Summary
Citrus huanglongbing (HLB) is the most devastating citrus disease worldwide. ‘Candidatus Liberibacter asiaticus’ (Las) is the most prevalent HLB causal agent that is yet to be cultured. Here, we analysed the flagellar genes of Las and Rhizobiaceae and observed two characteristics unique to the flagellar proteins of Las: (i) a shorter primary structure of the rod capping protein FlgJ than other Rhizobiaceae bacteria and (ii) Las contains only one flagellin‐encoding gene flaA (CLIBASIA_02090), whereas other Rhizobiaceae species carry at least three flagellin‐encoding genes. Only flgJAtu but not flgJLas restored the swimming motility of Agrobacterium tumefaciens flgJ mutant. Pull‐down assays demonstrated that FlgJLas interacts with FlgB but not with FliE. Ectopic expression of flaALas in A. tumefaciens mutants restored the swimming motility of ∆flaA mutant and ∆flaAD mutant, but not that of the null mutant ∆flaABCD. No flagellum was observed for Las in citrus and dodder. The expression of flagellar genes was higher in psyllids than in planta. In addition, western blotting using flagellin‐specific antibody indicates that Las expresses flagellin protein in psyllids, but not in planta. The flagellar features of Las in planta suggest that Las movement in the phloem is not mediated by flagella. We also characterized the movement of Las after psyllid transmission into young flush. Our data support a model that Las remains inside young flush after psyllid transmission and before the flush matures. The delayed movement of Las out of young flush after psyllid transmission provides opportunities for targeted treatment of young flush for HLB control.
Keywords: citrus, flagella, HLB control, huanglongbing, Liberibacter, movement, psyllid
Introduction
Citrus huanglongbing (HLB, also called citrus greening) is the most devastating citrus disease worldwide. The most prevailing HLB pathogen in the world is ‘Candidatus Liberibacter asiaticus’ (Las), an unculturable bacterium vectored by Asian citrus psyllid (ACP, Diaphorina citri), which is an invasive pest for citrus‐producing areas outside of Asia (Bové, 2006; Wang and Trivedi, 2013). Currently, HLB management strategies mainly rely on psyllid control and numerous horticultural approaches (Blaustein et al., 2018; Li et al., 2016, 2017, 2018) since all commercial citrus varieties are susceptible to HLB (Folimonova et al., 2009). Current HLB and ACP management has not prevented HLB from spreading worldwide (Wang, 2019). For example, HLB has spread throughout Florida, to other major citrus‐producing states (e.g. California and Texas) in the USA, and to neighbouring countries including Mexico and countries in the Caribbean and Central America (Wang et al., 2017). It is paramount to understand the biology and virulence mechanism of Las to design a suitable and efficient HLB control strategy. However, its unculturability has hampered our investigation of the biology and virulence mechanism of Las. Its close relatives, such as Liberibacter crescens, Agrobacterium and Sinorhizobium, have been used as surrogates in previous studies (Andrade and Wang, 2019; Jain et al., 2019; Naranjo et al., 2019; Vahling‐Armstrong et al., 2012).
Las has been observed to be distributed throughout the whole plant, including leaf, stem, root, flower and seed coat (Tatineni et al., 2008). However, the details of Las movement in planta are still unknown. Most bacteria possess more than one organelle for motility. For most bacteria, flagella are the major motility organelles responsible for swimming and swarming, whereas type IV pili are responsible for the twitching motility. Las encodes the type IVc tight adherence‐pili (Tad) that are usually not associated directly with motility. Instead, they are involved in adherence (Andrade and Wang, 2019). Flagellum‐mediated motility allows bacterial cells to move toward and explore nutrient‐rich habitats and move away from unfavourable environments (Anderson et al., 2010; Berry and Armitage, 1999; Moens and Vanderleyden, 1996; Zhu et al., 2013). Flagella also play a key role in surface attachment and host–bacteria interactions (Heindl et al., 2014; Rossez et al., 2015). It has been known that some flagellated plant pathogenic bacteria suppress expression of flagella in the plant, probably to avoid triggering plant defence (Chatnaparat et al., 2016; Yu et al., 2013). Las contains 30 flagellar genes located in three clusters on the chromosome (Fig. 1) (Duan et al., 2009). However, it remains to be determined whether Las synthesizes flagella during within‐plant movement. This is a fundamental question that has clear implications for the rate at which disease symptoms appear in both new flush and old leaves.
Figure 1.
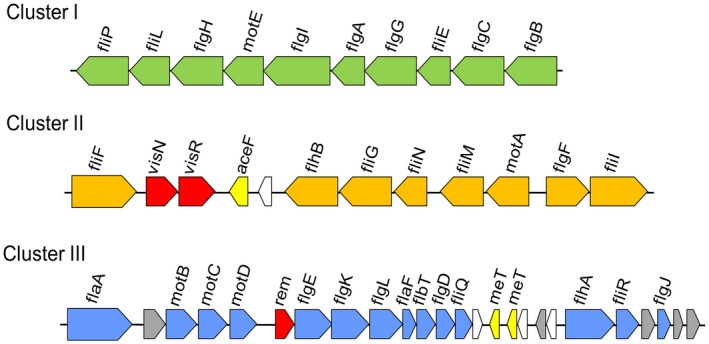
Genetic organization of the three clusters encoding flagellar genes in the ‘Candidatus Liberibacter asiaticus’ genome. The names of the coding regions shown in the diagram are based on their homology with flagellar genes in Rhizob i aceae bacteria. Grey arrows, conserved hypothetical protein; white arrows, hypothetical proteins.
The flagellar machinery can be divided into five parts called the basal body, the hook, the hook–filament junction zone, the filament and the filament cap (DeRosier, 2006; Evans et al., 2014; Ghosh, 2004; Minamino and Namba, 2004). The basal body is composed of the cytoplasmatic C‐ring (FliG, FliM and FliN), the inner or MS‐ring (FliF), the periplasmatic P‐ring (FlgI), the outer membrane L‐ring (FlgH), the proximal rod (FlgB, FlgC, FlgF and FliE) and the distal rod (FlgG) (Jones and Aizawa, 1991; Park et al., 2006). The basal body is embedded in the cell surface and plays a role in flagella rotation together with the Mot proteins (Minamino et al., 2008). The hook and filament, which are tubular structures composed of subunits of hook (FlgE) and flagellin (FliC), respectively, extend outwards from the cell (Macnab, 2003). The hook length and the switch of specificity from substrates hook‐to‐filament are tightly regulated by FliK (Erhardt et al., 2010; Waters et al., 2007). Deletion of fliK leads to formation of prolonged flagellar hooks (polyhooks), lack of filament structures and nonmotile phenotypes (Minamino et al., 2009). The hook–filament junction zone (FlgK and FlgL) and the filament cap (FliD) are located between the hook and filament and at the tip of the filament, respectively (Ikeda et al., 1996). Flagellar assembly begins with the basal body, proceeds with the hook and finishes with the filament. Capping proteins (FlgJ, FlgD and FliD) are needed to permit and regulate the polymerization of rod, hook and filament, respectively, at different stages of flagellar assembly (Minamino and Namba, 2004). FlgJ, FlgD and FliD exist at the tip of the growing rod, hook and filament structures, respectively (Evans et al., 2014).
The flagellar export apparatus is built into the central pore of the basal body MS‐ring, and it is formed by interaction of six membrane proteins: FlhA, FlhB, FliP, FliO, FliQ and FliR, and three cytoplasmic proteins FliH, FliI and FliJ. FliI is the ATPase that provides the energy for the translocation of proteins across the cytoplasmic membrane (Macnab, 2003). FliH acts as an ATPase regulator, coupling the energy of ATP hydrolysis to flagellar protein export (Evans et al., 2014; Macnab, 2003). Also, the chaperone, FliJ, prevents premature aggregation of substrates in the cytoplasm, and FlgA, assists the P‐ring assembly in the periplasm (Evans et al., 2006; Macnab, 2003).
Flagellins are the subunits that compose the flagellar filament and represent the major flagellar structural protein (Altegoer et al., 2014; Rossez et al., 2015). The number of flagellin‐encoding genes in bacterial genomes varies and may range between one (Escherichia coli) and seven (Vibrio parahaemolyticus) (Fedorov and Kostyukova, 1984; Kim and McCarter, 2000; Stewart and McCarter, 2003). The flagellar filaments in E. coli K‐12 are built up from only one flagellin protein (FliC) (Turner et al., 2012), whereas Bdellovibrio bacteriovorus (Thomashow and Rittenberg, 1985), Campylobacter coli (Guerry et al., 1990), Caulobacter crescentus (Driks et al., 1989), Agrobacterium tumefaciens (Deakin et al., 1999; Mohari et al., 2018), Rhizobium lupini strain H13‐3 and Sinorhizobium meliloti have at least two flagellin proteins forming their flagellar filaments (Scharf et al., 2001).
Young flush has been shown to be critical for ACP and Las infection. ACP preferentially feeds and exclusively reproduces on young, newly emerged flush shoots of citrus. ACP nymphs feed and complete their life stages on young flush shoots. This observation appears to be universal across different citrus‐producing regions worldwide (Bové, 2006; Hall et al., 2013; Setamou et al., 2016; Sétamou and Bartels, 2015; Tomaseto et al., 2016). Not surprisingly, citrus trees are normally infected when new flush is present (Hall et al., 2016). In addition, ACPs that acquire Las as adults are poor vectors of Las compared with ACPs that acquire the pathogen as nymphs (Pelz‐Stelinski et al., 2010). A threshold infection level (c. 106 Las/psyllid) is required for successful transmission to citrus plants (Ukuda‐Hosokawa et al., 2015). How Las moves after psyllid transmission remains largely unexplored.
Here, we investigated the flagella of Las. Our results indicate that Las flagellar genes express at low levels in planta. Las moves to other sink tissues with phloem sap when young leaves mature into sources. This delayed Las movement after ACP transmission into young flush might present opportunities to develop new strategies for HLB management.
Results
Comparison of flagellar genes of ‘Ca. Liberibacter’ and other members of Rhizobiacea e
The analysis of the flagellar components encoded by the Las genome (Fig. 1 and Table 1) showed the following characteristics that are shared by all bacterial species in Rhizobiaceae: (i) lack of cytoplasmic chaperone FliJ and the components of export apparatus FliH/FliO; (ii) the function of FliK is performed by MotD (Eggenhofer et al., 2006); (iii) the rod‐capping FlgJ protein lacks the muramidase domain in its carboxy‐terminus (Herlihey et al., 2014); and (iv) lack of FliD, the filament cap (Table 1). It is important to note that all the traits cited above are shared by members of Rhizobiaceae that are known to contain flagellar apparatus and show swimming behaviour, such as A. tumefaciens and S. meliloti (Attmannspacher et al., 2005; Deakin et al., 1999; Mohari et al., 2018).
Table 1.
Identity of residues of flagellar proteins encoded by Rhizobiaceae bacteria (as percentage values)
| Gene | Las | Atu | Sme | Lcc | Laf | Lso |
|---|---|---|---|---|---|---|
| Basal body | ||||||
| FliG | + | 46 | 45 | 54 | 81 | 82 |
| FliM | + | 30 | 30 | 42 | 70 | 66 |
| FliN | + | 47 | 50 | 50 | 77 | 74 |
| FliF | + | 49 | 50 | 56 | 78 | 75 |
| FlgI | + | 64 | 64 | 68 | 85 | 87 |
| FlgH | + | 54 | 56 | 63 | 80 | 76 |
| FlgB | + | 45 | 47 | 57 | 76 | 75 |
| FlgC | + | 59 | 62 | 68 | 79 | 85 |
| FlgF | + | 44 | 46 | 49 | 77 | 75 |
| FliE | + | 37 | 37 | 42 | 67 | 63 |
| FlgG | + | 59 | 62 | 69 | 88 | 90 |
| FlgJ | +* | 29 | 32 | 37* | 61* | 68* |
| MotA | + | 62 | 58 | 64 | 60 | 63 |
| MotB | + | 31 | 32 | 36 | 65 | 75 |
| MotC | + | 29 | 27 | 34 | 65 | 60 |
| Hook | ||||||
| FlgE | + | 50 | 41 | 58 | 75 | 74 |
| FliK/MotE | + | 32 | 28 | 29 | 44 | 48 |
| FlgD | + | 43 | 44 | 57 | 85 | 75 |
| Junction zone | ||||||
| FlgK | + | 36 | 36 | 47 | 76 | 69 |
| FlgL | + | 27 | 30 | 39 | 61 | 57 |
| Filament | ||||||
| FlaA | + | 50 | 42 | 49 | 67 | 61 |
| FlaB | † | + | + | − | − | − |
| FlaC | † | + | + | − | − | − |
| FlaD | † | + | + | − | − | − |
| Capping filament | ||||||
| FliD | † | † | † | † | † | † |
| Export apparatus | ||||||
| FlhA | + | 66 | 65 | 68 | 86 | 82 |
| FlhB | + | 44 | 45 | 54 | 80 | 74 |
| FliP | + | 60 | 57 | 67 | 90 | 86 |
| FliO | † | † | † | † | † | † |
| FliQ | + | 57 | 61 | 65 | 90 | 90 |
| FliR | + | 40 | 43 | 51 | 81 | 78 |
| FliH | † | † | † | † | † | † |
| FliI | + | 62 | 63 | 68 | 87 | 84 |
| Chaperones | ||||||
| FliJ | † | † | † | † | † | † |
| FlgA | + | 42 | 46 | 53 | 70 | 69 |
Comparative analysis of flagellar proteins encoded by different species of R h izob i aceae bacteria. Las, ‘Candidatus Liberibacter asiaticus’ (CP001677); Laf, ‘Candidatus Liberibacter africanus’ (CP004021); Lso, ‘Candidatus Liberibacter solanacearum’ (CP002371); Lcc, Liberibacter crescens strain BT‐1 (CP003789); Atu, Agrobacterium tumefaciens (AE007869); Sme, Sinorhizobium meliloti strain 1041 (AL591688).
Indicates a shorter primary structure for FlgJ homologues when compared to A. tumefaciens and S. meliloti.
No homologues were identified. + and − indicate presence and absence of a homologue in the analysed genome, respectively.
We observed two characteristics unique to the flagellar proteins of Las and Liberibacter crescens BT‐1, a culturable bacterium, that may determine the fate of the Las flagellar machinery:
It has a shorter primary structure of the rod‐capping protein FlgJ than other Rhizobiaceae bacteria (Fig. 2). In previous work, it was shown that the minimal functional region required of FlgJ for promoting the rod assembly consists of 151 residues in its amino terminus (Hirano et al., 2001). The primary structure of FlgJ in Las (CLIBASIA_01980) contains 112 amino acid residues, whereas in A. tumefaciens (Atu0584) and S. meliloti (SMc03071), FlgJ homologues consist of 175 and 184 residues, respectively.
Las contains only one flagellin‐encoding gene flaA (CLIBASIA_02090), whereas other Rhizobiaceae species carry at least three flagellin‐encoding genes (Fig. 1, Table 1). For example, Rhizobium lupini H13‐3 and S. meliloti produce flagellar filaments composed of three and four flagellin subunits, respectively (Scharf et al., 2001). These flagellin‐encoding genes are named flaA through flaD. Indeed, the complex flagellar filaments in Rhizobiaceae bacteria are composed of a majority flagellin and at least one secondary flagellin. Mutational analysis of flagellin genes revealed that, in both R. lupini and S. meliloti, FlaA is the principal flagellin and that FlaB, FlaC and FlaD are secondary (Deakin et al., 1999). In this way, FlaA and at least one secondary flagellin are required for assembling a functional flagellar filament in R. lupini and S. meliloti (Scharf et al., 2001). Similar results were demonstrated in A. tumefaciens (Mohari et al., 2018). Thus, the unique features of Las in FlgJ and FlaA are probably critical to functionality of its flagella.
Figure 2.

Sequence alignment of the rod cap proteins (FlgJ) encoded by Rhizob i aceae bacteria showing the rod‐binding domain highlighted in green. Atu, Agrobacterium tumefaciens (AE007869); Sme, Sinorhizobium meliloti strain 1041 (AL591688); Rhf, Sinorhizobium fredii NGR234 (CP001389); CLas, ‘Candidatus Liberibacter asiaticus’ (CP001677); CLaf, ‘Candidatus Liberibacter africanus’ (CP004021); CLso, ‘Candidatus Liberibacter solanacearum’ (CP002371); Lcc, Liberibacter crescens strain BT‐1 (CP003789); CLam, ‘Candidatus Liberibacter americanus’ (CP006604). Alignment result was obtained by ClustalW and protein domain was identified with Pfam.
Characterization of the function of FlgJ and FlaA of Las using Agrobacterium as a surrogate
We first evaluated whether FlgJ and FlaA of Las are functional by assessing whether they can complement corresponding mutants of A. tumefaciens (Atu). For this purpose, we constructed a nonpolar mutant of flgJ in A. tumefaciens. As expected, the flgJ mutant of A. tumefaciens became immobile, which is similar to the ∆flaABCD mutant with all four flagellin genes deleted (Fig. 3). Only flgJAtu but not flgJLas restored the swimming motility of A. tumefaciens flgJ mutant (Fig. 3A,B), indicating flgJLas might not be functional or works differently in Las. To verify whether FlgJLas can physically interact with the proximal rod proteins FlgB and FliE as shown in other well‐studied systems (Hirano et al., 2001), we performed a pull‐down experiment with those proteins. Interestingly, FlgJLas interacted with FlgB but not with FliE (Fig. 4).
Figure 3.
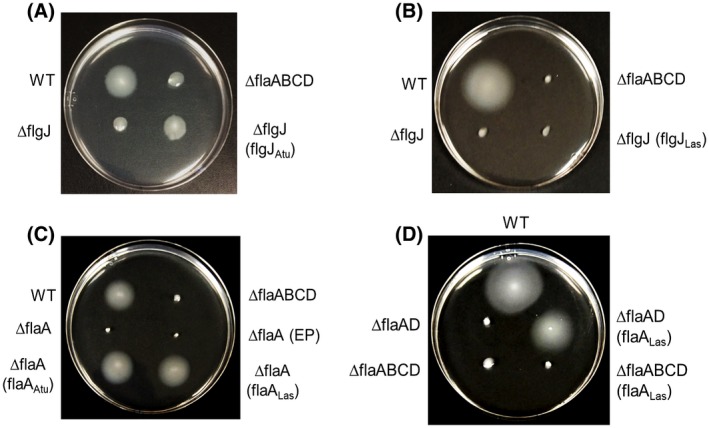
Functional analysis of FlgJ and FlaA of ‘Candidatus Liberibacter asiaticus’ (Las) via complementation analysis of their corresponding mutants of Agrobacterium tumefaciens (Atu) with swimming assay. (A) Complementation of A. tumefaciens flgJ mutant with flgJAtu.. (B) Complementation of A. tumefaciens flgJ mutant with flgJLas. (C) Complementation of A. tumefaciens flaA mutant with flaALas and flaAAtu. (D) Complementation of A. tumefaciens flaAD and flaABCD mutants with flaALas . WT, A. tumefaciens strain C58; ∆flaABCD, flagellum null mutant; ∆flgJ, A. tumefaciens C58 with deletion of flgJ; ∆flaA, A. tumefaciens C58 with in‐frame deletion in flaA; ∆flgJ (flgJAtu), flgJ mutant complemented with A. tumefaciens flgJ; ∆flgJ (flgJLas ), flgJ mutant complemented with Las flgJ; ∆flaA (flaAAtu), flaA mutant complemented with A. tumefaciens flaA; ∆flaA (flaALas), flaA mutant complemented with Las flaA; ∆flaA (EP), ∆flaA carrying empty plasmid; ∆flaAD, A. tumefaciens C58 with in‐frame deletion for the flagellin‐encoding genes flaA and flaD; ∆flaABCD, A. tumefaciens C58 carrying deletion for all flagellin genes flaA, flaB, flaC and flaD; ∆flaAD (flaALas) and ∆flaABCD (flaALas), mutants ∆flaAD and ∆flaABCD harbouring the flagellin‐encoding gene flaA from Las. Bacterial cells were incubated at 28 °C and photographed after 3 days.
Figure 4.
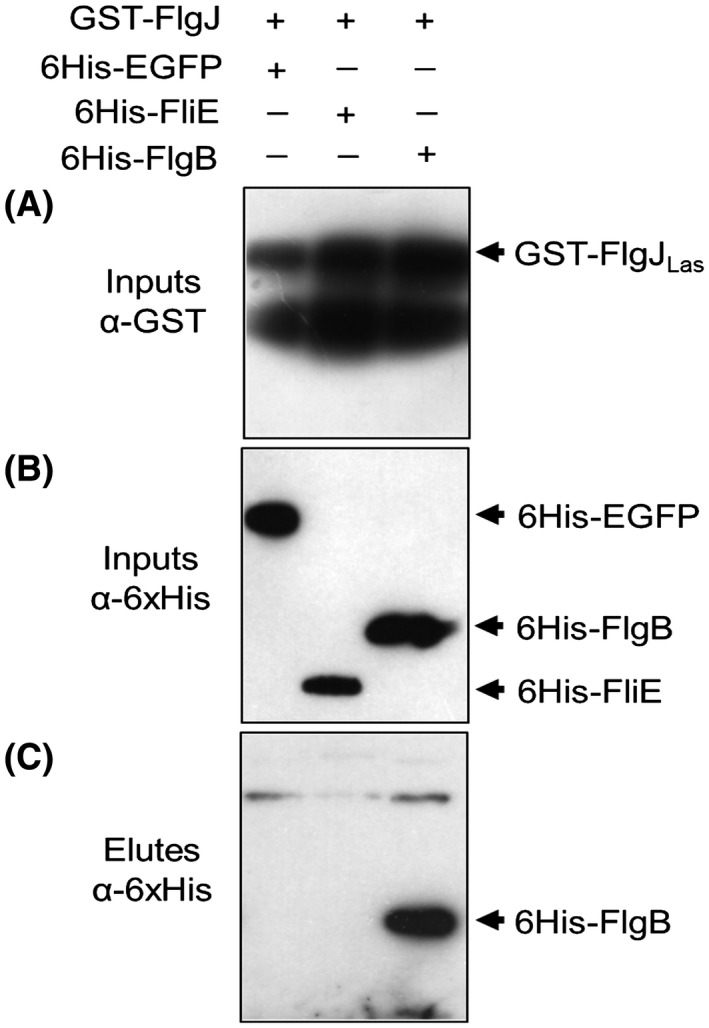
Pull‐down assay showed a physical interaction between FlgJLas and FlgB. (A) The protein GST‐FlgJLas was used as a bait for the pull‐down assay. GST‐FlgJLas was expressed in Escherichia coli, immobilized and washed on glutathione sepharose beads, and incubated with E. coli lysates containing 6×HisEGFP, 6×HisFlgB or 6×HisFliE. Pull‐down inputs containing the supernant of total cell extracts (A) and (B) were immunobloted using the anti‐GST and anti‐6×His antibodies, respectively. Eluted protein fractions (Elutes) were probed with anti‐6×His antibodies.
To test the function of flagellin protein FlaALas, we generated mutant strains of A. tumefaciens carrying deletion in flaA alone (∆flaA), in flaA and flaD genes (∆flaAD), and in all flagellin‐encoding genes (∆flaABCD). Interestingly, ectopic expression of flaALas in A. tumefaciens mutants restored the swimming motility of ∆flaA mutant (Fig. 3C) and ∆flaAD mutant (Fig. 3D), but not that of the null mutant ∆flaABCD (Fig. 3D). The swimming halos for the tested strains were calculated and the means and standard deviation (n = 3) values are shown in Fig. S2. These results indicate that in the A. tumefaciens flagellar system FlaALas functions as the majority flagellin interacting with other secondary flagellins such as FlaB or FlaC, which remain intact in the ∆flaAD mutant, to form a fully functional flagellar filament. However, FlaALas alone could not form a fully functional flagellar filament in A. tumefaciens.
Observation of Las flagellum in psyllids and in planta
To verify whether Las forms a functional flagellum, we visualized Las cells isolated from Las‐infected plants and psyllids using transmission electron microscopy (TEM). For the visualization, Las cells were confirmed using Las‐specific antibody against Las OmpA and the second antibody anti‐rabbit IgG‐gold. Las cells isolated from grapefruit (Citrus paradisi) seed coats (Fig. 5A) and parasitic dodder stems (Cuscuta sp.) and psyllids are all filament‐shaped or long‐rod‐shaped. Previous observation of Las under TEM suggested that it is polymorphic with Las cells being mostly filamentous in psyllids, but in different shapes in plants with diameter 0.33–1.5 μm and length 2.6–6.3 μm (Hartung et al., 2010). No flagellum was observed for Las cells isolated from grapefruit seed coats (Fig. 5A) and parasitic dodder stems (Fig. 5B). For Las isolated from psyllid guts, no flagellum was observed on most Las cells. A few Las cells showed thread‐like structures from which we could not make a conclusive claim whether they are indeed flagella (Fig. 5C‐H). However, L. crescens, which shares the same flagellar genes as Las (Table 1), was observed to contain flagella under TEM (Fig. 6).
Figure 5.
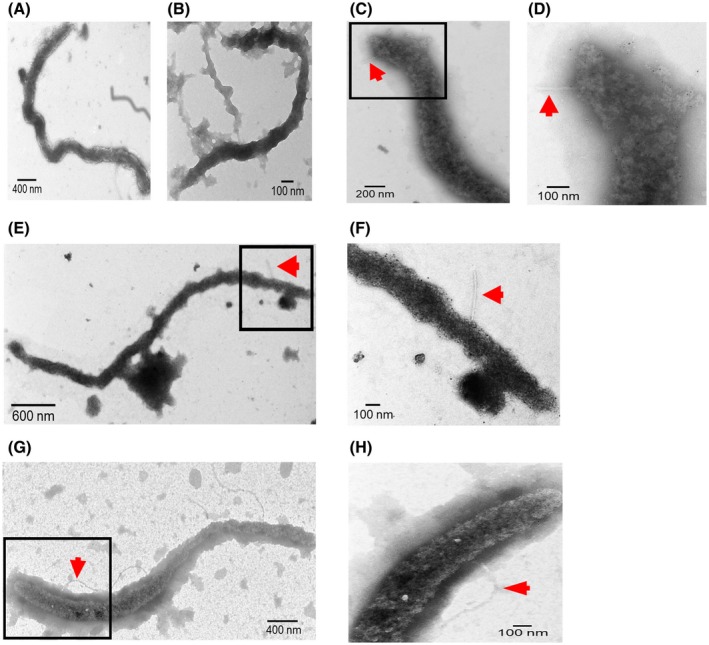
Transmission electron microscopy analysis of ‘Candidatus Liberibacter asiaticus’ (Las). (A) Las cell isolated from Las‐infected grapefruit seed coat. (B) Las cell isolated from dodder stem. (C)–(H) Las isolated from psyllid guts. (D), (F) and (H) are enlarged views of the indicated area in (C), (E) and (G), respectively. Las cells were visualized by using negative staining and antibody against Las OmpA and second antibody anti‐rabbit IgG‐gold. Arrows indicate thread‐like structures. Size bars are indicated in the images.
Figure 6.
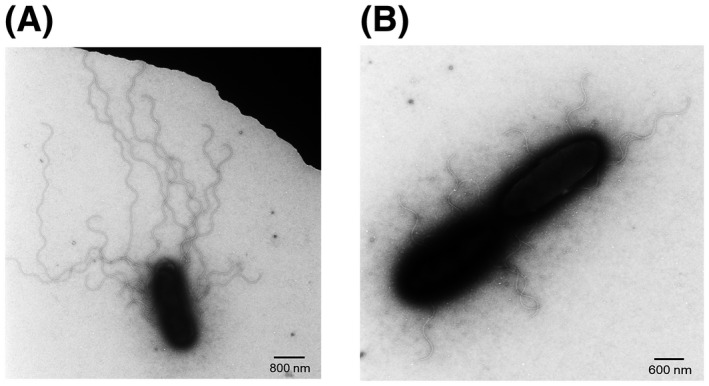
Visualization of the flagellar structures in Liberibacter crescens BT‐1 cells. Negatively stained cells were analysed by transmission electron microscopy. Size bars are indicated in the images.
Expression of flagellar genes in psyllids and in planta
To further assess whether Las synthesizes flagella in psyllids and plants, we tested the expression of flagellar genes at both transcription and translation levels using quantitative reverse transcription PCR (RT‐qPCR). We found that flagellar genes were up‐regulated in psyllids compared to in planta (Fig. 7). Specifically, the flagellin gene flaA is highly up‐regulated in psyllids, which is consistent with our previous study (Yan et al., 2013). Likewise, flagellin protein was detected for Las inside psyllids, but not inside plants based on western blot (Fig. 8). Taken together, our data suggest Las lacks flagella in planta or flagellar genes express at low level in planta.
Figure 7.
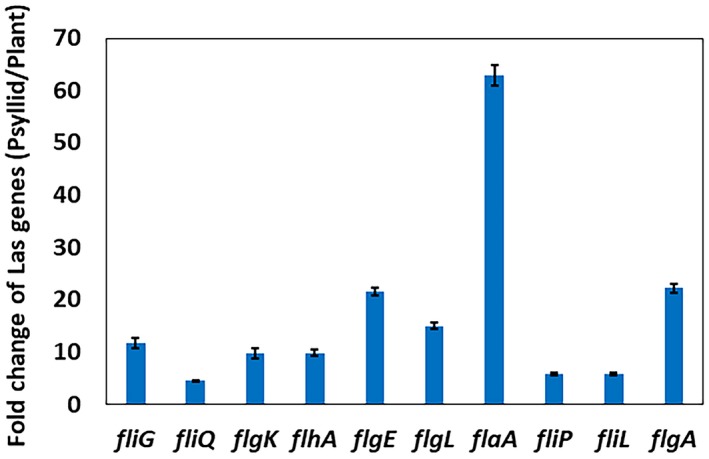
RT‐qPCR analysis of selected ‘Candidatus Liberibacter asiaticus’ (Las) genes encoding flagellum apparatus. Fold change is the relative gene expression (in psyllid versus in planta) of Las.
Figure 8.
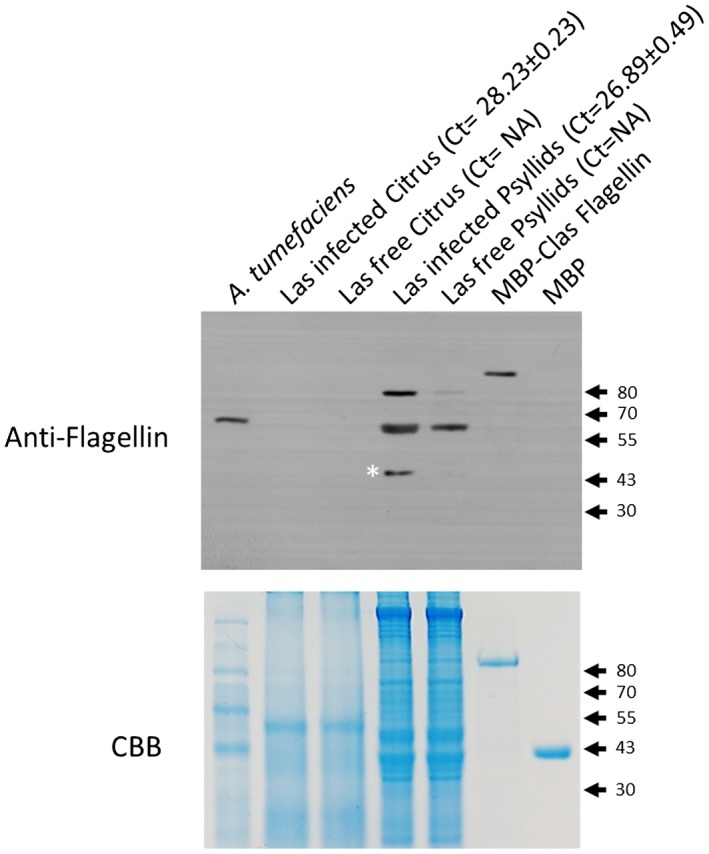
Flagellin detection. The antibody produced using the peptide of ‘Candidatus Liberibacter asiaticus’ (Las) flagellin protein (sequence: DRVSSGLRVSDAAD) was used to detect flagellin protein in different bacterial samples (left to right): Agrobacterium tumefaciens C58 derivative strain; Las‐infected citrus (Ct = 28.23 ± 0.23); Las‐free citrus control; Las‐infected psyllids (Ct = 26.89 ± 0.49); Las‐free psyllid control; positive MBP‐Las flagellin fusion protein control and negative MBP protein control. The band marked with an asterisk is the flagellin protein of Las with MW 48 kDa. Coomassie brilliant blue (CBB) served as a loading control. The experiment was repeated twice with similar results.
Movement of Las in planta after ACP transmission of Las into young flush
Las has been known to translocate through the sieve pore to move from the infection sites to tissues throughout the plants (Kim et al., 2009). Since Las does not produce flagella or its flagellar gene expression is low in planta, its movement to other parts (e.g. roots) of the plant from the infection site is unlikely to be mediated by flagella. Instead, we hypothesize that Las moves with phloem sap to sink tissues when young flush matures. To test this hypothesis, we caged ten Las‐positive ACPs/flush (Valencia sweet orange) for each plant for different durations and tested for Las at 54 days post‐psyllid feeding. For each treatment, we used five plants with one plant as a biological replicate. After 54 days, 40%, 100%, 100% and 80% of tested leaves were Las‐positive following 6, 11, 18 and 29 days of ACP feeding, respectively. However, none of the root samples were Las‐positive, indicating that Las had not moved to roots.
To further understand Las movement inside plants after ACP transmission, we trimmed nine citrus plants (Hamlin sweet orange on Cleo) to trigger young flush. We caged 15 psyllids/plant together with young flush and five mature leaves. Las was detected in young flush‐developed leaves in five of the nine plants at 4 weeks after ACP transmission, and 7, 9 and 15 weeks after ACP transmission. Las was detected in the roots in five of the infected plants at 15 weeks after ACP transmission, but not at other time points. Las was not detected in all the old leaf samples inside or outside of the cage at the aforementioned time points. Taken together, our data are consistent with our hypothesis that Las moves from source to sink passively with phloem sap and will likely stay in the young flush after ACP transmission and before flush matures.
Discussion
Las flagella have multiple important features. First, FlgJ of Las is not able to complement the flgJ mutant of A. tumefaciens. FlgJ plays an important role in flagellar assembly (Hirano et al., 2001). FlgJLas, as its homologues in several other bacterial phyla, including Alphaproteobacteria, Deltaproteobacteria, Epsilonproteobacteria and Spirochaetes (Nambu et al., 2006), contains only the rod‐capping domain involved in protein–protein interactions, but lacks the peptidoglycan hydrolase domain, which allows the elongating flagellar rod to penetrate through the peptidoglycan (PG) layer. The peptidoglycan hydrolase domain is commonly found in FlgJ from other bacteria but its function can be developed by other peptidoglycan hydrolase protein not related to flagellar proteins. Liberibacter genomes code for one peptidoglycan hydrolase, which contains a soluble lytic transglycosylase (SLT) domain and a periplasmic signal peptide (Fig. S1). In enteric bacteria such as Salmonella enterica serovar Typhimurium, flgJ null mutants fail to produce the flagellar rods, hooks and filaments, and are nonmotile (Hirano et al., 2001; Nambu et al., 1999). Instead, the flgJ mutant of Borrelia burgdorferi, which contains a single‐domain FlgJ homolgue as Las, can form intact flagellar basal bodies but had fewer and disoriented flagellar hooks and filaments (Zhang et al., 2012). Accordingly, the motility of the flgJ mutant of B. burgdorferi was partially deficient. In addition, FlgJ seems to be functional because it was able to interact with FlgB. Thus, Liberibacter spp. may have evolved some novel means involving FlgJ to assemble the rod. Second, even though Las encodes a functional FlaA that complements the corresponding mutant of A. tumefaciens, the complex flagellar filament in Rhizobiaceae bacteria is formed by the interaction of FlaA with at least one secondary flagellin, such as FlaB, C or D, that is absent in Las. Ectopic expression of FlaALas in ΔflaABCD did not restore the motility of the null mutant, suggesting that FlaALas is incapable of forming a fully functional filament. Alternatively, the observation of flagella in L. crescens suggests synthesis of Las flagella related to FlgJ and FlaA might involve some novel mechanisms different from known examples in Rhizobiaceae. Lastly, even though we could not totally rule out the possibility that Las might contain flagella in certain conditions (such as in psyllids, as implied in Fig. 8), expression of the flagellar genes is repressed in planta. Indeed, western blotting was not able to detect flagellin in planta.
The flagellar features of Las in planta might help Las avoid the elicitation of plant defence responses since flagellin is known to trigger plant immunities as typical pathogen‐associated molecular patterns (Thomma et al., 2011). Interestingly, mammalian cells use different receptors for recognition of extracellular flagellin or intracellular flagellin. For sensing flagellin outside the mammalian cell, the immune system uses Toll‐like receptor TLR‐5 (Yoon et al., 2012). If flagellin protein is detected within the cytosol of a cell, it is detected by the Nod‐like receptor (NLR) Ipaf (Franchi et al., 2006). We reason that flagellin might activate some early immune responses when Las is transmitted into the phloem by psyllids, during which the flagellin expressed in psyllids remain available in planta. It is likely the recognition is not mediated by FLS2 (Gómez‐Gómez and Boller, 2002), but instead by some unknown intracellular receptors. However, it is expected flagellar expression is turned off soon after transmission. Interestingly, multiple plant pathogenic bacteria, such as Pseudomonas syringae (Cheng et al., 2018; Markel et al., 2016) and Xanthomonas axonopodis pv. glycines (Chatnaparat et al., 2016), have been known to use a similar strategy by reducing expression of flagellar genes and losing flagella once reaching their habitats inside the plant. On the other hand, the higher expression of flagellar genes in psyllids suggests flagella might be important for its movement inside the psyllid body to different organs and cell organelles (Ghanim et al., 2017). The induction of flagellar genes in psyllids probably results from differential physiochemical conditions inside psyllids from the phloem sap. In addition, the ACP genome encodes a deficient innate immune system lacking a number of genes that encode for antimicrobial peptides and the Imd pathway associated with defence against Gram‐negative bacteria (Arp et al., 2017).
We have proposed the following movement model for Las after psyllid transmission into young flush (Fig. 9). In Phase 1 (lag phase), Las initially undergoes no or slow growth to adapt to the environmental changes from psyllids to phloem, then begins multiplication inside the young flush after an undefined period (Phase 2/multiplication phase). In the first two phases, Las stays inside the young flush before it turns into a source. With the maturation of the young flush, the infected leaf transits from a sink into a source. Meanwhile, Las begins to move from the infected leaf to other parts of the plant, such as roots, following phloem sap from source to sink (Phase 3/movement phase). The Las flagellar features probably suggest that Las moves with phloem sap rather than relying on flagella‐mediated movement. This model is consistent with the data that newly infected young leaves become infectious within 10–15 days after an inoculum of Las from an adult psyllid (Lee et al., 2015). This movement pattern also promotes acquisition and transmission of Las by nymphs. Las has been suggested to move at approximately 2–3 cm/day in citrus after graft inoculation (S. Lopes, personal communication). However, we do not think that detecting Las in the leaves but not in the root results from the slow movement of Las. The average height (top to root) of the trimmed trees used for our Las movement assay is approximately 20 cm and we conducted the qPCR analysis at 54 days after psyllid feeding, which is supposed to be enough for Las to reach the root if moving right after feeding. In addition, microscopic analysis of Las in planta showed that Las cells float freely in the phloem sap without attaching to the sieve tube cell walls or each other (i.e. without forming biofilms) (Ding et al., 2018; Folimonova and Achor, 2010; Hartung et al., 2010; Hilf et al., 2013; Kim et al., 2009). A similar phenomenon has been observed for Lso and ‘Ca. Liberibacter americanus’ in planta (Liefting et al., 2009; Secor et al., 2009; Wulff et al., 2014). The bacteria move primarily in the vertical direction along the sieve tubes through the sieve pores, rather than horizontally to adjacent sieve tubes. This hypothesis was first suggested by observations of the uneven colonization of Las in dodder, where adjacent phloem vessel elements were observed to be completely full of Las or free of the pathogen (Hartung et al., 2010). Even though Las has delayed movement out of young flush, its population is highest in the seed coat compared to other tissues based on our experience. Dodder also hosts a high density of Las based on our previous experience. We used different tissues, including young flush, in our microscopy studies and could only observe Las when seed coats and dodder were used. Indeed, the procedure used to observe flagella under TEM is quite challenging for unculturable bacteria. More robust evidence for the Las movement model can be acquired by fluorescent microscopical analysis of green fluorescent protein (GFP)‐labelled Las, as previously conducted for xylem‐inhibiting Xylella (Newman et al., 2003), provided Las can be cultured or be labelled by GFP in vivo in the future.
Figure 9.
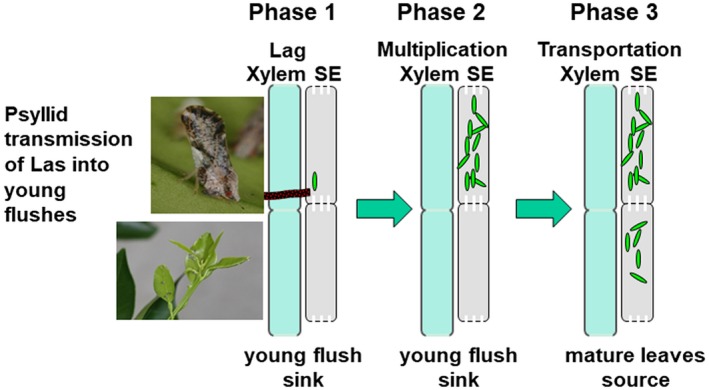
Schematic representation of ‘Candidatus Liberibacter asiaticus’ (Las) movement with phloem sap after psyllid transmission. Phase 1: After psyllid feeding and injecting Las into the phloem of young flush, Las initially undergoes a lag phase with no or slow growth to adapt to the environmental changes. Phase 2: Las begins multiplication inside the young flush after an undefined period. Phase 3: With the maturation of the young flush, the infected leaf transits from a sink into a source. Meanwhile, Las begins to move from the infected leaf to other parts of the plant, such as roots, following phloem sap from source to sink.
Those unique features of the HLB pathosystem centring on young flush suggest it represents the Achilles' heel for Las, and is thus the key point for HLB/ACP management. The psyllids preferentially feed and exclusively reproduce on young, newly emerged flush shoots of citrus, and their nymphs feed and complete their life stages on young flush shoots. Young flush is critical for psyllids acquiring and transmitting Las (Bové, 2006; Hall et al., 2013; Setamou et al., 2016; Sétamou and Bartels, 2015; Tomaseto et al., 2016). The importance of controlling psyllids during flush stages has been recognized (Hall et al., 2016). The delayed movement of Las out of the young flush provides opportunities to develop novel control strategies. Interestingly, traditional citrus growers in China not only rely on high‐frequency psyllid control of young flush using insecticides, but also remove flush (e.g. physical removal or spray with young flush killer) during summer or winter (for certain areas). This flush removal practice probably contributes to reducing the ACP population, preventing ACP from acquiring Las from young flush of HLB‐positive trees and transmitting Las to young flush. The delayed movement of Las in young flush also helps the development of targeted early detection of Las after psyllid transmission and before HLB symptom appearance (Pandey and Wang, 2019). In addition, targeted treatment of young flush with bactericides might prevent Las from establishing in planta after psyllid transmission. The minimum concentration of oxytetracycline required to suppress Las populations in planta has been suggested to be 0.68 and 0.86 µg/g under greenhouse and field conditions, respectively (Li et al., 2019). Depending on application method, oxytetracycline concentrations in leaf tissues have been reported to range from 0 to 0.53 µg/g (Li et al., 2019; Vincent et al., 2019), below the concentrations required to suppress Las. It seems that successful prevention of Las establishment after psyllid transmission requires optimized delivery methods to increase bactericidal concentration to the minimum concentration of bactericides required to suppress Las populations in planta. The bactericidal effect needs to cover the whole flush period until leaf maturation. Additionally, it is interesting to know whether the targeted treatment effect can be obtained with non‐antibiotic bactericides such as generally recognized as safe (GRAS) antimicrobial compounds.
In summary, the flagellar characteristics of Las support a delayed movement of Las from young flush after psyllid transmission. The movement pattern of Las provides unique opportunities for early diagnosis of HLB and targeted treatment of young flush to control HLB.
Experimental Procedures
Bacterial strains, plasmids and growth conditions
The plasmids, oligonucleotides and bacterial strains used in this study are listed in Tables S1 and S2. Escherichia coli cells were grown at 37 °C in Luria–Bertani (LB) medium. The A. tumefaciens strain C58 and mutant strains were grown in LB medium at 28 °C. Plasmids were introduced into E. coli by heat‐shock at 42 °C and into A. tumefaciens by electroporation. A grobacterium tumefaciens deletion mutants were generated using the suicide vectors pNPTS138 as described elsewhere (Hibbing and Fuqua, 2011). Cells of L. crescens strain BT‐1 were grown at 28 °C at 150 rpm in BM7 medium. Antibiotics were used at the following concentrations: kanamycin 50 mg/mL, ampicillin 100 mg/mL, gentamicin 10 mg/mL.
Deletion of flagellar genes and construction of complemented strains of A. tumefaciens
DNA manipulations and PCR were performed according to standard procedures (Sambrook et al., 2006). To construct the flaA, flaD and flgJ deletion mutants, approximately 1 kb of the upstream and downstream regions of those flagellar genes were amplified by PCR from A. tumefaciens genomic DNA (oligonucleotides are listed in Table S2), and the two fragments were ligated to produce an in‐frame deletion. The approximately 2 kb fragments that resulted were then cloned into the HindIII site of the pNPTS138 suicide vector, generating the plasmids pNPTS‐FlaA, pNPTS‐FlaD and pNPTS‐FlgJ (Andrade and Wang, 2019; Andrade et al., 2014). These constructs were introduced into A. tumefaciens cells by electroporation, and the wild‐type copies were replaced by the deleted version after two recombination events. For the first and second recombination events, A. tumefaciens cells were selected in LB medium without NaCl (LBON) with kanamycin and in LBON plus 5% sucrose, respectively. The single mutants ∆flaA, ∆flaD and ∆flgJ and the double mutant ∆flaAD were confirmed by PCR. To produce the flagellin null mutant, two fragments of 1 kb of the upstream and downstream regions of flaA and flaC were amplified and ligated. A fragment of 2 kb was generated and cloned into the HindIII site of pNPTS138. This resulting construct pNTPS‐FlaABC, carrying in‐frame deletion of flaA‐flaB‐flaC, was introduced into A. tumefaciens flaD mutant strain by electroporation. After selections in appropriate media, the null mutant ∆flaABCD was confirmed by PCR. To complement ∆flgJ, ∆flaA, the double mutant ∆flaAD and the null mutant strains, fragments including the coding region of flaA or flgJ genes were amplified by PCR from Las genomic DNA. Also, the coding region of flgJ was amplified from A. tumefaciens genome. Those three fragments were inserted into the BamHI site of the pTF53 vector that contains the constitutive promoter Trp (Andrade and Wang, 2019; De Feyter et al., 1993), creating the plasmids pF53‐Las_FlaA, pF53‐Las_FlgJ and pF53‐Atu_FlgJ. The construct pF53‐FlaA_Las was used to transform ∆flaA, ∆flaAD and ∆flaA BCD mutant strains. ∆flgJ mutant strain was transformed with pF53‐Las_FlgJ or pF53‐Atu_FlgJ constructs. After electroporation, the transformed A. tumefaciens cells were selected on LB medium with gentamicin. All constructs were sequenced for confirmation.
Motility assay
The swimming motility of A. tumefaciens was assayed on semisolid agar as described previously (Merritt et al., 2007). Briefly, bacteria cells grown in LB media for 48 h at 28 °C were stabbed in 0.3% (w/v) agar ATGN modified medium plates (20 mM NaCl, 10 mM (NH4)2SO4, 5 mM MgSO4, 1 mM CaCl2, 0.16 mM KH2PO4, 0.32 mM K2HPO4, 0.01 mM FeSO4, 10 mM fructose, 10 mM sucrose, 0.03% casamino acid). Motility was evaluated and photographed after 3 days of incubation at 28 °C. The means of three replicates and standard deviation values were calculated. Statistical analysis was conducted by Student’s t‐test (P < 0.05).
Pull‐down assay
The constructs used in the pull‐down assay are listed in Table S1. Those constructs were used to transform E. coli strain BL21(DE3). The transformants were precultured overnight in 3 mL of LB medium plus specific antibiotics (Table S1) at 37 °C and 200 rpm. Then, precultured cells were diluted 1:100 in 100 mL of LB medium and grown at 37 °C until OD600nm = 0.6. The expression of the recombinant proteins was induced by addition of 1 mM IPTG to the growth medium and the induction process occurred at 30 °C and 200 rpm for 6 h. After induction, E. coli cells were pelleted by centrifugation at 2348 g and 4 °C for 10 min. Pelleted E. coli cells expressing GST‐FlgJLas protein were washed in phosphate‐buffered saline (PBS, pH 7.4) and suspended into lysis buffer (PBS, 100 µg/mL lysozyme and 40 µg/mL DNAse I) and sonicated to generate the cell lysates. After centrifugation, the cell lysates were incubated with 0.3 mL of glutathione sepharose high‐performance beads for 1 h then beads mixed with cell lysates were loaded on a 10 mL column, according to the manufacturer’s instructions (GE Healthcare, Pittsburgh, PA, USA). The beads were washed four times with PBS to remove the unbound proteins and incubated with E. coli cell lysates containing 6×HisEGFP, 6×HisFLgB or 6×HisFliE for 2 h at 4 °C. After washing three times, the proteins were eluted with PBS plus reduced glutathione (7 mg/mL). Eluted samples were mixed with 4× SDS‐PAGE loading buffer and boiled for 5 min. The controls (Inputs) and eluted samples were subjected to SDS 12% polyacrylamide gel electrophoresis and immunoblotting by using anti‐GST (1:5000) and anti‐6×His (1:3000) (Sigma, St Louis, MO, USA) antibodies followed by secondary antibodies raised against rabbit (1:10 000) (Sigma).
Las‐infected psyllids and plants
Las‐infected psyllid colonies were reared in laboratory cultures maintained on Las‐infected sweet orange trees at the University of Florida, Citrus Research and Education Center (CREC‐UF, Lake Alfred, FL, USA). Psyllid colonies were maintained in insect‐proof cages inside environmentally controlled growing chambers with a 12‐h photoperiod. Las‐infected seeds were obtained from lopsided fruits from HLB‐positive grapefruit (Citrus paradisi). Las‐infected dodder plants (Cuscuta sp.) on HLB‐positive periwinkle (Catharanthus roseus) were used for TEM assay. Plants were grown in a greenhouse with a 12‐h photoperiod and controlled temperature. Las‐positive dodder tendrils were a gift from Dr Ed Etxeberria (University of Florida, CREC, Lake Alfred, FL, USA). Las in the insects, citrus plants, seed coats and dodder plants was verified by qPCR (Hartung et al., 2010; Hilf et al., 2013; Yan et al., 2013). A sleeve net (80 mesh) was used to cage Las‐positive psyllids on young flush only. The caged plants were kept within a bigger cage to prevent interference from other psyllids.
Isolation of Las from psyllid guts and plant materials
About 80 Las‐infected adult psyllids (Ct value = 26 to 32) were collected from the psyllid room (CREC‐UF) and used to dissect midguts in sterile PBS under a dissecting stereomicroscope by using depressed glass wells and fine entomological needles. After tissue samples were dissected, the tissues were washed three times with PBS and transferred to a sterile Eppendorf tube containing 0.3% bacitracin in PBS (Ghanim et al., 2016). Samples were pipetted up and down slowly to release the bacteria cells attached to the midgut.
Selected seeds isolated from infected and symptomatic citrus plants (Ct value about 27 to 29) were washed five times in sterile water, dried in filter paper and the coats were removed by using high‐precision straight fine tweezers. Seed coats were divided into very small pieces and suspended with 0.3% bacitracin (Thermo Fisher, Waltham, MA, USA) in PBS.
Individual dodder tendrils positive for Las (Ct = 24 to 26) were removed intact from parasitized plants. A segment of 10 mm was removed from the centre of dodder stem pieces, washed five times in sterile water, dried in paper filter, transferred to 0.3% bacitracin in PBS and cut into very small pieces with razor blades. The mixtures were pipetted up and down slowly to release the bacteria. Solution containing Las was transferred to a sterile Eppendorf tube.
Transmission electron microscopy and antibody‐labelled Las cells
Bacterial cultures of L. crescens were grown at 28 ºC in BM7 medium with 5 mM putrescine (Sigma) to OD600nm of 0.5. One droplet of suspended bacteria was placed on formvar‐ and carbon‐coated 400‐mesh copper grids and allowed to settle for 1 min. The excess water was wicked away with filter paper. The samples were then stained, before drying, with 1% aqueous ammonium molybdate (Sigma) to which a few drops of bacitracin water (20 μg of bacitracin dissolved in 10 mL water) was added as a spreading agent. The stains were added to the grids and wicked away with filter paper immediately. The grids were allowed to dry for 1 h before being viewed and photographed on a Morgagni 268 transmission electron microscope (FEI Company, Eindhoven, Netherlands). Similar procedures were repeated for bacterial suspension diluted 1:5 in PBS obtained from Las‐infected psyllid midguts, grapefruit seed coats and dodder stems.
The labelling of Las bacteria absorbed in the grids was done as follows. Grids with absorbed Las cells were incubated with polyclonal antibody against Las anti‐OmpA raised in rabbit (Abcam, Cambridge, MA, USA). Rabbit antisera against Las OmpA was diluted 1:250 in PBST buffer [1x PBS, 1% (w/v) bovine serum albumen and 1% (v/v) Triton X‐100]. After 1 h incubation, the grids were washed three times for 5 min each in PBST buffer. Grids were incubated with anti‐rabbit IgG (whole molecule)‐gold (Sigma) diluted 1:5 in PBST buffer and incubated for 30 min. After incubation, the grids were washed three times in PBST buffer and twice in sterile water. Finally, grids were stained with 1% aqueous ammonium molybdate and after drying for 1 h they were visualized and photographed on a Morgagni 268 TEM (FEI Company).
Flagellin antibody and detection of flagellin via western blotting
To detect the flagellin protein in different samples, the peptide DRVSSGLRVSDAAD of Las flagellin was synthesized as an antigen to produce Las flagellin antibody from rabbits. The total proteins were isolated from A. tumefaciens strain GV2260, Las‐infected citrus and Las‐infected psyllids. Agrobacterium tumefaciens cells were grown in YEP medium to OD600 nm = 0.6. Cells were pelleted by centrifugation, resuspended in 2× SDS‐PAGE loading buffer and boiled for 5 min. Protein extracts from both Las‐infected and Las‐free psyllids (ten insects) and citrus plants (pieces of petioles) were obtained by snap freezing the samples in liquid nitrogen following maceration of the tissues in tissue Lyser (Qiagen, Germantown, MD, USA). Macerated psyllid samples were resuspended in I‐PER Insect Cell protein extraction reagent (Thermo Fisher), and macerated citrus samples were resuspended in CelLytic P Cell lysis reagent (Sigma). Resuspended samples were centrifuged at 13 523 g for 15 min at 4 °C, then the supernatants were diluted in 2× SDS‐PAGE loading buffer. Ten microlitres of protein of each sample was loaded for 10% SDS‐PAGE electrophoresis and western blot analysis. Full‐length flagellin fused with a maltose‐binding protein (MBP) tag was used as a positive control. Las‐free citrus and psyllids, and free MBP protein were used as negative controls.
RT‐qPCR analysis of Las flagellar genes
Comparative analysis of expression of the selected flagellum genes encoded by the Las genome was verified in Las‐infected plants of Valencia sweet orange (C. sinensis) and in Las‐infected psyllids as described previously (Yan et al., 2013). All RT‐qPCRs were performed using a 7500 Fast Real‐time PCR system (Applied Biosystems, Foster City, CA, USA) with a QuantiTect SYBR Green RT‐PCR kit (Qiagen). The primers were designed from the sequence of the Las genome using IDT SciTolls (http://www.idtdna.com/pages/scitools). The total reaction volume of one‐step RT‐qPCR was 25 μL and contained 2 × QuantiTect SYBR Green RT‐PCR Master Mix (12.5 μL), 10 μM gene‐specific primers (1.25 μL), QuantiTect RT Mix (0.5 μL) and 50 ng of RNA template (1 μL). 16S rRNA was used as the endogenous control. Reactions were incubated at 50 °C for 30 min and at 95 °C for 15 min, then cycled (40 times) at 94 °C for 15 s, 54–56 °C for 30 s and 72 °C for 30 s. Melting curve analysis was conducted to verify the specificity of the RT‐qPCR products. Two technical replicates and three biological replicates were used for each of the genes. The relative fold change in target gene expression was calculated using the formula 2− ΔΔCt (Livak and Schmittgen, 2001). Statistical analysis of all data was conducted by Student’s t‐test.
Supporting information
Fig. S1 Sequence alignment of the peptidoglycan hydrolase encoded by ‘Candidatus Liberibacter’ species was generated by ClustalW. The signal peptide sequence found by PrediSi is highlighted in yellow. The SLT domain is indicated in green. CLas, ‘Ca. Liberibacter asiaticus’ (CLIBASIA_00965); CLaf, ‘Ca. Liberibacter africanus’ (G293_01215); CLam, ‘Ca. Liberibacter americanus’ (Lam_376); CLso, ‘Ca. Liberibacter solanacearum’ (CKC_02595); Lcc, Liberibacter crescens BT‐1 (B488_10490).
Fig. S2 Swimming motility assay of the Agrobacterium tumefaciens wild‐type, ΔflaAD and ΔflaABCD mutant strains, and the mutant strains complemented with ‘Candidatus Liberibacter asiaticus’ (Las) flaA. The mean values ± the standard deviations (n = 3) are plotted. Mean values were compared to the wild‐type, * indicate statistically significant difference (P < 0.05, Student t test). WT, A. tumefaciens wild‐type strain carrying the empty vector. ΔflaAD and ΔflaABCD, A. tumefaciens mutant strains carrying the empty vector. ΔflaAD + flaALas and ΔflaABCD + flaALas, mutant strains carrying the Las flaA gene.
Table S1 Bacterial strains and plasmids used in this study.
Table S2 Oligonucleotides used in this study.
Acknowledgements
This work has been supported by Florida Citrus Initiative Program, Citrus Research and Development Foundation, and the USDA MAC program. We thank Dr Ed Etxeberria for the Las‐positive dodder materials. This project was also supported by the National Key Research and Development Program of China (2018YFD0201500) to T.S.Y.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
- Altegoer, F. , Schuhmacher, J. , Pausch, P. and Bange, G. (2014) From molecular evolution to biobricks and synthetic modules: a lesson by the bacterial flagellum. Biotechnol. Genet. Eng. Rev. 30, 49–64. [DOI] [PubMed] [Google Scholar]
- Anderson, J.K. , Smith, T.G. and Hoover, T.R. (2010) Sense and sensibility: flagellum‐mediated gene regulation. Trends Microbiol. 18, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M. and Wang, N. (2019) The Tad pilus apparatus of ‘Candidatus Liberibacter asiaticus’ and its regulation by VisNR. Mol. Plant–Microbe Interact. 32, 1175–1187. [DOI] [PubMed] [Google Scholar]
- Andrade, M.O. , Farah, C.S. and Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5′ UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas . PLoS Pathog. 10, e1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arp, A.P. , Martini, X. and Pelz‐Stelinski, K.S. (2017) Innate immune system capabilities of the Asian citrus psyllid, Diaphorina citri . J. Invertebr. Pathol. 148, 94–101. [DOI] [PubMed] [Google Scholar]
- Attmannspacher, U. , Scharf, B. and Schmitt, R. (2005) Control of speed modulation (chemokinesis) in the unidirectional rotary motor of Sinorhizobium meliloti . Mol. Microbiol. 56, 708–718. [DOI] [PubMed] [Google Scholar]
- Berry, R.M. and Armitage, J.P. (1999) The bacterial flagella motor. Adv. Microb. Physiol. 41, 291–337. [DOI] [PubMed] [Google Scholar]
- Blaustein, R.A. , Lorca, G.L. and Teplitski, M. (2018) Challenges for managing ‘Candidatus Liberibacter’ spp. (huanglongbing disease pathogen): current control measures and future directions. Phytopathology, 108, 424–435. [DOI] [PubMed] [Google Scholar]
- Bové, J.M. (2006) Huanglongbing: a destructive, newly‐emerging, century‐old disease of citrus. J. Plant Pathol. 88, 7–37. [Google Scholar]
- Chatnaparat, T. , Prathuangwong, S. and Lindow, S.E. (2016) Global pattern of gene expression of Xanthomonas axonopodis pv. glycines within soybean leaves. Mol. Plant–Microbe Interact. 29, 508–522. [DOI] [PubMed] [Google Scholar]
- Cheng, F. , Ma, A. , Zhuang, G. and Fray, R.G. (2018) Exogenous N‐acyl‐homoserine lactones enhance the expression of flagella of Pseudomonas syringae and activate defence responses in plants. Mol. Plant Pathol. 19, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin, W.J. , Parker, V.E. , Wright, E.L. , Ashcroft, K.J. , Loake, G.J. and Shaw, C.H. (1999) Agrobacterium tumefaciens possesses a fourth flagelin gene located in a large gene cluster concerned with flagellar structure, assembly and motility. Microbiology, 145, 1397–1407. [DOI] [PubMed] [Google Scholar]
- DeRosier, D. (2006) Bacterial flagellum: visualizing the complete machine in situ . Curr. Biol. 16, R928–R930. [DOI] [PubMed] [Google Scholar]
- Ding, F. , Allen, V. , Luo, W. , Zhang, S. and Duan, Y. (2018) Molecular mechanisms underlying heat or tetracycline treatments for citrus HLB control. Hortic. Res. 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driks, A. , Bryan, R. , Shapiro, L. and DeRosier, D.J. (1989) The organization of the Caulobacter crescentus flagellar filament. J. Mol. Biol. 206, 627–636. [DOI] [PubMed] [Google Scholar]
- Duan, Y. , Zhou, L. , Hall, D.G. , Li, W. , Doddapaneni, H. , Lin, H. , Liu, L. , Vahling, C.M. , Gabriel, D.W. , Williams, K.P. , Dickerman, A. , Sun, Y. and Gottwald, T. (2009) Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plant–Microbe Interact. 22, 1011–1020. [DOI] [PubMed] [Google Scholar]
- Eggenhofer, E. , Rachel, R. , Haslbeck, M. and Scharf, B. (2006) MotD of Sinorhizobium meliloti and related alpha‐proteobacteria is the flagellar‐hook‐length regulator and therefore reassigned as FliK. J. Bacteriol. 188, 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt, M. , Hirano, T. , Su, Y. , Paul, K. , Wee, D.H. , Mizuno, S. , Aizawa, S. and Hughes, K.T. (2010) The role of the FliK molecular ruler in hook‐length control in Salmonella enterica . Mol. Microbiol. 75, 1272–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L.D. , Stafford, G.P. , Ahmed, S. , Fraser, G.M. and Hughes, C. (2006) An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. USA. 103, 17474–17479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L.D. , Hughes, C. and Fraser, G.M. (2014) Building a flagellum outside the bacterial cell. Trends Microbiol. 22, 566–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov, O.V. and Kostyukova, A.S. (1984) Domain structure of flagellin. FEBS Lett. 171, 145–148. [DOI] [PubMed] [Google Scholar]
- De Feyter, R. , Yang, Y. and Gabriel, D.W. (1993) Gene‐for‐genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant–Microbe Interact. 6, 225–237. [DOI] [PubMed] [Google Scholar]
- Folimonova, S.Y. and Achor, D.S. (2010) Early events of citrus greening (huanglongbing) disease development at the ultrastructural level. Phytopathology, 100, 949–958. [DOI] [PubMed] [Google Scholar]
- Folimonova, S.Y. , Robertson, C.J. , Garnsey, S.M. , Gowda, S. and Dawson, W.O. (2009) Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology, 99, 1346–1354. [DOI] [PubMed] [Google Scholar]
- Franchi, L. , Amer, A. , Body‐Malapel, M. , Kanneganti, T.D. , Ozören, N. , Jagirdar, R. , Inohara, N. , Vandenabeele, P. , Bertin, J. , Coyle, A. , Grant, E.P. and Núñez, G. (2006) Cytosolic flagellin requires Ipaf for activation of caspase‐1 and interleukin 1beta in salmonella‐infected macrophages. Nat. Immunol. 7, 576–582. [DOI] [PubMed] [Google Scholar]
- Ghanim, M. , Fattah‐Hosseini, S. , Levy, A. and Cilia, M. (2016) Morphological abnormalities and cell death in the Asian citrus psyllid (Diaphorina citri) midgut associated with ‘Candidatus Liberibacter asiaticus’. Sci. Rep. 6, 33418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim, M. , Achor, D. , Ghosh, S. , Kontsedalov, S. , Lebedev, G. and Levy, A. (2017) ‘Candidatus Liberibacter asiaticus’ accumulates inside endoplasmic reticulum associated vacuoles in the gut cells of Diaphorina citri . Sci. Rep. 7, 16945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, P. (2004) Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68, 771–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez‐Gómez, L. and Boller, T. (2002) Flagellin perception: a paradigm for innate immunity. Trends Plant Sci. 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Guerry, P. , Logan, S.M. , Thornton, S. and Trust, T.J. (1990) Genomic organization and expression of Campylobacter flagellin genes. J. Bacteriol. 172, 1853–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, D.G. , Gottwald, T.R. , Stover, E. and Beattie, G.A.C. (2013) Evaluation of management programs for protecting young citrus plantings from huanglongbing. HortScience, 48, 330–337. [Google Scholar]
- Hall, D.G. , Albrecht, U. and Bowman, K.D. (2016) Transmission rates of ‘Ca. Liberibacter asiaticus’ by Asian citrus psyllid are enhanced by the presence and developmental stage of citrus flush. J. Econ. Entomol. 109, 558–563. [DOI] [PubMed] [Google Scholar]
- Hartung, J.S. , Paul, C. , Achor, D. and Brlansky, R.H. (2010) Colonization of dodder, Cuscuta indecora, by ‘Candidatus Liberibacter asiaticus’ and ‘Ca. L. americanus’. Phytopathology, 100, 756–762. [DOI] [PubMed] [Google Scholar]
- Heindl, J.E. , Wang, Y. , Heckel, B.C. , Mohari, B. , Feirer, N. and Fuqua, C. (2014) Mechanisms and regulation of surface interactions and biofilm formation in Agrobacterium . Front. Plant Sci. 5, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlihey, F.A. , Moynihan, P.J. and Clarke, A.J. (2014) The essential protein for bacterial flagella formation FlgJ functions as a β‐N‐acetylglucosaminidase. J. Biol. Chem. 289, 31029–31042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing, M.E. and Fuqua, C. (2011) Antiparallel and interlinked control of cellular iron levels by the Irr and RirA regulators of Agrobacterium tumefaciens . J. Bacteriol. 193, 3461–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf, M.E. , Sims, K.R. , Folimonova, S.Y. and Achor, D.S. (2013) Visualization of ‘Candidatus Liberibacter asiaticus’ cells in the vascular bundle of citrus seed coats with fluorescence in situ hybridization and transmission electron microscopy. Phytopathology, 103, 545–554. [DOI] [PubMed] [Google Scholar]
- Hirano, T. , Minamino, T. and Macnab, R.M. (2001) The role in flagellar rod assembly of the N‐terminal domain of Salmonella FlgJ, a flagellum‐specific muramidase. J. Mol. Biol. 312, 359–369. [DOI] [PubMed] [Google Scholar]
- Ikeda, T. , Oosawa, K. and Hotani, H. (1996) Self‐assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J. Mol. Biol. 259, 679–686. [DOI] [PubMed] [Google Scholar]
- Jain, M. , Munoz‐Bodnar, A. and Gabriel, D.W. (2019) ‘Candidatus Liberibacter asiaticus’ peroxiredoxin (LasBCP) suppresses oxylipin‐mediated defense signaling in citrus. J. Plant Physiol. 236, 61–65. [DOI] [PubMed] [Google Scholar]
- Jones, C.J. and Aizawa, S. (1991) The bacterial flagellum and flagellar motor: structure, assembly and function. Adv. Microb. Physiol. 32, 109–172. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K. and McCarter, L.L. (2000) Analysis of the polar flagellar gene system of Vibrio parahaemolyticus . J. Bacteriol. 182, 3693–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.S. , Sagaram, U.S. , Burns, J.K. , Li, J.L. and Wang, N. (2009) Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: microscopy and microarray analyses. Phytopathology, 99, 50–57. [DOI] [PubMed] [Google Scholar]
- Lee, J.A. , Halbert, S.E. , Dawson, W.O. , Robertson, C.J. , Keesling, J.E. and Singer, B.H. (2015) Asymptomatic spread of huanglongbing and implications for disease control. Proc. Natl. Acad. Sci. USA. 112, 7605–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Trivedi, P. and Wang, N. (2016) Field evaluation of plant defense inducers for the control of citrus huanglongbing. Phytopathology, 106, 37–46. [DOI] [PubMed] [Google Scholar]
- Li, J. , Pang, Z. , Trivedi, P. , Zhou, X. , Ying, X. , Jia, H. and Wang, N. (2017) ‘Candidatus Liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol. Plant–Microbe Interact. 30, 620–630. [DOI] [PubMed] [Google Scholar]
- Li, J. , Li, L. , Pang, Z. , Kolbasov, V. , Ehsani, R. , Carter, E. and Wang, N. (2018) Developing citrus huanglongbing management strategies based on the severity of symptoms in HLB‐endemic citrus‐producing regions. Phytopathology. 109, 582–592. [DOI] [PubMed] [Google Scholar]
- Li, J. , Pang, Z. , Duan, S. , Lee, D. , Kolbasov, V. and Wang, N. (2019) The in planta effective concentration of oxytetracycline against ‘Candidatus Liberibacter asiaticus’ for suppression of citrus huanglongbing. Phytopathology. 10.1094/PHYTO-06-19-0198-R. [DOI] [PubMed] [Google Scholar]
- Liefting, L.W. , Sutherland, P.W. , Ward, L.I. , Paice, K.L. , Weir, B.S. and Clover, G.R.G. (2009) A new ‘Candidatus Liberibacter’ species associated with diseases of solanaceous crops. Plant Dis. 93, 208–214. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(−ΔΔC(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Macnab, R.M. (2003) How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100. [DOI] [PubMed] [Google Scholar]
- Markel, E. , Stodghill, P. , Bao, Z. , Myers, C.R. and Swingle, B. (2016) AlgU controls expression of virulence genes in Pseudomonas syringae pv. tomato DC3000. J. Bacteriol. 198, 2330–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt, P.M. , Danhorn, T. and Fuqua, C. (2007) Motility and chemotaxis in Agrobacterium tumefaciens surface attachment and biofilm formation. J. Bacteriol. 189, 8005–8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino, T. and Namba, K. (2004) Self‐assembly and type III protein export of the bacterial flagellum. J. Mol. Microbiol. Biotechnol. 7, 5–17. [DOI] [PubMed] [Google Scholar]
- Minamino, T. , Imada, K. and Namba, K. (2008) Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18, 693–701. [DOI] [PubMed] [Google Scholar]
- Minamino, T. , Moriya, N. , Hirano, T. , Hughes, K.T. and Namba, K. (2009) Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol. Microbiol. 74, 239–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens, S. and Vanderleyden, J. (1996) Functions of bacterial flagella. Crit. Rev. Microbiol. 22, 67–100. [DOI] [PubMed] [Google Scholar]
- Mohari, B. , Thompson, M.A. , Trinidad, J.C. , Setayeshgar, S. , and Fuqua, C. (2018) Multiple flagellin proteins have distinct and synergistic roles in Agrobacterium tumefaciens motility. J. Bacteriol. 200:e00327-18 10.1128/JB.00327-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu, T. , Minamino, T. , Macnab, R.M. and Kutsukake, K. (1999) Peptidoglycan‐hydrolyzing activity of the FlgJ protein, essential for flagellar rod formation in Salmonella typhimurium . J. Bacteriol. 181, 1555–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambu, T. , Inagaki, Y. and Kutsukake, K. (2006) Plasticity of the domain structure in FlgJ, a bacterial protein involved in flagellar rod formation. Genes Genet. Syst. 81, 381–389. [DOI] [PubMed] [Google Scholar]
- Naranjo, E. , Merfa, M.V. , Ferreira, V. , Jain, M. , Davis, M.J. , Bahar, O. , Gabriel, D.W. and De La Fuente, L. (2019) Liberibacter crescens biofilm formation in vitro: establishment of a model system for pathogenic ‘Candidatus Liberibacter’ spp. Sci. Rep. 9, 5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, K.L. , Almeida, R.P. , Purcell, A.H. and Lindow, S.E. (2003) Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera . Appl. Environ. Microbiol. 69, 7319–7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, S.S. and Wang, N. (2019) Targeted early detection of citrus HLB causal agent ‘Candidatus Liberibacter asiaticus’ before symptom expression. Phytopathology. 109, 952–959. [DOI] [PubMed] [Google Scholar]
- Park, S.Y. , Lowder, B. , Bilwes, A.M. , Blair, D.F. and Crane, B.R. (2006) Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. USA. 103, 11886–11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelz‐Stelinski, K.S. , Brlansky, R.H. , Ebert, T.A. and Rogers, M.E. (2010) Transmission parameters for ‘Candidatus liberibacter asiaticus’ by Asian citrus psyllid (Hemiptera: Psyllidae). J. Econ. Entomol. 103, 1531–1541. [DOI] [PubMed] [Google Scholar]
- Rossez, Y. , Wolfson, E.B. , Holmes, A. , Gally, D.L. and Holden, N.J. (2015) Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 11, e1004483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. , Russell, D.W. , Rudolph, G. Schmieder Fund (2006) The Condensed Protocols from Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Scharf, B. , Schuster‐Wolff‐Buhring, H. , Rachel, R. and Schmitt, R. (2001) Mutational analysis of the Rhizobium lupini H13–3 and Sinorhizobium meliloti flagellin genes: importance of flagellin A for flagellar filament structure and transcriptional regulation. J. Bacteriol. 183, 5334–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secor, G.A. , Rivera, V.V. , Abad, J.A. , Lee, I.M. , Clover, G.R.G. , Liefting, L.W. , Li, X. and De Boer, S.H. (2009) Association of ‘Candidatus Liberibacter solanacearum’ with zebra chip disease of potato established by graft and psyllid transmission, electron microscopy, and PCR. Plant Dis. 93, 574–583. [DOI] [PubMed] [Google Scholar]
- Setamou, M. , Alabi, O.J. , Kunta, M. , Jifon, J.L. and da Graca, J.V. (2016) Enhanced acquisition rates of ‘Candidatus Liberibacter asiaticus’ by the Asian citrus psyllid (Hemiptera: Liviidae) in the presence of vegetative flush growth in citrus. J. Econ. Entomol. 109, 1973–1978. [DOI] [PubMed] [Google Scholar]
- Sétamou, M. and Bartels, D.W. (2015) Living on the edges: spatial niche occupation of Asian citrus psyllid, Diaphorina citri Kuwayama (Hemiptera: Liviidae), in citrus groves. PLoS ONE, 10, e0131917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, B.J. and McCarter, L.L. (2003) Lateral flagellar gene system of Vibrio parahaemolyticus . J. Bacteriol. 185, 4508–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatineni, S. , Sagaram, U.S. , Gowda, S. , Robertson, C.J. , Dawson, W.O. , Iwanami, T. and Wang, N. (2008) In planta distribution of ‘Candidatus Liberibacter asiaticus’ as revealed by polymerase chain reaction (PCR) and real‐time PCR. Phytopathology, 98, 592–599. [DOI] [PubMed] [Google Scholar]
- Thomashow, L.S. and Rittenberg, S.C. (1985) Waveform analysis and structure of flagella and basal complexes from Bdellovibrio bacteriovorus 109J. J. Bacteriol. 163, 1038–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P. , Nurnberger, T. and Joosten, M.H. (2011) Of PAMPs and effectors: the blurred PTI‐ETI dichotomy. Plant Cell, 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaseto, A. , Krugner, R. and Lopes, J. (2016) Effect of plant barriers and citrus leaf age on dispersal 639 of Diaphorina citri (Hemiptera: Liviidae). J. Appl. Entomol. 140, 91–102. [Google Scholar]
- Turner, L. , Stern, A.S. and Berg, H.C. (2012) Growth of flagellar filaments of Escherichia coli is independent of filament length. J. Bacteriol. 194, 2437–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukuda‐Hosokawa, R. , Sadoyama, Y. , Kishaba, M. , Kuriwada, T. , Anbutsu, H. and Fukatsu, T. (2015) Infection density dynamics of the citrus greening bacterium ‘Candidatus Liberibacter asiaticus’ in field populations of the psyllid Diaphorina citri and its relevance to the efficiency of pathogen transmission to citrus plants. Appl. Environ. Microbiol. 81, 3728–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahling‐Armstrong, C.M. , Zhou, H. , Benyon, L. , Morgan, J.K. and Duan, Y. (2012) Two plant bacteria, S. meliloti and ‘Ca. Liberibacter asiaticus’, share functional znuABC homologues that encode for a high affinity zinc uptake system. PLoS ONE, 7, e37340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, C. , Pierre, M. , Li, J. and Wang, N. (2019) Implications of heat treatment and systemic delivery of foliar‐applied oxytetracycline on citrus physiological management and therapy delivery. Front. Plant Sci. 10, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, N. (2019) The citrus huanglongbing crisis and potential solutions. Mol. Plant, 12, 607–609. [DOI] [PubMed] [Google Scholar]
- Wang, N. and Trivedi, P. (2013) Citrus huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology, 103, 652–665. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Pierson, E.A. , Setubal, J.C. , Xu, J. , Levy, J.G. , Zhang, Y. , Li, J. , Rangel, L.T. and Martins, J. (2017) The Candidatus Liberibacter‐host interface: insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 55, 451–482. [DOI] [PubMed] [Google Scholar]
- Waters, R.C. , O’Toole, P.W. and Ryan, K.A. (2007) The FliK protein and flagellar hook‐length control. Protein Sci. 16, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulff, N.A. , Zhang, S. , Setubal, J.C. , Almeida, N.F. , Martins, E.C. , Harakava, R. , Kumar, D. , Rangel, L.T. , Foissac, X. , Bove, J.M. and Gabriel, D.W. (2014) The complete genome sequence of ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing. Mol. Plant–Microbe Interact. 27, 163–176. [DOI] [PubMed] [Google Scholar]
- Yan, Q. , Sreedharan, A. , Wei, S. , Wang, J. , Pelz‐Stelinski, K. , Folimonova, S. and Wang, N. (2013) Global gene expression changes in ‘Candidatus Liberibacter asiaticus’ during the transmission in distinct hosts between plant and insect. Mol. Plant Pathol. 14, 391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S.I. , Kurnasov, O. , Natarajan, V. , Hong, M. , Gudkov, A.V. , Osterman, A.L. and Wilson, I.A. (2012) Structural basis of TLR5‐flagellin recognition and signaling. Science, 335, 859–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Lund, S.P. , Scott, R.A. , Greenwald, J.W. , Records, A.H. , Nettleton, D. , Lindow, S.E. , Gross, D.C. and Beattie, G.A. (2013) Transcriptional responses of Pseudomonas syringae to growth in epiphytic versus apoplastic leaf sites. Proc. Natl. Acad. Sci. USA. 110, E425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Tong, B.A. , Liu, J. and Li, C. (2012) A single‐domain FlgJ contributes to flagellar hook and filament formation in the Lyme disease spirochete Borrelia burgdorferi . J. Bacteriol. 194, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. , Kojima, S. and Homma, M. (2013) Structure, gene regulation and environmental response of flagella in Vibrio . Front. Microbiol. 4, 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequence alignment of the peptidoglycan hydrolase encoded by ‘Candidatus Liberibacter’ species was generated by ClustalW. The signal peptide sequence found by PrediSi is highlighted in yellow. The SLT domain is indicated in green. CLas, ‘Ca. Liberibacter asiaticus’ (CLIBASIA_00965); CLaf, ‘Ca. Liberibacter africanus’ (G293_01215); CLam, ‘Ca. Liberibacter americanus’ (Lam_376); CLso, ‘Ca. Liberibacter solanacearum’ (CKC_02595); Lcc, Liberibacter crescens BT‐1 (B488_10490).
Fig. S2 Swimming motility assay of the Agrobacterium tumefaciens wild‐type, ΔflaAD and ΔflaABCD mutant strains, and the mutant strains complemented with ‘Candidatus Liberibacter asiaticus’ (Las) flaA. The mean values ± the standard deviations (n = 3) are plotted. Mean values were compared to the wild‐type, * indicate statistically significant difference (P < 0.05, Student t test). WT, A. tumefaciens wild‐type strain carrying the empty vector. ΔflaAD and ΔflaABCD, A. tumefaciens mutant strains carrying the empty vector. ΔflaAD + flaALas and ΔflaABCD + flaALas, mutant strains carrying the Las flaA gene.
Table S1 Bacterial strains and plasmids used in this study.
Table S2 Oligonucleotides used in this study.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.


