Abstract
Objective
This article presents our approaches to extraction of medications and associated adverse drug events (ADEs) from clinical documents, which is the second track of the 2018 National NLP Clinical Challenges (n2c2) shared task.
Materials and Methods
The clinical corpus used in this study was from the MIMIC-III database and the organizers annotated 303 documents for training and 202 for testing. Our system consists of 2 components: a named entity recognition (NER) and a relation classification (RC) component. For each component, we implemented deep learning-based approaches (eg, BI-LSTM-CRF) and compared them with traditional machine learning approaches, namely, conditional random fields for NER and support vector machines for RC, respectively. In addition, we developed a deep learning-based joint model that recognizes ADEs and their relations to medications in 1 step using a sequence labeling approach. To further improve the performance, we also investigated different ensemble approaches to generating optimal performance by combining outputs from multiple approaches.
Results
Our best-performing systems achieved F1 scores of 93.45% for NER, 96.30% for RC, and 89.05% for end-to-end evaluation, which ranked #2, #1, and #1 among all participants, respectively. Additional evaluations show that the deep learning-based approaches did outperform traditional machine learning algorithms in both NER and RC. The joint model that simultaneously recognizes ADEs and their relations to medications also achieved the best performance on RC, indicating its promise for relation extraction.
Conclusion
In this study, we developed deep learning approaches for extracting medications and their attributes such as ADEs, and demonstrated its superior performance compared with traditional machine learning algorithms, indicating its uses in broader NER and RC tasks in the medical domain.
Keywords: adverse drug events, electronic health records, deep learning, named entity recognition, relation extraction
INTRODUCTION
Electronic health record (EHR) systems have been widely used in clinical practice and have generated a mass of clinical data which has become an enabling resource for clinical research including pharmacovigilance. The goal of pharmacovigilance is to detect, monitor, characterize, and prevent adverse drug events (ADEs) associated with pharmaceutical products.1 Traditionally, spontaneous reporting systems were developed to collect safety information, but they were passive and reactive. The imperative to shift the paradigm towards a more proactive approach has resulted in an exploration of accessible data resources including EHRs, biomedical literature, product labels, and content from social media.2,3 Some previous studies have attempted to detect ADEs using EHRs in pharmacovigilance.4,5 For example, ICD-9 encoded data can be used for ADE identification; however, it has low sensitivity.6 But the rich information in clinical narratives can help to improve ADE detection, since more details of the diseases (such as signs and symptoms, disease status, and severity) are all typically recorded in clinical text.1 Although manual chart-review is an effective tool for ADE detection, it is also very expensive7 and time-consuming. To address this problem, natural language processing (NLP) serves as a potential solution to detect ADEs accurately and automatically from free text, which makes it possible to build a real-time sustainable and large-scale pharmacovigilance system in clinical practice.
Many studies have attempted to extract medication, ADEs, and their relations using NLP. Some representative systems and studies extracting medication and ADEs from EHRs include MedEx,8 USyd’s system,9 and MedXN.10 Moreover, the clinical NLP community has launched a series of challenges during past years to promote the system development for recognizing medication and ADEs.11 Some community challenges that focused on identifying medications and ADEs from EHRs and drug labels included 1) the 2009 Informatics for Integrating Biology and the Bedside,12 2) the 2017 Text Analysis Conference,13 3) the Medication, Indication, and Adverse Drug Events from Electronic Health Record Notes,14 and 4) The 2018 National NLP Clinical Challenges (n2c2).15
Early systems mainly used regular expressions and rule-based methods to extract medications.8,10 With the increasing availability of annotated data sets from community challenges, machine learning (ML)-based approaches have become predominant. Usually, the clinical NLP pipeline for this task consists of 2 steps: 1) concepts of medications and attributes are recognized first and 2) their relations are identified. Concept extraction is a typical sequence labeling problem in ML, and conditional random field (CRF) was the most commonly used method in previous studies.9,16,17 On the other hand, relation recognition can be represented as a classification problem, and support vector machine (SVM) was commonly used in some previous studies.9,17 With the rapid development of deep learning methods in recent years, some studies used deep learning models to extract medication, ADEs, and their relations. Typically, recurrent neural networks (RNN) and their variants such as bi-directional LSTM CRF (BI-LSTM-CRF)18,19 were used for clinical named entity recognition (NER) tasks.20–22 For example, Dandalal et al.23 developed a system to recognize medications, ADEs, and indications for the Medication, Indication, and Adverse Drug Events challenge, etc. In their system, a sequence labeling model was used to recognize all medications and symptoms first, and then a classifier was used to classify whether a pair of medication and symptom has a relation of ADE or indication.
However, there is an inevitable error propagation issue with the pipeline-based method: 1) the inaccurately identified concepts will directly affect the end-to-end performance of relation recognition. Therefore, some studies in the open domain have investigated joint-learning methods which can recognize concepts and relations simultaneously, so that the information from recognition of concepts and relations can benefit each other.24,25 However, very few studies in the clinical domain have worked on this problem. 2) In addition to the issue of error propagation, the traditional 2-step pipeline method faces another major problem, namely, omitted annotations of attribute entities. In the challenge of 2018 n2c2 Track 2, entities of ADE and reason were not to be annotated in a gold-standard corpus if they are not associated with any drug. This makes the corpus unable to train an effective NER model and misses a majority of negative attribute–concept candidate pairs that are required to train an effective relation classifier.
To address the above challenges, we proposed a joint learning model for our participation in the 2018 n2c2 Track 2 in which the entities of ADE, the reason, and their relations with medications are recognized simultaneously in a single-sequence labeling model. There are 3 subtasks in this challenge: 1) NER: identifying drugs and their attributes; 2) relations classification: identifying relations between drugs and their attributes given gold-standard concepts; and 3) end-to-end: between drugs and their attributes given gold-standard concepts. More details of the challenge, such as annotation guidelines and data set information, can be found in Ozlem et al.15 Here we describe our entry to the challenge, which is mainly based on new deep learning approaches and was top-ranked among all participating teams.
MATERIALS AND METHODS
Data set
The n2c2 corpus included 505 discharge summaries, which came from the MIMIC-III (Medical Information Mart for Intensive Care III) clinical care database.26 The corpus contained 9 types of clinical concepts including drug and its 8 attributes (reason, ADE, frequency, strength, duration, route, form, and dosage). The relations between drug and the 8 attributes were also annotated. The training data set included 303 discharge summaries and the test set included 202 discharge summaries.
Study overview
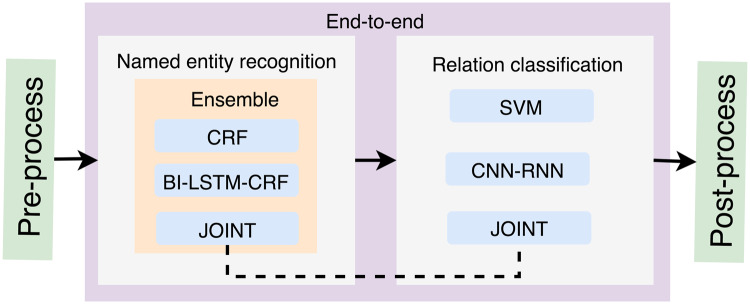
Figure 1 shows an overview of our study design. Our system consists of 4 components: pre-processing, NER, RC, and post-processing. The subtask 1 is a NER task, for which 3 ML-based methods were investigated, including CRF, BI-LSTM-CRF, and a joint-learning method. Ensemble methods were also employed to combine the outputs from the 3 single models to obtain a better performance. Furthermore, the subtask 2 is a RC task, for which we examined SVM, a deep learning method with a parallel neural network of Convolution Neural Network and Recurrent Neural Network (CNN-RNN), and the joint–learning-based method as in subtask 1. In addition, rule-based postprocessing was applied to the outputs of the RC component. The details of the system components are described below.
Figure 1.
Overview of our system for the n2c2 challenge.
Preprocessing
The preprocessing module includes basic steps such as sentence boundary detection, tokenization, and POS tagging, which were done using the Clinical Language Annotation, Modeling, and Processing toolkit.27
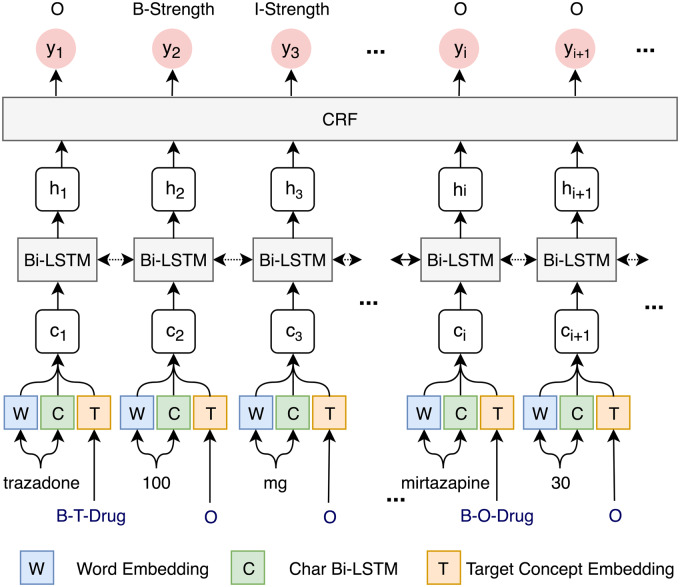
Named entity recognition
Annotated data were transformed into the BIO format, where “B” represents the beginning of an entity, “I” represents other words inside an entity, and “O” represents all other nonentity words. Figure 2 showed a sample sentence using the BIO format to represent the label of each word. The NER models will predict the BIO labels for the input sentences, which will be transformed back to named entities. The 3 NER algorithms, CRF, BI-LSTM_CRF, and a new approach that jointly recognizes attribute entities and their relations to drugs (JOINT), were individually built to recognize medication and attribute entities and then combined using different ensemble strategies.
Figure 2.
An example of BIO tags for named entity representation in the sequence labeling task.
CRF
CRF is known for good performance in NER, including in medication information extraction.9,16,17 Therefore, we chose it as a strong baseline for comparison with deep learning approaches. We used the CRF implementation in the Clinical Language Annotation, Modeling, and Processing toolkit27 to recognize all the entities, including drugs and their attributes, simultaneously (with features of word shape, n-gram, prefix-suffix, orthographic, and discretized word embeddings, etc). The Supplementary MaterialTable 1 shows the descriptions of the features and their values for the word “trazadone” (in Figure 2) as an example.
Table 1.
The lenient F1-score of single machine learning models and ensemble methods for the named entity recognition task. The boldface represents the best performance on each type of entities among different single methods and different ensemble methods, respectively
| Single model |
Ensemble method |
|||||
|---|---|---|---|---|---|---|
| CRF | BI-LSTM-CRF | JOINT | Committee | Stacking | Category | |
| Drug | 0.9495 | 0.9545 | 0.9556 | 0.9556 | 0.9514 | 0.9545 |
| Strength | 0.9783 | 0.9822 | 0.9737 | 0.9832 | 0.9818 | 0.9805 |
| Duration | 0.7865 | 0.8559 | 0.8193 | 0.8624 | 0.862 | 0.8604 |
| Route | 0.9474 | 0.9556 | 0.94 | 0.9562 | 0.9559 | 0.9551 |
| Form | 0.9551 | 0.9583 | 0.9442 | 0.9575 | 0.9585 | 0.9538 |
| ADE | 0.2453 | 0.4224 | 0.4306 | 0.5295 | 0.3876 | 0.4306 |
| Dosage | 0.9328 | 0.9427 | 0.9325 | 0.9482 | 0.9477 | 0.9444 |
| Reason | 0.5743 | 0.6796 | 0.6638 | 0.6749 | 0.6724 | 0.6812 |
| Frequency | 0.9693 | 0.9743 | 0.9522 | 0.9748 | 0.974 | 0.9746 |
| Overall | 0.9194 | 0.933 | 0.9237 | 0.9345 | 0.9323 | 0.9326 |
Abbreviations: ADE, adverse drug event; BI-LSTM-CRF, bi-directional LSTM CRF;
CRF, conditional random field.
BI-LSTM-CRF
Among different deep learning approaches, BI-LSTM-CRF is widely used and shows good performances in different NER tasks.20–22,28 BI-LSTM-CRF uses a BI-LSTM to give scores of all possible labels for each token in a sequence, and predicts a token’s label using its neighbor’s information using a CRF layer.18,19 Here we followed the architecture of the BI-LSTM-CRF in the work of Lample et al.18 The input embedding included word embedding and character embedding, where the word embedding was trained on the MIMIC-III corpus word2vec package with a dimension size of 50.29 The method in Lample et al.’s work was used to generate character embeddings by employing a BI-LSTM neural network, in which the input were all characters of a word represented by random initialized vectors (with a dimension size of 35), and the output were the character embeddings for the word (with a dimension size of 200).
JOINT
As previously mentioned, the 2-step pipeline method of NER and relation classification suffers from the error propagation issue. The omitted annotations of attribute entities (eg, ADE and reason) make it more challenging to train both the attribute NER and relation classification models effectively. To address the previously outlined issues, we proposed a joint method for attribute detection, which identifies their mentions and classifies their relations with medications in 1 step. For NER, the JOINT method still consists of 2 steps: 1) we first recognized drugs using a BI-LSTM-CRF model, and 2) each identified drug was then used as a feature for another BI-LSTM-CRF model to recognize attribute entities such as ADEs. All attribute entities identified in this way were considered to have relations with the target drug. More details of relation recognition are described in the Relation Classification section later.
Ensemble strategies
For this work, 3 ensemble methods were proposed: 1) Committee: the final label for a token is the majority of output labels of the single models, 2) Stacking30: a meta-model was trained to combine output from single models in which its input was labels from single models and output was combined labels. Here, a CRF model was trained as a meta-model whose features were the output “BIO” from 3 single models. 3) Category-level best: The predictions of the single model that outperformed its peers on each category were chosen.
Relation classification
Once entities were recognized, we fed them into 1 of the following 3 relation classification methods.
SVM
Given drugs and other entities, the recognition of relations can be transformed into classification problems. A classifier identifies true relation pairs (d, e) from all possible candidate pairs of a drug d and a nondrug entity e within a sentence. SVM has been used in previous relation classification tasks on clinical text and achieved a good performance.9,17,31 We used SVM as a baseline method to compare it with other deep learning methods in the end-to-end and relation classification tasks. The features used here are described in the Supplementary MaterialTable 2. For example, there are 2 drugs and 5 attributes in the sentence example in Figure 2, and totally 10 candidate relation pairs were generated for classification. In “trazadone 100 mg QHS and mirtazapine 30 mg PO QHS”, there is a candidate relation pair between “trazadone” and “100 mg”, and its features (see details in Supplementary MaterialTable 2) are extracted as the input for the SVM method.
Table 2.
The lenient F1-score of machine learning models and rule-based post-processing for relation classification task. All entities used by these methods are gold-standard entities. The boldface represents the best performance for each type of relation
| SVM | SVM + postprocessing | CNN-RNN | CNN-RNN + postprocessing | JOINT | JOINT + postprocessing | |
|---|---|---|---|---|---|---|
| Strength → Drug | 0.9704 | 0.9792 | 0.9760 | 0.9853 | 0.9865 | 0.9916 |
| Dosage → Drug | 0.9637 | 0.9798 | 0.9642 | 0.9818 | 0.9720 | 0.9860 |
| Duration → Drug | 0.84 | 0.8947 | 0.8519 | 0.9125 | 0.8829 | 0.9292 |
| Frequency → Drug | 0.9525 | 0.9735 | 0.9592 | 0.9810 | 0.9692 | 0.9873 |
| Form → Drug | 0.9728 | 0.9867 | 0.9713 | 0.9864 | 0.9765 | 0.9890 |
| Route → Drug | 0.9581 | 0.9742 | 0.9668 | 0.9805 | 0.9736 | 0.9858 |
| Reason → Drug | 0.7328 | 0.8364 | 0.7464 | 0.8466 | 0.7579 | 0.8488 |
| ADE → Drug | 0.7604 | 0.8221 | 0.7528 | 0.8112 | 0.7946 | 0.8502 |
| Overall | 0.9256 | 0.9521 | 0.9304 | 0.9574 | 0.9399 | 0.9630 |
Abbreviations: ADE, adverse drug event; CNN, convolution neural network; RNN, recurrent neural network; SVM, support vector machine.
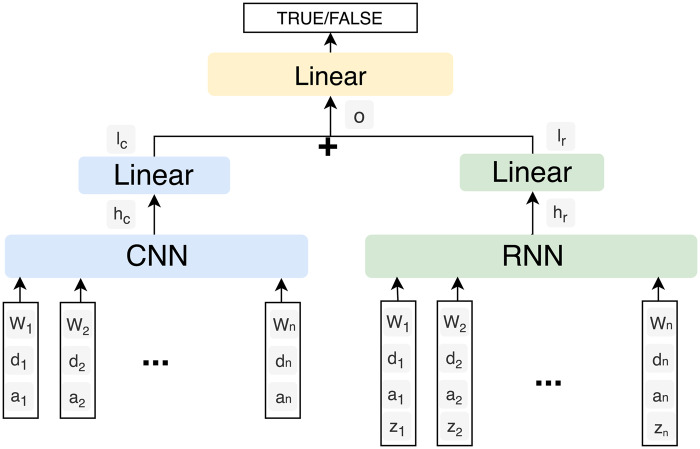
CNN-RNN
CNN and RNN are the 2 major deep learning based architectures which can be used in relation classification tasks.32,33 In order to achieve a better performance, we used a parallel combination of CNN and RNN. Figure 3 shows the architecture of the CNN-RNN model. We generated all possible candidate relation pairs in the same way as we did in the SVM method, and the input features for each relation pair included word embeddings (“w”), POS embeddings (“z”) and position embeddings (“d” and “a”). Specifically, word embeddings and position embeddings were sent to CNN, whereas word embeddings, position embeddings, and POS embeddings were sent to RNN. The output vectors (“hc” and “hr”) of CNN and RNN were passed to the linear layers, respectively. The outputs (“lc” and “lr”) from the linear layers were then concatenated (“o”) and a final linear layer was used to generate predictions. The vector names are marked in Figure 3, and their dimension sizes are provided in Supplementary MaterialTable 4. We used the position embedding method proposed by Zeng et al.33 to represent positions of an entity e in a sentence s. More details of the position embeddings are shown in Equation (1), where L is a hyper-parameter, m and n are the beginning and ending indices of e in s, SL is the length of a sentence s, and N is a predefined value. L denotes the maximum number of words in a sentence to be considered, which is 250 in the task. N is the number for marking entities in a sentence. The following example which illustrates the position of embeddings has been included in Supplementary Material Table 3.
| (1) |
Figure 3.
Architecture of the CNN-RNN model. “w” and “z” represent word embedding and POS embedding, respectively. “d” and “a” represent position embeddings. “hc,” “hr,” “lc,” “lr,” and “o” are outputs of layers in the architecture.
Table 4.
Examples of sentences for error analysis. The bold represents entities
| Examples of sentences |
|---|
| Example 1. “After discharge her fever resolved, but the rash did not. She used Sarna lotion and Benadryl at home…” |
| Example 2. “The rash might worsen before…” |
| Example 3. “… received Benadryl for extremity erythema and itching during peri-administration with vancomycin…” |
JOINT
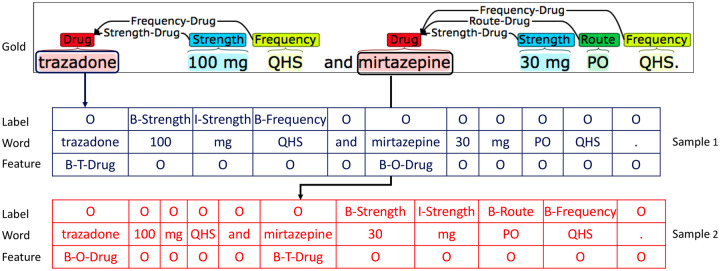
The JOINT approach relies on a novel transformation, which takes annotated sentences and produces drug-focused sequences. That is, given a sentence with identified drug entities, we generated a sequence which labels attributes for each drug in the sentence. If there were multiple drugs, we generated multiple attribute-labeled sequence samples. For instance, in Figure 4, the sentence has 2 drugs, “trazadone” and “mirtazapine”. Thus, we generated 2 labeled sequences: 1 with “trazadone” as the target concept, and the other with “mirtazapine” as the target. In each sequence, we labeled the attribute entities that are associated with the target drug using a BIO scheme. For example, in the sample generated for “trazadone”, only “100 mg” and “QHS” were labeled as “B-Strength I-Strength” and “B-Frequency”, respectively. Other attributes entities like “30 mg”, “PO” and the second “QHS” that are not associated with “trazadone,” were labeled as “O” in this sample. Notably, we provided the information of the target drug to the model during the training and prediction stages as the form of features.
Figure 4.
An illustration of the drug-focused sequence transformation, where each separate sequence encodes all attributes for each concept (drug).
However, there are no sequence labeling methods that can directly take the generated sequence along with the target drug information as input. In this study, we propose a neural architecture (Figure 5) that models the target drug information alongside its corresponding sequence with labeled attributes. The proposed neural architecture is an extension of the BI-LSTM-CRF model proposed by Lample et al.,18 with the important distinction that the input layer contains not only the vector representations of individual words, but also the vector representation of the target concept. We generated a sequence of semantic tags to represent the target concept information using a BIO scheme. As shown in Figure 5, we also use “B-T-Drug” and “B-O-Drug” to distinguish the target drug from other nontarget drugs. A semantic tag lookup table initialized at random contains an embedding for every tag. The semantic tag embeddings were then learned directly from the data during the training of the model. The word embeddings were trained on the MIMIC-III corpus using the word2vec package.29 The character embeddings were generated using the same method as that in the BI-LSTM-CRF model for NER. Their dimension sizes together with other model parameters are provided in Supplementary Material Table 5. The character, word, and semantic tag representations were then concatenated into . This helps the network to keep track of both sentential and positional context information of the target concept. The BI-LSTM layer was used for encoding long input sequences to, which performs conditioning of the concatenated representations on the left and right contexts. Finally, the CRF layer provided output tag predictions , which are the predicted attribute labels of the target concept.
Figure 5.
The architecture of BI-LSTM-CRF neural network for attribute detection of a given drug. We concatenated word, character, and target concept (semantic tag) embeddings as input.
In the NER and end-to-end tasks, the drugs predicted from the committee method were used as features for the JOINT approach, while gold-standard drugs were used for the single relation classification task.
Postprocessing
We conducted error analysis on the training data and summarized obvious errors and proposed some rules to fix them and further enhance the recognition performance. The 2 major rules are:
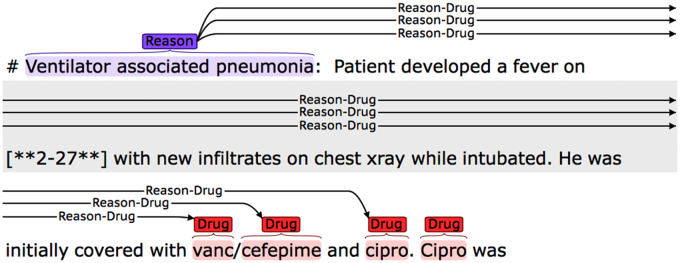
1) Recover the relations for isolated nondrug entities. According to the annotation guideline, any attribute should have at least 1 relation with a drug, so we linked the isolated nondrug entities with their nearest drugs. Figure 6 shows an example in which the isolated reason entity, “Ventilator associated pneumonia”, was linked to its neighbor vanc. Because the 3 drugs vanc, cefepime, and cipro were in parallel, so it was also linked to cefepime and cipro.
Figure 6.
Rule-based postprocessing for the relation classification and end-to-end tasks.
2) Remove relations that spanned across sections. In some relations predicted by the ML model, the 2 entities were located in different sections. Such relations were removed from the final output.
Evaluation
In the NER subtask, CRF was used as the baseline method and was compared with both the BI-LSTM-CRF and the JOINT methods. In the relation classification and end-to-end subtasks, SVM was used as the baseline method and compared with the CNN-RNN and the JOINT methods.
The primary evaluation metric is the lenient micro F1 score in the challenge (equation 4). We used a 5-fold cross-validation on the training data set (303 documents) to optimize parameters for the models. However, we noticed that parameters and performance for each of the 5 models were quite different, which makes it difficult to decide on optimal parameters for a single model that uses all training data. Therefore, we kept all 5 models and used them to predict the test data set, and then combined outputs from the 5 models to make the final decision using the committee method.
| (2) |
| (3) |
| (4) |
RESULTS
Named entity recognition
Table 1 displays the F1-score of NER using the single models and the ensemble methods. In single models, the BI-LSTM-CRF outperformed the CRF and the JOINT method on all categories except the ADE, whereas the JOINT method performed best on the ADE. The committee method was the optimal ensemble method. It outperformed single models on 7 of the 9 categories and improved the overall performance by 0.15%. In contrast, the stacking and category-levels’ best methods failed to improve the performance over single models.
Relation classification
Table 2 shows the lenient F1-score of ML models and performance after their integration with rule-based postprocessing for the relation classification task, respectively. Both of the CNN-RNN and JOINT methods outperformed the baseline SVM method. The JOINT method beat the CNN-RNN method on all categories. F1-scores of the JOINT method were 0.0418 and 0.0115 higher than the CNN-RNN on the recognition of ADE and reason, and 0.0095 higher than the CNN-RNN on the whole. Notably, rule-based postprocessing could improve the performances of all 3 ML-based relation classification methods. For example, the F1-score of the CNN-RNN was enhanced 0.0463 for duration, 0.0909 for reason, and 0.0556 for ADE, respectively.
End-to-end task
Table 3 illustrates the micro lenient F1-score of the JOINT method and 2 pipeline methods that used the committee method to recognize named entities, and the SVM and the CNN-RNN to recognize relations, respectively. In the pipeline methods, the CNN-RNN outperformed the SVM method. Unlike the results in the relation classification task, the JOINT method performed worse than the CNN-RNN method on the whole, which was slightly better on 3 of the 9 types of relations (relations between drug and strength, dosage and frequency). The JOINT method was 0.0928 lower than the CNN-RNN on the relation between drug and the ADE. The reason for this result could most likely be due to the fact that in the end-to-end task, the JOINT method used predicted drugs instead of gold-standard drugs in the relation classification task. The postprocessing showed an improved performance over the CNN-RNN method (0.8792 vs 0.8905). However, it did not improve the performance of the JOINT method.
Table 3.
The lenient F1-score of different machine learning models and rule-based postprocessing for the end-to-end task. The boldface represents the best performance on each type of relation
| SVM | SVM + postprocessing | CNN-RNN | CNN-RNN + postprocessing | JOINT | JOINT + postprocessing | |
|---|---|---|---|---|---|---|
| Strength → Drug | 0.9574 | 0.9646 | 0.9637 | 0.9720 | 0.9644 | 0.9644 |
| Dosage → Drug | 0.9218 | 0.9337 | 0.9231 | 0.9353 | 0.9245 | 0.9245 |
| Duration → Drug | 0.7395 | 0.7735 | 0.7400 | 0.7861 | 0.7366 | 0.7366 |
| Frequency → Drug | 0.9361 | 0.9522 | 0.9405 | 0.9582 | 0.9425 | 0.9425 |
| Form → Drug | 0.941 | 0.9510 | 0.9404 | 0.9516 | 0.9363 | 0.9363 |
| Route → Drug | 0.9228 | 0.9350 | 0.9299 | 0.9415 | 0.9287 | 0.9287 |
| Reason → Drug | 0.5626 | 0.5756 | 0.5722 | 0.5792 | 0.5637 | 0.5630 |
| ADE → Drug | 0.4734 | 0.4718 | 0.4749 | 0.4755 | 0.3821 | 0.3790 |
| Overall | 0.8750 | 0.8853 | 0.8792 | 0.8905 | 0.8775 | 0.8774 |
Abbreviations: ADE, adverse drug event; CNN, convolution neural network; RNN, recurrent neural network; SVM, support vector machine.
DISCUSSION
In this study, we investigated deep learning methods to recognize drug names, attribute entities, and their relations from clinical narratives. By combining ML models with rule-based postprocessing, our system achieved the second-best performance among 28 participating teams and 60 submitted runs on the NER task, and the best performance on the relation classification and end-to-end tasks among 48 and 50 runs, respectively in the challenge of the 2018 n2c2 Track 2. Our results demonstrated the feasibility of the JOINT method for relation extraction, with the goal to alleviate the issue of rare attributes such as ADEs and reasons in the data set. Based on the experiences acquired during this study, future research directions to improve the performance may be derived.
Variability of deep learning models across folds. In the 5-fold cross validation during the training, the optimal parameters and performance of the trained model in each fold varied a lot, which made it difficult to pick 1 as the optimal model. Supplementary Material Table 6 shows the performance of the BI-LSTM-CRF model on each fold. The overall performance ranged from 0.9252 to 0.9299, and the performance on ADE recognition ranged from 0.3358 to 0.4946. It indicates that the sample selection bias of the training data set had an influence on the performance of the ML method. Moreover, the hyper-parameters (eg, best epoch in deep learning) of each model were also different, making it difficult to choose 1 optimal set for training a model on the entire training data set. To resolve this problem, the committee method was employed to combine models from all the 5 folds. The overall performance of the combined prediction was 0.0015 higher than the best performance on an individual fold (fold 5), which achieved the optimal performance on 5 of the 9 categories (Supplementary Material Table 6). Therefore, the ensemble method could be a solution to address the issue of the sampling bias due to the random partitioning of the training data set. Another potential solution is to calculate the average performance of models from 5 folds on each epoch and pick the best one.
The JOINT method. The JOINT method outperformed the CNN-RNN in the RC subtask (0.9399 vs. 0.9304, and 0.9630 vs. 0.9574 after postprocessing, respectively), demonstrating that it’s a promising architecture for problem representation. However, it obtained lower performance in the end-to-end subtask (0.8775 vs. 0.8792, and 0.8774 vs. 0.8905 after postprocessing, respectively). One potential reason is that the JOINT method is highly dependent on the target drugs and was more sensitive to the accuracy of drug recognition, in comparison with the CNN-RNN. Drug predictions with a high recall will be examined in the JOINT model in our future work. Further, the postprocessing step had little influence on the performance of the JOINT method in the end-to-end task, because the post-processing aimed to recover relations between isolated attributes and their target drugs (ie, the nearest drugs as used in the rule). However, there’s no isolated nondrug entities in the predictions of the JOINT method for further improvement. In contrast, in the relation classification task, some gold-standard nondrug entities were not recognized by the JOINT method. Such entities were linked with their target drugs by the postprocessing, leading to an improved performance. Besides, the current architecture of the JOINT model also needs to be expanded in the future to represent inter-sentence relations, which accounts for ∼30% of reasons and ADEs in the whole data set.
Integration of external data sets. In order to improve the performance of drug recognition, we also conducted an experiment by adding an external corpus with drug annotation, the MTsamples corpus,34 into the training set. However, the BI-LSTM-CRF model achieved an F1 score of 0.9200, which was lower than the original model trained without the external data set. It may be because the annotation guidelines were slightly different between these 2 data sets. Instead of adding samples into the training set directly, a better way may be to learn transferable representation from external data sets. Additionally, the performance may be improved by reweighting samples from different data sets. In some studies, a base model is first trained on external data sets, and then fine-tuned using the destination data set.35–37
Error analysis. As shown in Table 1, although our system achieved a higher performance on most attributes, recognition of ADE and reason are still challenging (with F1-score 0.5295 and 0.6749, respectively). One potential reason is that the majority of medical problems present in the clinical text are negative samples that are not labeled as ADE or reason. By estimation, only a small proportion (2% and 8%, respectively) of medical problems in the training set were ADEs and reasons. Such a data distribution and annotation scheme make it difficult for ML methods to learn general patterns. For example, there are 202 “rash” in the whole training set, but only 43 of them are labeled as ADE and 18 of them are labeled as reason. Table 4 presents an illustration of this problem: In Example 1, “rash” is the reason for drugs “Sarna” and “Benadryl”, but it is neither a reason or an ADE in Example 2. Another challenge is that some reasons/ADEs and their related drugs are not located in the same sentence. In the training set, as mentioned before ∼30% reasons and ADEs and their related drugs are not located in the same sentences. In the NER task, our ML models were only trained on single sentences, missing the information from the whole context (eg, drug “Sarna” and “Benadryl”) in Example 1 on their neighbor sentences. Similarly, relation classification models trained with single sentences cannot predict relations across multiple sentences. Furthermore, some medical problems are labeled both as ADE and reason. In Example 3, “extremity erythema and itching” is the reason for “Benadryl”, while it is also the ADE of “vancomycin”. Our methods can only output 1 label for each word, hence some ADEs or reasons were missed.
Limitations and future work. Our study has limitations which we outline here. First, although we tried to use the external corpus to improve the performance on the NER task, exactly how to integrate existing medical data sets/knowledge into the deep learning framework has not been fully explored yet. For example, knowledge bases with existing drug indications, such as the MEDication Indication resource,38 has the potential to further improve the performance. In the future, we will use existing domain knowledge of drug indications and ADEs to reduce recognition errors by using them as features in ML models and as rules in postprocessing. Second, as discussed previously, methods to leverage existing corpora from other similar tasks are still challenging and need further investigation. In addition, the deep learning field is evolving quickly. Recently, novel deep learning architectures based on improved language models like ELMo,39 BERT,37 etc. generated better performances in the open domain on some classical NLP tasks including NER and relation classification.36,37,39 In the future, we plan to evaluate and adapt these methods to the biomedical domain.
CONCLUSION
In this study, we developed deep learning approaches for extracting medications and their attributes from clinical text and demonstrated its superior performance compared with traditional ML algorithms. Our approach may inform development of novel clinical NLP methods on biomedical information extraction and benefit applications of EHRs in clinical practice such as pharmacovigilance.
FUNDING
This work was supported by National Library of Medicine grant number 5R01LM010681, National Cancer Institute grant number U24 CA194215, National Institute of General Medical Sciences grant number 5U01TR002062 and Cancer Prevention and Research Institute of Texas grant number RP160015.
AUTHOR CONTRIBUTIONS
HX, CT, YZ, SW, QW, ZJ, JD, ZL, JW, and YX designed the study. QW, ZJ, JD, JW, ZL, and YZ performed the research and analyzed the data under HX’s supervision. QW, JX, YZ, and HX wrote the manuscript. All authors revised and approved the manuscript.
CONFLICT OF INTEREST STATEMENT
Dr. Xu and The University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
Supplementary Material
REFERENCES
- 1. Luo Y, Thompson WK, Herr TM, et al. Natural language processing for EHR-based pharmacovigilance: a structured review. Drug Saf 2017; 40 (11): 1075–89. [DOI] [PubMed] [Google Scholar]
- 2. Coloma PM, Trifirò G, Patadia V, et al. Postmarketing safety surveillance. Drug Saf 2013; 36 (3): 183–97. [DOI] [PubMed] [Google Scholar]
- 3. Harpaz R, Callahan A, Tamang S, et al. Text mining for adverse drug events: the promise, challenges, and state of the art. Drug Saf 2014; 37 (10): 777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang X, Hripcsak G, Markatou M, et al. Active computerized pharmacovigilance using natural language processing, statistics, and electronic health records: a feasibility study. J Am Med Inform Assoc 2009; 16 (3): 328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haerian K, Varn D, Vaidya S, et al. Detection of pharmacovigilance-related adverse events using electronic health records and automated methods. Clin Pharmacol Ther 2012; 92 (2): 228–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nadkarni PM. Drug safety surveillance using de-identified EMR and claims data: issues and challenges. J Am Med Inform Assoc 2010; 17 (6): 671–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phansalkar S, Hoffman JM, Hurdle JF, et al. Understanding pharmacist decision making for adverse drug event (ADE) detection. J Eval Clin Pract 2009; 15 (2): 266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010; 17 (1): 19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patrick J, Li M.. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc 2010; 17 (5): 524–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sohn S, Clark C, Halgrim SR, et al. MedXN: an open source medication extraction and normalization tool for clinical text. J Am Med Inform Assoc 2014; 21 (5): 858–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C-C, Lu Z.. Community challenges in biomedical text mining over 10 years: success, failure, and the future. Brief Bioinform 2016; 17 (1): 132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uzuner O, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 17 (5): 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roberts K, Demner-Fushman D, Tonning JM. Overview of the TAC 2017 adverse reaction extraction from drug labels track. In: Proceedings of the Tenth Text Analysis Conference Gaithersburg, MD, USA; 2017. https://tac.nist.gov/publications/2017/additional.papers/TAC2017.ADR_overview.proceedings.pdf (accessed 6 Jan 2019).
- 14. Jagannatha A, Liu F, Liu W, et al. Overview of the first natural language processing challenge for extracting medication, indication, and adverse drug events from electronic health record notes (MADE 1.0). Drug Saf 2019; 42 (1): 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Uzuner O. National NLP Clinical Challenges (n2c2). https://portal.dbmi.hms.harvard.edu/projects/n2c2-t2/ (accessed 3 May 2019).
- 16. Chapman AB, Peterson KS, Alba PR, et al. Detecting adverse drug events with rapidly trained classification models. Drug Saf 2019: 42 (1): 147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aramaki E, Miura Y, Tonoike M, et al. Extraction of adverse drug effects from clinical records. Stud Health Technol Inform 2010; 160 (Pt 1): 739–43. [PubMed] [Google Scholar]
- 18. Lample G, Ballesteros M, Subramanian S, et al. Neural architectures for named entity recognition. In: Proceedings of NAACL-HLT San Diego, CA; 2016: 260–70.
- 19. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF models for sequence tagging. CoRR abs/1508.01991; 2015.
- 20. Jagannatha AN, Yu H.. Structured prediction models for RNN based sequence labeling in clinical text. Proc Conf Empir Methods Nat Lang Process 2016; 2016: 856–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu Z, Yang M, Wang X, et al. Entity recognition from clinical texts via recurrent neural network. BMC Med Inform Decis Mak 2017; 17 (Suppl 2): 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dandala B, Joopudi V, Devarakonda M, et al. IBM Research System at MADE 2018: Detecting Adverse Drug Events from Electronic Health Records. In: Proceedings of Machine Learning Research; 2018: 39–47.
- 23. Dandala B, Joopudi V, Devarakonda M.. Adverse drug events detection in clinical notes by jointly modeling entities and relations using neural networks. Drug Saf 2019; 42 (1): 135–46. [DOI] [PubMed] [Google Scholar]
- 24. Li Q, Ji H. Incremental Joint Extraction of Entity Mentions and Relations. In: Proceedings of the 52nd Annual Meeting of the Association for Computational Linguistics Baltimore, MD: Association for Computational Linguistics; 2014: 402–12.
- 25. Miwa M, Bansal M. End-to-end relation extraction using LSTMs on sequences and tree structures. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics Berlin, Germany; 2016: 1105–16.
- 26. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3: 160035.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soysal E, Wang J, Jiang M, et al. CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Informatics Assoc 2017; 25(3):331–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Z, Tang B, Wang X, et al. De-identification of clinical notes via recurrent neural network and conditional random field. J Biomed Inform 2017; 75: S34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mikolov T, Chen K, Corrado G, et al. Distributed representations of words and phrases and their compositionality. In: NIPS 2013 Proceedings of the 26th International Conference on Neural Information Processing Systems Lake Tahoe, Nevada; 2013: 3111–9.
- 30. Zhi-Hua Z. Ensemble Methods: Foundations and Algorithms. Boca Raton, FL: Chapman & Hall/CRC; 2012. [Google Scholar]
- 31. Lee H-J, Zhang Y, Xu J, et al. UTHealth at SemEval-2016 task 12: an end-to-end system for temporal information extraction from clinical notes. In: Proceedings of SemEval-2016 San Diego, California; 2016: 1292–7.
- 32. Zhang D, Wang D. Relation classification via recurrent neural network. CoRR arXiv:1508.01006; 2015.
- 33. Zeng D, Liu K, Lai S, et al. Relation classification via convolutional deep neural network. In: Proceedings of COLING 2014, the 25th International Conference on Computational Linguistics. Dublin, Ireland: 2014. 2335–44.
- 34.Transcribed Medical Transcription Sample Reports and Examples – MTSamples. https://www.mtsamples.com/ (accessed 25 Jan 2019).
- 35. Lin BY, Lu W. Neural adaptation layers for cross-domain named entity recognition. In: Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing Brussels, Belgium: Association for Computational Linguistics; 2018: 2012–22.
- 36. Openai AR, Openai KN, Openai TS, et al. Improving Language Understanding by Generative Pre-Training. openai Prepr; 2018. https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf (accessed 25 Jan 2019).
- 37. Devlin J, Chang M-W, Lee K, et al. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. CoRR arXiv1810.04805; 2018. https://arxiv.org/abs/1810.04805 (accessed 12 Oct 2018).
- 38. Wei W-Q, Cronin RM, Xu H, et al. Development and evaluation of an ensemble resource linking medications to their indications. J Am Med Inform Assoc 2013; 20 (5): 954–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peters ME, Ammar W, Bhagavatula C, et al. Semi-supervised sequence tagging with bidirectional language models. CoRR arXiv:1705.00108; 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.