Abstract
Clonal heterogeneity and evolution of mantle cell lymphoma (MCL) remain unclear despite the progress in our understanding of its biology. Here, we report a 71-yr-old male patient with an aggressive MCL and depict the clonal evolution from initial diagnosis of typical MCL to relapsed blastoid MCL. During the course of the disease, the patient was diagnosed with classic Hodgkin lymphoma (CHL) and received a CHL therapeutic regimen. Molecular analysis by next-generation sequencing of both MCL and CHL demonstrated clonally related CHL with characteristic immunophenotype and PDL1/2 gains. Moreover, our data illustrate the clonal heterogeneity and acquisition of additional genetic aberrations including a rare fusion of SEC22B-NOTCH2 in the process of clonal evolution. Evidence obtained from our comprehensive immunophenotypic and genetic studies indicates that MCL and CHL can originate from a common precursor by divergent clonal evolution, which may pose a therapeutic challenge.
Keywords: B-cell lymphoma, Hodgkin lymphoma
INTRODUCTION
Clonal evolution of mantle cell lymphoma (MCL) is still poorly understood. Intratumoral heterogeneity, subclonal architecture, and clonal dynamic changes after chemotherapy have been demonstrated (Bea et al. 2013). The presence of Hodgkin/Reed–Sternberg (HRS)-like cells has been reported in rare patients suggesting clonal evolution HRS-like cells from MCL (Schneider et al. 2014; Murray et al. 2017; Kramer et al. 2019). Interestingly, all these cases had scattered HRS-like cells in a background of typical MCL (Caleo et al. 2003; Tinguely et al. 2003; Hayes et al. 2006; Schneider et al. 2014; Giua et al. 2015; Murray et al. 2017; Kramer et al. 2019). The mixed inflammatory microenvironment seen in classic Hodgkin lymphoma (CHL) was seldom observed, mimicking the so-called type I morphology with HRS-like cells in chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) (de Leval et al. 2004; Mao et al. 2007; Xiao et al. 2016). Current recommendations are against diagnosing or managing these cases as bona fide CHL (Xiao et al. 2016; Swerdlow et al. 2017). Therefore, it is still unclear if MCL can clonally evolve to CHL. Here, we report an exceedingly rare event in a patient with MCL, who developed a clonally related CHL after eradication of MCL and then relapsed again as blastoid MCL with additional genetic aberrations. Comprehensive genetic evidence indicates a divergent clonal evolution from a common precursor to MCL and CHL.
CASE PRESENTATION
A 70-yr-old male with a stage IV MCL (Fig. 1A–F) received four cycles of rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) and lenalidomide, followed by two cycles of rituximab and high dose cytarabine (R-HiDAC), and 6 mo of lenalidomide maintenance. A year after the initial diagnosis, restaging PET showed new FDG avid hilar, abdominal, left inguinal nodes, and a new FDG avid focus in the right rib. Biopsy of the left inguinal lymph node and bilateral thoracic hilar nodes showed typical CHL and no evidence of MCL by both immunohistochemical and flow cytometric studies (Fig. 1G–M; Supplemental Fig. S1). He was diagnosed with stage IV CHL, nodular sclerosing subtype, and treated with brentuximab vedotin, doxorubicin, vinblastine, and dacarbazine (BV-AVD). Treatment was poorly tolerated and was complicated by anemia and thrombocytopenia requiring transfusions; therefore, only four cycles were administered. PET imaging at the end of the fourth cycle showed the persistence of disease in the left acetabulum, which was biopsied and showed blastoid MCL without pathologic evidence of CHL (Fig. 1N–S). The patient received radiation therapy to the localized disease. However, the patient relapsed postradiation in multiple bony sites. Currently, the patient is being treated with rituximab and ibrutinib and awaiting assessment of response.
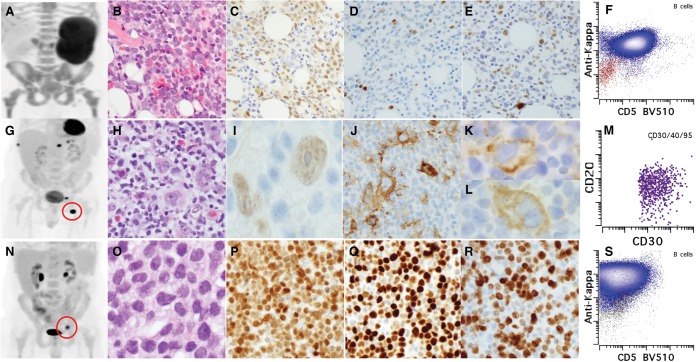
Figure 1.
Pathologic features of MCL and CHL. (A) PET imaging showed diffuse involvement in lymph nodes, spleen, and bone marrow. (B) H&E morphology of MCL in bone marrow (×200). (C–E) Immunohistochemical stains of MCL including Cyclin D1 (C), Ki-67 (D), and TP53 (E). (F) Immunophenotype of MCL by flow cytometry (gated CD19+ B cells are shown). (Blue) κ restriction B cells, (red) λ restriction B cells. (G) PET imaging showed localized uptake in left inguinal node and ribs (red circle represents biopsied left inguinal node). (H) H&E morphology of CHL in the node (note the mixed inflammatory background including eosinophils and plasma cells; also see Supplemental Fig. S1A). (I–L) Immunohistochemical stains of CHL including Cyclin D1 (I, weak expression in HRS cells), CD30 (J), PDL1 (K), and PDL2 (L). (M) Immunophenotype of HRS cells by flow cytometry (FSChiSSChiCD30+CD40+CD95+ cells are gated). (N) PET imaging showed relapsed blastoid MCL (red circle represents the biopsied acetabulum lesion). (O) H&E morphology of blastoid MCL (×200). (P–R) Immunohistochemical stains of blastoid MCL including Cyclin D1 (P), Ki-67 (Q), and TP53 (R). (S) Immunophenotype of blastoid MCL by flow cytometry (CD19+ B cells are gated). (Blue) κ restricted population, (red) λ restricted population.
RESULTS
The findings of immunohistochemical studies on the three lymphomas are summarized in Supplemental Table S2. Cyclin D1 was expressed in MCL, CHL (HRS cells), and blastoid MCL (Fig. 1C,I,P). Ki-67 proliferation index and TP53 expression were low in MCL (Fig. 1D,E), whereas both were high in the blastoid MCL (Fig. 1O,R). HRS cells in CHL showed characteristic phenotype with the absence of CD20 (Supplemental Fig. S1), strong expression of MUM-1 (Supplemental Fig. S1) and strong CD30 (Fig. 1J), and up-regulation of PDL1 and PDL2 (Fig. 1K,L).
Chromosome analysis of the bone marrow aspirate involved by MCL revealed a complex karyotype with subclones in 12 of 20 metaphase cells. The chromosome abnormalities included t(11;14), the hallmark of MCL, and multiple numerical and structural chromosome abnormalities, including deletions of the short arms of Chromosomes 1, 8, 9, 12, and 17, and in the long arm of Chromosome 6, loss of Chromosomes Y, 9, and 13 and 21, three to four marker chromosomes, and a ring chromosome (not shown).
FISH analysis confirmed the IGH/CCND1 fusion with a typical signal pattern in the bone marrow aspirate specimen with MCL (Supplemental Fig. S2A). FISH analysis using the CCND1 break-apart probes confirmed the CCND1 rearrangements in the HRS cells of the groin lymph node biopsy with CHL and in the acetabulum bone biopsy tissue with blastoid MCL (Supplemental Fig. S2B,C), with a complex signal pattern in both the HRS cells and in blastoid MCL cells. Thus, all three specimens with MCL, CHL, and blastoid MCL carried the t(11;14), with additional abnormalities in CHL and blastoid MCL.
FISH analysis using PDL1/PDL2 probes, along with a centromere probe for Chromosome 9 as an internal control for copy-number assessment, showed a high level gain of PDL1 and PDL2 in 25%, and polysomy 9 in 60% of the HRS cells analyzed in groin tissue with CHL (Supplemental Fig. S2D). FISH analysis of the acetabulum tissue with blastoid MCL showed no evidence of PDL1/2 amplification or translocations. However, loss of PDL1/2 and one CEP9 copy was observed in 86% cells, consistent with loss of one Chromosome 9, which was also detected in the bone marrow specimen with MCL (Supplemental Fig. S2D,E).
A clonal IGHV gene rearrangement was identified in the original MCL (Fig. 2A) and was further characterized by NGS, which revealed a unique clonal sequence (IGHV3-23 IGHJ4) with a mutation rate of 3.5%, consistent with a mutated somatic hypermutation status. Comparative analysis of the CHL and blastoid MCL revealed an identical IGHV clonal sequence, also with a mutation rate of 3.5%, confirming clonal relatedness (Fig. 2B,C). NGS study using a targeted panel containing 400 genes identified somatic mutations in TP53 p.R248Q, NSD2 p.E1099K, and IDH1 p.E262K in all three lymphomas (Table 1; Supplemental Table S3). A subclonal KMT2D p.L3542Vfs*13 (c.10624_10625delCT) mutation was detected in the initial MCL and was subsequently found in the CHL at a comparable allele fraction to the shared TP53 variant, suggesting that this became the predominant clone; however, this KMT2D variant was not detected in the blastoid MCL. Instead, the blastoid MCL had additional SETBP1 p.S568I and a different KMT2D variant p.F3966_Q3971 delins* mutation that was not present in the initial MCL or CHL. In addition, SEC22B-NOTCH2 gene fusion was detected in blastoid MCL but not reported in MCL or CHL.
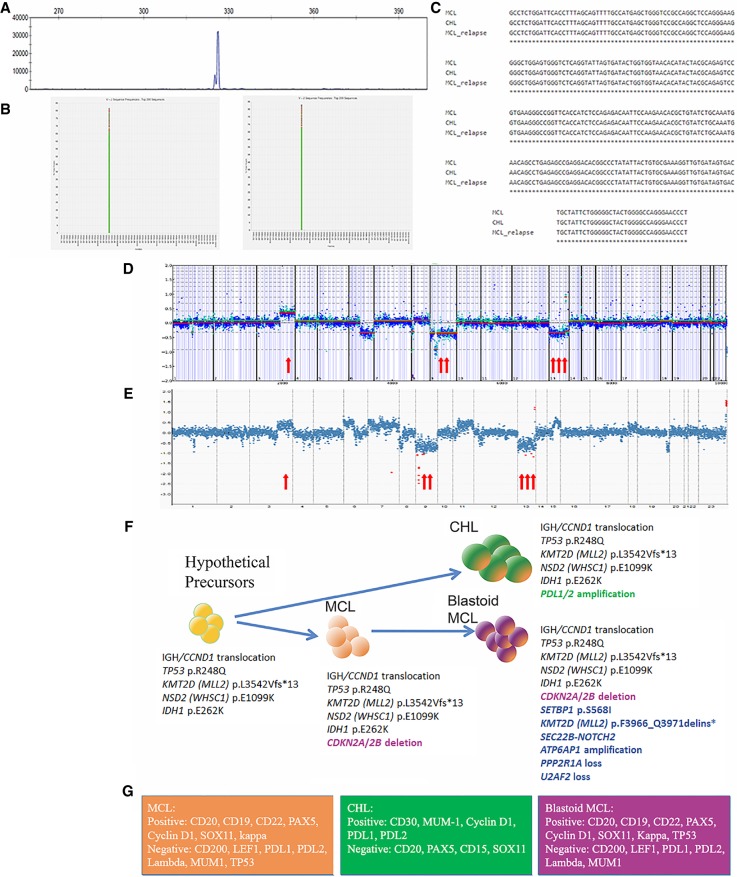
Figure 2.
Molecular profiles of MCL and CHL. (A) Fragment analysis by PCR studies showed a clonal peak in IGH rearrangements (identical peaks are shared by all three lymphomas). Shown here is the initial MCL. (B) Next-generation sequencing studies (LymphoTrack assay) showed identical clones between MCL (left) and CHL (right). The same clone was also identified in blastoid MCL (not shown). (C) Alignment of identical IGHV sequences between all three lymphomas. (D,E) Copy-number plots for MCL (D) and blastoid MCL (E) showing shared broad copy-number gains on Chromosome arms 3q and broad copy-number losses on 8p, 9, and 13. In the original MCL, broad copy-number loss of 6q is noted; however, relapsed MCL showed gains on 6p, 7, 11q, and 15q25–26, which were not appreciated in the original MCL. (F) Graphic illustration of divergent clonal evolution of a common precursor to CHL and MCL. Genetic aberrations are shown accordingly. (G) Immunophenotype of the three lymphomas.
Table 1.
Somatic variants detected in the patient
| Gene | Chromosome | HGVS DNA reference | HGVS protein reference | Variant type | Predicted effect | dbSNP ID | Genotype |
|---|---|---|---|---|---|---|---|
| TP53 | 17p13.1 17:7577538 |
NM_000546.5: c.743G>A | p.Arg248Gln | SNV; missense | Substitution | rs11540652 | Heterozygous |
| IDH1 | 2q34 2:209106784 |
NM_005896: c.784G>A | p.Glu262Lys | NA | |||
| NSD2 (WHSC1) | 4p16.3 4:1962801 |
NM_001042424; c.3295G>A | p.Glu1099Lys | SNV; missense | Substitution | rs772470710 | Heterozygous |
| KMT2D (MLL2) | 12q13.12 12: 49427964 |
NM_003482; c.10624_10625delCT | p.Leu3542Valfs*13 | Indel; nonsense | Frameshift | NA | Heterozygous |
| KMT2D (MLL2) | 12q13.12 12:49426576 |
NM_00348; c.11897_11911delTTCAACAGCAGCAGC | p.Phe3966_Gln3971delins* | Indel; nonsense | Nonsense | NA | Heterozygous |
| SETBP1 | 18q12.3 18:42531008 |
NM_015559; c.1703G>T | p.Ser568Ile | SNV; missense | Substitution | NA | Heterozygous |
(SNV) Single-nucleotide variant, (indel) insertion/deletion.
Copy-number alterations (CNAs) detected by NGS testing were compatible with the karyotyping results of the initial MCL (see above), including loss of Chromosomes 9 and 13, Chromosome arms 6q and 8p, and deletion of 9p21, which includes the CDKN2A/B genes. NGS analysis also revealed gain of the long arm of Chromosome 3, which was most likely represented as the marker chromosome in karyotyping analysis. Comparison of the copy-number profiles of the original MCL and blastoid MCL also showed similar CNAs as described above (deletion of 8p, losses of Chromosomes 9 and 13) and additional genomic imbalances in the blastoid MCL, including gain of Chromosome 6, which override the net copy level of Chromosome 6, gain of Chromosome 7, 11q with CCND1 involvement (see FISH above) and 15q25–26 (Fig. 2D,E; Supplemental Table S3). As CNA analysis is highly dependent on tumor purity, it was not informative in the groin lymph node biopsy involved by CHL because of the rarity of HRS cells (<20% of total cells; data not shown).
Clonally related HRS-like cells in MCL have only been reported in three prior cases (Supplemental Table S4; Schneider et al. 2014; Murray et al. 2017; Kramer et al. 2019). In these studies, the clonal relationship was established by PCR studies of the IGHV regions after microdissection (Schneider et al. 2014; Murray et al. 2017) and confirmed by FISH studies showing IGH/CCND1 translocation in both MCL and HRS-like cells (Schneider et al. 2014; Murray et al. 2017; Kramer et al. 2019). Our case is exceptional as a bona fide CHL was diagnosed rather than MCL with HRS c-like cells, and a MCL component was not present in the biopsy with CHL either by flow cytometric study (with a sensitivity approaching 0.01%) or immunohistochemical studies (Supplemental Fig. S1; Supplemental Table S2). Our comprehensive analysis provided convincing evidence at multiple levels demonstrating clonal relatedness between MCL and CHL including shared IGH/CCND1 rearrangements, identical IGHV clonal sequence, and shared mutations of TP53 p.R248Q, NSD2 p.E1099K and IDH1 p.E262K. However, the additional differential genetic aberrations seen between MCL and CHL, such as loss of CDKN2A/B and Chromosome 9 in MCL, PDL1/2 gain, and Chromosome 9 polysomy in CHL, suggest a common precursor clone with a divergent clonal evolution to MCL and CHL. Notably, a subclonal KMT2D p.L3542Vfs*13 (c.10624_10625delCT) mutation was present in the initial MCL, and this mutation became the predominant clone in CHL but was not detected in blastoid MCL, suggesting the clone harboring KMT2D p.L3542Vfs*13 (c.10624_10625delCT) mutation might have evolved to CHL. The blastoid MCL likely originated from a different subclone, which acquired mutations in SETBP1, a KMT2D variant that was different from the initial variant (p.F3966_Q3971delins*), and a SECB22-NOTCH2 fusion. These findings implicate the presence of a common precursor harboring IGH/CCND1 rearrangements and mutations of TP53 p.R248Q, NSD2 p.E1099K, and IDH1 p.E262K. KMT2D p.L3542Vfs*13 (c.10624_10625delCT) mutation is likely to present in the common precursor, albeit at a subclonal level (Fig. 2F). The possibility of a linear evolution from the initial MCL to CHL is not favored (Fig. 2F).
Discussion
MCLs are considered to arise from a subset of CD5+ naive B cells, whereas a small proportion of MCLs appear to be of post–germinal center (post-GC) origin (Jares et al. 2012). In this case, NGS studies demonstrated an identical rate of IGHV hypermutation in all three lymphomas in our patient indicating a post-GC origin (Schneider et al. 2014). The characteristic genetic aberrations of IGH/CCND1 translocations occur at the pre-B stage (Jares et al. 2012). The hypothetical precursors also had mutations in several genes including TP53 p.R248Q, NSD2 p.E1099K, IDH1 p.E262K, and likely KMT2D p.L3542Vfs*13 (c.10624_10625delCT), a typical MCL mutational profile. These results suggest a common precursor at GC or post-GC stage in our case. Interestingly, the NOTCH2 inversion event leading to a SEC22B-NOTCH2 fusion was detected in blastoid MCL. NOTCH2 mutations and NOTCH2’s signaling pathway have been implicated in the pathogenesis of aggressive MCL (Bea et al. 2013). This fusion, initially discovered in triple-negative aggressive breast cancer (Robinson et al. 2011), is highly active in up-regulating NOTCH signaling and the expression of downstream targets including CMYC and CCND1 (Robinson et al. 2011). The identification of this fusion provides an alternative mechanism of aberrant NOTCH2 signaling in MCL and might also explain the aggressive behavior in this patient.
The underlying mechanisms of a divergent clonal evolution to HRS-like cells and/or CHL in MCL include EBV infection, acquisition of additional genetic aberrations such as PDL1/2 amplification, and certain mutations in MCL that might contribute to this process such as TP53. Interestingly, in our case deregulation of PDL1/2 in the setting of multiple prior genetic aberrations, including TP53 mutations, might be sufficient to generate the CHL phenotype. The clinical significance of HRS-like cells in MCL remains to be clarified. The reported cases were rarely managed with CHL appropriate regimens (Caleo et al. 2003; Tinguely et al. 2003; Hayes et al. 2006; Schneider et al. 2014; Giua et al. 2015; Murray et al. 2017; Kramer et al. 2019), which is consistent with the current recommendations for management of CLL/SLL with “type I” HRS-like (Swerdlow et al. 2017). Once a diagnosis of bona fide CHL is established, management with CHL appropriate regimen would be reasonable as demonstrated by control of CHL component in this case. It is unclear if CHL changes the clinical course of MCL although our patient had an aggressive clinical course.
METHODS
Biopsies were formalin-fixed paraffin-embedded (FFPE) and processed for standard H&E staining and immunohistochemistry. Single-cell suspension was prepared from submitted fresh tissues and subjected to flow cytometric analysis. The bone marrow aspirate specimens were studied with conventional chromosome and FISH analysis. FFPE tissue blocks were also submitted for FISH tests, clonality analysis by IGH gene rearrangement, and targeted next-generation sequencing analysis (see Supplemental Table S1).
ADDITIONAL INFORMATION
Data Deposition and Access
The variants reported in this study have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000930027–SCV000930032.
Ethics Statement
The patient has consented to MSK protocol 09-141: Collection of Human Biological Specimens from Patients for Research Studies. This protocol requires written informed consent. This protocol allows for the collection of biological specimens from patients to perform laboratory studies. The use of the data collected is covered under MSK protocol 16-1289: Clinicopathological Features of Hematological Malignancies. This protocol allows for the retrospective analysis of hematopathology-related data, data previously reviewed for research purposes, and clinical records in MSKCC. Findings from these analyses can be used in peer-reviewed publications to disseminate knowledge. Protocol 16-1289 has been approved under a waiver of informed consent, because the research involves no more than minimal risk to the participants or their privacy.
Acknowledgments
This study was supported in part through the National Institutes of Health/National Cancer Institute (NIH/NCI) Cancer Center Support Grant P30 CA008748 and Lymphoma Spore grant P50 CA192937. W.X. is supported by a start-up fund from the Department of Pathology at MSKCC.
Author Contributions
W.X. and H.T. conceived the study, collected and analyzed the data, and wrote the manuscript. K.P.-D. edited the manuscript and analyzed and annotated all the molecular data, including the critical comparative analysis. C.L.B., A.B., J.S., S.F., and A.K. provided critical clinical information. M.E.A., M.R., S.F., J.Y., and A.D. interpreted data. J.B. coordinated the project. R.A. and Y.Z. performed and reviewed cytogenetic studies. All the authors approved the final version of the manuscript.
Competing Interest Statement
C.L.B. has received research support from Janssen, Novartis, Epizyme, and Xynomics and an honorarium from Dava Oncology. A.D. has received personal fees from Roche, Corvus Pharmaceuticals, Physicians’ Education Resource, Seattle Genetics, Peerview Institute, Oncology Specialty Group, Pharmacyclics, Celgene, and Novartis and research grants from National Cancer Institute, and Roche/Genentech.
Referees
Mahesh Swaminathan
Anonymous
Supplementary Material
Footnotes
[Supplemental material is available for this article.]
REFERENCES
- Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martin-Garcia D, Jares P, Gine E, Pinyol M, Royo C, Nadeu F, et al. 2013. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci 110: 18250–18255. 10.1073/pnas.1314608110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caleo A, Sánchez-Aguilera A, Rodríguez S, Dotor AM, Beltrán L, de Larrinoa AF, Menárguez FJ, Piris MA, García JF. 2003. Composite Hodgkin lymphoma and mantle cell lymphoma: two clonally unrelated tumors. Am J Surg Pathol 27: 1577–1580. 10.1097/00000478-200312000-00012 [DOI] [PubMed] [Google Scholar]
- de Leval L, Vivario M, De Prijck B, Zhou Y, Boniver J, Harris NL, Isaacson P, Du MQ. 2004. Distinct clonal origin in two cases of Hodgkin's lymphoma variant of Richter's syndrome associated with EBV infection. Am J Surg Pathol 28: 679–686. 10.1097/00000478-200405000-00018 [DOI] [PubMed] [Google Scholar]
- Giua R, Fontana D, Deda G, Bianchi A, Rabitti C, Antonelli Incalzi R. 2015. Composite mantle-cell lymphoma and classical Hodgkin lymphoma in a very old adult. J Am Geriatr Soc 63: 824–826. 10.1111/jgs.13355 [DOI] [PubMed] [Google Scholar]
- Hayes SJ, Banerjee SS, Cook Y, Houghton JB, Menasce LP. 2006. Composite mantle-cell lymphoma and classical Hodgkin lymphoma. Histopathology 48: 621–623. 10.1111/j.1365-2559.2005.02296.x [DOI] [PubMed] [Google Scholar]
- Jares P, Colomer D, Campo E. 2012. Molecular pathogenesis of mantle cell lymphoma. J Clin Invest 122: 3416–3423. 10.1172/JCI61272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Uppal G, Wang ZX, Gong JZ. 2019. Mantle cell lymphoma with Hodgkin and Reed–Sternberg cells: review with illustrative case. Appl Immunohistochem Mol Morphol 27: 8–14. 10.1097/PAI.0000000000000527 [DOI] [PubMed] [Google Scholar]
- Mao Z, Quintanilla-Martinez L, Raffeld M, Richter M, Krugmann J, Burek C, Hartmann E, Rudiger T, Jaffe ES, Müller-Hermelink HK, et al. 2007. IgVH mutational status and clonality analysis of Richter's transformation: diffuse large B-cell lymphoma and Hodgkin lymphoma in association with B-cell chronic lymphocytic leukemia (B-CLL) represent 2 different pathways of disease evolution. Am J Surg Pathol 31: 1605–1614. 10.1097/PAS.0b013e31804bdaf8 [DOI] [PubMed] [Google Scholar]
- Murray C, Quinn F, Illyes G, Walker J, Castriciano G, O'Sullivan P, Grant C, Vandenberghe E, Bird B, Flavin R. 2017. Composite blastoid variant of mantle cell lymphoma and classical Hodgkin lymphoma. Int J Surg Pathol 25: 281–286. 10.1177/1066896916672556 [DOI] [PubMed] [Google Scholar]
- Robinson DR, Kalyana-Sundaram S, Wu Y-M, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. 2011. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med 17: 1646–1651. 10.1038/nm.2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider S, Crescenzi B, Schneider M, Ascani S, Hartmann S, Hansmann ML, Falini B, Mecucci C, Tiacci E, Küppers R. 2014. Subclonal evolution of a classical Hodgkin lymphoma from a germinal center B-cell-derived mantle cell lymphoma. Int J Cancer 134: 832–843. 10.1002/ijc.28422 [DOI] [PubMed] [Google Scholar]
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J. 2017. World Health Organization classification of tumours of haematopoietic and lymphoid tissues. IARC, Lyon. [Google Scholar]
- Tinguely M, Rosenquist R, Sundström C, Amini RM, Küppers R, Hansmann ML, Bräuninger A. 2003. Analysis of a clonally related mantle cell and Hodgkin lymphoma indicates Epstein–Barr virus infection of a Hodgkin/Reed–Sternberg cell precursor in a germinal center. Am J Surg Pathol 27: 1483–1488. 10.1097/00000478-200311000-00014 [DOI] [PubMed] [Google Scholar]
- Xiao W, Chen WW, Sorbara L, Davies-Hill T, Pittaluga S, Raffeld M, Jaffe ES. 2016. Hodgkin lymphoma variant of Richter transformation: morphology, Epstein–Barr virus status, clonality, and survival analysis—with comparison to Hodgkin-like lesion. Hum Pathol 55: 108–116. 10.1016/j.humpath.2016.04.019 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The variants reported in this study have been submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) under accession numbers SCV000930027–SCV000930032.