Abstract
The tumor genome of a patient with advanced pancreatic cancer was sequenced to identify potential therapeutic targetable mutations after standard of care failed to produce any significant overall response. Matched tumor-normal whole-genome sequencing revealed somatic mutations in BRAF, TP53, CDKN2A, and a focal deletion of SMAD4. The BRAF variant was an in-frame deletion mutation (ΔN486_P490), which had been previously demonstrated to be a kinase-activating alteration in the BRAF kinase domain. Working with the Novartis patient assistance program allowed us to treat the patient with the BRAF inhibitor, dabrafenib. The patient's overall clinical condition improved dramatically with dabrafenib. Levels of serum tumor marker dropped immediately after treatment, and a subsequent CT scan revealed a significant decrease in the size of both primary and metastatic lesions. The dabrafenib-induced remission lasted for 6 mo. Preclinical studies published concurrently with the patient's treatment showed that the BRAF in-frame mutation (ΔNVTAP) induces oncogenic activation by a mechanism distinct from that induced by V600E, and that this difference dictates the responsiveness to different BRAF inhibitors. This study describes a dramatic instance of how high-level genomic technology and analysis was necessary and sufficient to identify a clinically logical treatment option that was then utilized and shown to be of clinical value for this individual.
Keywords: neoplasm of the pancreas
CASE PRESENTATION
A 65-yr-old male presented with a 2-wk history of jaundice preceded by a 3-wk history of abdominal discomfort and diarrhea, accompanied by a 15–20-pound weight loss in 1 mo. He had a prior medical history of morbid obesity and sleep apnea. Initial computed tomography (CT) revealed dilation of both the common bile duct and the pancreatic duct and a poorly defined hyperdense region in the pancreatic head. Upon referral to a gastroenterologist, the patient underwent endoscopic retrograde cholangiopancreatography with sphincterotomy. Brush cytology revealed pancreatic adenocarcinoma cells, and the patient subsequently underwent a Whipple procedure to remove the tumor. His tumor was moderately to poorly differentiated pancreatic ductal adenocarcinoma, T3N1, Stage IIB (tumor growing outside of the pancreas but not into major blood vessels/nerves, and in 3/21 lymph nodes). Given his relatively poor prognosis, he was offered adjuvant chemotherapy. Two months after the Whipple procedure, he started gemzar and abraxane that was continued for five cycles and received his last cycle with gemzar only because of weakness and anemia. Both the patient and treating physician decided on the doublet of gemzar and abraxane because of a better toxicity profile, simpler logistics, and improved clinical outcome in a metastatic setting. The patient was subsequently referred to radiation oncology for adjuvant chemoradiation therapy; however, a restaging CT scan performed 5 mo after adjuvant treatment showed multiple liver metastases, and therefore he did not receive any radiation treatment. A liver biopsy performed at the time of disease recurrence was positive for metastatic pancreatic adenocarcinoma, indicating that despite the initial staging of the patient's tumor as stage IIB it is likely that he had distant metastasis. He received a second line of combinatorial chemotherapy (5-FU/liposomal irinotecan) for 2 mo and third-line chemotherapy with FOLFOX (5-FU and oxaliplatin) with progression of his disease. His disease was monitored by serum tumor marker carbohydrate antigen (CA) 19-9 throughout his treatment. In addition, the patient underwent CT monitoring of the chest, abdomen, and pelvis. Based on his clinical status the patient was eligible for tumor genome profiling by the IBM Watson Health sponsored, New York Genome Center (NYGC) Cancer Alliance pilot study. The study was approved by Stony Brook University and Biomedical Research Alliance of New York institutional review boards.
METHODS
Whole-Genome, Whole-Exome, and Transcriptome Sequencing
The specimens were received as OCT (Optimal Cutting Temperature compound)-embedded tumor tissue from a liver metastasis and peripheral blood as normal. DNA and RNA purification, extraction, and library preparations were performed as previously reported (Wrzeszczynski et al. 2018). Whole-genome DNA sequencing was performed on the Illumina HiSeq X 2 × 150 bp paired-end sequencing (Illumina). The libraries were loaded at a 2:1 tumor:normal ratio to reach coverage of 110× (average read depth) for the tumor sample and 64× for the normal sample. As part of the study, whole-exome DNA sequencing was additionally performed using SureSelectXT Human All Exon V6 + COSMIC capture kit (Agilent) and 2 × 125 bp paired-end sequencing was performed on a Illumina HiSeq 2500 instrument to reach an average read depth of 238× for the tumor and 105× for the normal. The RNA sample was prepared using an mRNA protocol as previously reported (Wrzeszczynski et al. 2018), and the library was sequenced on the Illumina HiSeq 2500 2 × 125 bp rapid run platform obtaining approximately 57 million reads. Somatic single-nucleotide variants (SNVs), insertions and deletions (indels), structural variants (SVs), copy-number variants (CNVs), and fusions were called from whole-genome and transcriptome sequencing as previously reported (Wrzeszczynski et al. 2017). Whole-exome sequencing was used only to identify SNVs and indels. RNA sequencing was used to call fusions and confirm the expression of SNVs and indels.
Microarray Genotyping
Genotyping was performed as an internal quality control measure and to determine copy number (ploidy) and tumor content (purity). Extracted DNA was normalized, denatured, and neutralized. After an overnight amplification step, DNA was fragmented, precipitated, resuspended, and hybridized to Illumina HumanOmni2.5M BeadChips (Illumina WG-313-2511). BeadChips were loaded onto the Illumina HiScan microarray scanner, which yields fluorescence intensity data files that are interpreted in the context of biological information about the SNVs on the Bead Chips to generate genotype calls. Copy number, tumor purity, and ploidy are calculated using the ASCAT tool (Van Loo et al. 2010).
Sanger Sequencing
DNA derived from tumor was independently used as a template to PCR amplify BRAF exon 12 using the following primer sequences: exon 12F 5′-AATGGTATGGAGTTAGGGCTATG-3′ and exon 12R 5′-CTGGGAACCAGGAGCTAATAAA-3′. The resulting 258 bp product was purified using ExoSAP-IT (Thermo Fisher Scientific, 78201.1.ML) and Sanger sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific 4337455). The samples were loaded onto the ABI 3500 Dx machine and AB1 files were analyzed using Mutation Surveyor version 5.0 (SoftGenetics).
Variant Interpretation
Initial variant detection was performed as part of the NYGC whole genome and transcriptome clinical assay (Wrzeszczynski et al. 2018). SNV and indels were annotated via snpEff, snpSift (Cingolani et al. 2012), and GATK VariantAnnotator using annotation from ENSEMBL, COSMIC (Tate et al. 2019), Gene Ontology, and 1000 Genomes. All variants were annotated based on an in-house clinical classification system in which variants in targetable and COSMIC cancer census (Forbes et al. 2011) genes were identified and prioritized. Initial matching of each variant to drug(s) was performed by (i) identifying the tumor-specific gene variants, relative to normal germline DNA, based on SNV, CNV, and RNA sequencing data and (ii) matching to the expert-curated NYGC drug-to-gene database (June 2016). Our internal drug-to-gene database was assembled by manual curation of publicly available data from the National Comprehensive Network (NCCN; https://www.nccn.org/), the U.S. Food and Drug Administration (https://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs), Clinical Interpretations of Variants in Cancer (CIVIC) (civic.genome.wustl.edu), Precision Cancer Therapy-MD Anderson (https://pct.mdanderson.org/), OncoKB (oncokb.org), canSar (https://cansar.icr.ac.uk), Pharmacogenomics Knowledgebase (PharmGKB) (www.pharmgkb.org), ClinicalTrials.gov (clinicaltrials.gov), and directed literature searches.
RESULTS
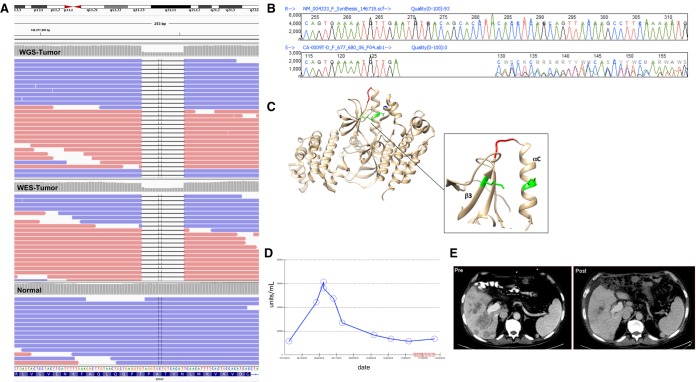
Whole-genome sequencing revealed a BRAF in-frame deletion of five amino acids within the kinase domain (Table 1). This deletion was also detectable by whole-exome sequencing, transcriptome sequencing and was confirmed by Sanger sequencing (Fig. 1A,B). Activating mutations in the KRAS signaling pathway are common in pancreatic cancers, with ∼90% of pancreatic ductal adenocarcinomas (PDACs) containing mutations in KRAS (Jones et al. 2008; Waddell et al. 2015). In one large cohort, BRAF mutations were detected in 1.4% of pancreatic carcinomas and were mutually exclusive with KRAS mutations (Foster et al. 2016). About half of these were deletions in the β3-αC loop of five residues, including the Asn486_Pro490 deletion seen in this specimen. The deletion potentially shifts the αC helix into an active conformation such that the catalytic residue Lys483 of β3 forms a salt bridge interaction with Glu501 of the αC helix (Fig. 1C). In preclinical studies this variant was shown to be sensitive to the BRAF inhibitor dabrafenib and a sorafenib-related compound, but resistant to vemurafenib in cell line assays (Foster et al. 2016). BRAF inhibitor sensitivity is lost in longer term assays, although the mutant remains sensitive to MEK inhibitors in the same time-frame. A dabrafenib/trametinib combination phase I/II clinical trial (NCT01989585) was identified.
Table 1.
Genomic findings
| Gene | CHR:POS | HGVS cDNA | HGVS protein | Variant type | Predicted effect | Variant allele frequency (Alt/Ref allele read count) | Read depth of variant |
|---|---|---|---|---|---|---|---|
| BRAF | 7:140477836 | NM_004333.4:c.1457_1471del ATGTGACAGCACCTA | NP_004324.2: p.Asn486_ Pro490del | In frame deletion | Activating mutation (Foster et al. 2016) | WGS: 66% (68/35) WES: 64% (77/44) |
WGS: 103× WES: 121× |
| CDKN2A | 9:21971120 | NM_001195132.1:c.238C>T | NP_001182061.1: p.Arg80a | Nonsense | Variant of uncertain significance likely loss of function (Rachakonda et al. 2013; Yarbrough et al. 1999) | WGS: 62% (48/29) WES: 66% (165/84) |
WGS: 77× WES: 249× |
| TP53 | 17:7579380 | NM_000546.5:c.299_306del AGAAAACC | NP_000537: p.Gln100Leufs Ter46 | Frame shift | Variant of uncertain significance likely oncogenic (Chakravarty et al. 2017) | WGS: 56% (42/33) WES: 52% (82/76) |
WGS: 75× WES: 158× |
| SMAD4 | 18q21.2 | NM_005359.5 | NM_005350.1 | Homozygous deletion | Loss of function (Jia et al. 2017) | Copy-number log2(T/N) = −1.624 | WGS (18q mean coverage): 96× |
aTermination (stop) codon (https://www.hgvs.org/mutnomen/standards.html).
Figure 1.
(A) BRAF Chr 7:140477836 (c.1457_1471delATGTGACAGCACCTA_p.Asn486_Pro490del) as visualized in whole-genome sequencing (top track, WGS-Tumor), whole-exome sequencing (center track, WES-Tumor), and blood normal sample (lower track, Normal). (B) Sanger sequencing electropherogram confirming the BRAF ΔNVTAP variant. (C) Location of the deletion variant in BRAF protein (red) and residues involved in a potential salt bridge (green) (PDB:1UWH). (D) CA 19-9 marker serum levels (units/mL) over time. (E) Patient CT scan of pre- (left) and post- (right) dabrafenib treatment demonstrating decrease in the hepatic metastatic tumor burden posttreatment.
Additional variants of clinical significance identified were a CDKN2A stop codon, a TP53 frameshift mutation and homozygous loss of SMAD4 (Table 1). Inactivating mutations in CDKN2A, TP53, and SMAD4 occur in >50% of pancreatic cancers (Waddell et al. 2015; Kamisawa et al. 2016). The CDKN2A mutation likely affected all copies of CDKN2A based on variant allele frequency of 62% and an estimated tumor purity of 61%, which would result in complete loss of CDKN2A function. CDKN2A (p16/INK4a) binds to CDK4/6 regulating the cell cycle. Loss of CDKN2A can result in activation of CDK4 that in turn phosphorylates RB1. RB1-P can no longer inhibit E2F transcription factor resulting in cellular proliferation. Combinatorial therapy with CDK4/6 inhibitors, palbociclib and ribociclib, would be a potential therapeutic option (Franco et al. 2014). A TP53 early frameshift mutation, also likely to be homozygous (Table 1), would be predicted to lead to loss of p53 growth-inhibitory effects. The whole-genome copy-number profile suggested a highly aberrant genome with tumor ploidy of 2.46. SMAD4 two-copy homozygous loss at chromosome position 18q21.2 (log2(T/N) = −1.624) was detected resulting in possible loss of function of the tumor suppressor. SMAD4 has been identified as a key regulator in prostate adenocarcinoma progression (Ding et al. 2011).
SUMMARY
A patient with advanced pancreatic cancer was enrolled in the NYGC Cancer Alliance pilot study for tumor genome sequencing, which revealed somatic mutations in BRAF, TP53, CDKN2A, and a focal deletion of SMAD4. The case was presented at two molecular tumor boards—one at NYGC with scientists and physicians conducting the sequencing pilot study and one at Stony Brook Medical Center with representatives from clinical oncology and pathology. Given no further standard-of-care options and no nearby therapeutic clinical trials, the board recommended participation in the NCI-MATCH clinical trial. However, because the patient did not have the standard BRAF V600E mutation, he was not eligible for NCI-MATCH. The board decided that the demonstrated clinical utility of BRAF inhibition and combined BRAF/MEK inhibition in phase II clinical results in BRAF-mutant melanoma made a compelling case for trying this combination in this patient. Subsequently, the combination of these drugs was approved for BRAF-mutant lung, melanoma, and thyroid cancers (Long et al. 2017; Planchard et al. 2017; Subbiah et al. 2018). We therefore requested and obtained both the BRAF inhibitor dabrafenib and the MEK inhibitor trametinib from Novartis for off-label use. Because of the patient's poor performance status and liver dysfunction, the BRAF inhibitor dabrafenib was started at 75 mg po bid and gradually increased to full dose. The patient's overall clinical condition improved dramatically with dabrafenib. Since initiating therapy the patient had showed a clinical response in palpable liver disease, levels of the serum tumor marker CA 19-9 dropped immediately after treatment (Fig. 1D), and a subsequent CT scan revealed significant decrease in the size of both primary (not shown) and hepatic metastatic lesions (Fig. 1E). The patient responded well for ∼4 mo with no significant side effects but was noted to have increasing abdominal pain and weakness. With rising CA 19-9, trametinib was added but the patient did not respond to further therapy. Eventual CT scans showed progression of disease and the patient decided on palliative and hospice care. A second liver biopsy to better understand the potential mechanism of escape post–dabrafenib treatment was not possible because of the patient's deteriorating clinical condition. Somatic BRAF mutations occur in 3% of pancreatic cancers and are often inversely correlated with KRAS variants (Cancer Genome Atlas Research Network 2017; Guan et al. 2018). The ΔNVTAP BRAF has been observed in <0.5% pancreatic cases (Guan et al. 2018). Here, we show evidence of partial response and direct targetability of dabrafenib in a patient with an identified BRAF in-frame pathogenic deletion that had previously only been described in preclinical studies.
ADDITIONAL INFORMATION
Data Deposition and Access
The data (in vcf format) for this case can be obtained from ftp://ftp.nygenome.org/CA/9/ or by request to the corresponding author until it is made available in the EGA-archive.org public repository (ega-box-1298) as part of the larger study submission. The BRAF variant has been deposited into the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000666267.1.
Ethics Statement
This study was reviewed and approved by the Biomedical Research Alliance of New York Institutional Review Board (IRB), Rockefeller University and the Stony Brook University IRB. Written consent for genomic sequencing was obtained. The protocol allowed for the return of research sequencing results after confirmation in a clinical laboratory.
Acknowledgments
We thank the patient for participation in this study and the research use of his specimens.
Author Contributions
K.O.W. provided data analysis, interpretation, and initial drafting and revision of the final manuscript; S.R. provided data analysis and interpretation; M.O.F. designed the study, acquired data, and revised the manuscript; K.A. performed data analysis; M.S. and H.G. performed data analysis and revised the manuscript; V.F. and D.M. performed laboratory protocols, data acquisition, and data analysis and revised the manuscript; E.D. performed data analysis and laboratory protocols; D.K. performed data and quality review; A.R.C. performed the radiological analysis and interpretation; V.V.M. and V.J. performed data interpretation and revised the manuscript; R.B.D. conceived and designed the study, led the tumor board, performed data interpretation, and revised the manuscript; S.P. performed data interpretation and the initial drafting and revision of the manuscript; and M.C. performed data acquisition, patient care, interpretation, and initial drafting and revision of the manuscript.
Funding
This study was supported in part by a grant from the IBM corporation (IBM Watson Health) to the New York Genome Center, New York Genome Center philanthropic funds and Rockefeller University grant #UL1 TR000043 from the National Center for Advancing Translational Sciences (NCATS), and the National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. R.B.D. is a Howard Hughes Medical Institute Investigator.
Competing Interest Statement
The authors have declared no competing interest.
REFERENCES
- Cancer Genome Atlas Research Network. 2017. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell 32: 185–203 e113. 10.1016/j.ccell.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. 2017. OncoKB: a precision oncology knowledge base. JCO Precis Oncol 2017 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6: 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. 2011. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470: 269–273. 10.1038/nature09677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, et al. 2011. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–D950. 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster SA, Whalen DM, Özen A, Wongchenko MJ, Yin J, Yen I, Schaefer G, Mayfield JD, Chmielecki J, Stephens PJ, et al. 2016. Activation mechanism of oncogenic deletion mutations in BRAF, EGFR, and HER2. Cancer Cell 29: 477–493. 10.1016/j.ccell.2016.02.010 [DOI] [PubMed] [Google Scholar]
- Franco J, Witkiewicz AK, Knudsen ES. 2014. Cdk4/6 inhibitors have potent activity in combination with pathway selective therapeutic agents in models of pancreatic cancer. Oncotarget 5: 6512–6525. 10.18632/oncotarget.2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan M, Bender RJ, Pishvaian MJ, Halverson DC, Tuli R, Klempner SJ, Wainberg ZA, Singhi AD, Petricoin E, Hendifar AE. 2018. Molecular and clinical characterization of BRAF mutations in pancreatic ductal adenocarcinomas (PDACs). J Clin Oncol 36: 214 10.1200/JCO.2018.36.4_suppl.214 [DOI] [Google Scholar]
- Jia X, Shanmugam C, Paluri RK, Jhala NC, Behring MP, Katkoori VR, Sugandha SP, Bae S, Samuel T, Manne U. 2017. Prognostic value of loss of heterozygosity and sub-cellular localization of SMAD4 varies with tumor stage in colorectal cancer. Oncotarget 8: 20198–20212. 10.18632/oncotarget.15560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806. 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisawa T, Wood LD, Itoi T, Takaori K. 2016. Pancreatic cancer. Lancet 388: 73–85. 10.1016/S0140-6736(16)00141-0 [DOI] [PubMed] [Google Scholar]
- Long GV, Hauschild A, Santinami M, Atkinson V, Mandalà M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, et al. 2017. Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N Engl J Med 377: 1813–1823. 10.1056/NEJMoa1708539 [DOI] [PubMed] [Google Scholar]
- Planchard D, Smit EF, Groen HJM, Mazieres J, Besse B, Helland A, Giannone V, D'Amelio AM, Zhang P, Mookerjee B, et al. 2017. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non-small-cell lung cancer: an open-label, phase 2 trial. Lancet Oncol 18: 1307–1316. 10.1016/S1470-2045(17)30679-4 [DOI] [PubMed] [Google Scholar]
- Rachakonda PS, Bauer AS, Xie H, Campa D, Rizzato C, Canzian F, Beghelli S, Greenhalf W, Costello E, Schanne M, et al. 2013. Somatic mutations in exocrine pancreatic tumors: association with patient survival. PLoS One 8: e60870 10.1371/journal.pone.0060870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, Wen PY, Zielinski C, Cabanillas ME, Urbanowitz G, et al. 2018. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600–mutant anaplastic thyroid cancer. J Clin Oncol 36: 7–13. 10.1200/JCO.2017.73.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, et al. 2019. COSMIC: the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 47: D941–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loo P, Nordgard SH, Lingjaerde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, et al. 2010. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci 107: 16910–16915. 10.1073/pnas.1009843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al. 2015. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 518: 495–501. 10.1038/nature14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Frank MO, Koyama T, Rhrissorrakrai K, Robine N, Utro F, Emde AK, Chen BJ, Arora K, Shah M, et al. 2017. Comparing sequencing assays and human-machine analyses in actionable genomics for glioblastoma. Neurol Genet 3: e164 10.1212/NXG.0000000000000164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzeszczynski KO, Felice V, Abhyankar A, Kozon L, Geiger H, Manaa D, London F, Robinson D, Fang X, Lin D, et al. 2018. Analytical validation of clinical whole-genome and transcriptome sequencing of patient-derived tumors for reporting targetable variants in cancer. J Mol Diagn 20: 822–835. 10.1016/j.jmoldx.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarbrough WG, Buckmire RA, Bessho M, Liu ET. 1999. Biologic and biochemical analyses of p16INK4a mutations from primary tumors. J Natl Cancer Inst 91: 1569–1574. 10.1093/jnci/91.18.1569 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data (in vcf format) for this case can be obtained from ftp://ftp.nygenome.org/CA/9/ or by request to the corresponding author until it is made available in the EGA-archive.org public repository (ega-box-1298) as part of the larger study submission. The BRAF variant has been deposited into the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under accession number VCV000666267.1.