Abstract
Epithelial ovarian cancer (OvCa) is the most lethal female reproductive tract malignancy. A major clinical hurdle in patient management and treatment is that when using current surveillance technologies 80% of patients will be clinically diagnosed as having had a complete clinical response to primary therapy. In fact, the majority of women nonetheless develop disease recurrence within 18 mo. Thus, without more accurate surveillance protocols, the diagnostic question regarding OvCa recurrence remains framed as “when” rather than “if.” With this background, we describe the case of a 61-yr-old female who presented with a 3-mo history of unexplained whole-body rash, which unexpectedly led to a diagnosis of and her treatment for OvCa. The rash resolved immediately following debulking surgery. Nearly 1 yr later, however, the rash reappeared, prompting the prospect of tumor recurrence and requirement for additional chemotherapy. To investigate this possibility, we undertook a genomics-based tumor surveillance approach using a targeted 56-gene NGS panel and biobanked tumor samples to develop personalized ctDNA biomarkers. Although tumor-specific TP53 and PTEN mutations were detectable in all originally collected tumor samples, pelvic washes, and blood samples, they were not detectable in any biosample collected beyond the first month of treatment. No additional chemotherapy was given. The rash spontaneously resolved. Now, 2 yr beyond the patient's original surgery, and in the face of continued negative ctDNA findings, the patient remains with no evidence of disease. As this single case report suggests, we believe for the first time that ctDNA can provide an additional layer of information to avoid overtreatment.
Keywords: ovarian neoplasm
INTRODUCTION
Epithelial ovarian cancer (OvCa) is the most lethal female reproductive tract malignancy with an estimated 22,530 new cases and 13,980 deaths predicted for the United States in 2019 (SEER Cancer Statistics Review et al. 2019; Siegel et al. 2019). Surgical tumor debulking followed by platinum-based therapy represents the gold-standard treatment (Della Pepa et al. 2015). Unfortunately, and despite perceived clinical response to initial treatment, between 40% and 60% of all patients will have disease recurrence, and these rates increase for women with late-stage disease (75%). Overall, nearly 50% of women will have tumor recurrence within the first 18 mo following their initial treatment (Podratz and Cliby 1994; Rubin et al. 1999; Rahaman et al. 2005). Most patients ultimately die of their disease within the first 5 yr after initial cancer diagnosis (Paik et al. 2016).
Research efforts have focused on improving upon current surveillance strategies, which often include a combination of serial physical examinations, serum CA125 measurements, and radiologic examination with CT and PET scans (Niloff et al. 1985; Gu et al. 2009; Bell and Pannu 2011). Without accurate diagnostic and prognostic tests, and given the overwhelming rate of recurrence in this cancer, each symptom leads to the question of “when” rather than “if” regarding cancer recurrence and treatment. The use of circulating tumor DNA (ctDNA) has presented a promising tool to improve diagnostic, prognostic, and predictive value in many types of solid malignancies (Merker et al. 2018; Chin et al. 2019). A number of studies have demonstrated the use of liquid biopsy and ctDNA analysis in OvCa (Martignetti et al. 2014; Pereira et al. 2015; Cheng et al. 2017). The clinical relevance of these research-based procedures has not yet been demonstrated.
On the other end of the spectrum is early detection; there is no screening test nor is any recommended for this cancer. Currently, the overwhelming majority of women are diagnosed at a late stage of disease. Symptoms are often nonspecific. Most often, patients report abdominal pain or discomfort and mild digestive disturbances (Armstrong et al. 2010). Intriguingly, a rare number of women present with a paraneoplastic syndrome with dermatologic symptoms such as dermatomyositis. This idiopathic inflammatory myopathy is associated with malignancy (Field and Goff 2018). A cancer diagnosis is most commonly made simultaneously or within the first year of a diagnosis of dermatomyositis. OvCa was associated with just >8% of dermatomyositis patients and was often late stage at the time of diagnosis (Davis and Ahmed 1997; Dobloug et al. 2015). Dermatomyositis typically presents as photosensitivity of the upper trunk, face, ears, and hands as well as Gottron's papules, a heliotrope rash, and periungual telangiectasia. Patients may also report symmetrical proximal muscle weakness that occurs simultaneously with or independently of the dermatologic presentation (Christie et al. 2013).
We describe the case of a 61-yr-old female who originally presented following a 3-mo history of unexplained whole-body rash, which ultimately led to a diagnosis of late-stage OvCa. The rash resolved within days following her initial debulking surgery. Of central relevance to this report, the rash recurred 1 yr later. Although the traditional surveillance tests including the serum-based protein CA125 and CT/PET scans were within normal limits, the immediate clinical question, given the knowledge of a high rate of disease recurrence in this cancer even in the face of negative laboratory and radiology findings, was whether the reappearance of the rash heralded an early sign of cancer recurrence. We hypothesized that a liquid biopsy–based approach, using personalized ctDNA biomarkers based on her specific tumor signature, might provide diagnostic clarity.
RESULTS
Clinical Presentation
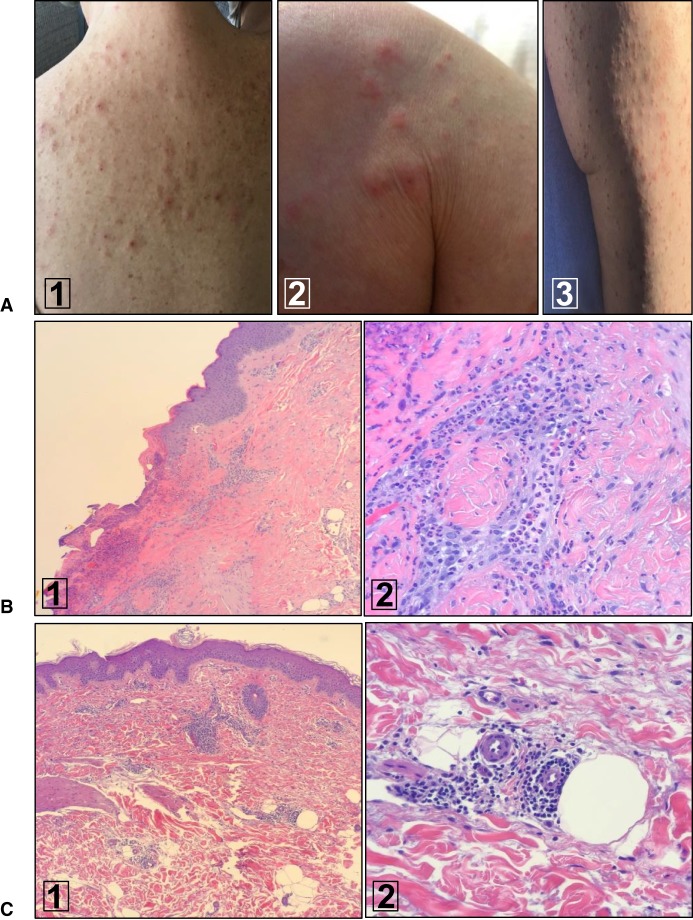
The patient was a 61-yr-old female whose last menstrual period was >10 yr prior to presentation and who had a family history of ovarian cancer. She originally sought medical care for an unexplained pruritic whole-body rash (Fig. 1A). Over a 3-mo period, she was evaluated by a total of seven different dermatologists and prescribed a number of different oral and topical treatments for presumed diagnoses that included scabies, interstitial dermatitis, and dermal hypersensitivity. The rash did not resolve and indeed continued to spread. Two independent skin biopsies performed during this time period revealed an underlying inflammatory process (Fig. 1B) but no definitive diagnosis. There was no evidence of any muscle weakness or other relevant findings on physical examination and in patient history. Despite multiple treatments, the unexplained rash did not resolve and raised the suspicion of a paraneoplastic syndrome; a diagnostic workup was initiated.
Figure 1.
Pruritic whole-body rash of unexplained origin. (A) Evidence of marked dermatographism and eczematous dermatitis on back (1), shoulder (2), and thigh (3). (B) Histologic findings of rash biopsied prior to OvCa diagnosis and treatment. (1) (H&E; 10×) Low-power view of right upper thigh excoriation with extensive parakeratosis. (2) (H&E; 40×) High-power view demonstrating acute and chronic inflammation surrounding a blood vessel. (C) Similar histologic findings of rash 1 yr following initial OvCa surgery and chemotherapy. (1) (H&E; 10×) Punch biopsy from the right lateral upper thigh demonstrating both deep and superficial dermatitis. (2) (H&E; 40×) Deep dermal blood vessels surrounded by inflammatory cells. The endothelial cells are rounded and consistent with a generalized reactivity phenotype.
A whole-body CT scan revealed the presence of multiple soft tissue masses along the left anterior peritoneum, the largest measuring 1.6 cm, and soft tissue prominence on the right ovary suggestive of ovarian carcinoma. The patient was then referred to our group for further evaluation and treatment.
A transvaginal ultrasound was performed. This revealed the presence of a 4.7 × 2.5 × 2.5-cm elongated heterogeneous solid mass consistent with a fallopian tube malignancy. There was also a 1.0 × 0.7 × 1.0-cm mass to the right anterolateral aspect of the uterine fundus and a 1.4-cm solid vascular implant in the cul de sac with evidence of peritoneal thickening. Serum CA125 levels were elevated (117.4 U/mL). The patient underwent debulking surgery.
A total abdominal hysterectomy, bilateral salpingo-oophorectomy, omentectomy, and rectosigmoid resection with primary anastomosis were performed. No residual disease was noted at the completion of surgery. Pathologic examination of the tumor confirmed a high-grade serous ovarian carcinoma, stage IIIC, arising from the right fallopian tube (Fig. 2). Most notably, at the time of discharge 2 d following her surgery, the patient's rash had almost completely resolved.
Figure 2.
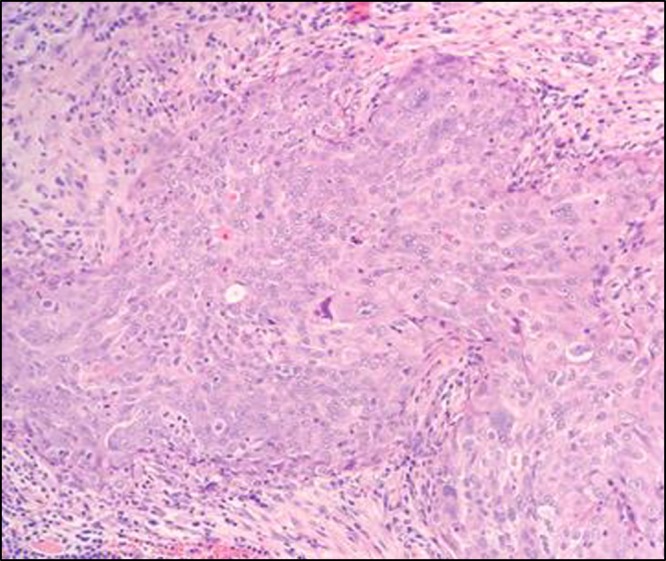
Stage IIIC ovarian cancer. High-grade serous nuclei displaying significant pleomorphism, prominent nucleoli, and apoptosis (H&E; 20×).
The patient was started on primary platinum-based chemotherapy with carboplatin and paclitaxel. Because of an allergic reaction to paclitaxel (facial flushing, lower back pain, and mild, scratchy throat) the treatment was switched to and completed with carboplatin and paclitaxel protein-bound (Abraxane). Bevacizumab (Avastin) was added later once approved by her insurance provider. After eight cycles of chemotherapy and based on PET/CT scan imaging and serum CA125 levels, all within normal limits, the patient was diagnosed as having had a complete clinical response. A second look laparoscopy was negative for cytologic evidence of disease. The patient was started on maintenance treatment with oral cyclophosphamide (Cytoxan) (Fig. 3, top). The patient continued with routine surveillance follow-up with clinical examinations, imaging, and CA125 levels. Serum CA125 levels were all within normal limits, and PET scans were negative (Fig. 3, bottom).
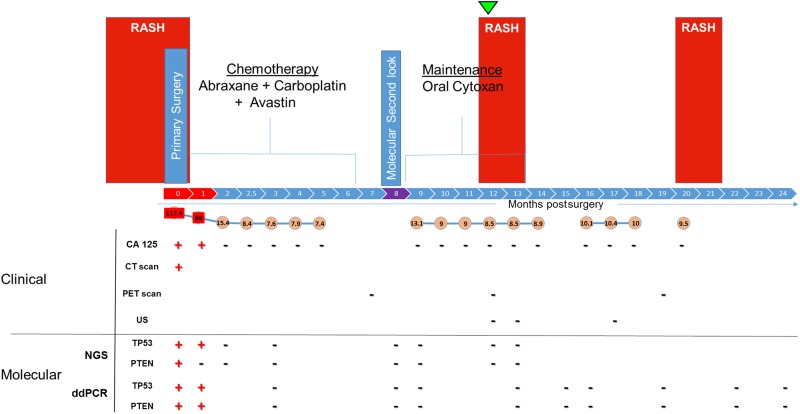
Figure 3.
Timeline of patient clinical history, testing, and results. Schematic demonstrating the patient's overall presentation and clinical course highlighted with both traditional and precision medicine–directed molecular results. The patient developed a whole-body rash of unexplained origin that was resistant to multiple topical and systemic treatments 3 mo prior to her diagnosis of ovarian cancer. The original rash disappeared within days of tumor debulking at the time of primary surgery and initiation of chemotherapy. The rash reappeared ∼1 yr later and suggested the possibility of cancer recurrence. Results of serum CA125 levels, CT and PET scans, and ultrasound (US) throughout her care are shown. The patient's tumor was sequenced at the timepoint shown (green arrowhead), with the express intent to identify somatic mutations and develop patient-specific ctDNA biomarkers. These tumor-specific mutations were simultaneously screened for in blood samples, second-look biopsies, and a pelvic wash sample (purple), all of which had been previously collected in real time and biobanked. In the upper and lower panels, the clinical presentation, surgical history, and chemotherapy events are shown as well as the clinical and molecular results including both next-generation sequencing (NGS) and droplet digital polymerase chain reaction (ddPCR) assay results, respectively. Results are shown as positive (+) or negative (−).
Twelve months following her initial surgery, the whole-body rash returned. It appeared similar in character and distribution to the original rash. Dermatologic evaluation and biopsy was again performed (Fig. 1C). The histologic features associated with this rash appeared similar to at the time of the original presentation. Radiologic and serum CA125 testing were within normal limits.
Given the reappearance of the rash, its shared presentation, distribution, and histologic findings, and the patient's initial presentation history, coupled with the known high recurrence rate of this disease even in the face of negative serum and radiologic findings, recommendations were raised to treat the patient with a second line of chemotherapy for a suspected OvCa recurrence.
Genomic Analyses
Based on previous studies demonstrating that ctDNA can represent a powerful tool in detecting otherwise occult cancers (e.g., Pereira et al. 2015; Kato et al. 2017; de Melo and Jardim 2018; Shen et al. 2018), we sought to generate a tumor-specific mutation profile and targeted liquid biopsy ctDNA probes for this patient. As the patient was already enrolled in our longitudinal precision medicine study, which includes longitudinal biobanking of tumor and blood samples, we isolated germline DNA from blood, genomic DNA from the patient's primary and disseminated tumors, and a pelvic wash (second-look laparoscopy) and cell-free DNA from blood samples being collected for research purposes throughout her period of care (Fig. 3, bottom panel).
Using a targeted, pan-cancer 56-gene panel (Swift Biosciences) and ultra-deep sequencing (>2000× coverage) of the three original tumor samples, somatic mutations were identified and then validated by Sanger sequencing. From this panel, mutations were identified in two genes, PTEN (p.P248S) and TP53 (p.R249M) (Table 1). Using these two mutations as tumor-specific guides for this patient, panel-based sequencing was also performed on all additional blood and pelvic wash samples collected throughout the course of her care. To validate tumor-specific mutations in plasma and pelvic wash samples custom TaqMan assays were designed using the Life Technologies Web-based design tool (http://www.thermofisher.com/order/custom-genomic-products/tools/genotyping/) and digital droplet polymerase chain reaction (ddPCR; RainDance Technologies) was performed.
Table 1.
PTEN and TP53 variant information
| Gene | Chr:Pos (hg19) | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (ACMG) | dbSNP | Genotype | ClinVar ID |
|---|---|---|---|---|---|---|---|---|
| PTEN | 10:89717717-89717717 | c.151C > T, c.742 C > T, c.1261C > T | p.P51S, p.P248S, p.P421S | Missense | Uncertain significance | - | Heterozygous | - |
| TP53 | 17:7577535-7577535 | c.269G > T, c.350G > T, c.629G > T, c.713G > T, c.746G > T | p.R90M, p.R117M, p.R210M, p.R238M, p.R249M | Missense | Pathogenic | 587782329 | Heterozygous | RCV000428988.1 |
Tumor-specific somatic mutations were identified in the pelvic wash and a number of plasma samples obtained at the time of surgery and within the first month of treatment (Table 2). Notably, and using a combination of panel-based and ddPCR-based assays, tumor-specific PTEN and TP53 mutations were detected in plasma collected only at day of surgery and 8 d postsurgery (Table 2). In contrast, neither of these tumor-specific PTEN and TP53 mutations were detected in any other of the subsequent 10 plasma samples collected over the next 22 mo. In addition, molecular analysis of peritoneal washes that had been obtained by laparoscopic examination at the completion of chemotherapy (Fig. 3; highlighted in purple at month 8 of the timeline), a procedure we have named the “molecular second look” (Schwartz et al. 2018), also failed to detect either of these two mutations. Sensitivity and linearity test results for each ddPCR assay, performed as we have previously described (Pereira et al. 2015), established that both of the assays can detect several copies of mutant target and can discriminate between wild-type and mutant sequence for both probes to as low as 0.02% (Supplemental Fig. S1).
Table 2.
Detection of tumor-specific next-generation sequencing (NGS) mutations by ddPCR in samples obtained within the first month following surgery
| Sample origin | Sample type | NGS PTEN | NGS TP53 | ddPCR PTEN | ddPCR TP53 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele frequency | Major allele (FW, RV) | Minor allele (FW, RV) | Allele frequency | Major allele (FW, RV) | Minor allele (FW, RV) | Allele frequency | Allele frequency | ||||||
| Right fallopian tube | tDNA | 76.3 | 173 | 276 | 585 | 959 | 75.0 | 498 | 597 | 1605 | 1908 | 78.0 | 75.0 |
| Colonic tumor | tDNA | 4.8 | 979 | 2052 | 54 | 103 | 4.2 | 3924 | 6031 | 181 | 274 | 3.3 | 5.1 |
| Sigmoid tumor | tDNA | 35.4 | 598 | 1010 | 350 | 549 | 35.8 | 1629 | 1885 | 917 | 1087 | 24.3 | 36.5 |
| Pelvic wash | cfDNA | 23.2 | 1184 | 1790 | 393 | 525 | 28.2 | 3553 | 4137 | 1443 | 1631 | 25.5 | 29.0 |
| Plasma day of surgery | cfDNA | 0.6 | 1201 | 2295 | 11 | 11 | 0.8 | 3963 | 4114 | 30 | 35 | 1.9 | 1.4 |
| Plasma 8 d postsurgery | cfDNA | neg. | n/a | n/a | n/a | n/a | 0.8 | 6744 | 7918 | 63 | 71 | 0.2 | 0.4 |
(NGS) Next-generation sequencing, (ddPCR) droplet digital polymerase chain reaction, (FW) forward, (RV) reverse, (tDNA) transfer DNA, (cfDNA) circulating free DNA.
Taken together, the clinical and molecular data strongly supported the conclusion that the patient remained in remission. The decision was made not to start the patient on chemotherapy. Within weeks of making that decision, the rash spontaneously resolved.
DISCUSSION
In this study, we used a liquid biopsy–based approach to monitor on the molecular level for the possibility of tumor recurrence in a patient whose initial presentation for OvCa was an unexplained rash and whose rash reappeared 1 yr after her initial treatment. As part of our Gynecologic Cancer Translational Research Program, biobanked tumor specimens, peritoneal wash samples, and longitudinally collected blood samples allowed us to define tumor-specific mutations, generate highly sensitive, patient-specific ctDNA biomarkers, and then track them during the natural history of the disease. In this patient, we could demonstrate that despite the unique dermatologic presentation that lead to the diagnosis of her cancer, the rash's recurrence was not associated with OvCa recurrence. The negative molecular findings were consistent with the other surveillance findings, including CA125, ultrasound, and PET scans, and so no new chemotherapy regimen was initiated. Intriguingly, and as shown in Figure 3, the patient developed a rash once again, 20 mo after initial treatment and 8 mo after the time frame described in Results. Again, this whole-body rash emerged, lasted 2 wk, and resolved. The patient, now 2 yr after initial surgery and primary chemotherapy treatment, remains otherwise well and without evidence of OvCa recurrence.
The recurrence of this patient's rash presented a diagnostic dilemma. Given that this patient's rash first brought her to medical attention and her eventual cancer diagnosis and then disappeared immediately following her original surgery suggested a possible biologic link between the two. Although rare, paraneoplastic syndromes are systemic manifestations that can lead to the diagnosis of an occult malignancy. A known paraneoplastic syndrome that has been established to be associated with OvCa is dermatomyositis, a rare inflammatory myopathy (Zerdes et al. 2017; Field and Goff 2018). Symptoms include a rash on the upper trunk, face, ears, arms, and hands, as well as symmetric proximal muscle weakness that may occur simultaneously or independently (Christie et al. 2013). Patients diagnosed with dermatomyositis have a general increased risk of developing cancers such as ovarian, lung, pancreas, breast, and colorectal in the 5 years after the appearance of a rash. The risk of OvCa is eight times higher in patients with dermatomyositis compared to the general population (Dobloug et al. 2015; Field and Goff 2018). Therefore, patients older than 40 who present with this condition are recommended to undergo further screening for OvCa with CA125 every 6 mo for 5 yr (Arshad and Barton 2016). Although no formal neuromuscular examination was performed, our patient had no evidence of myopathy.
In general, and following chemotherapy, up to 50% of patients with OvCa and normal CA125 levels nonetheless have a persistent disease (Bast 2010; Parkinson et al. 2016). We and others have described the use of ctDNA in serum (Rubin et al. 1999; Rahaman et al. 2005; Pereira et al. 2015) and pelvic washes, a so-called “molecular second look” (Schwartz et al. 2018), as a highly sensitive method of detecting OvCa recurrence and minimal residual disease. For these reasons, we decided to analyze all longitudinally collected and stored blood samples from this patient and to use ctDNA analysis to evaluate the status of her disease Through targeted NGS, we have identified tumor-specific mutation in TP53 and PTEN genes in cfDNA collected on the day of surgery and 8 d postsurgery. Neither of the mutations were then detected in any of the blood samples collected thereafter including during the second look and recurrence of the rash. Taken together, the clinical and molecular results obtained during each recurrence of the unexplained rash provided confidence in the decision to not treat the patient and not causally link the rash recurrences with recurrences of the patient's cancer. It should be noted that there is no evidence that early treatment of recurrence in ovarian cancer correlates with improved prognosis. As per current ovarian cancer treatment guidelines, this patient would not have had an indication to receive chemotherapy regardless of the results of ctDNA analysis.
In conclusion, and as this single case report suggests, a personalized molecular approach toward OvCa surveillance offers not only an opportunity for the earliest detection of recurrence for treatment but can also assist in avoiding overtreatment.
METHODS
Patient and Patient-Derived Samples
The patient was provided written consent and enrolled and all samples used in this study were collected in accordance with the Institutional Review Board of the Icahn School of Medicine at Mount Sinai (GCO# 10-1166). All experimental protocols were approved by the Icahn School of Medicine at Mount Sinai in accordance with our research protocol (GCO# 10-1166). All clinical investigations were conducted according to the principles expressed in the Declaration of Helsinki. Patient samples were collected over a 2-yr period between 2017 and 2019.
Blood and tumor samples were collected and biobanked at the Icahn School of Medicine at Mount Sinai. For plasma separation, blood samples were collected in BD Vacutainer SST Plus Blood Collection Tubes (BD Biosciences) and processed between 4 and 6 h after collection. Blood samples were centrifuged at 1200 rpm for 10 min at 4°C for separation of plasma. All samples were aliquoted and stored at −130°C until use.
DNA Extraction Protocols
Circulating free (cfDNA) was extracted from 1 mL of plasma (Circulating Nucleic Acid Kit, QIAGEN) and eluted with 105 µL of AVE buffer as we have previously described (Nair et al. 2016). Germline DNA was extracted from 10 mL of whole blood (ArchivePure DNA Kit, 5' Gaithersburg) according to manufacturer's protocol. Tumor DNA was extracted from ∼25–50 mg of tissue (DNeasy Blood and Tissue Kit, QIAGEN) according to the manufacturer's instructions. Quantification of cfDNA, genomic DNA, and tumor DNA was performed by Qubit fluorometry.
Next-Generation Sequencing
As we have previously described these methods in great detail (Nair et al. 2016), we describe them briefly herein. All of the samples from germline PBMC DNA, tumor DNA, plasma cfDNA, and pelvic wash DNA samples were sequenced to an average of 2000× coverage using a targeted amplicon panel (Accel-Amplicon 56G V2 Oncology Panel, Swift Biosciences).
Resulting targeted NGS libraries were sequenced on an Illumina MiSeq using V2 chemistry. Somatic variant calling was performed using LoFreq and GATKhc algorithms. A target of 2000× coverage and 10-ng inputs enabled the lower limit of detection to be set to the 1% fraction. All germline mutations were filtered using QIAGEN's CLC Genomics Workbench 12.0.2, and overlap analysis between GATKhc and LoFreq was performed. All somatic mutations were annotated to determine the functional impact, and then mutations considered as common SNPs (MAF > 0.1) using QIAGEN's Ingenuity Variant Analysis (IVA) were removed. Remaining mutations were then analyzed using QCI to determine potential clinical significance.
PCR Assay Design and Validation
Custom TaqMan Assays were designed using the Life Technologies Web-based design tool (www.lifetechnologies.com/order/custom-genomic-products). Assays contained VIC or FAM-labeled probes, which probed for the wild-type and mutant variants, respectively. The specificity of each assay was first validated by quantitative PCR. Next, sensitivity and lower limits of detection were established by digital droplet PCR (RainDance Technologies), as we have previously described (Pereira et al. 2015).
ADDITIONAL INFORMATION
Data Deposition and Access
The variants described in this paper have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000902407 and SCV000902408.
Ethics Statement
This study was approved by the Icahn School of Medicine at Mount Sinai's Program for the Protection of Human Subjects. Written informed consent was obtained from the patient prior to sample collection.
Acknowledgments
The authors thank the patient for her participation in this study and members of the Department of Obstetrics, Gynecology, and Reproductive Sciences, Icahn School of Medicine at Mount Sinai, New York, and Department of Gynecologic Oncology, Western Connecticut Health Network for their assistance in sample collection and annotation.
Author Contributions
D.P., P.D., and J.A.M. conceptualized the project. Data D.P., S.C.C., A.-M.B., P.D., and J.A.M. curated the data. D.P., S.C.C., P.D., and J.A.M. performed the formal analysis. P.D. and J.A.M. acquired funding. D.P., S.C.C., M.M.P., O.C.-V., J.-N.B., A.-M.B., J.I., L.Y., K.T., J.R., P.D., and J.A.M. investigated. D.P., P.D., and J.A.M. administered the project. D.P., S.C.C., M.M.P., A.-M.B., P.D., and J.A.M. wrote the original draft. D.P., S.C.C., M.M.P., O.C.-V., J.-N.B., A.-M.B., J.I., L.Y., T.K., J.R., P.D., and J.A.M. reviewed and edited the manuscript.
Funding
This study was supported in part by generous funding from the Gordon family, the Ruttenberg family, the Goldstone family, the Mannheimer family, the Varadi Ovarian Initiative in Cancer Education (VOICE) and support in recognition of Margaret Heller. The funders had no role in study design, data collection, or its analysis.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Armstrong D, Axilbund J, Bell J, Bristow R, Chi D, Diaz-Montes T, Eitan R, Ferriss J, Giuntoli R, Gross A, et al. 2010. Epidemiology and clinical presentation of ovarian cancer. In Early diagnosis and treatment of cancer series: ovarian cancer (ed. Bristow R, Armstrong D), pp. 1–15. Saunders, Philadelphia. [Google Scholar]
- Arshad I, Barton D. 2016. Dermatomyositis as a paraneoplastic phenomenon in ovarian cancer. BMJ Case Rep 2016: bcr2016215463 10.1136/bcr-2016-215463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bast RC Jr. 2010. CA 125 and the detection of recurrent ovarian cancer: a reasonably accurate biomarker for a difficult disease. Cancer 116: 2850–2853. 10.1002/cncr.25203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DJ, Pannu HK. 2011. Radiological assessment of gynecologic malignancies. Obstet Gynecol Clin North Am 38: 45–68. 10.1016/j.ogc.2011.02.003 [DOI] [PubMed] [Google Scholar]
- Cancer Stat Facts: Ovarian Cancer. 2019. SEER Cancer Statistics Review. National Cancer Institute, Bethesda, MD. [Google Scholar]
- Cheng X, Zhang L, Chen Y, Qing C. 2017. Circulating cell-free DNA and circulating tumor cells, the “liquid biopsies” in ovarian cancer. J Ovarian Res 10: 75 10.1186/s13048-017-0369-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, Azad TD, Dudley JC, Chaudhuri AA. 2019. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Mol Diagn Ther 23: 311–331. 10.1007/s40291-019-00390-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie A, McKay N, Nussey F. 2013. Dermatomyositis as presenting feature of ovarian cancer, treated with neo-adjuvant chemotherapy and interval debulking surgery. Gynecol Oncol Case Rep 6: 13–15. 10.1016/j.gynor.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MD, Ahmed I. 1997. Ovarian malignancy in patients with dermatomyositis and polymyositis: a retrospective analysis of fourteen cases. J Am Acad Dermatol 37: 730–733. 10.1016/S0190-9622(97)70109-9 [DOI] [PubMed] [Google Scholar]
- Della Pepa C, Tonini G, Pisano C, Di Napoli M, Cecere SC, Tambaro R, Facchini G, Pignata S. 2015. Ovarian cancer standard of care: are there real alternatives? Chin J Cancer 34: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo Gagliato D, Jardim DLF. 2018. Noninvasive cancer biomarkers in solid malignancies: circulating tumor DNA-clinical utility, current limitations and future perspectives. Ann Transl Med 6: 233 10.21037/atm.2018.05.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobloug GC, Garen T, Brunborg C, Gran JT, Molberg O. 2015. Survival and cancer risk in an unselected and complete Norwegian idiopathic inflammatory myopathy cohort. Semin Arthritis Rheum 45: 301–308. 10.1016/j.semarthrit.2015.06.005 [DOI] [PubMed] [Google Scholar]
- Field C, Goff BA. 2018. Dermatomyositis—key to diagnosing ovarian cancer, monitoring treatment and detecting recurrent disease: case report. Gynecol Oncol Rep 23: 1–3. 10.1016/j.gore.2017.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P, Pan LL, Wu SQ, Sun L, Huang G. 2009. CA 125, PET alone, PET–CT, CT and MRI in diagnosing recurrent ovarian carcinoma: a systematic review and meta-analysis. Eur J Radiol 71: 164–174. 10.1016/j.ejrad.2008.02.019 [DOI] [PubMed] [Google Scholar]
- Kato S, Krishnamurthy N, Banks KC, De P, Williams K, Williams C, Leyland-Jones B, Lippman SM, Lanman RB, Kurzrock R. 2017. Utility of genomic analysis in circulating tumor DNA from patients with carcinoma of unknown primary. Cancer Res 77: 4238–4246. 10.1158/0008-5472.CAN-17-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignetti JA, Camacho-Vanegas O, Priedigkeit N, Camacho C, Pereira E, Lin L, Garnar-Wortzel L, Miller D, Losic B, Shah H, et al. 2014. Personalized ovarian cancer disease surveillance and detection of candidate therapeutic drug target in circulating tumor DNA. Neoplasia 16: 97–103. 10.1593/neo.131900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merker JD, Oxnard GR, Compton C, Diehn M, Hurley P, Lazar AJ, Lindeman N, Lockwood CM, Rai AJ, Schilsky RL, et al. 2018. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol 36: 1631–1641. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- Nair N, Camacho-Vanegas O, Rykunov D, Dashkoff M, Camacho SC, Schumacher CA, Irish JC, Harkins TT, Freeman E, Garcia I, et al. 2016. Genomic analysis of uterine lavage fluid detects early endometrial cancers and reveals a prevalent landscape of driver mutations in women without histopathologic evidence of cancer: a prospective cross-sectional study. PLoS Med 13: e1002206 10.1371/journal.pmed.1002206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niloff JM, Bast RC Jr, Schaetzl EM, Knapp RC. 1985. Predictive value of CA 125 antigen levels in second-look procedures for ovarian cancer. Am J Obstet Gynecol 151: 981–986. 10.1016/0002-9378(85)90678-7 [DOI] [PubMed] [Google Scholar]
- Paik ES, Kim TJ, Lee YY, Choi CH, Lee JW, Kim BG, Bae DS. 2016. Comparison of survival outcomes after recurrence detected by cancer antigen 125 elevation versus imaging study in epithelial ovarian cancer. J Gynecol Oncol 27: e46 10.3802/jgo.2016.27.e46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson CA, Gale D, Piskorz AM, Biggs H, Hodgkin C, Addley H, Freeman S, Moyle P, Sala E, Sayal K, et al. 2016. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med 13: e1002198 10.1371/journal.pmed.1002198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira E, Camacho-Vanegas O, Anand S, Sebra R, Catalina Camacho S, Garnar-Wortzel L, Nair N, Moshier E, Wooten M, Uzilov A, et al. 2015. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One 10: e0145754 10.1371/journal.pone.0145754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podratz KC, Cliby WA. 1994. Second-look surgery in the management of epithelial ovarian carcinoma. Gynecol Oncol 55: S128–S133. 10.1006/gyno.1994.1351 [DOI] [PubMed] [Google Scholar]
- Rahaman J, Dottino P, Jennings TS, Holland J, Cohen CJ. 2005. The second-look operation improves survival in suboptimally debulked stage III ovarian cancer patients. Int J Gynecol Cancer 15: 19–25. 10.1136/ijgc-00009577-200501000-00004 [DOI] [PubMed] [Google Scholar]
- Rubin SC, Randall TC, Armstrong KA, Chi DS, Hoskins WJ. 1999. Ten-year follow-up of ovarian cancer patients after second-look laparotomy with negative findings. Obstet Gynecol 93: 21–24. 10.1016/S0029-7844(98)00334-2 [DOI] [PubMed] [Google Scholar]
- Schwartz M, Camacho-Vanegas O, Wood AM, Dashkoff M, Whitelock C, Harkins TT, Cohen CJ, Beddoe AM, Dottino P, Martignetti JA. 2018. Applying precision medicine to ovarian cancer: proof-of-principle for a “molecular second look”. Int J Gynecol Cancer 28: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen SY, Singhania R, Fehringer G, Chakravarthy A, Roehrl MHA, Chadwick D, Zuzarte PC, Borgida A, Wang TT, Li T, et al. 2018. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563: 579–583. 10.1038/s41586-018-0703-0 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. 2019. Cancer statistics, 2019. CA Cancer J Clin 69: 7–34. 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- Zerdes I, Tolia M, Nikolaou M, Tsoukalas N, Velentza L, Hajiioannou J, Mitsis M, Kyrgias G. 2017. How can we effectively address the paraneoplastic dermatomyositis: diagnosis, risk factors and treatment options. J BUON 22: 1073–1080. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variants described in this paper have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000902407 and SCV000902408.