Abstract
Objective
The study sought to describe the literature describing clinical reasoning ontology (CRO)–based clinical decision support systems (CDSSs) and identify and classify the medical knowledge and reasoning concepts and their properties within these ontologies to guide future research.
Methods
MEDLINE, Scopus, and Google Scholar were searched through January 30, 2019, for studies describing CRO-based CDSSs. Articles that explored the development or application of CROs or terminology were selected. Eligible articles were assessed for quality features of both CDSSs and CROs to determine the current practices. We then compiled concepts and properties used within the articles.
Results
We included 38 CRO-based CDSSs for the analysis. Diversity of the purpose and scope of their ontologies was seen, with a variety of knowledge sources were used for ontology development. We found 126 unique medical knowledge concepts, 38 unique reasoning concepts, and 240 unique properties (137 relationships and 103 attributes). Although there is a great diversity among the terms used across CROs, there is a significant overlap based on their descriptions. Only 5 studies described high quality assessment.
Conclusion
We identified current practices used in CRO development and provided lists of medical knowledge concepts, reasoning concepts, and properties (relationships and attributes) used by CRO-based CDSSs. CRO developers reason that the inclusion of concepts used by clinicians’ during medical decision making has the potential to improve CDSS performance. However, at present, few CROs have been used for CDSSs, and high-quality studies describing CROs are sparse. Further research is required in developing high-quality CDSSs based on CROs.
Keywords: clinical reasoning ontology, clinical decision support, clinical ontology, clinical concepts, ontology properties
INTRODUCTION
Clinical decision support systems (CDSSs), when integrated with electronic health record (EHR) systems, are an integral part of health information technology.1,2 CDSSs assist clinicians during the health-related decision-making process by presenting situation-specific clinical knowledge and patient information, in an appropriate format, at the appropriate time of the care process.2 Barriers to CDS development include lack of incentives, lack of standardized clinical terminology, outdated legacy EHR, lack of transferability of clinical decision support (CDS) logic from one system to another, lack of experts needed to translate medical knowledge into a CDS knowledge base (KB), and the low computer literacy of the end user.3
Clinicians encounter a significant number of alerts every day, and the usefulness of these alerts is questionable. Van der Sijs et al4 conducted a systematic review to assess physician response to drug safety alerts and found that 49%-96% of alerts were overridden. Studies have noted that clinicians often override alerts that are considered clinically irrelevant, reveal information that is already known by the clinician, or do not take into account other relevant information pertinent to the case.5,6 An unfortunate unintended consequence of CDSSs is “alert fatigue,” due to their high false positive rate.7 Traditionally, alerts have been designed to follow a rigid decision tree accessing only specific and limited patient information.8 Hence, alert logic often misses important relevant patient information, leading to inappropriate alerting. Other factors contributing to high false positive rates include low alerting threshold, lack of personalization, lack of clinical importance, and inaccuracy per updated guidelines.4,9,10
Alert-based CDSSs usually are comprised of 3 components: a KB (encompassing scientific and medical information, patient information from the EHR and CDS logic), a user interface that allows the user to communicate with the system, and an inference engine that provides the platform for the functionality of the CDSS.8 Currently, much of the patient data within EHRs, especially reasons for clinicians’ decisions, are in unstructured text format. Most logic-based CDSSs that rely on structured data are unable to utilize data related to clinical reasoning because the clinical data present within the EHR and the data structure of the KB are insufficient for the effective function of traditional alert-based CDSSs.
One approach that developers have employed to improve CDSSs is to model clinical reasoning through ontologies to simulate the decision-making processes carried out by clinicians.11–14 Clinical reasoning is the process used by clinicians to obtain and analyze data to reach a decision regarding a patient.15 It requires general understanding of evidence-based medical knowledge and the ability to isolate relevant medical information related to the specific case, based on a specific patient’s information.16 In treating patients, clinicians are faced with questions such as “What is the patient’s diagnosis?” and” When did symptoms start?” They are also faced with more complex questions related to reasoning such as “Why was a particular medication given over another?” or “What were the other diagnoses considered?” The data structures currently used within EHRs do not lend themselves readily to identifying answers to questions regarding clinical reasoning. This limitation also cripples the KBs used by current CDSSs. An ontology that details clinical reasoning will allow us to categorize and organize these reasons, thereby making them available for CDSS, and forms the basis for a more sophisticated system that utilizes previous patient-specific clinician reasoning when alerting.
An ontology is a formal representation of knowledge within a domain; typically, a hierarchically arranged set of unique terms known as concepts, their attributes, and the semantic relationships between those concepts.17 Ontologies organize domain knowledge into structures that computers can read, and humans can understand. Clinical reasoning ontologies (CROs) represent the concepts used by clinicians reasoning about diagnostic and therapeutic interventions and making diagnoses.18,19 Patient-specific clinical data are mapped into these CROs to make them usable in clinical reasoning axioms and to allow for the description of clinical decisions. CROs capture clinicians’ reasoning process by defining clinical concepts, mapping patient data to these concepts, and the defining the semantic relationships between them. This data structure will enable the creation of a more personalized KB for CDSSs. For example, clinicians can indicate, when prescribing, that a certain medication should be prescribed to the patient even though the patient is on a medication that could potentially interact with the prescribed drug, because the patient has previously tolerated the medication combination. A CDSS could be designed to access this information and learn that although generally there is a drug-drug interaction, it is irrelevant for this patient, and therefore, do not alert. Thus, in utilizing CRO-based CDSSs, one could decrease the pernicious phenomena of overalerting, and mitigate alert fatigue by creating more personalized and smarter CDSSs.
The ability to reuse existing ontologies would reduce some of the barriers to the development of CDSSs and could possibly speed the development process. The Open Biological and Biomedical Ontology (OBO) Foundry, a collective that provides access to biological, biomedical, and clinical related ontologies, could be a potential source for a CRO.20 However, the ontologies in OBO tend to focus on a specific aspect of clinical entities rather than cognitive processes. For example, the Human Disease Ontology classifies human-related diseases according to their etiology and provides a standardized ontology of disease and phenotypic terms that allow for semantic mapping of diseases across existing vocabularies.21 Other ontologies, such as the Cardiovascular Disease Ontology, focus on specific disease processes.22 Although OBO lists several such ontologies, an ontology encapsulating the “reasoning concepts” behind the clinical decision across overall patient-clinician encounter without restricting to a specific disease entity does not exist.23
In the absence of an existing standard, researchers are developing their own CROs to represent specific disease processes or different aspects of clinical workflows. The purpose of these ontologies includes improving interoperability,24 improving information gathering,25 aiding medical education,26 administrative support,27 and improving CDSSs.11 At least some of the ontologies that are used in CDSSs appear to map some reasoning axioms creating partial CROs.11–14
Given the clinical importance of CRO-based CDSSs and lack of a comprehensive literature review of current research on CROs in CDSSs, we believe that a systematic review is needed that provides an overview of the existing CRO-based CDSSs, with a compilation and classification of the concepts and properties present within these ontologies. This paper represents such a review to identify and summarize published works that describe CDSSs based on clinical ontologies with a focus on ontologies that contain clinical reasoning concepts and semantic relationships. We included a catalogue of the concepts and properties used within these ontologies and identify the current practices for developing and applying CROs to CDSSs. The results of our summary provide a resource for researchers and developers working on CRO-based CDSSs to select characteristics applicable to their efforts and can be used as a reference to guide future research and potential synergies of current practices in CRO-based CDSSs.
The objective of this systematic review is to describe the literature outlining clinical reasoning ontologies used to empower CDSSs and identify and classify the concepts (medical knowledge concepts and reasoning concepts) and their properties (semantic relationships and attributes) within these ontologies to guide future research.
MATERIALS AND METHODS
We reviewed the literature with the objective of answering the following study questions:
What are the existing CROs used to empower CDSSs?
How are the CROs and the CDSSs evaluated by their developers?
What are the characteristics of the existing CROs that are used by researchers and developers working on CRO-based CDSSs (ie, medical knowledge concepts, reasoning concepts, semantic relationships, and attributes)?
We followed the Preferred Reporting Items for Systematic Reviews and Meta-Analysis guidelines as far as appropriate for this review, to minimize the selection bias of included studies.28 A study protocol was written before the investigation (the study protocol was written in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analysis and published systematic reviews before investigation and was submitted to PROSPERO to be registered; the study was deemed as outside PROSPERO’s scope).
Data sources and search strategy
We searched databases including PubMed, PubMed Central, and Scopus from their inception to January 30, 2019. Multiple search terms and combinations of search terms were tested to determine the search strategy that identified the broadest results possible. Consensus among the authors was reached before deciding on the search strings. MeSH (Medical Subject Headings) terms were not used in the search strings as they were found to identify many irrelevant studies. We found that including both singular and plural forms within the second query broadened the search and identified studies that would have otherwise been missed. We used the following search strings:
-
PubMed and PubMed Central search terms:
“Clinical cognition” OR “Clinical Reasoning” OR (“Ontology” AND “Evidence Based Medicine”)
-
Scopus and Google Scholar (GS) search terms:
(“Decision support system” OR “Decision support systems”) AND (ontology OR terminology)
We included GS as an additional source to capture any relevant “grey” literature. Grey literature comprises nonformal scholarly publications produced by organizations outside of traditional academic publishers and can include dissertations, technical reports, conference proceedings articles from nongovernmental organizations and policy institutions.29 Many innovations in technology are initially published in these forms. There are some limitations to GS (eg, the search algorithm can personalize the search to the user, thus hindering replicability).30 Additionally, studies on GS have suggested the search should be limited to the first few pages due to diminishing returns.31 Indeed, we found that the relevancy of the articles greatly diminished after 10 pages; hence, we confined our search results to first 10 pages. The final search was conducted on February 2, 2019.
Study selection
The identified studies were evaluated according to the inclusion criteria: (1) studies exploring terminologies related to clinical reasoning and CDS, (2) studies exploring application or development of CDSSs that use CROs or clinical ontologies with reasoning axioms, and (3) studies exploring computerized methodology to draw relationships between clinical concepts.
The study selection was performed in stages. In stage 1, eligibility criteria were refined by 2 authors (P.I.D., J.J.C.) who independently reviewed subsets of 100 titles. The percent agreement was calculated following the independent review. Disagreements were discussed with the aim of revising and fine-tuning the eligibility criteria. This process was repeated with the revised criteria and another subset of 100 titles until a 94% agreement was reached. In stage 2, the titles were assessed for inclusion by a single reviewer (P.I.D.). The abstracts of all selected articles during stage 2 were then evaluated in stage 3 independently by the 2 reviewers (P.I.D., J.J.C.). Articles accepted, based on abstracts, by either reviewer advanced to the fourth stage of screening, in which 2 authors (P.I.D., J.J.C.) screened the full text of each article. The final article list is a compilation of articles accepted by both reviewers during stage 4.
Data extraction and synthesis
Data related to CDSS purpose, medical domain, computational methods, ontology scope and purpose, knowledge source, and characteristics such as concepts (medical knowledge and reasoning) and properties (relationship and attributes) were extracted from the study articles. The information provided within the articles was abstracted using an iteratively structured form by one of the authors (P.I.D.). The ontologies were categorized as new, existing, or revised based on whether the study article described using an ontology newly created by the CDS development team, used an existing ontology without modification, or used an existing ontology but modified to better fit CDSS scope, respectively. The other authors were consulted, as needed, for data extraction, and any conflicts were resolved via discussion and consensus.
We compiled concepts and properties used within the CRO. We reached group consensus about the classification of properties as either “relationships” or “attributes” and concepts as either “reasoning concepts” or “medical knowledge concepts.” We combined the concepts and removed duplicates based on the descriptions provided within the text, tables, and concept maps provided in the publications. When necessary, a more descriptive term was used to identify the final concept based on its description. The same methodology was performed for properties. When a definition of a concept or property was unavailable within the article, we inferred the definition using the informed assessment of the 2 medical expert authors.
Last, we extracted data regarding the CDSSs, and any ontology evaluations performed by the development team (internal validity and usability testing). See Supplementary List 1 for definitions of characteristic terms.
Quality assessment
The ontology evaluation comprises intrinsic (ie. technical) and extrinsic (ie. usability) testing. We defined intrinsic evaluation as an assessment of the ontology based on a set of criteria: accuracy, clarity, internal consistency, completeness, conciseness, expandability, and efficiency.32,33 Extrinsic evaluation relates to function and is defined as measurement of effectiveness of the CRO-based CDSS and its ease of use.34 We based our definitions of evaluation criteria established by Gomez-Perez.32
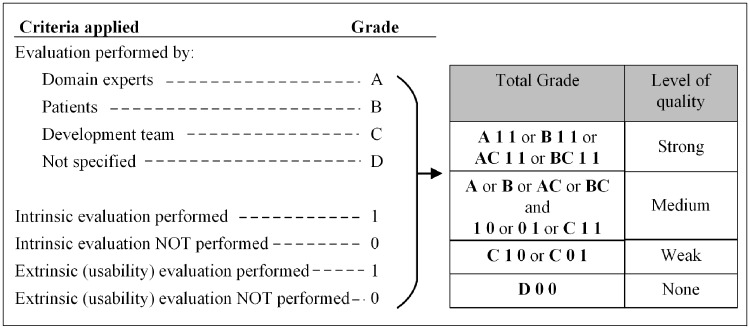
We conducted the quality assessment by evaluating the quality related data described in the publications. Any mention of performance of accuracy, clarity, internal consistency, completeness, conciseness, expandability, or efficiency were grouped under intrinsic evaluation as per our definition, and any mention of user testing were categorized as extrinsic. We conducted our evaluation based on predefined criteria as indicated in Figure 1. The CDSSs were then categorized as high, moderate, or low level of quality. Owing to the descriptive nature of the included studies, the Cochrane risk of bias is not applicable.
Figure 1.
Criteria used for study quality assessment.
RESULTS
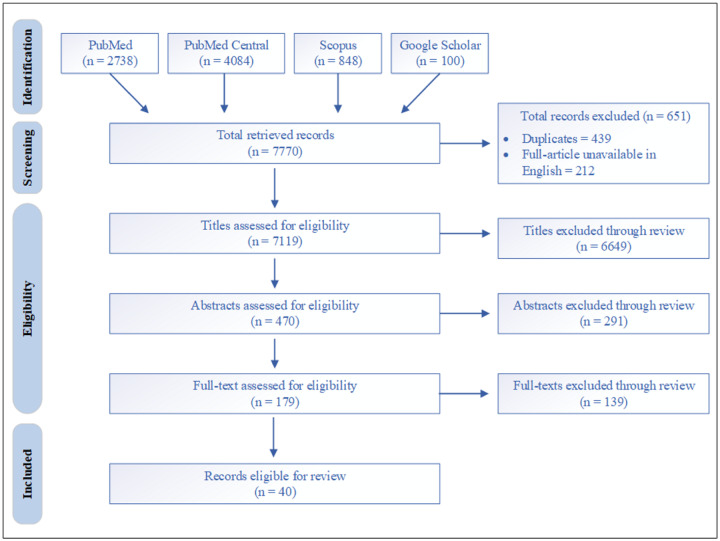
The database searches yielded a total of 7770 results. After excluding duplicates and articles in which the full-text version was not available in English, we reviewed 7119 titles. Of these, 470 articles met eligibility criteria for abstract review, which led to 179 articles for full-text review. Forty studies met the inclusion criteria and were reviewed in detail. The selection of articles is outlined in Figure 2.
Figure 2.
Search results.
Characteristics of CDSSs
The characteristics of the CRO-based CDSSs are summarized in Table 1. The articles by Farrish and Grando56 and by Grando et al57 were identified as describing the same CRO-based CDSS; therefore, they were merged. Similarly, articles by Abidi63 and Abidi et al64 described the same CRO-based CDSS; hence, they were combined, resulting in 38 CRO-based CDSSs. All of the final 40 articles were found in either MEDLINE or Scopus. None of the final articles were exclusive to GS.
Table 1.
Summary of studies included (n = 38)
| Author | Computational methods | Medical domain | CDSS purpose | Associated ontologies |
|---|---|---|---|---|
| Mohammed and Benlamri11 | RB, proximity-based, machine learning | DM2 and HTN | Provides differential diagnosis recommendation based on patient's data and CPGs | Patient ontology, disease symptoms ontology |
| Sene et al12 | RB, pattern-matching algorithm, NLP | Geriatric oncology | Assist during telemedicine based on CBR process and the conventional medical reasoning | Medical ontology |
| Denekamp and Peleg13 | Multiphase, anchor-based, Bayesian | Diagnosis | Assist physicians in the process of MCM-oriented diagnosis | TiMeDDx - Knowledge model |
| Uciteli et al35 | RB | Perioperative risk | Identify and analyze risks in perioperative treatment process to aid in avoiding errors | Risk identification ontology (RIO) |
| Bau et al36 | RB | Diabetic management during surgery | Assist with the management of diabetic patients during surgery | Domain ontology |
| Merlo et al37 | OB | Functional behavioral problems | Provide an evidence-based approach to behavioral experts in diagnosing behavioral problems | FBA ontology |
| Jimenez-Molina et al38 | OB, fuzzy logic, algorithm | Chronic disease | Manage all stages of chronic patient diagnosis and treatment based on business process management approach | MCCS ontology, process ontology, actors ontology |
| Shen et al39 | OB, machine learning | Infectious diseases | Diagnose infectious diseases based on patient entered data and provide antibiotic treatment recommendations | Domain ontology |
| El-Sappagh et al40 | OB, RB | DM2 | Assists with the treatment of DM2 | DM2 Treatment Ontology (DMTO) |
| Abidi41 | OB, RB, algorithm | Comorbidity conditions | A CPG integration framework to provide primary care physicians, institutional specific CPG medicated CDSs for comorbidities | Comorbidity CPG ontology |
| Beierle et al42 | OB | BC | Support treatment decisions in cancer therapy by revising co-medications and drug interactions | Ontology for Cancer Therapy Application |
| Shang et al43 | RB | Chronic disease (HTN and DM2) | Service oriented sharable CDSS that integrate multiple CPGs, for chronic diseases | Infrastructure ontology, special ontology |
| Berges44 | OB | GHJ rehabilitation | Assist physiotherapists during the treatment processes related to GHJ | Telerehabilitation Ontology (TrhOnt) |
| Qi et al45 | RB | SpA | Provides patients with a personalized home-based self-management system for SpA | SpA ontology |
| Alsomali et al46 | RB | Penicillin-related adverse events | Alert clinicians of possible adverse drug events related to penicillin during drug prescription | Ontology of penicillin allergy |
| Zhang et al47 | RB | CPG | A sharable CDSS for management of clinical pathways that integrates into hospital CDS applications and fits into existing workflows | Decision support knowledge base generic ontology |
| Wilk et al27 | OB, RB | IHTs | Assist with formation of the IHTs to manage patients based on presentation-specific clinical workflows and team dynamics | IHT ontology |
| Zhang et al48 | RB, OB | DM2 | Provides patient specific recommendations on the management of inpatients with DM2 | Semantic healthcare knowledge ontology |
| Rosier et al49 | RB, OB | Cardiology | Improve AF-related CIED alert triage | Cardio-vascular disease ontology |
| Jafarpour et al50 | RB, OB, algorithm | CPG | Provide computerized CDS based on CPGs using an OWL-based execution engine | CPG ontology |
| Alharbi et al51 | RB | Diabetes | Decision support for diagnosis and treatment of diabetes based on CPG | Diabetes Ontology, Patient ontology |
| Shen et al14 | OB, machine learning, NLP, fuzzy logic | Disease diagnosis and treatment | Provides clinicians and patients with an optimal personalized diagnostic and treatment plan | Knowledge Model Agent Type (KMAT) ontology |
| El-Sappagh et al52 | RB | Diabetes | Assist with the diagnosis and management of diabetes | Case base ontology |
| Budovec et al26 | RB | Radiology | Provides radiology differential diagnosis in an interactive website and an educational tool | Radiology Gamuts Ontology (RGO) |
| Wang et al53 | RB, probability | General medical CPGs | Personalized CPGs for disease specific treatment to be used by individual hospitals. | Local ontology |
| Eccher et al54 | RB, OB | Cancer therapy | Facilitate the interoperability between a CPG-based DSS for cancer treatment and an oncological EPR | Therapies ontology |
| Martínez-Romero et al55 | RB, OB | CICU | Provides supervision and treatment assistance for critical patients in CICU with acute cardiac disorders | Critical Cardiac Care Ontology (C3O) |
| Farrish and Grando56; Grando et al57 | RB | Medication | Assists with management of polypharmacy prescriptions for patients with MCC to reduce the overall treatment complexity | Drug ontology |
| Omaish et al58 | RB, OB | ACS | Assists ED physicians with treatment of ACS patients based on computerized ACS CPGs | CPG ontology |
| Riaño et al59 | OB, ranking of weighted options | Home care of chronic diseases | Assists with the management of chronically ill patients including development of personalized treatment plans | Case profile ontology |
| Adnan et al (2010)60 | OB, NLP, RB | High risk discharge medications | provides advice recommendations for high risk discharge medications, to be used in the Electronic Discharge Summary | Medication information ontology |
| Prcela et al61 | RB | Heart failure | provides CDS for heart failure | Heart failure ontology |
| Hussain and Abidi62 | RB | CPGs in Imaging studies | Provides a framework to computerize CPGs and to execute modeled CPGs based on patient data to deliver recommendations | CPG ontology, domain ontology, patient ontology |
| Abidi63; Abidi et al64 | RB | BC | An interactive BC follow-up CDSS for family physicians to assist with BC management and to provide educational material to patients | CPG ontology, patient ontology, BC ontology |
| Fox et al65 | OB | BC | Supports complex care pathways in BC | PROforma Task ontology, Goal ontology |
| Achour et al66 | OB, RB | Blood transfusion | Assists clinicians with the prescription of blood products for transfusion | Domain ontology |
| Wheeler et al67 | OB | HTN | A mobile self-management App to assists patients with the management of HTN | HTN management ontology |
| Sadki et al25 | OB, RB, algorithm | BC | Allows structured patient data acquisition for the management of BC patients | BC Knowledge Model |
ACS: acute coronary syndrome; App: application; BC: breast cancer; CBR: case-based reasoning; CDSS: clinical decision support system; CICU: cardiac intensive care unit; CPG: clinical pathway guideline; DM2: diabetes mellitus type 2; ED: emergency department; EPR: electronic patient record; FBA: functional behavioral assessment; GHJ: glenohumeral joint; HTN: hypertension; IHT: interdisciplinary healthcare team; MCC: multiple chronic conditions; MCCS: medical context and contextual services; MCM: main clinical manifestation; NLP: natural language processing; OB: ontology based; RB: rule based; SpA: spondylarthritis; TiMeDDx: name of the ontology.
Rule-based computational methods use IF/THEN logic rules for inferencing. Ontology-based methods make inferences by following the relationships within the ontology. In addition, “algorithm” was used to describe when an inference was based on a specific calculation. Thirty CDSSs (79%) used rule-based computation for inferencing, 22 (58%) used an ontology-based method, 6 (16%) used algorithms, 3 (8%) used natural language processing, 3 (8%) used machine learning, and 2 (5%) used fuzzy logic. Other computational methods included probability, proximity-based, anchor-based, and ranking of weighted option. Twenty (5 ontology-based and 15 rule-based) CDSSs used only 1 computational method.
A wide range of medical domains were addressed by the CDSSs: 12 dealt with management of chronic diseases (5 diabetes, 1 hypertension, 1 heart failure, and 5 multiple chronic diseases), 6 with cancer management (4 breast cancer and 2 general cancer treatment), 3 with cardiac-related conditions, 3 with medication management and adverse events, 3 with general clinical guidelines, 2 with radiology, 2 with diagnosis, and 7 with others (1 each of preoperative risk, infectious disease, glenohumeral joint rehabilitation, spondylarthritis treatment, healthcare teams, diagnosis and treatment, blood transfusion).
Characteristics of CROs
All the CROs were used as the KB for their respective CDSS. A total of 34 CDSSs (90%) used only 1 ontology, 4 CDSSs used 2 ontologies, and 2 CDSSs used 3 ontologies (Table 2). The ontology scope correlated with the medical domain. The types of knowledge sources employed during the ontology development (with the corresponding number of ontologies) included domain experts (n = 23), clinical pathway guidelines (CPGs) (n = 22), literature (n = 20), existing ontologies or terminologies (n = 14), EHR (n = 11), clinical workflows (n = 2), and software including websites (n = 1). Most CDSSs (81%) employed multiple sources with only 7 studies using 1 type of knowledge sources (4 using CPG only, 2 using existing ontology, 1 using literature). The size of the ontologies appears to vary significantly, although most publications did not mention the actual number of concepts and properties.
Table 2.
Description of the ontologies identified within the CDSSs
| Author | Ontology scope | Sources of knowledge | Ontology—source(s)a | Ontology sizeb |
|
|---|---|---|---|---|---|
| Concepts | Properties | ||||
| Mohammed and Benlamri11 | Patient parameter; diseases and symptoms | Existing ontologies | Multiple existing plus new | >241b | 13 ** |
| Sene et al12 | Medical concepts in geriatric oncology | Lit, domain experts | New | 61b | ND |
| Denekamp and Peleg13 | Clinical data items related to diagnosis | Lit, CPG, domain experts | New | 5b | 6 ** |
| Uciteli et al35 | Perioperative risk | CPG, domain experts, existing ontology | Multiple existing | 19b | 13b |
| Bau et al36 | Medical knowledge related to DM2 management | Domain expert, EHR, hospital clinical workflow | New | 31b | 13b |
| Merlo et al37 | Structure and the semantics of functional behavioral assessment methods | Domain experts, lit | New | 15b | 15b |
| Jimenez-Molina et al38 | Medical context; clinical pathways; healthcare professionals | CPG, domain experts, EHR | New | 24b | 24b |
| Shen et al39 | Infectious disease | Existing ontologies, lit, CPG, websites | New | 1 267 004 | 12b |
| El-Sappagh et al40 | DM2 | Lit, CPG, domain experts, EHR, existing ontologies | Multiple existing | >10 700 | 279 |
| Abidi41 | CPG | CPG, domain experts | New | 102 | 58 |
| Beierle et al42 | Cancer drugs: active ingredients, interactions, drug regimens | Lit, EHR, existing software | Revised existing | 40b | 18b |
| Shang et al43 | HTN and DM2 CPGs; disease concepts related to HTN and DM2 | CPG | New | 47 | 121 |
| Berges44 | Physiotherapy process related to glenohumeral joint | Existing ontologies and databases, EHR treatment protocol, domain experts | Multiple existing | 2351 | 100 |
| Qi et al45 | Spondylarthritis and definitions for alert type | Lit, CPG, domain experts | New | 22b | 22b |
| Alsomali et al46 | Penicillin allergy related adverse events | Lit, existing ontologies | New | 52 | 15 |
| Zhang et al47 | Patient data, CDS related domain knowledge, CDS rules | CPG | New | 62 | 94b |
| Wilk et al27 | Clinical workflow, interdisciplinary healthcare team member and patient specific concepts | Lit, domain experts | Revised existing | 21b | 19b |
| Zhang et al48 | DM2 | Lit, CPG, EHR, domain experts, existing terminologies | New | 127 | 196 |
| Rosier et al49 | AF and CIED alerts | Lit | New | 252 | 25 |
| Jafarpour et al50 | Nursing, CHF, and AF CPGs | Existing ontology | Revised existing | 12b | 13b |
| Alharbi et al51 | Diabetes | CPG, domain experts | New | 7b | 19 |
| Shen et al14 | Diagnosis, prognosis, and treatment (example: gastric cancer) | Lit, EHR | New | 92b | 58b |
| El-Sappagh et al52 | Case base reasoning context in diabetes; patient attributes | Domain experts, lit, CPG, existing ontology, EHR | Multiple existing | 132 | 48b |
| Budovec et al26 | Radiology information needed for diagnosis | Lit, domain experts | New | 4b | 3b |
| Wang et al53 | CPG | EHR, CPG, domain experts | New | 88b | 11b |
| Eccher et al54 | Cancer treatment | Domain experts, oncological workflows, existing ontologies | New | 82b | 9b |
| Martínez-Romero et al55 | Medical care related to acute cardiac disorder in cardiac-ICU | Lit, domain experts | New | 40b | 7b |
| Farrish and Grando56; Grando et al57 | Generic drugs and related information | Lit, existing ontologies, CPG, domain experts | Multiple existing | 16b | 35b |
| Omaish et al58 | CPG related to ACS management | CPG, domain experts | New | 29b | 1b |
| Riaño et al59 | Chronic disease management, home care | CPG, lit, EHR, domain experts, ICD10 | New | 143b | 8b |
| Adnan et al60 | Medication knowledge specific to post discharge patient information | EHR, lit, existing websites and terminologies | New | 40b | 7b |
| Prcela et al61 | Heart failure | CPG (congestive and acute HF) | New | 200 | > 100 |
| Hussain and Abidi62 | Imaging CPG; patient health parameters | CPG (EU Radiation Protection 118 Referral Guideline for Imaging) | New | 30b | 7b |
| Abidi63; Abidi et al64 | Structure of BC follow-up CPG; patient parameter; medical knowledge related to BC found within the CPG | CPG, domain experts | New | 12b | 45b |
| Fox et al65 | BC (diagnosis, treatment, management) | Lit, CPG, existing ontologies | Multiple existing plus new | 79b | ND |
| Achour et al66 | blood transfusion | Domain experts, existing terminologies | New | 17b | 2b |
| Wheeler et al67 | CPGs, behavior change theories, and associated behavior change strategies related to HTN | CPG, Lit, domain experts | New | 50 | 71 |
| Sadki et al25 | Patient data in BC stage and management | CPG | New | 4b | 6b |
AF: atrial fibrillation; BC: breast cancer; CDSS: clinical decision support system; CHF: congestive heart failure; CIED: cardiac implant electronic devices; CPG: clinical pathway guideline; DM2: diabetes mellitus type 2; EHR: electronic health record; HF: heart failure; HTN: hypertension; ICU: intensive care unit; Lit: literature; ND: not discernable.
aIdentify if the clinical reasoning ontology discussed is new, existing, or revised; new—if it is a new ontology created by the development team specifically for the CDSS; existing—if the development team used an ontology that is already in existence without altering it; revised—if the development team used an already existing ontology but with some alterations to suit the CDSS purpose.
bOntology size is not explicitly stated. The size is determined by adding the number of concepts and properties described within the article (in body or in images).
Quality assessment data
Our quality assessment revealed that 30 (79%) studies described the evaluation of the CRO-based CDSS. In 29 (76%) cases, intrinsic evaluations were performed and 20 (53%) studies employed test cases or comparison studies. A test case was defined as a set of variables under which the system’s function is tested. For example, the accuracy of TiMeDDx was tested by analyzing the diagnosis inferred for patient vignettes describing multiple symptoms.13 Comparison studies compared the outcome of the CDSS with a gold standard, domain expert, or another CDSS. For example, in the article by Shen et al,39 the system’s diagnostic capability was tested by comparing the diagnosis of the CDS to that of the clinician.
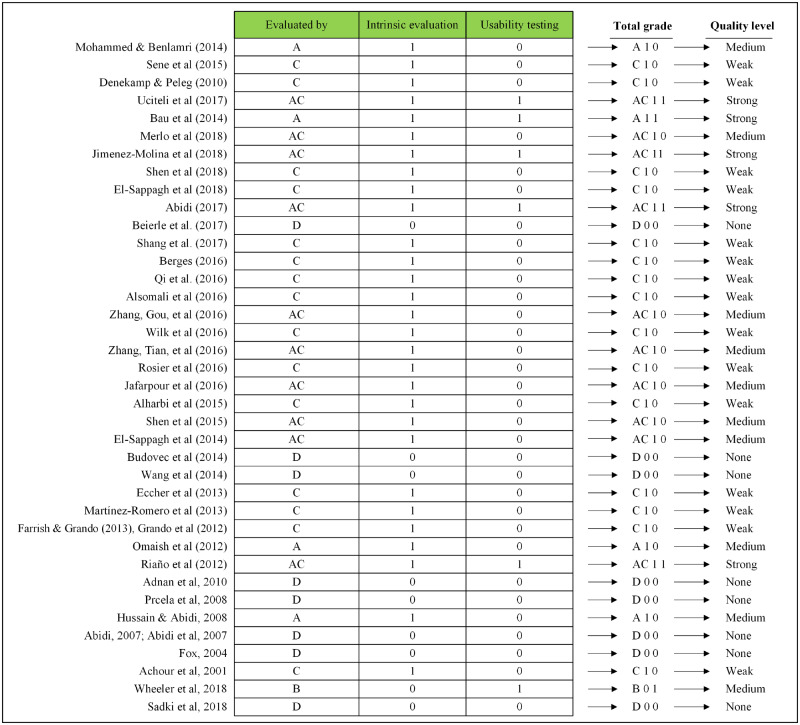
Nine of the publications mentioned performing intrinsic evaluation but did not elaborate the purpose. Usability testing was only performed in 6 CDSSs. Only 5 studies achieved a high quality level, while 10 had a medium quality level, and 23 had a weak quality level. Our assessment revealed that 8 studies did not report a formal evaluation of their CDS or CRO. The CRO-based CDSSs in our study set did not discuss testing related to clinical salience in practice or effects on clinical outcomes. Figure 3 summarizes the quality assessment of included studies.
Figure 3.
Quality assessment of the clinical decision support systems and their ontologies.
Concepts and properties extracted from CROs
A total of 1315 concepts and 603 properties were identified from the study articles. We then removed duplicates and combined concepts with similar descriptions, producing a final list of 567 concepts. These were then categorized into 339 medical knowledge and 228 reasoning concepts. We considered concepts that describe medical information related to patient, disease processes, clinical workflows, and clinic function such as history, symptoms, assessment, treatment plan, lab tests, administration process, and risk factors, as medical knowledge concepts. The medical knowledge concepts from all the studies were grouped, duplicates were removed, and concepts with the same definition were combined, resulting in 126 unique medical knowledge concepts and 31 subconcepts. For example, we combined concepts patient history46 and history14,40 under the concept history; concepts route of administration,40,59delivery option,12 and application route42 under the concept route of administration; and concepts rule,47logic,62 and SWRL: Rule52 under the concept Logic. We determined that the concepts comprised 15 medical domains. See Supplementary Table S1 for full list of the medical knowledge concepts.
Reasoning concepts were also categorized by removing duplicates and combining the concepts with the same definition. For example, we grouped concepts ActDocumentation48 and Make record of data65 under the concept Data documentation; concepts task67 and enact tasks65 under the concept enact tasks; and concepts Application_purpose,12Therapeutic purpose,14 and Treatment_intent54 under the concept Treatment_purpose. Thirty-eight unique reasoning concepts with 86 subconcepts were identified. The reasoning concepts expanded over 5 medical domains. See Table 3 for full list of reasoning concepts and Supplementary Table S2 for their definitions.
Table 3.
List of reasoning concepts (see Supplementary Table S2 for reasoning concepts definitions)
| Medical domain | Reasoning concept | Reasoning subconcepts |
|---|---|---|
| Action | Inform patient or colleague about | Process information, appointment, results, management, risk |
| Data documentation | ||
| Enquiry to acquire information | Family history, personal history, current problem and background, past problem and associated information, availability of services, appointments | |
| Enquiry to recall for service | Arrange service | |
| Enquiry to request with response | Appointment, results, second opinion, specialist services, investigations | |
| Enquiry to confirm action has been done | ||
| Decision | Eligibility for participation in trails, eligibility for service, need for referral, diagnosis, detection, etiology, pathology, need for follow-up, investigation, prophylaxis, risk assessment, choice of therapy | |
| Assessment | COMB, automatic motivation, physical capability, psychological capability, reflective motivation, social opportunity, behavioral change technique | |
| Comparison of… . | Comparison of behavior, comparison of outcomes | |
| Plan | Referral for service, follow-up, manage treatment pathway, arrange/rearrange services | |
| Acquire information/knowledge about specific setting | Acquire information about setting, acquire comparison data in setting | |
| Detect | ||
| Classify | Staging | |
| Eligibility | Investigations, referral, therapy, research trail | |
| Assess level of some parameter | Urgency, risk, need, quality | |
| Predict | ||
| Diagnosis | ||
| Prognosis | ||
| Action_description | Decisional_action_description, Drug_prescription_description, Clinical_action_description, Drug_administration_description, Surgical_action_description, Laboratory_exam_action_description | |
| Enact tasks | Communicate, Educate, Inform, Act_Observation, Act_Patient_Encounter, Act_Procedure, Act_Substance_Administration, Act_Registration, Act_Working_List, Act_Care_Plan, Feedback and monitoring | |
| Goals | Achieve some state of world | Limit changes to current state, bring about required future state, empower staff, prevent unwanted future state, ensure compliance with plan |
| Goal type | Cessation goal, acquisition goal, shapeable goal, intervention goal | |
| Treatment | Treatment decision | Decide between alternative interventions, decide whether to carry out intervention or not, decide type of investigation, Decide scheduling of intervention |
| Treatment_purpose | ||
| Dose modification | Add serum, decrease dose, increase dose, continue, finish | |
| Influential factors | Motivation, opportunity, obstacle, reward and threat | |
| Intervention function | ||
| CPG | Similarity measure | Exact, difference, complex |
| Confidence | ||
| Antecedents | ||
| Guideline_Step | Decision_Option, Diagnostic_Step, Discharge_Step, Admission_Step, Transfer_Step, Control_of_disease | |
| Associations | ||
| Repetition and substitution | ||
| Regulation | ||
| Covert learning | ||
| Scheduled consequences | ||
| Tip | ||
| TDFDomain (Theoretical Domains Framework) |
COMB: capability, opportunity, motivation, and behavior model; CPG: clinical pathway guideline.
Properties were also analyzed in similar fashion leading to 240 unique properties: 103 attributes and 137 relationships. The properties comprised relationships and attributes across 17 domains. Table 4 displays a sample list of properties, their facets, and their designation as attribute or property (see Supplementary Table S3 for the full list).
Table 4.
List of properties (see Supplementary Table S3 for full list)
| Domain | Property | Facet | Range | R vs. A |
|---|---|---|---|---|
| Record | has_Patient | Medical record | A | |
| hasHighLevelContext | High-level context | R | ||
| Patient | has_patient_profile | Patient properties | R | |
| has_patient_ID | Patient ID | A | ||
| has_lab_test | has_Part, has_Unit, has_Status | Lab test details | R | |
| has_Lab_test_value | Test value | A | ||
| has_diagnosis | hasSide | Diagnosis, location | R | |
| has_diagnosis_severity | Disease severity | A | ||
| has_history | EndingDate | Patient's history | R | |
| has_Family_History | isRelativeOf | Family history | R | |
| has_treatment_plan | Treatment plan | R | ||
| has_symptom_or_sign | Symptoms and sign | R | ||
| has_presentation | Chief presentation | R | ||
| has_measurement | has_UpperLimitValue, has_ExactValue | Value | A | |
| Disease_since_date | Date | A | ||
| has_complication | Complication | R | ||
| has_previous_treatment_plan | Treatment plan | R | ||
| has_HealthcareProvider | hasSpecialty, plays_role_of, actorName | Healthcare provider | R | |
| has Alarm | Alarm types | R | ||
| has_demographic | hasName, Sex, has Age, Ethnicity | Demographic data | R | |
| Diagnostic process | observationMethod | Observation method | R | |
| observed_data | Data value | A | ||
| Assessment_Reason | Reason | R | ||
| has_pain | Pain level | A | ||
| has_device | hasMedicalDevice, hasTool | Medical device | R | |
| has_Assessment | Assessment | R | ||
| has_patient_reported_findings | has_VAS_value, has_ASDAS, etc | Questionnaire value | A | |
| has_Recommendation | Recommendation | R | ||
| Signs and symptoms | Is_assessed_by | Assessment name | R | |
| has_RecoveryRate | Recovery rate | A | ||
| has_MortalityRate | Mortality rate | A | ||
| is_not_caused_by | Factors | R | ||
| cause_by | Causing factor | R | ||
| is_symptom_of | Disease | R | ||
| Diagnosis and disease | hasSyndrome | Syndrome name | R | |
| has_severity | Severity level | A | ||
| has_treatment | antibiotic2bacteria | Treatment | R | |
| has_causing_factors | bacteria2infection | Causing factor | R | |
| hasRisk | Risk factor | R | ||
| affected_Body_Site | Body part | R | ||
| hasLabTest | Lab test name | R | ||
| hasStatus | Status | A | ||
| hasSyndromeDuration | Time | A | ||
| has_new_stage | Cancer stage | A | ||
| is_transmitted_by | Vector | R | ||
| has_complication | Complication list | R | ||
| occurs_with | Disease, symptom | R | ||
| hasExperimentalData | Experimental data | R | ||
| Treatment | hasHealthRecord | hasEHR_ID | Health record ID | A |
| has_education_program | has_provider, has_section | Education program | R | |
| has_next_evaluation_date | Date | A | ||
| part_of | part_of | Treatment plan | R | |
| has_intervention_goal | isAppropriateForInterventionGoal | Intervention goal | R | |
| has_pharmacological_plan | Medication list | R | ||
| is_recommended_for_illness | Recommendation | R | ||
| Medication | Can_be_combined_with | Medication | R | |
| Contradict_with | Contradict_with_drug, _with_drug | Drug ingredient | R | |
| has_treatment_target | has_A1C_lowering_level, etc | Treatment target | A | |
| has_active_ingredient | Active ingredient | A | ||
| has_administrationProcess | Administration process | R | ||
| has_cost | Medication cost | A | ||
| has_order_start_date | Date | A | ||
| has_order_stop_date | date | A | ||
| has_dose | hasPatientDrugUnRec, etc | Dose | R | |
| dosage_Measurement_Unit | measurement unit | A | ||
| has_cumulative_dose | accumulative dose | A | ||
| has_maximum_dose | maximumDrugUnits, maximumDosage | medication dosage | R | |
| has_frequency (freq) | maximum_Freq, minimum_Freq | Drug frequency | A | |
| has_application_route | Drug application route | A | ||
| has_explanation | Explanation | R | ||
| has_toxicity | Toxicity | A | ||
| has_Therapy_description | withSpecificFluids | Drug therapy direction | A | |
| Nutrition | has_amount | has_calcium, has_carbohydrate_grams, | Quantity | A |
| has_calories | has_total_calories, | Amount of calories | A | |
| Time | has_time | number_of_times, hasExerciseTime | Time | A |
| has_temporal_entity | Temporal data | A | ||
| has_temporal_relation | equals, before, after, hasBeginning | Temporal relation | R | |
| Trend_in_TimePeriod | Time period | A | ||
| Alert | has_Alert | hasLow-, hasHigh- hasMedium-Alert | Alert level | A |
| AssociatedToDynamicContext | Dynamic context | R | ||
| Anatomy | nerve_supply | nerve | R | |
| has_location | Anatomic location | R | ||
| CDS/CPG | has_input | CDS input | A | |
| has_Outcome | Outcome specification | A | ||
| hasDecisionRule | CDS function, logic | R | ||
| has_Trigger | hasTriggerSource, triggersException | CDS trigger | R | |
| has_logic_component | has Arc, hasEndNode, hasStartNode | Arc, Node | R | |
| hasInformationReturn | Treatment information | R | ||
| Risk | risk_for_adverse_situation | Risk situation | R | |
| Risk_related_recommendation | Diagnostic test | R | ||
| Clinical Team | executes | Clinical workflow | R | |
| hasPractitionerStatus | Practitioner status | R | ||
| has_Action | has_directive, hasPatientAction, etc | Action | R | |
| Task | Evokes | Diagnosis | R | |
| Synergistically_evokes | Diagnosis | R | ||
| hasCondition | Medical condition | R | ||
| has_status | hasTaskState, hasWorkFlowStatus | Task status | R | |
| is_followed_by | Task | R | ||
| has_decision_option | Decision option | R | ||
| has_act_relations | hasActPtn, hasPtnAct, hasActRelTarget | Relationship type | R | |
| is_assigned | is_responsible_for, managesPatient, | Medical team member | R | |
| Universal | Priority | Priority level | A | |
| Reason | isWarrantedBy | Reason | R | |
| hasFunction | Function | R | ||
| isInputOf | Indicator | R | ||
| isOutputOf | Output | R | ||
| Functional terms | description | Rule description, model | R | |
| attribute | Attribute of model | A | ||
| hasDataCategory | subclass, hasScenario | Subclass, scenario | R | |
| terminologyName | Name string | A | ||
| code | procedureCode, DisplayName | Code | A | |
| hasStructuredData | Data type | A | ||
| translation | Translating code | A |
A: attribute; ASDAS: Ankylosing Spondylitis Disease Activity Score; CDS: clinical decision support; CPG: clinical pathway guideline; R: semantic relationship; VAS: visual analog scale.
DISCUSSION
In this systematic review, we investigated the literature exploring CROs used to empower CDSSs. We assessed the characteristics of the existing CDSSs that use CROs and determined the current practices used by the developers in creating the CROs. Tables 1 and 2 list the key findings. In summary, although there are many clinical ontologies in existence, we only identified 38 studies that used them in CDSSs. Moreover, these CROs restricted themselves to a specific clinical workflow. Ontologies such as the Breast Cancer Ontology42 and DMTO53 only contain concepts related to a specific disease, whereas ontologies like RIO36 and C3O56 are restricted to specific workflows within a specific subspecialty. These limitations are understandable considering the enormity of the medical field. The restricted scope of the ontologies limits their applicability across the full medical domain.
Medical decisions involve complex inferential processes, some, if not all, at least in part use “reasoning.” The difficulty in developing a sophisticated CDSSs that only alerts the clinician when appropriate, reducing the need for overrides, or assists with complex decision-making processes such as providing a differential diagnosis that is personalized to each patient, lies with the difficulties associated with decoding what constitutes clinical reasoning. Many researchers have proposed different approaches for utilizing ontologies to decrypt clinical reasoning especially for the betterment of CDSSs.11–14,35–67 We noted that even when CDSSs use CROs, most of them do so in combination with other inferencing methods such as rule-based inferencing to adequately represent the knowledge needed for the CDSS. This finding is expected given the complexity associated with clinical reasoning and KBs.
Our analysis also revealed that most developers referred to multiple data sources during ontology development, including existing ontologies, domain experts, literature, clinical guidelines, and the EHR. Currently, however, there is neither a standard format to identify appropriate sources for an ontology nor a standard document to which developers can refer to as a starting point. CROs and CRO-based CDSSs are generally being developed and studied in isolation. We believe that the broader informatics community will benefit from knowing the best practices used by existing systems. More importantly, our study provides a list of concepts and properties for an initial starting point, as is found in other research fields such as drug development or genetic research. We note, for example, that there are multiple ontologies developed by different groups for clinical workflows related to breast cancer25,42,63–65 and diabetes.11,36,40,48,52 As such, we believe that our lists of medical knowledge concepts, clinical reasoning concepts, and properties will provide a foundation for starting the development process of future ontologies. Furthermore, our findings could be used as the basis for a standard to improve access to data by CDSS developers, implementers, or evaluators to improve the function and interoperability of EHR and CDSS.
Implications for EHR improvement and future research
Clinical ontologies are increasingly used as a means for improving various aspects of health care.68–70 CDS is one such area in medicine in which clinical ontologies are being used to develop more efficient and accurate systems. Most CROs focused on a specific disease process, workflow, or subspecialty; hence, they tend to only map clinical reasoning concepts and relationships related those aspects. Thus, most CROs create only a partial representation of clinical knowledge used by clinicians. A more comprehensive CRO will facilitate better structuring of the KB and allow CDSSs to access a wider range of information that can both complement and improve extant CDSSs without being restrictive to only one aspect of patient care.71 This inclusiveness would allow for the development of more complex CDSSs that can incorporate and act upon data related to the whole patient. In turn, CDSSs could be better personalized to provide alerts only when they are clinically relevant to the patient. This would lead to significantly fewer alerts and alleviate alert fatigue.
Developers of clinical ontologies and CDSSs should consider expanding the number and the types of reasoning concepts mapped in CROs. In our study, we identified 38 unique reasoning concepts that belonged to 5 medical domains. An expanded CRO can be used to identify and store reasoning behind many medical decisions that currently are only present in the free-text clinical notes (ie, history and physical examination, progress notes, consult notes, pathology reports, and radiology reports). There is a significant gap in existing CROs in mapping the data related to decisions one of the most important aspects of medical care. Clinicians are faced with many questions when reviewing a patient’s records regarding the actions taken by others in the past. Unfortunately, the clinical reasoning for decisions regarding patient care in many cases is often buried in free-text notes.72 A comprehensive CRO that captures the “why” of a decision will greatly assist clinicians in quickly accessing data and improving efficiency, and lead to better patient care.73
A CRO can also be used to improve the reuse of data for learning health systems. The Agency for Healthcare Research and Quality defines learning health systems as a healthcare system in which “internal data and experience are systematically integrated with external evidence and that knowledge is put into practice.”74 A CRO can assist in mapping the reasoning behind clinical decisions to be used for quality improvements, consensus of cases, case discussions in morning rounds, and use during multidepartmental conferences held to discuss complex patient cases. Moreover, easy access to reasoning can be a useful tool for the education of medical and nursing students and young clinicians, and as a component of continue education for clinicians.
Although we believe that our methods have been successful in identifying most or all ontology-based CDSSs, our efforts to summarize the ontologies used by these systems is limited, primarily because the foci of the articles we found generally dwelled more on the details of the logic and systems and less on cataloging the concepts and relations used. To the extent possible, we have compiled names and definitions provided in the articles, but given the limited details available, our ability to identify commonalities across systems was modest. However, now that the systems have been identified, along with their developers and general domains of interest, our study can provide a “starter set” of subsequent efforts to engage interested stakeholders to build a more comprehensive, well-defined ontology.
CONCLUSION
This review summarizes existing literature on CRO-based CDSSs. It identifies the current practices used within the development of the CROs and formulates lists of medical knowledge concepts, reasoning concepts, and properties (relationships and attributes) used by these CDSSs. The use of CROs, which map concepts used by clinicians’ during medical decision making, can significantly improve CDSS functionality. Although many CDSSs have been developed using clinical ontologies, few use CROs. As a result, high-quality studies describing CROs are sparse. Further research is required in developing high quality CROs-based CDSSs.
FUNDING
The work was supported, in part, by funds from the UAB Informatics Institute, as well as by National Center for Advancing Translational Sciences, of the National Institutes of Health, award number UL1TR003096 (to JJC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
PID and JJC conceptualized, designed, and conducted the study including study selection, data extraction, and data analysis. PID drafted the manuscript with significant intellectual input form JJC, and JJC and TKC assisted with creating and editing the article. All authors approved the final version of the article.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Osheroff JA, Teich JM, Levick DL, et al. Improving Outcomes with Clinical Decision Support: An Implementer’s Guide. 2nd ed. Chicago, IL: Healthcare Information and Management Systems Society; 2012. [Google Scholar]
- 2. Berner ES, La Lande TJ. Overview of clinical decision support systems In Berner ES, ed. Clinical Decision Support Systems Theory and Practice. 2nd ed. New York, NY: Springer; 2007: 3–18. [Google Scholar]
- 3. Middleton B, Sittig DF, Wright A.. Clinical decision support: a 25 year retrospective and a 25 year vision. Yearb Med Inform 2016; Suppl 1: S103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van der Sijs H, Aarts J, Vulto A, et al. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc 2006; 13 (2): 138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCoy AB, Thomas EJ, Krousel-Wood M, et al. Clinical decision support alert appropriateness: a review and proposal for improvement. Ochsner J 2014; 14 (2): 195–202. [PMC free article] [PubMed] [Google Scholar]
- 6. Taylor L, R T.. Reasons for physician non-adherence to electronic drug alerts. MedInfo 2004: 1101–5. http://moxxi.mcgill.ca/sites/moxxi.mcgill.ca/files/PDF-MEDINFO.pdf. Accessed March 12, 2019. [PubMed] [Google Scholar]
- 7. Kesselhem AS, Cresswell K, Phansalkar S, et al. Clinical decision support systems could be modified to reduce ‘Alert Fatigue’ while still minimizing the risk of litigation. Health Aff 2011; 30 (12): 2310–7. [DOI] [PubMed] [Google Scholar]
- 8. Dissanayake PI, Kochendorfer KM.. Clinical decision support systems in medicine In: Brown GD, Pasupathy KS, Patrick TB, eds. Health Informatics, a Systems Perspective. 2nd ed. Chicago, IL: Health Administration Press; 2018: 121–46. [Google Scholar]
- 9. Nanji KC, Slight SP, Seger DL, et al. Overrides of medication-related clinical decision support alerts in outpatients. J Am Med Inform Assoc 2014; 21 (3): 487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weingart SN, Toth M, Sands DZ, Aronson MD, Davis RB, Phillips RS.. Physicians' decisions to override computerized drug alerts in primary care. Arch Intern Med 2003; 163 (21): 2625–31. [DOI] [PubMed] [Google Scholar]
- 11. Mohammed O, Benlamri R.. Developing a semantic web model for medical differential diagnosis recommendation. J Med Syst 2014; 38 (10): 79.. [DOI] [PubMed] [Google Scholar]
- 12. Sene A, Kamsu-Foguem B, Rumeau P.. Telemedicine framework using case-based reasoning with evidences. Comput Methods Programs Biomed 2015; 121 (1): 21–35. [DOI] [PubMed] [Google Scholar]
- 13. Denekamp Y, Peleg M.. TiMeDDx–a multi-phase anchor-based diagnostic decision-support model. J Biomed Inform 2010; 43 (1): 111–24. [DOI] [PubMed] [Google Scholar]
- 14. Shen Y, Colloc J, Jacquet-Andrieu A, et al. Emerging medical informatics with case-based reasoning for aiding clinical decision in multi-agent system. J Biomed Inform 2015; 56: 307–17. [DOI] [PubMed] [Google Scholar]
- 15. Elstein A, Bordage J.. Psychology of clinical reasoning In: Dowie J, Elstein A, eds. Professional Judgment: A Reader in Clinical Decision-Making. New York, NY: Cambridge University Press; 1988. [Google Scholar]
- 16. Benner P, Hughes RG, Sutphen M.. Clinical reasoning, decisionmaking, and action: thinking critically and clinically In: Hughes RG, ed. Patient Safety and Quality: An Evidence-Based Handbook for Nurses. Rockville, MD: Agency for Healthcare Research and Quality; 2008. [PubMed] [Google Scholar]
- 17. Gruber TR. A translation approach to portable ontology specification. Knowl Acquis 1993; 5 (2): 199–220. [Google Scholar]
- 18. Musen MA. Scalable software architectures for decision support. Methods Inf Med 1999; 38 (4–5): 229–38. [PubMed] [Google Scholar]
- 19. Bright TJ, Yoko Furuya E, Kuperman GJ, Cimino JJ, Bakken S.. Development and evaluation of an ontology for guiding appropriate antibiotic prescribing. J Biomed Inform 2012; 45 (1): 120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.OBO Foundry. http://www.obofoundry.org/ Accessed April 5, 2019.
- 21.OBO Foundry. Human Disease Ontology. http://www.obofoundry.org/ontology/doid.html Accessed April 5, 2019.
- 22.OBO Foundry. Cardiovascular Disease Ontology. http://www.obofoundry.org/ontology/cvdo.html Accessed April 5, 2019.
- 23. Colicchio TK, Cimino JJ.. Clinicians’ reasoning as reflected in electronic clinical note-entry and reading/retrieval: a systematic review and qualitative synthesis. J Am Med Inform Assoc 2019; 26 (2): 172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corral MA, Antonelli L, Sánchez LE.. Health ontology and information systems: a systematic review. IEEE Latin Am Trans 2017; 15 (1): 103–20. [Google Scholar]
- 25. Sadki F, Bouaud J, Guézennec G, et al. Semantically structured web form and data storage: a generic ontology-driven approach applied to breast cancer. Stud Health Technol Inform 2018; 84: 50–63. [PubMed] [Google Scholar]
- 26. Budovec JJ, Lam CA, Kahn Jr CE.. Informatics in radiology: radiology gamuts ontology: differential diagnosis for the semantic web. Radiographics 2014; 34 (1): 254–64. [DOI] [PubMed] [Google Scholar]
- 27. Wilk S, Kezadri-Hamiaz M, Rosu D, et al. Using semantic components to represent dynamics of an interdisciplinary healthcare team in a multi-agent decision support system. J Med Syst 2016; 40 (2): 42.. [DOI] [PubMed] [Google Scholar]
- 28. Moher D, Liberati A, Tetzlaff J; The PRISMA Group, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med 2009; 6 (7): e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cornell University Library. Tutorial: grey literature. https://guides.library.cornell.edu/graylit Accessed June 6, 2019.
- 30. Piasecki J, Waligora M, Dranseika V.. Google search as an additional source in systematic reviews. Sci Eng Ethics 2018; 24 (2): 809–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Haddaway NR, Collins AM, Coughlin D, Kirk S.. The role of google scholar in evidence reviews and its applicability to grey literature searching. PLoS One 2015; 10 (9): e0138237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gomez-Perez A. Evaluation of ontologies. Int J Intell Syst 2001; 16: 391–409. [Google Scholar]
- 33. Hlomani H, S D.. Approaches, methods, metrics, measures, and subjectivity in ontology evaluation: a survey. Semantic Web J 2014; 15 http://www.semantic-web-journal.net/system/files/swj657.pdf Accessed Mar 10, 2019. [Google Scholar]
- 34. García-Barriocanal E, Sicilia MA, Sánchez-Alonso S.. Usability evaluation of ontology editors. Knowl Org 2005; 32 (1): 1–9. [Google Scholar]
- 35. Uciteli A, Neumann J, Tahar K, et al. Ontology-based specification, identification and analysis of perioperative risks. J Biomed Semant 2017; 8 (1): 36.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bau CT, Chen RC, Huang CY.. Construction of a clinical decision support system for undergoing surgery based on domain ontology and rules reasoning. Telemed J E Health 2014; 20 (5): 460–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Merlo G, Chiazzese G, Taibi D, Chifari A.. Development and validation of a functional behavioural assessment ontology to support behavioural health interventions. JMIR Med Inform 2018; 6 (2): e37.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jimenez-Molina A, Gaete-Villegas J, Fuentes J.. ProFUSO: business process and ontology-based framework to develop ubiquitous computing support systems for chronic patients’ management. J Biomed Inform 2018; 82: 106–27. [DOI] [PubMed] [Google Scholar]
- 39. Shen Y, Yuan K, Chen D, et al. An ontology-driven clinical decision support system (IDDAP) for infectious disease diagnosis and antibiotic prescription. Artif Intell Med 2018; 86: 20–32. [DOI] [PubMed] [Google Scholar]
- 40. El-Sappagh S, Kwak D, Ali F, et al. DMTO: A realistic ontology for standard diabetes mellitus treatment. J Biomed Semant 2018; 9 (1): 8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Abidi S. A knowledge-modeling approach to integrate multiple clinical practice guidelines to provide evidence-based clinical decision support for managing comorbid conditions. J Med Syst 2017; 41 (12): 193.. [DOI] [PubMed] [Google Scholar]
- 42. Beierle C, Sader B, Eichhorn C, et al. On the ontological modelling of co-medication and drug interactions in medical cancer therapy regimens for a clinical decision support system. In: proceedings IEEE Symposium on Computer Based Medical Systems; 2017: 105–110. [Google Scholar]
- 43. Shang Y, Wang Y, Gou L, et al. Development of a service-oriented sharable clinical decision support system based on ontology for chronic disease. Stud Health Technol Inform 2017; 245: 1153–7. [PubMed] [Google Scholar]
- 44. Berges I, Antón D, Bermúdez J, et al. TrhOnt: building an ontology to assist rehabilitation processes. J Biomed Semant 2016; 7 (1): 60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Qi J, Chen L, Leister W, et al. Towards knowledge driven decision support for personalized home-based self-management of chronic diseases. In: 2015 IEEE 12th International Conference on Ubiquitous Intelligence and Computing and 2015 IEEE 12th International Conference on Autonomic and Trusted Computing and 2015 IEEE 15th International Conference on Scalable Computing and Communications and Its Associated Workshops (UIC-ATC-ScalCom); 2015: 1724–9.
- 46. Alsomali W, Razzak I, Alshammari R.. Development of ontology for penicillin-Related adverse events. J Med Imaging Health Inform 2016; 6 (3): 620–6. [Google Scholar]
- 47. Zhang YF, Gou L, Tian Y, et al. Design and development of a sharable clinical decision support system based on a semantic web service framework. J Med Syst 2016; 40 (5): 118.. [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y, Tian Y, Zhou TS, et al. Integrating HL7 RIM and ontology for unified knowledge and data representation in clinical decision support systems. Comput Methods Programs Biomed 2016; 123: 94–108. [DOI] [PubMed] [Google Scholar]
- 49. Rosier A, Mabo P, Temal L, et al. Remote monitoring of cardiac implantable devices: Ontology driven classification of the alerts. Stud Health Technol Inform 2016; 221: 59–63. [PubMed] [Google Scholar]
- 50. Jafarpour B, Abidi SR, Abidi S.. Exploiting semantic web technologies to develop owl-based clinical practice guideline execution engines. IEEE J Biomed Health Inform 2016; 20 (1): 388–98. [DOI] [PubMed] [Google Scholar]
- 51. Alharbi RF, Berri J, El-Masri S. Ontology based clinical decision support system for diabetes diagnostic. In: 2015 Science and Information Conference (SAI); 2015: 597–602.
- 52. El-Sappagh SH, El-Masri S, Elmogy M, et al. An ontological case base engineering methodology for diabetes management systems-level quality improvement. J Med Syst 2014; 38 (8): 67.. [DOI] [PubMed] [Google Scholar]
- 53. Wang HQ, Zhou TS, Tian LL, et al. Creating hospital-specific customized clinical pathways by applying semantic reasoning to clinical data. J Biomed Inform 2014; 52: 354–63. [DOI] [PubMed] [Google Scholar]
- 54. Eccher C, Scipioni A, Miller AA, et al. An ontology of cancer therapies supporting interoperability and data consistency in EPRs. Comput Biol Med 2013; 43 (7): 822–32. [DOI] [PubMed] [Google Scholar]
- 55. Martínez-Romero M, Vázquez-Naya JM, Pereira J, et al. The iOSC3 system: Using ontologies and SWRL rules for intelligent supervision and care of patients with acute cardiac disorders. Comput Math Methods Med 2013; 2013: 650671.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Farrish S, Grando A.. Ontological approach to reduce complexity in polypharmacy. AMIA Annu Symp Proc 2013; 2013: 398–407. [PMC free article] [PubMed] [Google Scholar]
- 57. Grando A, Farrish S, Boyd C, et al. Ontological approach for safe and effective polypharmacy prescription. AMIA Annu Symp Proc 2012; 2012: 291–300. [PMC free article] [PubMed] [Google Scholar]
- 58. Omaish M, Abidi S, Abidi S.. Ontology-based computerization of acute coronary syndrome clinical guideline for decision support in the emergency department. Stud Health Technol Inform 2012; 180: 437–41. [PubMed] [Google Scholar]
- 59. Riaño D, Real F, López-Vallverdú JA, et al. An ontology-based personalization of health-care knowledge to support clinical decisions for chronically ill patients. J Biomed Inform 2012; 45 (3): 429–46. [DOI] [PubMed] [Google Scholar]
- 60. Adnan M, Warren J, Orr M.. Ontology based semantic recommendations for discharge summary medication information for patients. In: proceedings of the IEEE International Symposium on Computer Based Medical System; 2010: 456–61. [Google Scholar]
- 61. Prcela M, Gamberger D, Jovic A.. Semantic web ontology utilization for heart failure expert system design. Stud Health Technol Inform 2008; 136: 851–6. [PubMed] [Google Scholar]
- 62. Hussain S, Abidi SSR.. An ontology-based framework for authoring and executing clinical practice guidelines for clinical decision support systems. J Inform Technol Healthcare 2008; 6 (1): 8–22. [Google Scholar]
- 63. Abidi SR. Ontology-based modeling of breast cancer follow-up clinical practice guideline for providing clinical decision support. In: proceedings of the IEEE International Symposium on Computer Based Med System; 2007: 542–7. [Google Scholar]
- 64. Abidi SR, Abidi SS, Hussain S, et al. Ontology-based modeling of clinical practice guidelines: a clinical decision support system for breast cancer follow-up interventions at primary care settings. Stud Health Technol Inform 2007; 129 (Pt 2): 845–9. [PubMed] [Google Scholar]
- 65. Fox J, Alabassi A, Black E, et al. An ontological approach to modelling tasks and goals. Stud Health Technol Inform 2004; 101: 31–45. [PubMed] [Google Scholar]
- 66. Achour SL, Dojat M, Rieux C, et al. A UMLS-based knowledge acquisition tool for rule-based clinical decision support system development. J Am Med Inform Assoc 2001; 8 (4): 351–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wheeler TS, Vallis TM, Giacomantonio NB, et al. Feasibility and usability of an ontology-based mobile intervention for patients with hypertension. Int J Med Inform 2018; 119: 8–16. [DOI] [PubMed] [Google Scholar]
- 68. Kostopoulos K, Chouvarda I, Koutkias V, et al. An ontology-based framework aiming to support personalized exercise prescription: application in cardiac rehabilitation. In: 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2011: 1567–70. [DOI] [PubMed]
- 69. Schulz S, Martínez-Costa C.. How ontologies can improve semantic interoperability in health care In: Riaño D, Lenz R, Miksch S, Peleg M, Reichert M, ten Teije A, eds. Process Support and Knowledge Representation in Health Care. ProHealth 2013, KR4HC 2013. Lecture Notes in Computer Science. Cham, Switzerland: Springer; 2013: 8268. [Google Scholar]
- 70. Daughton AR, Priedhorsky R, Fairchild G, et al. An extensible framework and database of infectious disease for biosurveillance. BMC Infect Dis 2017; 17 (1): 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rospocher M, Serafini L. Ontology-centric decision support. In: proceedings of the 2012 International Conference on Semantic Technologies Meet Recommender Systems and Big Data; 2012: 61–72.
- 72. Hripcsak G, Johnson SB, Clayton PD.. Desperately seeking data: knowledge base-database links. In: proceedings of the Annual Symposium on Computer Applications in Medical Care; 1993: 639–43. [PMC free article] [PubMed] [Google Scholar]
- 73. Cimino JJ. Putting the “why” in EHR: capturing and coding clinical cognition. J Am Med Inform Assoc2019. [DOI] [PMC free article] [PubMed]
- 74. Turning AM. Computing machinery and intelligence. Mind 1950; 49: 433–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.