Summary
One of the most devastating fungal diseases of soybean in the southern USA is Cercospora leaf blight (CLB), which is caused mainly by Cercospora cf. flagellaris. Recent studies found that the fungal effector AVR4, originally identified in Cladosporium fulvum as a chitin‐binding protein, is highly conserved among other Cercospora species. We wanted to determine whether it is present in C. cf. flagellaris and, if so, whether it plays a role in the pathogen infection of soybean. We cloned the Avr4 gene and created C. cf. flagellaris ∆avr4 mutants, which produced little cercosporin and significantly reduced expression of cercosporin biosynthesis genes. The ∆avr4 mutants were also more sensitive to chitinase and showed reduced virulence on soybean compared to the wild‐type. The observed reduced virulence of C. cf. flagellaris ∆avr4 mutants on detached soybean leaves is likely due to reduced cercosporin biosynthesis. The phenotypes of reduced cercosporin production and cercosporin pathway gene expression, similar to those of the ∆avr4 mutants, were reproduced when wild‐type C. cf. flagellaris was treated with double‐stranded RNA targeting Avr4 in vitro. These two independent approaches demonstrated for the first time the direct involvement of AVR4 in the biosynthesis of cercosporin.
Keywords: C. kikuchii, Cercospora leaf blight, cercosporin biosynthesis, double‐stranded RNA, fungal virulence, gene disruption, gene silencing
Introduction
Soybean (Glycine max) is one of the most important agricultural crops and plays an important role in human and animal consumption. However, soybean is susceptible to various pathogens and pests throughout the growing season. Among fungal diseases, Cercospora leaf blight (CLB) and purple seed stain (PSS) are mainly caused by Cercospora kikuchii, C. cf. sigesbeckiae and C. cf. flagellaris (Albu et al., 2016; Matsumoto and Tomoyasu, 1925; Suzuki, 1921; Walters, 1980), with C. cf. flagellaris being the dominant causal agent of CLB in the southern USA (Albu et al., 2016). In the USA, CLB was first reported in 1978 (Walters, 1980). The first symptoms of CLB are usually observed in the late R5 (beginning of seed filling) and early R6 (end of seed filling) soybean growth stages on upper soybean leaves exposed to sunlight as reddish purple lesions, which become leathery, darken and gain bronze highlights as the disease progresses (Walters, 1980). At late soybean growth stages, angular‐to‐irregular lesions on infected leaf surfaces coalesce and eventually lead to premature defoliation.
In recent years, CLB was found in up to 70% of soybean fields surveyed in Alabama (Sikora et al., 2011). The damage caused by CLB has been a serious concern (Moore and Wolcott, 2000) because the disease has spread from the southern states to as far north as Iowa and has caused as high as 15–30% yield losses (Hartman et al., 1999; Wrather et al., 1997, 2001). The yield losses caused by CLB in Louisiana and Mississippi were estimated to be 2.38 and 2.07 million bushels in 2016, respectively (Allen et al., 2017). CLB is now among the top five most important diseases of soybean in the South. There is no effective management for CLB in soybean due to the long latent disease period, the lack of host resistance and effective fungicides.
During infection of soybean, C. cf. flagellaris produces a reddish‐purple pigment called cercosporin. It was first extracted from dry mycelia of C. kikuchii in 1957 and was identified as a perylenequinone (Kuyama and Tamura, 1957). It can absorb light energy and be converted to an energetically activated triplet state, which can react with oxygen and result in the generation of toxic reactive oxygen species, such as singlet oxygen and superoxide (Daub and Chung, 2007). The photosensitized cercosporin can cause peroxidation of membrane lipids, leading to membrane breakdown and cell death (Daub and Briggs, 1983), which contributes to leakage of nutrients and promotes growth and spread of fungal hyphae intracellularly (Daub, 1982; Daub and Ehrenshaft, 2000). Cercosporin is therefore considered a virulence factor and has been associated with lesion formation on soybean leaves (Upchurch et al., 1991a).
The genes involved in cercosporin biosynthesis have been identified in Cercospora nicotianae and consist of at least eight cercosporin toxin biosynthesis (CTB) genes with a possible four additional open reading frames (ORFs) (ORF9–12) that are organized in a cluster (Chen et al., 2007). The importance of some of these genes has been further investigated through gene disruption studies, which found that the expression of the eight CTB genes was coordinated and regulated by CTB8, a zinc finger transcription factor (Chen et al., 2007). In addition to CTB8, cercosporin biosynthesis is also regulated by CRG1, a transcription factor involved in cercosporin resistance (Chung et al., 2003) and by a MAP kinase kinase kinase (CZK3) (Shim and Dunkle, 2003). Several studies further demonstrated the importance of a cercosporin facilitator protein (CFP) and an ABC transporter (ATR1) in the biosynthesis or efflux of cercosporin and in fungal pathogenesis (Amnuaykanjanasin and Daub, 2009; Callahan et al., 1999; Choquer et al., 2005; Chung et al., 2003; Daub et al., 2005). A biosynthetic pathway for cercosporin was also proposed (Chen et al., 2007). However, recent studies presented alternative biosynthetic pathways obtained by the characterization of metabolites from a series of biosynthetic pathway gene knockouts, as well as the discovery of several additional genes required for cercosporin biosynthesis through a comparative genomic and conserved gene cluster analysis (de Jonge et al., 2017, 2018; Newman and Townsend, 2016; Shim and Dunkle, 2003).
Other than cercosporin production, there is little information on C. cf. flagellaris pathogenesis. One interesting observation made by Stergiopoulos et al. (2010) was that several Cercospora species, such as C. beticola, C. apii, C. nicotianae and C. zeina, contain a homologue of AVR4, a well‐studied fungal effector and a virulence factor of Cladosporium fulvum (Cf), on its host tomato (van Esse et al., 2007). Cf‐AVR4 was demonstrated to bind chitin on the fungal cell wall and shield it from digestion by host‐derived chitinases during infection (Joosten et al., 1994; van den Burg et al., 2006). The exact function of AVR4 is still unclear. However, the broadly conserved nature of AVR4 sequences across fungi with diverse lifestyles indicates that this effector may have conserved virulence functions that deregulate host immunity and facilitate infection on a wide range of hosts (Kohler et al., 2016).
If present, AVR4 from C. cf. flagellaris (Cfla‐AVR4) also may contribute to its virulence on soybean. Therefore, the current study aimed to clone the Cfla‐AVR4, disrupt it or suppress its expression through double‐stranded RNA (dsRNA) treatment, and then compare the changes in CLB disease development, expression of the CTB genes and cercosporin production both in vitro and in vivo to determine the possible roles played by AVR4 in C. cf. flagellaris. Cercosporin production was drastically reduced in ∆avr4 mutants compared to the wild‐type fungus in our in vitro and in planta assays. In addition, the ∆avr4 disruption mutants had lower expression levels of the CTB genes under light and dark conditions. Chitinase inhibited mycelial growth of ∆avr4 mutants in vitro, whereas the wild‐type fungus was not affected by the enzyme. Moreover, the growth and virulence of ∆avr4 mutants were clearly compromised on soybean leaves, suggesting that the mutants were more vulnerable to the deleterious effects of the host chitinases. Interestingly, cercosporin was also found to inhibit chitinase activity in vitro. These results suggest that AVR4 contributes to the virulence of C. cf. flagellaris on soybean through protecting fungal hyphae and affecting cercosporin biosynthesis. Wild‐type C. cf. flagellaris also exhibited reduced cercosporin production and reduced cercosporin biosynthesis gene expression when it was grown in liquid culture in the presence of Avr4‐specific dsRNA. The knowledge gained from the present study can lead to the development of more efficient strategies to manage CLB in soybean.
Results
Cloning and sequence analysis of C. cf. flagellaris Avr4 gene
A 0.35 kb Avr4 fragment was amplified from C. cf. flagellaris gDNA using primers designed based on conserved sequences of Avr4 homologues from other Cercospora species. The full‐length Avr4 gene was obtained by polymerase chain reaction (PCR) of previously constructed C. cf. flagellaris genomic libraries using primers designed to walk upward and downward into unknown genomic regions. In total, six overlapping fragments were amplified, sequenced and assembled into a 1.6 kb DNA fragment (Fig. S1, Supporting information), which has been deposited into GenBank under accession number MH673051. This Cfla‐Avr4 gene has an ORF from 686 to 1075 bp with no introns and encodes a protein of 129 amino acids with a putative transcriptional start site predicted at −15 bp upstream of the ATG start codon (Fig. S1). Sequence analysis showed high identity (97–99%) at the deduced amino acid level to AVR4 from C. beticola, C. nicotianae, C. zeina and C. apii, (Fig. S2). A BLAST search of NCBI databases revealed the presence of a putative chitin‐binding peritrophin‐A domain (NCBI accession: pfam01607) that is conserved in all five Cercospora species (Fig. S2). Furthermore, the nine cysteine residues reported in C. beticola, C. nicotianae, C. zeina and C. apii (Stergiopoulos et al., 2010) were also present in the C. cf. flagellaris Avr4 gene (Fig. S2).
Target gene disruption and confirmation of ∆avr4 mutants
To elucidate the role of AVR4 in C. cf. flagellaris virulence and fitness, ∆avr4 disruption mutants were created using the strategies outlined in Fig. 1A. Two DNA fragments containing 5ʹ Avr4 fused with 3ʹ of hygromycin phosphotransferase B gene (HYG) and 3ʹ Avr4 fused with 5ʹ HYG were obtained by fusion PCR and transformed directly into protoplasts of the wild‐type isolate. All 17 selected hygromycin‐resistant fungal transformants showed the presence of the 466 bp fragment from HYG by PCR using HYG‐specific primers NLC37 and NLC38 (Fig. 1B). Site‐specific integration of the split marker DNA fragments into the Avr4 gene locus was also verified through PCR with an upstream and downstream pair of primers. Selected mutants were confirmed to contain the expected extra 2.4 kb insertion of HYG compared to the wild‐type (1.6 kb) when its full length was amplified with primers Avr4F1 and Avr4R1(Fig. 1C). Three mutants (∆avr4M1, ∆avr4M2 and ∆avr4M3) were selected for further studies.
Figure 1.
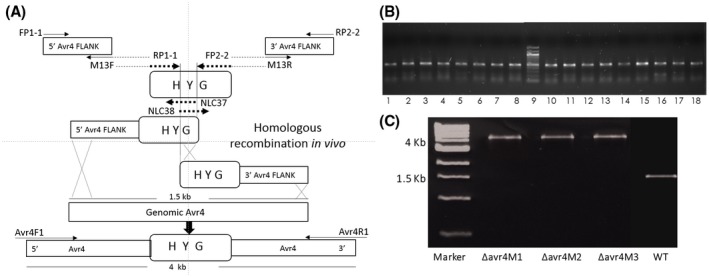
Targeted disruption of the Avr4 gene in C. cf. flagellaris. (A) The 5ʹ and 3ʹ fragments of Avr4 were amplified separately with primer pairs indicated and fused with the hygromycin cassette (HYG) as described in Experimental procedures. The resulting two PCR fragments were used to transform Cercospora cf. flagellaris protoplasts. Note: drawing is not to scale. (B) Sixteen selected ∆avr4 mutants (lines 1 to 7 and 10 to 18) contained the 466 bp HYG fragment when amplified by PCR using primer pair NLC37/NLC38. Lane 8 shows the 466 bp HYG fragment amplified from pUCATPH vector. Lane 9 shows the DNA size marker. (C) The presence of 4 kb PCR fragment using external primers Avr4F1/Avr4R1 in ∆avr4 mutants compared to the 1.6 kb Avr4 gene in C. cf. flagellaris wild‐type (WT) confirming the HYG insertion.
∆avr4 mutants grew faster on solid media, produced little cercosporin and had reduced expression of CTB genes in vitro
The ∆avr4 mutants grew faster on solid media than the wild‐type (Fig. S3). The difference in mycelium mat diameter became clear and significant 9 days after inoculation and it was about 50% larger in diameter than the wild‐type 13 days after inoculation (Fig. S3). In addition, the ∆avr4 mutants produced very little dark red pigment (a typical colour of concentrated cercosporin) under the fungal colony, which was strikingly different from the colour under the wild‐type colony (Figs S3 and 2). Cercosporin production of ∆avr4 mutants on potato dextrose agar (PDA) plates was approximately 10% of the amount of cercosporin produced by the wild‐type (Fig. 2). The difference in cercosporin production among the three ∆avr4 mutants was not significant.
Figure 2.
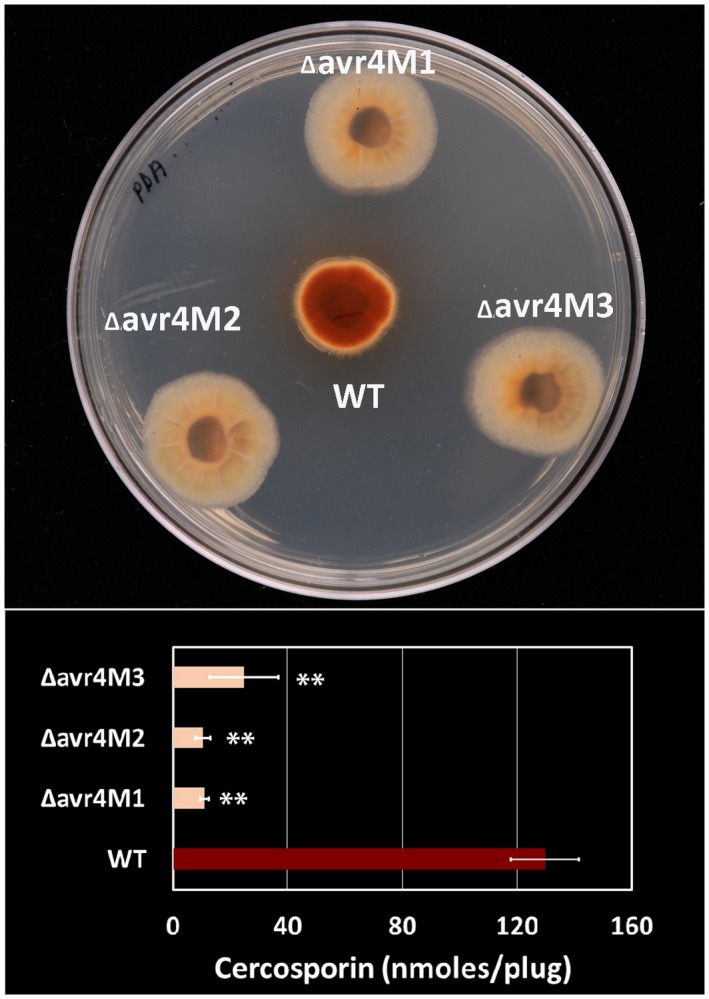
Cercosporin production of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants in vitro. The fungus was grown on potato dextrose agar plates under light for 5 days (top) and cercosporin was extracted with 5 M KOH and quantified by absorbance at 480 nm (bottom). Data are the mean and standard errors of three different experiments with five replicates of each fungal isolate. Asterisks (**) represent significant difference between WT and the mutants at P < 0.01.
When grown in liquid potato dextrose broth (PDB) or complete medium (CM) the same phenotype could be observed in which the wild‐type fungal culture became dark red in colour after 5 days of incubation at room temperature, whereas the mutant cultures exhibited a light yellow colour (Fig. S4). ∆avr4 mutants produced only 3–7% of the level of cercosporin compared to the wild‐type when grown in either liquid CM or PDB medium based on HPLC analysis (Fig. S5). Cercosporin was confirmed to be the main constituent in the red pigment produced by C. cf. flagellaris wild‐type in liquid media based on mass spectrometry analysis of the mass/charge (m/z) ratio of the dark red pigment and the cercosporin standard (Fig. S6).
Considering the reduction in cercosporin production by the ∆avr4 mutants in liquid and solid media compared to the wild‐type, the expression of cercosporin biosynthesis pathway genes was examined using real‐time reverse transcription (RT)‐PCR. It was found that disruption of Avr4 significantly reduced the expression of both CTB1 and CTB8 genes relative to β‐tubulin under light and dark conditions (about two‐ to three‐fold) (Fig. 3).
Figure 3.
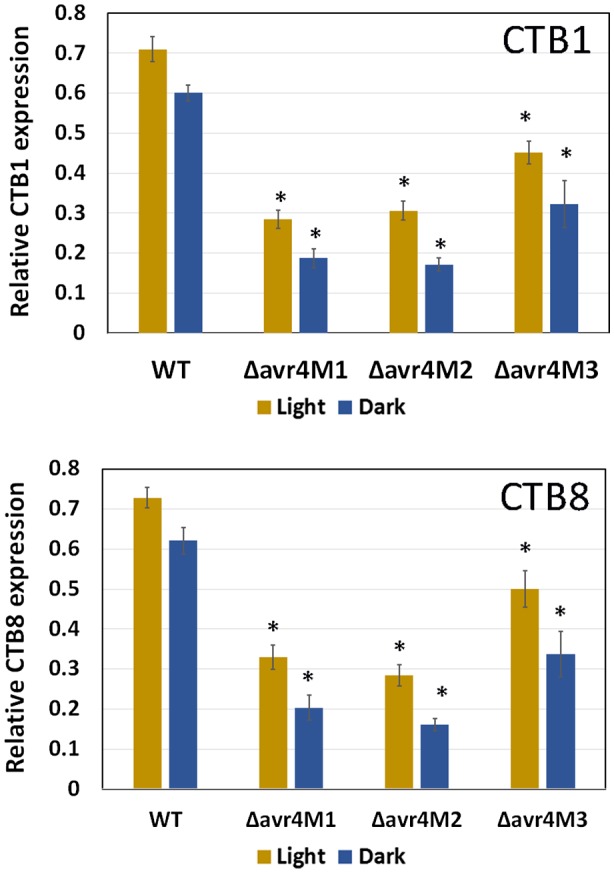
The expression of CTB1 and CTB8 relative to β‐tubulin in Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants. Fungal RNA was isolated from cultures grown in potato dextrose broth under light or darkness for 5 days. The expressions of CTB1 and CTB8 genes were normalized to that of β‐tubulin from the same sample. Data are the mean and standard errors of three different experiments with three biological replicates of each fungal isolate and three technical replicates of real‐time reverse transcription PCRs. Asterisks (*) indicate significant difference between the wild‐type and the mutants when grown under light or dark conditions (P < 0.05).
Chitinase reduced the growth of ∆avr4 mutants and chitinolytic activity is inhibited by cercosporin in vitro
AVR4 from C. fulvum was reported to protect the fungal cell wall from degradation by host chitinase during infection of tomato (van den Burg et al., 2006), therefore the effect of chitinase on the growth of ∆avr4 mutants compared to the wild‐type fungus was also examined. Inhibition of hyphal growth was observed when the mutants were grown next to wells containing 1 unit (U) of chitinase on CM agar plates (Fig. 4). Such suppression of growth by chitinase was not observed in the wild‐type (Fig. 4), demonstrating that AVR4 from C. cf. flagellaris plays a similar role in protecting the pathogen against host chitinases.
Figure 4.
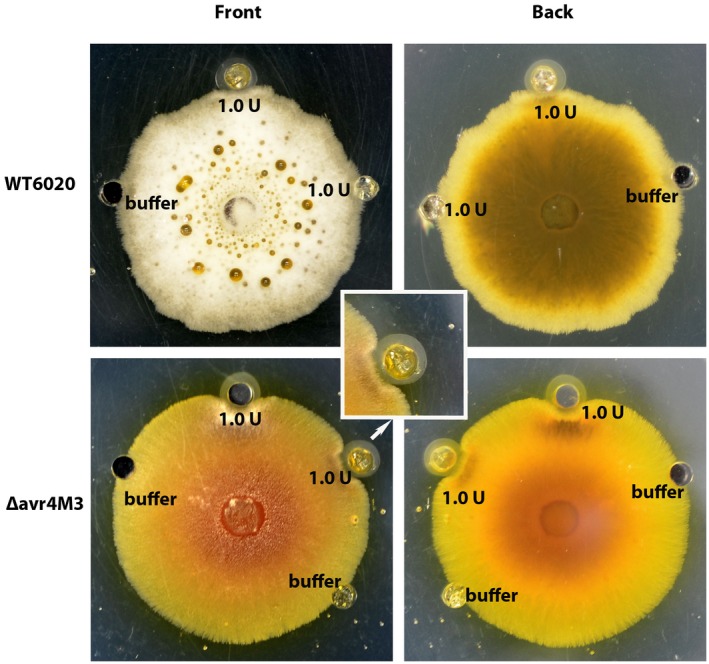
Growth of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutant in the presence of chitinase. One unit (U) of chitinase dissolved in 30 µL of 0.1 M potassium phosphate buffer (pH 6.5) was pipetted into wells of 1‐week‐old complete medium plates inoculated with C. cf. flagellaris WT or ∆avr4 mutants. Wells filled with 0.1 M phosphate buffer only were used as controls. Plates were sealed and kept at 25 °C. Fungal growth was visually assessed 2 days later.
A cup‐plate assay was also performed to determine whether cercosporin has a direct effect on chitinolytic activity. Reductions of 19% and 30% in chitinase activity were observed in the presence of 15 µM of cercosporin in the plate after 2 and 3 days, respectively, when the wells contained 0.25 U of chitinase (Fig. 5A,B). A slightly lower reduction in chitinolytic activity (12% and 16%, respectively) was observed after 2 and 3 days when 0.5 U of chitinase was used under the same conditions (Fig. 5A,B). The difference among the agarose plates with and without cercosporin became very clear after 5 days, when 60% and 70% reductions in enzyme activity were observed using 0.25 and 0.5 U of chitinase, respectively (Fig. 5B). However, the same concentration of cercosporin did not significantly inhibit the enzyme activity of the α‐amylase from Aspergillus oryzae on starch agarose cup plates compared to the control plates (Fig. 5C), suggesting that the inhibition of chitinase activity by cercosporin is enzyme‐specific.
Figure 5.
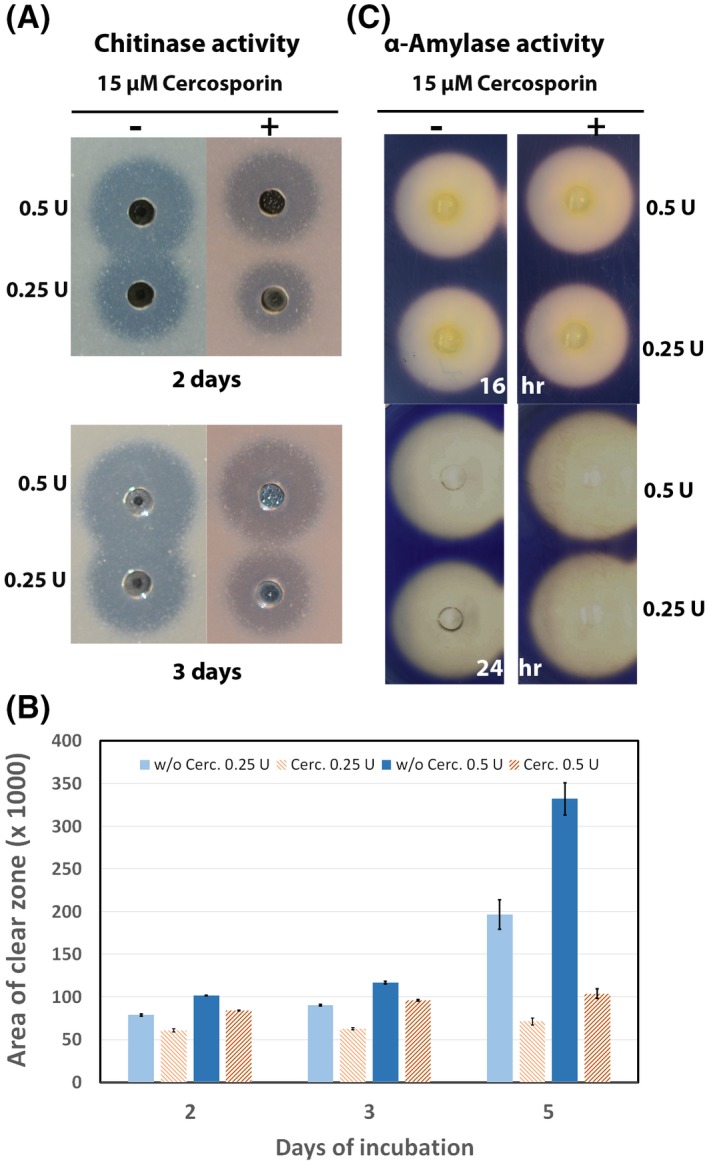
Chitinase and α‐amylase activity in the presence of cercosporin. (A) Agarose plates containing chitin and 15 µM or no cercosporin were used to assess chitin hydrolysis visualized as clearing zones around the wells containing 0.5 or 0.25 units (U) of chitinase after 48 and 72 h of incubation at 25 °C. (B) Area of clear zone assessed after 48, 72 and 120 h using Progenesis Software. (C) Enzyme activity of the α‐amylase from Aspergillus oryzae on 0.25% soluble starch agarose plates containing 15 µM or no cercosporin after 16 and 24 h of incubation at 25 °C.
Cercospora cf. flagellaris ∆avr4 mutants showed reduced virulence and growth on soybean leaves
To determine whether the loss of AVR4 alters the virulence of C. cf. flagellaris on soybean, the mycelial mats of the wild‐type isolate and ∆avr4 mutants from PDA were inoculated onto detached soybean leaves. The mutants grew twice as slowly as the wild‐type on the detached soybean leaves based on visual assessment, which is the opposite of what was observed when they were grown on PDA plates (Fig. S3). In addition, the mutants incited smaller necrotic and chlorotic lesions compared to the wild‐type at the site of mycelial inoculation (Fig. 6A,B), indicating a clear reduction in fungal virulence. This reduced virulence of ∆avr4 mutants on soybean leaves was confirmed by a 50% reduction in fungal growth inside the leaves inoculated with the mutants compared to those inoculated with the wild‐type pathogen (Fig. 6C).
Figure 6.
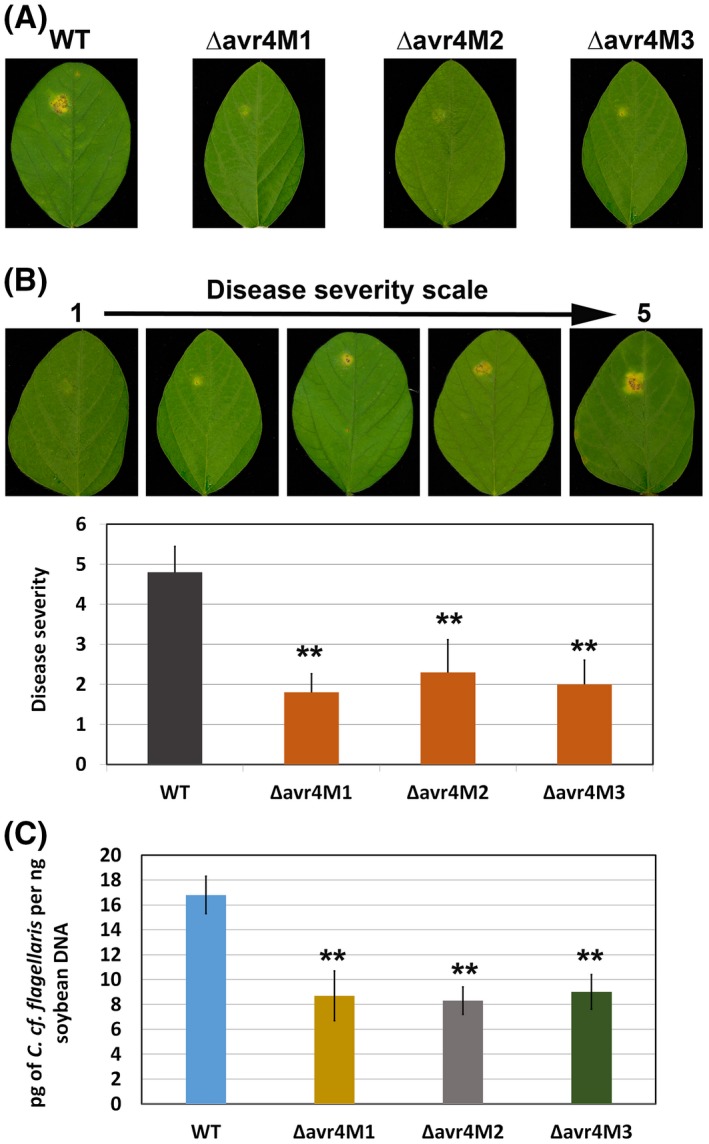
Virulence of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants on detached soybean leaves. (A) Detached soybean leaves were inoculated with mycelia plugs of the WT or ∆avr4 mutants and disease severity was determined at 14 days after inoculation. (B) Disease severity was determined based on the 1–5 rating scale (1 = less and 5 = more severe). Data are the mean and standard errors of three different experiments with three biological replicates (eight leaves) within each fungal isolate. Asterisks (**) indicate significant disease severity difference between the wild‐type and the mutants (P < 0.01). (C) Fungal growth was quantitatively assessed by determining the fungal DNA levels in soybean leaves through real‐time PCR. Data are the mean and standard errors of three different experiments with three biological replicates (eight leaves) for each wild‐type or mutant.
The development of minor necrotic and chlorotic lesions on soybean leaves inoculated with the mutants indicates that ∆avr4 mutants still produce a small amount of cercosporin during infection of soybean leaves. This was confirmed through HPLC analysis, which detected low levels of cercosporin ranging from 0.31 to 0.55 µg/g of leaf tissue in soybean leaves inoculated with C. cf. flagellaris mutants compared to 3.9 µg/g in leaves inoculated with the wild‐type fungus (Fig. S5).
Droplet digital PCR detected the presence of additional copies of HYG cassette in the mutants
To address the concern that ectopic integration of the HYG cassette could have contributed to any of the phenotypes observed above, genomic DNA from the wild‐type and the three mutants was analysed through droplet digital PCR using primers and probe for HYG as a target and the known single‐copy aminoadipate reductase gene (LYS2) as a reference. The results indicated that all three mutants have more than one copy of HYG in their genome (Table S1), which suggests that the observed phenotypes of the mutants could be due to knockout of unknown gene(s) at non‐target location(s). An independent study of suppressing Avr4 expression of the wild‐type C. cf. flagellaris in culture using dsRNA targeting the Avr4 gene was therefore conducted.
Avr4 dsRNA suppressed cercosporin production of the wild‐type C. cf. flagellaris grown in CM
Wild‐type C. cf. flagellaris produced 6.0 and 6.1 µg/mL cercosporin in half‐strength CM after 4 days in the absence of any dsRNA, or in the presence of the lysate prepared from Escherichia coli HT115 cells with L4440 plasmid only (EV), respectively (Fig. 7A,B). However, the cercosporin production in the wild‐type was reduced by 88% to 0.7 µg/mL when the culture was treated with Avr4 dsRNA prepared from bacterial cells containing the L4440‐Avr4 construct (Fig. 7A,B). This is consistent with the visual colour differences among the cercosporin extracts following different treatments (Fig. 7C). After 4 days of culturing in the presence of Avr4‐specific dsRNA, the expression of Avr4 was suppressed by 78% compared to the EV control (Fig. 7D). In addition, the expression of cercosporin biosynthesis pathway genes CTB1 and CTB8 was also significantly reduced (75% and 58%, respectively) in the mycelia that had been exposed to the extract containing Avr4‐specific dsRNA compared to the control that was treated with dsRNA prepared from E. coli cells that contained only the EV (Fig. 7D). This Avr4 dsRNA treatment study independently confirmed the involvement of Avr4 in cercosporin biosynthesis in C. cf. flagellaris, possibly through affecting the expression of cercosporin biosynthesis pathway genes such as CTB1 and CTB8.
Figure 7.
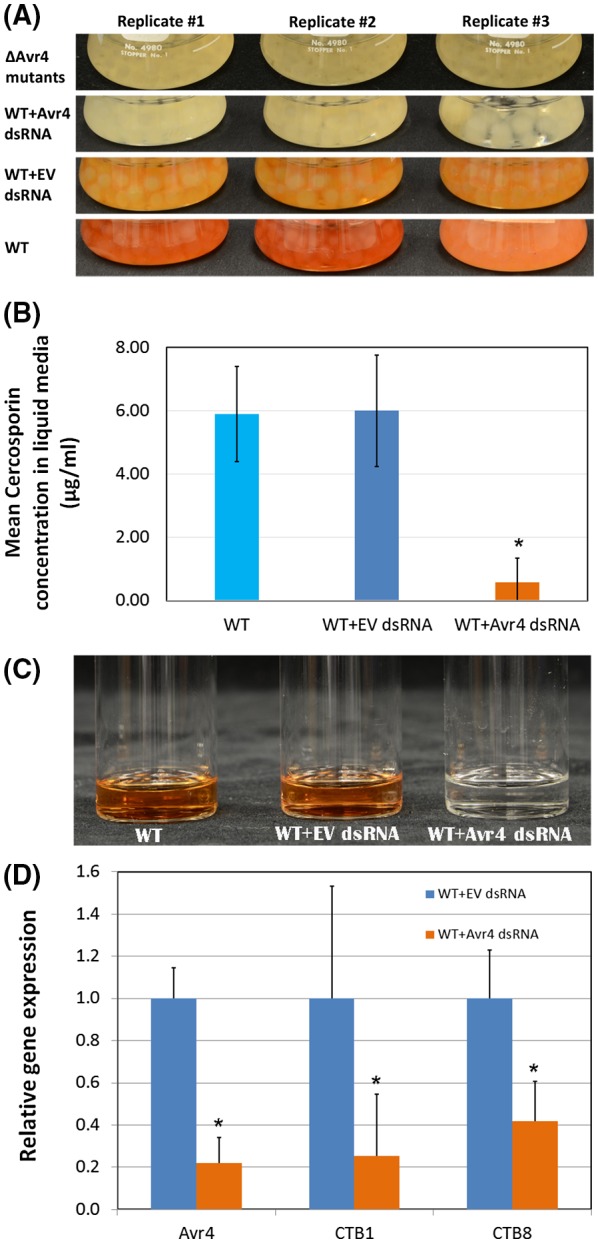
The effect of Avr4 dsRNA on cercosporin production and target gene expression. (A) Visual phenotype changes of the wild‐type (WT) Cercospora cf. flagellaris grown in half‐strength complete medium (CM) alone or treated with total nucleic acid extracted from Escherichia coli containing L4440 vector (WT + EV dsRNA) or containing Avr4 gene‐specific dsRNA (WT + Avr4 dsRNA) for 4 days in comparison to that of ∆avr4 mutant. The final concentration in the culture was 16 µg/mL for either EV dsRNA or Avr4 dsRNA. (B) Bar graph of cercosporin production in non‐treated WT C. cf. flagellaris grown in half‐strength CM or treated with total nucleic acid extract containing L4440 vector (EV) or containing Avr4 gene‐specific dsRNA (Avr4 dsRNA). (C) Visual comparison of cercosporin extracted from the WT C. cf. flagellaris under different treatments for 4 days. (D) Relative expressions of Avr4, CTB1 and CTB8 genes in the WT C. cf. flagellaris after different treatments.
Discussion
Cercospora cf. flagellaris is the causal agent of CLB disease in Louisiana and the interest in studying the AVR4 effector arose from the study by Stergiopoulos et al. (2010), which showed the presence of homologues of Avr4 in different species of the Dothideomycetes, including Cercospora spp., indicating its possible function as a virulence factor on distantly related host plants. Silencing of this gene in C. fulvum resulted in reduction of disease symptoms and fungal growth on tomato leaves (van Esse et al., 2007). AVR4 was found to be a chitin‐binding protein (van den Burg et al., 2006) and the presence of AVR4 in the apoplast of Arabidopsis and tomato plants has also been shown to enhance the susceptibility of these plants to several fungal pathogens (Cai et al., 2009; van Esse et al., 2007).
In the present study, the 1.6 kb fragment cloned from C. cf. flagellaris was identified as Avr4 based on its nucleotide and deduced amino acid sequence homologies with the Avr4 sequences from other Dothideomycete fungi (C. beticola, C. nicotianae, C. apii, C. zeina, Mycosphaerella fijiensis and C. fulvum). In addition, the Avr4 gene was detected in 150 isolates of C. cf. flagellaris collected from several locations throughout Louisiana. It is possible that Cercospora species maintain the conserved Avr4 to target a diverse range of hosts (Crous and Braun, 2003; Groenewald et al., 2013). Certainly, AVR4 as a chitin‐binding protein can be very useful in assisting fungi to enter and colonize different host plants that use chitinases as a defence mechanism to combat fungal invasion. Considering all the information above, we hypothesized that the presence of this effector in C. cf. flagellaris plays an important role in the virulence of this pathogen on soybean.
Avr4 disruption mutants were created from C. cf. flagellaris wild‐type isolate MRL 6020 2B, a well‐known cercosporin‐producing isolate, to elucidate the role of AVR4 in C. cf. flagellaris fitness and virulence. The first noticeable change in the phenotype of the ∆avr4 mutants was the reduced production of the dark‐red pigmentation on solid and liquid media, a characteristic of cercosporin that was verified through mass spectrometry (Fig. S6). Further investigation revealed that reduced cercosporin production was associated with the down‐regulation of CTB1 and CTB8 gene expression in the ∆avr4 mutants (Fig. 3). This result agreed with earlier studies showing that expression of CTB genes was correlated with toxin production (Chen et al., 2007).
The ∆avr4 mutants were found to contain more than one copy of the HYG cassette, indicating the presence of possible ectopic integrations besides the intended insertion in the Avr4 target gene (as determined through PCR). However, all three mutants shared the same phenotype and it is unlikely that random, unintended gene knockouts in three separately generated mutants contributed to those phenotypes without causing any other noticeable differences among them. Moreover, the above phenotypes, such as reduction in cercosporin production (Fig. 2) and significant suppression of cercosporin pathway genes (Fig. 3), were independently confirmed by in vitro dsRNA‐mediated suppression of Avr4 in the wild‐type (Fig. 7). These combined data conclusively demonstrated a direct role of AVR4 in affecting cercosporin biosynthetic pathway gene expression and cercosporin production. It is reasonable to speculate that the reduced virulence of the ∆avr4 mutants was most likely due to the reduced cercosporin production, since cercosporin was repeatedly reported as a virulence factor in Cercospora species, as well as a critical factor in the development of many plant diseases (Choquer et al., 2005; Gunasinghe et al., 2016; Staerkel et al., 2013; Upchurch et al., 1991a; Weiland et al., 2010). One possible explanation as to why different mutants with multiple copies of a HYG insertion exhibited the same phenotype is that the integrations may not be ectopic. Instead, they might have been integrated into the genome of the mutants as a concatemer at the intended target location (Avr4), which has been reported to occur (Kamisugi et al., 2006).
Apart from the altered cercosporin production, fungal growth was also different between C. cf. flagellaris ∆avr4 mutants and the wild‐type. Mutants were found to grow more rapidly on solid media compared to the wild‐type based on radial growth measurements (Fig. S3). This finding agrees with previous studies showing a negative correlation between fungal growth and cercosporin production (Jenns et al., 1989; Upchurch et al., 1991a). Compared to ∆avr4 mutants, the wild‐type strain grew more, not only saprophytically on the surface of inoculated soybean leaves based on our visual assessment, but also more abundantly inside the soybean leaves based on real‐time PCR quantification of fungal biomass (Fig. 6C), which clearly reflects the differences in virulence between the wild‐type pathogen and the mutants. Moreover, fungal growth was positively correlated with disease severity on soybean leaves, as wild‐type fungus caused more severe symptoms than the ∆avr4 mutants (Fig. 6).
Previous studies showed that the small conserved cysteine‐rich AVR4 protein binds chitin and protects C. fulvum against plant chitinases during infection of tomato and it also protects Trichoderma viride and Fusarium solani f. sp. phaseoli in the presence of chitinase and β‐1,3‐glucanase in vitro (van den Burg et al., 2006). This kind of protection against plant chitinases is essential for fungal development and colonization of its host. Recently, AVR4 from C. apii and C. beticola has been shown to bind chitin as well (Mesarich et al., 2016). In order to verify whether AVR4 from C. cf. flagellaris has the same protective function, a chitinase assay was also performed. As expected, ∆avr4 mutants were found to be more sensitive to chitinase than wild‐type C. cf. flagellaris (Fig. 4).
In order to verify if cercosporin could inhibit chitinase activity, a cup‐plate assay was performed and a clear reduction of the chitinolytic activity was observed in the presence of cercosporin, suggesting that this inhibition by cercosporin is protein‐specific, since the activity of an α‐amylase was not inhibited in the presence of cercosporin (Fig. 5). Therefore, the observed reduced colonization of soybean leaves by ∆avr4 mutants could be the combined result of lacking a functional AVR4 effector and the reduced production of cercosporin, which can not only cause peroxidation of membrane lipids leading to membrane breakdown and cell death (Daub and Chung, 2007; Daub and Hangarter, 1983), but can also directly inhibit chitinolytic activity.
Taken together, our findings suggest that this fungal effector protein suppresses host defence by affecting the expression of cercosporin pathway genes and toxin production. To the best of our knowledge, this is the first report concerning the roles of an AVR4 effector homologue in C. cf. flagellaris and the first report in linking AVR4 to cercosporin production. Further studies will be needed to understand how AVR4 affects cercosporin production in C. cf. flagellaris.
Experimental Procedures
Cloning of Avr4 from C. cf. flagellaris
Cercospora cf. flagellaris wild‐type isolate MRL 6020‐2B, a well‐known cercosporin‐producing isolate (Cai and Schneider, 2008), was grown for 5 days in PDB (Difco Laboratories Inc., Detroit, MI, USA) with constant shaking (200 rpm) at 25 °C under continuous light. Fungal dried mycelia were ground in liquid nitrogen and DNA was extracted with a GenElute Plant Genomic DNA Miniprep Kit (Sigma‐Aldrich, St Louis, MO, USA). Based on the Avr4 sequences available in GenBank (GU574324, GU574325, GU574326 and GU574327), Avr4F and Avr4R2 primers (see Table 1 for all primer sequences) were designed to confirm the presence of Avr4 in C. cf. flagellaris. The amplified fragment was cloned into pCR2.1‐TOPO TA Cloning vector (Invitrogen, Carlsbad, CA, USA) and sequenced. Genomic libraries of C. cf. flagellaris previously constructed using the Universal GenomeWalker kit (Clontech Laboratories Inc., Mountain View, CA, USA) by Chanda (2012) were used to clone the full‐length Avr4 gene according to the manufacturer’s guidelines. Adapter primers (AP1 and AP2) and gene‐specific primers (GSP1F and GSP1R, GSP2F and GSP2R) were used in two rounds of PCR to walk upwards and downwards into unknown genomic regions. The resulting DNA fragments were cloned into pCR2.1‐TOPO and sequenced.
Table 1.
List of primers used in this study
| Primer name | Oligonucleotide sequence (5ʹ→3ʹ) |
|---|---|
| Avr4F | AAGGATCCATGTACGGCCTCTTCCACCTC |
| Avr4R2 | AAGGTACCTTGGTGCAGGTGCAGGTGCTGA |
| AP1 | GTAATACGACTCACTATAGGGC |
| AP2 | ACTATAGGGCACGCGTGGT |
| GSP1F | AGACATGAAACGAACACACTGTATGG |
| GSP1R | AGTGCGTAGAGGCAGTCGAATGGAC |
| GSP2F | ACTACCAGGGAACTGCCTCGGATACAT |
| GSP2R | CCATACAGTGTGTTCGTTTCATGTCT |
| M13R | AGCGGATAACAATTTCACACAGGA |
| M13F | CGCCAGGGTTTTCCCAGTCACGAC |
| FP1‐1 | CCCGATGCTTGTCCGCAATA |
| RP1‐1* | TCCTGTGTGAAATTGTTATCCGCTTCTTGTTGTTGAGGCAGTGC |
| FP2‐2* | GTCGTGACTGGGAAAACCCTGGCGTGCTGGTCAATGGTGGAACG |
| RP2‐2 | CACAGTCTAACGCCTTCCGT |
| NLC37 | GGATGCCTCCGCTCGAAGTA |
| NLC38 | CGTTGCAAGAACTGCCTGAA |
| Avr4F1 | GCATTGCCCTAACAGACCATC |
| Avr4R1 | AGAATAGTGGGCGTTGCGT |
| Avra4sh_F* | GAATTAATACGACTCACTATAGGGAGACCATGTACGGCCTCTTCC |
| Avra4sh_R* | GAATTAATACGACTCACTATAGGGAGAGCATGTTGGAGGCTCCAC |
| CTB1 2F | ACCTTGCCTCAACTGTCTTAC |
| CTB1 2R | TGAAGCGACGAACGGATTT |
| CTB8 1F | GACAGCAGGTTATCTTCCAGAG |
| CTB8 1R | GTACTTATGCATCCACCACCA |
| Avr4WT‐F | CCGGTATCGCGTATGAAAGG |
| Avr4WT‐R | GAGAAGAAGTACTGCGACACGGT |
| Avr4WT‐PRB$ | FAM‐ATGCCGTGCTGGTCAATGGTGGA‐TAMRA |
| CKCTB6‐2F | CACCATGCTAGATGTGACGACA |
| CKCTB6‐2R | GGTCCTGGAGGCAGCCA |
| CKCTB6‐PRB$ | FAM‐CTCGTCGCACAGTCCCGCTTCG‐TAMRA |
| Hyg B qPCR F1 | GCT TTC AGC TTC GAT GTA GGA |
| Hyg B Prb 1$ | 5HEX/TAG CTG CGC /ZEN/CGA TGG TTT CTA CAA /3IABkFQ |
| Hyg B qPCR R1 | CGA TGC AAA GTG CCG ATA AAC |
| Lys2 qPCR Fl | GGT CAG TCG AAA TGG GTA TCT G |
| Lys2 qPCR Prb 1$ | 6‐FAM/TTG TTC GTT /ZEN/CCG GCT ACG TGA TGG /3IABkFQ |
| Lys2 qPCR Rl | TTG CTC ACG CCA GTC TTT |
The underlined are M13Foward and M13Reverse primer sequences (RP1‐1 and FP2‐2) or T7 promoter sequences needed for dsRNA production (Avr4sh_F and Avr4sh_R).
FAM and HEX are two different fluorescent dyes used to label the probes; TAMERA, ZEN and 3IABkFQ are different fluorescent quenchers.
Disruption of Avr4 in C. cf. flagellaris
Avr4 was disrupted by double homologous recombination using fusion PCR according to Yu et al. (2004). The hygromycin (HYG) cassette (2.5 kb) was amplified from pUCATPH (Lu et al., 1994) with the primers M13R and M13F. Cercospora cf. flagellaris genomic DNA was used to amplify the 5′ Avr4 fragment (0.4 kb) with FP1‐1 and RP1‐1, and the 3ʹ Avr4 fragment (0.5 kb) with FP2‐2 and RP2‐2. The underlined sequence in the primers RP1‐1 and FP2‐2 is complementary to the sequence of primers M13R and M13F, respectively. A second round of PCR was performed and a 1.5 kb fragment containing 5ʹ Avr4 fused with 3ʹ HYG was amplified using primers FP1‐1 and NLC37. A 2.5 kb fragment containing 5′ HYG fused with 3ʹ Avr4 was amplified with primers RP2‐2 and NLC38. The two PCR fragments, overlapping within the HYG region, were purified using a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany). These purified DNA fragments were used to transform the protoplasts of the C. cf. flagellaris wild‐type isolate as described below.
Protoplasts were prepared as previously described (Chung et al., 2002; Upchurch et al., 1991a). Fungal transformation was carried out as described by Turgeon et al. (2010). The colonies that appeared between 5 and 8 days were selected and transferred to CM plates containing 300 μg/mL hygromycin B (Roche Diagnostics, Indianaplis, IN, USA). Hygromycin‐resistant colonies were screened by PCR by using HYG‐specific primers NLC37 and NLC38. The primers Avr4F1 and Avr4R1 were used to validate site‐specific integration.
Fungal isolates, growth conditions and growth measurements
Cercospora cf. flagellaris MRL 6020‐2B (Cai and Schneider, 2005) and mutants (∆avr4M1, ∆avr4M2 and ∆avr4M3) were maintained on CM (Jenns et al., 1989) and the CM amended with 300 μg/mL hygromycin B, respectively. Fungal growth was assessed by placing a 7‐mm diameter mycelial plug in the centre of a PDA (IBI Scientific, Peosta, IA, USA) plate and colony radial growth measured every 2 days to determine the colony size increase over a period of 13 days. For liquid culture, the mycelial suspension prepared by grinding three 7‐mm mycelial plugs of 1‐week‐old fungal colony on solid CM in 2 mL of sterile water was used to inoculate 100 mL of liquid CM. Cultures were incubated at 25 °C with constant shaking (200 rpm) under light (240 μmol/m2/s) or dark (achieved by wrapping the flasks with two layers of aluminium foil) for 5 days.
Cercosporin extraction and quantification
From solid media, cercosporin was extracted from three 7‐mm mycelial plugs of 5‐day‐old fungal colony on PDA by soaking in 4 mL of 5 M KOH in the dark for 4 h and cercosporin was quantified using a spectrophotometer by measuring absorbance at 480 nm (Jenns et al., 1989). From liquid culture, cercosporin was extracted from 25 mL of 5‐day‐old fungal cultures grown in CM with 20 mL of ethyl acetate for 8 h as described by Gunasinghe et al. (2016). Extraction of cercosporin from soybean leaves infected with C. cf. flagellaris was also performed according to Gunasinghe et al. (2016). Briefly, soybean leaves (0.2 g) showing typical CLB symptoms were ground in 2 mL of ethyl acetate and the mixtures were kept overnight at 4 °C. Following centrifugation, the supernatant was collected and cercosporin was identified and quantified by high‐performance liquid chromatography (HPLC) as described below.
HPLC and mass spectrometry analyses of cercosporin
Cercosporin identification and quantification were achieved through comparing the retention time and area under the target peak of the pigment extracted from infected leaves or fungal cultures to the cercosporin standard (Sigma‐Aldrich). HPLC analysis of cercosporin was performed using a Waters (Milford, MA, USA) 2695 Separations Module with a Waters Atlantis C18 column (150 mm × 4.6 mm with 5 µm pore size) and detected using a Waters 2475 fluorescence detector with the mobile phases as described by Gunasinghe et al. (2016). In addition, cercosporin standard and the red pigment extracted from fungal cultures as well as C. cf. flagellaris inoculated soybean leaves were analysed by LC‐mass spectrometry through a fee‐based service at LSU Chemistry Department.
RNA isolation and real‐time PCR analysis of CTB1 and CTB8 gene expressions
Fungal cultures were grown for 5 days in CM with constant shaking (200 rpm). Cultures were incubated at 25 °C under either continuous white fluorescent light or in darkness, attained by wrapping flasks with two layers of aluminium foil. Dried fungal mycelia were ground to a fine powder in liquid nitrogen and RNA was isolated using RNeasy Plant Mini Kit (Qiagen). Subsequently, cDNA was synthesized from 500 ng of total RNA using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. RNase‐free DNase (Qiagen) was used to remove possible residual DNA contamination. Real‐time PCR experiments were performed using SYBR Green Master Mix (Applied Biosystems), CTB1 (2F and 2R) and CTB8 (1F and 1R) gene‐specific primers, and the ABI PRISM 7000 Sequence Detection System (Applied Biosystems) under standard conditions. The expression of CTB genes was normalized to β‐tubulin in the same culture to correct for template variations from one sample to another using the ∆C t method. The data presented here are a relative expression of target genes to β‐tubulin. Three biological replicates and three technical replicates within each biological replicate were used for each sample and a negative (template‐free) control.
Growth of C. cf. flagellaris wild‐type and ∆avr4 mutants in the presence of chitinase and agarose plate assay for chitinase and α‐amylase activity in the presence of cercosporin
A plate assay was performed to verify if growth of C. cf. flagellaris wild‐type and ∆avr4 mutants were affected by chitinase. Chitinase (Sigma‐Aldrich, #C8241‐25UN, ≥600 units/g) was dissolved in 0.1 M phosphate buffer. CM plates were inoculated with C. cf. flagellaris wild‐type or ∆avr4 mutants using 7‐mm diameter mycelial plugs and kept at 25 °C. After 14 days 4‐mm diameter wells were cut with a cork borer in the agar 1 mm away from the edge of the fungal colony and filled with 30 µL of 0.1 M phosphate buffer containing 1 U of chitinase or with 0.1 M phosphate buffer only as a control. Plates were sealed and kept at 25 °C for two additional days before fungal growth was visually assessed to determine whether mycelial development was affected around the wells or filter‐paper discs containing chitinase.
To verify if chitinolytic activity is influenced by cercosporin, a chitinase cup‐plate assay was performed. Glycol chitin substrate was synthesized according to Roberts and Selitrennikoff (1988) from shrimp‐shell chitin (Sigma‐Aldrich). To prepare chitin plates, agarose was dissolved (1.6% [w/v]) in 50 mM potassium phosphate buffer (pH 6.5). The dissolved agarose/buffer solution (20 mL) was cooled to 50–60 °C and 16.5 mg of glycol chitin and 30 µL of cercosporin (10 mM in acetonitrile) were added. Plates containing 30 µL acetonitrile only were used as controls. Four 7‐mm diameter wells were cut in the plates with a cork borer and 10 µL of 50 mM phosphate buffer only or buffer containing 0.25 or 0.5 U of chitinase enzyme were added to individual wells in the agarose plates. The plates were incubated at 25 °C and chitin hydrolysis, visualized as clear zones around the wells, was assessed after 48, 72 and 120 h. In order to determine whether the inhibition of activity by cercosporin is enzyme specific, 0.25 and 0.5 units of an α‐amylase from Aspergillus oryzae (Sigma, cat log #10065‐10G; 30 units/mg) in 10 µL of 50 mM potassium phosphate buffer (pH 6.5) was added to duplicated wells in 1% agarose plates containing 0.25% soluble starch in the above phosphate buffer with or without cercosporin. Four sets of plates (one control plate plus one cercosporin plate comprise a set) were used and one set of plates was stopped by adding 5 mM KI‐I2 solution at 16, 24, 48 and 72 h.
Fungal virulence assay on soybean
Soybean detached leaf assays were performed to examine the differences in fungal growth and virulence among the C. cf. flagellaris wild‐type and ∆avr4 mutants. Soybean plants cv. Syngenta 02JR423003 were grown in the greenhouse until R1 (beginning bloom) stage when leaves were collected and placed inside transparent plastic boxes containing moist paper towels. Mycelial agar plugs (7‐mm diameter) containing 2‐week‐old C. cf. flagellaris wild‐type or ∆avr4 mutants grown on PDA were placed on the adaxial surface of soybean leaves with the mycelial side touching the leaf and gently pressed. Mycelial plugs or macerated mycelia instead of spores are often used to study C. kikuchii infection on soybean due to the poor sporulation of the pathogen in vitro, a common issue for many Cercospora species (Almeida et al., 2005; Callahan et al., 1999; Upchurch et al., 1991b). The inoculated leaves were incubated under fluorescent light at room temperature. Disease severity was assessed 14 days post‐inoculation (dpi) using a 1–5 rating scale. The photos for the rating scale were selected from a large pool of plug‐inoculated soybean leaf photographs 2 weeks after inoculation, and assigned a number of 1 for least and 5 for most severe based on size and severity of lesion. Three boxes each containing eight trifoliate leaves were used for each treatment. Leaves from individual boxes were considered as one sample and individual boxes was considered as a replicate. The experiment was repeated three times.
Quantification of fungal growth using real‐time PCR
Fourteen days post‐inoculation, soybean leaves infected with C. cf. flagellaris wild‐type or ∆avr4 mutants were washed with sterile distilled water to remove agar plugs and fungal mycelia on the leaf surfaces before 22‐mm leaf discs were cut with a cork borer from inoculated leaves and ground in liquid nitrogen. Leaf genomic DNA was extracted as described above. Fungal biomass of C. cf. flagellaris in inoculated soybean leaves was quantified using real‐time PCR with primers CKCTB6‐2F/CKCTB6‐2R and fluorescent probe CKCTB6‐PRB in the ABI 7000 equipment (Applied Biosystems) according to Chanda et al. (2014).
Droplet digital PCR to determine the number of integrations of the Avr4 deletion cassette in the mutants
To determine whether there is any ectopic integration of the HYG cassette in non‐targeted areas in the mutants, a phenomenon that appears to occur frequently in C. fulvum even when using Agrobacterium tumefaciens‐mediated transformation (Ökmen et al., 2014), droplet digital PCR was performed at the Core Research Facility, at the Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, USA. The HYG gene copy number (target) was quantified using genomic DNA samples extracted from the wild‐type and mutants as templates, with a known single‐copy gene encoding aminoadipate reductase (LYS2) (An et al., 2002) as a reference. The primer and probe sequences used for droplet digital PCR are presented in Table 1. The accuracy of using droplet digital PCR to determine target gene copy number in comparison to Southern blot analysis has been well documented in earlier studies (Collier et al., 2017; Głowacka et al., 2016).
Avr4 dsRNA production in E. coli
To independently verify the role of AVR4 on cercosporin production, a 309 bp fragment of the Avr4 gene was amplified with primer pair Avr4sh_F and Avr4sh_R (Table 1), cloned into pCR2.1‐TOPO, and then into pL4440 between XhoI and SacI sites to form L4440‐Avr4 for dsRNA production in E. coli HT115 (DE3) cells according to Tenllado et al. (2003). dsRNA synthesis was induced with isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) (0.4 mM) for 5 h before the bacterial cells were harvested, washed twice with distilled water, resuspended in RNase‐free water to 1/20 of original culture volume and lysed by passing through a French press twice at 1000–1250 psi. After centrifugation for 10 min at 4 °C and 10 000g, the supernatant of the lysate was saved as crude dsRNA extract. To further purify the dsRNA, the crude extract was vigorously mixed with an equal volume of chloroform and pelleted by centrifugation at 10 000g for 10 min at 4 °C. The aqueous phase was transferred to a new 50 mL centrifuge tube and the dsRNA was ethanol precipitated and finally resuspended in 50 mL RNase‐free water for the in vitro culture study below.
Culturing C. cf. flagellaris in the presence of dsRNA
Freshly macerated 2‐week‐old C. cf. flagellaris wild‐type mycelia (0.5 mL) was added to 50 mL flasks containing 20 mL of half‐strength CM only (control) or 20 mL of half‐strength CM with dsRNA (at a final concentration of 16 µg/mL of culture) prepared from bacterial cells containing L4440‐Avr4 construct or half‐strength CM with cell lysate prepared from HT115 bacterial culture with the empty L4440 vector only (at a final concentration of 16 µg dsRNA per millilitre of culture) to serve as an additional control. Twenty millilitres of half‐strength CM inoculated with ground mycelia of 2‐week‐old C. cf. flagellaris Avr4 mutant was included as a positive control. After 4 days of culturing at 25 °C with constant light (240 μmol/m2/s), cercosporin production in liquid media was measured as described above using HPLC. Each treatment had five replicates and this experiment was repeated three times.
Supporting information
Fig. S1 Cercospora cf. flagellaris Avr4 gene sequence. The putative transcription start site at –15 relative to the putative start codon ATG (highlighted and italicized) is highlighted in bold and larger font size on top of the amino acid sequences.
Fig. S2 Alignment of AVR4 homologues from several Cercospora species. Alignment of AVR4 from C. cf. flagellaris, C. nicotianae, C. zeina, C. beticola and C. apii. was produced using Clustal Omega. Conserved amino acid residues are indicated with an asterisk. Cysteine residues (C) are shown in dark grey boxes as vertical lines. The chitin‐binding peritrophin‐A domain is shown in light grey boxes as a horizontal underline.
Fig. S3 Differences in phenotype and growth of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants. Colony radial growth of fungal cultures on potato dextrose agar plates was measured every 2 days to determine the colony size increase over a period of 13 days. Error bars indicate standard error of the mean of three different experiments with four biological replicates for each fungal isolate.
Fig. S4 Phenotypes of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants grown in liquid complete medium and potato dextrose broth. Cultures were incubated at 25 °C with constant shaking (200 rpm) under light (240 μmol/m2/s) for 5 days.
Fig. S5 HPLC analysis of red pigments extracted from Cercospora cf. flagellaris wild‐type (W)T and ∆avr4 mutants grown in liquid complete medium, potato dextrose broth and soybean leaves that had been inoculated with the pathogen. The peak height and retention time of the cercosporin standard (10 µg/mL) and the putative cercosporin in the extracts are indicated in the chromatograph for each of the samples.
Fig. S6 Mass spectrometry analysis of cercosporin standard and the red pigment extracted from Cercospora cf. flagellaris wild‐type (WT) grown in liquid complete medium. The major peak in cercosporin standard (A) has a measured mass to charge ratio (m/z) of 535.1603 and that of the major peak in the red pigment (B) is 535.1517. The measured m/z of both are very close to the calculated m/z of cercosporin standard (535.1599), confirming the red pigment is indeed composed mainly of cercosporin.
Table S1 Gene copy number analysis through droplet digital PCR of genomic DNA from the wild‐type Cercospora cf. flagellaris (MRL 6020‐2B) and the ∆avr4 mutants.
Acknowledgements
Cercospora cf. flagellaris isolate MRL 6020 2B was provided by Dr Raymond Schneider. The genomic libraries used for cloning Avr4 were constructed by Dr Ashok Chanda. The HYG gene encoding a phosphotransferase conferring hygromycin resistance in the pUCATPH vector (Lu et al., 1994) was kindly provided by Dr Gillian Turgeon (Department of Plant Pathology and Plant–Microbe Biology, Cornell University, USA). L4440 vector and E. coli strain HT115 (DE3) were kindly provided by Dr Tenllado (Departamento de Biología de Plantas, Centro de Investigaciones Biológicas, CSIC, Madrid, Spain). The research was supported by the Louisiana State Soybean and Small Grain Promotion Board. Published with the approval of the Director of the Louisiana State University Agricultural Center Agricultural Experiment Station as manuscript number 2017‐240‐31440.
References
- Albu, S. , Schneider, R.W. , Price, P.P. and Doyle, V.P. (2016) Cercospora cf. flagellaris and Cercospora cf. sigesbeckiae are associated with cercospora leaf blight and purple seed stain on soybean in North America. Phytopathology, 106, 1376–1385. [DOI] [PubMed] [Google Scholar]
- Allen, T.W. , Bradley, C.A. , Damicone, J.P. , Dufault, N.S. , Faske, T.R. , Hollier, C.A. , et al. (2017) Southern United States soybean disease loss estimates for 2016. SSDW Proceedings, 3–9. [Google Scholar]
- Almeida, Á.M. , Piuga, F.F. , Marin, S.R. , Binneck, E. , Sartori, F. , Costamilan, L.M. , Teixeira, M.R.O. and Lopes, M. (2005) Pathogenicity, molecular characterization, and cercosporin content of Brazilian isolates of Cercospora kikuchii . Fitopatol .Bras. 30, 594–602. [Google Scholar]
- Amnuaykanjanasin, A. and Daub, M.E. (2009) The ABC transporter ATR1 is necessary for efflux of the toxin cercosporin in the fungus Cercospora nicotianae . Fungal Genet. Biol. 46, 146–158. [DOI] [PubMed] [Google Scholar]
- An, K.‐D. , Nishida, H. , Miura, Y. and Yokota, A. (2002) Aminoadipate reductase gene: a new fungal‐specific gene for comparative evolutionary analyses. BMC Evol. Biol. 2, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg, H.A. , Harrison, S.J. , Joosten, M.H.A.J. , Vervoort, J. and de Wit, P.J.G.M. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant–Microbe Interact., 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Cai, G. and Schneider, R.W. (2005) Vegetative compatibility groups in Cercospora kikuchii, the causal agent of cercospora leaf blight and purple seed stain in soybean. Phytopathology, 95, 257–261. [DOI] [PubMed] [Google Scholar]
- Cai, G. and Schneider, R. (2008) Population structure of Cercospora kikuchii, the causal agent of Cercospora leaf blight and purple seed stain in soybean. Phytopathology, 98, 823–829. [DOI] [PubMed] [Google Scholar]
- Cai, G. , Schneider, R.W. and Padgett, G.B. (2009) Assessment of lineages of Cercospora kikuchii in Louisiana for aggressiveness and screening soybean cultivars for resistance to cercospora leaf blight. Plant Dis. 93, 868–874. [DOI] [PubMed] [Google Scholar]
- Callahan, T.M. , Rose, M.S. , Meade, M.J. , Ehrenshaft, M. and Upchurch, R.G. (1999) CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant–Microbe Interact. 12, 901–910. [DOI] [PubMed] [Google Scholar]
- Chanda, A.K. (2012) Molecular approaches to detect and control Cercospora kikuchii in soybeans In: Plant Pathology and Crop Physiology. LSU Doctoral Dissertations. Louisiana State University. [Google Scholar]
- Chanda, A.K. , Ward, N.A. , Robertson, C.L. , Chen, Z.Y. and Schneider, R.W. (2014) Development of a quantitative polymerase chain reaction detection protocol for Cercospora kikuchii in soybean leaves and its use for documenting latent infection as affected by fungicide applications. Phytopathology, 104, 1118–1124. [DOI] [PubMed] [Google Scholar]
- Chen, H. , Lee, M.H. , Daub, M.E. and Chung, K.R. (2007) Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae . Mol. Microbiol. 64, 755–770. [DOI] [PubMed] [Google Scholar]
- Choquer, M. , Dekkers, K.L. , Chen, H.‐Q. , Cao, L. , Ueng, P.P. , Daub, M.E. and Chung, K.R. (2005) The CTB1 gene encoding a fungal polyketide synthase is required for cercosporin biosynthesis and fungal virulence of Cercospora nicotianae . Mol. Plant–Microbe Interact. 18, 468–476. [DOI] [PubMed] [Google Scholar]
- Chung, K.‐R. , Shilts, T. , Li, W. and Timmer, L.W. (2002) Engineering a genetic transformation system for Colletotrichum acutatum, the causal fungus of lime anthracnose and postbloom fruit drop of citrus. FEMS Microbiol. Lett. 213, 33–39. [DOI] [PubMed] [Google Scholar]
- Chung, K.‐R. , Daub, M.E. , Kuchler, K. and Schüller, C. (2003) The CRG1 gene required for resistance to the singlet oxygen‐generating cercosporin toxin in Cercospora nicotianae encodes a putative fungal transcription factor. Biochem. Biophys. Res. Commun. 302, 302–310. [DOI] [PubMed] [Google Scholar]
- Collier, R. , Dasgupta, K. , Xing, Y.‐P. , Hernandez, B.T. , Shao, M. , Rohozinski, D. , Kovak, E. , Lin, J. , de Oliveira, M.L.P. , Stover, E. , McCue, K.F. , Harmon, F.G. , Blechl, A. , Thomson, J.G. and Thilmony, R. (2017) Accurate measurement of transgene copy number in crop plants using droplet digital PCR. The Plant Journal, 90, 1014–1025. [DOI] [PubMed] [Google Scholar]
- Crous, P.W. and Braun, U. (2003) Mycosphaerella and its anamorphs: 1. Names published in Cercospora and Passalora. Utrecht, Netherlands: Centraalbureau voor Schimmelcultures (CBS). [Google Scholar]
- Daub, M.E. (1982) Peroxidation of tobacco membrane lipids by the photosensitizing toxin, cercosporin. Plant Physiol. 69, 1361–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, M.E. and Briggs, S.P. (1983) Changes in tobacco cell membrane composition and structure caused by cercosporin. Plant Physiol. 71, 763–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, M.E. and Chung, K.‐R. (2007) Cercosporin: a photoactivated toxin in plant disease. St. Paul, MN: The American Phytopathological Society. [Google Scholar]
- Daub, M.E. and Ehrenshaft, M. (2000) The photoactivated cercospora toxin cercosporin: contributions to plant disease and fundamental biology. Annu. Rev. Phytopathol. 38, 461–490. [DOI] [PubMed] [Google Scholar]
- Daub, M.E. and Hangarter, R.P. (1983) Light‐induced production of singlet oxygen and superoxide by the fungal toxin, cercosporin. Plant Physiol. 73, 855–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daub, M.E. , Herrero, S. and Chung, K.R. (2005) Photoactivated perylenequinone toxins in fungal pathogenesis of plants. FEMS Microbiol. Lett. 252, 197–206. [DOI] [PubMed] [Google Scholar]
- van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , de Wit, P.J. and Thomma, B.P. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant–Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- Głowacka, K. , Kromdijk, J. , Leonelli, L. , Niyogi, K.K. , Clemente, T.E. and Long, S.P. (2016) An evaluation of new and established methods to determine T‐DNA copy number and homozygosity in transgenic plants. Plant Cell Environ. 39, 908–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenewald, J.Z. , Nakashima, C. , Nishikawa, J. , Shin, H.D. , Park, J.H. , Jama, A.N. , Groenewald, M. , Braun, U. and Crous, P. (2013) Species concepts in Cercospora: spotting the weeds among the roses. Stud. Mycol. 75, 115–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasinghe, N. , You, M.P. , Cawthray, G.R. and Barbetti, M.J. (2016) Cercosporin from Pseudocercosporella capsellae and its critical role in white leaf spot development. Plant Dis. 100, 1521–1531. [DOI] [PubMed] [Google Scholar]
- Hartman, G.L. , Sinclair, J.B. and Rupe, J.C. (1999) Compendium of soybean diseases. St. Paul, MN: American Phytopathological Society. [Google Scholar]
- Jenns, A.E. , Daub, M.E. and Upchurch, R.G. (1989) Regulation of cercosporin accumulation in culture by medium and temperature manipulation. Phytopathology, 79, 213–219. [Google Scholar]
- de Jonge, R. , Ebert, M.K. , Huitt‐Roehl, C.R. , Pal, P. , Suttle, J.C. , Neubauer, J.D. , Jurick, W.M. , Secor, G.A. , Thomma, B.P. , Van de Peer, Y. and Townsend, C.A. (2017) Ancient duplication and horizontal transfer of a toxin gene cluster reveals novel mechanisms in the cercosporin biosynthesis pathway. bioRxiv, 100545. [Google Scholar]
- de Jonge, R. , Ebert, M.K. , Huitt‐Roehl, C.R. , Pal, P. , Suttle, J.C. , Spanner, R.E. , Neubauer, J.D. , Jurick, W.M. , Stott, K.A. , Secor, G.A. and Thomma, B.P. (2018) Gene cluster conservation provides insight into cercosporin biosynthesis and extends production to the genus Colletotrichum . Proc. Natl. Acad. Sci. USA. 115, E5459–E5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten, M.H.A.J. , Cozijnsen, T.J. and De Wit, P.J.G.M. (1994) Host resistance to a fungal tomato pathogen lost by a single base‐pair change in an avirulence gene. Nature, 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Kamisugi, Y. , Schlink, K. , Rensing, S.A. , Schween, G. , von Stackelberg, M. , Cuming, A.C. , Reski, R. and Cove, D.J. (2006) The mechanism of gene targeting in Physcomitrella patens: homologous recombination, concatenation and multiple integration. Nucleic Acids Res. 34, 6205–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, A.C. , Chen, L.‐H. , Hurlburt, N. , Salvucci, A. , Schwessinger, B. , Fisher, A.J. and Stergiopoulos, I. (2016) Structural analysis of an Avr4 effector ortholog offers insight into chitin binding and recognition by the Cf‐4 Receptor. Plant Cell, 28, 1945–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyama, S. and Tamura, T. (1957) Cercosporin. A pigment of Cercosporina kikuchii Matsumoto et Tomoyasu. II. Physical and chemical properties of cercosporin and its derivatives. J. Am. Chem. Soc. 79, 5726–5729. [Google Scholar]
- Lu, S. , Lyngholm, L. , Yang, G. , Bronson, C. , Yoder, O.C. and Turgeon, B.G. (1994) Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme‐mediated integration. Proc. Natl. Acad. Sci. USA. 91, 12649–12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto, T. and Tomoyasu, R. (1925) Studies on the purple speck of soybean seed. Ann. Phytopathol. Soc. Jpn. 1, 1–14. [Google Scholar]
- Mesarich, C.H. , Stergiopoulos, I. , Beenen, H.G. , Cordovez, V. , Guo, Y. , Karimi Jashni, M. , Bradshaw, R.E. and de Wit, P.J. (2016) A conserved proline residue in Dothideomycete Avr4 effector proteins is required to trigger a Cf‐4‐dependent hypersensitive response. Mol. Plant Pathol. 17, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, S.H. and Wolcott, M.C. (2000) Using yield maps to create management zones in field crops. Louisiana Agriculture, 43, 12–13. [Google Scholar]
- Newman, A.G. and Townsend, C.A. (2016) Molecular characterization of the cercosporin biosynthetic pathway in the fungal plant pathogen Cercospora nicotianae . J. Am. Chem. Soc. 138, 4219–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ökmen, B. , Collemare, J. , Griffiths, S. , van der Burgt, A. , Cox, R. and de Wit, P.J.G.M. (2014) Functional analysis of the conserved transcriptional regulator CfWor1 in Cladosporium fulvum reveals diverse roles in the virulence of plant pathogenic fungi. Mol. Microbiol. 92, 10–27. [DOI] [PubMed] [Google Scholar]
- Roberts, W.K. and Selitrennikoff, C.P. (1988) Plant and bacterial chitinases differ in antifungal activity. J. Gen. Microbiol. 134, 169–176. [Google Scholar]
- Shim, W.‐B. and Dunkle, L.D. (2003) CZK3, a MAP kinase kinase kinase homolog in Cercospora zeae‐maydis, regulates cercosporin biosynthesis, fungal development, and pathogenesis. Mol. Plant–Microbe Interact. 16, 760–768. [DOI] [PubMed] [Google Scholar]
- Sikora, E.J. , Murphy, J.F. , Lawrence, K.S. and Mullen, J.M. (2011) Survey of fungal, nematode and viral diseases of soybean in Alabama. Plant Health Progress. 10.1094/PHP-2011-1227-01-RS. [DOI] [Google Scholar]
- Staerkel, C. , Boenisch, M.J. , Kröger, C. , Bormann, J. , Schäfer, W. and Stahl, D. (2013) CbCTB2, an O‐methyltransferase is essential for biosynthesis of the phytotoxin cercosporin and infection of sugar beet by Cercospora beticola . BMC Plant Biol. 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiopoulos, I. , van den Burg, H.A. , Ökmen, B. , Beenen, H.G. , van Liere, S. , Kema, G.H.J. and de Wit, P.J. (2010) Tomato Cf resistance proteins mediate recognition of cognate homologous effectors from fungi pathogenic on dicots and monocots. Proc. Natl. Acad. Sci. USA. 107, 7610–7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K. (1921) Studies on the cause of purple seed of soybeans. Chosen Agric. Assoc. Rep. 16, 24–28. [Google Scholar]
- Tenllado, F. , Martínez‐García, B. , Vargas, M. and Díaz‐Ruíz, J.R. (2003) Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 3, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon, B.G. , Condon, B. , Liu, J. and Zhang, N. (2010) Protoplast transformation of filamentous fungi. Methods Mol. Biol. 638, 3–19. [DOI] [PubMed] [Google Scholar]
- Upchurch, R.G. , Ehrenshaft, M. , Walker, D.C. and Sanders, L.A. (1991a) Genetic transformation system for the fungal soybean pathogen Cercospora kikuchii . Appl. Environ. Microbiol. 57, 2935–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upchurch, R.G. , Walker, D.C. , Rollins, J.A. , Ehrenshaft, M. and Daub, M.E. (1991b) Mutants of Cercospora kikuchii altered in cercosporin synthesis and pathogenicity. Appl. Environ. Microbiol. 57, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, H. (1980) Soybean leaf blight caused by Cercospora kikuchii . Plant Dis. 64, 961–962. [Google Scholar]
- Weiland, J. , Chung, K. and Suttle, J. (2010) The role of cercosporin in the virulence of Cercospora spp. to plant hosts. St. Paul, MN: APS Press. [Google Scholar]
- Wrather, J.A. , Anderson, T. , Arsyad, D. , Gai, J. , Ploper, L. , Porta‐Puglia, A. , Ram, H.H. and Yorinori, J.T. (1997) Soybean disease loss estimates for the top 10 soybean producing countries in 1994. Plant Dis. 81, 107–110. [DOI] [PubMed] [Google Scholar]
- Wrather, J. , Anderson, T. , Arsyad, D. , Tan, Y. , Ploper, L. , Porta‐Puglia, A. , Ram, H. and Yorinori, J. (2001) Soybean disease loss estimates for the top ten soybean‐producing countries in 1998. Can. J. Plant Pathol. 23, 115–121. [DOI] [PubMed] [Google Scholar]
- Yu, J.‐H. , Hamari, Z. , Han, K.‐H. , Seo, J.‐A. , Reyes‐Domínguez, Y. and Scazzocchio, C. (2004) Double‐joint PCR: a PCR‐based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Cercospora cf. flagellaris Avr4 gene sequence. The putative transcription start site at –15 relative to the putative start codon ATG (highlighted and italicized) is highlighted in bold and larger font size on top of the amino acid sequences.
Fig. S2 Alignment of AVR4 homologues from several Cercospora species. Alignment of AVR4 from C. cf. flagellaris, C. nicotianae, C. zeina, C. beticola and C. apii. was produced using Clustal Omega. Conserved amino acid residues are indicated with an asterisk. Cysteine residues (C) are shown in dark grey boxes as vertical lines. The chitin‐binding peritrophin‐A domain is shown in light grey boxes as a horizontal underline.
Fig. S3 Differences in phenotype and growth of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants. Colony radial growth of fungal cultures on potato dextrose agar plates was measured every 2 days to determine the colony size increase over a period of 13 days. Error bars indicate standard error of the mean of three different experiments with four biological replicates for each fungal isolate.
Fig. S4 Phenotypes of Cercospora cf. flagellaris wild‐type (WT) and ∆avr4 mutants grown in liquid complete medium and potato dextrose broth. Cultures were incubated at 25 °C with constant shaking (200 rpm) under light (240 μmol/m2/s) for 5 days.
Fig. S5 HPLC analysis of red pigments extracted from Cercospora cf. flagellaris wild‐type (W)T and ∆avr4 mutants grown in liquid complete medium, potato dextrose broth and soybean leaves that had been inoculated with the pathogen. The peak height and retention time of the cercosporin standard (10 µg/mL) and the putative cercosporin in the extracts are indicated in the chromatograph for each of the samples.
Fig. S6 Mass spectrometry analysis of cercosporin standard and the red pigment extracted from Cercospora cf. flagellaris wild‐type (WT) grown in liquid complete medium. The major peak in cercosporin standard (A) has a measured mass to charge ratio (m/z) of 535.1603 and that of the major peak in the red pigment (B) is 535.1517. The measured m/z of both are very close to the calculated m/z of cercosporin standard (535.1599), confirming the red pigment is indeed composed mainly of cercosporin.
Table S1 Gene copy number analysis through droplet digital PCR of genomic DNA from the wild‐type Cercospora cf. flagellaris (MRL 6020‐2B) and the ∆avr4 mutants.


