Summary
Zymoseptoria tritici is a filamentous fungus causing Septoria tritici blotch in wheat. The pathogen has a narrow host range and infections of grasses other than susceptible wheat are blocked early after stomatal penetration. During these abortive infections, the fungus shows a markedly different gene expression pattern. However, the underlying mechanisms causing differential gene expression during host and non‐host interactions are largely unknown, but likely include transcriptional regulators responsible for the onset of an infection programme in compatible hosts. MoCOD1, a member of the fungal Zn(II)2Cys6 transcription factor family, has been shown to directly affect pathogenicity in the rice blast pathogen Magnaporthe oryzae. Here, we analyse the role of the putative transcription factor Zt107320, a homologue of MoCOD1, during infection of compatible and incompatible hosts by Z. tritici. We show for the first time that Zt107320 is differentially expressed in host versus non‐host infections and that lower expression corresponds to an incompatible infection of non‐hosts. Applying reverse genetics approaches, we further show that Zt107320 regulates the dimorphic switch as well as the growth rate of Z. tritici and affects fungal cell wall composition in vitro. Moreover, ∆Zt107320 mutants showed reduced virulence during compatible infections of wheat. We conclude that Zt107320 directly influences pathogen fitness and propose that Zt107320 is involved in the regulation of growth processes and pathogenicity during infection.
Keywords: fungal dimorphism, gene expression, incompatible host, pycnidia formation, Septoria leaf blotch
Introduction
The fungus Zymoseptoria tritici (synonym Mycosphaerella graminicola) infects wheat and causes the disease Septoria tritici blotch. The pathogen is found worldwide where wheat is grown and can cause severe reduction in yield (Fones and Gurr, 2015). During infection, the fungus enters the leaf through stomata and establishes a hyphal network in the mesophyll. It propagates without causing visual symptoms for 7–14 days before inducing necrosis and producing asexual fructifications, pycnidia, in which pycnidiospores are produced to be dispersed via leaf–leaf contact or rain‐splash to neighbouring leaves (Brading et al., 2002; Kema et al. 1996b; Ponomarenko et al., 2011). Zymoseptoria tritici has a heterothallic mating system and meiosis leads to the production of wind‐borne ascospores that are considered to be the main primary inoculum in wheat fields (Kema et al., 1996a; Morais et al., 2016; Ponomarenko et al., 2011; Suffert et al., 2011). Under experimental conditions, the fungus has a narrow host range infecting wheat and shows abortive infections on closely related non‐host grass species like Triticum monococcum (Jing et al., 2008) and Brachypodium distachyon (Kellner et al., 2014; O'Driscoll et al., 2015). However, the underlying determinants of host specialization and host specificity of Z. tritici are largely unknown.
A previous study comparing the expression profiles of Z. tritici between early infection (4 days post‐infection) of the compatible host Triticum aestivum and the non‐host B. distachyon revealed 289 genes that were similarly expressed in the two hosts, but differentially expressed compared to growth in axenic culture (Kellner et al., 2014). These genes are likely crucial for Z. tritici during stomatal penetration, which occurs in the same way in both hosts. However, 40 genes showed differential expression between host and non‐host infections (Kellner et al., 2014) and are possibly involved in the discrimination of compatible and non‐compatible host–pathogen interactions. The signalling and regulatory networks responsible for these differential expression patterns are unknown, however. One of the differentially expressed genes encodes the putative transcription factor Zt107320. Expression of Zt107320 was significantly increased during infection of T. aestivum compared to the early infection of B. distachyon (Kellner et al., 2014), suggesting a host‐dependent regulation of the gene.
Zt107320 encodes a putative transcription factor belonging to the Zn(II)2Cys6 family. This gene family of transcription factors is exclusive to fungi (MacPherson et al., 2006; Pan and Coleman, 1990) and many members play an important role in the regulation of fungal physiology. For example, Zn(II)2Cys6 transcription factors in Magnaporthe oryzae, Fusarium oxysporum, Leptosphaeria maculans, Parastagonospora nodorum and Pyrenophora tritici‐repentis are involved in the regulation of fungal growth and pathogenicity (Fox et al., 2008; Galhano et al., 2017; Imazaki et al., 2007; Lu et al., 2014; Rybak et al., 2017). Interestingly, the homologue of Zt107320 in the rice blast pathogen M. oryzae, MoCOD1, was shown to affect conidiation and pathogenicity (Chung et al., 2013). MoCOD1 was found to be up‐regulated during conidiation and appressorium formation at 72 h post‐infection. Furthermore, the deletion mutant ∆MoCOD1 showed defects in conidial germination and appressorium formation. In planta, the mutant ∆MoCOD1 was attenuated in extending growth from the first‐invaded cells and caused markedly reduced symptoms when compared to the wild type (Chung et al., 2013).
Transcription factors, in general, regulate expression by integrating various signalling pathways and represent interesting targets for dissecting causes and mechanisms of pathogenicity and host specificity. In M. oryzae, a systemic approach was applied to characterize all 104 members of the Zn(II)2Cys6 family of transcription factors. Of these, 61 were shown to be involved in fungal development and pathogenicity (Lu et al., 2014). Similarly, in the head blight‐causing fungus Fusarium graminearum 26% of 657 tested transcription factors showed a phenotypic effect during mycelial growth, conidia production and toxin production (Son et al., 2011). In summary, a number of transcription factors, which play important roles in host infection and pathogenesis, have been identified for several important crop pathogens (Chen et al., 2017; Okmen et al., 2014; Xiong et al., 2015; Zhang et al., 2018; Zhuang et al., 2016). In Z. tritici, however, only a few regulatory genes have been characterized. Recently, two transcription factors, ZtWor1 (Mirzadi Gohari et al., 2014) and ZtVf1 (Mohammadi et al., 2017), were shown to be important regulators of development and virulence of Z. tritici during compatible infections of wheat, highlighting how transcription factors can be used to identify and dissect aspects of pathogenicity. ZtWor1 has been functionally characterized and appears to be involved in the cAMP‐dependent pathway, up‐regulated during the initiation of colonization and involved in regulating effector genes (Mirzadi Gohari et al., 2014). ZtVf1, a transcription factor belonging to the C2‐H2 subfamily, is required for virulence and its deletion leads to lower pycnidia density within lesions. Decreased virulence appears to be due to a reduced penetration frequency and impaired pycnidia development (Mohammadi et al., 2017). Further regulatory elements that affect pathogenicity in Z. tritici encompass several members of mitogen‐activated protein kinase pathways and G‐protein subunits [reviewed in (Rudd, 2015)].
Based on the close homology of Zt107320 to MoCOD1 and its significantly different expression profile during host and non‐host infections, we hypothesized that Zt107320 plays an important role in Z. tritici during early wheat infection. We show here that Zt107320 affects virulence of Z. tritici during compatible infections and regulates the dimorphic switch as well as the growth rate and cell wall properties of this important fungal plant pathogen.
Results
Phylogenetic analysis of Zt107320
In order to determine the distribution of homologues of Zt107320 we performed a similarity search on the protein level among fungal sequences. A total of 30 sequences were retrieved, which included orthologous sequences as well as the non‐orthologous Zt111096, which shows the highest similarity to Zt107320 in Z. tritici, and the well‐described transcription factor AmyR and MalR in Aspergillus niger and Aspergillus oryzae, respectively. These sequences were aligned and a phylogenetic analysis was conducted (Fig. 1a,b). Among these highly conserved proteins two protein domains are shared: Zn(2)‐C6 fungal‐type DNA‐binding domain superfamily (IRP036864) and a transcription factor domain (IRP007219) supporting a functional role of the putative transcription factors (see Fig. 1a). The retrieved sequences contained several proteins that have already been functionally dissected: the M. oryzae orthologue MoCod1, the Alternaria brassicola orthologue AbPf2 (AB06533.1), the A. niger homologue AmyR and the A. oryzae homologue MalR. Deletion of AbPf2 resulted in nonpathogenic strains (Cho et al., 2013). In the wild type, expression of AbPf2 decreased after initial colonization of host tissues and the authors concluded that AbPf2 regulates pathogenesis (Cho et al., 2013). AmyR in A. niger is considered to be a regulator of starch degradation and to regulate the expression of enzymes involved in polysaccharide degradation (Suzuki et al., 2015; vanKuyk et al., 2012; Zhang et al., 2016). MalR in A. oryzae is considered to regulate the expression of the maltose‐utilizing cluster genes (Suzuki et al., 2015). Constructing a phylogenetic tree revealed a monophyletic origin of Zt107320 with 27 of the 30 homologues, whereas AmyR, MalR and Zt111096 are non‐monophyletic in regard to Zt107320.
Figure 1.

Homology of Zt107320 among fungal transcription factors. (a) MUSCLE alignment (Edgar, 2004) of sequences homologous to Zt107320 that indicates regions according to their identity with the consensus sequence. The localizations of two functional domains, identified using InterProScan, in the consensus sequence are highlighted in orange and blue. (b) Phylogenetic tree based on 30 sequences showing the highest similarity with Zt107320 on the protein level, including the orthologues in the sister species Zymoseptoria pseudotritici and Z. ardabiliae. The phylogenetic tree was constructed using MUSCLE alignments and neighbour‐joining analysis with 1000 bootstrapping iterations. Shown is the consensus tree. Support of nodes by percentage of bootstrapping iterations is indicated.
Infections of B. distachyon by Z. tritici are blocked in the substomatal cavities during incompatible interaction coinciding with reduced expression of Zt107320
To study compatible and incompatible infections of Z. tritici in more detail, we inoculated leaves of 12–14‐day‐old seedlings of T. aestivum (cultivar Obelisk) and B. distachyon (ecotype Bd21), and analysed the infection development by confocal microscopy. Fungal cells germinated upon contact with the leaf surface and developed infection hyphae. In contrast to previous observations (O'Driscoll et al., 2015), we found that Z. tritici infection hyphae entered into open B. distachyon stomata at 4 days post‐inoculation (dpi). However, further infection development of Z. tritici was blocked in the substomatal cavities of B. distachyon leaves (Fig. 2a), similar to phenotypes previously observed in incompatible interactions with einkorn wheat (Jing et al., 2008). Consequently, Z. tritici hyphae did not colonize the mesophyll tissue of B. distachyon, no necrotic lesions developed and no asexual pycnidia formed. Interestingly, fungal growth was completely halted and no further growth was observed, even when leaves were examined 5 weeks post‐inoculation (Fig. 2a). In contrast, Z. tritici successfully infected and colonized the mesophyll tissue of the susceptible wheat cultivar Obelisk during different infection stages, as already described in detail (Haueisen et al., 2019). Penetration of leaf stomata at 4 dpi was followed by the establishment of a hyphal network in the mesophyll tissue until 7–11 dpi and subsequently the switch to necrotrophy when biomass increased substantially, resulting in the formation of pycnidia (Fig. 2a).
Figure 2.
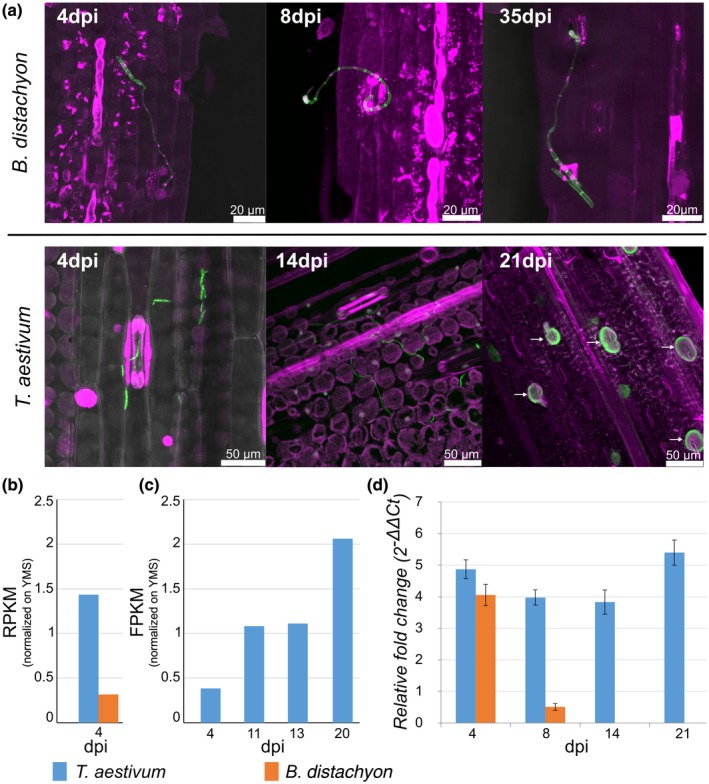
Zymoseptoria tritici infection hyphae are blocked in the substomatal cavities of Brachypodium distachyon, which coincides with reduced expression of Zt107320. (a) Micrographs showing Z. tritici cells and emerging hyphae during penetration of B. distachyon stomata and infection of Triticum aestivum. On T. aestivum, spores of Z. tritici germinated and hyphae penetrated the stomata at 4 days post‐inoculation (dpi). Intercellular mesophyll colonization of Z. tritici was observed until 14 dpi, resulting in the formation of pycnidia (indicated by arrows) at 21 dpi. In contrast, Z. tritici spores germinated on B. distachyon and hyphae penetrated stomata, but further growth was halted below the stomata with no subsequent leaf colonization (8–35 dpi). Maximum projections of confocal image z‐stacks are shown. Nuclei and grass cells displayed in purple and fungal hyphae or septae, respectively, in green. (b) Transcription of Zt107320 in compatible (T. aestivum) and incompatible (B. distachyon) hosts at 4 dpi normalized to the expression in YMS medium. Data from Kellner et al. (2014). (c) Transcription of Zt107320 in T. aestivum at indicated time points during in planta infection normalized to expression of the gene during growth in YMS medium. Data from Haueisen et al. (2019). (d) In planta expression of Zt107320 is displayed relative to expression during axenic growth. Values are normalized to the expression of the gene encoding the housekeeping protein GAPDH. Error bars indicate the standard error of the mean (SEM) of three independent biological replicates per sample.
Previously, comparative transcriptome analyses during Z. tritici infection of the host T. aestivum and the non‐host B. distachyon identified 40 differentially expressed genes at 4 dpi (Kellner et al., 2014) (Fig. 2b). During the course of the infection of a compatible host, a separate study based on transcriptome data showed that the expression of Zt107320 increases over time, with the highest expression at 20 dpi (Haueisen et al., 2019). This expression pattern is consistent with a putative role of Zt107320 in growth regulation (Fig. 2c). Building on this expression analysis, we focused on the putative transcription factor Zt107320 and validated the expression kinetics of Zt107320 by RT‐qPCR. We confirmed differential expression at 8 dpi (Fig. 2d). Expression in the non‐host B. distachyon was greatly reduced at 8 dpi compared to expression in axenic cultures, whereas the expression of Zt107320 during a compatible infection of the host T. aestivum was strongly up‐regulated during the course of the infection until 21 dpi.
Zt107320 is located with the nucleus during yeast‐like growth
The localization of Zt107320 within fungal cells was analysed using complementation strains that expressed a Zt107320‐eGFP fusion protein regulated by the native promoter. During yeast‐like growth on YMS medium, the green fluorescent fusion protein (GFP‐fusion) appeared to be located in the nucleus or in its immediate vicinity (see Fig. 3). This localization is similar to the reported nuclear localization of the homologue AbPf2 (Cho et al., 2013). The nuclear localization of the Zt107320‐eGFP fusion protein, however, was restricted to a fraction of the cells, which could indicate that the protein is also located in the cytoplasm. The observed localization of the Zt107320‐eGFP fusion protein is in concordance with the predicted nuclear localization of Zt107320 using the program WoLFPsort, which implements an algorithm for prediction of subcellular locations of proteins based on sequence composition and content (Horton et al., 2007) and therefore could support the putative functional role of Zt107320 as a transcription factor.
Figure 3.
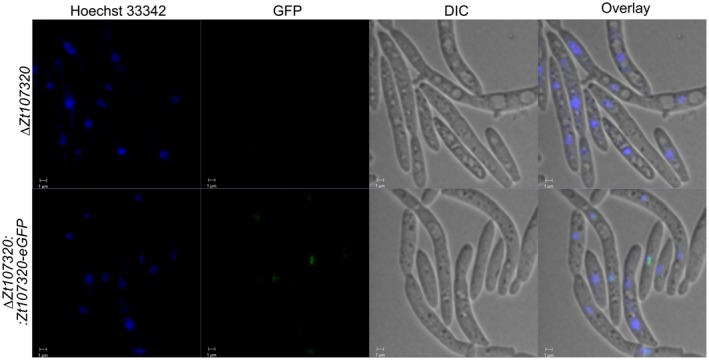
Localization of the Zt107320‐eGFP fusion protein detected by fluorescence microscopy. Nuclei were counterstained using the DNA‐specific dye Hoechst 33342. eGFP fluorescence co‐localized with the Hoechst 33342 signal indicating nuclear localization of the Zt107320‐eGFP fusion protein. eGFP fluorescence was restricted to the nuclei. DIC, differential interference contrast. Scale bars = 1 µm.
Zt107320 regulates growth and affects cell wall properties
Based on the coinciding halted growth and development and down‐regulation of Zt107320 in incompatible infections, we next asked whether Zt107320 is involved in the regulation of growth of Z. tritici in vitro. We determined the maximum growth rate r and the carrying capacity k of Z. tritici in liquid cultures using rich complex medium (YMS) and chemically defined minimal medium (MM) containing different carbohydrates. Growth was determined by OD600 measurements and data fitted assuming a logistic growth curve model. In YMS the two independently generated Zt107320 deletion mutants achieved a significantly lower maximum growth rate r compared to the wild type (median of 65% and 80% for strains 1 and 2, respectively), whereas growth rates were restored to the level of the wild type strain in the two complementation strains (median of 95% and 100%) (Fig. 4a). However, the maximum cell density, which equates to the carrying capacity k, was not different from the wild type for the two deletion strains (median of 104% and 96%) or the two complementation strains (median of 95% and 100%). Hence Zt107320 appears to affect the growth rate but not the carrying capacity in rich complex medium. In order to assess the effect of Zt107320 on the growth in conditions that promote hyphal growth and are therefore similar to the growth in planta we additionally used MM that included different carbohydrates as energy and carbon sources. Interestingly, in MM the maximal growth rate r was not significantly different from the wild type for two deletion strains (median for all MM: 102% and 107% of wild type) and the two complementation strains (median for all MM: 103% and 93% of wild type). However, the carrying capacity was lower for the two deletion strains (median for all MM: 79% and 83%) whereas the complementation strains reached the carrying capacity of the wild type (median for all MM 92% and 94%). The carrying capacity k for ∆Zt107320 was significantly reduced in MM that included fructose, glucose, sucrose and xylose, which are the carbohydrates that showed high carrying capacities (Figs 4a and S1). We thereby confirmed the relevance of Zt107320 as a putative regulator for growth that affects growth rates and carrying capacities. The effect on the carrying capacity but not growth rate in MM could indicate that Zt107320 affects the efficiency with which resources are utilized in MM and thereby affects growth during conditions when resources are limited. In contrast, in rich complex medium a higher overall growth rate can be achieved, with Zt107320 affecting the maximum growth rate but not the carrying capacity.
Figure 4.
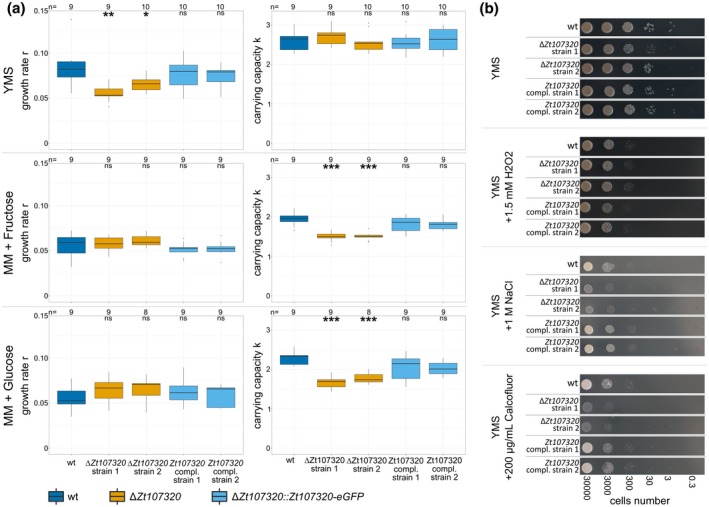
Zt107320 regulates growth and affects cell wall properties of Zymoseptoria tritici. (a) Maximum growth rate r and carrying capacity k measured during fungal growth in liquid culture, in rich complex YMS liquid medium and in liquid minimal medium MM containing fructose and glucose. Statistical significance inferred through an ANOVA and subsequent post hoc Tukey’s HSD comparing the deletion and complementation strains to the wild type is indicated as *P < 0.05, **P < 0.005, ***P < 0.0005. (b) In vitro growth of the wild type (wt), two deletion strains (∆Zt107320) and two complementation strains (∆Zt107320::Zt107320‐eGFP) on YMS medium including the supplemented compounds to assess the role of osmotic stress (1 M NaCl), reactive oxygen species (1.5 mM H2O2) and cell wall stressors (200 µg/mL Calcofluor).
The effect of Zt107320 deletion was not restricted to the growth rate but also included cell wall properties. Compared to the wild type, we observed reduced growth in vitro when challenging the deletion mutants with high osmotic stresses (0.5 M and 1 M NaCl; 1 M, 1.5 M and 2 M sorbitol) as well as with cell wall stress agents (300 µg/mL and 500 µg/mL Congo red; 200 µg/mL Calcofluor). Again, the wild type phenotype was restored in both complementation strains (Figs 4b and S2). Interestingly, temperature stress (28 °C) did not affect the wild type and the deletion mutants differently, as well as exposure to H2O2 (1.5 mM and 2 mM). This indicates a specific effect of Zt107320 on the cell wall properties but not on the ability of the fungus to counteract reactive oxygen species, which are produced by the plant during activation of immune responses (Jones and Dangl, 2006).
The dimorphic switch from yeast‐like to hyphal growth is considered to be central for pathogenicity during the early stages of infection (Kema et al. 1996b; Motteram et al., 2011; Yemelin et al., 2017). We therefore next asked whether Zt107320 is involved in regulating this morphological switch of Z. tritici. To promote hyphal growth we used solid minimal medium and supplied different carbon sources in order to compare carbon utilization and growth between the mutants and wild type. Initially, after 24 h of incubation on the solid media, no differences between the wild type, deletion strains and complementation strains were apparent (data not shown). However, after 14 days of incubation on the respective medium a prominent effect of medium composition and the deletion of Zt107320 on the growth pattern became apparent. The wild type Z. tritici strain showed markedly increased yeast‐like growth in the presence of all tested carbohydrates (Figs 5, S3 and S4). Interestingly, xylose or fructose as sole carbon source led to predominant hyphal growth in the wild type strain, which contrasts with the mainly yeast‐like growth observed in the presence of all other tested carbohydrates. The deletion of Zt107320 affected fungal growth morphology on all tested carbon sources. Compared to the wild type and the complementation strains, we observed increased hyphal growth for the ΔZt107320 strains on all carbon sources after 14 days of incubation (Figs 5, S3 and S4). Microscopic analysis showed the presence of cells with hyphal morphology and yeast‐like morphology in all conditions for the wild type strains, Zt107320 deletion and complementation strains. Yet, more hyphal growth—and in particular more branching from hyphae that resulted again in hyphal growth—was observed when Zt107320 was deleted. In contrast, less hyphal growth was observed for the wild type strain or complementation strains and branching of hyphal cells that resulted again in hyphal cells was only observed for these strains in the presence of xylose and fructose but no other carbohydrate (Figs 5, S3 and S4). For fructose and xylose a further increase in hyphal growth and branching compared to the wild type was observed in the two ∆Zt107320 deletion strains (Fig. S4). Under all conditions the wild type phenotype was restored in both complementation strains, confirming that Zt107320 plays a role in the regulation of growth in Z. tritici.
Figure 5.
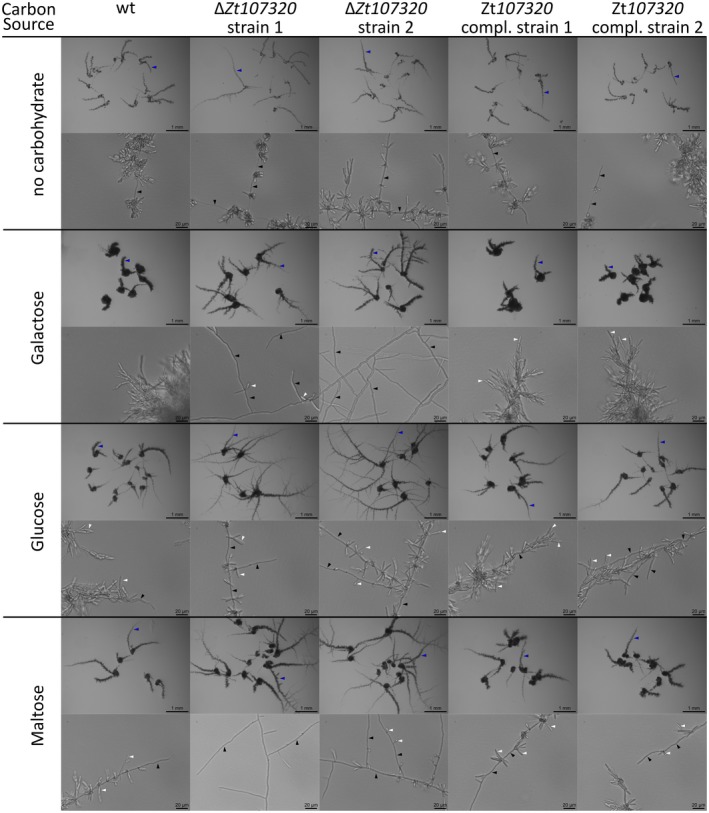
Zt107320 is involved in the regulation of the dimorphic switch of Zymoseptoria tritici. Pictures showing morphologies of colonies originating from single cells after 14 days of growth at 18 °C on minimal medium (MM) containing the indicated carbohydrates as carbon sources. Upper row for each carbon source: micrographs of colony morphologies. The two independent ∆Zt107320 strains showed an increase in number and degree of hyphal‐like protrusions (blue arrow marks one example for each condition) from colonies compared to the wild type, while in the two complementation strains the wild type colony morphology was restored. Lower row for each carbon source: micrographs showing cell morphologies in hyphal‐like protrusions from colonies. Under all conditions hyphae (examples are indicated by black arrows) and cells with yeast‐like morphologies (examples are indicated by white arrows) were present. The two ∆Zt107320 strains showed an increase in the hyphal growth compared to cells with yeast‐like morphologies. Branching of hyphae resulting in hyphal growth was increased in the two ∆Zt107320 strains whereas in the wild type (wt) and the two complementation strains branching from hyphae produced cells with yeast‐like morphologies (see also Figs S3 and S4).
Zt107320 affects the ability of Z. tritici to produce pycnidia
We next addressed whether the impact of Zt107320 deletion on growth rate, the dimorphic switch and cell wall properties also influences the ability of Z. tritici to infect its host, T. aestivum. We inoculated a predefined area of the second leaf of the susceptible wheat cultivar Obelisk and measured the number of pycnidia at 21 dpi. We observed a pronounced reduction in the production of pycnidia for the Zt107320 deletion strains. The density of pycnidia per cm2 of infected leaf area was reduced significantly for both independent deletions of Zt107320 (ANOVA, P < 1 × 10−7, P = 0.002) (Fig. 6a,b) compared to the wild type. Complementing the Zt107320 in locus fully restored the wild type phenotype in planta. The median for density of pycnidia per cm2 of leaf was reduced to 55% (range 30–65%) for the two independent deletion strains and restored to 95% (range 58–116%) for the two independent complementation strains. No significant effect was detected on the ability of the wild type, the deletion and complementation strains to induce necrosis. As the density of pycnidia is considered to be a quantitative measure for virulence (Stewart et al., 2016), significantly decreased pycnidia production indicates reduced virulence of mutants. Thus, we conclude that the putative transcription factor Zt107320, by its effect on the growth rate, the dimorphic switch and the cell wall properties, affects the fitness of Z. tritici during infection of the compatible wheat host.
Figure 6.
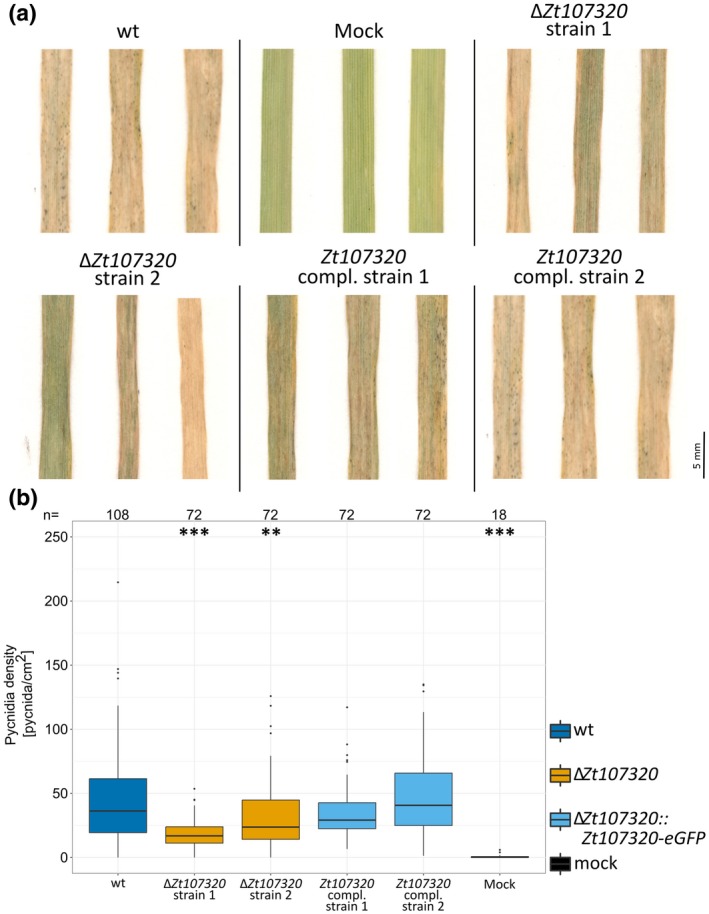
Zt107320 deletion affects pycnidia production during a compatible infection of wheat. (a) Pictures of representative leaves infected with the Zymoseptoria tritici wild type (wt), two independent deletion strains (∆Zt107320), two independent complementation strains (∆Zt107320::Zt107320‐eGFP) and mock‐treated leaves. (b) Boxplot depicting the pycnidia density (pycnidia per cm2 leaf) pooled from two independent experiments. Number of leaves (n) for each strain is indicated along the top. Statistical significance, inferred through an ANOVA and subsequent post hoc Tukey’s HSD comparing the deletion and complementation strains to the wild type, is indicated as *P < 0.05, **P < 0.005, ***P < 0.0005.
Discussion
We show that the putative transcription factor Zt107320, belonging to the fungal Zn(II)2Cys6 family, is involved in the infection programme of Z. tritici during a compatible interaction with the host plant T. aestivum. Transcription of Zt107320 is specifically induced during infection of wheat but not during the infection of the non‐host B. distachyon. This suggests a functional role of this gene in the regulation of the infection programme during a compatible host–pathogen interaction that allows the fungus to overcome host defences and propagate in the mesophyll tissue.
To date only a small number of genes have been shown to be involved in the virulence of this important wheat pathogen (Motteram et al., 2011; Orton et al., 2011; Poppe et al., 2015; Rudd, 2015; Yemelin et al., 2017). These genes encode the chitin‐binding LysM effector Mg3LysM (Lee et al., 2014; Marshall et al., 2011), two transcription factors ZtWor1 and ZtVf1 (Mirzadi Gohari et al., 2014; Mohammadi et al., 2017), three rapidly evolving small proteins (Hartmann et al., 2017; Poppe et al., 2015; Zhong et al., 2017) and members of more general processes in metabolism and cell signalling [reviewed in Rudd (2015)]. Indeed, in several studies multiple effector candidate genes have been deleted but for most mutants no or small effects on pathogenicity were observed (Gohari et al., 2015; Rudd et al., 2015). Therefore, a high level of functional redundancy seems to be present in Z. tritici. The deletion of Zt107320 results in a relatively small but significant reduction (by approximately 45%) in the pycnidia density, which contrasts to the fully or partially avirulent phenotype demonstrated for the two previously deleted transcription factors ZtWor1 and ZtVf1, respectively (Mirzadi Gohari et al., 2014; Mohammadi et al., 2017). Similar to our observation in the Zt107320 deletion mutant, deletion of MoCOD1, the rice blast homologue of Zt107320, led to a quantitative reduction in lesion size and number (Chung et al., 2013). This partial, but incomplete, quantitative reduction in virulence of ∆Zt107320 strains corresponds with the reduced, but still substantial, growth rate of these deletion mutants in vitro. Therefore, other mechanisms independent of Zt107320 are involved in the regulation of growth and development in vitro and in planta. Similarly, a partial, but incomplete, reduction of invasive growth in planta is observed in ∆MoCOD1 (Chung et al., 2013). The impact of Zt107320 on fungal growth and pathogenicity is similar to the effect caused by other members of the Zn(II)2Cys6 transcription factor family: TPC1 regulates invasive, polarized growth and virulence in the rice blast fungus M. oryzae (Galhano et al., 2017), and the transcription factor FOW2 is known to control the ability of F. oxysporum to invade roots and colonize plant tissue (Imazaki et al., 2007).
Zt107320 appears to be involved in the morphological switch from yeast‐like to hyphal growth as deletion of Zt107320 led to an increase in hyphal growth. More hyphal growth can be observed on MM and MM containing different carbon sources for the Zt107320 deletion strains compared to the wild type (Figs 5, S3 and S4). In particular the deletion mutants appear to not only grow more hyphae but also branch more from hyphae. The switch to filamentous growth is considered to be essential for plant infection (Kema et al., 1996b; Yemelin et al., 2017). For several factors that affect the dimorphic switch, an effect on the pathogenicity has been described. One of the first identified factors that regulates the dimorphic switch in Z. tritici was ZtHog1 (synonym: MgHog1), which is required for the dimorphic switch. Deleting ZtHog1 results in failure to infect wheat leaves (Mehrabi et al., 2006). Glycosylation also affects the switch to hyphal growth. Deletion of ZtAlg2, which encodes a putative mannosyltransferase in Z. tritici, prevents this switch and results in nonpathogenic strains (Motteram et al., 2011). In addition, the deletion of a putative glycosyltransferase, ZtGt2, was shown to reduce the ability to extend hyphae on solid surfaces and pathogenicity (King et al., 2017). Similarly, the deletion of MCC1, which encodes a putative c‐type cyclin, reduces the ability to produce aerial mycelia and impairs pathogenicity (Choi and Goodwin, 2011). This frequently described correlation between the dimorphic switch and pathogenicity was used as means to identify factors involved in pathogenicity in studies that screened for mutants with reduced hyphal growth. Many of these mutants were indeed also reduced in pathogenicity (Yemelin et al., 2017). Therefore, the involvement of Zt107320 in regulating this dimorphism underscores its importance for pathogenicity. Interestingly, in contrast to all earlier reports linking the dimorphic switch and pathogenicity in Z. tritici, the deletion of Zt107320 increases hyphal growth but decreases pathogenicity. This might be due to the effect of Zt107320 on the carrying capacity k in MM. The reduction of the carrying capacity in the Zt107320 deletion mutants, but not on the growth rate r, may indicate an effect of the transcription factor on carbon utilization once the carbon source becomes limited when approaching stationary phase. The differences in carrying capacity for the ∆Zt107320 strains could be indicative of an earlier nutrient limitation due to lower efficiency in nutrient usage. This nutrient limitation could therefore be responsible for the more pronounced switch to hyphal growth, in particular, since carbon limitation increases mycelial growth (Francisco et al., 2019). This is further supported by the fact that 24 h after inoculation onto solid MM no differences in growth pattern were observable between the wild type, the Zt107320 deletion and complementation strains (data not shown). At this time point nutrients could be considered to be non‐limiting. It is therefore interesting to speculate that the effect of the deletion of Zt107320 on the pathogenicity of Z. tritici could be due to the less efficient nutrient utilization during the course of the plant infection resulting in reduced pycnidia density.
Based on RT‐qPCR data we observed that Zt107320 is differentially expressed between non‐host and host infections and is further highly expressed during infections of compatible hosts. Although previous RNA‐Seq‐based transcriptome studies found Zt107320 to be expressed at different relative levels compared to liquid culture, both studies showed that it is up‐regulated during later stages of wheat infection and associated with necrotrophic host colonization, indicating a possible function for pycnidia formation (Haueisen et al., 2019; Rudd et al., 2015). Together, these findings support a role of Zt107320 in regulation of growth and pathogenicity of Z. tritici.
Interestingly, xylose, the main product of hemicellulose degradation by fungal xylanases, had a pronounced effect on the growth pattern of Z. ritici. Xylose supplementation not only increased overall growth compared to pure minimal medium but also led to an increase in hyphal growth. In Z. tritici, the switch to necrotrophic growth after an initial phase of symptomless infection, overall disease severity and quantitative pycnidiospore production are associated with the activity of endo‐β‐1,4‐xylanase (Siah et al., 2010; Somai‐Jemmali et al., 2017). During the switch to necrotrophy Z. tritici rapidly develops large hyphal networks and uses plant‐derived nutrients (Haueisen et al., 2019; Rudd et al., 2015). The observed effect of xylose on the growth morphology suggests that xylose—next to its role as a carbon source—may also direct growth and promote the spatial expansion of the intrafoliar hyphal network during the fungal lifestyle switch to necrotrophic growth. Indeed, genes encoding xylanases were shown to evolve under positive selection (Brunner et al., 2013), indicating an important functional role of this class of enzymes for adaptation to wheat infection. Although functional analyses of several xylanases in other plant pathogenic fungi resulted in no direct phenotypic effect (Brunner et al., 2013; Douaiher et al., 2007), xylanases have been proposed as virulence factors in Z. tritici (Douaiher et al., 2007). The results presented here indicate a possible role of xylose as a host infection‐associated signal molecule for Z. tritici and should warrant further analysis.
It is important to note that on resequencing of the wild type IPO323 isolate used in this study, we detected a 405 bp in‐frame deletion in the first exon of the Z. tritici homologue of white collar 1 (wc‐1, gene_ID: Zt09_chr_11_00064, Grandaubert et al. 2015) compared to the IPO323 reference genome. Such spontaneous genetic changes were previously reported to occur at an elevated frequency in Z. tritici during in vitro propagation (Möller et al., 2018). WC‐1 together with WC‐2 composes the White Collar complex (WCC) and acts as a transcription factor whose activity is regulated by light (Corrochano, 2019; He et al. 2002; Linden and Macino, 1997). In Neurospora crassa, the WCC was shown to affect the transcription of 5.6% of all expressed genes (Chen et al., 2009). It is therefore conceivable that the wc‐1 partial deletion has pleiotropic effects and may be responsible for the lower pathogenicity of the IPO323 isolate used in this study as compared to work published by other research groups. This would be in line with the observation that in M. oryzae the WC‐1 orthologue is involved in the regulation of virulence traits (Kim et al., 2011). Because WC‐1 is an important transcriptional regulator that can affect virulence, it is conceivable that the effects we observed for the putative transcription factor Zt107320 were influenced by epistatic effects between wc‐1 and Zt107320. In particular, the regulatory networks that comprise Zt107320 and WC‐1 might overlap or interfere with each other. Therefore, although all strains in this study had the wc‐1 partial deletion, the ∆Zt107320 phenotypes could have been affected by the wc‐1 partial deletion in a non‐additive manner (i.e. positive/negative epistasis). As a consequence, the phenotype of ∆Zt107320 in a strain that does not have the wc‐1 partial deletion may differ from the phenotype that we describe here. Nevertheless, with the complementation test included in our study, we can conclude that Zt107320 influences the dimorphic switch, growth and virulence of Z. tritici. Future studies should address the role WC‐1 might play in the regulation of virulence in Z. tritici and the potential interaction between Zt107320 and WC‐1.
In conclusion, we showed that Zt107320 affects the fitness of the wheat pathogen Z. tritici. Zt107320 is differentially expressed in host and non‐host environments, and is up‐regulated during the early stages of infection on compatible hosts and down‐regulated on non‐hosts. This down‐regulation corresponds to a considerably reduced growth and halted infections of Z. tritici after stomatal penetration of the non‐host B. distachyon. In addition, we can confirm that Zt107320 has a nuclear localization, consistent with its putative function as a transcription factor, and that it further regulates the dimorphic switch between yeast‐like and hyphal growth that is considered to be essential for pathogenicity. We therefore hypothesize that the putative transcription factor Zt107320 is part of the regulatory network that controls growth and development, and could be involved in integrating signals that differ between compatible and non‐compatible infections. Future studies should address the specific target genes of Zt107320 and their expression pattern during compatible and non‐compatible interactions. Furthermore, the transcriptional regulation of Zt107320 suggests that specific signals in the compatible host–pathogen interaction in wheat are responsible for the up‐regulation of this particular transcription factor‐encoding gene. Identification of these host‐derived signals will provide a fundamental insight into the molecular basis of host–pathogen interaction and host specificity in Z. tritici.
Experimental Procedures
Fungal strains and plant material
The Dutch isolate IPO323 was kindly provided by Gert Kema (Wageningen, Netherlands) and is available from the Westerdijk Institute (Utrecht, Netherlands) with the accession number CBS 115943. The isolate used in our experiments lacked accessory chromosome 18, presumably lost during culture maintenance in vitro (Kellner et al., 2014), and contains a 405 bp in‐frame deletion in gene Zt09_chr_11_00064 (chr 11:223,109‐223,513), a homologue of the white collar 1 (wc‐1) gene involved in controlling the expression of light‐regulated genes in N. crassa (Linden and Macino, 1997). Strains were maintained in either liquid yeast malt sucrose (YMS) broth (4 g/L yeast extract, 4 g/L malt extract, 4 g/L sucrose) at 18 °C on an orbital shaker or on solid YMS (+20 g/L agar) at 18 °C. The T. aestivum cultivar Obelisk was obtained from Wiersum Plantbreeding BV (Winschoten, Netherlands). Brachypodium distachyon inbred line Bd21 was kindly provided by Thierry Marcel (Bioger, INRA, France).
Sequence analysis
Phylogenetic analysis of Zt107320 was conducted using the software Geneious Prime v. 2019.0.4 (https://www.geneious.com). Protein sequences with homology to the translated protein sequence of Zt107320 were retrieved from the Ensemble Fungi database (Kersey et al., 2018). Furthermore, we included Zt107320 and its homologues from the sister species Z. pseudotritici and Z. ardabiliae, and the non‐orthologous sequences of the well‐described transcription factor AmyR from A. niger and MalR from A. oryzae, and the closely related gene MYCGRDRAFT_111096 from Z. tritici. A total of 30 matches were retrieved from the Ensemble Fungi database. Alignments were constructed using MUSCLE (Edgar, 2004) and trees constructed using the neighbour‐joining algorithm, building a consensus tree with 1000 bootstrapping replicates. Protein domains of the consensus sequence were identified using InterProScan (Quevillon et al., 2005). Prediction of the nuclear localization of the Zt107320 protein was conducted using the WoLF PSORT predictor (Horton et al., 2007).
Analysis of Z. tritici during its compatible and non‐compatible infections by confocal microscopy
Morphology and development of Z. tritici within and on the surface of leaves of B. distachyon inbred line Bd21 were analysed by confocal laser‐scanning microscopy (CLSM) as described previously (Haueisen et al., 2019). Analyses of compatible Z. tritici infections on T. aestivum 'Obelisk' were conducted by combining microtomy and CLSM as previously described (Rath et al., 2014). Distinct areas of the second leaf of 12‐day‐old (Bd21) and 14‐day‐old (wheat) seedlings were brush‐inoculated with 1 × 107 cells/mL in 0.1% Tween 20. Plants were incubated at 22 °C (day)/ 20 °C (night) and 100% humidity with a 16‐h light period for 48 h. Subsequently, humidity was reduced to 70%. Microscopy was conducted using a Leica TCS SP5 and analysis of imaging of z‐stacks was done using Leica Application Suite Advanced Fluorescence (Leica Microsystems, Germany) and AMIRA (FEITM Visualization Science Group, Germany).
Analysis of Zt107320‐eGFP expression using confocal microscopy
Cells were grown on solid YMS medium for 7 days before being scraped off the medium surface and introduced into 10 mM phosphate buffer (pH 7.2) containing 1 µg/µL Hoechst 33342 (Sigma‐Aldrich Chemie GmbH, Munich, Germany). Cells were incubated for 15–30 min in the dark and then transferred to a microscope slide and analysed by CLSM.
RNA isolation and RT‐qPCR
We analysed gene expression patterns of Zt107320 of Z. tritici using a quantitative reverse transcription PCR (RT‐qPCR) experiment. Total RNA was extracted from fungal axenic cultures (grown for 72 h in YMS medium at 18 °C and 200 rpm) and from snap‐frozen leaf tissue infected with Z. tritici (4, 8, 14 and 21 dpi) using the TRIzol reagent (Invitrogen, Karlsruhe, Germany), following the manufacturer’s instructions. Three biological replicates were included in the experimental set‐up. The cDNA samples were used in a RT‐qPCR experiment employing the iQ SYBR Green Supermix Kit (Bio‐Rad, Munich, Germany). PCR was conducted in a CFX96 RT‐PCR Detection System (Bio‐Rad) with the constitutively expressed control gene glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH). All primers are listed in Table S1.
Generation of Zt107320 deletion and complementation mutants by gene replacement
Fungal transformations and the creation of deletion and complementation mutants of Zt107320 were conducted as previously described (Poppe et al., 2015). In brief, gene deletions were generated by amplifying an approximately 1 kb region of the 5’ and 3’ flanking regions of Zt107320 using PCR. The amplified flanking sequences were fused to a hygromycin resistance (H ygR) cassette and an EcoRV cut vector backbone (pES61) using Gibson assembly (Gibson et al., 2009). Electrocompetent cells of the Agrobacterium tumefaciens strain AGL1 were transformed using standard protocols. These transformed A. tumefaciens cells were used for the transformation of Z. tritici as previously described (Zwiers et al., 2001). The same strategy was applied for the generation of the complementation strains by a C‐terminal fusion of the Zt107320 gene with an eGFP tag and a geneticin resistance cassette (NeoR) as a selection marker (Fig. S5). After transformation and homologous recombination in the ∆Zt107320 strain 1, the hygromycin resistance cassette was replaced by Zt107320‐eGFP and the geneticin resistance cassette (Fig. S5). Homologous recombination and integration were confirmed using PCR and Southern blot analysis by standard protocols. In short, genomic DNA was isolated using phenol‐chloroform isolation (Sambrook and Russell, 2001). Restriction digestion was performed using PvuII, followed by gel electrophoresis, blotting and detection using digoxygenin‐labelled probes binding to the upstream and downstream flank of Zt107320 (Fig. S5). In total four ∆Zt107320 strains and two ΔZt107320::Zt107320‐eGFP strains were generated.
In vitro phenotyping
The Z. tritici strains were grown on YMS solid medium for 5–7 days at 18 °C before the cells were scraped off the plate surface. For the determination of the growth rates, the cells were resuspended and counted. The cell density was adjusted to 50 000 cells/mL and 175 µL of the cell suspension was added to a well of a 96‐well plate. Plates were inoculated at 18 °C at 200 rpm with the OD600 being measured twice daily on a Multiskan Go plate reader (Thermo Scientific, Dreieich, Germany) (Table S2). Estimation of growth rates was done by employing the logistic growth equation as implemented in the growthcurver package v. 0.2.1 in R v. R3.4.1 (R Core Team, 2015). For the determination of the in vitro phenotypes, the cell number was adjusted to 107 cells/mL in ddH2O and serially diluted to 103 cells/mL. Three microlitres of each cell dilution was transferred onto YMS agar containing the tested compounds and incubated for 7 days at 18 or 28 °C. To test high osmotic stresses, 0.5 M NaCl, 1 M NaCl, 1 M sorbitol, 1.5 M sorbitol and 2 M sorbitol (Carl Roth GmbH, Karlsruhe, Germany) were added to the YMS solid medium. To test cell wall stresses, 300 µg/mL and 500 µg/mL Congo red and 200 µg/mL Calcofluor (Sigma‐Aldrich Chemie GmbH, Munich, Germany) were added to the YMS solid medium. Finally, to determine the effect of reactive oxygen species on mutant growth morphology, 2 mM H2O2 (Carl Roth GmbH) was added to the YMS solid medium.
To test whether Zt107320 affects the hyphal growth of Z. tritici and the ability of the fungus to use different carbon sources, we used MM as described in Barratt et al. (1965). Glucose, fructose, xylose, maltose, galactose, sorbitol, mannitol, starch and sucrose were added at a final concentration of 10 g/L and 20 g/L agar was included. Strains were grown on YMS solid medium for 7 days and resuspended into ddH2O. Cell number was adjusted and 3 µL of 5 × 103 cells/mL were added to the surface of the MM plates. Plates were incubated at 18 °C in the dark and growth was monitored using an inverse microscope and a stereomicroscope after 24 h and 14 days.
In planta phenotyping
For the in planta phenotypic assays, we germinated seeds of the wheat cultivar Obelisk on wet sterile Whatman paper for 4 days before potting using the soil Fruhstorfer Topferde (Hermann Meyer GmbH, Rellingen, Germany). Wheat seedlings were further grown for 7 days before inoculation. Zymoseptoria tritici strains were grown on YMS solid medium for 5 days at 18 °C before the cells were scraped from the plate surface. The cell number was adjusted to 108 cells/mL in H2O + 0.1% Tween 20, and the cell suspension was brushed onto approximately 5 cm on the abaxial and adaxial sides of the second leaf of each seedling. Inoculated plants were placed in sealed bags containing water for 48 h to facilitate infection through stomata. Plants were grown under constant conditions with a day/night cycle of 16 h light (c. 200 µmol/m2/s) and 8 h darkness in growth chambers at 20 °C. Plants were grown for 21 dpi at 90% relative humidity. At 21 dpi the infected leaves were cut, taped to sheets of paper and pressed for 5 days at 4 °C before being scanned at a resolution of 2400 dots per inch (dpi) using a flatbed scanner (HP Photosmart C4580, HP, Böblingen, Germany). Scanned images were analysed using an automated image analysis in Image J (Schneider et al., 2012) adapted from Stewart et al. (2016). The read‐out pycnidia/cm2 leaf surface was used for all subsequent analyses. See Table S3 for a summary of the in planta results.
Statistical analysis
Statistical analyses were conducted in R v. R3.4.1 (R Core Team, 2015) using the suite R Studio v. 1.0.143 (R Studio Team, 2015). Data inspection showed a non‐normal distribution for all datasets, including the measured pycnidia densities (pycnidia/cm2). Therefore, we performed an omnibus analysis of variance using rank‐transformation of the data (Conover and Iman, 1981) employing the model: pycnidia density ~ strain × experiment, r ~ strain and k ~ strain, respectively. Post hoc tests were performed using Tukey's HSD (Tukey, 1949).
Supporting information
Fig. S1 Zt107320 impacts maximum carrying capacity k but not the growth rate r of Zymoseptoria tritici in minimal medium containing sucrose and xylose. Maximum growth rate r and carrying capacity k of fungal cultures grown in liquid minimal medium (MM) containing the indicated carbohydrates as carbon sources. For sucrose and xylose as carbon source a significant effect of the deletion of Zt107320 on the carrying capacity was discernible, whereas the complementation strains were not statistically significantly different from the wild type (wt). All other carbon sources and the maximum growth rate were not significantly different for the two independent deletion strains. Statistical significance inferred through an ANOVA and subsequent post hoc Tukey’s HSD comparing the deletion and complementation strains to the wild type, is indicated as *P < 0.05; **P < 0.005; ***P < 0.0005.
Fig. S2 In vitro phenotype of Zt107320 wild type and mutants. In vitro growth of the wild type (wt), two independent deletion strains (∆Zt107320), two independent complementation strains (∆Zt107320::Zt107320‐eGFP) on YMS medium including the indicated compounds to assess the effect of osmotic stress (NaCl, sorbitol), reactive oxygen species (H2O2), cell wall stressors (Calcofluor, Congo Red) and increased temperature (28 °C) on growth and morphology of Zymoseptoria tritici.
Fig. S3 Growth morphologies of Zymoseptoria tritici wild type and ∆Zt107320 and ∆Zt107320::Zt107320‐eGFP strains on solid minimal medium in the presence of different carbon sources. Micrographs depicting growth morphologies after 14 days at 18 °C on media. Upper row and lower row: Two independent examples of colony morphology for each condition. Hyphal‐like protrusions originated from primary colonies. Hyphal‐like protrusions appeared more pronounced, more branched and covering a larger area for the two ∆Zt107320 strains compared to wild type (wt). The two Zt107320::Zt107320‐eGFP strains showed wild type phenotype. Scale bar represents 1 mm.
Fig. S4 Details of growth morphologies of Zymoseptoria tritici wild type and ∆Zt107320 and ∆Zt107320::Zt107320‐eGFP strains on solid minimal medium in the presence of different carbon sources. Detailed micrographs depicting growth morphologies after 14 days at 18 °C on media. Upper row and lower row: Details of one hyphal‐like protrusion depicted at differed magnification. Hyphal cells and yeast‐like cells can be seen in all conditions. In the two ∆Zt107320 strains more hyphal growth and more hyphal branching of hyphae resulting in hyphae occurred in comparison to the wild type (wt). The two ∆Zt107320::Zt107320‐eGFP complementation strains showed wild type morphologies. Scale bar represents 50 µm.
Fig. S5 Generation of Zt107320 mutants in Zymoseptoria tritici. (a) Schematic illustration of the gene replacement strategy used to delete the gene Zt107320. Generation of the ∆Zt107320 mutants by homologous recombination between the upstream (UF) and downstream flanking regions (DF) of Zt107320 in its genomic locus and a plasmid carrying the hygromycin resistance cassette (HygR) located between the UF and DF. Homologous recombination results in the integration of the hygromycin‐resistance gene cassette (HygR) in the locus of Zt107320. ∆Zt107320::Zt107300‐eGFP were generated by homologous recombination between UF and DF of the ∆Zt107320 strains and a transformed plasmid containing a C‐terminal fusion of Zt107320 and eGFP and a geneticin resistance cassette (NeoR) located between the UF and DF. Yellow bars indicate the position of the probes used in the Southern blot analyses. (b) Confirmation of Zt107320 mutants by Southern blot analyses.
Table S1 List of all primers used within this study.
Table S2 Summary of in vitro growth data.
Table S3 Summary of in planta phenotype.
Acknowledgements
The authors would like to thank all members of the Environmental Genomics group for critical comments and discussions. The study was funded by a personal grant to E.H.S. from the State of Schleswig Holstein and a fellowship from the Max Planck Society, Germany. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The authors have no conflict of interest.
References
- Barratt, R.W. , Johnson, G.B. and Ogata, W.N. (1965) Wild‐type and mutant stocks of Aspergillus nidulans . Genetics, 52, 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brading, P.A. , Verstappen, E.C.P. , Kema, G.H.J. and Brown, J.K.M. (2002) A gene‐for‐gene relationship between wheat and Mycosphaerella graminicola, the Septoria tritici blotch pathogen. Phytopathology, 92, 439–445. [DOI] [PubMed] [Google Scholar]
- Brunner, P.C. , Torriani, S.F.F. , Croll, D. , Stukenbrock, E.H. and McDonald, B.A. (2013) Coevolution and life cycle specialization of plant cell wall degrading enzymes in a hemibiotrophic pathogen. Mol. Biol. Evol. 30, 1337–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.‐H. , Ringelberg, C.S. , Gross, R.H. , Dunlap, J.C. and Loros, J.J. (2009) Genome‐wide analysis of light‐inducible responses reveals hierarchical light signalling in Neurospora . EMBO J. 28, 1029–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Le, X. , Sun, Y. , Li, M. , Zhang, H. , Tan, X. , Zhang, D. , Liu, Y. and Zhang, Z. (2017) MoYcp4 is required for growth, conidiogenesis and pathogenicity in Magnaporthe oryzae . Mol. Plant Pathol. 18, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, Y. , Ohm, R.A. , Grigoriev, I.V. and Srivastava, A. (2013) Fungal‐specific transcription factor AbPf2 activates pathogenicity in Alternaria brassicicola . Plant J. 75, 498–514. [DOI] [PubMed] [Google Scholar]
- Choi, Y.‐E. and Goodwin, S.B. (2011) Gene encoding a c‐type cyclin in Mycosphaerella graminicola is involved in aerial mycelium formation, filamentous growth, hyphal swelling, melanin biosynthesis, stress response, and pathogenicity. Mol. Plant–Microbe Interact. 24, 469–477. [DOI] [PubMed] [Google Scholar]
- Chung, H. , Choi, J. , Park, S.‐Y. , Jeon, J. and Lee, Y.‐H. (2013) Two conidiation‐related Zn(II)2Cys6 transcription factor genes in the rice blast fungus. Fungal Genet. Biol. 61, 133–141. [DOI] [PubMed] [Google Scholar]
- Conover, W.J. and Iman, R.L. (1981) Rank transformations as a bridge between parametric and nonparametric statistics. American Statistician, 35, 124–129. [Google Scholar]
- Corrochano, L.M. (2019) Light in the fungal world. From photoreception to gene transcription and beyond. Annu. Rev. Genet, 53 10.1146/annurev-genet-120417-031415. [DOI] [PubMed] [Google Scholar]
- Douaiher, M.‐N. , Nowak, E. , Durand, R. , Halama, P. and Reignault, P. (2007) Correlative analysis of Mycosphaerella graminicola pathogenicity and cell wall‐degrading enzymes produced in vitro: the importance of xylanase and polygalacturonase. Plant Pathol, 56, 79–86. [Google Scholar]
- Edgar, R.C. (2004) MUSCLE. Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fones, H. and Gurr, S. (2015) The impact of Septoria tritici blotch disease on wheat: an EU perspective. Fungal Genet. Biol. 79, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, E.M. , Gardiner, D.M. , Keller, N.P. and Howlett, B.J. (2008) A Zn(II)2Cys6 DNA binding protein regulates the sirodesmin PL biosynthetic gene cluster in Leptosphaeria maculans . Fungal Genet. Biol. 45, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco, C.S. , Ma, X. , Zwyssig, M.M. , McDonald, B.A. and Palma‐Guerrero, J. (2019) Morphological changes in response to environmental stresses in the fungal plant pathogen Zymoseptoria tritici . Sci. Rep. 9, 9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhano, R. , Illana, A. , Ryder, L.S. , Rodríguez‐Romero, J. , Demuez, M. , Badaruddin, M. , Martinez‐Rocha, A.L. , Soanes, D.M. , Studholme, D.J. , Talbot, N.J. and Sesma, A. (2017) Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 13, e1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, D.G. , Young, L. , Chuang, R.‐Y. , Venter, J.C. , Hutchison, C.A. and Smith, H.O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Gohari, A.M. , Ware, S.B. , Wittenberg, A.H.J. , Mehrabi, R. , M’Barek, S.B. , Verstappen, E.C.P. , van der Lee, T.A.J. , Robert, O. , Schouten, H.J. , de Wit, P.P.J.G.M. and Kema, G.H. (2015) Effector discovery in the fungal wheat pathogen Zymoseptoria tritici . Mol. Plant Pathol. 16, 931–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandaubert, J. , Bhattacharyya, A. and Stukenbrock, E.H. (2015) RNA‐seq based gene annotation and comparative genomics of four fungal grass pathogens in the genus Zymoseptoria identify novel orphan genes and species‐specific invasions of transposable elements. G3, 5, 1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, F.E. , Sanchez‐Vallet, A. , McDonald, B.A. and Croll, D. (2017) A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 11, 1189–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueisen, J. , Möller, M. , Eschenbrenner, C.J. , Grandaubert, J. , Seybold, H. , Adamiak, H. and Stukenbrock, E.H. (2019) Highly flexible infection programs in a specialized wheat pathogen. Ecol. Evol. 9, 275–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Q. , Cheng, P. , Yang, Y. , Wang, L. , Gardner, K.H. and Liu, Y. (2002) White collar‐1, a DNA binding transcription factor and a light sensor. Science, 297, 840–843. [DOI] [PubMed] [Google Scholar]
- Horton, P. , Park, K.‐J. , Obayashi, T. , Fujita, N. , Harada, H. , Adams‐Collier, C.J. and Nakai, K. (2007) WoLF PSORT. Protein localization predictor. Nucleic Acids Res. 35, W585–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imazaki, I. , Kurahashi, M. , Iida, Y. and Tsuge, T. (2007) Fow2, a Zn(II)2Cys6‐type transcription regulator, controls plant infection of the vascular wilt fungus Fusarium oxysporum . Mol. Microbiol. 63, 737–753. [DOI] [PubMed] [Google Scholar]
- Jing, H.‐C. , Lovell, D. , Gutteridge, R. , Jenk, D. , Kornyukhin, D. , Mitrofanova, O.P. , Kema, G.H.J. and Hammond‐Kosack, K.E. (2008) Phenotypic and genetic analysis of the Triticum monococcum–Mycosphaerella graminicola interaction. New Phytol. 179, 1121–1132. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kellner, R. , Bhattacharyya, A. , Poppe, S. , Hsu, T.Y. , Brem, R.B. and Stukenbrock, E.H. (2014) Expression profiling of the wheat pathogen Zymoseptoria tritici reveals genomic patterns of transcription and host‐specific regulatory programs. Genome Biol. Evol. 6, 1353–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kema, G.H. , Verstappen, E.C. , Todorova, M. and Waalwijk, C. (1996a) Successful crosses and molecular tetrad and progeny analyses demonstrate heterothallism in Mycosphaerella graminicola . Curr. Genet. 30, 251–258. [DOI] [PubMed] [Google Scholar]
- Kema, G.H.J. , Yu, D.Z. , Rijkenberg, F.H.J. , Shaw, M.W. and Baayen, R.P. (1996b) Histology of the pathogenesis of Mycosphaerella graminicola in wheat. Phytopathology, 86, 777–786. [Google Scholar]
- Kersey, P.J. , Allen, J.E. , Allot, A. , Barba, M. , Boddu, S. , Bolt, B.J. , Carvalho‐Silva, D. , Christensen, M. , Davis, P. , Grabmueller, C. , Kumar, N. , Liu, Z. , Maurel, T. , Moore, B. , McDowall, M.D. , Maheswari, U. , Naamati, G. , Newman, V. , Ong, C.K. , Paulini, M. , Pedro, H. , Perry, E. , Russell, M. , Sparrow, H. , Tapanari, E. , Taylor, K. , Vullo, A. , Williams, G. , Zadissia, A. , Olson, A. , Stein, J. , Wei, S. , Tello‐Ruiz, M. , Ware, D. , Luciani, A. , Potter, S. , Finn, R.D. , Urban, M. , Hammond‐Kosack, K.E. , Bolser, D.M. , de Silva, N. , Howe, K.L. , Langridge, N. , Maslen, G. , Staines, D.M. and Yates, A. (2018) Ensembl genomes 2018. An integrated omics infrastructure for non‐vertebrate species. Nucleic Acids Res. 46, D802–D808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S. , Singh, P. , Park, J. , Park, S. , Friedman, A. , Zheng, T. , Lee, Y.‐H. and Lee, K. (2011) Genetic and molecular characterization of a blue light photoreceptor MGWC‐1 in Magnaporth oryzae . Fungal Genet. Biol. 48, 400–407. [DOI] [PubMed] [Google Scholar]
- King, R. , Urban, M. , Lauder, R.P. , Hawkins, N. , Evans, M. , Plummer, A. , Halsey, K. , Lovegrove, A. , Hammond‐Kosack, K. and Rudd, J.J. (2017) A conserved fungal glycosyltransferase facilitates pathogenesis of plants by enabling hyphal growth on solid surfaces. PLoS Pathog. 13, e1006672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.‐S. , Rudd, J.J. , Hammond‐Kosack, K.E. and Kanyuka, K. (2014) Mycosphaerella graminicola LysM effector‐mediated stealth pathogenesis subverts recognition through both CERK1 and CEBiP homologues in wheat. Mol. Plant–Microbe Interact. 27, 236–243. [DOI] [PubMed] [Google Scholar]
- Linden, H. and Macino, G. (1997) White collar 2, a partner in blue‐light signal transduction, controlling expression of light‐regulated genes in Neurospora crassa . EMBO J. 16, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Cao, H. , Zhang, L. , Huang, P. and Lin, F. (2014) Systematic analysis of Zn2Cys6 transcription factors required for development and pathogenicity by high‐throughput gene knockout in the rice blast fungus. PLoS Pathog. 10, e1004432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacPherson, S. , Larochelle, M. and Turcotte, B. (2006) A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol. Mol. Biol. Rev. 70, 583–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, R. , Kombrink, A. , Motteram, J. , Loza‐Reyes, E. , Lucas, J. , Hammond‐Kosack, K.E. , Thomma, B.P.H.J. and Rudd, J.J. (2011) Analysis of two in planta expressed LysM effector homologs from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol. 156, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabi, R. , Zwiers, L.‐H. , de Waard, M.A. and Kema, G.H.J. (2006) MgHog1 regulates dimorphism and pathogenicity in the fungal wheat pathogen Mycosphaerella graminicola . Mol. Plant–Microbe Interact. 19, 1262–1269. [DOI] [PubMed] [Google Scholar]
- Mirzadi Gohari, A. , Mehrabi, R. , Robert, O. , Ince, I.A. , Boeren, S. , Schuster, M. , de Steinberg, G. , Wit, P.J.G.M. and Kema, G.H.J. (2014) Molecular characterization and functional analyses of ZtWor1, a transcriptional regulator of the fungal wheat pathogen Zymoseptoria tritici . Mol. Plant Pathol. 15, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi, N. , Mehrabi, R. , Gohari, A.M. , Mohammadi, G.E. , Safaie, N. and Kema, G.H.J. (2017) The ZtVf1 transcription factor regulates development and virulence in the foliar wheat pathogen Zymoseptoria tritici . Fungal Genet. Biol. 109, 26–35. [DOI] [PubMed] [Google Scholar]
- Möller, M. , Habig, M. , Freitag, M. and Stukenbrock, E.H. (2018) Extraordinary genome instability and widespread chromosome rearrangements during vegetative growth. Genetics, 210, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais, D. , Gélisse, S. , Laval, V. , Sache, I. and Suffert, F. (2016) Inferring the origin of primary inoculum of Zymoseptoria tritici from differential adaptation of resident and immigrant populations to wheat cultivars. Eur. J. Plant Pathol. 145, 393–404. [Google Scholar]
- Motteram, J. , Lovegrove, A. , Pirie, E. , Marsh, J. , van de Devonshire, J. , Meene, A. , Hammond‐Kosack, K. and Rudd, J.J. (2011) Aberrant protein N‐glycosylation impacts upon infection‐related growth transitions of the haploid plant‐pathogenic fungus Mycosphaerella graminicola . Mol. Microbiol. 81, 415–433. [DOI] [PubMed] [Google Scholar]
- O’Driscoll, A. , Doohan, F. and Mullins, E. (2015) Exploring the utility of Brachypodium distachyon as a model pathosystem for the wheat pathogen Zymoseptoria tritici . BMC Res. Notes, 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okmen, B. , Collemare, J. , van der Griffiths, S. , Burgt, A. , Cox, R. and de Wit, P.J.G.M. (2014) Functional analysis of the conserved transcriptional regulator CfWor1 in Cladosporium fulvum reveals diverse roles in the virulence of plant pathogenic fungi. Mol. Microbiol. 92, 10–27. [DOI] [PubMed] [Google Scholar]
- Orton, E.S. , Deller, S. and Brown, J.K.M. (2011) Mycosphaerella graminicola: from genomics to disease control. Mol. Plant Pathol. 12, 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, T. and Coleman, J.E. (1990) GAL4 transcription factor is not a” zinc finger” but forms a Zn (II) 2Cys6 binuclear cluster. Proc. Natl. Acad. Sci. USA. 87, 2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarenko, A. , Goodwin, S.B. , and Kema, G.H.J. (2011) Septoria tritici blotch (STB) of wheat. Plant Health Instructor. 10.1094/PHI-I-2011-0407-01. [DOI] [Google Scholar]
- Poppe, S. , Dorsheimer, L. , Happel, P. and Stukenbrock, E.H. (2015) Rapidly evolving genes are key players in host specialization and virulence of the fungal wheat pathogen Zymoseptoria tritici (Mycosphaerella graminicola). PLoS Pathog. 11, e1005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon, E. , Silventoinen, V. , Pillai, S. , Harte, N. , Mulder, N. , Apweiler, R. and Lopez, R. (2005) InterProScan. Protein domains identifier. Nucleic Acids Res. 33, W116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2015) R: A Language and Environment for Statistical Computing. Vienna, Austria: https://www.R-project.org. Accessed 5 November 2019. [Google Scholar]
- Rath, M. , Grolig, F. , Haueisen, J. and Imhof, S. (2014) Combining microtomy and confocal laser scanning microscopy for structural analyses of plant‐fungus associations. Mycorrhiza, 24, 293–300. [DOI] [PubMed] [Google Scholar]
- R Studio Team (2015) R Studio: Integrated Development Environment for R. https://support.rstudio.com/hc/en-us/articles/206212048-Citing-RStudio. Accessed 5 November 2019. [Google Scholar]
- Rudd, J.J. (2015) Previous bottlenecks and future solutions to dissecting the Zymoseptoria tritici–wheat host–pathogen interaction. Fungal Genet. Biol. 79, 24–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd, J.J. , Kanyuka, K. , Hassani‐Pak, K. , Derbyshire, M. , Andongabo, A. , Devonshire, J. , Lysenko, A. , Saqi, M. , Desai, N.M. , Powers, S.J. , Hooper, J. , Ambroso, L. , Bharti, A. , Farmer, A. , Hammond‐Kosack, K.E. , Dietrich, R.A. and Courbot, M. (2015) Transcriptome and metabolite profiling of the infection cycle of Zymoseptoria tritici on wheat reveals a biphasic interaction with plant immunity involving differential pathogen chromosomal contributions and a variation on the hemibiotrophic lifestyle definition. Plant Physiol. 167, 1158–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak, K. , See, P.T. , Phan, H.T.T. , Syme, R.A. , Moffat, C.S. , Oliver, R.P. and Tan, K.‐C. (2017) A functionally conserved Zn2 Cys6 binuclear cluster transcription factor class regulates necrotrophic effector gene expression and host‐specific virulence of two major Pleosporales fungal pathogens of wheat. Mol. Plant Pathol. 18, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. (2001) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schneider, C.A. , Rasband, W.S. and Eliceiri, K.W. (2012) NIH image to imageJ. 25 years of image analysis. Nat. Meth. 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siah, A. , Deweer, C. , Duyme, F. , Sanssené, J. , Durand, R. , Halama, P. and Reignault, P. (2010) Correlation of in planta endo‐β‐1,4‐xylanase activity with the necrotrophic phase of the hemibiotrophic fungus Mycosphaerella graminicola . Plant Pathol. 59, 661–670. [Google Scholar]
- Somai‐Jemmali, L. , Randoux, B. , Siah, A. , Magnin‐Robert, M. , Halama, P. , Reignault, P. and Hamada, W. (2017) Similar infection process and induced defense patterns during compatible interactions between Zymoseptoria tritici and both bread and durum wheat species. Eur. J. Plant Pathol. 147, 787–801. [Google Scholar]
- Son, H. , Seo, Y.‐S. , Min, K. , Park, A.R. , Lee, J. , Jin, J.‐M. , Lin, Y. , Cao, P. , Hong, S.‐Y. , Kim, E.‐K. , Lee, S.‐H. , Cho, A. , Lee, S. , Kim, M.‐G. , Kim, Y. , Kim, J.‐E. , Kim, J.‐C. , Choi, G.J. , Yun, S.‐H. , Lim, J.Y. , Kim, M. , Lee, Y.‐H. , Choi, Y.‐D. and Lee, Y.‐W. (2011) A phenome‐based functional analysis of transcription factors in the cereal head blight fungus, Fusarium graminearum . PLoS Pathog. 7, e1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, E.L. , Hagerty, C.H. , Mikaberidze, A. , Mundt, C.C. , Zhong, Z. and McDonald, B.A. (2016) An improved method for measuring quantitative resistance to the wheat pathogen Zymoseptoria tritici using high‐throughput automated image analysis. Phytopathology, 106, 782–788. [DOI] [PubMed] [Google Scholar]
- Suffert, F. , Sache, I. and Lannou, C. (2011) Early stages of septoria tritici blotch epidemics of winter wheat. Build‐up, overseasoning, and release of primary inoculum. Plant Pathol. 60, 166–177. [Google Scholar]
- Suzuki, K. , Tanaka, M. , Konno, Y. , Ichikawa, T. , Ichinose, S. , Hasegawa‐Shiro, S. , Shintani, T. and Gomi, K. (2015) Distinct mechanism of activation of two transcription factors, AmyR and MalR, involved in amylolytic enzyme production in Aspergillus oryzae . Appl. Microbiol. Biotechnol. 99, 1805–1815. [DOI] [PubMed] [Google Scholar]
- Tukey, J.W. (1949) Comparing individual means in the analysis of variance. Biometrics, 5, 99. [PubMed] [Google Scholar]
- vanKuyk, P.A. , Benen, J.A.E. , Wösten, H.A.B. , Visser, J. and de Vries, R.P. (2012) A broader role for AmyR in Aspergillus niger. Regulation of the utilisation of D‐glucose or D‐galactose containing oligo‐ and polysaccharides. Appl. Microbiol. Biotechnol. 93, 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, D. , Wang, Y. , Tang, C. , Fang, Y. , Zou, J. and Tian, C. (2015) VdCrz1 is involved in microsclerotia formation and required for full virulence in Verticillium dahliae . Fungal Genet. Biol. 82, 201–212. [DOI] [PubMed] [Google Scholar]
- Yemelin, A. , Brauchler, A. , Jacob, S. , Laufer, J. , Heck, L. , Foster, A.J. , Antelo, L. , Andresen, K. and Thines, E. (2017) Identification of factors involved in dimorphism and pathogenicity of Zymoseptoria tritici . PLoS ONE, 12, e0183065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Wang, S. , Zhang, X.X. , Ji, W. , Song, F. , Zhao, Y. and Li, J. (2016) The amyR‐deletion strain of Aspergillus niger CICC2462 is a suitable host strain to express secreted protein with a low background. Microb. Cell Fact. 15, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W.‐Q. , Gui, Y.‐J. , Short, D.P.G. , Li, T.‐G. , Zhang, D.‐D. , Zhou, L. , Liu, C. , Bao, Y.‐M. , Subbarao, K.V. , Chen, J.‐Y. and Dai, X.‐F. (2018) Verticillium dahliae transcription factor VdFTF1 regulates the expression of multiple secreted virulence factors and is required for full virulence in cotton. Mol. Plant Pathol. 19, 841–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, Z. , Marcel, T.C. , Hartmann, F.E. , Ma, X. , Plissonneau, C. , Zala, M. , Ducasse, A. , Confais, J. , Compain, J. , Lapalu, N. , Amselem, J. , McDonald, B.A. , Croll, D. and Palma‐Guerrero, J. (2017) A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 214, 619–631. [DOI] [PubMed] [Google Scholar]
- Zhuang, Z. , Lohmar, J.M. , Satterlee, T. , Cary, J.W. and Calvo, A.M. (2016) The master transcription factor mtfA governs aflatoxin production, morphological development and pathogenicity in the fungus Aspergillus flavus . Toxins, 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwiers, L.‐H. , Waard, De and Maarten, A. (2001) Efficient Agrobacterium tumefaciens‐mediated gene disruption in the phytopathogen Mycosphaerella graminicola . Curr. Genet. 39, 388–393. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Zt107320 impacts maximum carrying capacity k but not the growth rate r of Zymoseptoria tritici in minimal medium containing sucrose and xylose. Maximum growth rate r and carrying capacity k of fungal cultures grown in liquid minimal medium (MM) containing the indicated carbohydrates as carbon sources. For sucrose and xylose as carbon source a significant effect of the deletion of Zt107320 on the carrying capacity was discernible, whereas the complementation strains were not statistically significantly different from the wild type (wt). All other carbon sources and the maximum growth rate were not significantly different for the two independent deletion strains. Statistical significance inferred through an ANOVA and subsequent post hoc Tukey’s HSD comparing the deletion and complementation strains to the wild type, is indicated as *P < 0.05; **P < 0.005; ***P < 0.0005.
Fig. S2 In vitro phenotype of Zt107320 wild type and mutants. In vitro growth of the wild type (wt), two independent deletion strains (∆Zt107320), two independent complementation strains (∆Zt107320::Zt107320‐eGFP) on YMS medium including the indicated compounds to assess the effect of osmotic stress (NaCl, sorbitol), reactive oxygen species (H2O2), cell wall stressors (Calcofluor, Congo Red) and increased temperature (28 °C) on growth and morphology of Zymoseptoria tritici.
Fig. S3 Growth morphologies of Zymoseptoria tritici wild type and ∆Zt107320 and ∆Zt107320::Zt107320‐eGFP strains on solid minimal medium in the presence of different carbon sources. Micrographs depicting growth morphologies after 14 days at 18 °C on media. Upper row and lower row: Two independent examples of colony morphology for each condition. Hyphal‐like protrusions originated from primary colonies. Hyphal‐like protrusions appeared more pronounced, more branched and covering a larger area for the two ∆Zt107320 strains compared to wild type (wt). The two Zt107320::Zt107320‐eGFP strains showed wild type phenotype. Scale bar represents 1 mm.
Fig. S4 Details of growth morphologies of Zymoseptoria tritici wild type and ∆Zt107320 and ∆Zt107320::Zt107320‐eGFP strains on solid minimal medium in the presence of different carbon sources. Detailed micrographs depicting growth morphologies after 14 days at 18 °C on media. Upper row and lower row: Details of one hyphal‐like protrusion depicted at differed magnification. Hyphal cells and yeast‐like cells can be seen in all conditions. In the two ∆Zt107320 strains more hyphal growth and more hyphal branching of hyphae resulting in hyphae occurred in comparison to the wild type (wt). The two ∆Zt107320::Zt107320‐eGFP complementation strains showed wild type morphologies. Scale bar represents 50 µm.
Fig. S5 Generation of Zt107320 mutants in Zymoseptoria tritici. (a) Schematic illustration of the gene replacement strategy used to delete the gene Zt107320. Generation of the ∆Zt107320 mutants by homologous recombination between the upstream (UF) and downstream flanking regions (DF) of Zt107320 in its genomic locus and a plasmid carrying the hygromycin resistance cassette (HygR) located between the UF and DF. Homologous recombination results in the integration of the hygromycin‐resistance gene cassette (HygR) in the locus of Zt107320. ∆Zt107320::Zt107300‐eGFP were generated by homologous recombination between UF and DF of the ∆Zt107320 strains and a transformed plasmid containing a C‐terminal fusion of Zt107320 and eGFP and a geneticin resistance cassette (NeoR) located between the UF and DF. Yellow bars indicate the position of the probes used in the Southern blot analyses. (b) Confirmation of Zt107320 mutants by Southern blot analyses.
Table S1 List of all primers used within this study.
Table S2 Summary of in vitro growth data.
Table S3 Summary of in planta phenotype.


