Abstract
Intellectual disability (ID) is a clinically and genetically heterogeneous developmental brain disorder. The present study describes two male siblings, aged 7 and 1 yr old, with severe ID, spastic quadriplegia, nystagmus, and brain atrophy with acquired microcephaly. We used the exome sequencing to identify the causative gene in the patients and identified a hemizygous missense variant, c.1282T>A (p.W428R), in the p21-activated serine/threonine kinase 3 gene (PAK3), which is associated with X-linked ID. p.W428R is located within the highly conserved kinase domain and was predicted to induce loss of enzymatic function by three mutation prediction tools (SIFT, PolyPhen-2, and MutationTaster). In addition, this variant has not been reported in public databases (as of the middle of December 2018) or in the data from 3275 individuals of the Japanese general population analyzed using high-depth whole-genome sequencing. To date, only 13 point mutations and deletions in PAK3 in ID have been reported. The literature review illustrated a phenotypic spectrum of PAK3 pathogenic variant, and our cases represented the most severe form of the PAK3-associated phenotypes. This is the first report of a PAK3 pathogenic variant in Japanese patients with X-linked ID.
Keywords: intellectual disability, severe
CASE PRESENTATIONS
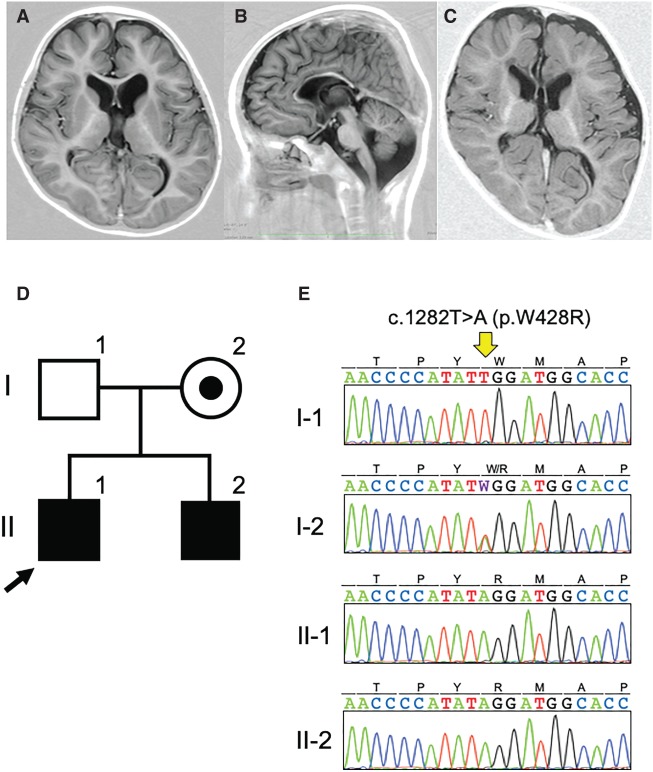
Patient MR94 II-1 was a 7-yr-old boy born to nonconsanguineous Japanese parents. He was born through normal delivery without asphyxia at 38 wk of gestation. His weight and head circumferences were 2720 g (−0.7 SD) and 31.5 cm (−1.3 SD), respectively. He acquired head control after age 3 yr, sat independently after age 6, but could not form words. He had a febrile convulsion at the age of 2 yr. When he was 5 yr old, his developmental quotient (DQ) was 12. When he was 6 yr 3 mo old, he suffered spastic quadriplegia and horizontal nystagmus. He showed no aggressive, hyperactive, or self-harming behavior but did show autistic stereotype movement. Hypotonia in his trunk, extremities, and facial muscles was noted. His height, weight, and head circumference were 100 cm (−3.0 SD), 14.6 kg (−1.8 SD), and 47 cm (−2.6 SD), respectively. Brain magnetic resonance imaging (MRI) showed cerebral white matter and midbrain atrophy, and a thin corpus callosum (Fig. 1AB; Table 1). His younger brother (MR94 II-2) also had X-linked intellectual disability (XLID). He was delivered by Caesarean section because of the breech presentation without asphyxia at 38 wk of gestation. His weight and head circumferences were 2878 g (−0.3 SD) and 30.5 cm (−2.0 SD), respectively. He rolled over after he was 1 yr old, but at the age of 1 yr and 2 mo, he had no head control, could not form words, and had a DQ < 10. He also showed spastic quadriplegia. His height, weight, and head circumference were 70 cm (−2.7 SD), 6.8 kg (−3.1SD), and 43 cm (−2.3 SD), respectively. No behavioral problems were noted, but autistic features were observed. Hypotonia in his trunk, extremities, and facial muscles was also noted. A brain MRI showed cerebral atrophy, myelination delay, and a thin corpus callosum (Fig. 1C).
Figure 1.
A PAK3 pathogenic variant in a Japanese family with X-linked intellectual disability (XLID). (A,B) Coronal and sagittal sections of T1-weighted magnetic resonance images of patient MR94 II-1. (C) A coronal section of T1-weighted magnetic resonance images of patient MR94 II-2. Reduced volume of the white matter and midbrain, as well as a thin corpus callosum, were noted. (D) Pedigree of family MR94. (E) Electrochromatogram of the PAK3 pathogenic variant, c.1282T>A, identified in two patients in family MR94.
Table 1.
Clinical features of patients with c.1282A>T (p.W428R) and reported patients with pathogenic variants in PAK3
| Phenotypic features | Patient 1 | Patient 2 | Sample number | Positive rate (%) |
|---|---|---|---|---|
| Variant | c.1282A>T (p.W428R) | |||
| Intellectual disability | Yes, severe | Yes, severe | 34/38 | 89.5 |
| Epileptic seizures | Yes | No | 9/39 | 23.1 |
| Autistic features | Yes | Yes | 6/38 | 15.8 |
| Aggressive behavior/self-injury | No | No | 10/38 | 26.3 |
| Global developmental delay | Yes | Yes | 32/41 | 78.0 |
| Language impairment | Yes | Yes | 36/37 | 97.3 |
| Spastic quadriplegia | Yes | Yes | 4/40a | 10.0 |
| Nystagmus | Yes | Yes | 4/40b | 10.0 |
| Hypotonia in the trunk and extremities | Yes | Yes | 12/35 | 34.3 |
| Hypotonia in the face | Yes | Yes | 12/39 | 30.8 |
| Microcephaly | Yes | Yes | 14/40 | 35.0 |
| Short stature | Yes | Yes | 6/38 | 15.8 |
| Low set ear | Yes | Yes | 6/38 | 15.8 |
| Long face | Yes | No | 10/40c | 25.0 |
| White matter atrophy | Yes | Yes | 2/8 | 25.0 |
| Midbrain atrophy | Yes | Yes | 3/10 | 30.0 |
| Hypoplasia of the corpus callosum | Yes | Yes | 6/9d | 66.7 |
aIncluding hemiplegia and lower limb spasticity.
bIncluding abnormality of eye movement.
cIncluding abnormal face shape.
dIncluding dysplastic corpus callosum and agenesis of corpus callosum.
VARIANT INTERPRETATION
We identified a hemizygous missense variant, c.1282T>A (p.W428R; GRch37. Chr X. 110439743), in the p21-activated serine/threonine kinase 3 gene (PAK3: NM_002578.3; OMIM300142) in MR94 II-1 and MR94 II-2 (Fig. 1D,E; Table 2). The variant was inherited from their mother, who is asymptomatic. The altered amino acid residue (p.W428) is located within the highly conserved kinase domain. The tryptophan residue is conserved from human to zebrafish (Supplemental Fig. 1). Hence, we expected that the variant would abolish the kinase activity of PAK3. A c.1282T>A variant in PAK3 has not been deposited in any database, including dbSNP build 151, ESP6500, 1000 Genomes, the Exome Aggregation Consortium, the Human Genetic Variation Database, the Genome Aggregation Database (gnomAD), ClinVar, or the Medical Genomics Japan Variant Database (as of the middle of December, 2018). We also searched the data from 3275 members of the Japanese general population acquired using high-depth whole-genome sequencing (Okada et al. 2018). However, not even a heterozygous carrier was found. Hence, the PAK3 variant identified in this study is considered to be “an extremely rare variant.” In addition, three mutation prediction tools, SIFT, PolyPhen-2, and MutationTaster, predicted the mutation to be “Damaging,” with a score of 0, “Probably damaging,” with a score 1, and “Disease causing,” with a score of 0.99, respectively. Finally, we classified and evaluated this variant using the guidelines for the classification of sequence variants from the American College of Medical Genetics and Genomics (Richards et al. 2015). We concluded that the variant identified in this family is “Class 4 (likely pathogenic)” (Supplemental Table 1). No pathogenic variants in other genes are identified. Array comparative genomic hybridization (aCGH) analyses in both parents and patients also identified no pathogenic copy number variants (Supplemental Table 2).
Table 2.
Genomic findings and variant interpretation
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (substitution, deletion, etc.) | dbSNP/dbVar ID | Genotype (heterozygous/homozygous) | ClinVar ID (optional) | Parent of origin (optional) | Observed effect (if shown to be different from the predicted effect) (optional) | Comments (optional) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PAK3 | X | NM_002578.3: c.1282T>A | p.W428R | Missense | Substitution | None | Hemizygous | SCV000914231 | X-linked recessive | None | Class 4, likely pathogenic |
SUMMARY
Families with a Chromosome X–linked inheritance pattern ID account for 5%–10% of all ID cases, suggesting an important role of the genes mutated on the X chromosome in males (Lubs et al. 2012). X-linked intellectual disability is divided into syndromic XLID (S-XLID) and nonsyndromic XLID (NS-XLID), the latter lacking dysmorphic features or other distinguishing symptoms (Ropers and Hamel 2005). To date, at least 141 genes responsible for S-XLID and NS-XLID have been reported (Neri et al. 2018). Among them, PAK3 was identified as the causative gene in patients with NS-XLID (OMIM300558; MRX30) (Allen et al. 1998). Currently, 10 types of point mutations, one splicing mutation, one small deletion, and a 90-kb deletion containing exons 6–18 are described in the Human Gene Mutation Database, Professional (Version: 2018.3) as of the middle of December, 2018. These are all found in non-Japanese individuals.
The clinical phenotypes of the patients with PAK3 mutations are variable, as summarized (Table 1; Supplemental Table 3). We noted several common features, including mild to severe ID (89.5%), language impairment (97.3%), and global developmental delay (78%), as also reported previously (Hertecant et al. 2017; Horvath et al. 2018). Microcephaly (35.0%) and hypotonia (trunk and extremities, 34.3%; face, 30.8%) may represent characteristic features of the patients with PAK3 mutations. Spastic paraplegia/hemiplegia (10%) and nystagmus (10%) were also found in multiple cases. Less frequent and nonspecific features include epileptic seizures (23.1%), autistic features (15.8%), aggressive behavior/self-injury (26.3%), and short stature (15.8%). Imaging findings were available only in a small number of patients, but hypoplasia of corpus callosum (66.7%), atrophy of midbrain (30.0%), and white matter (25%) were reported.
Our cases showed all features in Table 1 except for aggressive behavior. They also showed more severe ID compared with previously reported patients, suggesting that they had the most severe form of the disease caused by PAK3 mutations. Spastic quadriplegia and nystagmus appear to be rare phenotypes, but similar manifestations (i.e., lower limb spasticity and hemiplegia and oculomotor abnormalities) were also reported in other patients, suggesting that these features are possibly associated with PAK3 mutations. Hertecant et al. (2017) reported a monozygotic twin who presented with ID, behavioral problems, and unique macrocephaly caused by a p.Y427H mutation in PAK3, which is located one amino acid prior to the residue mutated in the present patients, who had acquired microcephaly, as seen in other patients. Reason for this opposite phenotypic expression remains unknown.
In conclusion, we report the first Japanese XLID family with a novel hemizygous PAK3 mutation. The literature review illustrated the phenotypic spectrum of PAK3-associated XLID and suggested that our cases represent a severe form of the disease.
METHODS
The biobank at the National Center of Neurology and Psychiatry is part of the National BioBank Network in Japan, which is a unique biorepository exclusively dedicated to collecting samples from patients with neuropsychiatric, muscular, and developmental diseases. The biobank contains DNA samples and clinical information from 583 families with neurodevelopmental diseases that were preserved and diagnosed from 2004 to 2016.
We performed exome sequencing to identify the causative gene in the family. Genomic DNAs were extracted from the patients and the parent's bloods by standard protocol. DNAs were processed by SureSelect XT Human All Exon V6 (Agilent Technologies) and BGISEQ DNA library preparation kit (BGI). Captured DNAs were sequenced using MGISEQ2000 (BGI) with 100-bp pair-end reads. Reads were mapped to the human genome reference (GRCh37/hg19) by BWA 0.7.5a-r405. Duplicated reads were removed by Picard 1.99. Variants were identified by Genome Analysis Toolkit v3.5 based on GATK Best Practice Workflow and annotated by ANNOVAR (2018 April 16). In all subjects, at least 98.1% of all coding regions were covered in 10 reads depth (Supplemental Table 4). The variant in PAK3 was confirmed by Sanger sequencing.
For the aCGH analysis, we utilized SurePrint G3 Human CGH microarray 8 × 60K (Agilent). The experimental procedures were performed according to the manufacturer's protocol (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis, Version 7.2, Agilent Technologies).
The following Web resources were used:
dbSNP build 151: https://www.ncbi.nlm.nih.gov/projects/SNP/
ESP6500: http://evs.gs.washington.edu/EVS/
1000 Genomes: http://grch37.ensembl.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000077264;r=X:110187513-110470589
Exome Aggregation Consortium: http://exac.broadinstitute.org
Human Genetic Variation Database: http://www.hgvd.genome.med.kyoto-u.ac.jp
gnomAD: http://gnomad.broadinstitute.org
Medical Genomics Japan Variant Database: https://mgend.med.kyoto-u.ac.jp
ADDITIONAL INFORMATION
Data Deposition and Access
PAK3 variant data was submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) and are available under accession number SCV000914231.
Ethics Statement
Clinical information and samples from patients and their parents were obtained with informed consent. The study was approved by the ethical committee of the National Center of Neurology and Psychiatry, Japan (IRB A2014-081 and XXXX-115 (20-9-6)).
Acknowledgments
We are very grateful to the patients and their parents who participated in this study.
Author Contributions
E.T., K.I., and Y.G. contributed to the conception and design of the study. A.I. and K.T. contributed to the genome analysis. A.I. contributed to the variant interpretation. E.T., C.A.-H., S.H., Y.I., E.N., K.I., and Y.G. managed the DNA samples and clinical information. S.K., Y.K., Y.M., and M.K. performed the whole genome sequencing and analyzed the data. A.I., E.T., C.A.-H., and K.I. wrote the manuscript and prepared the figures. Y.G. supervised the study.
Funding
This study was partially supported by the Program for an Integrated Database of Clinical and Genomic Information (17kk0205012h0002 to Y.G.); the Construction of integrated database of clinical and genomics information and sustainable system for promoting genomic medicine in Japan (18kk0205012s0303 to Y.G.); Practical Research Project for Rare/Intractable Diseases (18ek0109285h0002 and 19ek0109285h0003 to A.I. and Y.G.) from the Japan Agency for Medical Research and Development (AMED); and Intramural Research Grants (27-6 to Y.G.; 30-9 to A.I.) for Neurological and Psychiatric Disorders from the National Center of Neurology and Psychiatry, Japan.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. 1998. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet 20: 25–30. 10.1038/1675 [DOI] [PubMed] [Google Scholar]
- Hertecant J, Komara M, Nagi A, Al-Zaabi O, Fathallah W, Cui H, Yang Y, Eng CM, Al Sorkhy M, Ghattas MA, et al. 2017. A de novo mutation in the X-linked PAK3 gene is the underlying cause of intellectual disability and macrocephaly in monozygotic twins. Eur J Med Genet 60: 212–216. 10.1016/j.ejmg.2017.01.004 [DOI] [PubMed] [Google Scholar]
- Horvath GA, Tarailo-Graovac M, Bartel T, Race S, Van Allen MI, Blydt-Hansen I, Ross CJ, Wasserman WW, Connolly MB, van Karnebeek CDM. 2018. Improvement of self-injury with dopamine and serotonin replacement therapy in a patient with a hemizygous PAK3 mutation: a new therapeutic strategy for neuropsychiatric features of an intellectual disability syndrome. J Child Neurol 33: 106–113. 10.1177/0883073817740443 [DOI] [PubMed] [Google Scholar]
- Lubs HA, Stevenson RE, Schwartz CE. 2012. Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet 90: 579–590. 10.1016/j.ajhg.2012.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri G, Schwartz CE, Lubs HA, Stevenson RE. 2018. X-linked intellectual disability update 2017. Am J Med Genet A 176: 1375–1388. 10.1002/ajmg.a.38710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Momozawa Y, Sakaue S, Kanai M, Ishigaki K, Akiyama M, Kishikawa T, Arai Y, Sasaki T, Kosaki K, et al. 2018. Deep whole-genome sequencing reveals recent selection signatures linked to evolution and disease risk of Japanese. Nat Commun 9: 1631 10.1038/s41467-018-03274-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–423. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH, Hamel BC. 2005. X-linked mental retardation. Nat Rev Genet 6: 46–57. 10.1038/nrg1501 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
PAK3 variant data was submitted to the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) and are available under accession number SCV000914231.