Abstract
Low-grade serous ovarian cancer (LGSOC) is relatively chemoresistant, and no precision therapy is approved for this indication. Despite promising results in phase II trials, MEK inhibitors have failed to show improved progression-free survival in a phase III trial when compared to physician's choice chemotherapy. We report for the first time temporal changes in the tumor genome assessed in sequential tumor samples of a 48-yr-old patient with a KRAS-mutated LGSOC treated with the MEK inhibitor binimetinib. After an initial long-lasting partial response, rapidly progressive brain metastasis occurred, ultimately leading to patient death. Our study demonstrates that novel genomic alterations accumulated during the course of treatment as a result of therapeutic pressures led to MEK inhibitor resistance and, ultimately, disease evolution with an aggressive behavior observed in this patient. In particular, we describe the presence of ERBB3 amplification and aberrant ERBB3–MYC signaling as a potential mechanism of acquired MEK inhibitor resistance in a patient with LGSOC, which is similar to previous observations in KRAS-mutated colon and lung cancers. Our study highlights the need for an individualized approach to better understand tumor genome evolution and suggests that LGSOC patients may derive improved therapeutic benefit by using a combinatorial strategy used in other cancers in order to overcome emergent resistance to targeted therapies.
Keywords: ovarian neoplasm
INTRODUCTION
Low-grade serous ovarian cancer (LGSOC) accounts for ∼5%–8% of all ovarian cancers (OCs) and is characterized by slow growth pattern, younger age at diagnosis, poor response to chemotherapy, and longer overall survival when compared to high-grade serous OC (Gershenson et al. 2006). Optimal debulking surgery remains the cornerstone of upfront therapy for LGSOC and at the time of recurrence. Standard treatment includes the use of hormonal therapy and chemotherapy for which benefit is limited or unknown (Ledermann et al. 2013; Gourley et al. 2014). Despite the usual slow-growing pattern of disease, overall prognosis remains poor. Dysregulation of the KRAS/BRAF/MAPK/ERK pathway is commonly found in LGSOC (∼40%–60% harbor a RAS mutation), and activating mutations in KRAS, NRAS, or BRAF genes promote tumorigenesis through constitutive activation of the MAPK pathway (Cancer Genome Atlas Research 2011; Hunter et al. 2015). Inhibition of the MAPK pathway with the MEK inhibitor (MEKi) selumetinib has been investigated in a phase II trial, showing an overall response rate of 15% and a stable disease of 60% in patients with relapsed LGSOC, without correlation between MAPK pathway mutational status and response (Farley et al. 2013). However, interim analysis of the randomized phase III MILO study (NCT01849874) failed to demonstrate an improvement in progression-free survival with the MEK1/2i binimetinib (MEK162, ARRY-162) compared to physician choice chemotherapy (liposomal doxorubicin, paclitaxel, or topotecan) in patients with advanced LGSOC previously treated with platinum-based chemotherapy (Array BioPharma 2016 press-release). As such, the study was terminated early and targeted therapy approvals remain elusive in this disease. Further research is warranted to (i) improve our understanding of LGSOC biology; (ii) define predictive biomarkers; (iii) explore mechanisms of resistance to MEKi; and (iv) identify potential novel treatment strategies that are more effective while maintaining a good quality of life.
We report for the first time longitudinal genomic analyses of multiple and sequential tumor samples from a patient diagnosed with LGSOC who achieved a long-lasting partial response to binimetinib (17.5 mo) and subsequently developed unusually aggressive brain metastases (LGSOC pathology proven) leading to death shortly thereafter. We identified a potential mechanism of resistance to MEKi and molecular alterations associated with acquired aggressive clinical behavior in this patient.
RESULTS
Clinical Presentation
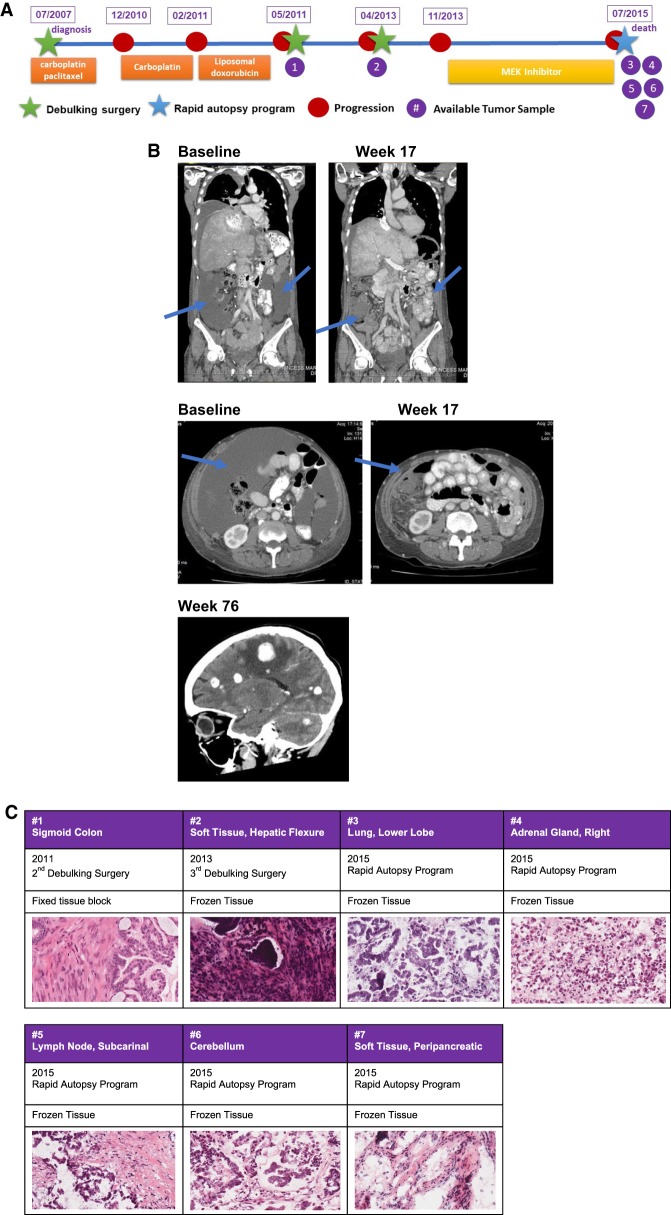
A 48-yr-old woman underwent total abdominal hysterectomy and bilateral salpingo-oophorectomy in July 2007 followed by six cycles of carboplatin and paclitaxel chemotherapy for a stage IIIC LGSOC (Fig. 1A; Mutch and Prat 2014). In December 2010, a chest–abdomen–pelvis computerized tomography (CT) documented recurrence with a pelvic mass and peritoneal carcinomatosis. She received three cycles of carboplatin with disease progression and a subsequent three cycles of liposomal doxorubicin with further growth of metastatic disease. In May 2011, she underwent secondary debulking surgery to no residual disease, and her pathology was consistent with LGSOC. In April 2013, because of further disease progression, she underwent a third debulking surgery with residual macroscopic disease >5 cm from an unresectable pelvic mass. Subsequent systemic treatment was delayed because of postsurgical complications with wound infection and incomplete healing. In November 2013, further disease progression occurred in the peritoneum, liver, right adrenal gland, abdominal and thoracic nodes, lungs, and pleura. In January 2014 (week 1), she was enrolled in the phase III MILO trial (NCT01849874) and randomized to receive binimetinib 45 mg orally twice a day. She achieved a partial response as per RECIST (Response Evaluation Criteria in Solid Tumor) 1.1 criteria at the first radiological assessment (9 weeks after treatment start) with best response at week 33 (47.7% reduction of the sum of the target lesions from baseline). From week 41, CT scans showed slow disease growth without reaching the criteria for progressive disease as per RECIST 1.1, and, thus, she continued to receive treatment with MEKi. In June 2015 (week 76), she developed acute left-side hemiparesis and a brain MRI showed multiple supra- and infratentorial metastasis (Fig. 1B). Despite whole-brain radiation, the patient had rapid neurological deterioration as a consequence of brain metastasis progression, ultimately leading to death. The patient consented to an ongoing Rapid Autopsy Program and tumor samples were collected from different disease sites (Table 1). The pathology report from all the disease sites, including brain metastasis, were reviewed by an expert Gynecology Pathologist and were all consistent with LGSOC (Fig. 1C).
Figure 1.
(A) Summary of patient clinical history since her diagnosis to her death and a timeline of samples available for genomic analysis. (B) Radiological images pre- (baseline) and on treatment (week 17) showing disease improvement during MEK inhibitor treatment (reduction of peritoneal effusion and improvement of liver involvement) and CT scan showing the onset of brain metastasis at week 76. (C) Hematoxylin and eosin sections (40×) showing diagnosis of low-grade serous ovarian carcinoma in all available samples.
Table 1.
Tumor samples collected
| # | Tissue site | Tissue type | Year |
|---|---|---|---|
| 1 | Sigmoid colon | Fixed tissue block | 2011 |
| 2 | Soft tissue, hepatic flexure | Frozen tissue | 2013 |
| 3 | Lower lobe, lung | Frozen tissue | 2015 |
| 4 | Right adrenal gland | Frozen tissue | 2015 |
| 5 | Lymph node, subcarinal | Frozen tissue | 2015 |
| 6 | Cerebellum | Frozen tissue | 2015 |
| 7 | Peripancreatic soft tissue | Frozen tissue | 2015 |
| Liver NORMAL | Frozen tissue | 2015 |
In this case study, we used comprehensive genomic analyses (whole-exome sequencing [WES], shallow whole-genome sequencing [s-WGS], and RNA sequencing [RNA-seq]) to explain the possible mechanism(s) leading to (a) acquired MEK resistance and (b) aggressive behavior with rapidly progressive brain metastasis in this patient.
Genomic Analyses
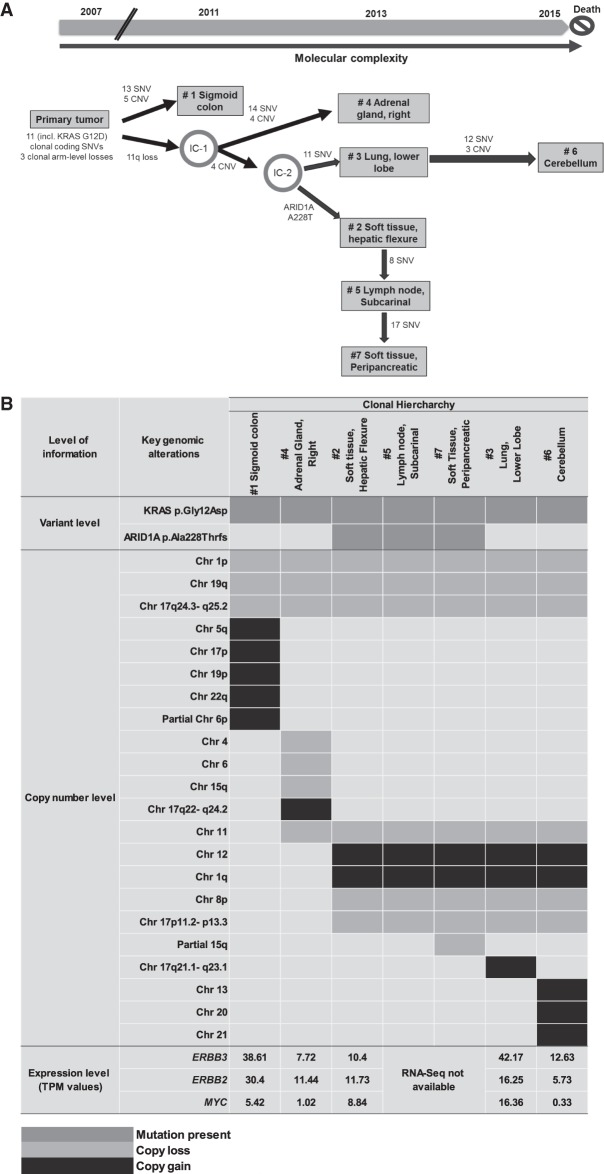
A longitudinal integrated analysis was performed across multiple tissues combining WES, s-WGS, and RNA-seq methods, which represented the complete disease course of this patient. This approach was used to examine and potentially explain the peculiar incidence of brain metastasis in this patient with LGSOC concurrent to the development of secondary resistance to MEKi. We reconstructed the trajectory of the disease evolution by comparing the tumor mutational spectrum and copy-number status of the different metastatic lesions alongside the clinical timing of their appearances. Our findings support the notion that the differential response seen to MEKi in this case is a consequence of tumor heterogeneity and clonal evolution over time (Fig. 2A,B).
Figure 2.
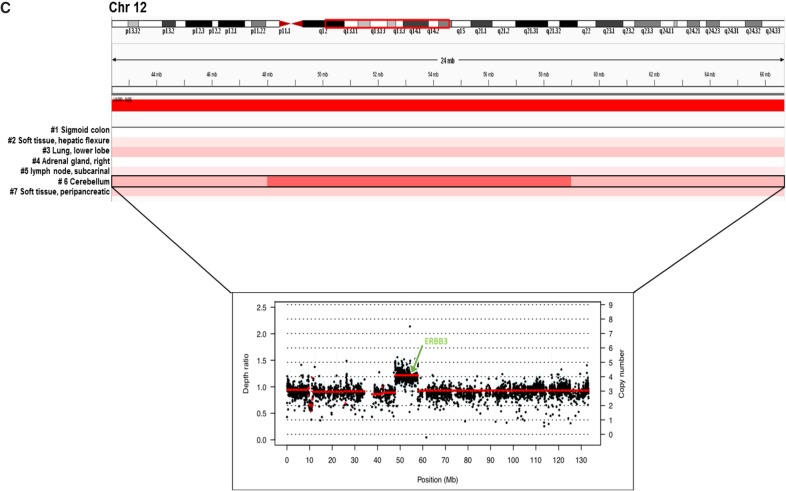
(A) Inferred spatial and temporal evolution of different lesions based on their clinical and shared molecular variants. Single-nucleotide variant (SNV) and copy-number variant (CNV) counts indicate somatic variants that are attributed to each branch along the proposed evolutionary tree. (Chr) Chromosome, (IC) intermediate clone, (IGV) Integrative Genomics Viewer. (B) Landscape of shared key genomic alterations between different lesions. (TPM) Transcripts per million. (C) IGV screenshot of Chr 12 across all lesions from s-WGS data. Inset shows exome depth ratios for the brain lesion, indicating focal amplification of the region encoding ERBB3. (Figure continued on next page.)
The presence of the driver mutation KRAS p.Gly12Asp detected in all metastatic clones along with the more recently described Chr 1p36.33 loss (Van Nieuwenhuysen et al. 2019) provided strong evidence of a model of branched evolution of the different metastatic subclones from a common LGSOC ancestry (Table 2). Notably, the sigmoid colon lesion was first observed in the 2011 CT scan, earlier than all the other lesions. The adrenal gland, soft-tissue hepatic flexure, lung, and soft-tissue peripancreatic lesions appeared in 2013 prior to MEKi treatment, whereas the cerebellar metastasis occurred in 2015 and its appearance corroborates with MEKi resistance and patient death caused by rapid disease progression (Fig. 2A).
Table 2.
Filtered variants of interest
| Gene | Chr | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP/dbVAR ID |
|---|---|---|---|---|---|---|
| KRAS | Chr 12 | c.35G > A | p.Gly12Asp | Missense_Mutation | Deleterious (0) | rs121913529 |
| ARID1A | Chr 1 | c.681_684delGGCC | p.Ala228ThrfsTer3 | Frame_Shift_Del | NA | NA |
(Chr) Chromosome, (HGVS) Human Genome Variation Society, (NA) not applicable, (dbSNP) Single-Nucleotide Polymorphism Database, (dbVAR) Database of Genomic Structural Variation.
Based on the timing of clinical interventions and the patterns of shared genomic aberrations, we inferred the hierarchy and interconnectedness of the different lesions. Our analyses revealed that the primary tumor might have carried within itself a subclonal architecture that underwent parallel selection during disease progression, giving rise to tumors in the sigmoid colon and an intermediate clone (IC-1). This IC-1 then acquired additional mutations and copy-number variations (CNVs) to give rise to (1) the adrenal gland metastasis and (2) an additional IC (IC-2). IC-2 further diverged, seeding the lung and the more complex brain lesion (via IC-3) and also branching off to acquire an ARID1A frameshift mutation (p.Ala288fs) common to a lineage of additional metastatic sites (hepatic flexure soft tissue, lymph node, and peripancreatic tissue) (Table 2; Fig. 2A,B; Supplemental Fig. S1).
Upon interrogating the genomic architecture of the brain lesion that led to MEKi failure, we identified unique gains in Chr 13, 20, and 21 and Chr 12q13.11–q14.1. Interestingly, the brain lesion exhibited novel focal amplification of ERBB3 that mapped onto this region of Chr 12 (Fig. 2C). Myc-dependent transcriptional up-regulation of ERBB3 has been previously implicated in the context of MEK inhibition in KRAS-mutated lung and colorectal cancers in preclinical studies (Sun et al. 2014). In our case, although ERBB3 was most highly expressed in lung and sigmoid colon lesions, the expression of its negative regulator MYC was also high in these two tissues (Fig. 2B). In contrast, MYC expression was lowest in the brain lesion (Fig. 2B), potentially allowing for the activation of ERBB3-dependent MEKi resistance exclusively in the brain.
Briefly, the sequence of events observed in this unique LGSOC patient highlights intratumor heterogeneity arising from tumor evolution and identifies unique genomic factors in the brain lesion that may contribute toward acquired resistance to targeted therapy and sustain the unusual aggressive behavior.
DISCUSSION
Clonal evolution resulting from persistent selective pressure that cancer cells undergo during treatment may lead to acquired drug resistance. Low-grade serous is a rare subtype of OC characterized by slow growth, early age of onset, resistance to chemotherapy, and lack of active treatments to improve patient outcome (Gershenson et al. 2006). Despite recent advances in defining the molecular characteristics of LGSOC, no targeted therapy is available for this disease, which represents an important unmet clinical need. Promising preliminary results of MEKi are not yet confirmed in a randomized trial in an unselected population (MILO trial, NCT01849874); however, signals of activity have been previously reported in genotype-selected patients carrying KRAS mutation and/or using combination strategies (Takekuma et al. 2016; Spreafico et al. 2017; Han et al. 2018). We conducted a comprehensive longitudinal analysis on a single patient with KRAS p.Gly12Asp-mutated LGSOC who had partial response to binimetinib for 17.5 mo (76 wk) after progression on several lines of treatment. The KRAS mutation was found unchanged across all tumor samples and this may explain the initial good response our patient achieved with binimetinib, suggesting that MEKi could still represent a relevant treatment option for KRAS-mutated LGSOC. To better define the role of MEKi in this tumor subtype, biomarker-driven trials are needed alongside complementary deep omics-based analyses of exceptional responders to provide rationale necessary for future targeted agent development in this population. Despite an initial radiological response, our patient developed a rapid progressive disease with uncommon brain metastasis rarely described in LGSOC (Marchetti et al. 2016). This is the first study to assess spatial and temporal evolution of molecular heterogeneity in LGSOC over 8 yr of therapy, comparing tumor samples pre- and post-MEKi. This includes comprehensive analysis of the underlying biology of late-stage brain metastasis to postulate putative mechanisms of acquired cellular aggression and resistance to MEK inhibition. As the prognosis of this patient was driven by the rapid evolution of brain metastasis, we interrogated genomic abnormalities present uniquely in the brain lesion when compared to other lesions that could explain the patient's phenotype.
In our patient, we observed the highest expression of the ERBB3 gene in the lung, followed by sigmoid colon, and brain metastasis (42.17, 38.61, and 12.63 transcripts per million [TPM], respectively). ERBB3 overexpression was previously described as a marker of acquired resistance to MEKi in KRAS-mutated lung and colon cancers in the preclinical setting (Sun et al. 2014). In this study, it was shown that MEKi of KRAS-mutated lung and colon cancer cells led to Myc degradation, which was concurrent with transcriptional up-regulation of ERBB2 and ERBB3 and the formation of kinase-active EGFR–ERBB3 and ERBB2–ERBB3 heterodimers (Sun et al. 2014). The authors also showed that combination therapy targeting EGFR and ERBB2 can reverse MEKi resistance and tumors with high ERBB3 expression levels are more likely to benefit from such combination treatment (Sun et al. 2014). Similar observation related to ERBB pathway involvement in MEKi resistance was also seen in a more recent study in LGSOC patient–derived cell lines (Fernández et al. 2019). In this study, a combined inhibition of MEK and EGFR led to tumor cell death in MEKi-resistant LGSOC cell lines.
In our patient, even though the highest expression levels of both ERBB2 and ERBB3 were observed in lung and sigmoid colon lesions, MYC expression was also relatively high in these two tissues (Fig. 2B). In contrast, the lowest MYC expression was observed in the brain lesion, suggesting that myc-induced transcriptional repression of ERBB genes was suppressed in the brain lesion, potentially explaining the unique phenomena of binimetinib resistance and rapid disease progression observed only in the brain lesion. Our study also identified unique amplifications in Chr 13, 20, and 21 in the MEKi resistant brain lesion. Chr 12 and 20 amplifications have been previously reported in recurrent LGSOC patient–derived cell lines following treatment with chemotherapy (Fernández et al. 2016). Further investigation of the role of genes located in these amplified chromosomal regions is warranted to understand potential mechanisms to bypass MEKi resistance. Notably, dual inhibition of SRC (located on Chr 20), along with MEK inhibition, has been shown to overcome MEK resistance and suppress cancer cells growth in preclinical models of high-grade serous ovarian cancer (Simpkins et al. 2018).
In conclusion, this study highlights the need for consistent monitoring of tumor evolution during the course of disease progression and the importance of applying an individualized approach to treatment based on the changing dynamics of a patient's tumor landscape. This approach may enhance the effectiveness of targeted treatment and, ultimately, lead to improvements in outcome. This study shows that common mechanisms of resistance (myc-induced transcriptional repression of ERBB genes) may exist across multiple tumor types, and as such, patients with a different histopathological diagnosis may benefit from similar genotype-matched combinatorial approaches. In addition, this study supports findings from previous preclinical studies and identifies unique chromosomal aberrations (amplifications in Chr 12, 13, 20, and 21) in LGSOC that, when probed in larger prospective translational studies, may provide novel insights into the biology of LGSOC. Future prospective trials are warranted to understand the generality of the ERBB3-driven MEKi resistance mechanism in LGSOC patients.
METHODS
Sample Collection
Freshly frozen or formalin-fixed paraffin-embedded (FFPE) tissue were obtained from the second and third debulking surgery and at the time of death and reviewed by an expert Gynecologist Pathologist to confirm the diagnosis of LGSOC in all the analyzed samples (Fig. 1C). Written informed consent for participation in the study and local Research Ethic Board approval for use of patient samples were obtained.
DNA Analyses
DNA and RNA samples were coisolated from frozen tissues at the Princess Margaret Genomics Center (PMGC) using QIAGEN All-Prep DNA/RNA/miRNA Universal CoIsolation kit. WES and s-WGS were performed from the DNA samples using Illumina HiSeq2500 and NextSeq500 sequencers, respectively. The mean depth of coverage across tumors samples was ∼250× and ∼0.15× for WES and sWGS, respectively (Table 3). WES and WGS were aligned to human genome reference (build hg38) using the Burrows–Wheeler aligner (bwa) mem (Li and Durbin 2009). For WES a normal liver tissue specimen was sequenced as a germline control with a mean depth of coverage of 53×. Somatic single-nucleotide variants (SNVs) and insertions and deletions (indels) were called using Strelka 1.01.14 (Saunders et al. 2012). SAMtools mpileup (Li et al. 2009) (v0.1.18), VarScan2 (v 2.3.6) (Koboldt et al. 2012), and Sequenza (v2.1.0) were used to determine allele-specific copy-number changes (Favero et al. 2014). Data was visualized and interpreted using University Health Network's cBioPortal instance (http://cbioportal.uhnresearch.ca) (Gao et al. 2013). Segmented copy number was generated from s-WGS data using ichorCNA (v0.1.0-17) (Adalsteinsson et al. 2017).
Table 3.
Sequencing coverage
| # | Sample | s-WGS coverage (×) | Exome coverage (×) | RNA-seq reads (in millions) | Tumor cellularity estimate (Sequenza) |
|---|---|---|---|---|---|
| 1 | Sigmoid colon | 0.16 | 254 | 405 | 0.88 |
| 2 | Soft tissue hepatic flexure | 0.09 | 260 | 214 | 0.34 |
| 3 | Lower lobe lung | 0.12 | 214 | 248 | 0.71 |
| 4 | Right adrenal gland | 0.21 | 241 | 320 | 0.63 |
| 5 | Lymph node, subcarinal | 0.18 | 244 | NA | 0.41 |
| 6 | Cerebellum | 0.15 | 245 | 199 | 0.83 |
| 7 | Peripancreatic soft tissue | 0.12 | 261 | NA | 0.58 |
| Normal (Liver) | 0.16 | 53 | NA | NA |
(NA) Not applicable, (s-WGS) shallow whole-genome sequencing.
RNA Analyses
RNA-seq libraries were prepared with 200 ng of total RNA from available pre- and post-MEKi samples using the TruSeq Stranded Total RNA kit with Ribo-Zero Gold from Illumina followed by sequencing using an Illumina NextSeq500 sequencer to generate ∼270 million reads/sample (Table 3). RNA-seq reads were aligned using STAR (v2.4.2a) (Dobin et al. 2013) to hg38 using the GENCODE v26 for transcript annotation. Gene expression analysis was performed using RSEM (v1.2.21) (Li and Dewey 2011). Fusions were detected in RNA-seq data using STAR-Fusion (Haas et al. 2017) and filtered to include only those with a least one spanning fragment and one junction read supporting the fusion (Supplemental Table 1).
ADDITIONAL INFORMATION
Data Deposition and Access
Interpreted variants have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000999192 and SCV000999193. According to the current version of the protocol approved by local Research Ethic Board, genomic data cannot be made publicly available and a specific patient consent cannot be obtained.
Ethics Statement
Written informed consent for participation in the study and local Research Ethic Board approval for use of patient samples were obtained.
Acknowledgments
This pilot project was supported by the Princess Margaret Cancer Foundation. We thank the Princess Margaret Genomics Centre for conducting all DNA extractions and genome sequencing. We acknowledge Katherine Karakasis for providing editorial support. We are also grateful to our patient and her family for allowing her tissues to be collected and used for this research project.
Author Contributions
This work was conducted at Princess Margaret Cancer Centre led by A.M.O., T.P., and S.L. I.C. and S.G. conceived and drafted the manuscript and performed data analyses and interpretation. A.D., J.B., and R.Q. performed bioinformatics analyses of sequencing data and assisted in data interpretation. P.S. reviewed and confirmed the histopathologic diagnosis of each tumor sample. Q.T. and M.B. coordinated the sample collection and management. A.M.O., T.P., and S.L. reviewed the data and the manuscript.
Funding
This project was funded by the Princess Margaret Cancer Foundation. All authors have approved the current version of the manuscript and its submission to CSH Molecular Case Studies.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, Gydush G, Reed SC, Rotem D, Rhoades J, et al. 2017. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 8: 1324 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Array BioPharma. 2016. Array BioPharma announces decision to discontinue MILO study in ovarian cancer [press-release]. Array BioPharma, Boulder, CO. [Google Scholar]
- Cancer Genome Atlas Research Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley J, Brady WE, Vathipadiekal V, Lankes HA, Coleman R, Morgan MA, Mannel R, Yamada SD, Mutch D, Rodgers WH, et al. 2013. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol 14: 134–140. 10.1016/S1470-2045(12)70572-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favero F, Joshi T, Marquard AM, Birkbak NJ, Krzystanek M, Li Q, Szallasi Z, Eklund AC. 2014. Sequenza: allele-specific copy number and mutation profiles from tumor sequencing data. Ann Oncol 26: 64–70. 10.1093/annonc/mdu479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández ML, DiMattia GE, Dawson A, Bamford S, Anderson S, Hennessy BT, Anglesio MS, Shepherd TG, Salamanca C, Hoenisch J, et al. 2016. Differences in MEK inhibitor efficacy in molecularly characterized low-grade serous ovarian cancer cell lines. Am J Cancer Res 6: 2235–2251. [PMC free article] [PubMed] [Google Scholar]
- Fernández ML, Dawson A, Hoenisch J, Kim H, Bamford S, Salamanca C, DiMattia G, Shepherd T, Cremona M, Hennessy B, et al. 2019. Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target. Cancer Cell Int 19: 10 10.1186/s12935-019-0725-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson DM, Sun CC, Lu KH, Coleman RL, Sood AK, Malpica A, Deavers MT, Silva EG, Bodurka DC. 2006. Clinical behavior of stage II–IV low-grade serous carcinoma of the ovary. Obstet Gynecol 108: 361–368. 10.1097/01.AOG.0000227787.24587.d1 [DOI] [PubMed] [Google Scholar]
- Gourley C, Farley J, Provencher DM, Pignata S, Mileshkin L, Harter P, Maenpaa J, Kim JW, Pujaide-Lauraine E, Glasspool RM, et al. 2014. Gynecologic Cancer InterGroup (GCIG) consensus review for ovarian and primary peritoneal low-grade serous carcinomas. Int J Gynecol Cancer 24: S9–S13. 10.1097/IGC.0000000000000257 [DOI] [PubMed] [Google Scholar]
- Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, Bankapur A, Ganote C, Doak TG, Pochet N, et al. 2017. STAR-Fusion: fast and accurate fusion transcript detection from RNA-seq. bioRxiv 10.1101/120295 [DOI] [Google Scholar]
- Han C, Bellone S, Zammataro L, Schwartz PE, Santin AD. 2018. Binimetinib (MEK162) in recurrent low-grade serous ovarian cancer resistant to chemotherapy and hormonal treatment. Gynecol Oncol Rep 25: 41–44. 10.1016/j.gore.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter SM, Anglesio MS, Ryland GL, Sharma R, Chiew YE, Rowley SM, Doyle MA, Li J, Gilks CB, Moss P, et al. 2015. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget 6: 37663–37677. 10.18632/oncotarget.5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledermann JA, Raja FA, Fotopoulou C, Gonzalez-Martin A, Colombo N, Sessa C, Group EGW. 2013. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 24: vi24–vi32. 10.1093/annonc/mdt333 [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti C, Ferrandina G, Cormio G, Gambino A, Cecere S, Lorusso D, De Giorgi U, Bogliolo S, Fagotti A, Mammoliti S, et al. 2016. Brain metastases in patients with EOC: clinico-pathological and prognostic factors. A multicentric retrospective analysis from the MITO group (MITO 19). Gynecol Oncol 143: 532–538. 10.1016/j.ygyno.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Mutch DG, Prat J. 2014. 2014 FIGO staging for ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol 133: 401–404. 10.1016/j.ygyno.2014.04.013 [DOI] [PubMed] [Google Scholar]
- Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. 2012. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 28: 1811–1817. 10.1093/bioinformatics/bts271 [DOI] [PubMed] [Google Scholar]
- Simpkins F, Jang K, Yoon H, Hew KE, Kim M, Azzam DJ, Sun J, Zhao D, Ince TA, Liu W, et al. 2018. Dual Src and MEK inhibition decreases ovarian cancer growth and targets tumor initiating stem-like cells. Clin Cancer Res 24: 4874–4886. 10.1158/1078-0432.CCR-17-3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreafico A, Oza AM, Clarke BA, Mackay HJ, Shaw P, Butler M, Dhani NC, Lheureux S, Wilson MK, Welch S, et al. 2017. Genotype-matched treatment for patients with advanced type I epithelial ovarian cancer (EOC). Gynecol Oncol 144: 250–255. 10.1016/j.ygyno.2016.12.002 [DOI] [PubMed] [Google Scholar]
- Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A. 2014. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell Rep 7: 86–93. 10.1016/j.celrep.2014.02.045 [DOI] [PubMed] [Google Scholar]
- Takekuma M, Wong KK, Coleman RL. 2016. A long-term surviving patient with recurrent low-grade serous ovarian carcinoma treated with the MEK1/2 inhibitor, selumetinib. Gynecol Oncol Res Pract 3: 5 10.1186/s40661-016-0026-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nieuwenhuysen E, Busschaert P, Laenen A, Moerman P, Han SN, Neven P, Lambrechts D, Vergote I. 2019. Loss of 1p36.33 frequent in low-grade serous ovarian cancer. Neoplasia 21: 582–590. 10.1016/j.neo.2019.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Interpreted variants have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession numbers SCV000999192 and SCV000999193. According to the current version of the protocol approved by local Research Ethic Board, genomic data cannot be made publicly available and a specific patient consent cannot be obtained.