Abstract
Objective
Academic medical centers and health systems are increasingly challenged with supporting appropriate secondary use of clinical data. Enterprise data warehouses have emerged as central resources for these data, but often require an informatician to extract meaningful information, limiting direct access by end users. To overcome this challenge, we have developed Leaf, a lightweight self-service web application for querying clinical data from heterogeneous data models and sources.
Materials and Methods
Leaf utilizes a flexible biomedical concept system to define hierarchical concepts and ontologies. Each Leaf concept contains both textual representations and SQL query building blocks, exposed by a simple drag-and-drop user interface. Leaf generates abstract syntax trees which are compiled into dynamic SQL queries.
Results
Leaf is a successful production-supported tool at the University of Washington, which hosts a central Leaf instance querying an enterprise data warehouse with over 300 active users. Through the support of UW Medicine (https://uwmedicine.org), the Institute of Translational Health Sciences (https://www.iths.org), and the National Center for Data to Health (https://ctsa.ncats.nih.gov/cd2h/), Leaf source code has been released into the public domain at https://github.com/uwrit/leaf.
Discussion
Leaf allows the querying of single or multiple clinical databases simultaneously, even those of different data models. This enables fast installation without costly extraction or duplication.
Conclusions
Leaf differs from existing cohort discovery tools because it does not specify a required data model and is designed to seamlessly leverage existing user authentication systems and clinical databases in situ. We believe Leaf to be useful for health system analytics, clinical research data warehouses, precision medicine biobanks, and clinical studies involving large patient cohorts.
Keywords: biomedical informatics, leaf, cohort discovery, observational health data sciences and informatics, data integration, cloud computing
INTRODUCTION
Healthcare organizations are challenged to manage ever increasing quantities of data, driven by the rise of electronic health record systems, patient-reported outcomes, registries, mobile health devices, genomic databases, and other clinical and nonclinical systems.1–3 Beyond importing these heterogeneous and complex data into enterprise data warehouses (EDWs), healthcare organizations are responsible for cleaning, integrating, and making the data accessible to appropriate users and consumers; maintaining organizational compliance; and protecting patient privacy and preventing security breaches. Even in cases in which data are successfully integrated and made available, their extraction can be time-consuming and difficult for consumers who may not have information technology (IT) or informatics backgrounds.
To advance the needs of our health system at the University of Washington and potentially other Clinical and Translational Science Awards (CTSA) centers, we developed Leaf, a lightweight drag-and-drop web application that enables direct querying of arbitrary clinical databases. Because Leaf is a “blank canvas” and does not require a particular clinical data model, making new data sources and biomedical concepts available in Leaf is often relatively simple, with no data extraction or transformation required. Utilizing an Agile development process with ongoing engagement of clinical users and stakeholders, we have incrementally improved Leaf functionality based on user feedback, adding capabilities such as real-time de-identification algorithms and direct REDCap4 export. We have prioritized the use of human-centered design5 techniques and modern web best practices to make Leaf’s user interface clear and intuitive. The result is a simple but powerful tool for our clinicians and researchers that requires fewer technical resources to maintain compared with other cohort discovery tools.
Background and significance
Cohort discovery tools have been demonstrated to be extremely useful for observational research, clinical trial recruitment, and hospital quality improvement programs.6–9 To find which cohort discovery tools were best suited to our institution, we investigated several, including Informatics for Integrating Biology and the Bedside (i2b2)6 and TriNetX.10 Our aim was to find a tool that was lightweight and easily deployable, adaptable to new data sources and use cases, and intuitive for our clinical and research users. As our institution operates a central EDW that is jointly used for quality improvement, analytics, and research, we also sought to avoid creating a costly data extraction process that would duplicate our EDW, instead working to leverage the EDW itself. The opportunities we perceived which drove the development of Leaf are summarized in the following sections.
Opportunity 1: a lightweight “sidecar” data service to the EDW
Data sources, schema, and row values in our EDW (or any clinical research biobank or databank) evolve with our electronic medical records (EMRs) and institutional use cases require new sources be ingested. Often this evolution of data cannot be predicted ahead of time, and any tools using EDW data must therefore be flexible and adaptable, ideally with minimal informatics effort. Our institution also made a significant investment in our EDW as a valuable resource available across our enterprise, and we sought a tool that did not attempt to recreate functionality in our EDW, but instead leveraged and complemented it. Finally, we envisaged a modular, modern, cloud-friendly tool that could be effectively deployed in either cloud architectures or on premises, interoperating alongside other tools in a service-oriented fashion (Figure 1).
Figure 1.
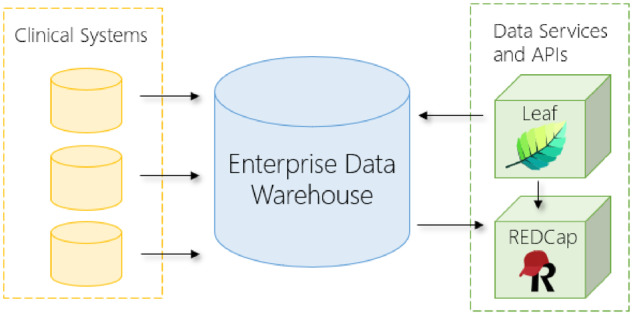
Diagram of the Leaf instance deployed at the University of Washington Medicine Enterprise Data Warehouse. Data from electronic medical records and other clinical systems flow into the enterprise data warehouse. Leaf, REDCap, and other tools are deployed in modular fashion to 41 query the enterprise data warehouse and interoperate for data extraction and analysis. API: application programming interface.
Opportunity 2: customized user interface for different sites or data
A second opportunity was the need for an intuitive user interface that could accommodate a wide array of data sources in a flexible manner and facilitate discoverability without overwhelming or frustrating users. We had learned from experience that an effective user interface needed to allow for both ontology-driven hierarchical data elements, such as diagnosis and procedure codes, and also local data presented in unique structure and textual descriptions. We also aimed to design an interface that could display not only aggregate patient counts, but also simple demographic visualizations and row-level cohort data available in a single click.
Opportunity 3: streamlined prep to research and data request processes
A third opportunity was a faster turnaround time for University of Washington (UW) researchers to generate and test hypotheses while reducing the burden of relatively simple cohort estimation requests on our research informatics team. As many UW researchers spent significant time requesting and waiting for cohort size estimates before submitting Institutional Review Board (IRB) requests, a tool that could fulfill many of those requests quickly and efficiently held great potential. To improve the timeliness of accessing clinical data after IRB approval as well, we believed that allowing users to enter IRB information directly and extensively logging their activity could provide a faster, secure, and more readily auditable workflow than previous manual processes.
Opportunity 4: better alignment with health system analytics
A fourth opportunity, related to the structure of UW Medicine informatics teams, was to reduce duplication of efforts between our research informatics team and other informatics teams involved in hospital operations and analytics. As each of these teams uses our EDW to query, catalog, and extract data, we perceived a unique opportunity to design a tool which served varied but often highly similar use cases for each. By allowing users to simply indicate their purpose (quality improvement or research) and de-identify data as needed, we believed that each informatics team and their users could greatly benefit.
Leaf began as a small innovative project by the first author to create a user-friendly tool that matched the powerful capabilities of the i2b2 web application but queried our EDW directly, leveraged the existing work of teams across our organization, and functioned with no data extraction required. After an initial proof-of-concept application was developed, Leaf was shown to leaders within our hospital system and CTSA, eventually becoming a flagship collaborative software project to improve self-service access to clinical data across research, analytics, and the biomedical informatics academic department at UW.
MATERIALS AND METHODS
Agile development and user engagement
Early in Leaf development, we sought to utilize Agile development methodology11 to ensure the Leaf user interface and features were driven by the needs of clinicians and researchers. The Leaf pilot project was formed to engage approximately 30 clinicians and researchers across various medical disciplines and organizations in our CTSA in weekly Agile sprints over approximately 6 months.
After an initial meeting was held to introduce the tool and discuss the scope of the project, weekly web-based meetings were conducted to iteratively introduce new features, gather feedback, and prioritize items for upcoming sprints. After each sprint, the development team worked to incorporate stakeholder ideas and coordinate with our EDW team to make additional data elements available.
After the pilot phase was complete, we planned and created a multitiered support system to triage user assistance and provide oversight of future Leaf development. In February 2018, a central instance of Leaf querying our EDW was approved as an institutionally-supported tool at UW.
System architecture
A Leaf instance is typically deployed with a 1:1 relationship to the clinical database or database server that it is intended to query (hereafter for simplicity we refer to clinical database in the singular, but multiple clinical databases on a single server can be queried in the same way provided that they share a Master Patient Index).12 A Leaf instance is composed of 3 core architectural elements common to many modern web applications:
A user-facing client application written in React and TypeScript.
A RESTful13 web application programming interface (API) deployed to a web server written in C# and .NET Core.
A small SQL Server application database for logging activity, tracking queries, and caching patient identifiers.
Additionally, a separate web server is typically used to filter web traffic and serve the client application to users in coordination with a SAML2-compliant identity provider, such as Shibboleth or Active Directory Federation Services. All components can be deployed on either Linux or Microsoft Windows operating systems.
Data model assumptions
Leaf makes minimal assumptions about any clinical database it queries. Indeed, one of the key insights of the Leaf application is that nearly all standard and nonstandard clinical databases conform to certain simple assumptions, which makes the largely model-agnostic nature of Leaf possible. The assumptions are the following:
The clinical database to be queried has a common identifying column name for patient identifiers across tables, such as PersonId, patient_id, or PAT_NUM.
Similarly, the database has a common identifying column name for clinical encounter identifiers, such as EncounterId or visit_occurrence_id.
These columns are defined globally for a given Leaf instance, and are expected to be present on every table, view, or subquery to be queried, though encounter identifier and date columns need only be present if a given SQL set has a 1:M relationship for a patient. In tables in which these columns are not present or are named differently, SQL views or subqueries can be used instead to derive them.
Login and access modes
Leaf is designed to be able to query clinical databases with identified patient information14 and hide or obfuscate that information when accessed in “de-identified mode.” The workflow for this is as follows:
On login users are presented with options for accessing data for the purposes of quality improvement or research. They must then enter quality improvement documentation (ie, project name, approving body) or IRB approval information, or alternatively indicate that they do not have this information. Users then select identified or de-identified access, with access limited to de-identified mode if the user has no approved quality improvement project or IRB application.
As users run queries and view row-level data in the “Patient List” screen, the Leaf REST API de-identifies the output SQL values depending on the data type (ie, a string, number, or date). If Leaf is in de-identified mode, de-identification is performed using an attribute-based schema tagging system that marks fields of supported Fast Healthcare Interoperability Resources (FHIR) templates with data about their sensitivity. Depending on the attributes of a given field, the underlying data will be left alone, masked, or excluded. For example, the FHIR Person name field is tagged as protected health information (PHI), and is removed entirely, while the Encounter admitDate is tagged as both PHI and mask-able, and so is shifted chronologically and included in the output to the client application.
For information on Patient List datasets based on FHIR templates, see the Extracting and exporting data for a cohort section.
De-identification methods
At a high level, PHI in Leaf is handled similarly to methods for secure password storage. When a query is run, the Leaf API generates 2 pieces of entropy:
A salt, unique to each patient in a given query in the form of a randomly generated global unique identifier.
A pepper, unique to each cohort extracted by a user in a single query in the form of a global unique identifier.
When row-level data are requested in de-identified mode, a unique identifier for each patient is generated by using a hash function on the combined salted and peppered value. Date-shifting is similarly performed as follows:
A random number generator is created using the hash of the patient’s query-specific salt.
The first random number value between –1000 and 1000 is extracted from the generator.
Each date for a patient is shifted by this value in hours.
For example, the Leaf “Basic Demographics” dataset (modeled on the FHIR Patient and Person resources) includes a personId field (a string) and a birthDate field (a date), both of which have been tagged as PHI elements in the Leaf source code. A given Leaf instance must configure an appropriate SQL SELECT statement for its respective clinical database which outputs required columns of these names and types, along with other columns not tagged as PHI (eg, gender).
When a user requests row-level data for a cohort, the Leaf API executes the configured SQL query on the clinical database, then proceeds to analyze the dataset metadata to develop a de-identification plan based on the fields tagged as PHI. After the de-identification plan is created for the query, it is executed by looping through each row and each PHI-tagged field of output, using the hash function on the salt, pepper, and row value to insert a random number (if a string) or a new date based on a randomly shifted value of the true date (if a date). If a given SQL query therefore returns identified values for the personId, birthDate, and gender columns of:
“MRN1234,” “1979-01-09 5: 00: 00,” “female”
The Leaf API will generate a de-identified output similar to:
“419858302,” “1978-11-25 13: 00: 00,” “female”
The de-identified data are then sent to the client application.
In practice, this ensures that each patient is given a different unique set of identifiers and date-shift values for every query they are included in, even if the criteria in the queries are identical. Though under Health Insurance Portability and Accountability Act of 1996 dates with more granular information than the year are considered PHI, the practice of using randomly shifted dates (rather than year alone) is similarly found in tools such as REDCap, and we have found it to sufficiently balance patient privacy concerns with analysis-related needs of Leaf users. For cohorts of a relatively small number of patients (eg, <10) Leaf does not currently remove or obfuscate the true count, though we intend to explore this in future work.
Concepts and mapping SQL objects
The Leaf application database has a central SQL table (app.Concept) which defines the concepts that comprise Leaf queries, their hierarchical relationship to other concepts, and textual representations seen by users. Each concept also contains a field for an arbitrary SQL WHERE clause which functions as a programmatic representation of the concept in the clinical database to be queried (example in Figure 2). The SQL FROM clause and corresponding date field (if any) for concepts are similarly found via a foreign key relationship to the app.ConceptSqlSet table, and thus all relevant SQL data for a concept can be returned in a simple JOIN. A given concept therefore requires only that the patient identifier field (and optionally, encounter identifier and date field) be present; any other fields or sets referenced in the WHERE clause are flexible and assumed to be present in the table or view.
Figure 2.
Example tabular values and Entity-Relation diagram illustrating hypothetical Leaf concepts for laboratory tests, hematology-based tests, and platelet count tests. Each concept has both textual and programmatic representation components. The “@” symbols within the SqlSetWhere field denote ordinal positions to insert the SQL Set alias at query compilation time. Additional fields in the tables have been omitted for brevity.
This structure allows concepts to be remarkably flexible in terms of programmatic representation (Figure 2) and presented visually to users in an intuitive fashion (Figure 3). Leaf’s concept table therefore functions as a programmatic and ontological metadata map of queries to a clinical database.
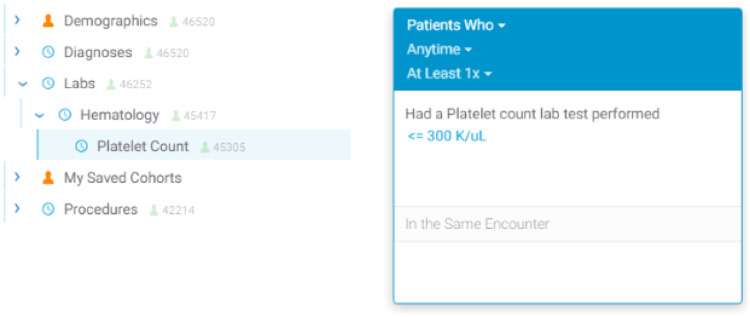
Figure 3.
User interface screenshots of the platelet count example concept as seen in Figure 1. The left image shows the concept within an intuitive nested hierarchy below “Hematology” and “Labs,” while the right image shows a simple visual query representation created after the user has dragged over the “Platelet Count” concept 43 and filtered by a value (≤300 K/µL).
To date, Leaf instances have been deployed and tested within our development environments and we have successfully validated this approach with a variety of data models, including the proprietary vendor-specified model of our EDW, the OMOP Common Data Model,15 i2b2,6 Epic Clarity,16 and MIMIC-III (Medical Information Mart for Intensive Care-III).17
Concept modifiers
Additional SQL logical for a concept—often called a “modifier”6—can be represented in Leaf by the addition of optional dropdown elements which appears after the user has dragged the concept into a panel. This can be represented visually to the user as:
Had a diagnosis of Type 2 diabetes mellitus without complications (ICD-10 E11.9) from any data source
If the user clicks “from any data source,” a dropdown will be shown with values such as:
From Billing
From Radiology
Each dropdown option contains both a textual representation and programmatic element. If the user clicks a dropdown option, the dropdown text changes to the selected option and the SQL logic representing the concept will have the selected dropdown option’s SQL (eg, @.data_source = “billing”) appended to the concept’s WHERE clause. Each concept can have multiple dropdowns, so it is possible to allow users to easily modify a diagnosis concept to specify both “From Billing” and “Primary Diagnosis,” for example.
In addition to dropdowns, Leaf also allows for numeric filters (demonstrated in Figure 3). Configuration metadata for additional concept elements, such as optional numeric data (ie, a numeric SQL field, default text, and units to display), dropdowns, and patient counts are stored in SQL tables in the Leaf application database and can be added programmatically or by using the application interface.
Query compilation
As users drag concepts over to define queries, the Leaf client creates an array of abstract syntax tree (AST) objects in JavaScript Object Notation representing the current user interface state and query definition. The AST objects themselves do not contain SQL, but instead contain ConceptId pointers to concepts in the Leaf app.Concept table included in the user’s query, along with additional metadata describing the query.
When the AST objects are sent to the Leaf REST API after the user clicks “Run Query,” the API performs the following steps:
Validates that the user is permitted to use each requested concept in the AST based on the ConceptId values in the Leaf application database.
Loads the SQL FROM and WHERE clauses and appropriate date and numeric fields (if applicable) for each concept.
Generates individual SQL queries for each of the 3 panels in the Leaf user interface that can be used to create queries. Empty panels are excluded.
Computes a summed estimated query cost per panel to the database using a simple heuristic determined by cached patient counts per concept as proxy for cost, with a reduction if concepts are constrained by dates or numeric values.
Generates a SQL Common Table Expression by ordering the panels by the estimated query cost values generated in (4). The queries are then linked by SQL INTERSECT and EXCEPT statements starting with the least estimated cost to optimize performance.
Runs the query against the clinical database, map-reducing the resulting patient identifiers in a hashset and caching them in the Leaf application database should the user later request row-level data for the cohort.
Returns a count of unique patients to the client application.
Building the concept tree using ontologies and the Unified Medical Language System
Because Leaf concept definitions are stored in a simple SQL table, relational database-based ontological mapping systems such as the Unified Medical Language System18 can be leveraged to dynamically build the Leaf concept tree programmatically, as has been demonstrated with i2b2.19 The Unified Medical Language System has been used extensively in our central EDW Leaf instance to build the concept tree sections for diagnosis and problem list codes (International Classification of Diseases-Ninth Revision [ICD-9], International Classification of Diseases-Tenth Revision [ICD-10], and SNOMED), procedure codes (Healthcare Common Procedure Coding System, Current Procedural Technology, Current Dental Technology, ICD-9, and ICD-10), and laboratory tests (Logical Observation Identifiers Names and Codes).
As the hierarchical nesting of concepts serves a largely visual rather than programmatic purpose (ie, descendent concepts do not automatically inherit the SQL logic of their parents) administrators must create this logic themselves when building concepts, typically using increasingly smaller ranges of codes via SQL BETWEEN statements as the tree descends. Interestingly, as Leaf concepts allow the use of virtually any database schema, it is also possible to alternatively use the i2b2 CONCEPT_DIMENSION or OMOP concept_ancestor tables to both build the concept tree and enforce this logic instead using IN or EXISTS statements, similar to i2b2.
Programmatically generated Leaf concepts can also be data-driven and unique to local clinical data. Scripts to generate unique concepts for clinics, services, hospital units, demographic characteristics, and other data types can be used to create arbitrary institution-specific concepts. Data such as those derived from natural language processing can similarly be made available in this way.
In terms of structure, Leaf allows for a variety of strategies for representing concepts depending on how best to present data to users and how clean the data are. For example, a hypothetical concept structure to query orders of ciprofloxacin for 50 mg may be:
a) Using a predefined nested structure where “50 mg” and other possible values are hard-coded as child concepts:
- Medication Orders
- Ciprofloxacin (SQL: WHERE @.med = “ciprofloxacin”)
- 50mg (SQL: WHERE @.med = ‘ciprofloxacin’ AND @.unit = ‘mg’ AND @.dosage = 50)
- …
b) Using a single ciprofloxacin concept with dropdowns such as that when dragged into a query the user would see:
Had a medication order of ciprofloxacin of any dose and strength
Whereupon clicking “of any dose and strength” they can select from a dropdown of values such as “50 mg,” “100 mg,” etc.).
c) Using a single ciprofloxacin numeric concept such that when dragged into a query the user would see:
Had a medication order of ciprofloxacin of any dose (mg)
Whereupon clicking “of any dose (mg)” they can specify a numeric value or range, and the unit is hard-coded as “mg.”
d) A combination of the above approaches.
Querying multiple Leaf instances simultaneously
Because Leaf AST-based query representations do not contain SQL but rather pointers to arbitrary concepts, mapping Leaf queries to various data models is notably straightforward, with a few important caveats.
Each Leaf concept contains an optional UniversalId field for the purpose of mapping the concept to a matching concept in a different Leaf instance, provided that the UniversalId values are the same. Concept UniversalId values must conform to the Uniform Resource Name specification20 and begin with the prefix urn: leaf: concept, but otherwise have no restrictions.
For example, 2 or more institutions with Leaf instances installed can choose to allow their users to query each other’s clinical databases using Leaf. After exchanging secure certificate and web address information, administrators must agree on common naming conventions for Leaf concept UniversalIds. A UniversalId of a concept for ICD-10 diagnosis codes related to type 2 diabetes mellitus could be defined as
urn : leaf : concept : diagnosis : coding = icd10 + code = e11.00 − e11.9
which serves as a human-readable identifier and includes the domain (diagnosis), the coding standard (ICD-10), and relevant codes to be queried (a range of diabetes mellitus type 2–related codes from E11.00 to E11.9).
Users from each institution continue to see the concepts defined by their local Leaf instance in the user interface. As users run queries across partner Leaf instances, however, each partner instance automatically translates the sender’s concepts to local concepts by UniversalIds. After concept translation, local instance-specific SQL queries are generated and executed, with results returned to the user. If a given Leaf instance does not have a local concept for any one of the concepts included in the user’s query, it responds with that information to the user and does not create a query.
Because each Leaf instance translates and runs queries independently, a missing concept in one instance does not affect others, which proceed to report results to the user. Importantly, Leaf server API instances do not communicate patient count results or row-level data directly to each other but instead send data only to the client after de-identification. Identified multi-instance queries are not allowed; if a user is in Identified mode all queries are restricted to their home institution.
Boolean logic and temporality constraints in cohort definition queries
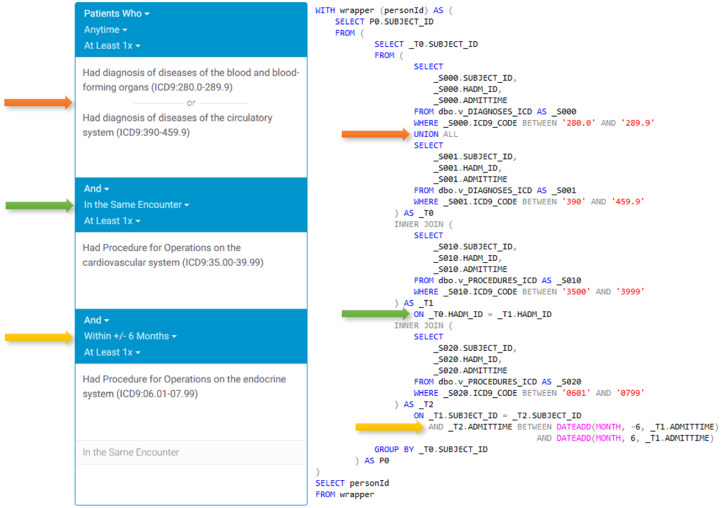
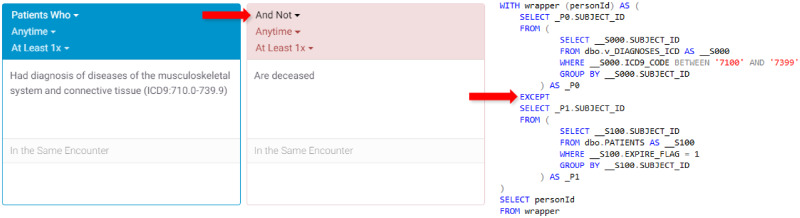
Leaf’s core functionality is as a flexible query constructor and execution engine. Queries can be configured to handle AND, OR, negation, and various temporality constraints, and include multiple visual cues to users. Temporal query relations are created vertically in the user interface (Figure 4), while additional inclusion or exclusion criteria with no temporal relations are represented horizontally in multiple panels (Figure 5). The query constructor user interface is designed to approximate natural language as much as possible to make query logic clear and readable to users.
Figure 4.
Visual representation of a single-panel query with the user interface on the left and corresponding compiled SQL representation on the right using the MIMIC-III (Medical Information Mart for Intensive Care-III) database. The example demonstrates the use of OR, same encounter, and temporal range logic.
Figure 5.
Visual representation of a 2-panel query with the user interface on the left and corresponding compiled SQL representation on the right. The second panel is used as an exclusion criteria and executed using a SQL EXCEPT statement.
Extracting and exporting data for a cohort
After executing a cohort estimation query, Leaf allows users to explore row-level data using the “Patient List” screen. Exportable datasets (hereafter simply “datasets”) are defined by an administrator and configured by dynamic SQL statements whose output must conform to a template of predefined column names. Dataset templates include “Encounter,” “Condition,” “Observation,” “Procedure,” and “Immunization,” and are based on denormalized tabular representations of FHIR resources.21
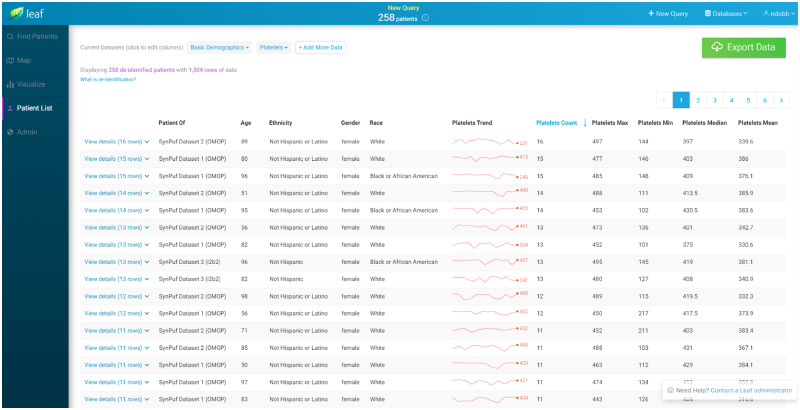
Similar to concepts, Leaf datasets include an optional UniversalId field which allows multiple Leaf instances to return predictable row-level FHIR-like clinical data (Figure 6), even if their underlying data models differ. After data are returned, users can quickly create and populate custom REDCap projects through the Leaf interface in both an identified and de-identified fashion. Leaf also employs an administrator-defined row export limit to ensure smooth operations and prevent large dataset exports without proper oversight.
Figure 6.
Screenshot of the Leaf user interface showing synthetic demographic and laboratory result data for a cohort from multiple Leaf instances, querying Observational Medical Outcomes Partnership and Informatics for Integrating Biology and the Bedside databases simultaneously. The Leaf client application automatically generates summary statistics for each patient. Granular row-level data can be accessed by clicking on a patient row in the table, and is directly exportable to REDCap via the REDCap application programming interface.
RESULTS
Since being released as a production tool at the University of Washington in February 2018, 302 users have run 18 791 queries against our EDW and created 854 REDCap projects populated with data by Leaf. During this time, our research informatics team has noted a relative increase in complex data extraction requests as usage of Leaf has grown, with a corresponding decrease in requests for simpler queries which we believe are likely being accomplished by clinicians and researchers using Leaf, though this is difficult to directly measure. Information on user roles and usage is listed in Table 1.
Table 1.
Leaf usage by role/position
| Role/position | Total users | Total queries |
|---|---|---|
| Attending physician or faculty | 80 (26.4) | 5250 (27.9) |
| Resident physician | 60 (19.8) | 4522 (24) |
| Research Coordinator or Scientist | 40 (13.2) | 4670 (24.8) |
| Central IT staff | 40 (13.2) | 429 (2.2) |
| Other or unknown | 27 (8.9) | 301 (1.6) |
| Departmental staff | 21 (6.9) | 1833 (9.7) |
| Student | 17 (5.6) | 937 (4.9) |
| Physician fellow | 8 (2.6) | 591 (3.1) |
| Nurse | 7 (2.3) | 135 (0.7) |
| Pharmacist | 2 (0.6) | 123 (0.6) |
Values are n (%).
IT: information technology.
Information on Leaf is available within our institution on the Institute of Translational Health Science’s website, internal intranet sites at the University of Washington, and through introductory presentations we have conducted. Many users continue to learn of Leaf by word of mouth, however, as we’ve pursued a “soft” production release to estimate informatics resources to support a growing user base going forward.
To assess impact, in spring 2019 a brief REDCap-based user feedback survey was sent to 232 Leaf users (excluding IT staff, users with access <3 months, and those believed to have left the institution), with a total response rate of 25.8% (60 of 232). Of the respondents, 48.3% (29 of 60) had at least 1 abstract, manuscript, poster, or presentation prepared using Leaf in some fashion, for a total of 68 abstracts and manuscripts (mean 1.1 per user) and 35 posters and presentations (mean 0.6 per user). Citations were voluntarily provided for 11 abstracts and manuscripts22–32 and 3 presentations,33–35 and there were links to usage for a summer internship research program36 and providing local context to national studies.37,38 Responses to the question “What purposes or projects have you used Leaf for?” are shown in Table 2.
Table 2.
Leaf usage by stated purpose(s)
| Purposes of use | Answered yes |
|---|---|
| IRB-approved research | 32 (53.3) |
| Data exploration | 25 (41.6) |
| Prep to research | 21 (35) |
| Hypothesis generation | 20 (33.3) |
| Quality improvement | 17 (28.3) |
| Testing | 11 (18.3) |
| Grant submissions | 9 (15) |
| Other | 1 (2.4) |
Values are n (%).
IRB: Institutional Review Board.
A total of 50% (30 of 60) responded using an open-ended comment field regarding their thoughts on Leaf in general. A total of 16.6% (6 of 30) mentioned the need for increased granularity of data and availability of historical data, and are specific to the data available in our EDW. A total of 63.3% (19 of 30) commented positively on Leaf’s value as a tool for research:
“I’m involved with several projects that use Leaf in a variety of ways… including all of our grant submissions and understanding how patients “flow” between EMS and trauma facilities in King County. For one recent successful grant proposal I think I was using Leaf every day for two months… It would have been incredibly difficult to get all the preliminary data if it wasn’t for Leaf.”
“The Leaf enterprise [query tool] is incredibly helpful in terms of determining number of patients in the EMR for particular research or QI queries and helping to extract variables that could be relevant to research. It has changed the way that I create and hypothesize my research projects.”
“I think Leaf is the most useful software to access [data generated by] the EHR [electronic health record]. I use it for resident projects, for case findings, for sample size estimates for grants, and to collect outcomes for IRB research projects.”
3.3% (1/30) mentioned confusion in using the user interface, and 10% (3/30) mentioned difficulty in validating data returned by Leaf queries:
“Stopped trying to use because of difficulty determining who is really in the cohort as intended.”
In terms of performance, using our central EDW production reporting database server with over 60 TB data, 144 cores, and 4194 GB memory, 97.5% of Leaf queries completed successfully with a mean execution time of 26.2 seconds, returning a mean 48 858 patients, and using a mean 9.1 concepts. The remaining 2.5% of queries failed largely due timeout issues (configured at 180 seconds). A total of 47.9% (9002 of 18 791) of queries utilized diagnosis or procedure code tables with over 320 million rows each. In general, we’ve found relatively simple queries to return results within 30 seconds, while more complex queries (ie, using more than 10 concepts or with temporal constraints) typically take 1-3 minutes to complete. We leave a more detailed examination of query performance and scalability to future work, but note that to date Leaf queries have been demonstrated to perform well using traditional relational database indexing methods on an EDW with nearly 5 million patients and over 2 decades of data.
DISCUSSION
Leaf represents a significant advancement in data-driven self-service cohort discovery tools by allowing CTSAs, large clinical research projects and enterprises to leverage existing clinical databases while reducing the burdens of data extraction and query construction on informatics teams. Because Leaf is a self-service tool, it removes the “informatics barrier” that many clinicians, trainees, and students face when trying to launch observational research or quality improvement initiatives and uncorks the potential of these faculty, fellows, residents, and students to implement their ideas.
Since becoming a formal project within the Center for Data to Health, we have refined Leaf to be readily accessible and valuable to the broader CTSA community. We are currently working with a growing number of CTSA sites to assist in piloting Leaf at other institutions.
Limitations
Despite its strengths, Leaf has a number of limitations that should be recognized. In terms of federated queries to multiple Leaf instances, the UniversalId model as a means of cross-institutional querying can require significant curation by administrators to ensure that the concept UniversalIds at each site match exactly, though this saves the costly effort of transforming and aligning data models. Also, while designed to be intuitive, the Leaf user interface requires some training to become proficient; many University of Washington users have requested assistance in crafting queries, even after receiving training. We suspect this is caused by the complexity of the underlying clinical data instead of the complexity of Leaf. While Leaf allows for highly flexible SQL queries, it is not designed to clean, integrate, or ensure semantic alignment among data, and is not a substitute for proper database maintenance, data quality testing, and refinement.
Last, while Leaf aims to securely de-identify patient data using current best practices, guaranteed prevention of patient reidentification remains elusive for data discovery tools like Leaf and an ongoing challenge in health care in general.39,40 To further tighten privacy protections, future Leaf development can focus on implementing greater configurability over date-shift increments, obfuscation, and anonymization algorithms to institutions deploying Leaf.
CONCLUSION
Leaf represents a successful example of a flexible, user-friendly cohort estimation and data extraction tool that can adapt to nearly any clinical data model. In this article, we describe how Leaf can be incorporated into existing enterprise workflows and infrastructure, allowing clinicians and researchers a window into clinical data while simultaneously reducing the informatics burden of supporting similar tools. As Leaf is now an open-source software project, we welcome discussion and collaboration and encourage other members of the CTSA community to join us in using and improving the tool.
FUNDING
This work was funded by the National Institutes of Health (NIH), the National Center for Advancing Translational Sciences (NCATS), NIH/NCATS UW-CTSA grant number UL1 TR002319, and NIH/NCATS CD2H grant number U24TR002306.
AUTHOR CONTRIBUTIONS
NJD wrote the majority of the article and is the creator and lead developer of Leaf. CS is the co-developer of Leaf and wrote the de-identification methods section. RB, JM, and BB led planning for data access, user management, and deployment at UW. BdV planned and advised on REDCap import and export functionality and user support tiers. EZ manages project feature requests and development plans. RH serves as faculty champion and has driven adoption and feature refinement. KS contributed to overall design and feature planning. AW and PT-H advise and provide implementation oversight. SDM directs overall development efforts and leads strategic planning.
ACKNOWLEDGMENTS
We gratefully acknowledge: our clinical and research users at the University of Washington for their suggestions and patience in testing Leaf; Robert Meizlik and Xiyao Yang for their development work on early versions of Leaf; Matthew Bartek (University of Washington); Nora Disis, Carlos De La Peña, Jennifer Sprecher, Marissa Konstadt (Institute of Translational Health Sciences); Sarah Biber and Melissa Haendel (Oregon Health and Science University, Center for Data to Health); and Shawn Murphy, Isaac Kohane, Diane Keogh, Douglas MacFadden, Griffin Weber, and other members of the i2b2 and SHRINE teams whose groundbreaking work we have been inspired by and remain indebted to.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Berner ES, Moss J.. Informatics challenges for the impending patient information explosion. J Am Med Inform Assoc 2005; 12 (6): 614–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shameer K, Badgeley MA, Miotto R, et al. Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief Bioinform 2017; 18 (1): 105–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011; 4 (1): 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42 (2): 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Norman DA, Draper SW.. User Centered System Design|New Perspectives on Human-Computer Interaction. Abingdon, United Kingdom: Taylor & Francis; 1986. https://www.taylorfrancis.com/books/e/9781482229639 Accessed April 6, 2019. [Google Scholar]
- 6. Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2). J Am Med Inform Assoc 2010; 17 (2): 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferranti JM, Gilbert W, McCall J, et al. The design and implementation of an open-source, data-driven cohort recruitment system: the Duke integrated subject cohort and enrollment research network (DISCERN). J Am Med Inform Assoc 2012; 19 (e1): e68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Penberthy L, Brown R, Puma F, Dahman B.. Automated matching software for clinical trials eligibility: measuring efficiency and flexibility. Contemp Clin Trials 2010; 31 (3): 207–17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson N, Abend A, Mandel A, et al. Implementation of a deidentified federated data network for population-based cohort discovery. J Am Med Inform Assoc 2012; 19 (e1): e60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.TriNetX. https://www.trinetx.com/ Accessed April 30, 2019.
- 11. Dybå T, Dingsøyr T.. Empirical studies of agile software development: a systematic review. Inform Softw Technol 2008; 50 (9–10): 833–59. [Google Scholar]
- 12. Carine F, Parrent N.. Improving patient identification data on the patient master index. Health Inf Manage 1999; 29 (1): 14–7. [DOI] [PubMed] [Google Scholar]
- 13. Pautasso C. RESTful web services: principles, patterns, emerging technologies In: Web Services Foundations. New York, NY: Springer; 2014: 31–51. [Google Scholar]
- 14.Office for Civil Rights (OCR). Methods for de-identification of PHI. https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html Accessed April 29, 2019.
- 15.OMOP Common Data Model–OHDSI. https://www.ohdsi.org/data-standardization/the-common-data-model/ Accessed April 6, 2019.
- 16.Epic. https://www.epic.com/ Accessed April 6, 2019.
- 17. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klann JG, Abend A, Raghavan VA, Mandl KD, Murphy SN.. Data interchange using i2b2. J Am Med Inform Assoc 2016; 23 (5): 909.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Klensin J, Saint-Andre P. Uniform Resource Names (URNs). https://tools.ietf.org/html/rfc8141 Accessed April 6, 2019.
- 21. Bender D, Sartipi K. HL7 FHIR: an agile and RESTful approach to healthcare information exchange; In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; 2013: 326–31.
- 22. Morcos M, Lood C, Hughes G.. Demographic, clinical and immunologic correlates among a cohort of 50 cocaine users demonstrating anti-neutrophil cytoplasmic antibodies. J Rheumatol 2019; 46 (9): 1151–6. [DOI] [PubMed] [Google Scholar]
- 23. Thomason J, Lood C, Hughes G.. The interferon gamma release assay is a novel predictor of disease activity in systemic Lupus erythematosus. Arthritis Rheumatol 2017; 69 (suppl 10). [Google Scholar]
- 24. Bartek MA, Kessler GL, Talbott JM, Nguyen J, Shalhub S.. Washington State abdominal aortic aneurysm mortality shows a steady decline between 1996 and 2016. J Vasc Surg 2019. Mar 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kang C, Bartek MA, Shalhub S, et al. PC040. disease-based observational cohort study of patients with thoracoabdominal aortic aneurysm. J Vasc Surg 2018; 67 (6): e183–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taylor AP, Freeman RV, Bartek MA, Shalhub S.. Left ventricular hypertrophy is a possible biomarker for early mortality after type B aortic dissection. J Vasc Surg 2019; 69 (6): 1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bartek MA, Aldea G, Nguyen J, et al. IP003. Aortic dissection-related mortality in Washington State remains unchanged from 1996 to 2016. J Vasc Surg 2018; 67 (6): e89–90. [Google Scholar]
- 28. Kang C, Bartek MA, Shalhub S, et al. Disease-based observation cohort study of patients with thoracoabdominal aortic aneurysm. J Vasc Surg 2018; 68 (3): e35–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bartek MA, Talbot J, Nguyen J, et al. Trends in Washington state aortic-related deaths over a 21-year period, 1996-2016. J Vasc Surg 2017; 66 (3): e54. [Google Scholar]
- 30. Thornblade LW, Verdial FC, Bartek MA, et al. The safety of expectant management for adhesive small bowel obstruction: a systematic review. J Gastrointest Surg 2019; 23 (4): 846–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang I, Liew J, Morcos B, Zuo S, Crawford C, Bays AM.. A pharmacist managed titration of urate-lowering therapy to streamline gout management. Arthritis Rheumatol 2019; 39 (9): 1637–41. [DOI] [PubMed] [Google Scholar]
- 32. Yang H, Veldtman G, Bouma B, et al. Non-vitamin K antagonist oral anticoagulants in adults with a Fontan circulation. Are they safe? Open Heart2019; 6 (1): e000985. [DOI] [PMC free article] [PubMed]
- 33. Mills B, Muller E, Feinstein B, et al. Differences in injury circumstances and outcomes between firearm injuries transported via EMS and private vehicle. In: proceedings from the Society for the Advancement of Violence and Injury Research Annual Meeting; April 1–3, 2019; Cincinnati, OH.
- 34. Powelson L. Chronic pain after trauma. In: proceedings from the UW Medicine Anesthesiology and Pain Medicine Grand Rounds; June 27, 2018; Seattle, WA.
- 35. Amin TM, Schorn M, Krashin D, et al. Frequency of opioid use for migraines in the ED. In: proceedings from the World Congress on Pain; September 12–16, 2018; Boston, MA.
- 36.INSIGHT Program Awarded Federal Research Education Grant. http://depts.washington.edu/hiprc/insight-program-awarded-federal-research-education-grant/ Accessed April 6, 2019.
- 37.HealthLink, UW Medicine. UW Medicine warns of danger to kids around a lawnmower. https://www.king5.com/article/news/health/uw-medicine-warns-of-danger-to-kids-around-a-lawnmower/281-576066862 Accessed April 6, 2019.
- 38. Malcolm K, Hurst A. More bike sharing, fewer helmets. Are head injuries on the rise? https://kuow.org/stories/more-bike-sharing-fewer-helmets-are-head-injuries-on-the-rise/ Accessed April 6, 2019.
- 39. Sweeney L. K-anonymity: a model for protecting privacy. Int J Uncertain Fuzz Knowl Based Syst 2002; 10 (5): 557–70. [Google Scholar]
- 40. El Elam K, Jonker E, Arbuckle L, Malin B.. A systematic review of re-identification attacks on health data. PLoS One 10 (4): e0126772. [DOI] [PMC free article] [PubMed] [Google Scholar]