Abstract
Whole-exome sequencing was used to identify the genetic etiology of a rapidly progressing neurological disease present in two of six siblings with early childhood onset of severe progressive spastic paraparesis and learning disabilities. A homozygous mutation (c.2005G>T, p, V669L) was found in VAC14, and the clinical phenotype is consistent with the recently described VAC14-related striatonigral degeneration, childhood-onset syndrome (SNDC) (MIM#617054). However, the phenotype includes a distinct clinical presentation of retinitis pigmentosa (RP), which has not previously been reported in association with VAC14 mutations. Brain magnetic resonance imaging (MRI) revealed abnormal magnetic susceptibility in the globus pallidus, which can be seen in neurodegeneration with brain iron accumulation (NBIA). RP is a group of inherited retinal diseases with phenotypic/genetic heterogeneity, and the pathophysiologic basis of RP is not completely understood but is thought to be due to a primary retinal photoreceptor cell degenerative process. Most cases of RP are seen in isolation (nonsyndromic); this is a report of RP in two siblings with VAC14-associated syndrome, and it is suggested that a connection between RP and VAC14-associated syndrome should be explored in future studies.
Keywords: moderate global developmental delay, retinitis pigmentosa inversa
CASE PRESENTATION
A 21-yr-old male (referred to hereafter as the proband), with developmental disabilities and retinitis pigmentosa (RP) (Table 1; Fig. 1) was referred to our clinic and agency for services and benefits evaluation from the Office of People with Developmental Disabilities in New York State.
Table 1.
Phenotypic features—HPO terminology
| VAC14 syndrome | Proband 1 (II-1) | Mother (I-1) | Father (I-2) | Sibling sister (II-6) |
|---|---|---|---|---|
| HP:0002376 Loss of developmental skills | Y | Y | N | Y |
| HP:0002317 Unsteady gait | Y | N | N | Y |
| HP:0002307 Drooling | Y | N | N | N |
| HP:0012179 Delayed speech | Y | N | N | Y |
| HP:0006957 Loss of ability to walk | Y | N | N | Y |
| HP:0006957 Hypotonia | Y | N | N | Y |
| HP:0000007 Autosomal recessive inheritance | Y | Y | Y | Y |
| HP:0001561 Polyhydramnios | N/A | Y | N | N |
| HP:0009381 Short fingers | Y | Y | N/A | Y |
| HP:0001263 Global developmental delay | Y | N/A | N/A | Y |
| HP:0000750 Delayed speech and language | Y | N/A | N/A | Y |
| HP:0002015 Dysphagia | Y | N/A | N/A | Y |
| HP:0001167 Abnormality of finger | N | Y | N/A | N/A |
| HP:0000518 Cataract | Y | N/A | N/A | Y |
(N/A) Not available, (Y) yes, present, (N) no, not present.
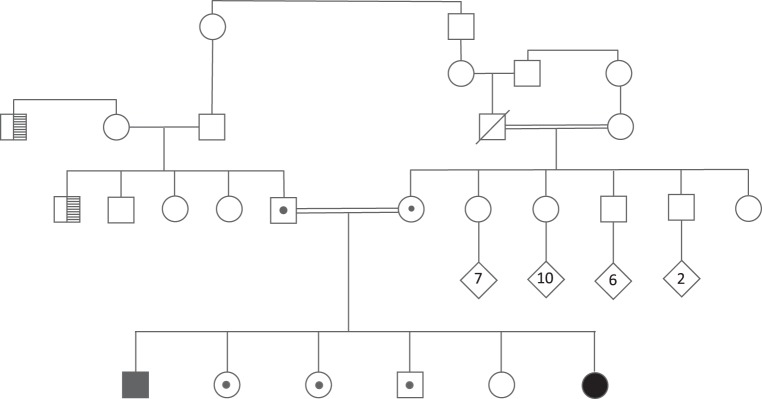
Figure 1.
Family pedigree: ancestry is Sephardic and Ashkenazim Syrian Jewish. Affected probands (confirmed to be homozygous for V669L/ V669L in VAC14) are marked with black-filled symbols. Carriers (verified by sequencing to be heterozygous WT/V669L in VAC14) are marked with a dot in the symbol. Proband 1: age 21, male, developmental delays, spastic paraplegia, RP, basal ganglia anomaly, speech problems, premature birth at 34 wk. Sibling 2: age 20, female; sibling 3: age 18, female; sibling 4: age 20, male; sibling 5: age 14, female; sibling 6 (Proband 2): age 4, mild developmental delays, hypotonia, nondysmorphic, RP; mother: age 44, clubfoot, abnormal fingers and toes, bruises easily, with five siblings: female age 40, female age 31 (has child with craniosynostosis), brother age 38, brother age 39, sister age 31 with clubfoot, hydrocephalus s/p shunt, abnormal finger and toe formation; father: age 51; the ages of the paternal siblings are not known. Parents are second cousins once removed. Consanguinity in the pedigree, as maternal grandparents are first cousins. History of night blindness with the paternal uncle and paternal great uncle of the probands (marked with cross-hatching in the pedigree).
Pregnancy
The proband's 23-yr-old mother's pregnancy was complicated with oligohydramnios, premature rupture of membranes at 34 wk, and maternal febrile illness due to a urinary tract Escherichia coli infection. Birth weight was 2355 g (3 lb, 13 oz). The baby remained in the NICU for 2.5 wk, where he experienced complications of respiratory distress, possible neonatal sepsis, and lung atelectasis
Development and Behavior
The proband's mother reported an apparent normal first 2 yr of life with postnatal early milestones being achieved (crawled at 8 mo, walked at 13 mo, and was toilet trained by 2.5 yr); however, by his second birthday, the proband was noted to have unclear speech and an unstable gait. He was evaluated at 25 mo for early intervention services and received speech, occupational, and physical therapies, along with special instruction. At age 15.6 yr, he was administered a Stanford–Binet Intelligence Scale, 5th edition, and his full-scale IQ was 71 (mildly delayed), with nonverbal IQ of 68 and verbal IQ of 73. At age 13, he had a psychological evaluation using the Wechsler Adult Intelligence Scale, in which he had a verbal comprehension score of 71 (third percentile rank) and was considered borderline low. His visual impairment interfered significantly with his performance on the perceptual reasoning and information processing (PRI) tasks, so the PRI score was extremely low at 55 (0.1 percentile rank). He is able to provide personal hygiene care for himself.
Medical Presentation
Both the proband and his 4-yr-old affected sister were evaluated at New York Presbyterian Medical Center for their history of vision and developmental differences. The proband is noted to have progressive spastic paraparesis with hyperreflexia, along with a spastic ataxic gait, bilateral Babinski signs, slurred speech, and hypophonia. He walks on his toes. At age 5, he developed peripheral and night vision problems and was formally diagnosed with RP, following a full electroretinogram (ERG). His vision appeared stable from 2005 to 2009, but then progressively worsened over the years. He also has low muscle tone and speech delays. He has a long face and pointed chin, myopathic appearance with bilateral ptosis, and arching eyebrows. The proband had no dystonia, ataxia, or drooling. At age 9, a brain MRI demonstrated decreased signal of the globus pallidi on T2* gradient echo sequences. The radiology report raised the question of the “eye-of-the-tiger” sign (a mixture of globus pallidus T2 prolongation and susceptibility associated with PANK2 associated NBIA, one of the few NBIA disorders known at the time of the initial interpretation [Hayflick et al. 2003]). The retrospective review did not demonstrate this finding but did demonstrate faint striatal T2 prolongation as well as abnormal substantia nigra susceptibility for age (Supplemental Fig. 1).
Brain MR spectroscopy was normal, with no abnormal lactate signal, and blood lactic acid levels were in the normal range.
Previous Genetic Testing
The fragile X syndrome test was negative, the proband was negative for gene RPGR-ORF15 mutations (Molecular Vision Laboratory, Hillsboro, OR, 2017), and he was negative for mutations in the gene PANK2. The chromosomal microarray was normal.
Family Background
The proband lives at home with his parents and is the oldest of six siblings born into a Syrian Sephardic Jewish family. The proband's father is of mixed Syrian and non-Syrian Jewish ancestry, and his mother is of Syrian Jewish ancestry. The maternal grandparents are first cousins, and the father's parents are second cousins (Fig. 1). The proband has a 4-yr-old sister with similar manifestations including homozygosity for the same VAC14 variant. Four other siblings are heterozygous for, or do not have, the VAC14 mutation (see Fig. 1). In 2006, clinical genetics evaluation discussed the fact that congenital stationary night blindness was also present in one uncle and one great uncle, but this presentation was deemed unrelated to the present condition. The proband's mother has a history of clubfoot, finger and toe deformities, unusual facial features, and easy bruising. The proband's three maternal aunts, two maternal uncles, and one fraternal aunt have similar medical issues to his mother.
The affected 4-yr-old sister has developmental disabilities and RP (see Table 1).
Pregnancy—Proband's Sister
This sister was the product of an uncomplicated pregnancy. Her birth weight was 6 lb, 7 oz. There were no neonatal complications.
Development and Behavior—Proband's Sister
Her parents first started noticing issues with her vision when she began walking and was bumping into things and using her hands to feel around. She was diagnosed with RP and mild horizontal nystagmus.
She has a history of hypotonia and developmental delays. She rolled over at 9–10 mo, and walked and said first words after 18 mo.
Medical Presentation—Proband's Sister
Other than hypotonia, developmental disabilities, and RP, it is currently too early in her life to determine whether she will develop other neurologic symptoms comparable to her older brother.
TECHNICAL ANALYSIS
The proband, his affected sister, and parents were fully sequenced as part of a whole-exome sequencing study at Hadassah University Hospital. The laboratory (Otogenetics Ltd) captured ∼50 Mb of genomic DNA and sequenced the library on a HiSeq 2500 instrument. The fragments were 100–125 bp, paired-end reads. The samples were uploaded onto DNAnexus software and 37.73 million reads were aligned to the reference human genome (Hg19) (mean on target coverage × 67.9) (see Table 2). Variants were removed if they were: off-target (>6 bp from splice site), synonymous, far from splice sites, or common (MAF > 0.1%). This left 523 variants remaining. There were no coverage differences when compared to the same day samples. One variant NM_018052.5:c.2005G>T (p. V669L) in VAC14 was identified as being homozygous in the proband and his affected sister, inherited once from each parent (Table 3). Segregation analysis via Sanger sequencing confirmed the variants and demonstrated segregation of this variant with the reported phenotype (Fig. 1). Four other siblings are heterozygous or do not have the VAC14 mutation (see Fig. 1).
Table 2.
Quality metrics of exome sequencing data from Otogenetics
| Affected brother | Affected sister | Father | Mother | |
|---|---|---|---|---|
| TOTAL_READS (Mb) | 37.73 | 43.9 | 43.0 | 39.2 |
| MEAN_COVERAGE (×) | 67.9 | 79.3 | 75.0 | 70.3 |
| PF_UNIQUE_READS | 37.7 | 43.9 | 43.0 | 39.2 |
| Average aligned reads (%) | 98.8 | 98.7 | 98.7 | 98.6 |
Table 3.
Genomic/exome findings
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect (substitution, deletion, etc.) | dbSNP/ dbVar ID | Genotype (heterozygous/homozygous) | ClinVar ID (optional) | Parent of origin (optional) |
|---|---|---|---|---|---|---|---|---|---|
| VAC14 | Chr 16: 70729477a | NM_018052 | c.2005G>T, p. V669L | Missense | Substitution | rs1363536856 | Homozygous | Pending, SUB5893230 | Biparentally inherited |
aUsing Human February 2009 (GRCh37/hg19) Assembly.
Clinical whole-exome sequencing was also performed at Ambry Genetics for the affected sister in 2016, and re-analysis in 2018 identified the mutation in VAC14, and targeted DNA analysis in 2018 of the proband confirmed the alteration c.2005G>T (p. V669L).
The following RP genes in the exome data from Otogenetics were specifically analyzed for the presence of variants that may be contributing to the eye phenotype in both affected siblings:
ABCA4 ABHD12 ADIPOR1 AGBL5 AIPL1 ARHGEF18 ARL2BP ARL3 ARL6 BBS2 C21orf2i4 C2orf71 C2ORF71 C5AR2 C8ORF37 CA4 CDH16 CDHR1 CERKL CLRN1 CNGA1 CNGB1 CRB1 CWC27 CYP4V2 DHDDS DHX38 DNAJC17 EXOSC2 FLVCR1 FSCN2 GPR125 GUCA1B HGSNAT IDH3A IDH3B IFT172 IMPDH1 IMPG2 KIAA1549 KIZ LGALS9B LRAT MAK MERTK MFRP NEK2 NR2E3 NRL OFD1 OR2W3 orf15 PANK2 PCARE PDE6A PDE6B PDE6G PLK1S1 POC5 POMGNT1 PRCD PROM1 PRPF3 PRPF31 PRPF4 PRPF6 PRPF8 PRPH2 RBP3 RDH12 REEP6 RGR RHBDD2 RHO RLBP1 ROM1 RP1 RP1L1 RP2 RP9 RPE65 RPGR rpgr11a RRH SAG SEMA4A SLC7A14 SNRNP200 SRPX TLR4 TRNS2 TTC8 ttc8r TULP1 ush1 cpst WDR19 ZNF408 ZNF513.
None were identified. No differences in coverage were observed in the RP genes in affected individuals as compared to healthy parents.
The exome data from Ambry Genetics for the 4-yr-old affected sister were also reanalyzed. The QC of FASTQ files was done with FASTQC. The sequencing reads were mapped to the human Hg19 reference genome using a Burrows–Wheeler Aligner (BWA) with standard parameters (Li and Durbin 2009). After alignment, base quality score recalibration (BQSR) and indel realignment were performed with GATK (best practices pipeline) (McKenna et al. 2010). Duplicate read pairs were marked using PICARD (http://broadinstitute.github.io/picard/). Germline SNPs and indels were identified using FreeBayes (Garrison and Marth 2012) in this trio. The variant annotation was done with ANNOVAR (Wang et al. 2010). The variants with low depth (reads <30), minor allele frequencies (>1% in gnomAD), and that were intronic, synonymous, and in-frame indels were excluded. A recessive inheritance pattern was assumed on the basis of the pedigree. The homozygous variants in the proband were retained. There was no evidence for any mutations in PANK2, consistent with prior genetic testing results. Given the consanguinity in the family, additional homozygous variants were examined, including in the genes ZFHX3, ARID5A, SULT1C2, PITRM1, INSR, C20orf96, and ZCCHC3 (see Supplemental Excel File 1), but none of these were deemed as likely contributing to the phenotypes in both siblings.
We next used a phenotype-driven approach (Yang et al. 2015) for disease gene finding without an assumption for homozygous disease-causal variants. Using the same VCF file generated above, we uploaded the file to the wANNOVAR server, together with the description of the observed clinical phenotypes in the form of human phenotype ontology (HP:0002376, HP:0002317, HP:0012179, HP:0006957, HP:0006957, HP:0009381, HP:0001263, HP:0000750, HP:0002015, HP:0000518). The wANNOVAR server combined genotype information and phenotype information to derive a ranked list of candidate genes from exome sequencing data. In the results, VAC14 is ranked as the top gene with a score of 0.8, yet the second-highest ranked gene has a prioritization score of only 0.057. Thus, the phenotype-driven analysis of exome data strongly suggests that VAC14 is the only gene with a strong evidence of contributing to the observed phenotypes.
Additional variant interpretation of the Ambry WES data for the 4-yr-old affected sister was also conducted by the U.K. state registered Clinical Scientists in Sapientia software (Congenica Limited). FASTQ data was aligned to GRCh37 using BWA-MEM (0.7.12) and variant calling performed using GATK HaplotypeCaller (version 3.4-46). QC of FASTQ files was done with FASTQC (0.11.5) and VCF variant-based QC done using VariantRecalibrator (GATK). Variant annotation against all RefSeq and Ensembl transcripts was done via Ensembl Variant Effect Predictor (VEP; version 81). Variants were prioritized/ranked using Exomiser's Gene Pheno Score (version 10). Rare variants were identified by filtering using a maximum allele frequency setting of 0.5% in the following population data sets: EXAC, exome sequencing project (ESP), UK10K, and 1000 genomes. Variant consequence filters were used to select variants predicted to impact coding sequence, consensus splice sites, and splice region of VAC14-associated RefSeq and Ensembl transcripts. Resulting variants were analyzed in Sapientia using a gene agnostic approach utilizing filtering by inheritance and phenotype driven variant prioritization (Smedley et al. 2015). A virtual gene panel approach was used for review of variants in genes known to be associated with RP.
VARIANT INTERPRETATION
The VAC14 c.2005G>T (p. V669L) variant is not reported in the Genome Aggregation Database (gnomAD), a collection of observed variants from individuals with no known severe pediatric disease or first-degree relatives with severe pediatric disease. The p.V669 amino acid is conserved in available vertebrate species and in many invertebrates including Drosophila melanogaster and Caenorhabditis elegans. The p.V669L alteration is predicted by in silico analyses to be possibly damaging by PolyPhen, deleterious by MutationTaster, damaging by SIFT, and damaging by LRT. This variant was seen only once (out of 125,568 alleles) in the TOPMED project (Natarajan et al. 2018) and is therefore reported as SNP-rs1363536856, with further information available at. https://www.ncbi.nlm.nih.gov/snp/rs1363536856.
The alteration is homozygous in both affected siblings (Proband 1 and his sister), with the proband having a phenotype consistent with VAC14-associated neurodegeneration (Fig. 1). The sister is much younger and is currently more mildly affected, with hypotonia, developmental delays, and RP. The variant is in a carboxy-terminal domain of VAC14 that binds to FIG4 (Jin et al. 2008) and is involved in dimerization of VAC14 (Alghamdi et al. 2013), both of which could be affected by this variant.
This variant is classified as of Uncertain Significance according to the American College of Medical Genetics (ACMG) guidelines (Kleinberger et al. 2016; Richards et al. 2015) for the following reasons: PM1_Supporting, Located in a well-established functional domain without benign variation, modified to supporting, because although the domain has been characterized as functionally important (Jin et al. 2008; Alghamdi et al. 2013), it is not without benign variation as specified in the ACMG/AMP guideline; PM2 (moderate), Absent from controls (or at extremely low frequency if recessive) in Exome Sequencing Project, 1000 Genomes Project, or Exome Aggregation Consortium; PP1, cosegregation with disease in multiple affected family members in a gene definitively known to cause the disease, applying the LOD scoring system according to Oza et al. (2018) and PP3, multiple lines of computational evidence support a deleterious effect on the gene or gene product (conservation, evolutionary, splicing impact, etc.).
SUMMARY
Neurodegeneration with brain iron accumulation (NBIA) is a broad term that describes a heterogeneous group of progressive and incapacitating neurologic disorders in which there is iron deposition in certain brain areas, including the basal ganglia (Di Meo and Tiranti 2018; Alvarez-Cordoba et al. 2019). Major clinical symptoms can include spasticity, progressive dystonia, Parkinson's disease–like symptoms, neuropsychiatric alterations, and retinal degeneration. Among the NBIA disorders, the most frequent subtype is pantothenate kinase–associated neurodegeneration (PKAN) caused by defects in the gene encoding the enzyme pantothenate kinase 2 (PANK2), which catalyzes the first reaction of the coenzyme A biosynthesis pathway (Alvarez-Cordoba et al. 2019). To date, 12 disease-associated genes leading to NBIA have been identified, but ∼20% of cases are still genetically undefined (Di Meo and Tiranti 2018). These authors write that “only two genes, namely FTL and ceruloplasmin are directly associated to iron homeostasis while, at first glance, the other ten genes (PANK2, COASY, PLA2G6, C19orf12, FA2H, ATP13A2, WDR45, DCAF17, SCP2, and GTPBP2) seem to be unrelated and are involved in different cellular pathways” (Di Meo and Tiranti 2018).
The FIG4 protein is a subunit of the PI(3,5)P2 biosynthetic complex that also contains PIKFYVE, a PI3P kinase, and VAC14, a scaffold protein. VAC14 encodes a scaffold protein that is a component of the PIKfyve protein kinase complex. This complex is responsible for the synthesis of phosphatidylinositol 3,5-bisphosphate, an important component of cellular membranes, from phosphatidylinositol 3-phosphate. Mice lacking a functional copy of this gene exhibit severe neurodegeneration (Zhang et al. 2007; Jin et al. 2008).
It was recently shown that mutations in VAC14 can also be associated with Yunis–Varón syndrome (Lines et al. 2017). Mutations in the lipid phosphatase gene FIG4 are also associated with Yunis–Varón syndrome, along with Charcot–Marie–Tooth disease Type 4J, a peripheral neuropathy. Yunis–Varón syndrome (MIM#216340) presents with profound neonatal onset of hypotonia, failure to thrive, skeletal anomalies, and feeding intolerance. This can also include profound developmental delay, absent reflexes, persistent ophisthotonus, roving eye problems, severe feeding intolerance, skeletal abnormalities, heart defects, limb defects, cataracts, structural brain anomalies, and episodes of irritability (Lines et al. 2017). A more recent study presented an additional four families with variants in FIG4, in which the ocular findings in each affected child in Family 4 were initially described as bull's eye maculopathy (Lenk et al. 2019). The more severely affected sibling (Patient 6) was shown to have optic atrophy, retinal atrophy, and vascular attenuation.
Striatonigral degeneration with childhood onset (SNDC) (MIM#617054) is a progressive neurological disorder associated with overall regression, speech regression, abnormal gait, impaired movement, striatal abnormalities, and progressive dystonia of the upper limbs, trunk, and face. This disorder has also been shown to be associated with recessive VAC14 mutations (Lenk et al. 2016; Stutterd et al. 2017). Age of onset is typically between 18 mo and 3 yr (Lenk et al. 2016).
We report here a family that we hypothesize might have NBIA (based on the brain MRI for the proband), in which a homozygous missense variant in VAC14 was identified in two of six siblings with RP and neurologic deterioration. This variant is consistently predicted by various bioinformatics algorithms to be pathogenic, and it occurs in a region of the protein that binds to FIG4 (Jin et al. 2008) and is involved in dimerization of VAC14 (Alghamdi et al. 2013). This mutation in VAC14 (c.2005G>T, p, V669L) is inherited in an autosomal recessive manner. VAC14 (OMIM#604632) is located on Chromosome 16q22.1-q22.2 and is essential for the development of the brain and periphery sensory neurons in mammals (Zhang et al. 2007; Jin et al. 2008). The phenotypic features present in this family are compared in Supplemental Excel File 2 to some of the phenotypes found in Yunis–Varón syndrome and SNDC.
The radiology report from the brain MRI performed in 2006 noted an “eye-of-the-tiger”; however, this was unusual as it has been reported that the “eye-of-the-tiger” sign has been shown to be strongly correlated with PANK2 mutations, thus leading to the term “pantothenate kinase-associated neurodegeneration,” yet our proband and his younger sister did not have any mutations involving PANK2. The “eye-of-the-tiger” sign reflects tissue necrosis and edema (seen on T2-weighted MRI as hyperintensity) within a region of iron deposition (seen as hypointensity) (Hayflick et al. 2003). It is notable in the prior study that PANK2-mutation negative individuals were found to have the only hypointensity of the pallidum, which was labeled as “neurodegeneration with brain iron accumulation” (Hayflick et al. 2003). Review of the brain MRI did not confirm the prior report of an “eye-of-the-tiger” sign in our proband, and instead confirmed hypointensity in the globus pallidus and substantia nigra. Neurological examination of the proband did not reveal any indication of dystonia, ataxia, or drooling, which can be components of pantothenate kinase–associated neurodegeneration. It was noted in a prior report (Lenk et al. 2016) that one of their probands (Proband 1) also had hypointensity of the pallidum.
Retinitis pigmentosa refers to a group of inherited retinal diseases with phenotypic and genetic heterogeneity. The pathophysiologic basis of the progressive visual loss in patients with RP is not completely understood but is felt to be due to a primary retinal photoreceptor cell degenerative process mainly affecting the rods of the peripheral retina. In most cases RP is seen in isolation (nonsyndromic), but in some other cases, it may be a part of a genetic, metabolic, or neurologic syndrome or disorder. Nyctalopia, or night blindness, is the most common symptom of RP. The retinal vessels are often narrowed or attenuated and there is a waxy pallor appearance of the optic nerve head.
The manifestations seen in Proband 1 closely match the symptoms seen in SNDC; but RP is not part of the manifestations reported in other (younger) patients with this gene mutation (Lenk et al. 2016). Because Proband 1 and his affected younger sister have very similar manifestations, and they are the only two related members to be homozygous for the VAC14 mutation, we suggest that it is possible that visual impairment with RP is associated with this VAC14 mutation. Further confirmation of this association and the pathogenicity of this variant, in general, awaits the identification of additional families with the exact same homozygous variant and similar phenotypic features.
ADDITIONAL INFORMATION
Database Deposition and Access
The exome sequencing data were generated as part of clinical testing, so the underlying raw data are not consented for deposition to a public database. The variant has been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and can be found under accession number SCV000925663.
Ethics Statement
Both oral and written patient consent were obtained for research and publication, with approval of protocol #7659 for the Jervis Clinic by the New York State Psychiatric Institute–Columbia University Department of Psychiatry Institutional Review Board. Family consent was not given for photography of the children.
Acknowledgments
Appreciation is extended to the family, and to Ezzat El-Akkad, of IBR's Graphic Art Department, for producing Figure 1. We thank Miriam Meisler for feedback on the manuscript. The authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at http://gnomad.broadinstitute.org/about.
Author Contributions
G.J.L. contributed to design, analysis, and manuscript; E.M. to the manuscript; M.V. to design, clinical evaluation, analysis, and manuscript; Q.L., K.W., A.H., V.M., J.E., Y.H., and S.S. to exome sequencing and analysis; R.M. and D.C.D.V. to clinical evaluation and brain MRI interpretation; I.C. to clinical evaluation and genetic counseling; W.K.C. to clinical evaluation, analysis, and manuscript; and E.Y. to brain MRI interpretation.
Funding
Funding for this report was provided by the George A. Jervis Clinic of the New York State Institute for Basic Research in Developmental Disabilities (IBR), the New York State Office for People with Developmental Disabilities, and a grant to W.K.C. from the JPB Foundation.
Competing Interest Statement
The authors have declared no competing interest.
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Alghamdi TA, Ho CY, Mrakovic A, Taylor D, Mao D, Botelho RJ. 2013. Vac14 protein multimerization is a prerequisite step for Fab1 protein complex assembly and function. J Biol Chem 288: 9363–9372. 10.1074/jbc.M113.453712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Cordoba M, Villanueva-Paz M, Villalón-García I, Povea-Cabello S, Suárez-Rivero JM, Talaverón-Rey M, Abril-Jaramillo J, Vintimilla-Tosi AB, Sánchez-Alcázar JA. 2019. Precision medicine in pantothenate kinase-associated neurodegeneration. Neural Regen Res 14: 1177–1185. 10.4103/1673-5374.251203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meo I, Tiranti V. 2018. Classification and molecular pathogenesis of NBIA syndromes. Eur J Paediatr Neurol 22: 272–284. 10.1016/j.ejpn.2018.01.008 [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv. http://arxiv.org/abs/1207.3907
- Hayflick SJ, Westaway SK, Levinson B, Zhou B, Johnson MA, Ching KHL, Gitschier J. 2003. Genetic, clinical, and radiographic delineation of Hallervorden–Spatz syndrome. N Engl J Med 348: 33–40. 10.1056/NEJMoa020817 [DOI] [PubMed] [Google Scholar]
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, Petersen JL, Zhang Y, Park S, Duex JE, et al. 2008. VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P2 in yeast and mouse. EMBO J 27: 3221–3234. 10.1038/emboj.2008.248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger J, Maloney KA, Pollin TI, Jeng LJB. 2016. An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genet Med 18: 1165 10.1038/gim.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk GM, Szymanska K, Debska-Vielhaber G, Rydzanicz M, Walczak A, Bekiesinska-Figatowska M, Vielhaber S, Hallmann K, Stawinski P, Buehring S, et al. 2016. Biallelic mutations of VAC14 in pediatric-onset neurological disease. Am J Hum Genet 99: 188–194. 10.1016/j.ajhg.2016.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenk GM, Berry IR, Stutterd CA, Blyth M, Green L, Vadlamani G, Warren D, Craven I, Fanjul-Fernandez M, Rodriguez-Casero V, et al. 2019. Cerebral hypomyelination associated with biallelic variants of FIG4. Hum Mutat 40: 619–630. 10.1002/humu.23720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lines MA, Ito Y, Kernohan KD, Mears W, Hurteau-Miller J, Venkateswaran S, Ward L, Khatchadourian K, McClintock J, Bhola P, et al. 2017. Yunis–Varón syndrome caused by biallelic VAC14 mutations. Eur J Hum Genet 25: 1049–1054. 10.1038/ejhg.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Peloso GM, Zekavat SM, Montasser M, Ganna A, Chaffin M, Khera AV, Zhou W, Bloom JM, Engreitz JM, et al. 2018. Deep-coverage whole genome sequences and blood lipids among 16,324 individuals. Nat Commun 9: 3391 10.1038/s41467-018-05747-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza AM, DiStefano MT, Hemphill SE, Cushman BJ, Grant AR, Siegert RK, Shen J, Chapin A, Boczek NJ, Schimmenti LA, et al. 2018. Expert specification of the ACMG/AMP variant interpretation guidelines for genetic hearing loss. Hum Mutat 39: 1593–1613. 10.1002/humu.23630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. 2015. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17: 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Jacobsen JOB, Jäger M, Köhler S, Holtgrewe M, Schubach M, Siragusa E, Zemojtel T, Buske OJ, Washington NL, et al. 2015. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc 10: 2004–2015. 10.1038/nprot.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutterd C, Diakumis P, Bahlo M, Fanjul Fernandez MF, Leventer RJ, Delatycki M, Amor D, Chow CW, Stephenson S, Meisler MH, et al. 2017. Neuropathology of childhood-onset basal ganglia degeneration caused by mutation of VAC14. Ann Clin Transl Neurol 4: 859–864. 10.1002/acn3.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Robinson PN, Wang K. 2015. Phenolyzer: phenotype-based prioritization of candidate genes for human diseases. Nat Methods 12: 841–843. 10.1038/nmeth.3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zolov SN, Chow CY, Slutsky SG, Richardson SC, Piper RC, Yang B, Nau JJ, Westrick RJ, Morrison SJ, et al. 2007. Loss of Vac14, a regulator of the signaling lipid phosphatidylinositol 3,5-bisphosphate, results in neurodegeneration in mice. Proc Natl Acad Sci 104: 17518–17523. 10.1073/pnas.0702275104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.