosphorylation33-36 (Figure 1d). However, in Salmonella enterica subsp. enterica serovar Typhimurium, a wide range of regulatory mechanisms fine-tune when and to what extent LPS is modified to accommodate a more diverse life style (Figure 2 and supplementary information S3).
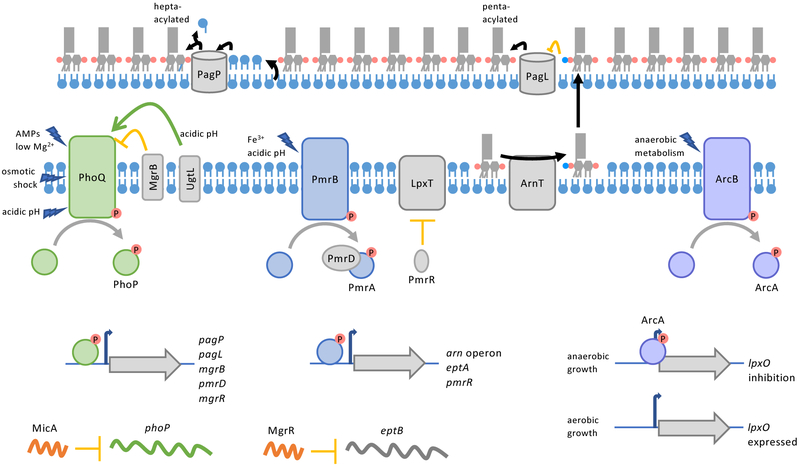
Figure 2: Regulation of LPS modifications.
(a) Regulation of LPS modification enzymes in Salmonella enterica subsp. enterica serovar Typhimurium. TCS PhoPQ, PmrAB, and ArcAB regulate genes that encode enzymes that alter the acylation (PagP, PagL), modify phosphates (aminoarabinose by ArnT and phosphoethanolamine by EptA), and hydroxylate an acyl chain (LpxO) of lipid A, respectively. PhoPQ also upregulates the protein PmrD which binds to and protects phosphorylated-PmrA, connecting these TCS. Small RNAs are connected to PhoPQ regulation; MicA inhibits translation of PhoP, and MgrR, when upregulated by PhoPQ, inhibits the gene eptB, encoding a core-oligosaccharide modifying phosphoethanolamine transferase. In addition, PhoPQ and PmrAB upregulate genes involved in negative feed-back loops for the respective TCS. PhoPQ upregulates MgrB that binds to and inhibits PhoQ. PmrAB upregulates the small protein PmrR and genes that for modifying lipid A with aminoarabinose (arn operon) and phosphoethanolamine (eptA). PmrR post-translationally inhibits the lipid A phosphotransferase LpxT. Decrease in LpxT activity and increased lipid A modification by ArnT and EptA alter the charge of the OM so that metal ions that activate PmrB are blocked from entering the cell. Finally, the activity of OM enzymes PagP and PagL are regulated by availability and modification of their substrates, respectively. When glycerophospholipids are mislocalized to the outer leaflet of the OM, PagP catalyzes the transfer of an acyl chain from donor glycerophospholipids to acceptor lipid A molecules. PagL deacylation of lipid A is inhibited by aminoarabinose modification of lipid A.
Many bacteria utilize two-component systems (TCS) to regulate LPS modification enzymes (supplementary information S3). At their core, these regulation systems utilize a sensor kinase that responds to a signal through autophosphorylation of a histidine residue. The phosphate is then transferred to a cognate response regulatory protein acting as a phosphorylation-mediated switch, turning on and off gene expression. PhoPQ and PmrAB are the most widely-spread TCS that affect lipid A modification genes37,38. Here we will briefly touch on the mechanism of these TCS with focus on recent advances.
PhoPQ is best studied in Salmonella enterica subsp. enterica serovar Typhimurium where it responds to many environmental stimuli including acidic pH, low concentrations of divalent cations, antimicrobial peptides (AMPs), and most recently reported osmotic shock38-41. The breadth of stimulating signals varies for PhoPQ of different bacteria38,42. However, work on PhoQ of Salmonella enterica subsp. enterica serovar Typhimurium and E. coli has revealed how various signals can feed into one sensor (Figure 2).
PhoQ contains cytoplasmic, transmembrane, and periplasmic domains. The periplasmic sensing domain contains a patch of acidic residues close to the membrane surface that is critical for sensing divalent cation concentrations and cationic AMPs43. Mg2+ and Ca2+ are thought to reduce repulsion between the acidic residues of PhoQ and acidic head group of glycerophospholipids in the IM and favor PhoQ in an off conformation43. Repulsion in the absence of divalent cations or disruption of this bridging by binding of cationic AMPs to this site causes a conformational change that favors autophosphorylation43. Initially, it was thought that pH changes may also activate PhoQ by affecting the periplasmic sensor domain44. However, recently it was suggested that pH changes are sensed by a cytoplasmic domain. When this cytoplasmic domain was altered as a result of mutations in the encoding gene, PhoQ is unable to sense pH changes but can still respond to Mg2+ limiting conditions45. Additionally, an IM protein in Salmonella enterica subsp. enterica serovar Typhimurium, UgtL, was proposed to bind PhoQ and amplify the autophosphorylation in response to acidic pH46. Finally, osmotic shock was proposed to activate PhoQ by a third mechanism whereby high osmolarity increases lateral pressure exerted by lipids on the transmembrane domains of PhoQ and causes a change in the protein conformation41. Altogether, work on Salmonella enterica subsp. enterica serovar Typhimurium PhoQ has demonstrated the intricacies of how a senor kinase can be adapted for multiple stimuli.
Activated-PhoQ phosphorylates PhoP, which in Salmonella enterica subsp. enterica serovar Typhimurium and other organisms transcriptionally regulates many virulence factors including lipid A modifications. PhoP directly regulates pagL and pagP genes, but also indirectly affects other LPS-modification genes through regulation of a small RNA (discussed later) and the protein PmrD22,47,48. While not present in all bacteria, Salmonella enterica spp. and E. coli encode the protein PmrD that mediates coupling between PhoPQ and PmrAB48,49. When upregulated by activation of PhoPQ, PmrD binds to phosphorylated PmrA and protects it from being deactivated by phosphatase activity of PmrB50. Thus, PhoPQ activation can indirectly feed into PmrAB regulation of genes like arnT and eptA37. To control the extent of PhoPQ activation, PhoP also upregulates expression of MgrB, an IM protein that provides negative feedback closing the PhoPQ regulatory circuit51.
PmrB is sensor kinase of the PmrAB TCS and is directly activated by binding of Fe3+, Al3+, and in some organisms Zn2+ to a periplasmic, metal-binding, ExxE motif52. PmrAB can also be activated by acidic pH, but full activation requires both PmrB and PmrD suggesting part of the activation is through PhoPQ activity53. PmrB’s proposed pH sensing domain consists of His and Glu residues in the periplasm that may react to altered protonation states53. Activation of PmrB causes it to phosphorylate PmrA which directly upregulates aminoarabinose (arn operon) and phosphoethanolamine (eptA) modifications. Activation also increases transcription of the small protein PmrR, which binds to and inhibits activity of the lipid A phosphotransferase, LpxT54. LpxT and EptA competitively modify the same site on LPS (Figure 1c), so LpxT inhibition enhances phosphoethanolamine modification of lipid A by EptA driving AMP resistance29.
Further, the combined effects of PmrR, EptA, and ArnT serve as a feedback loop controlling the amount to which PmrAB is activated54. Activation of PmrAB by metal ions like Fe3+ depends upon the charge of the OM. Upon encountering high levels of Fe3+, bacteria with a highly negatively-charged OM, due to the presence of bis- and tris-phosphorylated lipid A, experience high entry of Fe3+ into the cell where it activates PmrB. High activation of the PmrAB regulon produces the Arn proteins, EptA, and PmrR. PmrR quickly inhibits LpxT reducing the amount of tris-phosphorylated lipid A produced and somewhat reducing the negative charge of the OM. Production of aminoarabinose and phosphoethanolamine modified lipid A further reduces the negative charge of the OM. As the charge of the cell surface becomes more positive, less Fe3+ reaches PmrB and activation is fine-tuned54.
In addition to PhoPQ and PmrAB, organisms may have other TCS that allow them to respond to environmental signals (supplementary information S3). Demonstrated in Salmonella enterica serovar Enteritidis but likely more wide-spread, the ArcAB TCS is one of the regulators that responds to oxygen availability and regulates the gene encoding LpxO, the oxygenase responsible for hydroxylation of a lipid A acyl chain55. Pseudomonas aeruginosa, for example, encodes PhoPQ, PmrAB, and three additional TCS that regulate lipid A modification genes. The Pseudomonas ColRS system senses Zn2+ and specifically upregulates EptA, but not ArnT56. Whereas, ParRS and CprRS of Pseudomonas sense AMPs and specifically upregulates ArnT57,58. While both the sensor kinases of these systems, ParS and CprS, are activated by polymyxins, CprS is also activated by indolicidin and other peptides allowing Pseudomonas to respond to many environmental signals57,58. Vibrio cholerae was also recently shown to have an additional TCS, VprAB (also named CarRS), that senses bile, AMPs, and acidic pH to regulate its almEFG operon that modifies lipid A with glycine59,60. Finally, certain strains of Klebsiella pneumoniae have an additional TCS, CrrAB, that indirectly affects LPS modifications by upregulating the encoding gene for CrrC which in turn up regulates the genes that encode PmrAB61. Stimuli that activate CrrAB have yet to be determined. These examples demonstrate the breadth of which TCS are utilized to regulate LPS modifications.
Small RNAs commonly have connections to TCS and can either directly or indirectly regulate lipid A modification genes (supplementary information S3). MicA, a small RNA that represses synthesis of key OM proteins, also negatively regulates PhoPQ expression at the post-transcriptional level thereby impacting LPS structure as well62. The sRNA MgrR is upregulated by PhoPQ63. In turn, MgrR then inhibits transcription of the eptB gene, which encodes a phosphoethanolamine transferase that modifies the second Kdo sugar of the core oligosaccharide63. Since activation of PhoPQ typically leads to decoration of lipid A, MgrR inhibition of eptB expression is counter-intuitive, but further highlights the complex regulation of LPS modifications. The TCS and small RNAs described in brief here is not a comprehensive overview of all factors that affect transcription or translation of LPS modification genes. Other small RNAs and transcriptional regulators are listed in supplementary information S3.
Regulation of OM enzymes, like PagP, PagL, and LpxR, at the transcriptional level is a relatively slow process as it depends on assembly and turn-over. To more tightly control their activity, these enzymes are also regulated at the level of recognition or availability of their substrates (supplementary information S3). LpxR of Yersinia entercolitica and PagL of Salmonella enterica subsp. enterica serovar Typhimurium both poorly recognize lipid A that has been decorated with aminoarabinose64,65. These deacylases can modify lipid A with unmodified phosphates, but their activity is repressed under conditions where aminoarabinose modification at the IM is up-regulated64,65. PagP activity is instead regulated by availability of its donor substrate. PagP plays a role in maintaining the asymmetry of the OM by transferring an acyl chain from the glycerophospholipid phosphatidylethanolamine to lipid A66. In Salmonella enterica spp. and E. coli, the activity of PagP is normally low because the outer leaflet of asymmetric OM is lacking glycerophospholipids2. Upon OM disruption, such as in Mg2+ limiting conditions or exposure to AMPs, glycerophospholipids flip to the outer leaflet resulting in PagP-dependent acylation of lipid A67. These examples demonstrate some of the ways LPS modification enzymes are regulated post-translationally.
Consequences of LPS modifications
LPS modifications affect many physiological processes of Gram-negative bacteria. Here effects on permeability of the OM, recognition by immune cells, antimicrobial resistance, and OM vesiculation will be briefly reviewed (Figure 3).
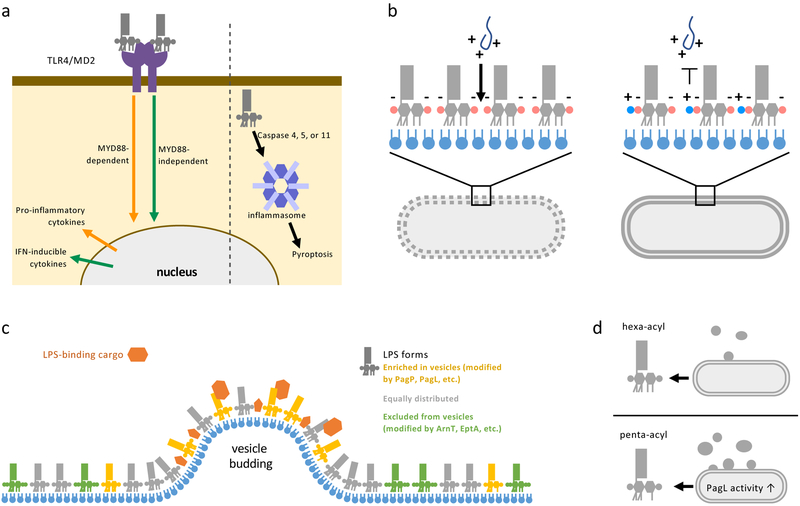
Figure 3: Consequences of LPS modifications.
(a) LPS can stimulate immune cell responses through recognition by surface receptors (left) or binding to the cytoplasmic inflammasome (right). TLR4/MD2 receptors on the surface of mammalian immune cells recognize lipid A and can activate two signaling pathways. The myeloid differentiation primary response protein 88 (MYD88)-dependent pathway upregulates proinflammatory cytokines that lead to inflammation and bacterial clearance. Alternatively, signaling through TIR domain-containing adaptor inducing IFNβ (TRIF), known as the MYD88-independent pathway, produces interferon (IFN) inducible cytokines that result in less inflammation, but are critical for adjuvanticity. Modifications of the phosphates and acyl chains of lipid A affect how well LPS is recognized by the TLR4/MD2 receptor and which signaling pathway is induced. Inflammasome recognition is mediates by caspases and lead to an inflammatory cell death pathway called pyroptosis. Modifications to acyl chains of LPS reduce stimulation of murine inflammasome response but does not affect stimulation of human cell-lines inflammasomes. (b) Cationic antimicrobial peptides (AMPs) produced by host immune cells or used as antibiotics (such as Polymyxin B and Colistin) to treat infectious bacteria act by first forming charge-charge interactions with the highly negatively-charged OM. AMPs then perforate the OM followed by the IM, leading to lysis of bacterial cells. LPS modification that reduce the negative charge or alter the acyl chains of lipid A provide resistance against AMPs by charge repulsion or decreasing the fluidity of the OM. (c) Gram-negative bacteria release vesicles that bud from the OM called outer membrane vesicle (OMVs). When the LPS in the OM and OMVs released are compared, certain chemical forms of LPS are enriched, equally distributed, or excluded (colored gold, grey, and green respectively) in OMVs. Some cargo proteins (orange) associated with OMVs such as heat-labile enterotoxin in enterotoxigenic E. coli, are recruited through specific interactions with LPS. Allowing the selective recruiting and secretion of certain proteins in OMVs. (d) Certain chemical forms of LPS, such as penta-acylated LPS produced by PagL activity, increase the size and number of OMVs released by bacteria, indicating that LPS modification can stimulate OMV formation.
Adaptation of OM permeability
LPS modifications are regulated in response to temperature, metal ion concentrations, pH, AMPs, and other conditions. While these are commonly described for their critical effects on pathogenesis, they also have a role in modulating OM permeability in many environments. Mg2+ bridging of the phosphates on lipid A and/or phosphates of the core oligosaccharide of LPS help stabilize the OM. In Mg2+ limiting conditions, however, the phosphates of LPS repel each other decreasing OM stability68. Through PhoPQ and PmrAB, bacteria like Salmonella enterica spp. respond by upregulating PagP, ArnT, and EptA that help to stabilize the membrane by producing more hepta-acylated lipid A (decreasing fluidity) and adding positively-charged moieties (aminoarabinose and phosphoethanolamine) to the 1 and 4′ phosphates68. It is not fully clear how having predominantly positively-charged lipid A, which would also charge clash, forms a more stable OM than having predominantly negatively-charged lipid A. Perhaps, because the positively-charged moieties are attached to phosphate groups, the zwitterionic nature of modified lipid A allows cross-bridging to occur between neighboring molecules in the membrane. Alternatively, since PagP activity indicates that some glycerophospholipids are mislocalized to the outer-leaflet, perhaps anionic glycerophospholipids (e.g. phosphatidylglycerol) contribute to cross-bridging positively-charged lipid A species. Still, when Mg2+ concentrations are high, constitutive expression of PagP, ArnT, and EptA increases OM permeability68 indicating that a negatively-charged lipid A domain increases bacterial fitness in the presence of divalent cations. Thus, bacteria employ different lipid A anchors in different environments to regulate OM permeability.
Evading immune system recognition
The human host immune system recognizes and responds to LPS as a common molecular signature of bacteria. LPS or LOS stimulate immune cells by two methods: binding to surface receptors and binding to a non-canonical inflammasome complex if internalized. Surface recognition starts with binding of LPS or LOS to LPS-binding proteins and CD14 (either soluble or membrane-bound) and then transfer of the molecule to toll-like receptor 4 (TLR4)-MD2 complexes on immune cells69. Hexa-acylated, bis-phosphorylated lipid A is highly recognized by the TLR4-MD2 co-receptor70,71 (Figure 3a). The signaling pathway stimulated upon TLR4-MD2 binding to lipid A depends on the adaptor proteins that are then recruited (Figure 3a). Recruitment of MYD88 (myeloid differentiation primary response protein 88) results in the MYD88-mediated pathway and production of proinflammatory cytokines. Meanwhile, recruitment of TRIF (TIR domain-containing adaptor inducing IFNβ) results in the MYD88-independent pathway and reduced inflammation72. LPS modifications, especially those that remove the phosphates or alter the acylation of lipid A, can reduce either recognition by TLR4-MD2 or alter which signaling pathway is triggered70,71,73. As such, many bacteria utilize lipid A modifications to evade recognition by the mammalian immune response. For example, H. pylori constitutively produces a dephosphorylated, tetra-acylated species of lipid A that promotes immune evasion and long-term carriage of the organism in the gastric mucosa74. Due to differences in modifications and acylation of lipid A, H. pylori is 100 to 1000-fold less immuno-stimulatory than E. coli74,75. In addition, many bacteria upregulate lipid A modifications during infection to evade TLR4-MD2 recognition. Intriguingly, the Bengoechea group76 detected upregulation of the lipid A modifications in Klebsiella pneumoniae directly extracted from infected tissue. This new approach allowed for the direct testing of what lipid A modifications are key for immune evasion in specific tissues. Altered LPS that have desired immune-stimulatory effects have also been explored as therapeutics (Box 2).
Box 2: Altered LPS as adjuvants and vaccines.
LPS can be utilized to stimulate the immune response, but high stimulation causes severe damage to tissues and organs, as occurs during sepsis72. LPS variants that stimulate the immune response without toxic effects have been explored as immunotherapeutics. In 1982, Ribi and colleagues136 described a method for chemically-altering LPS species from Salmonella minnesota to produce a mixture of mono-phosphorylated lipid A species (MPL) with reduced toxicity. After further development by the Corxia Corporation and GlaxoSmithKline Biologicals, this led to the FDA approval in 2009 of MPL, primarily 3-O-deacyl-4′-monophosphoryl lipid A, as an adjuvant in vaccines called MPL adjuvant™ 72. The MPL adjuvant™ shows reduced activation of the MYD88-dependent response and thus induces less toxic inflammation72, while providing key adjuvant properties. To further fine-tune the lipid A induced immune-response, our group137 and others138,139 have bioengineered non-pathogenic bacteria to produce various LPS glycoforms by combinations of overexpressing and knocking out of genes involved in lipid A synthesis and modification. In our work with E. coli137, altering the phosphates, number of acyl chains, position of acyl chains, and acyl chain hydroxylation of lipid A individually or in combination can give a range of TLR4-MD2 recognition and cytokine response. A similar range of immune stimulation was observed by the Van der ley139 and Ernst138 groups for LPS with various modifications produced by Neisseria meningitidis and Yersinia pestis, respectively. Interestingly, not all trends observed for TLR4-M2 recognition of lipid A species produced by E. coli held true for N. meningitidis, highlighting the importance of studying LPS-dependent immune modulation by various organisms.
Altering LPS has also been utilized to increase the efficacy of OMV vaccines. OMVs have been used as successful vaccine platforms, particularly for Neisseria meningitidis140, because they display molecular signatures of the pathogen, but are unable to replicate. LPS modifications that stimulate OMV formation or that alter the adjuvanticity have been combined with these platforms to have desired immune-stimulatory effects. In addition, the Feldman141 and DeLisa142,143 groups have developed methods to produce OMVs that contain LPS whose core oligosaccharide is modified with pathogen-associated glycans (capsule, heterologous O antigens, and other surface-displayed glycans). These approaches capitalize on the substrate promiscuity of the O-antigen ligase, WaaL, which can transfer a variety of sugar repeats from the undecaprenyl-pyrophosphate carrier to LPS3. The genes for producing a desired pathogen-associated glycan are heterologously expressed in non-pathogenic E. coli K-12 strains that do not produce O antigen141-143. The pathogen associated glycan is appended to LPS by WaaL and modified LPS is released in OMVs. Since LPS in OMVs is still endotoxic, the E. coli strains that produce these OMV vaccines can be further engineered to have modified lipid A with reduced endotoxicity, while retaining adjuvant properties142. These vaccine platforms have been used to produce OMVs displaying a variety of disease-associated glycans that upon immunization provide protection against the associated pathogen in disease models (e.g. Streptococcus pneumoniae, Campylobacter jejuni, Francisella tularensis subsp. Tularensis, and Neisseria meningitidis)141-143. Further, displaying glyco-engineered LPS on reduced-endotoxic and commensal strains of E. coli instead of OMVs could be promising as oral-administrable, cost-effective, whole-cell vaccines.
LPS that is internalized by host cells, especially from intracellular pathogens, is bound by caspases (caspases 4 and 5 in humans77, and caspase 11 in mice78,79) that then stimulate the inflammasome (Figure 3a). LPS stimulation of the inflammasome triggers an inflammatory cell lysis pathway called pyroptosis80. Similar to TLR4 recognition, lipid A is the minimal unit required for inflammasome stimulation77-79. Reduced acylation of lipid A reduces recognition by murine caspase 1178,79 but not by human caspases 4/581, indicating that these LPS modifications could be critical for pathogenesis of certain mammalian hosts. Further work is needed to determine if other LPS modifications, especially those employed by intracellular pathogens, allow for evasion of inflammasome recognition in their natural hosts.
Resistance to antimicrobial peptides
AMPs are produced and released by the immune response of the human host during inflammation to non-specifically clear bacteria. In addition, polymyxins are cationic AMPs naturally produced by Gram-positive bacteria and have been adapted for use as antibiotics to treat bacterial infections. AMPs are amphipathic; a typically cationic peptide mediates interaction with negatively-charged LPS with a hydrophobic domain that inserts into membranes forming pores82 (Figure 3b). Perforation of the OM allows entry and disruption of the IM resulting in cell lysis82. LPS modifications that remove or modify the phosphates of lipid A, modify the phosphates of the core oligosaccharide, and alter the acylation of lipid A can provide protection against AMPs (Figure 3b and supplementary information S2).
Recently it was demonstrated that one factor that contributes to long-term maintenance of commensal bacteria is LPS modifications that provide resistance to AMPs. The gut microbiota is a complex population of bacteria that are maintained long-term, in spite of the mammalian host inflammatory response83. Specifically, several Bacteroidetes thetaiotaomicron commensal isolates were found to have high resistance to polymyxin B mediated by expression of the lipid A phosphatase LpxF84. Further, when comparing the ability of isogenic strains with and without LpxF to colonize the mouse gut and be maintained after inflammation, both B. thetaiotaomicron populations could colonize, but the strain without LpxF was displaced by inflammation84. This finding indicated that lipid A modifications can contribute to long-term maintenance of commensal gut bacteria. While AMP resistance is a favorable trait for maintaining commensal bacteria, commensals can also become opportunistic pathogens and AMP resistance would make them more difficult to be cleared by the immune response.
Polymyxins are cationic AMPs naturally produced by bacteria that have been adapted for use as antibiotics. However, due to toxicity issues polymyxins are considered “last-resort” antibiotics. Still, with the increasing frequency of multi-drug resistant bacteria their use has also become necessary. Many pathogenic bacteria can become resistant to polymyxins through mutations that increase expression of chromosomally-encoded LPS-modifying enzymes like the Arn operon and EptA37. Fortuitously, these types of mutations do not easily spread between populations of bacteria. However, eptA homologs, named mcr genes for mobile colistin resistance, have been identified to be encoded in plasmids and phages that can be more easily spread85. In the few short years since the first report, mobile homologs of EptA have been identified in many pathogenic bacteria and have been detected worldwide, leading to concerns about spread of resistance to even last-resort antibiotics85. These findings only increase the insurmountable evidence that new therapeutics are desperately needed as other LPS modifications that require one enzyme and no special precursors, such as dephosphorylation by LpxF, could also be rapidly spread in a similar manner.
Effects on outer membrane vesicles
Gram-negative bacteria shed OM vesicles (OMVs) containing LPS as they grow. These OMVs are proposed to play roles in cell stress responses, nutrient acquisition, and pathogenesis86. Since LPS is a major constituent, altering LPS characteristics impact OMV processes, including recruiting cargo proteins and stimulating vesicle production. Pathogens like enterotoxigenic E. coli (ETEC) and Porphyromonas gingivalis produce virulence factors that bind to OMVs and are secreted into the environment87,88. Recruitment of these factors to OMVs is driven by binding of the cargo proteins to the sugars of LPS at the cell surface (Figure 3c). The pathogen ETEC produces a heat-labile enterotoxin that binds to the Kdo sugars of the core oligosaccharide88,89. Binding of labile toxin to LPS may be a mechanism of delivering the toxin on OMVs, but it also partially sequesters the toxin at the OM89.
Research on P. gingivalis has revealed increasingly elaborate interactions between OMV proteins and specific LPS species. Early on, it was noted that LPS glycoforms with a negatively-charged O antigen and a subset of key OM-associated proteins are enriched in OMVs87. Mutants that do not produce O antigen had OMVs with altered protein content supporting a role in recruitment87. Further, P. gingivalis encodes a proposed LPS modification gene, LptO, that affects OMV formation and secretion of gingipains. The activity of LptO is under debate, originally proposed as a lipid A deacylase and recently proposed to instead be a lipid A 1-phosphatase90,91. However, mutants with null alleles of IptO have 50% reduced OMVs and reduced secretion of proteins recognized by a type IX secretion systems, supporting LptO’s role in these processes90-92. Finally, a mechanism of recruitment of the disease-associated protein peptidylarginine deiminase (PPAD) to OMV LPS was recently proposed. PPAD citrullinate host proteins93 and is implicated in the autoimmune disease rheumatoid arthritis94,95. PPAD is produced in soluble and LPS-bound forms, but a single amino acid change (Gln 373 to Lys) in PPAD reduces the LPS-bound form of the protein96,97. Further, work is needed to clarify how PPAD and other P. gingivalis OMV proteins associate with LPS. Another excellent example of how LPS structure influences OMV formation is the interaction of the quorum sensing molecule PQS of P. aeruginosa with lipid A98. PQS binds to the 4′ phosphate group of LPS, inserts partially into the membrane, and induces curvature that stimulates OMV formation98,99. Thus, this signaling molecule stimulates its own secretion into the environment.
Recently chemical alterations to LPS have also been implicated in stimulation of vesicle formation. Specifically, the Feldman group saw that deacylation of LPS by overexpressing PagL increased both the number and size of OMVs released by Salmonella enterica subsp. enterica serovar Typhimurium100 (Figure 3d). Interestingly, the Kuehn group101 also recently found that when they closely monitored OMVs upon shifts between neutral/high Mg2+ and acidic/low Mg2+ media, PhoPQ and PmrAB activation increases the number and size of OMVs released. Aminoarabinose and phosphoethanolamine modified LPS were found to be under-represented, while hepta-acylated LPS (from PagP activity) was enriched in OMVs101. The cooccurrence of increased size and number of OMVs with enrichment of PagP-modified LPS could indicate this modification also stimulates OMV production101. The Kuehn group also proposed that exclusion of certain LPS forms from OMVs, like aminoarabinose and phosphoethanolamine-modified LPS upon PhoPQ and PmrAB activation, could be a mechanism of quickly enriching these modified forms in the OM during environmental transitions101. These two connections between LPS modifications and OMVs raise many questions for cell envelope biogenesis and bacterial pathogenesis. Under what conditions are PagP and PagL upregulated to stimulate OM vesiculation, to alter fluidity of the OM by altering LPS acylation, or both? Does the selective secretion of PagL- and PagP-modified forms of LPS in OMVs, which are less immune-stimulatory, impact the host immune response to pathogenic bacteria? As discussed earlier, aminoarabinose-modified LPS inhibits the activities of LpxR and PagL; does this serve as a regulatory feed-back to decrease OM vesiculation once the OM LPS content has been appropriately altered?
Inhibitors of LPS biogenesis and modification
Much focus has been put on developing inhibitors of LPS biogenesis and modification as it is an ideal target for antimicrobial treatment due to its uniqueness to bacteria, its essential nature, and its role in virulence. Inhibitors have been explored for many of the enzymes involved in Kdo synthesis and the Raetz pathway (Figure 4)102,103, focusing on enzymatic steps that are essential in most Gram-negative bacteria. The most promising inhibitors of LPS synthesis have been LpxC inhibitors due to their broad-activity, reviewed thoroughly by Erwin104. However, recent findings have suggested that more focus should be put on development of inhibitors of LpxK and LpxH, because, even in bacteria that can grow in the absence of LPS, inhibition of these enzymes results in build-up of toxic intermediates105,106. Such inhibitors could be particularly key for treating pathogenic Acinetobacter baumannii strains which are commonly multi-drug resistant and can survive in the absence of LOS (see Box 1). Inhibitors of the latter steps in lipid A/LPS synthesis could still prove useful in combination with drugs that normally cannot permeate the asymmetric OM. The most recent studies targeting LPS synthesis have attempted to increase the efficacy of LpxC inhibitors107, identify compounds that target LpxA through virtual screening methods108,and develop new high-throughput screening strategies109,110 for other Lpx enzymes (e.g. LpxH).
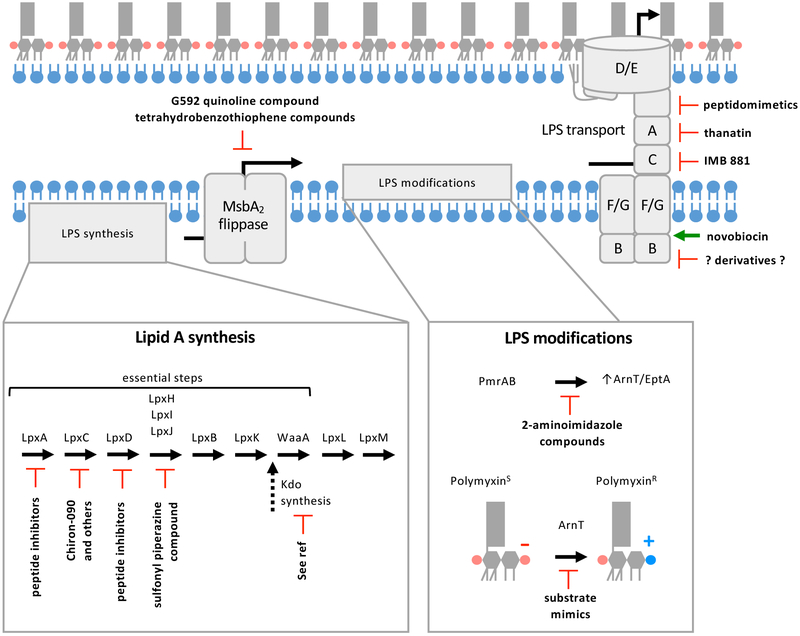
Figure 4: Summary of therapeutic strategies to target LPS biogenesis.
Compounds have been identified that inhibit (indicated by red blocked arrows) enzymes involved in lipid A synthesis, LPS flipping by MsbA2, regulators of LPS modifications, enzymes that modify LPS, and transport of LPS to the OM. The DNA gyrase novobiocin has also been demonstrated to bind the Lpt machinery and activate LPS transport (indicated by green arrow). While this activity does not inhibit growth, it does synergistically increase the efficacy of polymyxins.
Compounds that target transporters of LPS have also been explored as antimicrobials (Figure 4). Two classes of inhibitors were recently reported that target MsbA2. The first are quinolone-like compounds discovered by Genentech that bind to the transmembrane-pocket of MsbA2 and lock it in an LPS-bound, inward-facing conformation111. These inhibitors potently inhibit growth of E. coli and Klebsiella, but are less effective against MsbA2 from Pseudomonas, indicating they could pan out to be narrow-range antimicrobials111. Kahne and collaborators also recently reported a class of compounds that act as inhibitors of MsbA2 in Acinetobacter baumannii112. The compounds appear to stimulate wasteful ATP-hydrolysis of MsbA2 that is uncoupled from LPS flipping112.
A compound that affects coupling of ATP-hydrolysis and LPS transport has also been discovered for the Lpt complex (Figure 4). It was found that the already well-described DNA-gyrase inhibitor, novobiocin, is able to bind to the Lpt machinery113. However, novobiocin binding to the Lpt transporter increases LPS transport instead of inhibiting113. While novobiocin’s effect on LPS transport is not inhibitory, it could be a good starting point for the rationale design of compounds targeting Lpt. Further, novobiocin and novobiocin derivatives, including compounds unable to inhibit DNA gyrase, were found to work synergistically with polymyxins in strains that are naturally sensitive to polymyxins114. These findings indicate that increased LPS transport potentiates polymyxins114 and that combination therapies could be used to reduce the amount of polymyxin needed to treat bacterial infections. However, in strains displaying modified lipid A, novobiocin does not significantly synergize with polymyxin (unpublished, Trent laboratory), indicating that a combination therapy would have limitations in the clinic.
Three additional compounds have been reported that target the periplasmic domains of the Lpt transporter (Figure 4). LptC, LptA, and the N-terminus of LptD contain homologous β jellyroll domains that interact head-to-tail by strand addition to bridge the periplasm9. LPS molecules travel across the Lpt periplasmic bridge with acyl-chains protected in a hydrophobic groove formed by the β-jellyroll domains9. A class of macrocyclic peptidomimetics were found to specifically inhibit LPS transport of Pseudomonas species by binding to the periplasmic domain of LptD115,116. These peptidomimetics are proposed to block LPS as it traverses the periplasm115,117. In addition, two previously discovered antibacterial compounds, thanatin and IMB-881, were recently found to act by disrupting assembly of the LPS transporter subunits (Figure 4). Thanatin is capable of binding to the periplasmic domain of LptD and LptA118. Binding of thanatin to LptA occurs at the N-terminus β-strand in a way hypothesized to block interactions between LptA and LptC118. IMB-881 was also demonstrated to block interactions between LptA and LptC in a screen using a yeast two-hybrid system and subsequent surface plasmon resonance experiments119. Thanatin and IMB-881 have antibacterial activity against many Gram-negative bacteria118,119 and could be developed into useful wide-spectrum antimicrobials.
Finally, inhibitors that block the expression or directly inhibit the enzymatic activity of lipid A modifying enzymes have been reported (Figure 4). Since LPS modifications are critical for virulence of many pathogens and antibiotic resistance, these compounds could be used to directly treat many bacterial infections or in combination therapy to potentiate polymyxins. Inhibitors were discovered that caused down regulation of PmrAB and reduced lipid A modifications in Acinetobacter baumannii and Klebsiella pneumoniae120. Indeed, these compounds potentiated polymyxins120. Substrate mimics have also been reported that can directly inhibit ArnT which would be expected to have a similar effect121. These compounds could prove to be useful for treating bacteria with PmrAB-mediated resistance to polymyxins. However, they would be unable to reverse polymyxin resistance arising from the plasmid-encoded homologs of eptA (mcr genes) whose expression is PmrA-independent. No compounds have yet been identified that target EptA, but the recent full-length structure of the enzyme could serve as a starting point for rational design of inhibitors122. Altogether, much work has been devoted to designing or identifying compounds that inhibit LPS biogenesis and modification. More work is needed to explore which of these has potential as a clinical antimicrobial or to identify new compounds with promise.
Concluding remarks and future directions
Here we have touched on the striking diversity of the chemical alteration of LPS employed by pathogenic bacteria and the associated physiological consequences. Yet, it is clear that we do not have a complete story of all biological processes that LPS is involved in and many open questions remain. It is still unclear why LPS is essential in almost all Gram-negative bacteria. Further work on bacteria able to survive in the absence of LPS, like A. baumannii, will provide clues to what additional roles LPS holds as a major constituent of the OM. Also, from a bacterial physiology standpoint, LPS biogenesis is a complex process; LPS is synthesized on the cytoplasmic side of the IM, transported across multiple membranes, and modified in all compartments of the cell. While the major synthetic enzymes and transporters have been identified in many bacteria, how synthesis and transport are coordinated to ensure efficient assembly of the bacterial OM is unknown. Do LPS synthetic enzymes form large synthome complexes? In organisms that constitutively modify LPS, what are the checkpoints to make sure these have occurred? (Box 3) Further, LPS biogenesis and its modification likely have connections to other cellular processes in bacteria that have yet to be discovered. For example, the LPS modification enzyme LpxT utilizes undecaprenyl-pyrophosphate as a phosphate donor and contributes to recycling of this carrier molecule123. Conceptually, this also opens up the possibility that undecaprenyl-pyrophosphate could serve as phosphate donor for other processes in the periplasm, where no ATP available. In addition, EptC in Campylobacter jejuni is a phosphoethanolamine transferase that remarkably modifies lipid A, a heptose sugar of the core oligosaccharide, N-linked glycans of proteins and the rod of the C. jejuni flagellum124,125. Many bacteria contain multiple homologs of phosphoethanolamine transferases; how many of these are involved in LPS modifications, protein modifications or both? LPS modification enzymes could have a larger role in regulating activity of cell envelope proteins.
Box 3: Selecting which LPS to transport.
To assure LPS synthesis is complete before transport begins, the IM flippase, MsbA2, transports hexa-acylated LPS more efficiently than LPS with a tetra-acylated lipid A domain144. The molecular mechanism for selectively is still not understood, but mutations that alter a proline either in transmembrane 1 or 2 of MsbA resulted in increased transport of lipid IVA in strains with disrupted Kdo synthesis145. Recent cryo-EM structures of MsbA2 prior to and after flipping suggests a trap and flip model where lipid A binds in a deep cavity of MsbA2 (see figure) that allows it to translocate to the outer leaflet of the IM prior to flipping146. ATP hydrolysis then causes transmembrane rearrangement to flip and release lipid A146. Selectivity for hexa-acylated lipid A likely occurs prior to the “trap” state as it would be energetically unfavorable to release lipid A back into the inner leaflet once this step is reached.
Many modifications of LPS and attachment of O antigen, if present, to the coreoligosaccharide occur at the periplasmic leaflet of the IM3. Presumably, it would be advantageous to assure that desired core-lipid A modifications and O-antigen addition occur before transport by the Lpt machinery. Work in Burkholderia cenocepacia and E. coli suggests that the Lpt machinery recognizes charges at the 1 and 4′ positions of lipid A in order to control which molecules are transported. In B. cenocepacia, LPS is predominantly aminoarabinose-modified and this modification is essential for viability147. B. cenocepacia that produce unmodified, negatively-charged lipid A are only viable with a compensatory mutation that alters an Asp residue in the substrate cavity of LptFG (colored red on LptG in figure) to neutral His suggesting a charge interaction between transporter and substrate148. Further analysis of this residue in LptG suggested it may be coevolved with the charge of the 1 and 4′ position of lipid A produced by various Gram-negative bacteria149. In E. coli, LptG contained a positively-charged residue at this site and lipid A is predominantly unmodified at the 1 and 4′ phosphates. Further, mutations in E. coli that alter this residue in LptG to have a negative charge conferred defects that could be suppressed by mutations that increased the amount of lipid A modified with positively-charged phosphoethanolamine149. These findings suggest that charge-charge interactions between the GlcN disaccharide backbone of lipid A and LptG are one way that a bacterium selects which form of LPS is transported to the OM. For Burkholderia cenocepacia gating which LPS is transported to the OM is a convenient way to assure that a constitutive modification of lipid A occurs. However, it is clear that not all bacteria with constitutive modifications to LPS utilize similar regulation mechanisms because constitutive modifications are not essential in many organisms like H. pylori.
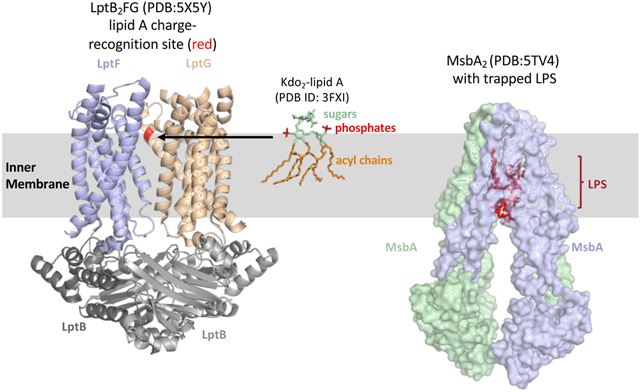
It has also become apparent that LPS has critical roles in interactions between bacteria and higher organisms. We have briefly reviewed how LPS can stimulate the human immune system, and both commensal and pathogenic bacteria have been implicated in immune system development impacting the incidence of asthma, allergies, and autoimmune diseases. Many studies have shown that the rate of autoimmune diseases is heightened in regions with improved sanitation and reduced childhood exposure to infectious agents126. Recently, in a study that followed gut microbiota development in infants from various regions, high abundance of Bacteroidetes in the gut during infancy was correlated with high incidence of autoimmune diseases127. Bacteroidetes species produce modified LPS that is less immuno-stimulatory, suggesting that LPS stimulation of the immune system is critical during development127. Finally, it is important to note that changes in lipid A/LPS structure also impact interaction between bacteria and plants with a number of LPS modifying enzymes characterized in Rhizobium species128. Our concepts of how bacteria contribute to the development of higher organisms are evolving and LPS is likely to have a major role in many more aspects of these processes.
Supplementary Material
Suggested Glossary terms
- Lipooligosaccharide
A form of LPS with an extended core oligosaccharide, but lacking O antigen
- TLR4/MD-2 (Toll-like receptor 4/myeloid differentiation factor 2)
A pattern-recognition receptor of the innate immune system that recognizes LPS/LOS initiating a robust signal cascade and inflammatory response in mammals
- Stringent response
A stress response of bacteria in reaction to nutrient limitation resulting in extreme physiological changes
- Small RNAs
Typically short, non-coding RNA molecules that interact with mRNAs to regulate gene expression or interact with proteins to regulate activity
- Outer membrane vesicles (OMVs)
Small, spherical outer-membrane blebs that are released from Gram-negative bacterial cells and contain membrane and periplasmic components
- Inflammasome
A intracellular, multiprotein complex in mammalian cells that recognizes microbial molecules and activates inflammatory response include pyroptosis and pro-inflammatory cytokines
- Pyroptosis
An inflammatory, programmed cell death that typically associated with infection of intracellular pathogens
- Capsule
A thick layer of polysaccharides that surrounds a bacterial cell, also referred to as capsular polysaccharide
References
- 1.Muhlradt PF & Golecki JR Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. European journal of biochemistry 51, 343–352 (1975). [DOI] [PubMed] [Google Scholar]
- 2.Nikaido H & Vaara M Molecular basis of bacterial outer membrane permeability. Microbiological reviews 49, 1–32 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitfield C & Trent MS Biosynthesis and export of bacterial lipopolysaccharides. Annual review of biochemistry 83, 99–128, doi: 10.1146/annurev-biochem-060713-035600 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Muhlradt PF, Menzel J, Golecki JR & Speth V Outer membrane of salmonella. Sites of export of newly synthesised lipopolysaccharide on the bacterial surface. European journal of biochemistry 35, 471–481 (1973). [DOI] [PubMed] [Google Scholar]
- 5.Osborn MJ, Gander JE & Parisi E Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. The Journal of biological chemistry 247, 3973–3986 (1972). [PubMed] [Google Scholar]
- 6.Young HE et al. Discovery of the Elusive UDP-Diacylglucosamine Hydrolase in the Lipid A Biosynthetic Pathway in Chlamydia trachomatis. mBio 7, e00090, doi: 10.1128/mBio.00090-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin EJ, O'Brien JP, Ivanov PL, Brodbelt JS & Trent MS Identification of a broad family of lipid A late acyltransferases with non-canonical substrate specificity. Molecular microbiology 91, 887–899, doi: 10.1111/mmi.12501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hankins JV et al. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxy-acyltransferase and its role in innate immune recognition. Molecular microbiology 81, 1313–1329, doi: 10.1111/j.1365-2958.2011.07765.x (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N & Kahne D Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nature reviews. Microbiology 14, 337–345, doi: 10.1038/nrmicro.2016.25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweet CR et al. Enzymatic synthesis of lipid A molecules with four amide-linked acyl chains. LpxA acyltransferases selective for an analog of UDP-N-acetylglucosamine in which an amine replaces the 3"-hydroxyl group. The Journal of biological chemistry 279, 25411–25419, doi: 10.1074/jbc.M400597200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Mourik A et al. Altered linkage of hydroxyacyl chains in lipid A of Campylobacter jejuni reduces TLR4 activation and antimicrobial resistance. The Journal of biological chemistry 285, 15828–15836, doi: 10.1074/jbc.M110.102061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gattis SG, Chung HS, Trent MS & Raetz CR The origin of 8-amino-3,8-dideoxy-D-manno-octulosonic acid (Kdo8N) in the lipopolysaccharide of Shewanella oneidensis. The Journal of biological chemistry 288, 9216–9225, doi: 10.1074/jbc.M113.453324 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belunis CJ & Raetz CR Biosynthesis of endotoxins. Purification and catalytic properties of 3-deoxy-D-manno-octulosonic acid transferase from Escherichia coli. The Journal of biological chemistry 267, 9988–9997 (1992). [PubMed] [Google Scholar]
- 14.Hankins JV & Trent MS Secondary acylation of Vibrio cholerae lipopolysaccharide requires phosphorylation of Kdo. The Journal of biological chemistry 284, 25804–25812, doi: 10.1074/jbc.M109.022772 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobau S, Mamat U, Brabetz W & Brade H Molecular cloning, sequence analysis, and functional characterization of the lipopolysaccharide biosynthetic gene kdtA encoding 3-deoxy-alpha-D-manno-octulosonic acid transferase of Chlamydia pneumoniae strain TW-183. Molecular microbiology 18, 391–399 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Mamat U, Baumann M, Schmidt G & Brade H The genus-specific lipopolysaccharide epitope of Chlamydia is assembled in C. psittaci and C. trachomatis by glycosyltransferases of low homology. Molecular microbiology 10, 935–941 (1993). [DOI] [PubMed] [Google Scholar]
- 17.Chung HS & Raetz CR Dioxygenases in Burkholderia ambifaria and Yersinia pestis that hydroxylate the outer Kdo unit of lipopolysaccharide. Proceedings of the National Academy of Sciences of the United States of America 108, 510–515, doi: 10.1073/pnas.1016462108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White KA, Lin S, Cotter RJ & Raetz CR A Haemophilus influenzae gene that encodes a membrane bound 3-deoxy-D-manno-octulosonic acid (Kdo) kinase. Possible involvement of kdo phosphorylation in bacterial virulence. The Journal of biological chemistry 274, 31391–31400 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Boll JM et al. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. mBio 6, e00478–00415, doi: 10.1128/mBio.00478-15 (2015).Identification of secondary acyltransferases in Acinetobacter baumannii including an unusual, dual-functioning LpxM that catalyzes transfer of acyl chains to two positions.
- 20.Eshghi A, Henderson J, Trent MS & Picardeau M Leptospira interrogans lpxD Homologue Is Required for Thermal Acclimatization and Virulence. Infection and immunity 83, 4314–4321, doi: 10.1128/iai.00897-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y et al. LPS remodeling is an evolved survival strategy for bacteria. Proceedings of the National Academy of Sciences of the United States of America 109, 8716–8721, doi: 10.1073/pnas.1202908109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo L et al. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95, 189–198 (1998). [DOI] [PubMed] [Google Scholar]
- 23.Reynolds CM et al. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3'-acyloxyacyl moiety of lipid A. The Journal of biological chemistry 281, 21974–21987, doi: 10.1074/jbc.M603527200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trent MS, Ribeiro AA, Lin S, Cotter RJ & Raetz CR An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-L-arabinose to lipid A: induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. The Journal of biological chemistry 276, 43122–43131, doi: 10.1074/jbc.M106961200 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Gibbons HS, Lin S, Cotter RJ & Raetz CR Oxygen requirement for the biosynthesis of the S-2-hydroxymyristate moiety in Salmonella typhimurium lipid A. Function of LpxO, A new Fe2+/alpha-ketoglutarate-dependent dioxygenase homologue. The Journal of biological chemistry 275, 32940–32949, doi: 10.1074/jbc.M005779200 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Hankins JV, Madsen JA, Giles DK, Brodbelt JS & Trent MS Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proceedings of the National Academy of Sciences of the United States of America 109, 8722–8727, doi: 10.1073/pnas.1201313109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Needham BD & Trent MS Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nature reviews. Microbiology 11, 467–481, doi: 10.1038/nrmicro3047 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanistanon D et al. A Francisella mutant in lipid A carbohydrate modification elicits protective immunity. PLoS pathogens 4, e24, doi: 10.1371/journal.ppat.0040024 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herrera CM, Hankins JV & Trent MS Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Molecular microbiology 76, 1444–1460, doi: 10.1111/j.1365-2958.2010.07150.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowicki EM, O'Brien JP, Brodbelt JS & Trent MS Characterization of Pseudomonas aeruginosa LpxT reveals dual positional lipid A kinase activity and co-ordinated control of outer membrane modification. Molecular microbiology 94, 728–741, doi: 10.1111/mmi.12796 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerouge I & Vanderleyden J O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS microbiology reviews 26, 17–47, doi: 10.1111/j.1574-6976.2002.tb00597.x (2002). [DOI] [PubMed] [Google Scholar]
- 32.Samuel G & Reeves P Biosynthesis of O-antigens: genes and pathways involved in nucleotide sugar precursor synthesis and O-antigen assembly. Carbohydrate research 338, 2503–2519 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Stead C et al. A novel 3-deoxy-D-manno-octulosonic acid (Kdo) hydrolase that removes the outer Kdo sugar of Helicobacter pylori lipopolysaccharide. Journal of bacteriology 187, 3374–3383, doi: 10.1128/jb.187.10.3374-3383.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stead CM, Beasley A, Cotter RJ & Trent MS Deciphering the unusual acylation pattern of Helicobacter pylori lipid A. Journal of bacteriology 190, 7012–7021, doi: 10.1128/jb.00667-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tran AX et al. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. The Journal of biological chemistry 279, 55780–55791, doi: 10.1074/jbc.M406480200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran AX, Stead CM & Trent MS Remodeling of Helicobacter pylori lipopolysaccharide. Journal of endotoxin research 11, 161–166, doi: 10.1179/096805105x37349 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Chen HD & Groisman EA The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annual review of microbiology 67, 83–112, doi: 10.1146/annurev-micro-092412-155751 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prost LR & Miller SI The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cellular microbiology 10, 576–582, doi: 10.1111/j.1462-5822.2007.01111.x (2008). [DOI] [PubMed] [Google Scholar]
- 39.Garcia Vescovi E, Soncini FC & Groisman EA Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174 (1996). [DOI] [PubMed] [Google Scholar]
- 40.Richards SM, Strandberg KL, Conroy M & Gunn JS Cationic antimicrobial peptides serve as activation signals for the Salmonella Typhimurium PhoPQ and PmrAB regulons in vitro and in vivo. Frontiers in cellular and infection microbiology 2, 102, doi: 10.3389/fcimb.2012.00102 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan J, Jin F, Glatter T & Sourjik V Osmosensing by the bacterial PhoQ/PhoP two-component system. Proceedings of the National Academy of Sciences of the United States of America 114, E10792–e10798, doi: 10.1073/pnas.1717272114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gooderham WJ et al. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology (Reading, England) 155, 699–711, doi: 10.1099/mic.0.024554-0 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Bader MW et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472, doi: 10.1016/j.cell.2005.05.030 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Prost LR et al. Activation of the bacterial sensor kinase PhoQ by acidic pH. Molecular cell 26, 165–174, doi: 10.1016/j.molcel.2007.03.008 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Choi J & Groisman EA Acidic pH sensing in the bacterial cytoplasm is required for Salmonella virulence. Molecular microbiology 101, 1024–1038, doi: 10.1111/mmi.13439 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi J & Groisman EA Activation of master virulence regulator PhoP in acidic pH requires the Salmonella-specific protein UgtL. Science signaling 10, doi: 10.1126/scisignal.aan6284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo L et al. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science (New York, N.Y.) 276, 250–253 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Kox LF, Wosten MM & Groisman EA A small protein that mediates the activation of a two-component system by another two-component system. The EMBO journal 19, 1861–1872, doi: 10.1093/emboj/19.8.1861 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubin EJ, Herrera CM, Crofts AA & Trent MS PmrD is required for modifications to escherichia coli endotoxin that promote antimicrobial resistance. Antimicrobial agents and chemotherapy 59, 2051–2061, doi: 10.1128/aac.05052-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kato A & Groisman EA Connecting two-component regulatory systems by a protein that protects a response regulator from dephosphorylation by its cognate sensor. Genes & development 18, 2302–2313, doi: 10.1101/gad.1230804 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lippa AM & Goulian M Feedback inhibition in the PhoQ/PhoP signaling system by a membrane peptide. PLoS genetics 5, e1000788, doi: 10.1371/journal.pgen.1000788 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wosten MM, Kox LF, Chamnongpol S, Soncini FC & Groisman EA A signal transduction system that responds to extracellular iron. Cell 103, 113–125 (2000). [DOI] [PubMed] [Google Scholar]
- 53.Perez JC & Groisman EA Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Molecular microbiology 63, 283–293, doi: 10.1111/j.1365-2958.2006.05512.x (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato A, Chen HD, Latifi T & Groisman EA Reciprocal control between a bacterium's regulatory system and the modification status of its lipopolysaccharide. Molecular cell 47, 897–908, doi: 10.1016/j.molcel.2012.07.017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fernandez PA et al. Fnr and ArcA Regulate Lipid A Hydroxylation in Salmonella Enteritidis by Controlling lpxO Expression in Response to Oxygen Availability. Frontiers in microbiology 9, 1220, doi: 10.3389/fmicb.2018.01220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nowicki EM, O'Brien JP, Brodbelt JS & Trent MS Extracellular zinc induces phosphoethanolamine addition to Pseudomonas aeruginosa lipid A via the ColRS two-component system. Molecular microbiology 97, 166–178, doi: 10.1111/mmi.13018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernandez L et al. Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrobial agents and chemotherapy 54, 3372–3382, doi: 10.1128/aac.00242-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fernandez L et al. The two-component system CprRS senses cationic peptides and triggers adaptive resistance in Pseudomonas aeruginosa independently of ParRS. Antimicrobial agents and chemotherapy 56, 6212–6222, doi: 10.1128/aac.01530-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bilecen K et al. Polymyxin B resistance and biofilm formation in Vibrio cholerae are controlled by the response regulator CarR. Infection and immunity 83, 1199–1209, doi: 10.1128/iai.02700-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Herrera CM et al. The Vibrio cholerae VprA-VprB two-component system controls virulence through endotoxin modification. mBio 5, doi: 10.1128/mBio.02283-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng YH, Lin TL, Lin YT & Wang JT Amino Acid Substitutions of CrrB Responsible for Resistance to Colistin through CrrC in Klebsiella pneumoniae. Antimicrobial agents and chemotherapy 60, 3709–3716, doi: 10.1128/aac.00009-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coornaert A et al. MicA sRNA links the PhoP regulon to cell envelope stress. Molecular microbiology 76, 467–479, doi: 10.1111/j.1365-2958.2010.07115.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon K & Gottesman S A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Molecular microbiology 74, 1314–1330, doi: 10.1111/j.1365-2958.2009.06944.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawasaki K, Ernst RK & Miller SI Inhibition of Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. Journal of bacteriology 187, 2448–2457, doi: 10.1128/jb.187.7.2448-2457.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reines M et al. Deciphering the acylation pattern of Yersinia enterocolitica lipid A. PLoS pathogens 8, e1002978, doi: 10.1371/journal.ppat.1002978 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bishop RE et al. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. The EMBO journal 19, 5071–5080, doi: 10.1093/emboj/19.19.5071 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia W et al. Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. The Journal of biological chemistry 279, 44966–44975, doi: 10.1074/jbc.M404963200 (2004). [DOI] [PubMed] [Google Scholar]
- 68.Murata T, Tseng W, Guina T, Miller SI & Nikaido H PhoPQ-mediated regulation produces a more robust permeability barrier in the outer membrane of Salmonella enterica serovar typhimurium. Journal of bacteriology 189, 7213–7222, doi: 10.1128/jb.00973-07 (2007).LPS modifications upregulated by PhoPQ and PmrAB alter the permeability properties of the OM that is more stable in Mg2+ limiting environments, but less stable in high Mg2+ environments.
- 69.Sassi N, Paul C, Martin A, Bettaieb A & Jeannin JF Lipid A-induced responses in vivo. Advances in experimental medicine and biology 667, 69–80, doi: 10.1007/978-1-4419-1603-7_7 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Kong Q et al. Palmitoylation state impacts induction of innate and acquired immunity by the Salmonella enterica serovar typhimurium msbB mutant. Infection and immunity 79, 5027–5038, doi: 10.1128/iai.05524-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kong Q et al. Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infection and immunity 80, 3215–3224, doi: 10.1128/iai.00123-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casella CR & Mitchell TC Putting endotoxin to work for us: monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cellular and molecular life sciences : CMLS 65, 3231–3240, doi: 10.1007/s00018-008-8228-6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamamoto M & Akira S Lipid A receptor TLR4-mediated signaling pathways. Advances in experimental medicine and biology 667, 59–68, doi: 10.1007/978-1-4419-1603-7_6 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Cullen TW et al. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS pathogens 7, e1002454, doi: 10.1371/journal.ppat.1002454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mandell L et al. Intact gram-negative Helicobacter pylori, Helicobacter felis, and Helicobacter hepaticus bacteria activate innate immunity via toll-like receptor 2 but not toll-like receptor 4. Infection and immunity 72, 6446–6454, doi: 10.1128/iai.72.11.6446-6454.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Llobet E et al. Deciphering tissue-induced Klebsiella pneumoniae lipid A structure. Proceedings of the National Academy of Sciences of the United States of America 112, E6369–6378, doi: 10.1073/pnas.1508820112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192, doi: 10.1038/nature13683 (2014). [DOI] [PubMed] [Google Scholar]
- 78.Hagar JA, Powell DA, Aachoui Y, Ernst RK & Miao EA Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science (New York, N.Y.) 341, 1250–1253, doi: 10.1126/science.1240988 (2013).Along with Kayagaki et al. 2013, discovered that murine caspase 11 interacts with cytoplasmic LPS and stimulates the Inflammasome independently of TLR4 stimulation.
- 79.Kayagaki N et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science (New York, N.Y.) 341, 1246–1249, doi: 10.1126/science.1240248 (2013).Along with Hagar et al. 2013, discovered that murine caspase 11 interacts with cytoplasmic LPS and stimulates the Inflammasome independently of TLR4 stimulation.
- 80.Guo H, Callaway JB & Ting JP Inflammasomes: mechanism of action, role in disease, and therapeutics. Nature medicine 21, 677–687, doi: 10.1038/nm.3893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lagrange B et al. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nature communications 9, 242, doi: 10.1038/s41467-017-02682-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peschel A & Sahl HG The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nature reviews. Microbiology 4, 529–536, doi: 10.1038/nrmicro1441 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Faith JJ et al. The long-term stability of the human gut microbiota. Science (New York, N.Y.) 341, 1237439, doi: 10.1126/science.1237439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cullen TW et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science (New York, N.Y.) 347, 170–175, doi: 10.1126/science.1260580 (2015).LPS modifications that mediate AMP resistance contribute to stable maintenance of gut microbiota.
- 85.Schwarz S & Johnson AP Transferable resistance to colistin: a new but old threat. The Journal of antimicrobial chemotherapy 71, 2066–2070, doi: 10.1093/jac/dkw274 (2016). [DOI] [PubMed] [Google Scholar]
- 86.Schwechheimer C & Kuehn MJ Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nature reviews. Microbiology 13, 605–619, doi: 10.1038/nrmicro3525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haurat MF et al. Selective sorting of cargo proteins into bacterial membrane vesicles. The Journal of biological chemistry 286, 1269–1276, doi: 10.1074/jbc.M110.185744 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Horstman AL & Kuehn MJ Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. The Journal of biological chemistry 275, 12489–12496 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Horstman AL, Bauman SJ & Kuehn MJ Lipopolysaccharide 3-deoxy-D-manno-octulosonic acid (Kdo) core determines bacterial association of secreted toxins. The Journal of biological chemistry 279, 8070–8075, doi: 10.1074/jbc.M308633200 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen YY et al. The outer membrane protein LptO is essential for the O-deacylation of LPS and the co-ordinated secretion and attachment of A-LPS and CTD proteins in Porphyromonas gingivalis. Molecular microbiology 79, 1380–1401, doi: 10.1111/j.1365-2958.2010.07530.x (2011). [DOI] [PubMed] [Google Scholar]
- 91.Rangarajan M et al. LptO (PG0027) Is Required for Lipid A 1-Phosphatase Activity in Porphyromonas gingivalis W50. Journal of bacteriology 199, doi: 10.1128/jb.00751-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sato K et al. Identification of Porphyromonas gingivalis proteins secreted by the Por secretion system. FEMS microbiology letters 338, 68–76, doi: 10.1111/1574-6968.12028 (2013). [DOI] [PubMed] [Google Scholar]
- 93.Gabarrini G et al. Conserved Citrullinating Exoenzymes in Porphyromonas Species. Journal of dental research 97, 556–562, doi: 10.1177/0022034517747575 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Konig MF, Paracha AS, Moni M, Bingham CO 3rd & Andrade F Defining the role of Porphyromonas gingivalis peptidylarginine deiminase (PPAD) in rheumatoid arthritis through the study of PPAD biology. Annals of the rheumatic diseases 74, 2054–2061, doi: 10.1136/annrheumdis-2014-205385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wegner N et al. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunological reviews 233, 34–54, doi: 10.1111/j.0105-2896.2009.00850.x (2010). [DOI] [PubMed] [Google Scholar]
- 96.Gabarrini G et al. Dropping anchor: attachment of peptidylarginine deiminase via A-LPS to secreted outer membrane vesicles of Porphyromonas gingivalis. Sci Rep 8, 8949, doi: 10.1038/s41598-018-27223-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gabarrini G et al. There's no place like OM: Vesicular sorting and secretion of the peptidylarginine deiminase of Porphyromonas gingivalis. Virulence 9, 456–464, doi: 10.1080/21505594.2017.1421827 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mashburn-Warren L et al. Interaction of quorum signals with outer membrane lipids: insights into prokaryotic membrane vesicle formation. Molecular microbiology 69, 491–502 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li A, Schertzer JW & Yong X Molecular conformation affects the interaction of the Pseudomonas quinolone signal with the bacterial outer membrane. The Journal of biological chemistry, doi: 10.1074/jbc.AC118.006844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Elhenawy W et al. LPS Remodeling Triggers Formation of Outer Membrane Vesicles in Salmonella. mBio 7, doi: 10.1128/mBio.00940-16 (2016).PagL-mediated deacylation of LPS in Salmonella stimulates production of OMVs.
- 101.Bonnington KE & Kuehn MJ Outer Membrane Vesicle Production Facilitates LPS Remodeling and Outer Membrane Maintenance in Salmonella during Environmental Transitions. mBio 7, doi: 10.1128/mBio.01532-16 (2016).During environmental transition that alter LPS modification, certain LPS species are selectively secreted and others under-represented in OMVs.
- 102.Cipolla L et al. New targets for antibacterial design: Kdo biosynthesis and LPS machinery transport to the cell surface. Current medicinal chemistry 18, 830–852 (2011). [DOI] [PubMed] [Google Scholar]
- 103.Zhou P & Zhao J Structure, inhibition, and regulation of essential lipid A enzymes. Biochimica et biophysica acta. Molecular and cell biology of lipids 1862, 1424–1438, doi: 10.1016/j.bbalip.2016.11.014 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Erwin AL Antibacterial Drug Discovery Targeting the Lipopolysaccharide Biosynthetic Enzyme LpxC. Cold Spring Harbor perspectives in medicine 6, doi: 10.1101/cshperspect.a025304 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richie DL et al. Toxic Accumulation of LPS Pathway Intermediates Underlies the Requirement of LpxH for Growth of Acinetobacter baumannii ATCC 19606. PloS one 11, e0160918, doi: 10.1371/journal.pone.0160918 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wei JR et al. LpxK Is Essential for Growth of Acinetobacter baumannii ATCC 19606: Relationship to Toxic Accumulation of Lipid A Pathway Intermediates. mSphere 2, doi: 10.1128/mSphere.00199-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Piizzi G et al. Design, Synthesis, and Properties of a Potent Inhibitor of Pseudomonas aeruginosa Deacetylase LpxC. Journal of medicinal chemistry 60, 5002–5014, doi: 10.1021/acs.jmedchem.7b00377 (2017). [DOI] [PubMed] [Google Scholar]
- 108.Pratap S et al. Acyl chain preference and inhibitor identification of Moraxella catarrhalis LpxA: Insight through crystal structure and computational studies. International journal of biological macromolecules 96, 759–765, doi: 10.1016/j.ijbiomac.2017.01.005 (2017). [DOI] [PubMed] [Google Scholar]
- 109.Nayar AS et al. Novel antibacterial targets and compounds revealed by a high-throughput cell wall reporter assay. Journal of bacteriology 197, 1726–1734, doi: 10.1128/jb.02552-14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Richie DL et al. A pathway-directed positive growth restoration assay to facilitate the discovery of lipid A and fatty acid biosynthesis inhibitors in Acinetobacter baumannii. PloS one 13, e0193851, doi: 10.1371/journal.pone.0193851 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ho H et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 557, 196–201, doi: 10.1038/s41586-018-0083-5 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Zhang G et al. Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proceedings of the National Academy of Sciences of the United States of America 115, 6834–6839, doi: 10.1073/pnas.1804670115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.May JM et al. The Antibiotic Novobiocin Binds and Activates the ATPase That Powers Lipopolysaccharide Transport. Journal of the American Chemical Society 139, 17221–17224, doi: 10.1021/jacs.7b07736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mandler MD et al. Novobiocin Enhances Polymyxin Activity by Stimulating Lipopolysaccharide Transport. Journal of the American Chemical Society 140, 6749–6753, doi: 10.1021/jacs.8b02283 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Srinivas N et al. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science (New York, N.Y.) 327, 1010–1013, doi: 10.1126/science.1182749 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Vetterli SU, Moehle K & Robinson JA Synthesis and antimicrobial activity against Pseudomonas aeruginosa of macrocyclic beta-hairpin peptidomimetic antibiotics containing N-methylated amino acids. Bioorganic & medicinal chemistry 24, 6332–6339, doi: 10.1016/j.bmc.2016.05.027 (2016). [DOI] [PubMed] [Google Scholar]
- 117.Werneburg M et al. Inhibition of lipopolysaccharide transport to the outer membrane in Pseudomonas aeruginosa by peptidomimetic antibiotics. Chembiochem 13, 1767–1775, doi: 10.1002/cbic.201200276 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Vetterli SU et al. Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli. Science advances 4, eaau2634, doi: 10.1126/sciadv.aau2634 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang X et al. Identification of an anti-Gram-negative bacteria agent disrupting the interaction between LPS transporters LptA and LptC. International journal of antimicrobial agents, doi: 10.1016/j.ijantimicag.2018.11.016 (2018). [DOI] [PubMed] [Google Scholar]
- 120.Harris TL et al. Small molecule downregulation of PmrAB reverses lipid A modification and breaks colistin resistance. ACS chemical biology 9, 122–127, doi: 10.1021/cb400490k (2014). [DOI] [PubMed] [Google Scholar]
- 121.Kline T et al. Synthesis of and evaluation of lipid A modification by 4-substituted 4-deoxy arabinose analogs as potential inhibitors of bacterial polymyxin resistance. Bioorganic & medicinal chemistry letters 18, 1507–1510, doi: 10.1016/j.bmcl.2007.12.061 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anandan A et al. Structure of a lipid A phosphoethanolamine transferase suggests how conformational changes govern substrate binding. Proceedings of the National Academy of Sciences of the United States of America 114, 2218–2223, doi: 10.1073/pnas.1612927114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Touze T, Tran AX, Hankins JV, Mengin-Lecreulx D & Trent MS Periplasmic phosphorylation of lipid A is linked to the synthesis of undecaprenyl phosphate. Molecular microbiology 67, 264–277, doi: 10.1111/j.1365-2958.2007.06044.x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cullen TW et al. EptC of Campylobacter jejuni mediates phenotypes involved in host interactions and virulence. Infection and immunity 81, 430–440, doi: 10.1128/iai.01046-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cullen TW & Trent MS A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proceedings of the National Academy of Sciences of the United States of America 107, 5160–5165, doi: 10.1073/pnas.0913451107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bach JF & Chatenoud L The hygiene hypothesis: an explanation for the increased frequency of insulin-dependent diabetes. Cold Spring Harbor perspectives in medicine 2, a007799, doi: 10.1101/cshperspect.a007799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vatanen T et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell 165, 1551, doi: 10.1016/j.cell.2016.05.056 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Raetz CR, Reynolds CM, Trent MS & Bishop RE Lipid A modification systems in gram-negative bacteria. Annual review of biochemistry 76, 295–329, doi: 10.1146/annurev.biochem.76.010307.145803 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Moffatt JH et al. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrobial agents and chemotherapy 54, 4971–4977, doi: 10.1128/aac.00834-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Peng D, Hong W, Choudhury BP, Carlson RW & Gu XX Moraxella catarrhalis bacterium without endotoxin, a potential vaccine candidate. Infection and immunity 73, 7569–7577, doi: 10.1128/iai.73.11.7569-7577.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Steeghs L et al. Meningitis bacterium is viable without endotoxin. Nature 392, 449–450, doi: 10.1038/33046 (1998). [DOI] [PubMed] [Google Scholar]
- 132.Beceiro A et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrobial agents and chemotherapy 58, 518–526, doi: 10.1128/aac.01597-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Boll JM et al. A penicillin-binding protein inhibits selection of colistin-resistant, lipooligosaccharide-deficient Acinetobacter baumannii. Proceedings of the National Academy of Sciences of the United States of America 113, E6228–e6237, doi: 10.1073/pnas.1611594113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Henry R et al. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrobial agents and chemotherapy 56, 59–69, doi: 10.1128/aac.05191-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Powers MJ & Trent MS Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proceedings of the National Academy of Sciences of the United States of America, doi: 10.1073/pnas.1806714115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qureshi N, Takayama K & Ribi E Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium. The Journal of biological chemistry 257, 11808–11815 (1982). [PubMed] [Google Scholar]
- 137.Needham BD et al. Modulating the innate immune response by combinatorial engineering of endotoxin. Proceedings of the National Academy of Sciences of the United States of America 110, 1464–1469, doi: 10.1073/pnas.1218080110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gregg KA et al. Rationally Designed TLR4 Ligands for Vaccine Adjuvant Discovery. mBio 8, doi: 10.1128/mBio.00492-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zariri A, Pupo E, van Riet E, van Putten JP & van der Ley P Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Sci Rep 6, 36575, doi: 10.1038/srep36575 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sanders H & Feavers IM Adjuvant properties of meningococcal outer membrane vesicles and the use of adjuvants in Neisseria meningitidis protein vaccines. Expert review of vaccines 10, 323–334, doi: 10.1586/erv.11.10 (2011). [DOI] [PubMed] [Google Scholar]
- 141.Price NL et al. Glycoengineered Outer Membrane Vesicles: A Novel Platform for Bacterial Vaccines. Sci Rep 6, 24931, doi: 10.1038/srep24931 (2016).Engineering of non-pathogenic E. coli to produce LPS modified with glycans from pathogenic bacteria, such as capsule from Streptococcus pneumoniae and heptasaccharide N-glycan Campylobacter jejuni, that can be isolated in OMVs and used as vaccines.
- 142.Chen L et al. Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proceedings of the National Academy of Sciences of the United States of America 113, E3609–3618, doi: 10.1073/pnas.1518311113 (2016).Engineering of non-pathogenic E.coli to produce LPS modified with glycans from pathogenic bacteria, such as O antigen from Francisella tularensis subsp. Tularensis, that can be isolated in OMVs and used as vaccines.
- 143.Valentine JL et al. Immunization with Outer Membrane Vesicles Displaying Designer Glycotopes Yields Class-Switched, Glycan-Specific Antibodies. Cell chemical biology 23, 655–665, doi: 10.1016/j.chembiol.2016.05.014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou Z, White KA, Polissi A, Georgopoulos C & Raetz CR Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. The Journal of biological chemistry 273, 12466–12475 (1998). [DOI] [PubMed] [Google Scholar]
- 145.Mamat U et al. Single amino acid substitutions in either YhjD or MsbA confer viability to 3-deoxy-d-manno-oct-2-ulosonic acid-depleted Escherichia coli. Molecular microbiology 67, 633–648, doi: 10.1111/j.1365-2958.2007.06074.x (2008). [DOI] [PubMed] [Google Scholar]
- 146.Mi W et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237, doi: 10.1038/nature23649 (2017).Structural snapshot of MsbA2 in complex with LPS that suggests a “trap and flip” model for LPS flipping across the IM.
- 147.Ortega XP et al. A putative gene cluster for aminoarabinose biosynthesis is essential for Burkholderia cenocepacia viability. Journal of bacteriology 189, 3639–3644, doi: 10.1128/jb.00153-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hamad MA, Di Lorenzo F, Molinaro A & Valvano MA Aminoarabinose is essential for lipopolysaccharide export and intrinsic antimicrobial peptide resistance in Burkholderia cenocepacia(dagger). Molecular microbiology 85, 962–974, doi: 10.1111/j.1365-2958.2012.08154.x (2012). [DOI] [PubMed] [Google Scholar]
- 149.Bertani BR, Taylor RJ, Nagy E, Kahne D & Ruiz N A cluster of residues in the lipopolysaccharide exporter that selects substrate variants for transport to the outer membrane. Molecular microbiology, doi: 10.1111/mmi.14059 (2018).In E.coli, LPS extraction from the IM by the LPS transporter requires interactions with the 1 and 4′ positions of lipid A.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.