Abstract
Introduction
CXCL14 was a significantly under-expressed mRNA in hepatocellular carcinoma tissues according to our microarray analysis, as well as head and neck squamous cell carcinoma and cervical squamous cell carcinoma. CXCL14 was considered a tumor suppressor in some studies; however, its role in HBV infection has not been identified.
Methods
CXCL14 mRNA expression was quantified from 20 male HCC patients, and the fold change in cancer tissues was calculated by comparisons with normal adjacent tissues. Overall, 212 patients with chronic HBV infection and 180 HBV-free controls were recruited to investigate the association between CXCL14 polymorphisms and HBV progression as well as liver function parameters. Serum CXCL14 levels were determined by enzyme-linked immunosorbent assay (ELISA), and comparisons were made between different HBV status and different CXCL14 genotypes.
Results
The mRNA expression of CXCL14 was 0.33-fold in HCC tissues when compared with adjacent tissues. The frequencies of rs2237062 and rs2547, but not rs2237061, were significantly different between patients with mild hepatitis and moderate-to-severe hepatitis. Moreover, rs2237062 and rs2547 polymorphisms correlated with impaired liver function parameters. ELISA results suggested that HBV-free controls had the highest level of CXCL14, while mild hepatitis patients had low levels, and patients with moderate-to-severe hepatitis had the lowest level. GA+AA genotypes of rs2547 were associated with reduced levels of serum CXCL14 because it introduced a stop codon at residue 109.
Conclusion
CXCL14 was significantly suppressed in HBV-related HCC tissues, and its polymorphisms were linked with advanced stage chronic HBV infection and impaired liver function.
Keywords: CXCL14, HBV infection, disease progression, polymorphism
Introduction
According to the latest statistics published by Bray et al,1 liver cancer qualifies as the sixth most common cancer globally, while its mortality ranked fourth for all types of cancers, and it is estimated that about 841,000 newly diagnosed cancer cases and 782,000 deaths occur annually worldwide. Its incidence also varies markedly in different regions; in particular, liver cancer has a high incidence in Eastern Asia, South-Eastern Asia, and Northern Africa. Hepatocellular carcinoma (HCC) is the most common type of liver cancer, comprising 70%–85% of all diagnosed liver cancer cases, and intrahepatic cholangiocarcinoma along with other rare types can be found in 10%–15% of total cases.2 It has been generally acknowledged that hepatitis B virus (HBV) and hepatitis C virus infection are the major risk factors for developing HCC, and a systematic review revealed that a high proportion of HBV infection is found in HCC cases originating from Eastern Asia, especially China, where approximately 76% of HCC cases were HBV positive.3 The Chinese Center for Disease Control and Prevention conducted a national-wide serological survey in 2014 involving 31,713 study participants aged from 1 to 29 years, and revealed a significant reduction of HBV surface antigen prevalence in samples compared with a prevalence of 10% in 1992.4 Such a significant reduction of HBV surface antigen prevalence can be attributed to the implementation of the Expanded Program on Immunization, offering HBV vaccination to every child after birth. Despite the rapid progress made in reducing HBV prevalence, especially in the young generation, its prevalence remains high in people aged above 50 years old because of the lack of a therapy that eradicates chronic HBV infection.5 China has the largest population in the world; therefore, large amounts of people with chronic HBV infection are at risk of developing HCC. However, among the large amount of chronic HBV patients, only a few eventually progress to HCC, indicating that other factors, including genetic factors, are involved in the development of HCC.
In our previous study, we used microarray analysis to investigate the genome-wide long non-coding RNA and mRNA expression profile of HCC tissues and adjacent normal tissues collected from male HBV-related HCC cases.6 The results indicated that chemokine ligand 14 (CXCL14) was the most under-expressed mRNA in HCC tissues with a fold change of 58.03, when compared with adjacent normal tissues. A previous study showed that CXCL14 expression was downregulated in HCC tissues, and an in vitro assay showed it induced tumor cell apoptosis.7 As a member of the CXC chemokine family, it was first identified in breast and kidney cells and was shown to be constitutively and widely expressed in normal tissues.8 An in situ mRNA hybridization study suggested that the expression of CXCL14 mRNA was absent in head and neck squamous cell carcinoma and cervical squamous cell carcinoma.9 However, publications have emerged that offer contradictory findings regarding CXCL14 expression in cancer. For example, its high expression in stroma was correlated with a shorter survival of breast cancer.10 To date, there has been little published data on CXCL14 expression in HCC. A genome-wide association study conducted in the Japanese population suggested that an intron polymorphism rs2237062 was associated with the elevated risk of HCC in patients with chronic HCV infection.11 We conducted the present study to confirm the expression of CXCL14 in HCC and to investigate its role in the progression of chronic HBV infection, which is an intermediate stage in the development of HCC.
Materials and Methods
HBV-Related HCC Subjects
During the study period of November 2015 to December 2017, 20 male HCC patients with chronic HBV infection were included from Zhongshan Hospital, Xiamen University. The inclusion criteria were as follows: (1) pathologically diagnosed with primary HCC (ICD10: C22.9); (2) male; (3) chronic HBV infection confirmed by ELISA prior to the onset of HCC; and (4) permanent residents who lived in Xiamen over 10 years and aging from 20 to 79 years. Patients were excluded if any of the following conditions were met: (1) presence of cancers other than HCC; (2) autoimmune hepatitis or toxic hepatitis; and (3) refusal to participate.
Sample Preparation
Primary HCC tissues and normal tissues 5 cm distant from the tumor edge were collected from each included patient during liver resection and placed in a liquid nitrogen pre-freezing RNase-free vial for 5 mins, then stored at −78°C prior to RNA extraction. Tissue samples were subjected to RNA extraction using Trizol reagent (Invitrogen, MA, USA). The purity and concentration of RNA were determined from OD260/280 readings using a NanoDrop ND-1000 (Invitrogen), and the integrity was evaluated using standard denaturing agarose gel electrophoresis. RNA extracts with a volume exceeding 8 μg underwent further analysis.
Quantitative Real-Time PCR Reaction
To quantify the expression of CXCL14, quantitative real-time PCR (qPCR) was performed using a CFX96 real-time PCR detection system (Bio-Rad, CA, USA). The following primer sequences were used for real-time RT-PCR analysis: (CXCL14): F-5′-CTGCGAGGAGAAGATGGTTA-′3, R-5′-CTTTGCACAAGTCTCCCAAC-′3.
PCR was performed in a final volume of 20 μL consisting of 10 μL of 2 × SYBR qPCR Mix, 1 μL of each primer, 2 μL cDNA, and 4 μL ddH2O. The real-time PCR conditions were as follows: 1 cycle of 1 min at 94°C, followed by 40 cycles of 30 s at 94°C for denaturation, 15 s at 57°C for annealing, and 15 s at 72°C for extension, followed by 10 mins for the final extension. All samples were run in triplicate. qPCR Human Reference cDNA (Takara Bio, Dalian, China) was used as an endogenous reference gene. CXCL14 mRNA expression was normalized to the expression of human reference cDNA. The 2−ΔΔCT method was used to evaluate the relative change of CXCL14 expression in HCC tissues when compared with normal adjacent tissues.
Patients with Chronic HBV Infection and HBV-Free Controls
Overall, 212 patients with chronic HBV infection were recruited from Zhongshan Hospital, Xiamen University between November 2015 to December 2017. Chronic HBV infection was confirmed by a commercial ELISA kit and results were quantified by real-time fluorescence quantitative PCR. To rule out confounding effects caused by previous anti-viral treatment so that we could study CXCL14 polymorphisms associated with chronic HBV patients in their natural state, only treatment-naïve patients were recruited in this study. Patients were included if they were aged between 20 and 79 years, had HBV DNA levels ≥107 copies/mL, and had no prior history of anti-viral treatment. The exclusion criteria were as follows: co-infection with HCV/human immunodeficiency virus (HIV), prior receipt of anti-viral treatment, and refusal to participate. Eligible patients were further assigned into two groups based on their hepatitis activity scores obtained using the METAVIR score. Subjects with activity scores of A0-A1 were categorized as having mild hepatitis, while subjects with activity scores of A2-A3 were categorized as having moderate-to-severe chronic hepatitis. Another 180 controls in this study who did not HBV, HCV, or HIV infections were recruited by medical professionals at the same study site. The present study was officially approved by the Ethic Committee of Zhongshan Hospital Xiamen University through a secret ballot and was in agreement with the Helsinki declaration. All study subjects signed a written informed consent form before enrollment.
Data Collection
On the date of entry, each subject was interviewed face-to-face by an extensively trained investigator, and the following information was collected including gender, age, weight, height, marital status, smoking status, alcohol consumption, and family history of hepatitis. Information on HBV infection (date of HBsAg detection, HBsAg, anti-HBs, HBeAg, anti-HBe, and anti-HBc status, HBV genotype, HBV titer), and the results of liver function parameters were recorded in a database by reviewing medical records with the permission of the subjects. The precise definitions of smoking and alcohol consumption were as follows: individuals who smoked at least one cigarette per day for over one year were defined as smokers, and those who consumed one or more alcoholic drink per week for over 6 months were categorized as alcohol drinkers. Fibrosis stage, hepatitis activity, and inflammation grade were established using the METAVIR system by liver biopsy in all included subjects with chronic HBV infection.
Genotyping Assay for CXCL14 Polymorphisms
We collected a venous blood sample from each enrolled subject. Blood samples were centrifuged at 3600 ×g for 10 mins to separate the serum and blood cells and then stored at −78°C prior to DNA extraction. Human genome DNA was extracted from blood cells using the Magna Pure LC 2.0 system (Roche Applied Science, Mannheim, Germany). CXCL14 polymorphisms including rs2237061, rs2237062, and rs2547 were genotyped by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/MS). All procedures were performed according to the manufacturer’s instructions (Sequenom, San Diego, CA, USA). A negative water control and reference DNA were used as quality control measures during the genotyping assay. Approximately 5% of the samples were randomly selected and genotyping was repeated as duplicated controls. The genotyping call rate was 100% according to the results from the platform.
Determination of Serum CXCL14 Levels
Serum CXCL14 levels were measured using a Human CXCL14/BRAK DuoSet ELISA kit (R&D Systems, MN, USA), and the lower detection limit for this kit was 62.5 pg/mL. All procedures were conducted in accordance with the manufacturer’s instructions. After the experiment, the results were calculated using a microplate reader set to 450 nm with a standard curve.
Statistical Analysis
The relative change of CXCL14 in tissues was compared by Student’s t-test directly on the –ΔΔCT value. The association between CXCL14 polymorphisms and chronic HBV progression was evaluated by logistic regression. Comparisons of serum CXCL14 levels between three groups were compared by one-way ANOVA. All statistical analyses were conducted with SPSS software version 25 (SPSS, Inc., IL, USA).
Results
CXCL14 mRNA Expression Levels Determined by qRT-PCR
We included 20 HBV-related male HCC cases, and HCC tissues and adjacent tissues were collected during hepatectomy. The median age among HCC cases was 46.00 years, and the mean tumor size and standard deviation were 6.36 cm and 1.91 cm, respectively. After RNA extraction from tissues, we used agarose gel electrophoresis to inspect the sample RNA’s integrity. Subsequently, the sample RNA was subjected to qRT-PCR as described above. CXCL14 in HCC tissues was significantly reduced at a ratio of 0.33 compared with adjacent tissues.
Demographic Characteristics of Patients with Chronic HBV Infection and HBV-Free Controls
As shown in Table 1, the present study included 212 patients (including 151 mild hepatitis patients and 61 moderate-to-severe hepatitis patients) with chronic HBV infection and 180 HBV-free controls. The mean age of patients with mild hepatitis was 30.70±7.09 years, 31.25±7.78 years in the moderate-to-severe hepatitis patients, and 29.50±6.52 years in the HBV-free controls, and the proportion of males was 66.2%, 67.2% and 66.7%, respectively. The statistical analysis found no significant difference in BMI, marital status, smoking status, alcohol consumption and family history of hepatitis B between the three groups, indicating they had comparable demographic characteristics. Regarding the HBV genotype, 81.1% of patients with chronic HBV infection were genotype B, and the rest were genotype C (18.9%), consistent with the genotype distribution data of Southern China.
Table 1.
Demographic Characteristics of Patients with Chronic HBV Infection and HBV-Free Controls
| Variables | Mild Hepatitis (n=151) Group 1 | Moderate to Severe Hepatitis(n=61) Group 2 | HBV-Free Controls (n=180) Group 3 | P value |
|---|---|---|---|---|
| Gender | ||||
| Male | 100 (66.2) | 41 (67.2) | 120 (66.7) | |
| Female | 51 (33.8) | 20 (32.8) | 60 (33.3) | 0.990 |
| Age | 30.70±7.09 | 31.25±7.87 | 29.50±6.52 | 0.239 |
| BMI | 21.39±2.94 | 21.45±3.41 | 21.10±3.30 | 0.160 |
| Marital Status | ||||
| Single | 40 (26.5) | 19 (31.1) | 62 (34.4) | |
| Married | 108 (71.5) | 42 (68.9) | 117 (65.0) | |
| Divorced | 3 (2.0) | 0 (0.0) | 1 (0.6) | 0.332 |
| Smoking Status | ||||
| Smokers | 17 (11.3) | 13 (21.3) | 28 (15.6) | |
| Never-smokers | 134 (88.7) | 48 (78.7) | 152 (84.4) | 0.162 |
| Alcohol Consumption | ||||
| No | 20 (13.2) | 14 (23.0) | 39 (21.7) | |
| Yes | 131 (86.8) | 47 (77.0) | 141 (78.3) | 0.094 |
| Family History of Hepatitis B | ||||
| No | 102 (67.5) | 40 (65.6) | 125 (69.4) | |
| Yes | 49 (32.5) | 21 (34.4) | 55 (30.6) | 0.839 |
| HBV Genotype | ||||
| Genotype B | 123 (81.5) | 49 (80.3) | – | |
| Genotype C | 28 (18.5) | 12 (19.7) | – | 0.280 |
CXCL14 Polymorphisms and Disease Progression of Chronic HBV Infection
In accordance with the METAVIR score system, we categorized 212 patients with chronic HBV infection into mild hepatitis (Group 1, n=151) and moderate-to-severe hepatitis (Group 2, n=61). Based on the results of MALDI-TOF/MS, we obtained the genotypes of rs2237061, rs2237062, and rs2547 for all study participants. The genotype distribution in each CXCL14 polymorphism was calculated and compared between the three groups to determine whether they were differentially distributed in patients with mild hepatitis, moderate-to-severe hepatitis patients and HBV-free controls. In addition, the association between the above-mentioned CXCL14 polymorphisms and chronic HBV progression was investigated by univariate logistic regression (Table 2). We found that the genotype distribution of rs2237062 was significantly different between group 1 and group 2, as well as between group 2 and group 3. Similarly, an identical trend was also observed in rs2547 when we compared the genotype distribution between the three groups. These data suggested that the genotype CC carriers of rs2237062 had a 3.87-fold (95% CI: 1.56–9.61) risk of developing to moderate or severe hepatitis compared with the wild type GG carriers. An elevated risk of chronic HBV progression was also found in genotype AA carriers with rs2547, with an OR of 16.17 (95% CI: 1.90–138.00). However, we did not detect any difference in the genotype distribution of rs2237061 or its ORs calculated by univariate logistic regression. An analysis of alleles in three CXCL14 polymorphisms was also conducted. Similarly, we observed an elevated risk for HBV progression in rs2237062 and rs2547 but not rs2237061.
Table 2.
The Genotype Distribution of CXCL14 Polymorphisms and Its Association with Chronic HBV Progression
| Genotype | Group 1 | Group 2 | Group 3 | P value | OR (95% CI)a |
|---|---|---|---|---|---|
| rs2237061 | |||||
| CC | 110 (72.8) | 43 (70.5) | 132 (73.3) | 0.129a | 1.00 (Ref) |
| CT | 33 (21.9) | 10 (16.4) | 39 (21.7) | 0.991b | 0.78 (0.35–1.71) |
| TT | 8 (5.3) | 8 (13.1) | 9 (5.0) | 0.081c | 2.56 (0.90–7.25) |
| C | 253 (83.8) | 96 (78.7) | 303 (84.2) | 0.214 a | 1.00 (Ref) |
| T | 49 (16.2) | 26 (21.3) | 57 (15.8) | 0.891 b | 1.40 (0.82–2.38) |
| 0.166 c | |||||
| rs2237062 | |||||
| GG | 76 (50.3) | 25 (41.0) | 97 (53.9) | 0.006a* | 1.00 (Ref) |
| GC | 64 (42.4) | 22 (36.1) | 79 (43.9) | 0.087b | 1.05 (0.54–2.03) |
| CC | 11 (7.3) | 14 (23.0) | 4 (2.2) | <0.001c* | 3.87 (1.56–9.61)* |
| G | 216 (71.5) | 72 (59.0) | 273 (75.8) | 0.012 a | 1.00 (Ref) |
| C | 86 (28.5) | 50 (41.0) | 87 (24.2) | 0.209 b | 1.74 (1.13–2.71) |
| <0.001 c | |||||
| rs2547 | |||||
| GG | 124 (82.1) | 46 (75.4) | 153 (85.0) | 0.004a* | 1.00 (Ref) |
| GA | 26 (17.2) | 9 (14.8) | 26 (14.4) | 0.773b | 0.93 (0.41–2.14) |
| AA | 1 (0.7) | 6 (9.8) | 1 (0.56) | 0.001c* | 16.17 (1.90–138.00)* |
| G | 274 (90.7) | 101 (82.8) | 332 (92.2) | 0.021 a | 1.00 (Ref) |
| A | 28 (9.3) | 21 (17.2) | 28 (7.8) | 0.492b | 2.04 (1.11–3.75) |
| 0.003c |
Notes: aGroup 1 vs Group 2. bGroup 1 vs Group 3. cGroup 2 vs Group 3. *P<0.05
Comparison of Liver Function Parameters Based on CXCL14 Polymorphisms
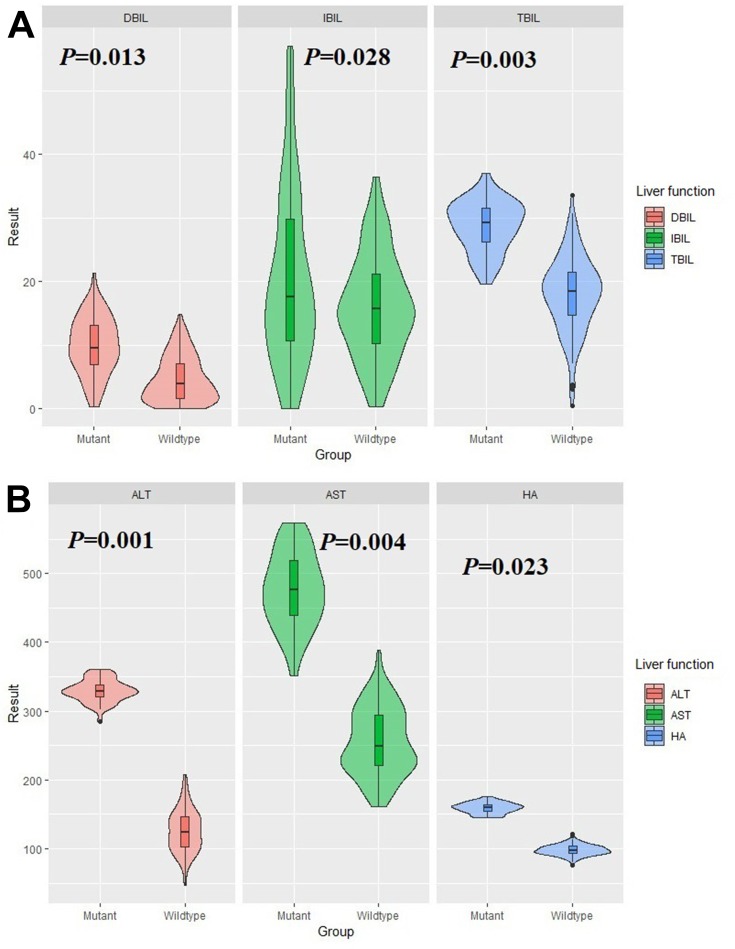
Univariate logistic regression indicated that the polymorphism on rs2237062 and rs2547, but not rs2237061, was positively correlated with chronic HBV progression among patients with chronic HBV infection. Therefore, we conducted a Student’s t-test to compare the liver function parameters based on the genotypes of rs2237062 and rs2547 to determine whether the CXCL14 polymorphisms affected liver function parameters. As shown in Table 3 and Figure 1, carriers of the GC+CC genotype in rs2237062 demonstrated a significantly elevated level of TBIL (29.79±4.06 vs 18.14±6.18), DBIL (9.92±4.91 vs 4.13±3.96) and IBIL (19.92±17.23 vs 15.72±10.19). No significant differences were found among the other liver function parameters between the different genotypes of rs2237062. Surprisingly, we also observed significant increases in ALT, AST, and HA among carriers of the GA+AA genotype in rs2547 when compared with GG wildtype carriers. However, there were no differences in bilirubin between the different genotypes of rs2547.
Table 3.
Comparison on Liver Function Parameters Among Patients with Chronic HBV Infection by CXCL14 Polymorphisms
| Liver Function Parameters | rs2237062 GG Genotype | GC+CC Genotype | P value | rs2547 GG Genotype | GA+AA Genotype | P value |
|---|---|---|---|---|---|---|
| TBIL (umol/L) | 18.14±6.18 | 29.79±4.06 | 0.003* | 24.61±7.02 | 25.33±4.84 | 0.901 |
| DBIL (umol/L) | 4.13±3.96 | 9.92±4.91 | 0.013* | 7.42±8.68 | 7.38±5.26 | 0.991 |
| IBIL (umol/L) | 15.72±10.19 | 19.92±17.23 | 0.028* | 18.13±14.85 | 17.95±14.27 | 0.947 |
| AST (U/L) | 277.57±74.91 | 304.66±56.24 | 0.530 | 251.27±52.12 | 489.78±57.56 | 0.004* |
| ALT (U/L) | 146.78±32.42 | 174.93±26.82 | 0.274 | 127.53±30.20 | 329.16±20.75 | 0.001* |
| TP (g/L) | 73.37±8.28 | 74.37±4.55 | 0.265 | 74.28±4.50 | 72.30±11.91 | 0.089 |
| GLOB (g/L) | 41.34±5.33 | 42.48±3.48 | 0.077 | 42.18±3.54 | 41.03±7.22 | 0.147 |
| ALB (g/L) | 32.43±4.44 | 31.89±3.84 | 0.351 | 32.10±4.01 | 32.24±4.64 | 0.852 |
| TF (g/L) | 226.42±31.25 | 231.77±32.92 | 0.230 | 228.07±32.10 | 235.95±32.57 | 0.178 |
| CP (g/L) | 24.77±3.64 | 23.94±4.43 | 0.138 | 24.07±3.84 | 25.36±5.15 | 0.086 |
| HA (ug/L) | 98.03±8.07 | 117.34±9.78 | 0.124 | 98.64±7.50 | 157.78±7.87 | 0.023* |
Note: * P<0.05.
Abbreviations: TBIL, total bilirubin; DBIL, direct bilirubin; IBIL, indirect bilirubin; ALT, alanine aminotransferase; AST, aspartate transaminase; TP, total protein; GLOB, globulin; ALB, albumin; TF, transferrin; CP, ceruloplasmin; HA, Hyaluronan.
Figure 1.
Comparison on liver function parameters by rs2237062 (A) and rs2547 (B) genotypes. (A) DBIL, IBIL and TBIL are significantly higher in GC+CC genotype carriers of rs2237062; (B) ALT, AST and HA are elevated in GA+AA genotype carriers of rs2547.
Comparison of Serum CXCL14 Levels Between Groups and Genotypes
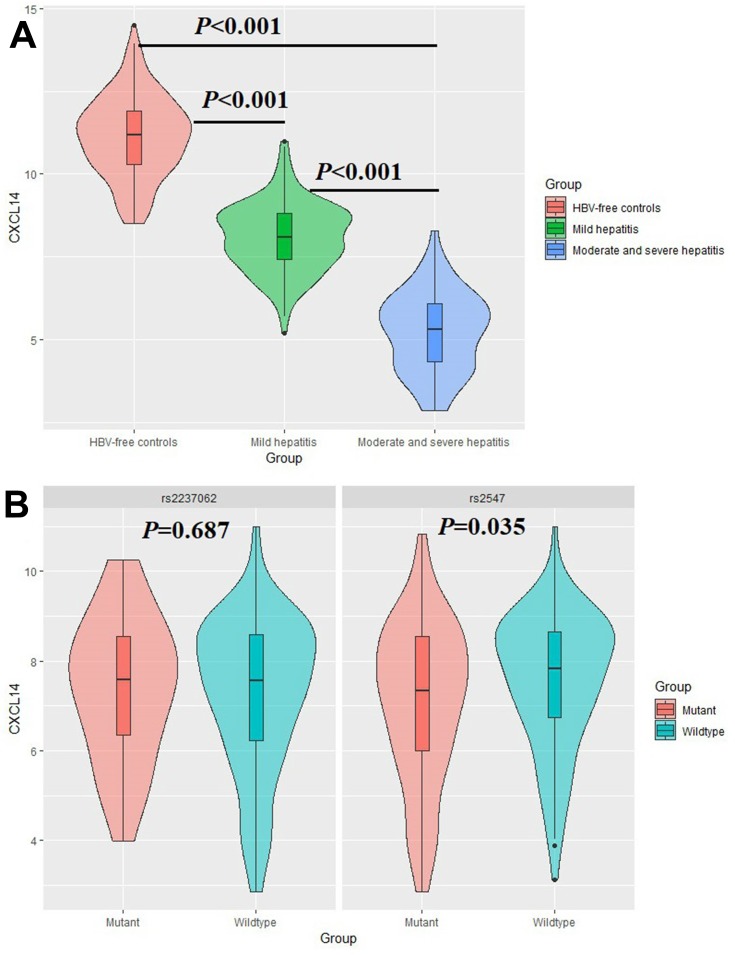
We conducted one-way ANOVA to compare CXCL14 levels between patients with mild hepatitis, moderate-to-severe hepatitis and HBV-free controls. As shown in Table 4 and Figure 2A, the CXCL14 level was 11.14±1.22 ng/mL in HBV-free controls, while a significant reduction was detected in mild hepatitis patients (8.10±1.03 ng/mL) and moderate-to-severe hepatitis patients (5.23±1.21 ng/mL) (P<0.05). To further investigate the association between CXCL14 polymorphisms and CXCL14 serum level, we compared the CXCL14 level between patients with chronic HBV infection by genotype. As shown in Table 5 and Figure 2B, there was no significant difference in CXCL14 level between rs2237062 genotypes; however, in contrast, a significant reduction was observed in carriers of the GA+AA genotype (7.55±1.53 vs 7.06±1.79) in rs2547 when compared with wild type carriers.
Table 4.
One-Way ANOVA of CXCL14 Level Between Three Groups
| Group | CXCL14 Level (ng/mL) | F | P value |
|---|---|---|---|
| Group 1 | 8.10±1.03 | <0.001a* | |
| Group 2 | 5.23±1.21 | <0.001b* | |
| Group 3 | 11.14±1.22 | 683.773 | <0.001c* |
Notes: aGroup 1 vs Group 2. bGroup 1 vs Group 3. cGroup 2 vs Group 3. *P<0.05
Figure 2.
Comparison on serum CXCL14 level by groups (A) and rs2237062 and rs2547 genotypes (B). (A) CXCL14 protein level is higher in HBV-free controls, and it decreases in patients with moderate and severe hepatitis; (B) CXCL14 protein level is significantly lower in carriers of rs2547 GA+AA genotype.
Table 5.
Comparison on CXCL14 Level Between Genotypes Among Patients with Chronic HBV Infection
| Genotype | CXCL14 Level (ng/mL) | t | P value |
|---|---|---|---|
| rs2237062 GG genotype | 7.37±1.60 | ||
| GC+CC genotype | 7.25±1.71 | 0.404 | 0.687 |
| rs2547 GG genotype | 7.55±1.53 | ||
| GA+AA genotype | 7.06±1.79 | 2.126 | 0.035* |
Note: *P<0.05.
Discussion
The present study confirmed that reduced CXCL14 mRNA expression in HCC tissues when compared with adjacent tissues. Furthermore, two polymorphisms, but not rs2237061, in CXCL14 were associated with more advanced progression of chronic HBV infection and compromised liver function. We also compared the serum CXCL14 level in patients with chronic HBV infection and HBV-free controls and found that the CXCL14 level was significantly higher in HBV-free controls, and significantly reduced when comparing patients with mild hepatitis and moderate-to-severe hepatitis patients. In addition, among patients with chronic HBV infection, carriers of rs2547 mutant had a lower CXCL14 level when compared with wild type carriers.
Generally, CXCL14 has been acknowledged to be a tumor suppressor in certain types of cancer, on the basis of its low expression in carcinoma tissues and abundant expression in normal tissues.10 Several anticancer mechanisms of CXCL14 have been proposed, for example, CXCL14 is a chemoattractant for monocytes, B cells, dendritic cells, and NK cells. A transwell chemotaxis assay showed that CXCL14 is involved with the trafficking of NK cells to sites of malignancy; therefore, the suppression of CXCL14 might prevent NK immune surveillance, consequently leading to the promotion of oncogenesis.12 Our results obtained from comparisons of CXCL14 mRNA expression between HCC tissues and normal tissues by qRT-PCR indicated that CXCL14 might have a tumor-suppressing effect, because its expression was 0.33-fold in HCC tissues compared with adjacent tissues.
We also investigated the impact of an intron polymorphism rs2237062 and a protein-coding polymorphism rs2547 on the progression of chronic HBV infection. We found that the CC genotype was more frequently observed in cases with a more advanced progression of HBV infection, and it was also correlated with elevated serum bilirubin. To date, only one publication has reported the association between CXCL14 polymorphisms and HBV progression; however, it failed to detect a significant difference in rs2237062 genotype distribution between patients with HCC, patients with chronic infection, and HBV-free controls.13 However, this previous publication did suggest that the CC genotype was more frequently observed in HCC patients with advanced TNM stages. Although our results of genotype distribution were not consistent with the report published by Gu et al,14 we both observed the involvement of CXCL14 in the development of HCC.
Another major finding was that the AA genotype of rs2547 was more frequent in HBV patients of advanced stage, and that mild hepatitis patients had a similar genotype distribution when compared with HBV-free controls. After we applied a dominant model to compare liver function parameters, we found that mutant genotypes, combining GA and AA genotypes, had increased levels of ALT, AST, and HA, which are important biomarkers for evaluating liver fibrosis, which is an important pathological step before carcinogenesis. The serum level of HA has generally been acknowledged to be a useful biomarker for diagnosing liver fibrosis with a sensitivity of 82.4% and a specificity of 78.2%.15 Based on the results obtained above, we assumed that rs2547 had a critical role in the progression of HBV infection, considering all elevated liver function parameters were of clinical significance. Currently, no known empirical research has focused on exploring relationships between rs2547 and HBV infection. One previous study found that rs2547 was associated with susceptibility to developing aggressive prostate cancer using a microarray technique.16 According to the statistical analysis, this supports the idea that the rs2547 polymorphism is correlated with a more severe liver impairment and fibrosis, and that AA genotype carriers have a 16.17-fold risk of progressing to the advanced stage of hepatitis. However, despite the large OR we observed in univariate analysis, the data must be interpreted with caution because of the rarity of the AA genotype in the included study participants. Because of the difference in frequency of the AA genotype between mild hepatitis (0.7%) and moderate-to-severe hepatitis (9.8%), the risk can be large in terms of mathematics. Therefore, validation of the association between rs2547 and HBV progression in a larger population is required.
In addition, the present study evaluated the serum CXCL14 level in patients with HBV infection and HBV-free controls. The results of one-way ANOVA showed a linear trend, where CXCL14 was highest in HBV-free controls, lower in patients with mild hepatitis, and lowest in patients with moderate-to-severe hepatitis. This indicated that CXCL14 was inversely correlated with HBV progression. A comparison was also made between the genotypes of rs2237062 and rs2547 among patients with HBV infection; however, there was no significant difference between rs2237062 genotypes, although the wild type GG carriers of rs2547 had a significantly higher level of CXCL14 when compared with the GA+AA genotype. Therefore, the gene-coding polymorphism, rs2547, might impact the serum CXCL14 level. The polymorphism on rs2547 introduced a stop-gain in the coding sequence to produce an incomplete protein. Thus, it can be assumed that the presence of the rs2547 polymorphism would lead to the reduction of the serum level of CXCL14 and would result in a more severe liver injury.
Conclusion
In this study, we determined an association between CXCL14 polymorphisms and HBV progression by logistic regression. In addition, the serum level of CXCL14 between different genotypes was compared. The current data indicated that the serum CXCL14 level was reduced in patients with chronic HBV infection when compared with healthy controls. Furthermore, CXCL14 polymorphisms were associated with degenerated liver function, especially rs2547, which was also associated with elevated HA level, an important indicator of liver cirrhosis. These findings provide important insights into the role of CXCL14 in HBV infection. However, these results should be considered with caution because of the small number of study participants. Therefore, further investigation should be undertaken to investigate how CXCL14 affects liver function and carcinogenesis.
Acknowledgments
This work was supported by the National Natural Science Foundation [Grant No. 81871729, 81802089, 81772260, 81771312, 81672094, 81471967, 81471231, 81401749, 81301501, 81201360, 81271335, 81101324, and 81171625], the Key Projects for Province Science and Technology Program of Fujian [Grant No. 2018D0014], the National Science Foundation for Distinguished Young Scholars of Fujian [Grant No. 2014D001], the Major Special Projects for Serious Illness of Xiamen [Grant No. 3502Z20179045], the Natural Science Foundation of Fujian Province [Grant No. 2016J01628, 2016J01626], and the Youth Scientific Program of Fujian Provincial Health and Family Planning Committee [Grant No. 2016-2-67]. The funders played no role in the study design, data collection or analyses, the decision to publish, or manuscript preparation.
Author Contributions
HCY, JJN, and SLL were mainly involved in experimental process, conceptualization and financial support. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Massarweh NN, El-Serag HB. Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Control. 2017;24(3):1073274817729245. doi: 10.1177/1073274817729245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Martel C, Maucort-Boulch D, Plummer M, Franceschi S. World-wide relative contribution of hepatitis B and C viruses in hepatocellular carcinoma. Hepatology. 2015;62(4):1190–1200. doi: 10.1002/hep.27969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui F, Shen L, Li L, et al. Prevention of chronic hepatitis B after 3 decades of escalating vaccination policy, China. Emerg Infect Dis. 2017;23(5):765–772. doi: 10.3201/eid2305.161477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng R, Qu C, Zhang S, et al. Liver cancer incidence and mortality in China: temporal trends and projections to 2030. Chin J Cancer Res. 2018;30(6):571–579. doi: 10.21147/j.issn.1000-9604.2018.06.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu J, Lin Y, Liu P, Yu Y, Su C, Wang X. Microarray analysis on the lncRNA expression profile in male hepatocellular carcinoma patients with chronic hepatitis B virus infection. Oncotarget. 2016;7(46):76169–76180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Huang P, Zhang L, et al. Antitumor efficacy of C-X-C motif chemokine ligand 14 in hepatocellular carcinoma in vitro and in vivo. Cancer Sci. 2013;104(11):1523–1531. doi: 10.1111/cas.12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hromas R, Broxmeyer HE, Kim C, et al. Cloning of BRAK, a novel divergent CXC chemokine preferentially expressed in normal versus malignant cells. Biochem Biophys Res Commun. 1999;255(3):703–706. doi: 10.1006/bbrc.1999.0257 [DOI] [PubMed] [Google Scholar]
- 9.Frederick MJ, Henderson Y, Xu X, et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol. 2000;156(6):1937–1950. doi: 10.1016/S0002-9440(10)65067-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sjöberg E, Augsten M, Bergh J, Jirström K, Östman A. Expression of the chemokine CXCL14 in the tumour stroma is an independent marker of survival in breast cancer. Br J Cancer. 2016;114(10):1117–1124. doi: 10.1038/bjc.2016.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, Ji G, Wang Y, et al. Large-scale search of single nucleotide polymorphisms for hepatocellular carcinoma susceptibility genes in patients with hepatitis C. Hepatology. 2005;42(4):846–853. doi: 10.1002/hep.v42:4 [DOI] [PubMed] [Google Scholar]
- 12.Ito S, Ozawa S, Ikoma T, Yajima N, Kiyono T, Hata R. Expression of a chemokine BRAK/CXCL14 in oral floor carcinoma cells reduces the settlement rate of the cells and suppresses their proliferation in vivo. Biomed Res. 2010;31(3):199–206. doi: 10.2220/biomedres.31.199 [DOI] [PubMed] [Google Scholar]
- 13.Starnes T, Rasila KK, Robertson MJ, et al. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: implications for the downregulation of CXCL14 in malignancy. Exp Hematol. 2006;34(8):1101–1105. doi: 10.1016/j.exphem.2006.05.015 [DOI] [PubMed] [Google Scholar]
- 14.Gu X, Wang H, Wang A, et al. An intronic polymorphism rs2237062 in the CXCL14 gene influences HBV-related HCC progression in Chinese population. Mol Biol Rep. 2012;39(2):797–803. doi: 10.1007/s11033-011-0801-7 [DOI] [PubMed] [Google Scholar]
- 15.Tangkijvanich P, Kongtawelert P, Pothacharoen P, Mahachai V, Suwangool P, Poovorawan Y. Serum hyaluronan: a marker of liver fibrosis in patients with chronic liver disease. Asian Pac J Allergy Immunol. 2003;21(2):115–120. [PubMed] [Google Scholar]
- 16.Williams KA, Lee M, Hu Y, et al. A systems genetics approach identifies CXCL14, ITGAX, and LPCAT2 as novel aggressive prostate cancer susceptibility genes. PLoS Genet. 2014;10(11):e1004809. doi: 10.1371/journal.pgen.1004809 [DOI] [PMC free article] [PubMed] [Google Scholar]