Abstract
Genetic rearrangements involving the anaplastic lymphoma kinase (ALK) gene confer sensitivity to ALK tyrosine kinase inhibitors (TKIs) and superior outcome in non-small-cell lung cancer (NSCLC). However, clinical courses vary widely, and recent studies suggest that molecular profiling of ALK+ NSCLC can provide additional predictors of therapy response that could assist further individualization of patient management. As repeated tissue biopsies often pose technical difficulties and significant procedural risk, analysis of tumor constituents circulating in the blood, including ctDNA and various proteins, is increasingly recognized as an alternative method of tumor sampling (“liquid biopsy”). Here, we report the case of a KLC1–ALK-rearranged NSCLC patient responding to crizotinib treatment and demonstrate how analysis of plasma and serum biomarkers can be used to identify the ALK fusion partner and monitor therapy over time. Results of ctDNA sequencing and copy-number alteration profiling as well as serum protein concentrations at various time points during therapy reflected the current remission status and could predict the subsequent clinical course. At the time of disease progression, we identified four distinct secondary mutations in the ALK gene in ctDNA potentially causing treatment failure, accompanied by rising levels of CEA and CYFRA 21–1. Moreover, several copy-number variations were detected at the end of the treatment, including an amplification of a region on Chromosome 12 encompassing the TP53 regulator MDM2. In summary, our findings illustrate the utility of noninvasive longitudinal molecular profiling for assessing remission status, exploring mechanisms of treatment failure, predicting subsequent clinical course, and dissecting dynamics of drug-resistant clones in ALK+ lung cancer.
Keywords: lung adenocarcinoma
CASE PRESENTATION
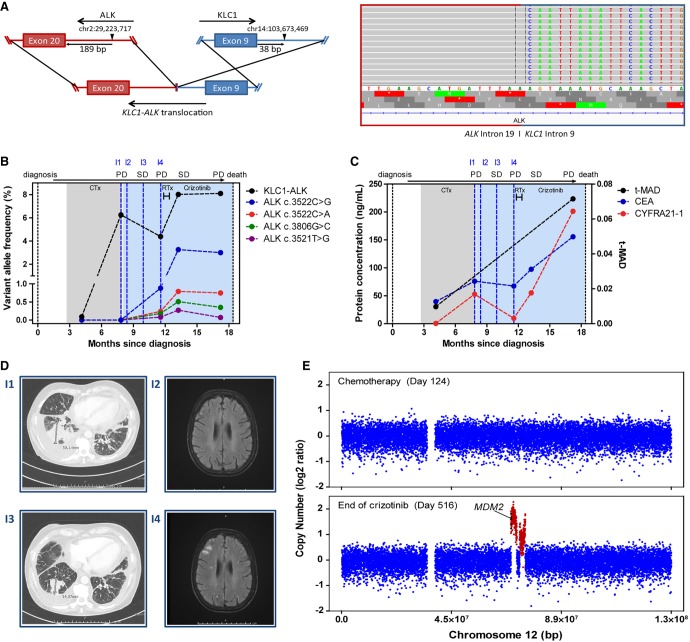
A 66-yr-old woman without significant comorbidities was diagnosed with metastatic adenocarcinoma of the right lung including ipsilateral pleural and diffuse bone spread (T2N0M1c). Initial treatment with carboplatin/pemetrexed chemotherapy resulted in a partial remission after two cycles and was followed by pemetrexed maintenance. Meanwhile, an ALK rearrangement was detected by fluorescence in situ hybridization (FISH). Subsequent RNA next-generation sequencing (NGS) of the tumor identified transcripts of KLC1 exon 9 fused with ALK exon 20 (K9–A20). Retrospective targeted deep sequencing of circulating tumor DNA (ctDNA) from several longitudinal blood plasma samples collected at different time points confirmed the translocation and identified the DNA breakpoints in KLC1 intron 9 and ALK intron 19 (Fig. 1A). The first plasma sample, collected at the time of partial remission after two cycles of carboplatin/pemetrexed chemotherapy, showed a variant allele frequency (VAF) of 0.09% for the ALK fusion and no single-nucleotide variants (SNVs) in the ALK gene (Fig. 1B). Analysis of a simultaneously collected serum sample revealed a low abundance of CEA (39.52 ng/µL) and CYFRA 21–1 (0.53 ng/µL; Fig. 1C). Two months after initiation of pemetrexed maintenance, the progression of the primary tumor and pleural effusion was noted by chest CT (Fig. 1D, I1), and new cerebral metastases were detected by brain MRI (Fig. 1D, I2). At this timepoint, the retrospectively determined KLC1–ALK fusion VAF rose to 6.25% and the tumor protein markers CEA, CYFRA 21–1 were also increased. Second-line treatment with crizotinib (250 bid) was initiated, and the patient initially responded with tumor shrinkage of ∼40%, as determined by radiological imaging (Fig. 1D, I3). During the next disease progression with multiple new brain lesions 50 d later (Fig. 1D, I4), the fusion VAF in retrospective analysis was 4.4% (i.e., lower than at the time of the previous systemic progression). However, four distinct secondary mutations in the kinase domain of the ALK gene were identified in ctDNA: ALK p.F1174C (c.35221T>G) with 0.08% VAF, ALK p.F1174L (c.3522C>G with 0.88% VAF and c.3522C>A with 0.24% VAF), and ALK p.G1269A (c.3806G>C) with 0.18% VAF (Fig. 1B). After whole-brain radiotherapy with 30 Gy (10 fractions of 3 Gy), clinical and radiological follow-up during the following three months indicated stable disease, wherefore crizotinib treatment was continued. However, a plasma sample taken only 3 wk after the last radiation dose retrospectively revealed rising VAFs of the fusion KLC1–ALK (8.01%) and all four secondary ALK variants, the most abundant of which was ALK p.F1174 (c.3522C>G with 3.26% VAF; Fig. 1B). At the same time, CEA and CYFRA 21–1 concentrations were also elevated (Fig. 1C). In accordance with the rising VAFs and protein levels, and despite the apparently stable radiologic findings, multilocular disease progression occurred just 1 mo later, with multiple new lung and bone metastases as confirmed by PET-CT. Retrospective molecular reassessment with liquid biopsy at that time point showed steadily high VAFs, whereas concentrations of protein markers rose further (CEA: >155 ng/µL, CYFRA 21–2: >200 ng/µL; Fig. 1B,C). Based on the radiologic progression, crizotinib treatment was discontinued and a switch to ceritinib was planned, but the patient died before the therapy could be started. We also performed retrospective shallow whole-genome sequencing (sWGS) of ctDNA from a plasma sample after two cycles of carboplatin/pemetrexed and from the last sample upon progression under crizotinib. Although sWGS of ctDNA during the first months after diagnosis revealed no copy-number variations (CNVs) with a trimmed median absolute deviation from copy-number neutrality (t-MAD) of 0.0098, noticeable alterations were detected at the end of the treatment (t-MAD = 0.0414), including an amplification of the region encompassing the TP53 regulator MDM2 on Chromosome 12 (Fig. 1C,E).
Figure 1.
(A) Schematic of the predicted genomic translocation of KLC1–ALK and aligned ctDNA reads in IGV spanning the rearrangement of KLC1 intron 8 and ALK intron 19. (B) Kinetics of mutated allele frequencies detected in ctDNA during treatment. Corresponding images (I1–I4) are shown in E. Phases of stable disease and progressive disease are indicated on top. (C) Kinetics of serum protein concentrations during treatment. Corresponding images (I1–I4) are shown in D. Phases of stable disease and progressive disease are indicated on top. (D) Corresponding chest CT of the primary tumor (on the left; I1, I3) and brain MRI images of cerebral metastases (on the right; I2, I4) taken during the therapy course. (E) Copy-number profiles of Chromosome 12 from sWGS of ctDNA. The upper panel shows the CNV profile before crizotinib treatment initiation; the bottom panel shows the CNV profile at the end of treatment. (SD) Stable disease, (PD) progressive disease, (CTx) chemotherapy, (RTx) whole-brain radiotherapy.
TECHNICAL ANALYSIS AND METHODS
The histological diagnosis of lung adenocarcinoma was performed by experienced pulmonary pathologists on a formalin-fixed and paraffin-embedded (FFPE) biopsy of the primary tumor. The presence of the ALK rearrangement was determined by fluorescent in situ hybridization (FISH) using ALK break-apart probes (ZytoLight SPEC ALK probe, ZytoVision). A fusion transcript with exon 9 of KLC1 fused to ALK exon 20 (K9–A20) was confirmed by RNA NGS using the AmpliSeq RNA Lung Cancer Fusion Panel (Thermo Fisher Scientific) as described previously (Pfarr et al. 2016; Volckmar et al. 2019).
For ctDNA analysis, plasma was isolated from blood samples, centrifuged within 30 min of collection, and processed with the AVENIO ctDNA Analysis Kit according to the manufacturer's instructions (Roche Diagnostics). Briefly, cell-free DNA was isolated from 2 mL of plasma using the AVENIO cfDNA Isolation Kit (Roche) and quantified with the Qubit dsDNA High Sensitivity Kit (Thermo Fisher). Targeted sequencing libraries were prepared from 39.9 ng DNA in median (range 24.5–50 ng) using the AVENIO ctDNA Library Preparation Kit with the AVENIO Targeted and Surveillance Panel (both from Roche) for hybridization-based enrichment of a 17-gene (81-kb) and a 197-gene (198-kb) panel, respectively. Among their target regions, both panels are designed to capture the exons 19 to 28 as well as intron 19 of the ALK gene. CtDNA libraries from all plasma samples were initially enriched and sequenced with the AVENIO Targeted panel. To validate variants detected with the Targeted panel and to potentially identify further mutations in an extended genomic target region, all plasma samples were repeatedly enriched and sequenced with the AVENIO Surveillance panel. All protocols were conducted according to the manufacturer's recommendations. Enriched libraries were sequenced on an Illumina NextSeq550 using the High Output Kit V2 (300 cycles) according to the manufacturer's protocol (Illumina) with a median unique target sequence coverage of 7760× (range 4177×–10,869×). Automated raw data processing and data analysis was performed with the AVENIO ctDNA analysis software (Roche). Manual QC of called variants and visualization of aligned reads spanning the DNA breakpoints in KLC1 intron 9 and ALK intron 19 was done using the Integrative Genomics Viewer (IGV) (Robinson et al. 2017).
For low-coverage sWGS, sequencing libraries were prepared from 2.5 ng cell-free DNA using the KAPA Hyper Prep Kit with KAPA Dual-Indexed Adapters for Illumina platforms (both Roche). Following sequencing adapter ligation for 15 h at 16°C to achieve high ligation efficiency, libraries were amplified in 11 PCR cycles and purified according to the manufacturer's protocol. Samples were sequenced in a multiplex of 48 sWGS libraries per lane on an Illumina HiSeq4000 with 100-bp paired-end reads (Illumina). Raw sequencing reads were processed and aligned using the automated pipeline OTP (Reisinger et al. 2017). Genome-wide copy number profiles were estimated from low-coverage sWGS data of ctDNA using ichorCNA (Adalsteinsson et al. 2017), and t-MAD scores were calculated to quantify CNVs (Mouliere et al. 2018).
For protein analysis, serum was isolated from blood samples and centrifuged 30 min after collection to ensure complete coagulation. Serum CEA and CYFRA 21–1 levels were determined from 10 µL of serum by multiplexed flow cytometry using the MILLIPLEX MAP Human Circulating Cancer Biomarker Magnetic Bead Panel (Merck) with the Luminex Bio-Plex 200 immunoassay platform (Bio-Rad). All protocols were conducted according to the manufacturer's recommendations.
Plasma and serum biosamples have been provided by Lungbiobank Heidelberg/BMBH in accordance with the regulations of the BMBH and the approval of the ethics committee of the University of Heidelberg. Analysis of clinical data and tissue and blood samples of the patient in this study was performed after informed consent and approval by the ethics committee of Heidelberg University (S-270/2001 and S-296/2016).
VARIANT INTERPRETATION
The median survival of ALK-rearranged non-small-cell lung cancer (ALK+ NSCLC) patients treated with tyrosine kinase inhibitors (TKIs) currently exceeds 5 yr, but clinical courses vary widely (Duruisseaux et al. 2017). Recent studies have demonstrated the association of this clinical heterogeneity with specific molecular tumor characteristics, such as the exact type of ALK fusion variant, including non-EML4 fusions, as well as the presence of TP53 and other mutations (Christopoulos et al. 2018a,b; Kang et al. 2018; Kron et al. 2018; Christopoulos et al. 2019a,c). Thus, broader molecular profiling of ALK+ NSCLC, using, for example, RNA-/DNA-NGS appears to provide information about several clinically relevant parameters that could be used to optimize therapies and personalized patient treatment beyond the readout obtainable from immunohistochemistry and FISH (Volckmar et al. 2019).
In the lung adenocarcinoma case presented here, a translocation of the ALK gene locus was initially detected by FISH analysis. Subsequent RNA sequencing identified fusion transcripts with exon 9 of KLC1 fused to ALK exon 20 (K9–A20), which has been detected only at the RNA level in the past, without any information about TKI sensitivity (Togashi et al. 2012). To the best of our knowledge, our study is the first report of the exact intronic double-strand breakpoints and the resulting interchromosomal rearrangement for a KLC1–ALK fusion, which we could identify using hybridization-based deep ctDNA sequencing (Table 1; Fig. 1B). Our results show that the KLC1–ALK fusion of our patient encompasses the DNA sequence encoding the 5′-end part of KLC1, including the promoter domain, together with the sequence coding for the intracellular part of the ALK protein, including the entire tyrosine kinase domain, suggesting sensitivity to treatment with ALK inhibitors. Indeed, the patient showed a good response to crizotinib with partial tumor remission and is—to the best of our knowledge—the first KLC1–ALK NSCLC case successfully treated with ALK TKI in the literature.
Table 1.
Genomic findings
| Gene | Chr | HGVS DNA ref | HGVS Protein ref | Variant type | Predicted effect | Allele frequency | Target coverage | Mutation read count |
|---|---|---|---|---|---|---|---|---|
| KLC1–ALK | 2 | t(2;14)(p23.1;32.33) (hg19 Chr 2:29,223,717:Chr 14:103,673,469) | n/a | KLC1–ALK fusion | Oncogenic, sensitivity to ALK inhibitor | 8.09% | 5860× | 474 |
| ALK | 2 | c.3521T>G | p.F1174C | Substitution | Crizotinib resistance | 0.27% | 10,316× | 28 |
| ALK | 2 | c.3522C>A | p.F1174L | Substitution | Crizotinib resistance | 0.79% | 10,192× | 81 |
| ALK | 2 | c.3522C>G | p.P1174L | Substitution | Crizotinib resistance | 3.26% | 10,192× | 332 |
| ALK | 2 | c.3806G>C | p.G1269A | Substitution | Crizotinib resistance | 0.51% | 16,190× | 83 |
At the time of failure for any treatment, be it chemotherapy, crizotinib, or after cerebral radiotherapy (Fig. 1D, I1), disease progression was detectable in liquid biopsies as increasing VAF of the KLC1–ALK fusion and rising protein tumor markers. Moreover, at the time of cerebral disease progression with multiple new brain lesions under crizotinib (Fig. 1E,I3), ctDNA assessment identified four distinct mutations in the ALK kinase domain, which potentially cause the progression (Table 1; Fig. 1B). Three out of the four (ALK c.3522C>G, ALK c.3522C>G/A, and c.3521T>G) encode the p.F1174C/L variants, which are associated with resistance to ceritinib and crizotinib (Gainor et al. 2016; Lin et al. 2017). The fourth mutation (ALK c.3806G>C) encodes the p.G1269A variant, which is associated with resistance to crizotinib (Gainor et al. 2016). Interestingly, the four secondary mutations show similar courses over time, suggesting a parallel evolution of multiple resistant clones with distinct molecular resistance mechanisms. Although Gainor et al. detected all these resistance mutations in tissue specimens obtained from progressing EML4–ALK-rearranged tumors, our report is the first to describe these mutations occurring simultaneously in ctDNA of a KLC1–ALK patient progressing under crizotinib. In line with this finding, recent proof-of-concept studies also demonstrated the feasibility of identifying and tracking resistance mutations to ALK inhibitors with serial liquid biopsies (Dagogo-Jack et al. 2018; De Carlo et al. 2018; McCoach et al. 2018), illustrating the potential utility of plasma ctDNA genotyping in the clinical patient management in ALK+ NSCLC (Rolfo et al. 2018). Besides ALK+ NSCLC, several studies demonstrated the potential of ctDNA typing for cancer detection, molecular profiling, therapy monitoring, and identification of treatment resistance in various tumor entities (Murtaza et al. 2013; Abbosh et al. 2017; Wan et al. 2017; Cohen et al. 2018). Finally, although imaging suggested stable disease after radiotherapy with remission of the cerebral lesions, rising ctDNA levels of the KLC1–ALK fusion and of all four secondary ALK point mutations suggested poor disease control. Importantly, this was accompanied by rising concentrations of both protein tumor markers CEA and CYFRA 21–1 in synchronous serum samples (Fig. 1C), and confirmed by the subsequent clinical course, which showed multifocal radiologic progression a few weeks later in conjunction with further increases in all ctDNA and protein markers.
Panel sequencing utilizes enrichment of specific loci and enables deep sequencing to detect recurrent hotspot or resistance mutations; however, it is limited to mutations within the target regions. In contrast, whole-genome or -exome sequencing of plasma DNA facilitates comprehensive molecular profiling, including de novo mutations and CNVs, but is only feasible for increased ctDNA fractions as the analytical sensitivity is limited by the relatively low coverage. Increasing the tumor-derived fraction of cell-free DNA could help to overcome this limitation and to improve the detection of somatic aberrations at lower sequencing depths (Mouliere et al. 2018). Here, we demonstrate a complementary use of targeted deep sequencing and low-coverage sWGS. In addition to mutation typing, we performed sWGS of cell-free DNA to assess CNV profiles at the genome-scale. Prior to crizotinib initiation, no CNVs were detected in the plasma sample of our patient. In contrast, remarkable alterations with fourfold t-MAD increase were detected at the end of treatment, including an amplification of a region on Chromosome 12 encompassing the TP53 regulator gene MDM2 (Fig. 1C,E). MDM2 amplifications have been directly associated with worse prognosis and TKI resistance in RET-rearranged lung cancers (Dworakowska et al. 2004; Somwar et al. 2016), whereas impairment of TP53 function, as would be expected with MDM2 overexpression, is an important determinant of poor survival in ALK+ NSCLC, either at baseline or if acquired during the course of the disease (Kron et al. 2018; Christopoulos et al. 2019b,c). Thus, although the presence of the amplification in the baseline cannot be excluded because of the overall low ctDNA fraction in this sample, the findings suggest that CNVs, including the MDM2 amplification, might contribute to the progression of ALK+ NSCLC patients beyond secondary mutations in the ALK kinase domain.
SUMMARY
We report the first case of KLC1–ALK-rearranged lung adenocarcinoma responding to treatment with crizotinib and demonstrate the utility of multiple plasma and serum biomarkers for identification of ALK fusion partners and longitudinal disease monitoring in ALK+ NSCLC. At the time of treatment, failure under chemotherapy, crizotinib and cerebral radiotherapy, rising allelic frequencies of the ALK fusion, distinct ALK resistance mutations (in case of TKI), emerging CNV, and increasing concentrations of protein markers reflected the poor disease control and predicted the subsequent clinical deterioration, in part earlier and more reliably than radiologic findings. Our findings illustrate the potential clinical utility of noninvasive longitudinal molecular profiling for assessing remission status, exploring mechanisms of treatment failure, predicting subsequent clinical course, and dissecting dynamics of drug-resistant clones in ALK+ lung cancer.
ADDITIONAL INFORMATION
Data Deposition and Access
The variants of this case have been submitted to COSMIC (https://cancer.sanger.ac.uk/cosmic) and are available under the accession number COSP46927.
Ethics Statement
Analysis of clinical data and tissue and blood samples of the patient in this study was performed after informed consent and approval by the ethics committee of Heidelberg University (S-270/2001 and S-296/2016).
Acknowledgments
The authors thank Ingrid Heinzmann-Groth and Saskia Östringer of Translational Research Unit (STF) of Thoraxklinik Heidelberg for assistance with the collection of patient samples, the Berlin Institute of Health Core Unit Genomics at the Charité and the Genomics and Proteomics Core Facility at the German Cancer Research Center (DKFZ) for sequencing analyses, and the Omics IT and Data Management Core Facility at the German Cancer Research Center (DKFZ) for data management and processing. The authors also thank Gregor Obernosterer (Roche Diagnostics) for logistic support.
Author Contributions
S.D., P.C., L.G., M.T., A.S., and H.S. designed the study. P.C., M.R., and M.T. were responsible for patient treatment. S.D., P.C., L.G., A.-L.V., C.-P.H., S.J.O., T.Z., M.A.S., M.M., T.M., M.R., M.T., A.S., and H.S. processed the samples. S.D., P.C., L.G., A.-L.V., V.E., Z.Y., T.Z., M.S., M.T., A.S., and H.S. analyzed the data. All authors wrote and revised the manuscript.
Funding
This work was supported by the German Center for Lung Research (DZL), by the German Cancer Consortium (DKTK), by the Heidelberg Center for Personalized Oncology at the German Cancer Research Center (DKFZ-HIPO), and by Roche Molecular Solutions.
Competing Interest Statement
S.D. reports speaker's honoraria from Roche; P.C. reports research funding from Novartis, Roche, AstraZeneca, and Takeda, as well as advisory board and/or lecture fees from Boehringer, Pfizer, and Chugai; A.-L.V. reports speaker's honoraria from Astra Zeneca; V.E. reports advisory board and lecture fees from AstraZeneca and Thermo Fisher; M.T. reports advisory board honoraria from Novartis, Lilly, BMS, MSD, Roche, Celgene, Takeda, AbbVie, and Boehringer, speaker's honoraria from Lilly, MSD, and Takeda, research funding from AstraZeneca, BMS, Celgene, Novartis, and Roche, and travel grants from BMS, MSD, Novartis, and Boehringer; A.S. reports advisory board honoraria from BMS, Bayer, AstraZeneca, Thermo Fisher, Novartis, and Seattle Genomics, speaker's honoraria from BMS, Bayer, Illumina, AstraZeneca, Novartis, Thermo Fisher, MSD, and Roche, as well as research funding from Chugai, Bayer, and BMS; and H.S. reports advisory board and speaker's honoraria from Roche.
Referees
Alexander Wyatt
Anonymous
REFERENCES
- Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, et al. 2017. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545: 446–451. 10.1038/nature22364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adalsteinsson VA, Ha G, Freeman SS, Choudhury AD, Stover DG, Parsons HA, Gydush G, Reed SC, Rotem D, Rhoades J, et al. 2017. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun 8: 1324 10.1038/s41467-017-00965-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos P, Endris V, Bozorgmehr F, Elsayed M, Kirchner M, Ristau J, Buchhalter I, Penzel R, Herth FJ, Heussel CP, et al. 2018a. EML4-ALK fusion variant V3 is a high-risk feature conferring accelerated metastatic spread, early treatment failure and worse overall survival in ALK+ non–small cell lung cancer. Int J Cancer 142: 2589–2598. 10.1002/ijc.31275 [DOI] [PubMed] [Google Scholar]
- Christopoulos P, Kirchner M, Endris V, Stenzinger A, Thomas M. 2018b. EML4-ALK V3, treatment resistance, and survival: refining the diagnosis of ALK+ NSCLC. J Thoracic Dis 10: S1989–S1991. 10.21037/jtd.2018.05.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos P, Budczies J, Kirchner M, Dietz S, Sültmann H, Thomas M, Stenzinger A. 2019a. Defining molecular risk in ALK+ NSCLC. Oncotarget 10: 3093–3103. 10.18632/oncotarget.26886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos P, Dietz S, Kirchner M, Volckmar AL, Endris V, Neumann O, Ogrodnik S, Heussel CP, Herth FJ, Eichhorn M, et al. 2019b. Detection of TP53 mutations in tissue or liquid rebiopsies at progression identifies ALK+ lung cancer patients with poor survival. Cancers (Basel) 11: E124 10.3390/cancers11010124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos P, Kirchner M, Bozorgmehr F, Endris V, Elsayed M, Budczies J, Ristau J, Penzel R, Herth FJ, Heussel CP, et al. 2019c. Identification of a highly lethal V3+TP53+ subset in ALK+ lung adenocarcinoma. Int J Cancer 144: 190–199. 10.1002/ijc.31893 [DOI] [PubMed] [Google Scholar]
- Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, Javed AA, Wong F, Mattox A, et al. 2018. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science (80- ) 359: 926–930. 10.1126/science.aar3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagogo-Jack I, Brannon AR, Ferris LA, Campbell CD, Lin JJ, Schultz KR, Ackil J, Stevens S, Dardaei L, Yoda S, et al. 2018. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis Oncol 2018 10.1200/PO.17.00160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlo E, Schiappacassi M, Urbani M, Doliana R, Baldassarre G, Da Ros V, Santarossa S, Chimienti E, Berto E, Fratino L, et al. 2018. Therapeutic decision based on molecular detection of resistance mechanism in an ALK-rearranged lung cancer patient: a case report. Onco Targets Ther 11: 8945–8950. 10.2147/OTT.S184745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duruisseaux M, Besse B, Cadranel J, Pérol M, Mennecier B, Bigay-Game L, Descourt R, Dansin E, Audigier-Valette C, Moreau L, et al. 2017. Overall survival with crizotinib and next-generation ALK inhibitors in ALK-positive non-small-cell lung cancer (IFCT-1302 CLINALK): a French nationwide cohort retrospective study. Oncotarget 8: 21903–21917. 10.18632/oncotarget.15746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworakowska D, Jassem E, Jassem J, Peters B, Dziadziuszko R, Żylicz M, Jakóbkiewicz-Banecka J, Kobierska-Gulida G, Szymanowska A, Skokowski J, et al. 2004. MDM2 gene amplification: a new independent factor of adverse prognosis in non–small cell lung cancer (NSCLC). Lung Cancer 43: 285–295. 10.1016/j.lungcan.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, Dagogo-Jack I, Gadgeel S, Schultz K, Singh M, et al. 2016. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov 6: 1118–1133. 10.1158/2159-8290.CD-16-0596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Zhang XC, Chen HJ, Zhou Q, Tu HY, Li WF. 2018. Uncommon ALK fusion partners in advanced ALK-positive non-small-cell lung cancer. J Clin Oncol 36: 8561 10.1200/JCO.2018.36.15_suppl.8561 [DOI] [Google Scholar]
- Kron A, Alidousty C, Scheffler M, Merkelbach-Bruse S, Seidel D, Riedel R, Ihle MA, Michels S, Nogova L, Fassunke J, et al. 2018. Impact of TP53 mutation status on systemic treatment outcome in ALK-rearranged non-small-cell lung cancer. Ann Oncol 29: 2068–2075. 10.1093/annonc/mdy333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Riely GJ, Shaw AT. 2017. Targeting ALK: precision medicine takes on drug resistance. Cancer Discov 7: 137–155. 10.1158/2159-8290.CD-16-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoach CE, Blakely CM, Banks KC, Levy B, Chue BM, Raymond VM, Le AT, Lee CE, Diaz J, Waqar SN, et al. 2018. Clinical utility of cell-free DNA for the detection of ALK fusions and genomic mechanisms of ALK inhibitor resistance in non-small cell lung cancer. Clin Cancer Res 24: 2758–2770. 10.1158/1078-0432.CCR-17-2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, Mair R, Goranova T, Marass F, Heider K, et al. 2018. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 10: eaat4921 10.1126/scitranslmed.aat4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaza M, Dawson SJ, Tsui DWY, Gale D, Forshew T, Piskorz AM, Parkinson C, Chin SF, Kingsbury Z, Wong ASC, et al. 2013. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497: 108–112. 10.1038/nature12065 [DOI] [PubMed] [Google Scholar]
- Pfarr N, Stenzinger A, Penzel R, Warth A, Dienemann H, Schirmacher P, Weichert W, Endris V. 2016. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosomes Cancer 55: 30–44. 10.1002/gcc.22297 [DOI] [PubMed] [Google Scholar]
- Reisinger E, Genthner L, Kerssemakers J, Kensche P, Borufka S, Jugold A, Kling A, Prinz M, Scholz I, Zipprich G, et al. 2017. OTP: an automatized system for managing and processing NGS data. J Biotechnol 261: 53–62. 10.1016/j.jbiotec.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. 2017. Variant review with the integrative genomics viewer. Cancer Res 77: E31–E34. 10.1158/0008-5472.CAN-17-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, Herbst RS, Mok TS, Peled N, Pirker R, et al. 2018. Liquid biopsy for advanced non–small cell lung cancer (NSCLC): a statement paper from the IASLC. J Thorac Oncol 13: 1248–1268. 10.1016/j.jtho.2018.05.030 [DOI] [PubMed] [Google Scholar]
- Somwar R, Smith R, Hayashi T, Ishizawa K, Charen AS, Khodos I, Mattar M, He J, Balasubramanian S, Stephens P, et al. 2016. MDM2 amplification (Amp) to mediate cabozantinib resistance in patients (Pts) with advanced RET-rearranged lung cancers. J Clin Oncol 34: 9068 10.1200/JCO.2016.34.15_suppl.9068 [DOI] [Google Scholar]
- Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, Nakajima T, Mano H, Takeuchi K. 2012. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One 7: e31323 10.1371/journal.pone.0031323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckmar AL, Leichsenring J, Kirchner M, Christopoulos P, Neumann O, Budczies J, de Oliveira CMM, Rempel E, Buchhalter I, Brandt R, et al. 2019. Combined targeted DNA and RNA sequencing of advanced NSCLC in routine molecular diagnostics: analysis of the first 3,000 Heidelberg cases. Int J Cancer 145: 649–661. 10.1002/ijc.32133 [DOI] [PubMed] [Google Scholar]
- Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N. 2017. Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17: 223–238. 10.1038/nrc.2017.7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The variants of this case have been submitted to COSMIC (https://cancer.sanger.ac.uk/cosmic) and are available under the accession number COSP46927.