Summary
Plants have evolved powerful immune systems to recognize pathogens and avoid invasions, but the genetic basis of plant susceptibility is less well‐studied, especially to oomycetes, which cause disastrous diseases in many ornamental plants and food crops. In this research, we identified a negative regulator of plant immunity to the oomycete Phytophthora parasitica, AtRTP5 (Arabidopsis thaliana Resistant to Phytophthora 5), which encodes a WD40 repeat domain‐containing protein. The AtRTP5 protein, which was tagged with green fluorescent protein (GFP), is localized in the nucleus and plasma membrane. Both the A. thaliana T‐DNA insertion rtp5 mutants and the Nicotiana benthamiana RTP5 (NbRTP5) silencing plants showed enhanced resistance to P. parasitica, while overexpression of AtRTP5 rendered plants more susceptible. The transcriptomic analysis showed that mutation of AtRTP5 suppressed the biosynthesis of endogenous jasmonic acid (JA) and JA‐dependent responses. In contrast, salicylic acid (SA) biosynthesis and SA‐dependent responses were activated in the T‐DNA insertion mutant rtp5‐3. These results show that AtRTP5 acts as a conserved negative regulator of plant immunity to Phytophthora pathogens by interfering with JA and SA signalling pathways.
Keywords: negative regulator, oomycetes, Phytophthora, plant hormone, plant immunity
Introduction
A class of proteins in plants has been uncovered to act as negative regulators of plant immunity by promoting infection and facilitating compatible interactions to evade plant immunity (Boevink et al., 2016; He et al., 2018). These proteins participate in plant immunity by interfering with different signalling pathways. The leucine‐rich repeat receptor‐like kinase (LRR‐RLK) BIR2 acts as a negative regulator of pathogen‐associated molecular pattern (PAMP) triggered immunity by preventing the interaction of LRR‐RLK BAK1 with its ligand‐binding protein flagellin sensing 2 (FLS2) (Halter et al., 2014). The bir2 mutants showed enhanced cell death in the attendance of PAMP and more resistance to bacterial pathogens (Halter et al., 2014). Some well‐studied negative regulators are also recruited by eukaryotic microorganism effectors, such as RXLR effectors secreted by Phytophthora pathogens, which cause destructive diseases resulting in enormous losses in agriculture and the economy (Jiang and Tyler, 2012; Wu et al., 2014). The plant K‐homology (KH) RNA‐binding protein, KRBP1, is recruited by the Phytophthora infestans RXLR effector Pi04089. Overexpression of KRBP1 enhances the colonization of P. infestans, while KRBP1 protein mutants at KH domains fail to interact with RNA and lose the function of promoting infection (Wang et al., 2015). In recent years, some other susceptibility (S) genes have been identified as negative regulators of plant immunity by modifying plant hormone networks. Arabidopsis RTP1 encodes a novel endoplasmic reticulum (ER)‐localized protein and negatively regulates Arabidopsis thaliana resistance to biotrophic pathogens, possibly by affecting the transcription of salicylic acid (SA) responsive genes (Pan et al., 2016).
Plant hormones play important roles in response to various abiotic or biotic stresses and in regulating the balance between growth and resistance. Jasmonic acid (JA) and SA are two hormones that participate in modulating the plant immunity response to different stimuli (Alazem and Lin, 2015). JA is required as a defence signal against necrotrophic pathogens and herbivorous insects, while SA often renders plants more resistant to biotrophic and hemibiotrophic pathogens (Cui et al., 2010; Pan et al., 2016). Activation or suppression of the JA and SA signalling pathways generally trigger expression of pathogenesis‐related (PR) genes harbouring a number of enzymatic and biological properties (Takahashi et al., 2004). Antagonistic cross‐talk between the JA and SA pathways has been identified in most studies. The bacterial pathogen Pseudomonas syringae pv. tomato (Pst) DC3000 uses the virulence factor coronatine (COR) to repress the expression of the key SA biosynthetic enzyme ICS1 (isochorismate synthase 1) and a methyltransferase BSMT1 by activating the transcription of NAC‐type TFs (Attaran et al., 2014). Two type III effector genes of P. syringae, HopZ1 and HopX1, which respectively encode a putative acetyltransferase and a cysteine protease, increase pathogenicity by promoting the COI1‐dependent degradation of jasmonate‐ZIM‐domain (JAZ) proteins (Gimenez‐Ibanez et al., 2014; Jiang et al., 2013).
Identification of the negative regulators in host plants is essential to understand the pathogenic mechanisms and to provide a possibility for broad‐spectrum, durable resistance breeding. However, little is known about the genetic basis of plant susceptibility to pathogens, especially the pathogenic oomycetes, or about the underlying mechanisms by which the plant factors participate in immune signalling pathways. In this report, we identified a new negative regulator of plant immunity in A. thaliana to Phytophthora parasitica, AtRTP5 (At5G43930), which encodes a WD40‐repeat domain‐containing protein. Both rtp5 mutants and the NbRTP5‐silencing plants showed enhanced resistance to Phytophthora pathogens compared to the control plants, while overexpression of AtRTP5 promoted the colonization of P. parasitica and P. infestans in the host plants, suggesting it plays a conserved role in regulating plant immunity. Whole‐genome transcription analysis and qRT‐PCR results suggested that AtRTP5 regulated the accumulation of endogenous JA and SA and downstream responses to JA and SA.
Results
T‐DNA insertion rtp5 mutants showed enhanced resistance to P. parasitica
Using the P. parasitica–A. thaliana compatible interaction system (Wang et al., 2011b), A. thaliana T‐DNA insertion mutants (Pan et al., 2016; Zuo et al., 2000) were screened for enhanced resistance to P. parasitica. The insertion sites of the stably resistant mutants were identified by whole‐genome resequencing using uninfected detached rosette leaves of 4‐week‐old A. thaliana. This led to the identification of AtRTP5 (Arabidopsis thaliana Resistant to P. parasitica 5). In the AtRTP5 protein sequence, a nuclear localization sequence (NLS) was predicted by cNLS Mapper (score > 5) and a WD40 repeat domain was predicted by the Conserved Domain Search Service (default parameters) of the National Center for Biotechnology Information (NCBI) (Fig. 1a). The phylogenetic tree was constructed in MEGA X with the neighbour‐joining method based on the alignment between the AtRTP5 protein sequence and its homologues in plants (Fig. S1 and Table S1). The results showed that AtRTP5 was ubiquitous in land plants (Fig. 1b).
Figure 1.
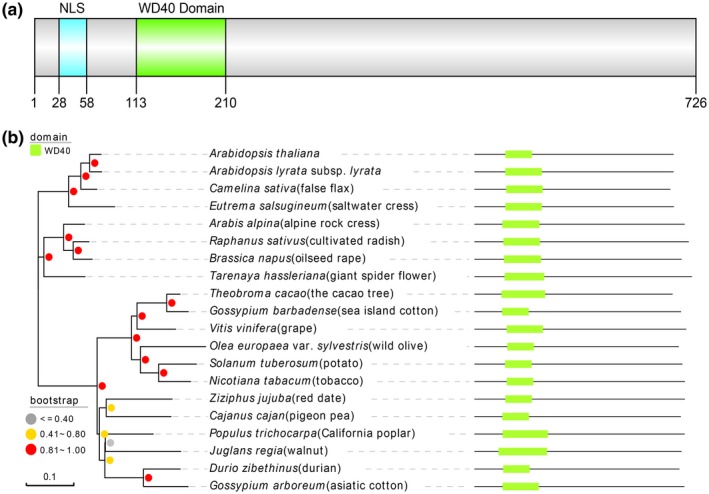
AtRTP5 encodes a WD40 domain‐containing protein and is ubiquitous in land plants. (a) Diagram of conserved domains in AtRTP5 protein sequence. (b) Phylogeny of AtRTP5 homologues in parts of plant species and their corresponding domain architecture. The evolutionary analysis was constructed in MEGA X with the neighbour‐joining method. The grey, yellow and red dots represent the percentages of the associated taxa clustered together in replicate trees in the bootstrap test (1000 replicates).
We obtained two T‐DNA insertion mutants of AtRTP5 from ABRC, rtp5‐2 (SALK_079027C) and rtp5‐3 (SALK_112605C). The T‐DNA insertion site of rtp5‐2 is located in the promoter region close to the ATG start codon and the T‐DNA insertion site of rtp5‐3 is located in the eighth exon (Fig. 2a). The AtRTP5 expression level is decreased by 40% in rtp5‐2 and nearly 100% in rtp5‐3 (Fig. 2b). We inoculated the detached rosette leaves of 4‐week‐old A. thaliana and roots of 2‐week‐old seedlings with P. parasitica zoospores. The detached leaves from the two T‐DNA mutants did not have obvious water‐soaked lesions at 3 days post‐inoculation (dpi), as did the detached leaves from wild‐type A. thaliana Col‐0 (Fig. 2c,d). The biomass measurement result and plant disease severity assessment showed fewer hyphae in rtp5‐2 and rtp5‐3 around the infection sites compared to Col‐0 (Fig. 2e,f). The seedling roots inoculation assay showed similar results. Twelve days after inoculation, while almost all of the Col‐0 seedlings collapsed, the rtp5‐2 and rtp5‐3 inoculated seedlings sprouted new leaves (Fig. S2a). The disease severity assessment also suggested seedlings of rtp5‐2 and rtp5‐3 were more resistant (Fig. S2b). These results imply that AtRTP5 is a negative regulator of plant immunity during the A. thaliana–P. parasitica compatible interaction.
Figure 2.
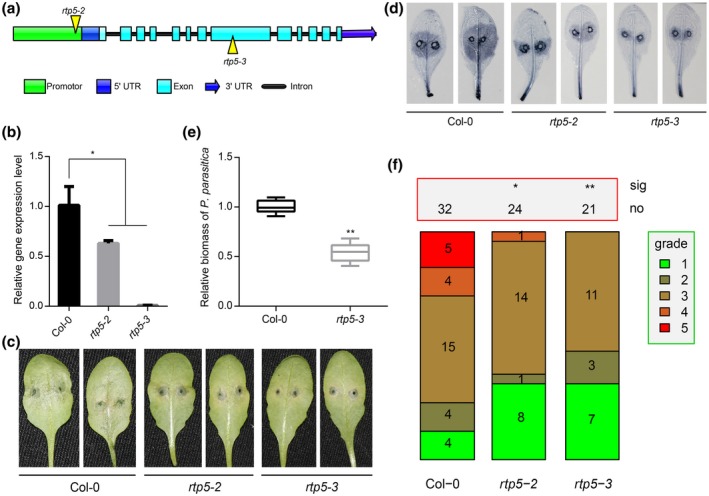
Arabidopsis thaliana T‐DNA insertion rtp5 mutants showed enhanced resistance to Phytophthora parasitica. (a) Diagram of the T‐DNA insertion sites in rtp5‐2 and rtp5‐3. (b) The qRT‐PCR results indicate that the expression level of AtRTP5 decreased in the T‐DNA insertion mutants. The relative expression levels of AtRTP5 were normalized to AtUBC9. Statistical analysis was determined by one‐way ANOVA followed by Tukey’s multiple comparison tests. (c) Leaf inoculation assay illustrated that the T‐DNA insertion mutant plants were resistant to P. parasitica, scored 3 days post‐inoculation (dpi). More than ten leaves from ten different plants were tested in each experiment, and three independent experiments were performed. (d) Inoculated leaves at 3 dpi were stained with trypan blue. (e) Relative DNA ratio of P. parasitica compared to A. thaliana at 3 dpi (PPTG_09948/AtUBC9) was determined by qPCR. Statistical analysis was determined by Student’s t‐test. (f) The plant disease severity assessment of the infected leaves shown in (c) and (d). All infected leaves were divided into five grades depending on the pathogen colonization area ratio. Grade 1, 0–10%; Grade 2, 10–30%; Grade 3, 30–50%; Grade 4, 50–80%; Grade 5, >80%. Error bars in (b) and (e) indicate SD of three biological replicates. All asterisks indicate significant differences (*P < 0.05, **P < 0.01).
Overexpression of AtRTP5 accelerated infection of Phytophthora pathogens
Transgenic plants overexpressing AtRTP5 in the Col‐0 background showed no obvious changes in development phenotypes (Fig. 3a). qRT‐PCR results confirmed the high transcription level of AtRTP5 in two homozygous overexpression lines, RTP5OE2 and RTP5OE9 (Fig. 3b). Detached rosette leaves from the 4‐week‐old RTP5OE2 and RTP5OE9 plants challenged with P. parasitica zoospores showed that growth of P. parasitica was greatly enhanced in RTP5OE2 and RTP5OE9 lines relative to Col‐0 (Fig. 3c,d). The inoculated leaves of RTP5OE2 and RTP5OE9 lines were more susceptible than Col‐0 (Fig. 3e). We validated these results by infiltrating Agrobacterium harbouring pART27::AtRTP5CFlag in N. benthamiana leaves and inoculating P. parasitica zoospores 2 days later. The lesion diameters measured at 2 to 3 dpi showed that AtRTP5 promoted P. parasitica infection (Fig. 3f).
Figure 3.
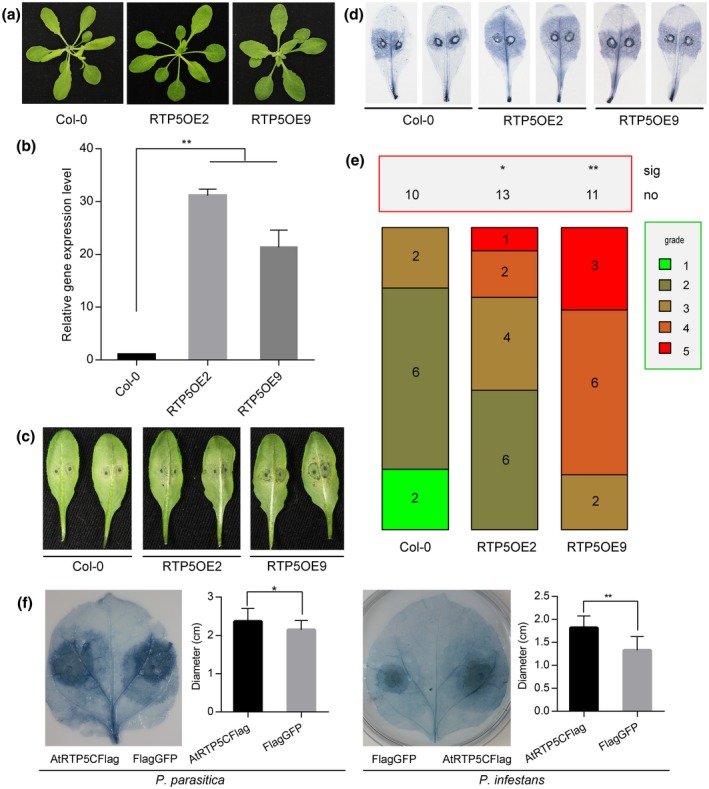
Overexpression of AtRTP5 in host plants accelerated infection of Phytophthora parasitica and P. infestans. (a) Col‐0 phenotype has no differences compared with RTP2OE2 and RTP2OE9 during the adult plant stage. (b) The qRT‐PCR results indicate that AtRTP5 was significantly up‐regulated in the overexpression plants. The relative expression levels of AtRTP5 were normalized to AtUBC9. Statistical analysis was determined by one‐way ANOVA followed by Tukey’s multiple comparison tests. (c) The detached leaves of overexpressing Arabidopsis thaliana plants were challenged with P. parasitica zoospores and were more susceptible compared to A. thaliana Col‐0. The photograph was taken at about 60 hours post‐inoculation (hpi). More than ten leaves from ten different plants were tested in each experiment, and three independent experiments were performed. (d) Infected leaves at 3 days post‐inoculation(dpi) were stained with trypan blue. (e) Statistical analysis of plant disease severity. The infected leaves were divided into five grades as described in Fig. 2. (f) Nicotiana benthamiana leaves infiltrated with Agrobacterium tumefaciens GV3101 harbouring pART27::AtRTP5CFlag construct and pART27::Flag‐GFP construct were challenged by P. parasitica and P. infestans, respectively. The statistical analysis by Student’s t‐test of lesion diameters showed that overexpression of AtRTP5 enhanced the infection of P. parasitica and P. infestans. Error bars in (b) and (f) indicate SD of three biological replicates. Asterisks indicate significant differences (*P < 0.05, **P < 0.01).
To investigate whether AtRTP5 affected plant resistance to other Phytophthora pathogens, we inoculated P. infestans zoospores on N. benthamiana leaves expressing an AtRTP5‐Flag fusion protein. Flag‐tagged green fluorescent protein (GFP) was expressed as a control for inoculation assays. Lesion diameters of leaves expressing AtRTP5‐Flag were significantly larger than those of the control group (Fig. 3f), indicating that AtRTP5 promoted infection of P. infestans and acted as a negative regulator of plant resistance against Phytophthora pathogens.
Silencing of NbRTP5 genes by VIGS enhanced plant resistance to P. parasitica
Virus‐induced gene silencing (VIGS) was used to investigate the function of AtRTP5 homologous genes of N. benthamiana in plant immunity. Four sequences, designated as NbRTP5‐1 (Niben101Scf00274g03001.1), NbRTP5‐2 (Niben101Scf11483g00012.1), NbRTP5‐3 (Niben101Scf05619g00010.1) and NbRTP5‐4 (Niben101Scf02842g01008.1), were identified in the N. benthamiana genome, encoding four proteins 50–80% identical to AtRTP5 (Fig. S3a). The expressions of four NbRTP5 genes (NbRTP5s) were also triggered by P. parasitica infection, determined by RNA‐Seq data of infected N. benthamiana leaves (Fig. S3c). One VIGS construct, pTRV2::NbRTP5s, was generated to specifically silence the four NbRTP5 genes simultaneously. The transcription levels of the NbRTP5 genes were reduced by 50–80% in the plants harbouring the pTRV2::NbRTP5s construct compared to the TRV‐GFP control plants (Fig. S3b). TRV‐NbRTP5s plants grew more strongly and were taller than TRV‐GFP plants (Fig. 4a). About 21 days post‐infiltration of constructs with virus, P. parasitica zoospores were inoculated on detached leaves. The TRV‐NbRTP5s plants showed enhanced resistance to P. parasitica, with significantly smaller disease lesions compared to the TRV‐GFP plants (Fig. 4b,c). Significantly, more P. parasitica colonization was detected in control plants (Fig. 4d), indicating that NbRTP5 genes accelerate P. parasitica infection.
Figure 4.
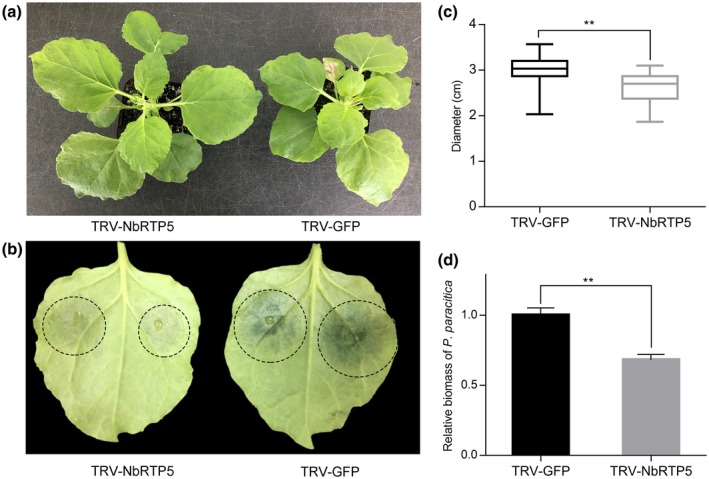
Silencing of NbRTP5 genes by virus‐induced gene silencing (VIGS) enhanced plant resistance to Phytophthora parasitica. (a) Silencing of NbRTP5 genes resulted in plants growing more robust. (b) Detached leaves of VIGS plants were challenged by P. parasitica zoospores and the lesion diameters were measured at 3 days post‐inoculation (dpi). Water‐soaked lesions of TRV‐NbRTP5s leaves were more visible compared to TRV‐GFP leaves. (c) Statistical analysis of lesion diameters showed that TRV‐NbRTP5s plants were more resistant to P. parasitica. (d) Relative DNA ratio of P. parasitica compared to Nicotiana benthamiana (PPTG_09948/NbActin) at 3 dpi was determined by qPCR. Amount of colonization in TRV‐NbRTP5s plants was less than that in TRV‐GFP plants. Student’s t‐test was conducted for statistical analysis in (c) and (d). Error bars in (c) and (d) indicate SD of three biological replicates, with more than ten leaves per replicate. **Indicates significant differences from control at P < 0.01.
AtRTP5 was induced by P. parasitica infection and the nucleus localization is essential for biological function of AtRTP5
Expression of AtRTP5 was induced by P. parasitica. We analysed the relative expression level of AtRTP5 at different time points post‐infection by P. parasitica. AtRTP5 was up‐regulated during the biotrophic phases of P. parasitica infection and peaked at 24 hours post‐inoculation (hpi) (Fig. 5d). The expression pattern was confirmed by RNA‐Seq of inoculated seedling root materials (Fig. S4).
Figure 5.
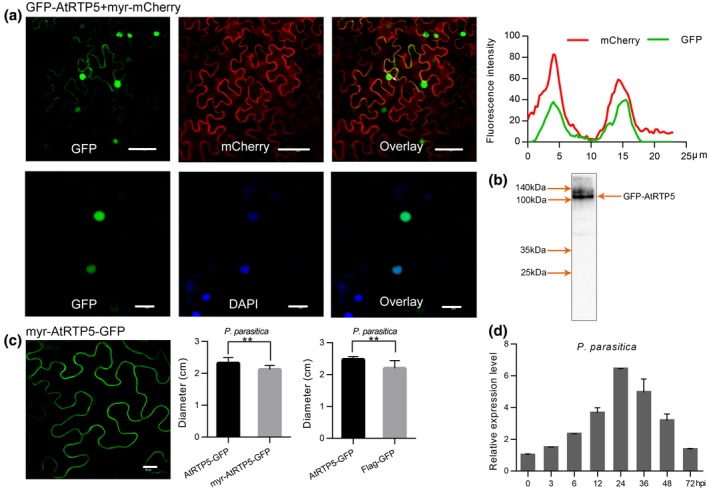
AtRTP5 transcription was induced by the infection of Phytophthora parasitica and nuclear localization of AtRTP5 protein was required for function in plant immunity. (a) Recombinant protein GFP‐AtRTP5 was localized in the nucleus and plasma membrane. The fluorescence of GFP‐AtRTP5 overlapped with that of Myr‐mCherry (upper panel). The blue fluorescence of the DAPI‐stained nucleus could merge with green fluorescence (lower panel). (b) The protein was extracted from Nicotiana benthamiana leaves expressing GFP‐AtRTP5 and a western blot showed the correct protein band of GFP‐AtRTP5 (c. 106 kDa). (c) Myr peptide was added to the N terminus of AtRTP5‐GFP, which resulted in the fusion protein transferring to the plasma membrane from the nucleus. Myr‐AtRTP5‐GFP had no function of promoting pathogen infection. Ten leaves were tested in each experiment and three independent experiments were performed. (d) The expression pattern of AtRTP5 determined by qRT‐PCR indicated that AtRTP5 was up‐regulated during the infection of P. parasitica. Total RNA was obtained from detached leaves infected by P. parasitica at different times. All scale bars in (a) and (c) are 40 μm.
GFP was fused to the N‐terminus of AtRTP5 driven by the CaMV 35S promoter. pART27::GFP‐AtRTP5 was transiently expressed in N. benthamiana leaves using the Agrobacterium‐mediated transient expression system. Strong green fluorescence signals were observed in the nucleus and a slight fluorescence signal was observed at the plasma membrane (PM) (Fig. 5a). pART27::GFP‐AtRTP5 was co‐expressed in N. benthamiana with a PM‐localized recombinant protein Myr‐mCherry, a myristoylation signal sequence MGCSVSK (Zheng et al., 2014) fused to the N‐terminus of the mCherry sequence. GFP‐AtRTP5 overlapped with Myr‐mCherry, indicating its localization at the PM (Fig. 5a). The nuclear localization was confirmed with 4',6‐diamidino‐2‐phenylindole (DAPI) staining (Otto, 1990), by staining the tobacco leaves expressing GFP‐AtRTP5. Blue fluorescence was observed in the nucleus, overlapping with the green fluorescence of GFP‐AtRTP5 (Fig. 5a). The protein integrity of GFP‐AtRTP5 was confirmed by western blot (Fig. 5b).
To test whether nuclear localization was required for biological function of AtRTP5 protein, a myristoylation signal sequence MGCSVSK (Zheng et al., 2014) was fused to the N‐terminus of the AtRTP5‐GFP recombinant protein, resulting in AtRTP5‐GFP being relocalized to the PM (Fig. 5c). Phytophthora parasitica zoospores were inoculated on two sides of each leaf expressing AtRTP5‐GFP and Myr‐AtRTP5‐GFP, which was respectively co‐expressed with p19 (Voinnet et al., 2003). The lesion diameters were measured at 48 to 72 hpi. The Myr‐AtRTP5‐GFP fusion protein lost the function of promoting pathogen infection (Fig. 5c), suggesting that nuclear localization was required for the AtRTP5 protein to promote pathogen infection.
AtRTP5 was associated with the plant hormone signalling pathway
We performed RNA‐Seq using uninfected leaves of Col‐0, T‐DNA insertion mutant rtp5‐3 and the overexpression transformant RTP5OE2. Applying q < 0.005 and twofold as a cut‐off, the transcriptome profile (accession number PRJNA562027) analysis showed that there were 415 transcripts down‐regulated and 523 transcripts up‐regulated in rtp5‐3 compared to Col‐0. The overexpression of AtRTP5 (RTP5OE2) resulted in increased expression of 591 genes and decreased expression of 353 genes. Among the down‐regulated transcripts in rtp5‐3, 17 were also up‐regulated in RTP5OE2, including AtRTP5. Only three of the up‐regulated transcripts in rtp5‐3 were down‐regulated in RTP5OE2 (Fig. 6b).
Figure 6.
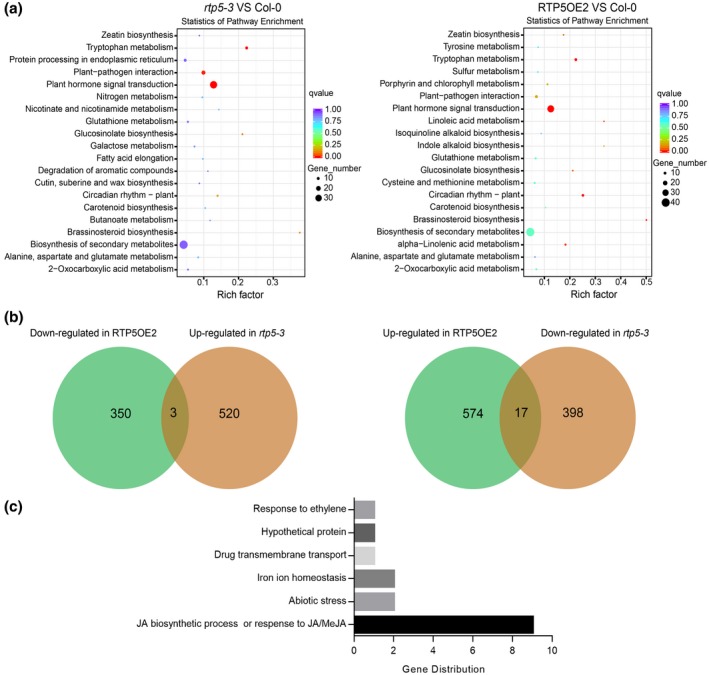
Analysis of the RNA‐Seq data of uninfected Arabidopsis thaliana Col‐0, rtp5‐3 and overexpression plant RTP5OE2. (a) The KEGG enrichment analysis showed that the differentially expressed genes (DEGs) of rtp5‐3 compared with Col‐0 and the DEGs of RTP5OE2 compared with Col‐0 were mainly involved in plant hormone signalling pathways. The sizes and the colours of the dots represent the numbers of DEGs and the q‐value, respectively. (b) Venn diagrams showing the overlap of genes down‐regulated in RTP5OE2 while up‐regulated in rtp5‐3 compared with Col‐0 (left), and the overlap of genes up‐regulated in RTP5OE2 while down‐regulated in rtp5‐3 compared with Col‐0 (right). (c) The distribution of the 17 overlapping genes up‐regulated in RTP5OE2 but down‐regulated in rtp5‐3; nine of them are involved in the JA signalling pathway.
We analysed these differentially expressed genes (DEGs) based on the KEGG (Kyoto Encyclopedia of Genes and Genomes) Orthology‐Based Annotation System (KOBAS 2.0, corrected P‐value < 0.05). The majority of the DEGs between rtp5‐3 and Col‐0 were involved in the plant hormone signalling pathway, and similar results were observed with RTP5OE2 compared to Col‐0 (Fig. 6a). Of the 17 genes down‐regulated in rtp5‐3 but up‐regulated in RTP5OE2 (Fig. 6b), nine of them (except AtRTP5) were involved in the JA signalling pathway or in the response to JA/methyl jasmonate (MeJA). One gene responded to auxin and ethylene. The other five genes performed different biological functions, such as cellular iron ion homeostasis or abiotic stress (Fig. 6c). We confirmed the results using qRT‐PCR, with the JA‐related genes (JARGs) except JARG9 (BT4) (Robert et al., 2009) being down‐regulated in rtp5‐3 without infection. The transcription of these JARGs was significantly suppressed by P. parasitica infection, especially during the early infection stages (Fig. S5), suggesting that AtRTP5 plays an important role in regulating the JA signalling pathway and that JA is not favourable for plant resistance against P. parasitica.
Biosynthesis of endogenous JA was significantly suppressed in rtp5‐3
As shown in RNA‐Seq data and qRT‐PCR results, the expression level of JARG6 (Oxophytodienoate‐Reductase 3, AtOPR3), which is involved in biosynthesis of JA (Schaller et al., 2000), was significantly down‐regulated in rtp5‐3 compared to Col‐0. In Arabidopsis, AtLox2 (A. thaliana Lipoxygenase 2) encodes a lipoxygenase localized in the chloroplast, catalysing the conversion of fatty acids to the hydroperoxyl derivatives required for JA accumulation (Liavonchanka and Feussner, 2006; Turner et al., 2002). AtAOS (A. thaliana Allene Oxide Synthase) encodes an allene oxide synthase that belongs to the cytochrome p450 CYP74 protein family. AOS catalyses the dehydration of 13‐(S)‐hydroperoxylinolenic acid and converts it to an unstable epoxide 12,13‐epoxy‐linolenic acid, which is converted to the jasmonate precursor 12‐oxophytodienoic acid (OPDA) (Devoto and Turner, 2003; Lyons et al., 2013). Similar expression patterns were shown in the transcriptome data and in the qRT‐PCR results. Both AtAOS and AtLox2 were drastically down‐regulated in rtp5‐3, demonstrating that AtRTP5 was associated with the biosynthesis of endogenous JA. The transcripts of all other genes, including AtJMT (jasmonic acid carboxyl methyltransferase) and AtJAR1 (jasmonate resistant 1), which are involved in the transformations from JA to MeJA and to jasmonoyl‐l‐isoleucine (JA‐Ile), respectively (Staswick et al., 2002; Suza and Staswick, 2008; Turner et al., 2002), and AtMYC2 (jasmonate insensitive 1, JIN1), AtJAZ1 (jasmonate‐zim‐domain protein 1) and AtPDF1.2 (plant defensin 1.2), which respond to JA downstream of the signalling pathway (Brown et al., 2003; Chini et al., 2009; Fernández‐Calvo et al., 2011; Niu et al., 2011; Thines et al., 2007), were reduced in rtp5‐3 (Fig. 7a). These results suggest that AtRTP5 affects not only the biosynthesis of endogenous JA but also the downstream responses of this signalling pathway.
Figure 7.
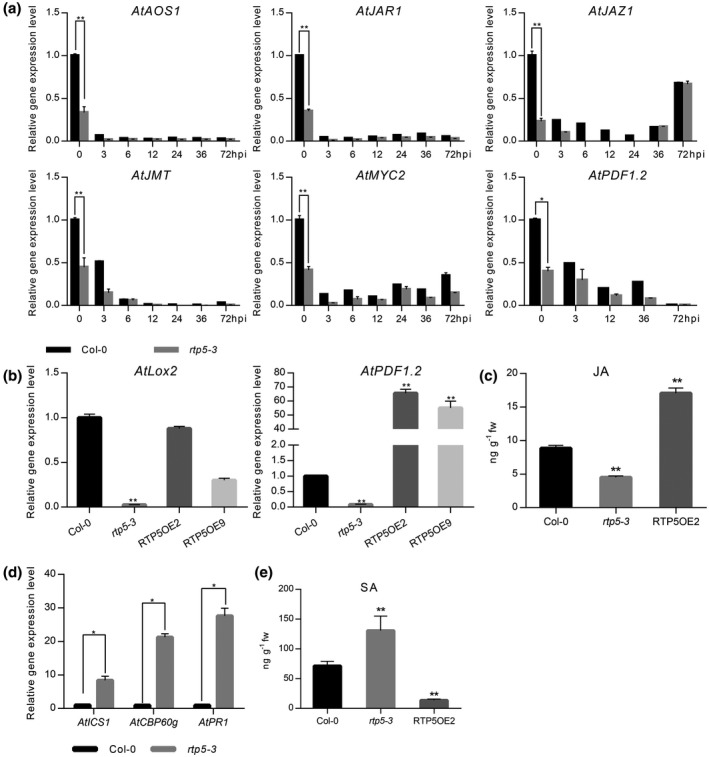
Deficiency of AtRTP5 suppressed jasmonic acid (JA) biosynthesis while activating the salicylic acid (SA) signalling pathway. (a) All examined JA signalling pathway marker genes were down‐regulated in rtp5‐3 compared with Arabidopsis thaliana Col‐0 during Phytophthora parasitica infection. Total RNA was obtained from detached leaves infected by P. parasitica at different times. (b) JA marker genes AtLox2 and AtPDF1.2 were down‐regulated in rtp5‐3 without infection, which were complemented by the overexpression of AtRTP5 (RTP5OE2 and RTP5OE9). (c) The endogenous JA contents of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 were determined by HPLC‐MS/MS with four biological replicates. The biosynthesis of JA was suppressed in rtp5‐3, but activated in RTP5OE2. (d) qRT‐PCR results showed that SA signalling pathway marker genes were up‐regulated in rtp5‐3. Total RNA was obtained from uninfected detached leaves of Col‐0 and rtp5‐3, respectively. (e) The endogenous SA contents of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 were determined by HPLC‐MS/MS with four biological replicates. SA was accumulated in rtp5‐3 and reduced in RTP5OE2, compared to Col‐0. The relative expression levels of tested genes in (a), (b) and (d) were normalized to AtUBC9. Three independent experiments were performed for each test of the gene expression levels. Statistical analysis was determined by Student’s t‐test between samples from two genotypes and determined by one‐way ANOVA followed by Tukey’s multiple comparison tests for samples from multiple genotypes.
We also examined the expression levels of AtLox2 and AtPDF1.2 in the uninfected overexpression transformant RTP5OE2 (Fig. 7b). The results show that AtPDF1.2 is up‐regulated in overexpression plants, while the expression level of AtLox2 is almost unchanged compared to Col‐0, but is higher than rtp5‐3. This suggests that overexpression of AtRTP5 complements the defect of the JA signal but does not result in its over‐accumulation. We tested the endogenous JA contents of 2‐week‐old seedlings of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 without infection using HPLC‐MS/MS. The JA content in rtp5‐3 decreased while it increased in RTP5OE2, compared to Col‐0 (Fig. 7c). AtRTP5 interfered with the JA signalling pathway by affecting the endogenous biosynthesis processes and the downstream gene responses.
SA signalling pathway was activated in the absence of AtRTP5
We tested the expression level of several SA signalling pathway‐related genes in rtp5‐3 and Col‐0 without infection (Fig. 7d). The isochorismate synthase ICS1, required to synthesize SA for plant immunity (Wildermuth et al., 2001), was highly up‐regulated in rtp5‐3. Similar expression patterns of CBP60g and PR1, which contributed to the production of SA in response to microbe‐associated molecular patterns (MAMPs) (Wang et al., 2011a) and responded to SA accumulation (Uknes et al., 1993), were observed (Fig. 7d). Taken together, the SA signalling pathway was activated in rtp5 mutant plants, which suggested that AtRTP5 plays a role as a suppressor of the SA signalling pathway.
We measured the endogenous SA contents of 2‐week‐old seedlings of Col‐0, rtp5‐3 and overexpression plant RTP5OE2 without infection using HPLC‐MS/MS. The content of endogenous SA was increased in rtp5‐3 while it was decreased in RTP5OE2 (Fig. 7e). In summary, SA significantly accumulated in the absence of AtRTP5 and the transcription of genes responding to SA was activated in rtp5‐3.
Discussion
AtRTP5 encodes a WD40 repeat (WDR) protein containing three WD40 repeat motifs, predicted by NCBI Conserved Domain Search Service (CD Search). We found that RTP5 (AtRTP5 and NbRTP5s) functions as a conserved susceptibility factor enhancing plant susceptibility to P. parasitica and P. infestans in A. thaliana and N. benthamiana, respectively. AtRTP5 also regulates plant hormone signalling pathways by interfering with the biosynthesis of JA and SA, and with the downstream responses.
The WDR domain is one of the most abundant protein interaction domains in eukaryotes (Miller et al., 2016; Ouyang et al., 2012), typically consisting of a seven‐bladed β‐propeller‐like domain with an overall doughnut shape (Schapira et al., 2017). WDR domain‐containing proteins participate in signal transduction, transcriptional regulation, plant organ development and innate immunity, providing platforms for protein–protein and protein–DNA interactions or constituting multiprotein complexes (Perfus‐Barbeoch et al., 2004; Schapira et al., 2017). A representative WDR‐containing protein family is the TRANSPARENT TESTA GLABRA (TTG) proteins family (Bouyer et al., 2008; Park et al., 2012). The Arabidopsis TTG1, which encodes a WDR protein containing four WD40 repeat motifs, interacts with bHLH proteins (GL3, EGL3 and TT8) to regulate TTG1‐dependent development pathways (Zhao et al., 2008). The TTG1‐GL3 protein complex can recruit the R2R3‐MYB proteins to form a TTG1‐bHLH‐MYB regulatory complex activating the downstream target transcriptions (Li et al., 2012; Zhao et al., 2008). Tobacco (Nicotiana tabacum) NtTTG1 is a target protein of the oomycete elicitin protein ParA1 (Li et al., 2012). An NtTTG1‐like gene, NtTTG2, suppresses the resistance to bacteria and viruses by repressing the localization of NPR1 and the transcription of pathogenesis‐related (PR) genes regulated by SA/NPR1 (Wang et al., 2009). Therefore, it is possible that proteins produced by Phytophthora pathogens are delivered into plant cells and facilitate infection by interacting with RTP5 directly or indirectly.
Genes involved in JA biosynthesis and JA responses, such as AtLox2, AtJAR1 and AtMYC2, were all down‐regulated in the AtRTP5 mutant rtp5‐3. The endogenous JA content in rtp5‐3 was significantly less than that in Col‐0, suggesting that AtRTP5 plays a role in JA biosynthesis. Plant hormones, especially JA and SA, fulfil essential roles in the regulation of plant development and innate immunity, but the mechanisms that plants use to regulate the hormone network are little known. Endogenous accumulation of SA antagonizes JA‐dependent responses and, conversely, JA can antagonize SA responses depending on the plant species and attacker strategies (Koornneef and Pieterse, 2008; Verhage et al., 2010). Both CBP60g, which contributes to production of SA in the presence of MAMPs (Wang et al., 2011), and the SA response gene PR1 were up‐regulated in the absence of AtRTP5, suggesting that the SA signalling pathway is activated in rtp5‐3. SA plays important roles in plant resistance to Phytophthora pathogens (Pan et al., 2016; Sugano et al., 2013; Wang et al., 2013); therefore, we suggest that accumulated SA in rtp5‐3 contributes to plant resistance against P. parasitica.
Our study uncovered a novel negative regulator of plant immunity, AtRTP5, that can promote the infection of Phytophthora pathogens by interfering with plant hormone signalling pathways. Absence of AtRTP5 triggers the accumulation of endogenous SA and enhances plant immunity. A simple working model of AtRTP5 that summarizes the content of this paper is shown in Fig. 8.
Figure 8.
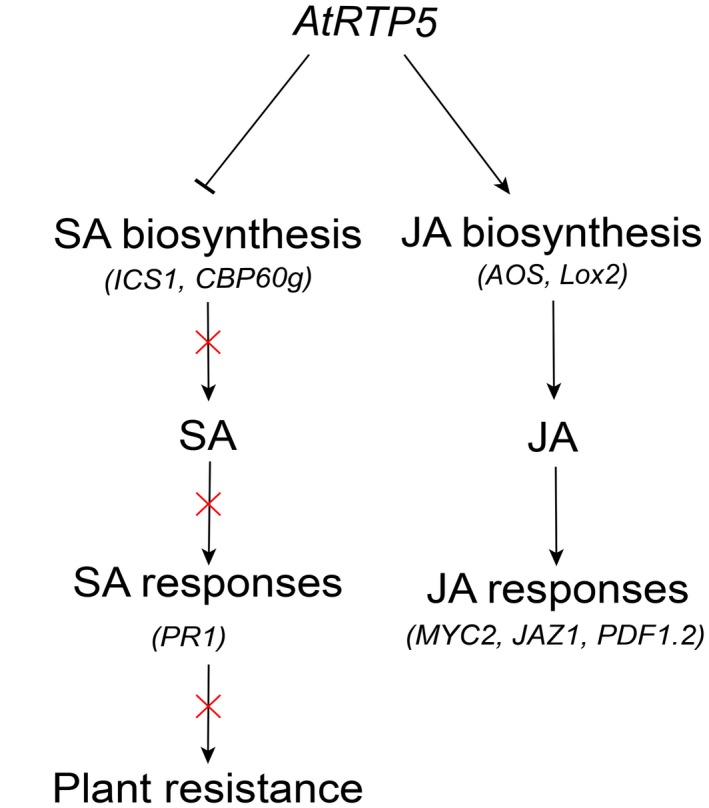
Schematic model for the role of AtRTP5 in plant immunity. Silencing of AtRTP5 promoted plant immunity by repressing the JA signalling pathway and activating SA biosynthesis and SA‐dependent responses. AtRTP5 suppressed the expression of SA biosynthesis‐related genes ICS1 and CBP60g, and the endogenous SA content was decreased in overexpression plants. The decreased SA suppressed PR1 expression and negatively affected plant resistance to Phytophthora pathogens. AtRTP5 accelerated the expression of JA biosynthesis‐related genes AOS and Lox2, and the endogenous JA content was increased in overexpression plants. The increased JA accelerated the expression of MYC2, JAZ1 and PDF1.2.
Experimental Procedures
Plant materials and growth condition
Arabidopsis seeds were surface‐sterilized before being planted on 1/2 MS medium and were incubated at 23 °C for about 10 days before being transplanted into soil. For the whole‐seedling inoculation assay, 2‐week‐old seedlings were used. For other experiments, 10‐day‐old seedlings were transferred to soil and grown at 23 °C, with a photoperiod of 13h:11h, light:dark. Ecotype Columbia‐0 (Col‐0) was used as wild‐type control plant in the infection assays. Arabidopsis T‐DNA insertion mutants in Col‐0 background, rtp5‐2 (SALK_079027C) and rtp5‐3 (SALK_112605C), were obtained from the Arabidopsis Biological Resource Center (ABRC). Homozygotes were identified by PCR using primers rtp5‐2‐LP or rtp5‐3‐LP, rtp5‐2‐RP or rtp5‐3‐RP, and LBb1.3, listed in Table S2.
The overexpression lines in Col‐0 background were obtained using the Agrobacterium tumefaciens‐mediated floral dipping transformation method (Zhang et al., 2006). The generated transformants were identified by screening and genotyping on 1/2 MS medium with kanamycin. At least three independent T3 homozygous transformants were used in subsequent experiments.
Nicotiana benthamiana was grown in the same conditions as previously described (Fan et al., 2018).
Plasmid constructs
For transient expression assays and A. thaliana genetic transformation, a full‐length coding sequence of AtRTP5 was cloned from A. thaliana cDNA using gene‐specific primers (Table S2). The amplified fragment was ligated into pKannibal (Wesley et al., 2001). The vector was digested at the NotI site and a 3 kb fragment including CaMV 35S promoter, AtRTP5 coding sequence and OCS terminator was ligated into the binary vector pART27 (Gleave, 1992). Fusion fragments GFP‐AtRTP5, AtRTP5‐GFP and AtRTP5C‐Flag were obtained by overlapping PCR or by specific primers containing Flag‐tag (Table S2), and were inserted into vector pKannibal and pART27.
For VIGS, two NbRTP5 sequences used in construct pTRV2:NbRTP5s were cloned from the N. benthamiana cDNA and were fused by overlapping PCR. The fusion fragment was ligated into the tobacco rattle virus (TRV) TRV2 vector (Hayward et al., 2011; Liu et al., 2002).
Agroinfiltration and inoculation assays
Agrobacterium‐mediated transient expression assay by infiltration was performed as described previously (Fan et al., 2018). Briefly, constructs were transformed into A. tumefaciens strain GV3101 (Rossi et al., 1993) by electroporation. The transformed cells were incubated in Luria–Bertani medium with appropriate antibiotics at 28 °C, 200 rpm, for about 30 h in the dark. Then the bacteria were pelleted and resuspended in infiltration buffer (10 mM 2‐(N‐morpholino) ethane sulphonic acid (MES), 10 mM MgCl2 and 200 mM acetosyringone, pH 5.6). The resuspended bacteria were mixed with the silencing suppressor p19 (Voinnet et al., 2003) in a 1:1 or 1:1:1 ratio, and adjusted to a final concentration of OD600 = 0.2 for each construct.
About two explanate leaves of each 6‐week‐old N. benthamiana plant were selected to be infiltrated with A. tumefaciens strains harbouring pART27::AtRTP5CFlag or pART27::AtRTP5‐GFP and p19 on one side of the leaf, and infiltrated with A. tumefaciens strains harbouring GFP and p19 as negative control on the other side of the same leaf. Two days later, a 8 µL droplet of zoospore suspension (50 000 zoospores/mL) of the P. parasitica strain, GFP‐tagged Pp016, was inoculated near the infiltrated sites.
Pathogen culture condition and infection assays
Phytophthora parasitica strain, GFP‐tagged Pp016 and P. infestans strain Pi21336 were used for plant infection assays in this paper. Phytophthora parasitica was grown on 5% (v/v) CA (cleared carrot juice and agar) medium with 0.005% (w/v) β‐sitosterol and 0.01% (w/v) CaCO3. Production of P. parasitica zoospores was conducted as previously described (Wang et al., 2011b). For infection of AtRTP5 mutant plants, the zoospore concentration was adjusted to 4 × 105/mL, and 10 μL droplets of zoospores were inoculated per leaf, which was slightly wounded by a toothpick only on one side. For infection of detached leaves of AtRTP5 overexpression plants, the zoospore concentration was adjusted to 2 × 105/mL, and 10 μL droplets were inoculated per leaf. More than ten leaves from different plants were tested in each experiment and three independent experiments were performed. In each experiment, all infected leaves were divided into five grades depended on the pathogen colonization area ratio, as described in Fig. 2. The statistical significance of the plant disease severity assessment was determined by the Wilcoxon Rank‐Sum test at 3 dpi. Leaf discs (around the inoculated site, diameter 1 cm) from all infected leaves were collected as one sample in each line. Genomic DNA was extracted with the CTAB (cetyltrimethylammonium bromide) method and used as a template in real‐time qPCR experiments for quantifying the pathogen biomass (the ratio between pathogen and plant genomic DNA). Three independent experiments were performed and the statistical significance was determined by Student’s t‐test. For root infection, 2‐week‐old A. thaliana seedlings were removed from 1/2 MS medium in aseptic conditions and the roots were dipped into 2 × 105/mL zoospore suspension for about 10 s. Subsequently, the infected seedlings were transferred to 1/2 MS medium without sugar and statistical analysis was performed after incubating at 23 °C, 5000 Lx, 16h:8h, light:dark for about 15 days. Ten seedlings were used in each experiment and three independent experiments were performed. All infected seedlings were divided into three grades depending on their growth situation. Statistical significance was determined by Wilcoxon Rank‐Sum test. For N. benthamiana leaves infection with P. parasitica, concentration of zoospores was adjusted to 30–50 zoospores/μL and 8–10 μL suspension was inoculated on both sides of the leaf. Ten leaves from different individual plants were used for each experiment and four independent experiments were performed. Lesion diameters were measured at 2–3 dpi. Student’s t‐test was conducted to statistical analysis.
P. infestans was cultured on rye agar medium at 18 °C for about 10 days before zoospore production. The hyphae were flooded with 5 mL cold dH2O, and were scraped to release sporangia (Wang et al., 2015). Plates with sporangia solution were placed at 4 °C for about 1 h to release zoospores and the concentration was adjusted to 100 zoospores/μL. Each N. benthamiana leaf infiltrated by A. tumefaciens with constructs was inoculated with 8–10 μL zoospore suspension on both sides. Ten leaves from different individual plants were used for each experiment and three independent experiments were performed. Lesion diameters were measured after incubating at 18 °C for about 5 days. Student’s t‐test was conducted to statistical analysis.
TRV‐based VIGS in N. benthamiana
As described previously (King et al., 2014), A. tumefaciens strain GV3101 harbouring pTRV2::NbRTP5s, pTRV2::GFP or pTRV2::PDS constructs was mixed with strains carrying pTRV1 vector in a 1:1 ratio to achieve final concentration of OD600 = 0.2 for each component. The largest leaves of 3‐week‐old plants were chosen for infiltration and the silenced plants were used to inoculate with P. parasitica 3 weeks later. The VIGS primers used in this study are shown in Table S2.
About 8 µL (50 000 zoospores/mL) zoospore suspension of GFP‐tagged Pp016 was inoculated near the infiltrated sites. Lesion diameters were measured at 3 dpi. Leaf discs (around the inoculated site, diameter 3 cm) from one side of all infected leaves were collected as one sample in each experiment, and three independent experiments were performed. Genomic DNA was extracted with the CTAB method and used as a template in real‐time qPCR experiments for quantifying the pathogen biomass (the ratio between pathogen and plant genomic DNA). Student’s t‐test was conducted as statistical analysis.
Gene expression
Plant total RNA was extracted using the TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) method. For qRT‐PCR, the first‐strand cDNA was synthesized from 700 ng total RNA using the PrimeScript RT Reagent Kit (TaKaRa, Dalian, China) according to the manufacturer’s recommendations. Quantitative analysis for the relative expression level of the tested genes was performed using the SYBR Premix Kit (Roche, Basel, Switzerland) on Q7 Real‐Time Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The Ct values of tested genes were normalized to AtUBC9 (At4G27960) in A. thaliana and NbActin (Niben101Scf09133g02006.1) in N. benthamiana. Statistical analysis was determined by Student’s t‐test between samples from two genotypes and determined by one‐way ANOVA followed by Tukey’s multiple comparison tests for samples from multiple genotypes.
RNA‐Seq and data analyses
Total RNA isolated from uninfected detached leaves of Col‐0, rtp5‐3 and RTP5OE2 were sequenced with an Illumina HiSeq4000 PE150 at Novogene Bioinformatics Technology Co., Ltd (Beijing, China). The clean data were mapped to the Tair10 genome release files using TopHat2 (mismatch = 2). The reads numbers mapped to each gene were counted using HTSeq (‐m union) and the reads count was used to calculate FPKM (expected number of fragments per kilobase of transcript sequence per million base pairs sequenced) (Trapnell et al., 2010). DEGSeq R package (q < 0.005 and |log2 (fold change)| > 1) was used for differential expression analysis. The P‐values were adjusted with the Benjamini and Hochberg method. The read number information of the RNA‐Seq data is shown in Table S3.
Author Contributions
W.S. and W.L. designed the experiments. W.L., D.Z., J.D., Y.M. and X.K. performed the experiments. W.L., Q.Z., T.L. and W.S. analysed the data. W.L. and W.S. wrote the manuscript. All authors reviewed the manuscript.
Supporting information
Fig. S1 Alignment results of AtRTP5 homologues in land plants. The homologous genes were identified by aligning the AtRTP5 amino acid sequence to the NCBI non‐redundant protein sequence database using the BLASTP algorithm. The multisequence alignment was performed with Clustal X2 and the alignment result was visualized with GeneDoc.
Fig. S2 The seedling root inoculation assays showed that rtp5‐2 and rtp5‐3 were resistant to Phytophthora parasitica infection. (a) Two‐week‐old seedlings roots of Arabidopsis thaliana Col‐0, rtp5‐2 and rtp5‐3 were dipped into zoospore suspension of P. parasitica and were transferred to 1/2 MS‐S medium (1/2 MS medium without saccharose) for about 2 weeks before assessment of disease severity. (b) The plant disease severity assessment of infected seedlings showed in (a). All infected seedlings were divided into three grades depending on their growth situation.
Fig. S3 Homologues of AtRTP5 in Nicotiana benthamiana. (a) Four sequences, designated as NbRTP5‐1 to NbRTP5‐4, were identified in the N. benthamiana genome, encoding four proteins that are 50–80% identical to AtRTP5. (b) The silencing efficiencies of four NbRTP5 genes were more than 80%. qRT‐PCR experiments were determined by using cDNA synthesized from total RNA of TRV‐NbRTP5 and TRV‐GFP plants. (c) Expression of the four NbRTP5 genes was triggered by P. parasitica infection. Total RNA was obtained from N. benthamiana leaves infected with P. parasitica zoospores.
Fig. S4 Expression pattern of AtRTP5 determined by RNA‐Seq data of infected seedling roots. Total RNA was obtained from seedling roots of Arabidopsis thaliana Col‐0 and rtp5‐3 during different infection stages.
Fig. S5 The jasmonic acid‐related genes (JARGs) were down‐regulated in mutant rtp5‐3 as determined by qRT‐PCR. The transcription profiles of JARGs were confirmed by qRT‐PCR. Total RNA was obtained from detached leaves infected by Phytophthora parasitica at 0, 24 and 48 hours post‐inoculation (hpi), respectively. The gene descriptions of these JARGs are shown in Table S4. JARG5 is JAZ1 and its transcription profile is shown in Fig. 7a. The transcription profile of JARG9 determined by qRT‐PCR is slightly different from the RNA‐Seq data.
Table S1 Accession number list of AtRTP5 homologues shown in Fig. 1b.
Table S2 Primers used in this study.
Table S3 The read counts of the RNA‐Seq data.
Table S4 Annotation for jasmonic acid‐related genes (JARGs).
Acknowledgements
We would like to thank Dr Jianru Zuo (Chinese Academy of Sciences, China) and the Arabidopsis Biological Resource Centre (ABRC, USA) for providing Arabidopsis mutant seeds, and Professors Jim Peacock and Liz Dennis (CSIRO Agriculture and Food, Canberra, Australia), and Gary Loake (University of Edinburgh, UK) for their helpful suggestions on the manuscript. This work was supported by the China Agriculture Research System (CARS‐09), the National Natural Science Foundation of China (31125022 and 31561143007), and the Programme of Introducing Talents of Innovative Discipline to Universities (project 111) from the State Administration of Foreign Experts Affairs (#B18042).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.
References
- Alazem, M. and Lin, N.S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaran, E. , Major, I.T. , Cruz, J.A. , Rosa, B.A. , Koo, A.J. , Chen, J. , Kramer, D.M. , He, S.Y. and Howe, G.A. (2014) Temporal dynamics of growth and photosynthesis suppression in response to jasmonate signaling. Plant Physiol. 165, 1302–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink, P.C. , McLellan, H. , Gilroy, E.M. , Naqvi, S. , He, Q. , Yang, L. , Wang, X. , Turnbull, D. , Armstrong, M.R. , Tian, Z. and Birch, P.R.J. (2016) Oomycetes seek help from the plant: Phytophthora infestans effectors target host susceptibility factors. Mol. Plant, 9, 636–638. [DOI] [PubMed] [Google Scholar]
- Bouyer, D. , Geier, F. , Kragler, F. , Schnittger, A. , Pesch, M. , Wester, K. , Balkunde, R. , Timmer, J. , Fleck, C. and Hulskamp, M. (2008) Two‐dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6, e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, R.L. , Kazan, K. , McGrath, K.C. , Maclean, D.J. and Manners, J.M. (2003) A role for the GCC‐box in jasmonate‐mediated activation of the PDF1.2 gene of Arabidopsis . Plant Physiol. 132, 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A. , Boter, M. and Solano, R. (2009) Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid‐signalling module. FEBS J. 276, 4682–4692. [DOI] [PubMed] [Google Scholar]
- Cui, H. , Wang, Y. , Xue, L. , Chu, J. , Yan, C. , Fu, J. , Chen, M. , Innes, R.W. and Zhou, J.M. (2010) Pseudomonas syringae effector protein AvrB perturbs Arabidopsis hormone signaling by activating MAP kinase 4. Cell Host Microbe, 7, 164–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devoto, A. and Turner, J.G. (2003) Regulation of jasmonate‐mediated plant responses in Arabidopsis . Ann. Bot. 92, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, G. , Yang, Y. , Li, T. , Lu, W. , Du, Y. , Qiang, X. , Wen, Q. and Shan, W. (2018) A Phytophthora capsici RXLR effector targets and inhibits a plant PPIase to suppress endoplasmic reticulum‐mediated immunity. Mol. Plant, 11, 1067–1083. [DOI] [PubMed] [Google Scholar]
- Fernández‐Calvo, P. , Chini, A. , Fernández‐Barbero, G. , Chico, J.M. , Gimenez‐Ibanez, S. , Geerinck, J. , Eeckhout, D. , Schweizer, F. , Godoy, M. and Franco‐Zorrilla, J.M. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell, 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Boter, M. , Fernandez‐Barbero, G. , Chini, A. , Rathjen, J.P. and Solano, R. (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis . PLoS Biol. 12, e1001792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992) A versatile binary vector system with a T‐DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Halter, T. , Imkampe, J. , Mazzotta, S. , Wierzba, M. , Postel, S. , Bücherl, C. , Kiefer, C. , Stahl, M. , Chinchilla, D. and Wang, X. (2014) The leucine‐rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 24, 134–143. [DOI] [PubMed] [Google Scholar]
- Hayward, A. , Padmanabhan, M. and Dinesh‐Kumar, S. (2011) Virus‐induced gene silencing in Nicotiana benthamiana and other plant species In:Methods in Molecular Biology, Plant Reverse Genetics. Vol. 678, (Pereira A. ed.), pp. 55–63. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- He, Q. , Naqvi, S. , McLellan, H. , Boevink, P.C. , Champouret, N. , Hein, I. and Birch, P.R.J. (2018) Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl Acad. Sci. USA. 115, E7834–E7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, R.H. and Tyler, B.M. (2012) Mechanisms and evolution of virulence in oomycetes. Annu. Rev. Phytopathol. 50, 295–318. [DOI] [PubMed] [Google Scholar]
- Jiang, S. , Yao, J. , Ma, K.W. , Zhou, H. , Song, J. , He, S.Y. and Ma, W. (2013) Bacterial effector activates jasmonate signaling by directly targeting JAZ transcriptional repressors. PLoS Pathog. 9, e1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, S.R. , McLellan, H. , Boevink, P.C. , Armstrong, M.R. , Bukharova, T. , Sukarta, O. , Win, J. , Kamoun, S. , Birch, P.R.J. and Banfield, M.J. (2014) Phytophthora infestans RXLR effector PexRD2 interacts with host MAPKKKε to suppress plant immune signaling. Plant Cell, 26, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, A. and Pieterse, C.M. (2008) Cross talk in defense signaling. Plant Physiol. 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, B. , Gao, R. , Cui, R. , Lu, B. , Li, X. , Zhao, Y. , You, Z. , Tian, S. and Dong, H. (2012) Tobacco TTG2 suppresses resistance to pathogens by sequestering NPR1 from the nucleus. J. Cell Sci. 125, 4913–4922. [DOI] [PubMed] [Google Scholar]
- Liavonchanka, A. and Feussner, I. (2006) Lipoxygenases: occurrence, functions and catalysis. J. Plant Physiol. 163, 348–357. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S. (2002) Virus‐induced gene silencing in tomato. Plant J. 31, 777–786. [DOI] [PubMed] [Google Scholar]
- Lyons, R. , Manners, J.M. and Kazan, K. (2013) Jasmonate biosynthesis and signaling in monocots: a comparative overview. Plant Cell Rep. 32, 815–827. [DOI] [PubMed] [Google Scholar]
- Miller, J.C. , Chezem, W.R. and Clay, N.K. (2016) Ternary WD40 repeat‐containing protein complexes: evolution, composition and roles in plant immunity. Front. Plant Sci. 6, 1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu, Y. , Figueroa, P. and Browse, J. (2011) Characterization of JAZ‐interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis . J. Exp. Bot. 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto, F. (1990) DAPI staining of fixed cells for high‐resolution flow cytometry of nuclear DNA In: Methods in Cell Biology, Flow Cytometry, Vol. 33, ( Zbigniew, D. , Harry, A.C. , eds.), pp. 105–110. Amsterdam: Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- Ouyang, Y. , Huang, X. , Lu, Z. and Yao, J. (2012) Genomic survey, expression profile and co‐expression network analysis of OsWD40 family in rice. BMC Genom. 13, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q. , Cui, B. , Deng, F. , Quan, J. , Loake, G.J. and Shan, W. (2016) RTP1 encodes a novel endoplasmic reticulum (ER)‐localized protein in Arabidopsis and negatively regulates resistance against biotrophic pathogens. New Phytol. 209, 1641–1654. [DOI] [PubMed] [Google Scholar]
- Park, C.H. , Chen, S. , Shirsekar, G. , Zhou, B. , Khang, C.H. , Songkumarn, P. , Afzal, A.J. , Ning, Y. , Wang, R. and Bellizzi, M. (2012) The Magnaporthe oryzae effector AvrPiz‐t targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen‐associated molecular pattern–triggered immunity in rice. Plant Cell, 24, 4748–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus‐Barbeoch, L. , Jones, A.M. and Assmann, S.M. (2004) Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7, 719–731. [DOI] [PubMed] [Google Scholar]
- Robert, H.S. , Quint, A. , Brand, D. , Vivian‐Smith, A. and Offringa, R. (2009) BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 58, 109–121. [DOI] [PubMed] [Google Scholar]
- Rossi, L. , Escudero, J. , Hohn, B. and Tinland, B. (1993) Efficient and sensitive assay for T‐DNA‐dependent transient gene expression. Plant Mol. Biol. Rep. 11, 220–229. [Google Scholar]
- Schaller, F. , Biesgen, C. , Müssig, C. , Altmann, T. and Weiler, E.W. (2000) 12‐Oxophytodienoate reductase 3 (OPR3) is the isoenzyme involved in jasmonate biosynthesis. Planta, 210, 979–984. [DOI] [PubMed] [Google Scholar]
- Schapira, M. , Tyers, M. , Torrent, M. and Arrowsmith, C.H. (2017) WD40 repeat domain proteins: a novel target class? Nat. Rev. Drug Discov. 16, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. , Tiryaki, I. and Rowe, M.L. (2002) Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole‐3‐acetic acids in an assay for adenylation. Plant Cell, 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S. , Sugimoto, T. , Takatsuji, H. and Jiang, C.J. (2013) Induction of resistance to Phytophthora sojae in soyabean (Glycine max) by salicylic acid and ethylene. Plant Pathol. 62, 1048–1056. [Google Scholar]
- Suza, W.P. and Staswick, P.E. (2008) The role of JAR1 in Jasmonoyl‐L‐isoleucine production during Arabidopsis wound response. Planta, 227, 1221–1232. [DOI] [PubMed] [Google Scholar]
- Takahashi, H. , Kanayama, Y. , Zheng, M.S. , Kusano, T. , Hase, S. , Ikegami, M. and Shah, J. (2004) Antagonistic interactions between the SA and JA signaling pathways in Arabidopsis modulate expression of defense genes and gene‐for‐gene resistance to cucumber mosaic virus. Plant Cell Physiol. 45, 803–809. [DOI] [PubMed] [Google Scholar]
- Thines, B. , Katsir, L. , Melotto, M. , Niu, Y. , Mandaokar, A. , Liu, G. , Nomura, K. , He, S.Y. , Howe, G.A. and Browse, J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature, 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Trapnell, C. , Williams, B.A. , Pertea, G. , Mortazavi, A. , Kwan, G. , van Baren, M.J. , Salzberg, S.L. , Wold, B.J. and Pachter, L. (2010) Transcript assembly and quantification by RNA‐Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G. , Ellis, C. and Devoto, A. (2002) The jasmonate signal pathway. Plant Cell, 14(Suppl), S153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S. , Dincher, S. , Friedrich, L. , Negrotto, D. , Williams, S. , Thompson‐Taylor, H. , Potter, S. , Ward, E. and Ryals, J. (1993) Regulation of pathogenesis‐related protein‐1a gene expression in tobacco. Plant Cell, 5, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhage, A. , van Wees, S.C. and Pieterse, C.M. (2010) Plant immunity: it’s the hormones talking, but what do they say? Plant Physiol. 154, 536–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O. , Rivas, S. , Mestre, P. and Baulcombe, D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Liu, R. , Chen, L. , Wang, Y. , Liang, Y. , Wu, X. , Li, B. , Wu, J. , Liang, Y. , Wang, X. , Zhang, C. , Wang, Q. , Hong, X. and Dong, H. (2009) Nicotiana tabacum TTG1 contributes to ParA1‐induced signalling and cell death in leaf trichomes. J. Cell Sci. 122, 2673–2685. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Tsuda, K. , Truman, W. , Sato, M. , le Nguyen, V. , Katagiri, F. and Glazebrook, J. (2011a) CBP60g and SARD1 play partially redundant critical roles in salicylic acid signaling. Plant J. 67, 1029–1041. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Meng, Y. , Zhang, M. , Tong, X. , Wang, Q. , Sun, Y. , Quan, J. , Govers, F. and Shan, W. (2011b) Infection of Arabidopsis thaliana by Phytophthora parasitica and identification of variation in host specificity. Mol. Plant Pathol. 12, 187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. , Bouwmeester, K. , Van de Mortel, J.E. , Shan, W. and Govers, F. (2013) A novel Arabidopsis–oomycete pathosystem: differential interactions with Phytophthora capsici reveal a role for camalexin, indole glucosinolates and salicylic acid in defence. Plant Cell Environ. 36, 1192–1203. [DOI] [PubMed] [Google Scholar]
- Wang, X. , Boevink, P. , McLellan, H. , Armstrong, M. , Bukharova, T. , Qin, Z. and Birch, P.R.J. (2015) A host KH RNA‐binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol. Plant, 8, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley, S.V. , Helliwell, C.A. , Smith, N.A. , Wang, M. , Rouse, D.T. , Liu, Q. , Gooding, P.S. , Singh, S.P. , Abbott, D. and Stoutjesdijk, P.A. (2001) Construct design for efficient, effective and high‐throughput gene silencing in plants. Plant J. 27, 581–590. [DOI] [PubMed] [Google Scholar]
- Wildermuth, M.C. , Dewdney, J. , Wu, G. and Ausubel, F.M. (2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature, 414, 562. [DOI] [PubMed] [Google Scholar]
- Wu, L. , Chen, H. , Curtis, C. and Fu, Z.Q. (2014) Go in for the kill: How plants deploy effector‐triggered immunity to combat pathogens. Virulence, 5, 710–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Henriques, R. , Lin, S.S. , Niu, Q.W. and Chua, N.H. (2006) Agrobacterium‐mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. [DOI] [PubMed] [Google Scholar]
- Zhao, M. , Morohashi, K. , Hatlestad, G. , Grotewold, E. and Lloyd, A. (2008) The TTG1‐bHLH‐MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development, 135, 1991–1999. [DOI] [PubMed] [Google Scholar]
- Zheng, X. , McLellan, H. , Fraiture, M. , Liu, X. , Boevink, P.C. , Gilroy, E.M. , Chen, Y. , Kandel, K. , Sessa, G. and Birch, P.R.J. (2014) Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22‐triggered immunity. PLoS Pathog. 10, e1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J. , Niu, Q.W. and Chua, N.H. (2000) An estrogen receptor‐based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Alignment results of AtRTP5 homologues in land plants. The homologous genes were identified by aligning the AtRTP5 amino acid sequence to the NCBI non‐redundant protein sequence database using the BLASTP algorithm. The multisequence alignment was performed with Clustal X2 and the alignment result was visualized with GeneDoc.
Fig. S2 The seedling root inoculation assays showed that rtp5‐2 and rtp5‐3 were resistant to Phytophthora parasitica infection. (a) Two‐week‐old seedlings roots of Arabidopsis thaliana Col‐0, rtp5‐2 and rtp5‐3 were dipped into zoospore suspension of P. parasitica and were transferred to 1/2 MS‐S medium (1/2 MS medium without saccharose) for about 2 weeks before assessment of disease severity. (b) The plant disease severity assessment of infected seedlings showed in (a). All infected seedlings were divided into three grades depending on their growth situation.
Fig. S3 Homologues of AtRTP5 in Nicotiana benthamiana. (a) Four sequences, designated as NbRTP5‐1 to NbRTP5‐4, were identified in the N. benthamiana genome, encoding four proteins that are 50–80% identical to AtRTP5. (b) The silencing efficiencies of four NbRTP5 genes were more than 80%. qRT‐PCR experiments were determined by using cDNA synthesized from total RNA of TRV‐NbRTP5 and TRV‐GFP plants. (c) Expression of the four NbRTP5 genes was triggered by P. parasitica infection. Total RNA was obtained from N. benthamiana leaves infected with P. parasitica zoospores.
Fig. S4 Expression pattern of AtRTP5 determined by RNA‐Seq data of infected seedling roots. Total RNA was obtained from seedling roots of Arabidopsis thaliana Col‐0 and rtp5‐3 during different infection stages.
Fig. S5 The jasmonic acid‐related genes (JARGs) were down‐regulated in mutant rtp5‐3 as determined by qRT‐PCR. The transcription profiles of JARGs were confirmed by qRT‐PCR. Total RNA was obtained from detached leaves infected by Phytophthora parasitica at 0, 24 and 48 hours post‐inoculation (hpi), respectively. The gene descriptions of these JARGs are shown in Table S4. JARG5 is JAZ1 and its transcription profile is shown in Fig. 7a. The transcription profile of JARG9 determined by qRT‐PCR is slightly different from the RNA‐Seq data.
Table S1 Accession number list of AtRTP5 homologues shown in Fig. 1b.
Table S2 Primers used in this study.
Table S3 The read counts of the RNA‐Seq data.
Table S4 Annotation for jasmonic acid‐related genes (JARGs).
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.


