Abstract
Osteoarthritis (OA) is the most common motor system disease in the elderly, with a high incidence and a huge social and economic burden. Therefore, it is urgent to study its potential pathogenesis to improve the therapeutic effect of the disease. In this study, we constructed a number of regulator-mediated OA dysfunction modules, and carried out in-depth analysis in order to examine the disease development process. Differential expression analysis, co-expression analysis and enrichment analysis were combined to screen genes related to disease progression. Subsequently, key regulatory factors in the process of OA were identified based on the pivotal regulators that may manipulate important parts of the module subnetwork. A total of 16 OA dysfunction modules were obtained, involving the aggregation of 3,239 module genes. Then, enrichment analysis showed that module genes were significantly involved in apoptosis, inflammation-related functions and signaling pathways. Finally, we revealed a series of regulators, including 842 ncRNA (miR-132-3p, miR-130a-3p and miR-590-3p), 59 transcription factors (NFKB1, RELA and STAT3). We consider that STAT3 is the core transcription factor and promotes the development of OA through the signal of NF-κB. Overall, our results provide biologists and pharmacists with a new way of thinking to reveal the disease process of OA, and provide a wider range of candidate targets for follow-up research.
Keywords: osteoarthritis, dysfunction module, STAT3, NF-κB signaling pathway
Introduction
Osteoarthritis (OA) is a degenerative joint disease involving cartilage and surrounding tissues. The disease usually progresses slowly, but may eventually result in joint failure, degeneration, pain and even disability (1). It is characterised by the focal area of articular cartilage loss in synovial joints and its related symptoms such as osteophyte formation, subchondral bone changes and synovitis (2). The risk of knee and hip diseases is high, followed by widespread lower limb, hand OA and hip diseases (3). The main symptoms are pain, stiffness, joint deformity and cracking (4). Women are more affected by OA than men, and the incidence of OA increases with age (5). So it is considered the most frequent chronic joint disease (6). In addition, OA is the leading cause of disability, and its incidence is increasing (7). In most cases, joint degeneration occurs, but the risk of OA increases with age, joint overload, joint abnormalities and collisions (8). In addition, risk factors for the disease include bone marrow edema, synovitis and joint effusion (9). Therefore, OA, as a common complex disease, is a prominent public health burden (10). Studies have shown that exercise therapy can effectively relieve pain in patients with OA of the hip or knee joint, but the cost is high (11). Non-surgical treatment is usually the best choice, mainly based on everyday life adaptation, weight loss and exercise, combined with drug therapy (12). In addition, conservative OA-specific therapy can also improve pain and function, and reduce the risks of surgery in patients with hip or knee OA (13). In addition to surgical treatment of severe OA, traditional treatments include non-steroidal anti-inflammatory drugs to relieve pain symptoms, anesthetic and non-anesthetic (limp) analgesics and physiotherapy (14). Viscoelastic supplementation with hypertonic acid (HA) injection is often used for local treatment of OA (15). OA has a potential inflammatory phenomenon which causes loss of chondrocytes, thus reducing the cartilage layer at the joint. Compounds with anti-inflammatory properties are potential therapeutic agents for OA (16). Some studies have shown that etoxib has a positive effect on OA inflammation, and shows good tolerance and low incidence of side effects (17). Collagen hydrolysate is a potential therapeutic agent for OA and osteoporosis (18). However, these drugs can only improve mild OA and have no good effect on serious diseases requiring surgical treatment. Therefore, how to reduce the clinical symptoms of severe OA patients by non-surgical methods is of particular importance.
Herein, we propose a comprehensive strategy for exploring the probable pathogenic process of OA based on functional dysfunction module. In this study, we identified that STAT3 may take part in these dysfunction modules and play an important role in mediating the NF-κB signaling pathway, thereby promoting OA. In conclusion, our integrated strategy based on functional modules not only helps to explore the hypothetical molecular mechanism of OA, but also provides rich resources and guidance for biologists to further design experiments.
Materials and methods
Differential expression analysis
We collected expression microarray data sets of OA-related disease samples from NCBI Gene Expression Omnibus (GEO) database (19), number GSE55235. Difference analysis was performed on the collected disease samples (patients with normal-OA) using the R language limma package (20).
The study was approved by the Ethics Committee of Second Hospital of Shanxi Medical University (Taiyuan, China).
Co-expression analysis
In order to study the co-expression of OA-related genes, we used weighted gene co-expression network analysis (WGCNA) (21) to analyze the RNA expression matrix of OA-related genes and to find the co-expression gene module. The weighted value of correlation coefficient, i.e., the N power of gene correlation coefficient, was utilized to calculate the correlation coefficient (Person coefficient) between any two genes. The connection between genes in the network obeys scale-free networks, which make the algorithm more biologically meaningful. Then, a hierarchical clustering tree was built by the correlation coefficient between genes. Different branches of the clustering tree represent separate gene modules, and different colors represent different modules. Finally, 16 significant OA-related gene co-expression modules were extracted.
Functional pathway enrichment analysis and identification of dysfunctional modules
Exploring the function and signaling pathway of genes is often an effective wany to study the molecular mechanism of diseases. R language Cluster Profiler package (22) was used to analyze gene enrichment in 16 modules (P-value cutoff = 0.01, q-value cutoff = 0.01) and KEGG pathway (P-value cutoff = 0.05, q value Cutoff = 0.2). Then, the functions and pathways related to the process of OA were screened and bubble maps were drawn.
Regulator analysis
Pivot regulator is defined as a regulator, which substantially regulates the dysfunction module of arthritis. Gene transcription and post-transcriptional regulation are often driven by non-coding genes (ncRNA) and transcription factors (TF). The transcription factor target data was downloaded from TRRUST V2 database (23), and 71 interaction pairs of 59 transcription factors were obtained. Then, ncRNA-RNA (protein) data were downloaded from RAID 2.0 database (24), and 1,661 interaction pairs involving 842 ncRNAs were obtained. Pivot analysis based on the interaction data was carried out to identify the regulatory effects of these transcription factors and ncRNA on the modules. Pivot analysis refers to search for at least two interacting drivers with the module in the target pair and calculating the significance of the interaction between the driver and the module according to the hypergeometric test. TF and ncRNA with P<0.01 are the pivots of the significant regulatory module. Finally, the core pivots were recognized by statistical analysis.
Results
Identification of time-series expression disorders in OA
In order to further explore the occurrence and progress of OA, data sets of gene expression profiles related to OA we downloaded from GEO database. Built on the differential analysis of genes, 3,239 differential genes were obtained to identify the key genes that play a continuous role in the regulation of OA in the disease process (Table SI).
Identification of staging related modules for functional OA
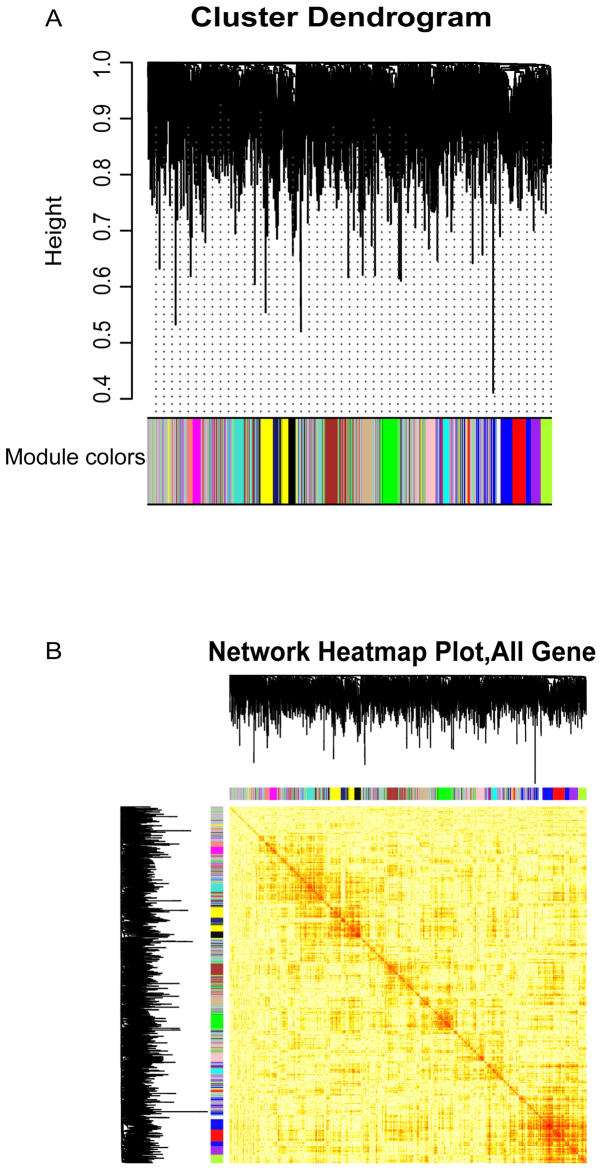
Based on WGCNA network, 16 co-expression modules (Fig. 1A and B) were generated for the analysis of persistent dysfunction genes. The key genes of each module were identified based on functional impairment modules, which showed significant clustering phenomena in the samples. These functional modules may participate in different functions and pathways, thus representing diverse regulatory mechanisms to mediate the occurrence and development of OA dysfunction.
Figure 1.
Clustering module of co-expression relations of OA-related genes. (A) According to the synergistic expression relationship of altered genes, 16 modules were clustered. A color represents a module. (B) Thermogram of modular gene expression in samples. In the OA disease sample, the phenomenon of grouping expression is intuitively presented. OA, osteoarthritis.
Functions and pathways of genes of interest
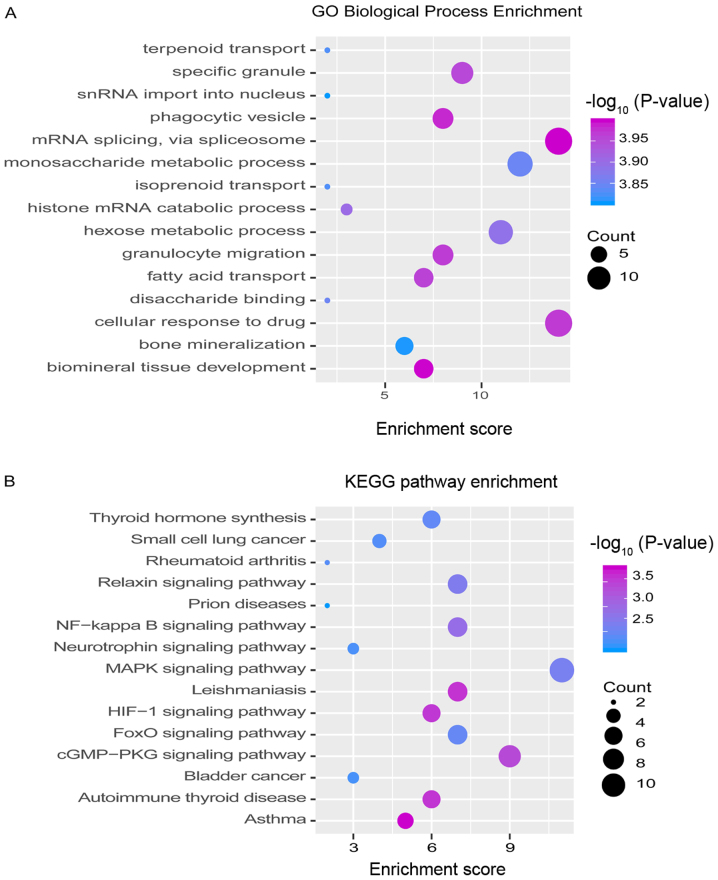
Studying the function and pathway of gene involvement is an essential means to identify the mediating pathogenesis. In order to study possible dysfunction caused by module gene disorder, the enrichment was analyzed of function and pathway of each module. The results showed that most of the functional modules added value in OA-related functions and pathways. GO function and KEGG pathway enrichment analysis on 16 functional modules were performed. A total of 10,759 functions and 263 KEGG pathway enrichment results were obtained. These include 1,576 molecular functions (MF), 852 cell components (CC) and 8,331 biological processes (BP) involving genes (Fig. 2 and Table SII).
Figure 2.
Modular genes participate in function and pathway identification of OA dysfunction module. (A) GO functional enrichment analysis involving module genes. The darker the color, the stronger the significance of enrichment. The larger the circle, the larger the proportion of module genes in GO functional entry genes. (B) Enrichment analysis of KEGG pathway involving modular genes. The darker the color, the stronger the significance of enrichment. The larger the circle, the larger the proportion of module genes to KEGG pathway entry genes. OA, osteoarthritis.
It is noteworthy that the AMPK/NF-κB signaling pathway in which they are substantially involved may be identified as the core signaling pathway to accelerate the disease progression of OA. As noted above, we found that the NF-κB signaling pathway may be closely related to the acceleration of OA.
TF and ncRNA driving OA
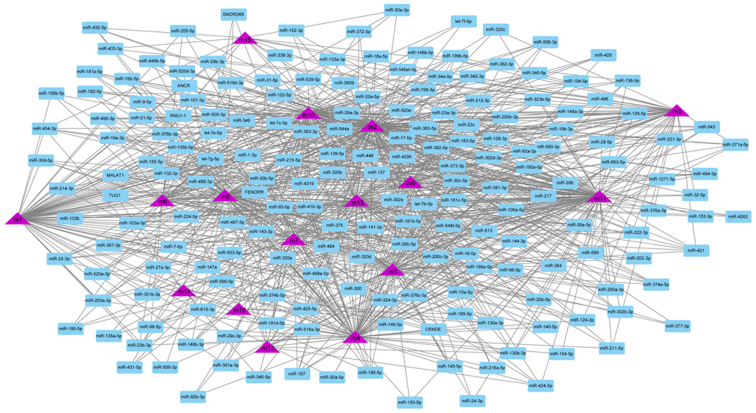
Although the regulation of single or several transcription factors and ncRNA on OA have been studied by biologists, few studies have focused on their comprehensive regulation of dysfunctional modules. According to the number of regulatory module genes and the significance, the potential regulatory effect of 901 pivot regulators on the module were determined. These include 842 ncRNA and 59 transcription factors, involving 1,661 ncRNA-module interaction pairs and 71 TF-module target pairs. Statistical analysis of the predicted results showed that miR-132-3p targeted as many as 11 dysfunction modules, respectively, and had a significant regulatory effect on OA (Fig. 3). In addition, miR-130a-3p and miR-590-3p regulated 10 dysfunction modules, respectively, and were identified as pivot regulators. Other ncRNAs also regulate multiple dysfunction modules to various degrees, and have potential effects on OA. The regulatory effect of transcription factors on diseases could be overlooked. Depending on statistics, NFKB1 and RELA have significant regulatory effects on four dysfunction modules, while STAT3 plays a role on three modules (Fig. 4). These transcription factors may mediate dysfunction modules to regulate the occurrence and progress of OA, and play a key role in the pathogenesis of OA. Other TFs also have a definite regulatory effect on functional modules, which may affect the progress of the disease. A thorough study of the regulatory role of pivot regulators on dysfunction modules will allow us to understand the underlying mechanisms of diseases. These pivot regulators can be used as candidates for further experimental studies.
Figure 3.
ncRNA regulatory network map of OA. Purple triangle represents a module, and blue box represents the ncRNA corresponding to the module. OA, osteoarthritis.
Figure 4.
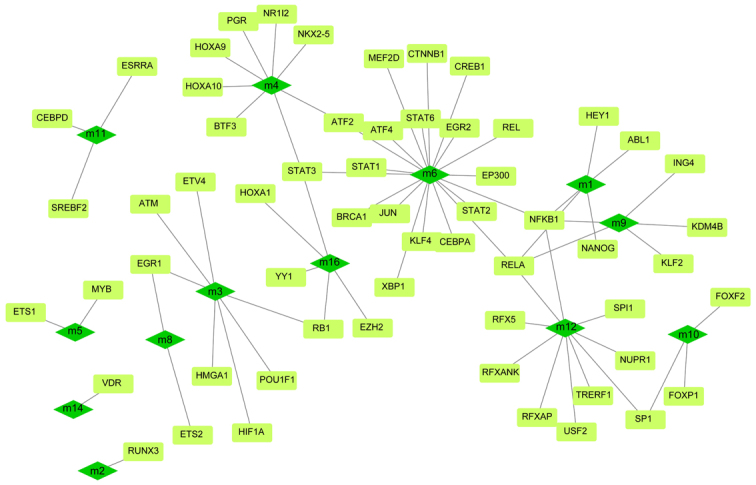
The regulatory network of transcription factors in OA includes dark green quadrilateral module and light green rectangle module. OA, osteoarthritis.
Discussion
OA is one of the most frequent chronic diseases. With the increase in life expectancy, the incidence of OA will continue to rise (25). The pathogenesis of OA involves many factors, including mechanics, effects of aging on the composition and structure of the cartilage matrix, and genetic factors (26). Among them, serum microRNA level has an important regulatory relationship with the development of severe OA of the knee joint and hip joint (27). Rousseau et al (28) found that serum periosteal protein was involved in the prevalence and development risk of knee OA in women. In order to fully explore the core pathways and regulators of OA, we first integrated related differential genes for differential analysis, and finally obtained 3,239 differential genes. The key gene statistics found that proteins play an important role in promoting the operative mechanism of OA. In view of the early stage of OA, including the effects of cell proliferation and chondrocytes in the synthesis of inflammatory mediators such as matrix proteins, proteases, growth factors and cytokines (26). Studies have revealed that in the late stage of OA, bone tissue blood flow and oxygen content significantly decreased. It also has a negative effect on bone cells, induces them to release protein (cytokines), promotes bone remodeling and cartilage destruction (29). We observed the co-expression behavior of the differentially expressed genes in the disease samples. From this, we obtained 16 co-expression modules. The genes contained in the modules were reckoned to have co-expression. Subsequently, in view of the consequences of enrichment analysis, we found that genes in 11 functional modules of OA mainly participate in the functional pathways of NF-κB signaling pathway. These include maintenance of protein localization in organelle, epidermal growth factor-activated receptor trans activation by G-protein coupled receptor signaling pathway and chondrocyte proliferation. The NF-κB signaling pathway is thought to be the core signaling pathway to promote the progression of OA. Through the review of Saito and Tanaka (30) we found two kinds of multifunctional signaling pathways in the development of OA: Notch and NF-κB in activated B cells, and determined that articular chondrocytes regulate the progress of OA through these pathways. miR-146 promotes the proliferation and inhibit apoptosis of OA chondrocytes by inhibiting the expression of TRAF6 and the inhibition of NF-κB signaling pathway (31). In addition, high expression of Sam68 can promote the activation of TNF-α-induced chondrocyte NF-κB signaling, the expression of decomposition genes and cell apoptosis, which provides a possible target for the pathophysiological study and treatment of OA (32).
To elucidate the transcriptional regulatory factors of OA, pivot regulators were analyzed based on transcriptional and post-transcriptional regulatory relationships. The results showed that microRNAs mainly composed of microRNAs such as miR-132-3p, miR-130a-3p and miR-590-3p played a major role in the regulation of OA. The results showed that miR-132-3p promoted cartilage formation and differentiation of rMSCs, possibly through targeting ADAMTS-5, which provided a new perspective for cartilage differentiation and pathology of OA (33). In addition, miR-130a plays an important role in regulating the expression of TNF-alpha in human chondrocytes, and it will be a novel therapeutic target for OA (34). Relevant studies have shown that elevated concentration of melatonin may lead to the inhibition of the expression of microRNA-590-3p and upregulation of the target gene of human osteoblast apoptosis (35). Building on our observations, we found that microRNAs that regulate multiple modules target genes and pathways related to OA, thus providing a comprehensive understanding of the molecular network underlying the pathogenesis of OA. Transcription factors such as NFKB1, RELA and STAT3 are key mediators in the process of OA-related potential diseases. Their interactions can regulate inflammation and metabolism in cells, and drive inflammatory cytokines and immune responses in OA microenvironment. In the process of OA, these three transcription factors mediate four and three dysfunction modules respectively, and play an important role in the pathogenesis of OA. Recent studies have demonstrated that upregulation of microRNA-9 or downregulation of NF-κB1 can promote cell proliferation and inhibit cell apoptosis, and downregulation of microRNA-9 can directly bind to NF-κB1, promote the proliferation and anti-apoptosis of knee OA chondrocytes (36). In addition, in vitro studies have shown that Rela is a fundamental subunit mediating the signal transduction of NF-κB, involved in cartilage formation and differentiation, cell survival and production of catabolic enzymes (37). These pivot regulators together mediate functional dysfunction modules, play an overall supervisory role, and represent the accelerating process of OA disease.
In conclusion, the study of gene involvement in signaling pathways will allow us to analyze the progress of disease. In view of the differential genes, we obtained, STAT3 speeds up the progression of OA through the NF-κB signaling pathway. Activation of nuclear factor NF-κB protein triggers the expression of a series of genes, leading to joint destruction and the occurrence and development of OA (OA) (38). ACY-1215, an inhibitor, mainly induces STAT3 in OA chondrocytes to downregulate the expression of matrix-degrading proteinase through NF-κB signaling pathway, thus improving cartilage degradation and exerting effective cartilage protection (39). In addition, chondroitin sulfate (CS) is a slow-acting disease regulator in the treatment of OA. Its beneficial effect is the anti-inflammatory properties caused by the inhibition of the signal transduction pathway of NF-κB or STAT3 (40). In general, functional module-based approaches cannot only explore the pathogenesis and development of diseases comprehensively and deeply, but also provide abundant resources for potential candidates of TF and pivot ncRNAs, and predict their potential therapeutic methods and mechanisms.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
FW wrote the manuscript, interpreted and analyzed the data. ZG designed the study and performed the experiments. YY was responsible for the analysis and discussion of the data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Second Hospital of Shanxi Medical University (Taiyuan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Litwic A, Edwards MH, Dennison EM, Cooper C. Epidemiology and burden of osteoarthritis. Br Med Bull. 2013;105:185–199. doi: 10.1093/bmb/lds038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dieppe PA, Lohmander LS. Pathogenesis and management of pain in osteoarthritis. Lancet. 2005;365:965–973. doi: 10.1016/S0140-6736(05)71086-2. [DOI] [PubMed] [Google Scholar]
- 3.Prieto-Alhambra D, Judge A, Javaid MK, Cooper C, Diez-Perez A, Arden NK. Incidence and risk factors for clinically diagnosed knee, hip and hand osteoarthritis: Influences of age, gender and osteoarthritis affecting other joints. Ann Rheum Dis. 2014;73:1659–1664. doi: 10.1136/annrheumdis-2013-203355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérez Martín Á. [Symptoms. Localizations: Knee, hip, hands, spine, other localizations] Aten Primaria. 2014;46(Suppl 1):11–17. doi: 10.1016/S0212-6567(14)70038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flugsrud GB, Nordsletten L, Reinholt FP, Risberg MA, Rydevik K, Uhlig T. Osteoarthritis. Tidsskr Nor Laegeforen. 2010;130:2136–2140. doi: 10.4045/tidsskr.09.1054. (In Norwegian) [DOI] [PubMed] [Google Scholar]
- 6.Bijlsma JW, Berenbaum F, Lafeber FP. Osteoarthritis: An update with relevance for clinical practice. Lancet. 2011;377:2115–2126. doi: 10.1016/S0140-6736(11)60243-2. [DOI] [PubMed] [Google Scholar]
- 7.Thomas AC, Hubbard-Turner T, Wikstrom EA, Palmieri-Smith RM. Epidemiology of posttraumatic osteoarthritis. J Athl Train. 2017;52:491–496. doi: 10.4085/1062-6050-51.5.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckwalter JA, Martin JA. Osteoarthritis. Adv Drug Deliv Rev. 2006;58:150–167. doi: 10.1016/j.addr.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Felson DT. An update on the pathogenesis and epidemiology of osteoarthritis. Radiol Clin North Am. 2004;42:1–9. doi: 10.1016/S0033-8389(03)00161-1. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg J, Zeggini E. Functional genomics in osteoarthritis: Past, present, and future. J Orthop Res. 2016;34:1105–1110. doi: 10.1002/jor.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kloek CJ, Bossen D, Veenhof C, van Dongen JM, Dekker J, de Bakker DH. Effectiveness and cost-effectiveness of a blended exercise intervention for patients with hip and/or knee osteoarthritis: Study protocol of a randomized controlled trial. BMC Musculoskelet Disord. 2014;15:269. doi: 10.1186/1471-2474-15-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grazina R, Andrade R, Bastos R, Costa D, Pereira R, Marinhas J, Maestro A, Espregueira-Mendes J. Clinical management in early OA. Adv Exp Med Biol. 2018;1059:111–135. doi: 10.1007/978-3-319-76735-2_5. [DOI] [PubMed] [Google Scholar]
- 13.Teoh LSG, Eyles JP, Makovey J, Williams M, Kwoh CK, Hunter DJ. Observational study of the impact of an individualized multidisciplinary chronic care program for hip and knee osteoarthritis treatment on willingness for surgery. Int J Rheum Dis. 2017;20:1383–1392. doi: 10.1111/1756-185X.12950. [DOI] [PubMed] [Google Scholar]
- 14.Adatia A, Rainsford KD, Kean WF. Osteoarthritis of the knee and hip. Part I: Aetiology and pathogenesis as a basis for pharmacotherapy. J Pharm Pharmacol. 2012;64:617–625. doi: 10.1111/j.2042-7158.2012.01458.x. [DOI] [PubMed] [Google Scholar]
- 15.Legré-Boyer V. Viscosupplementation: Techniques, indications, results. Orthop Traumatol Surg Res. 2015;101(Suppl):S101–S108. doi: 10.1016/j.otsr.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 16.Chin KY. The spice for joint inflammation: Anti-inflammatory role of curcumin in treating osteoarthritis. Drug Des Devel Ther. 2016;10:3029–3042. doi: 10.2147/DDDT.S117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivordova LE, Zavodovsky BV, Polyakova JV, Akhverdyan YR. Evidence of feasibility etoricoxib therapy in osteoarthritis in elderly patients. Adv Gerontol. 2016;29:286–290. (In Russian) [PubMed] [Google Scholar]
- 18.Moskowitz RW. Role of collagen hydrolysate in bone and joint disease. Semin Arthritis Rheum. 2000;30:87–99. doi: 10.1053/sarh.2000.9622. [DOI] [PubMed] [Google Scholar]
- 19.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets - update. Nucleic Acids Res. 2013;41(D1):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46D:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi Y, Zhao Y, Li C, Zhang L, Huang H, Li Y, Liu L, Hou P, Cui T, Tan P, et al. RAID v2.0: An updated resource of RNA-associated interactions across organisms. Nucleic Acids Res. 2017;45D:D115–D118. doi: 10.1093/nar/gkw1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pereira D, Ramos E, Branco J. Osteoarthritis. Osteoarthritis Acta Med Port. 2015;28:99–106. doi: 10.20344/amp.5477. [DOI] [PubMed] [Google Scholar]
- 26.Goldring MB, Goldring SR. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 27.Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, Distler A, Langley SR, Gelse K, Sesselmann S, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. 2015;74:e18. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 28.Rousseau JC, Sornay-Rendu E, Bertholon C, Garnero P, Chapurlat R. Serum periostin is associated with prevalent knee osteoarthritis and disease incidence/progression in women: The OFELY study. Osteoarthritis Cartilage. 2015;23:1736–1742. doi: 10.1016/j.joca.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 29.Racine J, Aaron RK. Pathogenesis and epidemiology of osteoarthritis. R I Med J. 2013;96:19–22. [PubMed] [Google Scholar]
- 30.Saito T, Tanaka S. Molecular mechanisms underlying osteoarthritis development: Notch and NF-κB. Arthritis Res Ther. 2017;19:94. doi: 10.1186/s13075-017-1296-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong JH, Li J, Liu CF, Liu N, Bian RX, Zhao SM, Yan SY, Zhang YB. Effects of microRNA-146a on the proliferation and apoptosis of human osteoarthritis chondrocytes by targeting TRAF6 through the NF-κB signalling pathway. Biosci Rep. 2017;37:37. doi: 10.1042/BSR20160578. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Xu L, Sun C, Zhang S, Xu X, Zhai L, Wang Y, Wang S, Liu Z, Cheng H, Xiao M, et al. Sam68 promotes NF-κB activation and apoptosis signaling in articular chondrocytes during osteoarthritis. Inflamm Res. 2015;64:895–902. doi: 10.1007/s00011-015-0872-3. [DOI] [PubMed] [Google Scholar]
- 33.Zhou X, Luo D, Sun H, Qi Y, Xu W, Jin X, Li C, Lin Z, Li G. miR-132-3p regulates ADAMTS-5 expression and promotes chondrogenic differentiation of rat mesenchymal stem cells. J Cell Biochem. 2018;119:2579–2587. doi: 10.1002/jcb.26421. [DOI] [PubMed] [Google Scholar]
- 34.Li ZC, Han N, Li X, Li G, Liu YZ, Sun GX, Wang Y, Chen GT, Li GF. Decreased expression of microRNA-130a correlates with TNF-α in the development of osteoarthritis. Int J Clin Exp Pathol. 2015;8:2555–2564. [PMC free article] [PubMed] [Google Scholar]
- 35.Meng X, Zhu Y, Tao L, Zhao S, Qiu S. miR-590-3p mediates melatonin-induced cell apoptosis by targeting septin 7 in the human osteoblast cell line hFOB 1.19. Mol Med Rep. 2018;17:7202–7208. doi: 10.3892/mmr.2018.8729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gu R, Liu N, Luo S, Huang W, Zha Z, Yang J. MicroRNA-9 regulates the development of knee osteoarthritis through the NF-kappaB1 pathway in chondrocytes. Medicine (Baltimore) 2016;95:e4315. doi: 10.1097/MD.0000000000004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi H, Chang SH, Mori D, Itoh S, Hirata M, Hosaka Y, Taniguchi Y, Okada K, Mori Y, Yano F, et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat Commun. 2016;7:13336. doi: 10.1038/ncomms13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigoglou S, Papavassiliou AG. The NF-κB signalling pathway in osteoarthritis. Int J Biochem Cell Biol. 2013;45:2580–2584. doi: 10.1016/j.biocel.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 39.Cheng C, Shan W, Huang W, Ding Z, Cui G, Liu F, Lu W, Xu J, He W, Yin Z. ACY-1215 exhibits anti-inflammatory and chondroprotective effects in human osteoarthritis chondrocytes via inhibition of STAT3 and NF-κB signaling pathways. Biomed Pharmacother. 2019;109:2464–2471. doi: 10.1016/j.biopha.2018.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Andrés RM, Payá M, Montesinos MC, Ubeda A, Navalón P, Herrero M, Vergés J, Terencio MC. Potential antipsoriatic effect of chondroitin sulfate through inhibition of NF-κB and STAT3 in human keratinocytes. Pharmacol Res. 2013;70:20–26. doi: 10.1016/j.phrs.2012.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.