Abstract
Retinal disease classification is a significant problem in computer-aided diagnosis (CAD) for medical applications. This paper is focused on a 4-class classification problem to automatically detect choroidal neovascularization (CNV), diabetic macular edema (DME), DRUSEN, and NORMAL in optical coherence tomography (OCT) images. The proposed classification algorithm adopted an ensemble of four classification model instances to identify retinal OCT images, each of which was based on an improved residual neural network (ResNet50). The experiment followed a patient-level 10-fold cross-validation process, on development retinal OCT image dataset. The proposed approach achieved 0.973 (95% confidence interval [CI], 0.971–0.975) classification accuracy, 0.963 (95% CI, 0.960–0.966) sensitivity, and 0.985 (95% CI, 0.983–0.987) specificity at the B-scan level, achieving a matching or exceeding performance to that of ophthalmologists with significant clinical experience. Other performance measures used in the study were the area under receiver operating characteristic curve (AUC) and kappa value. The observations of the study implied that multi-ResNet50 ensembling was a useful technique when the availability of medical images was limited. In addition, we performed qualitative evaluation of model predictions, and occlusion testing to understand the decision-making process of our model. The paper provided an analytical discussion on misclassification and pathology regions identified by the occlusion testing also. Finally, we explored the effect of the integration of retinal OCT images and medical history data from patients on model performance.
1. Introduction
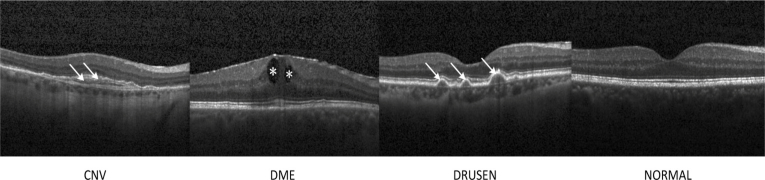
Nowadays, OCT has become a powerful imaging modality for non-invasive assessment of various retinal abnormalities [1–5], such as assisting the diagnosis of CNV, DME, and DRUSEN, as shown in Fig. 1. However, with the amount of image data produced by advanced OCT technology increasing, the feasibility of conventional manual OCT assessment in clinical practice has become largely unrealistic [6–8]. Likewise, the evaluation of retinal diseases on OCT images is subject to substantial inter- and intra-observer variability, when performed by experienced ophthalmologists, which can result in making inconsistency and unreliable interpretation, delaying accurate diagnosis, and creating a drain on healthcare resources [9–12]. Consequently, automated detection of retinal disorders on retinal OCT images would be of enormous benefit, which could help ophthalmologists to more efficiently evaluate and treat the eye diseases.
Fig. 1.
Representative optical coherence tomography (OCT) images. The arrows and the asterisks indicated the lesion sites. CNV: choroidal neovascularization; DME: diabetic macular edema.
Over the past several years, machine learning techniques have advanced the state-of-the-art in medical image detection. The results and details of the proposed and existing methods were provided in Table 1. Hussain et al. [13] adopted random forest algorithm to classify healthy and diseased retina using retinal features from spectral domain (SD)-OCT images, yielding mean accuracy of more than 96%. Lemaître et al. [14] proposed a method based on extracted local binary pattern features from OCT images and dictionary learning using bag of words models for DME detection, which achieved a sensitivity and a specificity of 81.2% and 93.7%, respectively. Alsaih et al. [15] introduced a linear support vector machine (SVM) to classify normal retina and DME, and obtained a 87.5% sensitivity and a 87.5% specificity. Srinivasan et al. [16] employed histogram of oriented gradients (HOG) descriptors and SVM classifiers for the detection of age-related macular degeneration (AMD), DME, and normal retina, which acquired 100%, 100%, and 86.67% accuracy at the OCT level, respectively. However, these classification approaches highly relied on features explicitly defined by ophthalmologists using their domain knowledge, resulting in time-consuming, weak generalization ability, and even unfeasibility in large datasets.
Table 1. The results and details of the proposed and existing methods.
| Author | Year | Database | Approach | Performance |
|---|---|---|---|---|
| Hussain et al. [13] | 2018 | Public (DHU, and Tian et al. Images) and proprietary | Random Forest algorithm | Classification of AMD, DME and NORMAL: Mean accuracy of more than 94%; Mean AUC: 0.99; Classification of diseased retina (AMD or DME) and healthy retina: The average accuracy, sensitivity and specificity were over 96%, 94% and 85%, respectively; Mean AUC: 0.99; N1; N2; N3 |
| Lemaître et al. [14] | 2016 | Proprietary | Local Binary Patterns | Sensitivity: 81.2%; Specificity: 93.7%; N1; N2; N3 |
| Alsaih et al. [15] | 2017 | Proprietary | Linear SVM | Sensitivity: 87.5%; Specificity: 87.5%; N1; N2; N3 |
| Srinivasan et al. [16] | 2014 | Public (DHU) | HOG descriptors and SVM classifiers | The accuracies of AMD, DME, and NORMAL were 100%, 100%, and 86.67%, respectively; N1; N2; N3 |
| Tan et al. [17] | 2018 | Public (Kasturba Medical College) | 14-layer deep CNN | Blindfold: Accuracy: 91.17%; Sensitivity: 92.66%; Specificity: 88.56%; Ten-fold: Accuracy: 95.45%; Sensitivity: 96.43%; Specificity: 93.75%; N1; N2; N3 |
| Gulshan et al. [18] | 2016 | Public (EyePACS-1 and Messidor-2) and proprietary | Combined the ten binary Inception V3 architecture as a deep neural network | On EyePACS-1 dataset: Mean sensitivity: 90.3%; Mean specificity: 98.1%; Mean AUC: 0.991; On Messidor-2 dataset: Mean sensitivity: 87.0%; Mean specificity: 98.5%; Mean AUC: 0.990; N1; N2; N3 |
| Lu et al. [19] | 2018 | Proprietary | Combined the four binary classifiers as a deep neural network | Ten-fold: Accuracy: 95.9%; Sensitivity: 94.0%; Specificity: 97.3%; AUC: 0.984; N1; N2; N3 |
| Li et al. [20] | 2019 | Proprietary | VGG-16 network | Accuracy: 98.6%; Sensitivity: 97.8%; Specificity: 99.4%; AUC: 100%; N1; N2; N3 |
| Karri et al. [21] | 2017 | Public (DHU) | Inception network | Mean accuracy: 99%, 89%, and 86%, respectively; N1; N2; N3 |
| Kermany et al. [22] | 2018 | Public (Mendeley database) | Inception V3 architecture | Accuracy: 96.6%; Sensitivity: 97.8%; Specificity: 97.4%; AUC: 0.999; N1; N2; Y3 |
| Fauw et al. [23] | 2018 | Proprietary | U-net and DenseNet | On the testing dataset 1 (Device Type 1): Accuracy: 94.5%; AUC: 0.9921; On the testing dataset 2 (Device Type 2): Accuracy: 96.6%; AUC: 0.9993; Y1; N2; N3 |
| Fang et al. [24] | 2019 | Public (UCSD dataset; NEH dataset) | LACNN | On UCSD dataset: Overall accuracy: 90.1%; Overall sensitivity 86.8%; Overall precision: 86.2%; On NEH dataset: Overall sensitivity: 99.33%; Overall precision: 99.39%; Overall AUC: 0.9940; Y1; N2; N3 |
| Rasti et al. [25] | 2018 | Public (NEH dataset; DHU dataset) | MCME model | On NEH dataset: Overall precision: 99.39%; Overall recall: 99.36%; Overall AUC: 0.998; On DHU dataset (optimal control parameter): Precision: 98.33%; Recall: 97.78%; AUC: 0.999; N1; N2; N3 |
| This study | 2019 | Proprietary | Multi-ResNet50 Ensembling | Ten-fold: Accuracy: 97.3%; Sensitivity: 96.3%; Specificity: 98.5%; AUC: 0.995; The best performance: Accuracy: 97.9%; Sensitivity: 96.8%; Specificity: 99.4%; AUC: 0.998; Kappa: 0.969; N1; Y2; Y3 |
DHU: Duke University, Harvard University, and University of Michigan; AMD: age-related macular degeneration; DME: diabetic macular edema; AUC: area under receiver operating characteristic; Y1: segmentation network and classification network for image classification tasks; N1: classification network for image classification tasks (no use of segmentation network); Y2: qualitative evaluation of model predictions; N2: no qualitative evaluation of model predictions; Y3: occlusion testing; N3: no occlusion testing; SVM: support vector machine; HOG: histogram of oriented gradient; CNN: convolutional neural network; curve; VGG: visual geometry group; UCSD: University of California San Diego; NEH: Noor Eye Hospital in Tehran; LACNN: lesion-aware convolutional neural network; MCME: multi-scale convolutional mixture of expert.
Tan et al. [17] utilized a deep convolutional neural network (CNN) to detect AMD, which achieved a mean accuracy of 91.17%, sensitivity of 92.66% and specificity of 88.56% using blindfold cross-validation strategy, and generated a mean accuracy of 95.45%, sensitivity of 96.43% and specificity of 93.75% with ten-fold cross-validation strategy. Gulshan et al. [18] applied deep learning to automatically detect diabetic retinopathy and DME in retinal fundus photographs, achieving a mean AUC of 0.991 with sensitivity of 90.3% and specificity of 98.1%, and a mean AUC of 0.990 with 87.0% sensitivity and 98.5% specificity on EyePACS-1 dataset and on Messidor-2 dataset, respectively. Although these methods could obtain promising results, they used the raw images to train the CNN from scratch requiring a large amount of training data and computation time to achieve a classification accuracy.
Lu et al. [19] developed a new intelligent system based on deep learning to automatically detect and differentiate multi-categorical abnormalities from OCT images, and a mean accuracy of 95.9% with 94.0% sensitivity, 97.3% specificity and a mean AUC of 0.984 was obtained. In their works, the image dataset was divided randomly into training, validation, and testing dataset at the image level. Multiple images from the same eye might include in the different partition (training, validation, or testing) so that the performance of model was biased. Li et al. [20] utilized deep transfer learning method based on the visual geometry group 16 (VGG-16) network to classify AMD and DME in retinal OCT images, which obtained a 98.6% accuracy, a 97.8% sensitivity, a 99.4% specificity, and a 100% AUC. Nevertheless, the construction of this model on an inadequate amount of data was susceptible to overfitting, and the validation dataset played the role of the testing dataset so that the reported performance could be biased. Karri et al. [21] described a transfer learning method based on Inception network to effectively identify retinal pathologies given retinal OCT images, and the mean of prediction accuracy across all validations for normal, AMD, and DME were 99%, 89%, and 86%, respectively. Nevertheless, gradient vanishing or gradient explode was prone to happen during the training process. The evaluation metric (only accuracy) was not extensive. In addition, this study focused only on addressing a “two diseases versus normal” task, and multiclass classifiers which could differentiate a specific abnormality among multi-categorical abnormalities, were more conformed to the clinical circumstances.
Recently, Kermany et al. [22] reported an accuracy of 96.6%, with a sensitivity of 97.8%, and a specificity of 97.4%, while the AUC was 0.999 in distinguishing urgent referrals (defined as CNV or DME) from drusen and normal exams. Furthermore, they also performed an occlusion testing to identify the areas contributing most to the neural network’s assignment of the predicted diagnosis. Yet, in this study, the validation dataset played the role of the testing dataset resulting in the biased reported performance. Meanwhile, 250 images from each category were chosen for the validation and testing while the numbers of images in each category in the training dataset were not the same (37,206 CNV, 11,349 DME, 8,617 DRUSEN, and 51,140 NORMAL), and this class imbalance might also affect the results. Furthermore, when the Inception V3 network with more layers used in this study was trained, gradient vanishing or gradient divergence was easier to happen. Also, the qualitative evaluation of model predictions and the detailed analysis of pathology regions identified by the occlusion testing were not performed. Fauw et al. [23] developed a deep learning architecture (an ensemble of segmentation and classification networks) to analyze clinical OCT scans and make referral suggestions, while its performance that reached or exceeded that of specialists on a range of sight-threatening retinal disorders was demonstrated. Although the performance increased benefiting from more segmentation model instances and more classification model instances, training time-complexity of the model was also increased and the performance of classification network heavily relied on that of segmentation network in this study. Fang et al. [24] proposed a lesion-aware convolutional neural network (LACNN) method to category retinal OCT images, and utilized retinal lesions detected by the lesion detection network (LDN) within OCT images to guide the CNN to focus on more discriminative information from local lesion-related regions for achieving more accurate classification (an overall accuracy of 90.1% and an overall sensitivity of 99.33% on the publicly UCSD dataset and NEH dataset, respectively). Rasti et al. [25] presented an ensemble model of multi-scale convolutional mixture of expert (MCME) to classify normal retina, dry AMD and DME, generating the precision rate of 98.86% and the AUC of 0.9985 with the MCME model of 4 scale-dependent experts. In these studies, the structure complexity of these deep learning networks was relatively high. The decision-making process of models was also not illustrated through qualitative evaluation of model predictions or occlusion testing excepting the study [22]. Moreover, the potential effect of integration of OCT images with the patients’ medical history data on model performance was not considered in these deep learning approaches [13–25].
Therefore, in order to address the afore-mentioned issues and maximize the clinical utility of automated detection, in this study we prepared and processed a relatively big dataset of retinal OCT images captured in real-world setting, and sought to explore the use of an ensemble of four improved ResNet50 to automatically classify CNV, DME, DRUSEN, and NORMAL for providing an accurate and timely detection of key pathology. Further, we performed qualitative evaluation of model predictions, and occlusion testing to help understand the decision-making process of the model. Also, we provided an analytical discussion on misclassification and pathology regions identified by the occlusion testing. Finally, we considered the potential benefit of integration of retinal OCT images with medical history data retrospectively collected from the corresponding patients. The proposed approach with high accuracy, high sensitivity, and high specificity, could potentially assist ophthalmologists to make a diagnostic decision.
2. Methods
2.1. Data collection
We conducted a retrospective study and collected a total of 21,357 retinal OCT images from 2,796 adult patients from the Shanghai Zhongshan Hospital and the Shanghai First People’s Hospital between 2014 and 2019. All images were deidentified to protect the privacy and security of patients’ health information prior to transfer to study investigators. These OCT images were used for the development and evaluation of the deep learning approach based on improved ResNet50. These OCT images were acquired with Heidelberg SD-OCT imaging systems (Spectralis software version 6.0a; Heidelberg spectralis, Heidelberg Engineering, Heidelberg, Germany). The baseline scan protocol consisted of: scan extent = volume scan (15°× 5°); scan sections = B-scans; 7 sections; and OCT automatic real time (ART) averaging, where ART was set to 20, which suggested that 20 SD-OCT images were averaged. This methodology obtained non-contact, cross-section frames in high resolution of the retina. Besides, we also retrospectively collected a short medical history of the patients like age, personal record of retinal disease, and personal retinal disorder related therapy. This study adhered to the tenets of the Declaration of Helsinki, and approved by the local ethics committee. Due to the anonymous and retrospective nature of this work, a waiver of informed consent was provided.
2.2. Image labeling and data augmentation
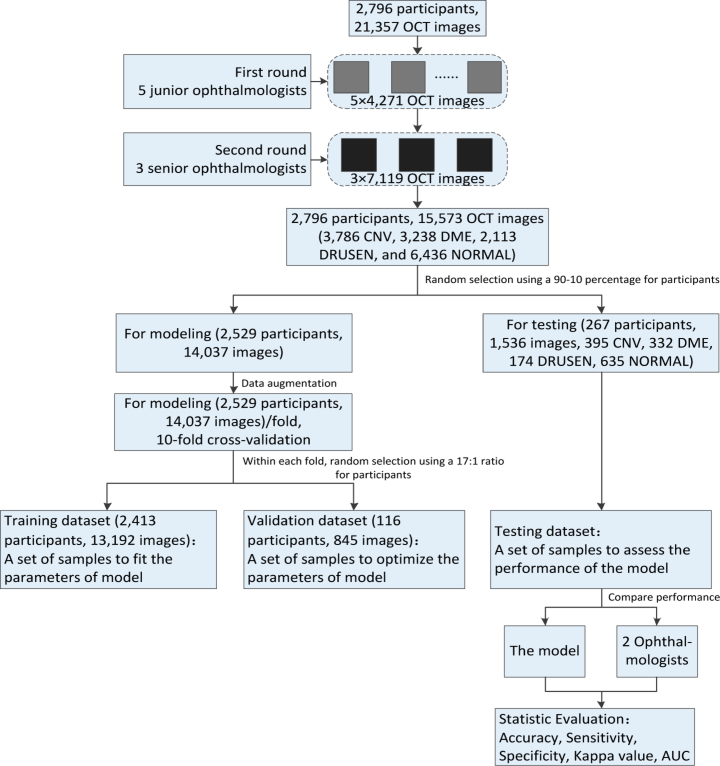
Prior to training the deep learning models, all OCT images were read and assessed by ophthalmologists, and graded for the present of CNV, DME, DRUSEN, and NORMAL. The gold standard annotation (image labeling) was performed by ophthalmologists. The current study invited 8 ophthalmologists to grade 21,357 OCT images. All images were reviewed masking other clinical information, and each ophthalmologist made a diagnosis independent to other ophthalmologists. In the first round, we randomly assigned these images to 5 junior ophthalmologists for screening and labeling, each of which reviewed about 4,271 OCT images. In the second round, 3 senior ophthalmologists were invited to verify and correct the labelling results, each of which reviewed about 7,119 OCT images. Senior ophthalmologists had approximately 10 to 15 years of experience, and junior ophthalmologists had up to 5 years of experience. The grading was performed on full-screen, high-resolution 27-in monitors. OCT images with severe artifacts causing misalignment and blurring of sections or significant image resolution reductions were removed directly. Only images with a clear consensus annotation between ophthalmologists were taken into the sample and imported into the database. For disagreement in image labels, they were adjudicated by an expert committee composed of senior ophthalmologists. The dataset selection and stratification process were displayed in Fig. 2. Eventually, the retinal OCT dataset for the experiment consisted of 15,573 images, of which 3,786 were affected by CNV, 3,238 were assigned images with DME appearances, 2,113 were related to DRUSEN cases, and the others represented healthy cases. We randomly selected 267 patients and obtained 1,536 images (395 CNV, 332 DME, 174 DRUSEN, 635 NORMAL) as testing dataset. The 14,037 images of another 2,529 patients were used as training dataset and validation dataset. We fitted and optimized the parameters of model on the training dataset and validation dataset, respectively, while the performance of our model was evaluated on the testing dataset.
Fig. 2.
Overall experimental design. OCT: optical coherence tomography; CNV: choroidal neovascularization; DME: diabetic macular edema; AUC: area under receiver operating characteristic curve.
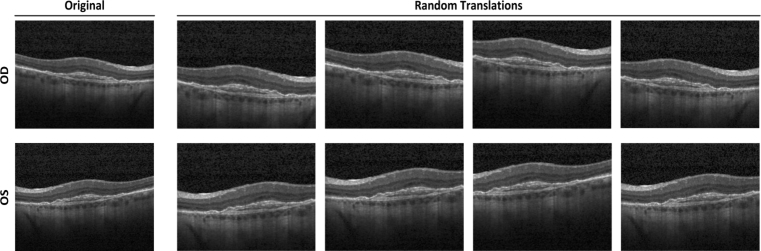
For the purpose of reflecting clinical care, increasing heterogeneity of OCT images within the training dataset, and reducing the possibility of overfitting, a data augmentation procedure was applied. Data augmentation can lead to better performing, more generalizable models which were invariant to certain types of image transformations and variations in image quality [26]. We first horizontally mirrored all images to mimic the inclusion of both oculus dexter (OD) and oculus sinister (OS) orientations of each image. Then, we applied a random cropping to all images including original and flipped images, and each cropped image was manually reviewed to ensure that each correctly retained pathological area. Through random cropping, the pathology areas of each image were randomly perturbed by a small amount, because the locations of these regions in the OCT images were not always identical in clinical care. The random cropping was repeated five times for each image. In total, 140,370 OCT images arising from (14,037 images) × (2 orientations) × (5 random crops) were obtained. We assigned each augmented image to the same label (CNV, DME, DRUSEN, and NORMAL) as the corresponding original input image. An example of input image and the corresponding augmentations was shown in Fig. 3. After augmentation, we used the Gaussian pyramid down sampling method [27] to establish a uniform size of 224 × 224 dimension for all images in the dataset as input to our model so that the computational requirements of our model could be satisfied.
Fig. 3.
Example of input CNV image (top left) and images arising from data augmentation. Each input image was augmented through horizontal mirroring to imitate alternative OD/OS orientation and random cropping to introduce small amount of perturbation. CNV: choroidal neovascularization; OD: oculus dexter; OS: oculus sinister.
2.3. Multi-ResNet50 ensembling for image recognition
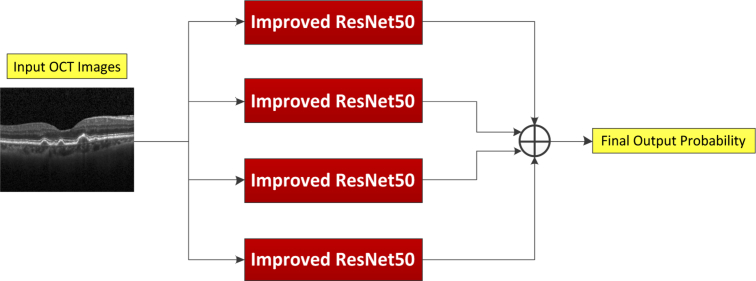
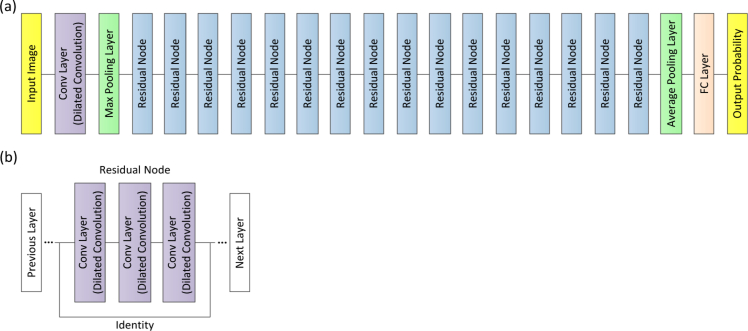
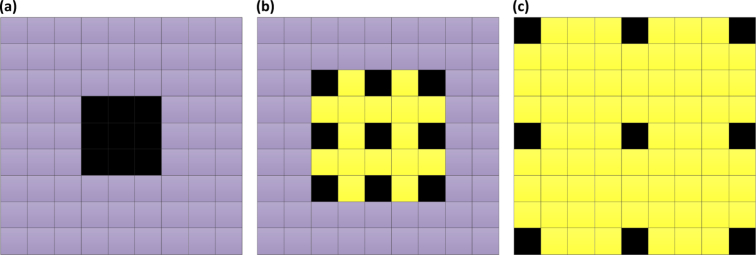
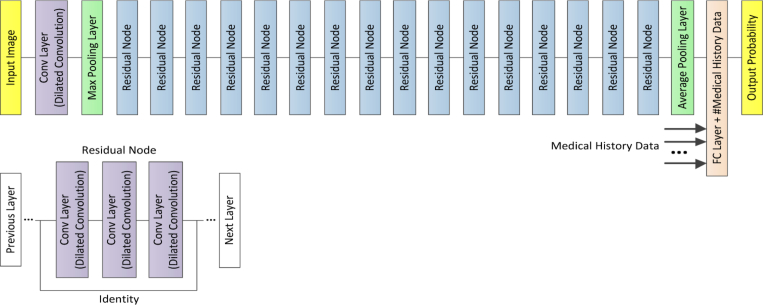
In the present work, our approach used an ensemble of four classification model instances as shown in Fig. 4, each of which was based on an improved ResNet50. The improved ResNet50 mainly consisted of convolutional layers, pooling layers, and fully connected layers, as shown in Fig. 5. The convolutional layers extracted features and transformed input images into hierarchical feature maps. The pooling layers (including max pooling and average pooling) merged semantically similar features into one to reduce the dimensionality of the extracted features. The fully connected layers combined these features and produced an image-level classification. In the improved ResNet50, the dilated convolution was introduced into the convolutional layers so that the constant resolution of the network could be guaranteed and the loss of resolution of the image space was minimized. The principle of dilated convolution was shown in Fig. 6. Figure 6(a) represented 3×3 convolution kernel with dilated rate of 1 corresponding to normal receptive field. Figure 6(b) displayed 3×3 convolution kernel with dilated rate of 2 corresponding to the receptive field of 5×5 normal convolution kernel. Figure 6(c) showed 3×3 convolution kernel with dilated rate of 4 equivalent to the receptive field of 9×9 normal convolution kernel. It was illustrated from Fig. 6 the dilated convolution supported increasing the receptive field of the convolution kernel without increasing the kernel parameters and avoiding excessive loss of resolution of the feature map. In addition, the batch normalization layer was added after each convolution layer to keep the input value of the nonlinear transformation function fall into the region sensitive to the input, and speed up the training by reducing internal covariate shift. Meanwhile, the improved ResNet50 could tackle the vanishing gradient problem and the gradient divergence problem by using the batch normalization and shortcut connections which skipped one or more layers when training deep networks (Fig. 5(b)). When performing image classification tasks, the same improved ResNet50 with a different order of the inputs and different random weight initalizations was trained. The final prediction was computed by the arithmetic average of class probabilities estimated by the constituent networks. Beside the benefits of uncertainty measure, the overall performance was also significantly improved using multi-ResNet50 ensembling by contrast to a single model instance.
Fig. 4.
Final predictions were yielded using an ensemble of four improved ResNet50 architectures, which was computed by the arithmetic average of class probabilities estimated by the constituent networks. OCT: optical coherence tomography.
Fig. 5.
(a) Schematics of the improved ResNet50 architecture. (b) The residual node was used as building block for the improved ResNet50 architecture. Conv: convolutional; FC: fully connected.
Fig. 6.
The receptive field with three different dilated rates.
Considering directly training each improved ResNet50 from scratch required a huge amount of data and computation time, we further utilized deep transfer learning technology which could apply features learned to perform one task to other tasks [28,29]. The improved ResNet50 used in this study was pre-trained on the ImageNet database containing thousands of different objects and scenes, and universal features learned from the pre-training could be reused for triage of retinal OCT images (CNV, DME, DRUSEN, and NORMAL) by transfer learning. Therefore, transfer learning not only greatly accelerated the training of improved ResNet50, but also made it possible to achieve a highly accurate model with a relatively small training dataset.
2.4. Model training
The image dataset for model training was separated randomly into multiple folds such that 10-fold cross-validation could be conducted to estimate and optimize our model. Within each fold, the dataset was split into independent training and validation datasets in a 17:1 ratio performed at the participant (not image) level, which meant that all images from a participant were contained in the same section (training or validation). Our model parameters were fitted and optimized on the corresponding training dataset and the validation dataset, respectively. The performances of the models were evaluated on the independent testing dataset consisting of only images of novel eyes that had not been encountered by the model during training. Among the 10 training models, the model with the best results was considered as the best model in this study. For training, a total of 10,000 steps with batch size of 200 images per step with a learning rate of 10−5 were performed, since the performance of validation dataset would not be further improved since then. Additionally, we also independently trained 3 ensembling binary classifiers to determine a breakdown of our model’s performance. Further, we selected the model with the highest performance on the testing dataset for comparing to results obtained by two ophthalmologists with rich clinical experience. The model was trained, validated, and tested on an Ubuntu 16.04 operation system with Intel Core i7-2700K 4.6 GHz CPU, 256 Gb RAM, Dual AMD Filepro 512 Gb PCIe-based flash storage, and NVIDIA GTX 1080 8 Gb GPU. Table 2 summarized training data and parameters.
Table 2. Summary of the parameters used in training the improved ResNet50.
| Parameter | Value |
|---|---|
| Weight Initialization | Pre-trained on ImageNet |
| Training dataset sample size/fold | 2,413 participants, 13,192 images (3,184 CNV, 2,731 DME, 1,825 DRUSEN, and 5,452 NORMAL) |
| Validation dataset sample size/fold | 116 participants, 845 images (207 CNV, 175 DME, 114 DRUSEN, and 349 NORMAL) |
| Testing dataset sample size/fold | 267 participants, 1,536 images (395 CNV, 332 DME, 174 DRUSEN, and 635 NORMAL) |
| Input image size | 224×224 pixels |
| Network output | Softmax probability of CNV, DME, DRUSEN, and NORMAL |
| Number of training steps | 10,000 |
| Learning rate | 10−5 |
CNV: choroidal neovascularization; DME: diabetic macular edema.
2.5. Visualizing model decisions
Considering the “black boxes” problem has been identified as an impediment to the application of deep learning in healthcare [30], we performed qualitative assessment of exemplar CNV, DME, DRUSEN, and NORMAL images along with model predictions, and occlusion testing on the entire set of testing images to help open this black box and understand the decision-making process of model. We invited an ophthalmologist to perform qualitative evaluation, by reviewing images on the testing dataset for which the model generated high confidence true predictions, relatively high confidence false predictions, and low confidence borderline predictions. Several random examples of each of these predictions from the test sample were selected to help illustrate model decisions and images that led to model errors. Through the use of occlusion testing [31], we could visualize the parts of the image which were most important to the deep learning classification. A blank 28×28 pixel window was overlaid on an image before applying the model to quantify the impact of each area on the model prediction. Then, this window was systematically slid over the entire image, and the probabilities of retinal disorders were recorded. The highest drop in the probability indicated the regions of OCT image with greatest impact on the deep learning algorithm classification. The interpretability of the predictive impact of features used in our model could help to assess whether the decision was based on key clinical features.
2.6. Statistical evaluation
We implemented overall accuracy, sensitivity, specificity, and AUC metrics with 95% CI to evaluate the performance of our model, and compared the performance of the best model to results obtained by ophthalmologists. Accuracy was calculated by dividing the number of correctly labeled images by the total number of test images. Sensitivity and specificity were derived by dividing the total number of correctly labeled abnormal and the total number of correctly labeled normal respectively by the total number of test images. Receiver operating characteristic (ROC) curves plotted by varying the operating threshold were used to assess the ability of our model on retinal OCT images in discriminating abnormality from normal. It provided the tradeoff between the sensitivity and 1-specificity. AUC was used to summarize the diagnostic accuracy of each parameter. The AUC of effective model ranging between 0.5 and 1 was higher, the performance of model was better. We also calculated a kappa value to quantify the degree of agreement between the best performance model and two ophthalmologists for each diagnostic category. The larger kappa value ranging from 0 to 1 meant better reliability. Statistical analyses were carried out using STATA version 14 and the Python package, SciPy version 0.19.1.
3. Results
3.1. Model evaluation on development dataset
Model performance was evaluated at the B-scan level on the independent testing dataset. In the multiclass comparison between CNV, DME, DRUSEN, and NORMAL, our model achieved an accuracy of 0.973 (95% CI, 0.971-0.975) with a sensitivity of 0.963 (95% CI, 0.960-0.966) and a specificity of 0.985 (95% CI, 0.983-0.987). A ROC curve was generated to access its capacity of separating normal from three abnormalities. A high AUC of 0.995 (95% CI, 0.994-0.996) was obtained. Table 3 summarized the performance of model at the B-scan level for detection of the three abnormalities.
Table 3. Performance of model at the B-scan level for detection of the three abnormalities.
| Accuracy (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | AUC (95% CI) |
|---|---|---|---|
| 0.973 (0.971–0.975) | 0.963 (0.960–0.966) | 0.985 (0.983–0.987) | 0.995 (0.994–0.996) |
CI: confidence interval; AUC: area under receiver operating characteristic curve.
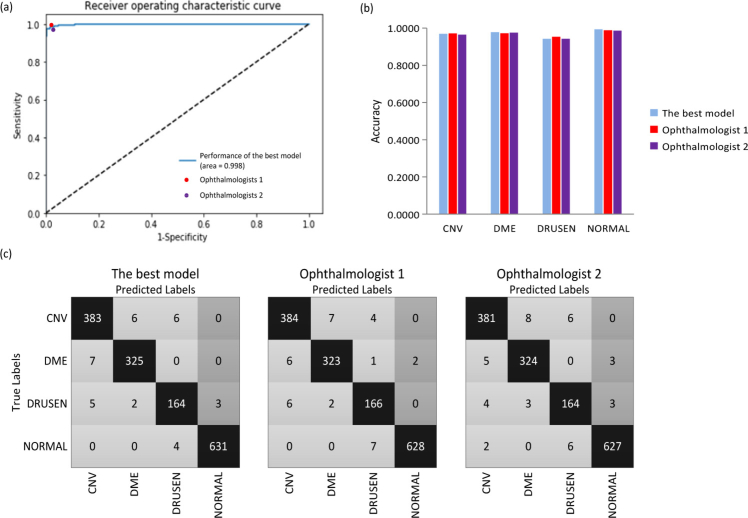
We further compared the performance of the best model to results obtained by two ophthalmologists with significant clinical experience at the B-scan level. The best model in Table 4 referred to the model with the best results obtained on the independent testing dataset using the 10-fold cross-validation strategy. The best model yielded an accuracy of 0.979, with a sensitivity of 0.968, a specificity of 0.994, while ophthalmologist 1 and ophthalmologist 2 got 0.977, 0.974 accuracy, 0.969, 0.964 sensitivity, and 0.989, 0.987 specificity, respectively (See Table 4). The kappa value for the best model was 0.969, slightly higher than those for the two ophthalmologists (0.968 and 0.963). The ROC curve for retinal abnormal detection with ophthalmologist performance was plotted for comparison at the B-scan level, where the AUC reached up to 0.998, as seen in Fig. 7(a). It was demonstrated from Fig. 7(a) the performance of our best model was similar to that of the two ophthalmologists.
Table 4. Performance of the best model and ophthalmologists at the B-scan level for detection of the three abnormalities.
| Accuracy | Sensitivity | Specificity | Kappa | |
|---|---|---|---|---|
| The best model | 0.979 | 0.968 | 0.994 | 0.969 |
| Ophthalmologist 1 | 0.977 | 0.969 | 0.989 | 0.968 |
| Ophthalmologist 2 | 0.974 | 0.964 | 0.987 | 0.963 |
Fig. 7.
(a) ROC curve for retinal diseases detected by the best model, with the operating points of the two ophthalmologists shown for comparison at the B-scan level. The curve showed that the best model demonstrated a performance that rivalled that of the two ophthalmologists. (b) A bar diagram comparing the specific accuracy of each category between the best model and two ophthalmologists at the B-scan level. (c) Three confusion matrixes for the best model and the two ophthalmologists’ predictions at the B-scan level, respectively. ROC: receiver operating characteristic; CNV: choroidal neovascularization; DME: diabetic macular edema.
The specific accuracy of each category obtained from the best model and the two ophthalmologists was reported at the B-scan level in Table 5. The results showed that the best model could correctly identify the three abnormalities and the normal control with 0.970, 0.979, 0.943, and 0.994 accuracy, respectively. At the B-scan level, the best model manifested superior compared to results derived from ophthalmologists in detecting DME and NORMAL, whereas the two ophthalmologists exhibited slightly better for DRUSEN. In the case of CNV, the best model’s accuracy was 0.005 better than ophthalmologist 2, with 0.002 poorer than ophthalmologist 1, as shown in Fig. 7(b).
Table 5. The specific accuracy of each category obtained from the best model and the two ophthalmologists at the B-scan level .
| Category | The best model | Ophthalmologist 1 | Ophthalmologist 2 |
|---|---|---|---|
| CNV | 0.970 | 0.972 | 0.965 |
| DME | 0.979 | 0.973 | 0.976 |
| DRUSEN | 0.943 | 0.954 | 0.943 |
| NORMAL | 0.994 | 0.989 | 0.987 |
CNV: choroidal neovascularization; DME: diabetic macular edema.
The specific assignment of each B-scan of different predictions regarding the correlation of their predicted labels with the true labels was depicted as confusion matrices in Fig. 7(c). As can be seen from Fig. 7(c), misclassification cases and types in the best model were comparable to those from the two ophthalmologists.
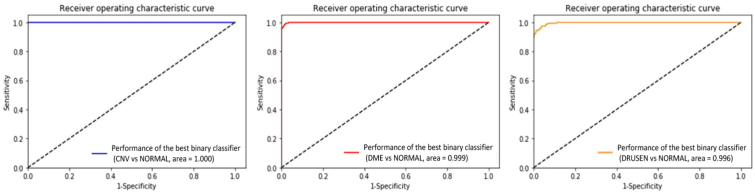
In addition, with the purpose of determining a breakdown of our model’s performance at the B-scan level, we implemented 3 ensembling binary classifiers to discriminate CNV/DME/DRUSEN from NORMAL using the corresponding OCT images. The 3 ensembling binary classifiers obtained 0.983 (95% CI, 0.981-0.985), 0.988 (95% CI, 0.987-0.989), 0.978 (95% CI, 0.976-0.980) accuracy, 0.984 (95% CI, 0.981-0.987), 0.986 (95% CI, 0.984-0.988), 0.981 (95% CI, 0.980-0.982) sensitivity, and 0.993 (95% CI, 0.992-0.994), 0.991 (95% CI, 0.990-0.992), 0.986 (95% CI, 0.984-0.998) specificity, while the AUC was 0.997 (95% CI, 0.996-0.998), 0.996 (95% CI, 0.995-0.997), 0.993 (95% CI, 0.991-0.995), respectively (See Table 6). The ROC curves of the best 3 ensembling binary classifiers at the B-scan level were as depicted in Fig. 8. The results of binary classification manifested a good breakdown of the model’s performance.
Table 6. Performance of the 3 ensembling binary classifiers at the B-scan level.
| CNV vs NORMAL | DME vs NORMAL | DRUSEN vs NORMAL | |
|---|---|---|---|
| Accuracy (95% CI) | 0.983 (0.981–0.985) | 0.988 (0.987–0.989) | 0.978 (0.976–0.980) |
| Sensitivity (95% CI) | 0.984 (0.981–0.987) | 0.986 (0.984–0.988) | 0.981 (0.980–0.982) |
| Specificity (95% CI) | 0.993 (0.992–0.994) | 0.991 (0.990–0.992) | 0.986 (0.984–0.998) |
| AUC (95% CI) | 0.997 (0.996–0.998) | 0.996 (0.995–0.997) | 0.993 (0.991–0.995) |
CI: confidence interval; CNV: choroidal neovascularization; DME: diabetic macular edema; AUC: area under receiver operating characteristic curve.
Fig. 8.
ROC curves of the best 3 ensembling binary classifiers to discriminate CNV, DME, and DRUSEN from NORMAL at the B-scan level. ROC: receiver operating characteristic; CNV: choroidal neovascularization; DME: diabetic macular edema.
3.2. Model evaluation on publicly DHU dataset and UCSD dataset
In this section, we firstly evaluated the performance of our model on the DHU dataset [16]. We adopted the same leave-three-out cross-validation as the study by Srinivasan et al., and conducted 45 experiments on the DHU dataset. In each experiment, our model was trained on 42 volumes, excluding one volume from each class, and tested the three volumes excluded from training. An entire volume is classified as the mode of the individual image classification results. We transferred our model pre-trained on our dataset and implanted the parameters of the pre-trained CNN model into the transferred CNN, while weight parameters of the output layer were initialized using Gaussian distribution. After fine-tuning, we investigated the performance of our model on the corresponding DHU dataset. Our approach correctly identified 93.3% cases with AMD, 100% cases with DME, and 100% cases of normal, which indicated that our approach manifested better performance compared to the method proposed by Srinivasan et al. (See Table 7). Additionally, in order to have a fair comparison to the study by Srinivasan et al. [16], we retrained and reevaluated our model without using the pre-trained weights on DHU dataset, in the same way as mentioned before. Our model still correctly detected 93.3% cases with AMD, 100% cases with DME, and 93.3% cases of normal, achieving a rather comparable performance to that by Srinivasan et al. [16] (See also Table 7).
Table 7. Fraction of volumes correctly classified during cross-validation.
| Methods | Classes | Fraction of volumes correctly classified |
|---|---|---|
| HOG descriptors and SVM classifiers [16] | AMD | 15/15 = 100% |
| DME | 15/15 = 100% | |
| NORMAL | 13/15 = 86.7% | |
| Our model (with the pre-trained weights on our development OCT dataset) | AMD | 14/15 = 93.3% |
| DME | 15/15 = 100% | |
| NORMAL | 15/15 = 100% | |
| Our model (without using the pre-trained weights) | AMD | 14/15 = 93.3% |
| DME | 15/15 = 100% | |
| NORMAL | 14/15 = 93.3% | |
HOG: histogram of oriented gradient; SVM: support vector machine; AMD: age-related macular degeneration; DME: diabetic macular edema; OCT: optical coherence tomography.
Then, we also investigated the performance of our deep learning approach on the UCSD dataset [24], and made a comparison with their work. The UCSD dataset consists of 84,484 OCT B-scans (8,866 DRUSEN, 37,455 CNV, 11,598 DME, and 26,565 NORMAL) obtained from 4,686 patients. Our model was trained in the same way as mentioned before. During our experiments, the whole UCSD datasets were sequentially split into six subsets. In each experiment, we trained our model on one subset and tested it on the remaining five subsets. We conducted repeatedly six times experiments with each of the six subsets used once as the training dataset. Finally, the experimental results were average values of all six experiments. Classification performance was assessed based on the same evaluation metrics [24]. The classification results were shown in Table 8. As can be observed from Table 8, our approach generated an overall accuracy (OA) of 90.4%, an overall sensitivity (OS) of 87.2%, and an overall precision (OP) of 86.7%, which indicated that the performance was comparable between our proposed approach and the LACNN method. In the time-complexity, the average training and test time for our approach was approximate 6.79 and 0.87 millisecond per image, respectively while the average training and test time for the LACNN network was about 9.5 and 1.2 millisecond per image, respectively. This illustrated that the time complexity of our approach was lower than that of LACNN. Further, for a fair comparison between our approach and LACNN method proposed by Fang et al. [24], we retrained our model without using the pre-trained weights on the UCSD dataset. In our experiments, the hyper-parameters for our approach were set to the basically same as that of LACNN (such as a batch of 24 images per step, an initial learning rate of 0.00001, and weight decay factor of 0.0002). We employed the same training setup and evaluation method as before. Our approach achieved an OA of 90.2%, with an OS of 86.9%, and an OP of 86.5% at the B-scan level (See Table 8). The result demonstrated that the performance was comparable between our approach and the LACNN method. In addition, we also measured the time-complexity of our approach. For our approach, the average training time was around 8.3 millisecond per image, while the average test time was approximate 0.88 millisecond per image. However, the average training and test time for the LACNN network was about 9.5 and 1.2 millisecond per image, respectively. Therefore, this showed that the time-complexity of our approach was still lower than that of LACNN.
Table 8. Classification results (in percentage) at the B-scan level on UCSD dataset.
| Methods | Classes | ACC | SE | PR | SP | AUC | OA | OS | OP |
|---|---|---|---|---|---|---|---|---|---|
| LACNN [24] | CNV | 92.7 ± 1.5 | 89.8 ± 4.5 | 93.5 ± 1.3 | 95.1 ± 1.6 | 97.7 ± 0.5 | 90.1 ± 1.4 | 86.8 ± 1.3 | 86.2 ± 2.3 |
| DME | 96.6 ± 0.2 | 87.5 ± 1.5 | 86.4 ± 1.6 | 98.0 ± 0.3 | 97.4 ± 0.4 | ||||
| DRUSEN | 93.6 ± 1.4 | 72.5 ± 7.9 | 70.0 ± 5.7 | 95.9 ± 2.1 | 93.4 ± 1.5 | ||||
| NORMAL | 97.4 ± 0.2 | 97.3 ± 1.0 | 94.8 ± 1.1 | 97.4 ± 0.5 | 99.2 ± 0.2 | ||||
| Our model (with the pre-trained weights on our development OCT dataset) | CNV | 92.3 ± 1.2 | 88.4 ± 3.1 | 93.7 ± 1.5 | 95.7 ± 1.5 | 97. 2 ± 0.4 | 90.4 ± 1.2 | 87.2 ± 1.4 | 86.7 ± 1.8 |
| DME | 97.1 ± 0.2 | 90.1 ± 1.2 | 94.3 ± 1.3 | 98.2 ± 0.4 | 97.8 ± 0.3 | ||||
| DRUSEN | 93.7 ± 1.3 | 73.1 ± 6.5 | 71.2 ± 4.3 | 96.3 ± 1.8 | 93.9 ± 1.1 | ||||
| NORMAL | 98.3 ± 0.2 | 98.1 ± 0.7 | 96.2 ± 1.0 | 98.4 ± 0.3 | 99.8 ± 0.08 | ||||
| Our model (without using the pre-trained weights) | CNV | 92.1 ± 1.3 | 88.1 ± 3.3 | 93.4 ± 1.6 | 95.5 ± 1.6 | 97. 1 ± 0.5 | 90.2 ± 1.3 | 86.9 ± 1.5 | 86.5 ± 2.0 |
| DME | 96.9 ± 0.2 | 89.2 ± 1.3 | 93.1 ± 1.4 | 98.1 ± 0.5 | 97.7 ± 0.4 | ||||
| DRUSEN | 93.5 ± 1.5 | 72.8 ± 7.1 | 70..7 ± 4.9 | 96.1 ± 1.9 | 93.7 ± 1.2 | ||||
| NORMAL | 97.9 ± 0.2 | 97.7 ± 0.8 | 95.6 ± 1.1 | 98.0 ± 0.4 | 99.5 ± 0.1 | ||||
UCSD: University of California San Diego; ACC: accuracy; SE: sensitivity; PR: precision; SP: specificity; AUC: area under receiver operating characteristic curve; OA: overall accuracy; OS: overall sensitivity; OP: overall precision; LACNN: lesion-aware convolutional neural network; CNV: choroidal neovascularization; DME: diabetic macular edema; OCT: optical coherence tomography.
3.3. Visualizing model decisions
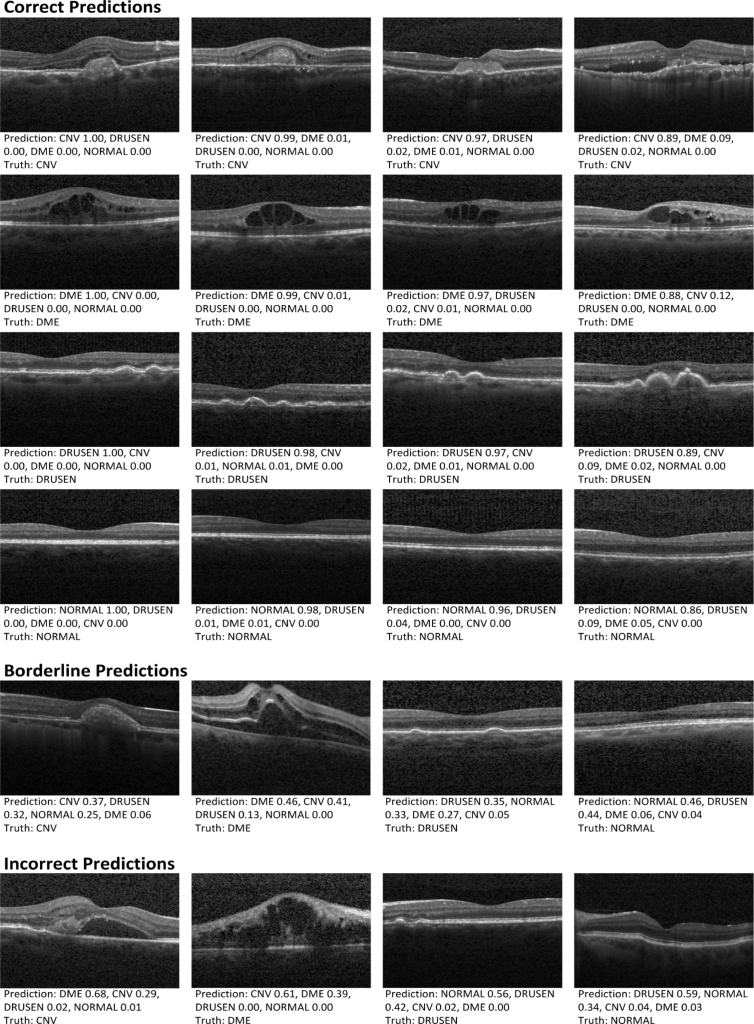
Figure 9 provided several random case examples from the test sample, which consisted of correct, incorrect, and borderline examples with the best model predictions and ophthalmologist-based truth. Among the correct retinal disorder predictions, hallmarks of CNV, DME, and DRUSEN were visible (CNV with neovascular membrane and associated subretinal fluid, DME with retinal-thickening-associated intraretinal fluid, and DRUSEN with undulations and elevations of retinal pigment epithelium (RPE) hyper-reflective band). In both the borderline and the incorrect cases, some examples confused CNV and DME as shown in Fig. 9. These examples with CNV and DME exhibited similar characteristics like subretinal fluid accumulation. Figure 9 also showed few mix-ups of DRUSEN and NORMAL. For the normal eye, RPE was usually evident as a highly concave backscattering layer posterior to the retina. Slightly convex in RPE may lead to confusion of DRUSEN and NORMAL in some conditions.
Fig. 9.
Predictions of the best model compared to ophthalmologist-based truth. Examples of correct, incorrect, and borderline predictions included the ophthalmologist truth and the best model prediction probabilities. The prediction probability value was between 0 and 1, and the category of an image identified by the algorithm was determined by the largest value. CNV: choroidal neovascularization; DME: diabetic macular edema.
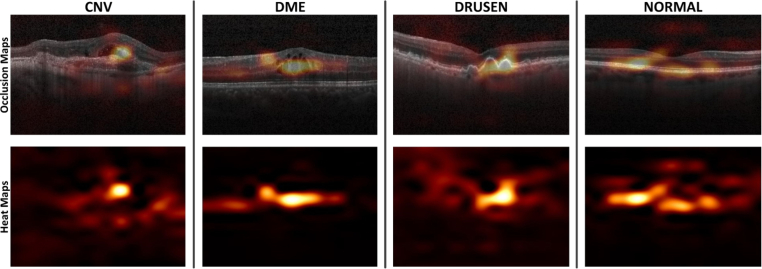
Occlusion testing was performed on the testing dataset to identify the regions which had the greatest impact on model decisions. As illustrated in Fig. 10, occlusion testing maps were shown for each group to illustrate regions most important to identifying symptoms. For the CNV eyes, subretinal/outer retinal hyper-reflective materials associated with the pigment epithelial detachment were identified as the most important regions and the intraretinal fluid contributed comparatively little to model decisions. For the DME eyes, the sub- and intra-retinal fluid accumulation were the key features for identification. The DRUSEN eyes were recognized as mound-like elevations with defined margins. In the normal eye, the RPE hyper-reflective band was identified as the region of interest. These regions recognized by occlusion testing were also validated by ophthalmologists to be the most clinically significant regions of pathology.
Fig. 10.
Occlusion testing maps showing most significant regions for detecting retinal diseases. In these images, golden regions indicated a large impact on model predictions while orange and red regions indicated a very limited impact on predictions. The heat map was created after prediction by assigning the softmax probability of the correct label to each occluded area. The occlusion map was generated by superimposing the heat map on the input image. CNV: choroidal neovascularization; DME: diabetic macular edema.
3.4. Integration of medical history data and multi-ResNet50 ensembling
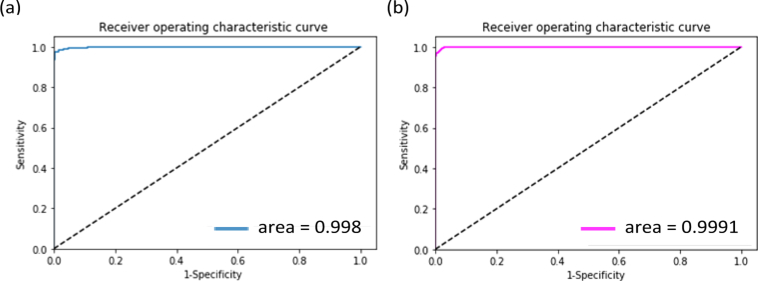
For the purpose of measuring the performance of the integration of retinal OCT images and medical history data of patients, we applied retinal OCT images and a short medical history data retrospectively collected from the corresponding patients to train the improved ResNet50 by introducing the raw data into the last fully connected layer of the network, as shown in Fig. 11. According to some major risk factors reported in the literature [23], we retrospectively selected some relevant medical history data of patients which was used in its raw format. The short medical history data utilized in this study mainly contained the age, personal retinal disease related therapy, and the personal record of retinal disorder. The values of the age were normalized to between 0 and 1. “Personal record of retinal disorder” contained patients’ historical diagnosis information relevant to three common retinal diseases (DME, CNV, and DRUSEN). “Personal retinal disease related therapy” included historical treatment courses and outcomes of the patients’ retinal disease (DME, CNV, and DRUSEN). When the patient suffered from DME, or CNV, or DRUSEN, the value of “Personal record of retinal disorder” was encoded as 1, otherwise its value was encoded as 0. If the patient with any of three retinal diseases had received prior treatment, the value of “Personal retinal disease related therapy” was encoded as 1. On the contrary, its value was set to 0. Although “Personal record of retinal disorder” and “Personal retinal disease related therapy” were available at the OCT level, we chose a part of B-scans relevant to three common retinal diseases including DME, CNV, and DRUSEN from the patient’s volume, and the corresponding information of “Personal record of retinal disorder” and “Personal retinal disease related therapy” from the same patient, resulting in a one-to-one correspondence between these B-scans and these metadata. Then, we integrated these B-scans and the corresponding information of “Personal record of retinal disorder” and “Personal retinal disease related therapy” to train our model. In this experiment, we still implemented 10-fold cross-validation to retrain the improved ResNet50 on the training dataset and validation dataset with the corresponding medical history data (including retinal OCT images from the previous training dataset and validation dataset plus medical history data of corresponding patients). After 10,000 steps, the training was stopped due to the absence of further improvement of the performance on the validation dataset since then. We carried out the experiment on the same independent testing dataset. Among the 10 training models, the model with the best results was regarded as the best model in this study. The performance of the best model was summarized in Table 9 and Fig. 12. It could be seen in Table 9 and Fig. 12 that the sensitivity and specificity had a slight improvement, without significant difference in the AUC by introducing medical history data. The results indicated that it could be of great value to improve the discriminative ability of retinal diseases by introducing medical information.
Fig. 11.
The improved ResNet50 architecture incorporating the medical history data and retinal OCT images, where the medical history data were integrated into the last fully connected layer of the network. Conv: convolutional; FC: fully connected.
Table 9. The comparison of AUC, sensitivity and specificity between considering only the retinal OCT images and integrating the images with medical data.
| Group | Sensitivity | Specificity | AUC |
|---|---|---|---|
| Only the retinal OCT images | 0.968 | 0.994 | 0.998 |
| Fusion age / personal record of retinal disorder / personal retinal disease related therapy | 0.979 | 0.997 | 0.9991 |
AUC: area under receiver operating characteristic curve; OCT: optical coherence tomography; CI: confidence interval.
Fig. 12.
ROC curves of the integration of retinal OCT images and medical history data. (a) displayed ROC curve with only retinal OCT images. (b) described ROC curve considering the integration of retinal OCT images with age, personal retinal disease related therapy, and the personal record of retinal disorder. ROC: receiver operating characteristic; OCT: optical coherence tomography.
4. Discussion
In the present study, we prepared and processed a relatively big dataset of retinal OCT images captured in real-world setting, and presented a novel ensemble of four classification model instances to detect three most common blinding retinal diseases from OCT images automatically and reliably, each of which was based on an improved ResNet50. The detection performance of this approach was validated using independent testing dataset and two publicly datasets (including DHU dataset and UCSD dataset). The results suggested that the approach described here had high accuracy, sensitivity, specificity, and AUC for implementing automated categorization of retinal disorders from OCT images, and achieved the performance at a level equivalent or better than that of ophthalmologists. Meanwhile, the results of binary classification validated a good breakdown of the model’s performance. Furthermore, we also further performed the qualitative assessment of exemplar CNV, DME, DRUSEN, and NORMAL images with model predictions, and occlusion testing to illustrate the decision-making process of the model. The results confirmed that our model made its decisions using accurate distinguishing features from OCT images which was in accordance with ophthalmologist assessment. Finally, we investigated the performance of the incorporation of retinal OCT images and medical history data of patients. The results showed a slight improvement in sensitivity and specificity with similar AUC values.
Although OCT could offer non-invasive real-time, high-resolution imaging on the structure of the retina and had become the mainstay of retinal diagnosis in clinical routine, subjective qualitative OCT image evaluation required significant clinical training and agreement was also limited, even among ophthalmologists with rich clinical experience [2,4,12,32]. Furthermore, it was inherently impractical to manually inspect large OCT datasets in busy clinics, and prone to errors [9]. An automated method in clinical practice was extremely useful to detect retinal diseases [33]. The proposed automated detection approach provided quick, objective and consistent image interpretation. There was no need to focus on the underlying retinal disorder process, and the results produced on the training dataset offered by historical diagnostic decisions determined their performances. The approach described here achieved a high level accuracy of 0.973 (95% CI, 0.971-0.975), with a sensitivity of 0.963 (95% CI, 0.960-0.966) and a specificity of 0.985 (95% CI, 0.983-0.987), while the AUC was 0.995 (95% CI, 0.994-0.996) at the B-scan level. In contrast to the previous studies (as displayed in Table 1), our model exhibited better performance than results obtained in [13–15,17,19,21], and achieved comparable performance to those in [16,18,20,22–25]. This suggested that it may be able to accurately detect CNV, DME, DRUSEN, and NORMAL while reducing the burden of false positives.
Most deep learning studies based on OCT images concentrated on the image segmentation involving complicated feature extraction or selection [10,11,34–37], which was time-consuming and required considerable skill and domain expertise to annotate the imaging data. Furthermore, a minor error introduced in segmentation would result in misalignment and misclassification [38,39]. Nonetheless, the deep learning approach adopted in this paper obviated such problems through learning predictive characteristics directly from the OCT images. Our model could automatically learn the richer and more distinct OCT image features for more accurate classification than CNN with shallower architectures [40–43]. This autonomous behavior could provide an opportunity to capture clinically important features or patterns of eye diseases which may not be detectable by the human eye. In addition, we adopted data augmentation method to increase the amount and type of variation of OCT images within the training data and reflect clinical care so that our model performance and generalizability could be improved.
Gradient vanishing or gradient explode may happen with the network depth increasing in deep learning. However, the improved ResNet50 in this study could tackle this problem by incorporating the batch normalization layers and the residual layers, which helped to aid the model achieve convergence during training, and gain significantly higher performance from a relatively deep network than obtained from shallower networks when conducting OCT image categorization tasks [40,44]. Sufficient data was the premise for good performance in deep learning. Unfortunately, it was difficult to collect an immense amount of retinal OCT images as the underlying datasets with the gold standard qualified by ophthalmologists in practical. Even if collected, the training would also require several days to adjust the huge parameters for model convergence, while a multi-class holdout model trained using transfer learning only spent about 3 hours in finishing training, validation, and testing on the corresponding datasets. Each binary classification could yield a high accuracy in under 1 hour. Therefore, initializing models via transfer learning was an important approach that should be considered whenever training a CNN to perform a new task, especially when limited data was a concern.
Deep learning commonly required a huge amount of data to increase the generalizability of the learnt model [45]. The construction of a model with data-driven feature quantifiers on an inadequate amount of data was susceptible to overfitting [46], which had a negative impact on performance during testing. In order to reduce the possibility of this problem and increase the robustness of the model, we applied two different strategies. First, the selection of the number of steps to complete the training process was stopped when the absence of further improvement in the performance on the validation dataset occurred. Second, we applied data augmentation method, such as horizontal flipping, and random cropping. These data augmentation techniques increased the amount and the diversity of OCT images within the training data, significantly improving our model performance and generalizability.
Deep learning based review of OCT images could also be useful in eye disease management as part of clinical decision support systems. The quantitative CNV, DME, DRUSEN, and NORMAL probabilities generated at the B-scan level by our model acted as a confidence score with respect to the eye disease prediction. When a score between 0.25 and 0.50 was produced, it indicated additional scrutiny compared to a score of 0.00 or 1.00. The quantitative analysis could also offer guidance to ophthalmologists on what specific image regions contributed to the model prediction. Clinicians could combine model predictions with all other patient-specific data to make patient management decisions. In addition, the approach developed in this study could be deployed on standard computing equipment anywhere in the world with a low cost, and provide reproducible evaluation of OCT images in patients with suspected eye diseases.
In order to insight into the decisions of multi-ResNet50 ensembling which have often been referred to as black boxes, a sampling of correct, incorrect, and borderline testing examples was reviewed by a ophthalmologist. Based on this additional review, our model did seem to correctly identify the related characteristics associated with CNV, DME and DRUSEN. In Fig. 9, CNV was related to neovascular membrane and associated subretinal fluid, DME was represented by retinal-thickening-associated intraretinal fluid, and DRUSEN was characterized by undulations and elevations of RPE hyper-reflective band. However, images with similar characteristics could cause low confidence predictions and model errors. In Fig. 9, these cases confused CNV and DME displayed the similar characteristics such as subretinal fluid accumulation. Also, for the normal eye, RPE was usually characterized as a highly concave backscattering layer posterior to the retina, and slightly convex in RPE may result in mix-up of DRUSEN and NORMAL in some cases. Further, occlusion testing was performed to identify image areas with the greatest importance on model predictions. In the case of CNV eyes, our model identified the subretinal/outer retinal hyper-reflective material as the most important part and the intraretinal fluid contributed comparatively little to model decisions. In the case of DME eyes, our model recognized the sub- and intra-retinal fluid accumulation as the key features for the deep learning classification. For the DRUSEN eyes, mound-like elevations with defined margins were identified as the greatest impact on model decisions. For the normal eye, the RPE hyper-reflective band was recognized as the regions of interest. Through occlusion testing, it was confirmed that the multi-ResNet50 ensembling made its decisions by accurate differentiating features from input OCT images, which was indeed identifying the areas of the image that was important for detecting eye disorders and demonstrated our model accessed retinal OCT images in a way similar to clinicians. This could recognize signs of eye diseases, increase opening opportunities for better training of clinicians on how to identify them in clinical practice, and potentially aid real-time clinical validation and future reviews or analysis for patients and physicians.
Given a broader view in the diagnosis, we prelimimarily investigated the effect of the integration of retinal OCT images and a short medical history data retrospectively acquired from the corresponding patients on model performance. The result indicated that there was no significant difference in the AUC, whereas the sensitivity and specificity improved slightly when introducing a short medical history data. The incorporation of medical history data from different sources could improve classification performance of the model. However, other methods such as qualitative evaluation and occlusion test, which could help understand and visualize the characteristics correlative with the final classification stood for a valuable option. Further tests with other new architectures and more medical history data should be investigated and evaluated.
Limitations in this study also should be considered. First, we collected retinal OCT images only from the Heidelberg Spectralis imaging system. Various of device settings, camera systems and population characteristics may have influence on retinal OCT images, and further affected model’s performance. In the future, we should entail the use of OCT images captured from different imaging systems in training, validation and testing datasets to evaluate our approach. Second, in the current study, we did not consider the longitudinal aspect of the data, and trained our model on the entire training dataset only considering each image individually. A future study was to use longitudinal images to train our model to improve the confidence of retinal disease predictions, and determine the extent (or rate) of retinal disease damage. Third, although a promising framework in this paper was provided for an automated detection of retinal diseases, the correct classification of a specific retinal disorders (such as diabetic retinopathy or glaucoma) could not be always guaranteed using only single OCT images in clinical practice. To this end, we should further investigate the artificial intelligence diagnosis of retinal diseases across multimodal data consisted of OCT angiography, visual field testing, and fundus photography and so on, in the future study.
5. Conclusion
In summary, a novel ensemble of four classification model instances to automatically and reliably detect retinal diseases from OCT images was developed in this study, each of which was based on an improved ResNet50. The diagnostic performance of this approach was verified on independent testing dataset and two publicly dataset (DHU dataset and UCSD dataset). In particular, the influence of integration of retinal OCT images and a short medical data retrospectively collected from the corresponding patients on model performance was also investigated. The results demonstrated that this approach was capable of discriminating between CNV, DME, DRUSEN, and NORMAL with high accuracy, sensitivity, specificity, and AUC. Moreover, in contrast to results obtained by ophthalmologists, our approach was equal to or superior to them. Further, the quantitative assessment of model predictions, and occlusion testing suggested our model indeed relied on regions commonly used by clinicians to detect CNV, DME, and DRUSEN. Finally, the results of integration of retinal OCT images with a short medical history data demonstrated a slight improvement in sensitivity and specificity with similar AUCs. Thereby, our approach could serve an important role in reliably guiding diseases and patient management in a field so intensively driven by imaging such as ophthalmology, and potentially help make large-scale automated screening programs for eye diseases by providing quick, objective and consistent image assessment.
Acknowledgements
The authors acknowledge the Shanghai Zhongshan Hospital and the Shanghai First People’s Hospital for our help and support. This work was also supported by the National Key Research and Development Program of China (2016YFF0101400), the National Natural Science Foundation of China (51675321), and the National Natural Science Foundation of China (61905144).
Funding
the National Key Research and Development Program of China (2016YFF0101400); the National Natural Science Foundation of China10.13039/501100001809 (51675321); the National Natural Science Foundation of China10.13039/501100001809 (61905144).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Li X., Shen L., Shen M., Qiu C. S., “Integrating handcrafted and deep features for optical coherence tomography based retinal disease classification,” IEEE Access 7, 33771–33777 (2019). 10.1109/ACCESS.2019.2891975 [DOI] [Google Scholar]
- 2.Samagaio G., Estévez A., Moura J. D., Novo J., Fernández M. I., Ortega M., “Automatic macular edema identification and characterization using OCT images,” Comput. Meth. Prog. Bio. 163, 47–63 (2018). 10.1016/j.cmpb.2018.05.033 [DOI] [PubMed] [Google Scholar]
- 3.Lee C. S., Baughman D. M., Lee A. Y., “Deep learning is effective for classifying normal versus age-related macular degeneration OCT images,” Ophthalmology Retina 1(4), 322–327 (2017). 10.1016/j.oret.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-López A., Ortega M., Penedo M. G., Charlon P., “A web-based framework for anatomical assessment of the retina using OCT,” Biosyst. Eng. 138, 44–58 (2015). 10.1016/j.biosystemseng.2015.04.001 [DOI] [Google Scholar]
- 5.Keane P. A., Patel P. J., Liakopoulos S., Heussen F. M., Sadda S. R., Tufail A., “Evaluation of age-related macular degeneration with optical coherence tomography,” Surv. Ophthalmol. 57(5), 389–414 (2012). 10.1016/j.survophthal.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 6.Hussain M. A., Bhuiyan A., Turpin A., Luu C. D., Smith R. T., Guymer R. H., Kotagiri R., “Automatic identification of pathology distorted retinal layer boundaries using SD-OCT imaging,” IEEE Trans. Biomed. Eng. 64(7), 1638–1649 (2017). 10.1109/TBME.2016.2619120 [DOI] [PubMed] [Google Scholar]
- 7.Sandhu H. S., Eltanboly A., Shalaby A., Keynton R. S., Schaal S., El-Baz A., “Automated diagnosis and grading of diabetic retinopathy using optical coherence tomography,” Invest. Ophthalmol. Visual Sci. 59(7), 3155–3160 (2018). 10.1167/iovs.17-23677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rasti R., Rabbani H., Mehridehnavi A., Hajizadeh F., “Macular OCT classification using a multi-scale convolutional neural network ensemble,” IEEE Trans. Med. Imaging 37(4), 1024–1034 (2018). 10.1109/TMI.2017.2780115 [DOI] [PubMed] [Google Scholar]
- 9.Toth C. A., Decroos F. C., Ying G. S., Stinnett S. S., Heydary C. S., Burns R., Maguire M., Martin D., Jaffe G. J., “Identification of fluid on optical coherence tomography by treating ophthalmologists versus a reading center in the comparison of age-related macular degeneration treatments trials,” Retina 35(7), 1303–1314 (2015). 10.1097/IAE.0000000000000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eltanboly A., Ismail M., Shalaby A., Switala A., El-Baz A., Schaal S., Gimel’farb G., El-Azab M., “A computer aided diagnostic system for detecting diabetic retinopathy in optical coherence tomography images,” Med. Phys. 44(3), 914–923 (2017). 10.1002/mp.12071 [DOI] [PubMed] [Google Scholar]
- 11.Fang L., Cunefare D., Wang C C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang L., Jin Y., Huang L., Guo S., Zhao G., Chen X., “Iterative fusion convolutional neural networks for classification of optical coherence tomography images,” J. Vis. Commun. Image R. 59, 327–333 (2019). 10.1016/j.jvcir.2019.01.022 [DOI] [Google Scholar]
- 13.Hussain M. A., Bhuiyan A., Luu C. D., Smith R. T., Guymer R. H., Ishikawa H., Schuman J. S., Ramamohanarao K., “Classification of healthy and diseased retina using SD-OCT imaging and random forest algorithm,” PLoS One 13(6), e0198281 (2018). 10.1371/journal.pone.0198281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemaître G., Rastgoo M., Massich J., Cheung C. Y., Wong T. Y., Lamoureux E., Milea D., Mériaudeau F., Sidibé D., “Classification of SD-OCT volumes using local binary patterns: Experimental validation for DME detection,” J. Ophthalmol. 2016, 3298606 (2016). 10.1155/2016/3298606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alsaih K., Lemaitre G., Rastgoo M., Massich J., Sidibé D., Meriaudeau F., “Machine learning techniques for diabetic macular edema (DME) classification on SD-OCT images,” Biomed. Eng. OnLine 16(1), 68 (2017). 10.1186/s12938-017-0352-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivasan P., Kim L. A., Mettu P. S., Cousins S. W., Comer G. M., Izatt J. A., Farsiu S., “Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images,” Biomed. Eng. OnLine 5(10), 3568–3577 (2014). 10.1364/BOE.5.003568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan J. H., Bhandary S. V., Sivaprasad S., Hagiwara Y., Bagchi A., Raghavendra U., Rao A. K., Raju B., Shetty N. S., Gertych A., Chua K. C., Acharya U. R., “Age-related macular degeneration detection using deep convolutional neural network,” Future Gener. Comp. Sy. 87, 127–135 (2018). 10.1016/j.future.2018.05.001 [DOI] [Google Scholar]
- 18.Gulshan V., Peng L., Coram M., Stumpe M. C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P. C., Mega J. L., Webster D. R., “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” JAMA 316(22), 2402–2410 (2016). 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 19.Lu W., Tong Y., Yu Y., Xing Y., Chen C., Shen Y., “Deep learning-based automated classification of multi-categorical abnormalities from optical coherence tomography images,” Trans. Vis. Sci. Techn. 7(6), 1–10 (2018). 10.1167/tvst.7.6.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F., Chen H., Liu Z., Zhang X., Wu Z., “Fully automated detection of retinal disorders by image-based deep learning,” Graefe's Arch. Clin. Exp. 257(3), 495–505 (2019). 10.1007/s00417-018-04224-8 [DOI] [PubMed] [Google Scholar]
- 21.Karri S. P. K., Chakraborty D., Chatterjee J., “Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration,” Biomed. Opt. Express 8(2), 579–592 (2017). 10.1364/BOE.8.000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kermany D. S., Goldbaum M., Cai W., Valentim C. C. S., Liang H., Baxter S. L., McKeown A., Yang G., Wu X., Yan F., Dong J., Prasadha M. K., Pei J., Ting M., Zhu J., Li C., Hewett S., Dong J., Ziyar I., Shi A., Zhang R., Zheng L., Hou R., Shi W., Fu X., Duan Y., Huu V. A. N., Wen C., Zhang E. D., Zhang C. L., Li O., Wang X., Singer M. A., Sun X., Xu J., Tafreshi A., Lewis M. A., Xia H., Zhang K., “Identifying medical diagnoses and treatable diseases by image-based deep learning,” Cell 172(5), 1122–1131.e9 (2018). 10.1016/j.cell.2018.02.010 [DOI] [PubMed] [Google Scholar]
- 23.Fauw J. D., Ledsam J. R., Romera-Paredes B., Nikolov S., Tomasev N., Blackwell S., Askham H., Glorot X., O’Donoghue B., Visentin D., Driessche G., Lakshminarayanan B., Meyer C., Mackinder F., Bouton S., Ayoub K., Chopra R., King D., Karthikesalingam A., Hughes C. O., Raine R., Hughes J., Sim D. A., Egan C., Tufail A., Montgomery H., Hassabis D., Rees G., Back T., Khaw P. T., Suleyman M., Cornebise J., Keane P. A., Ronneberger O., “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nat. Med. 24(9), 1342–1350 (2018). 10.1038/s41591-018-0107-6 [DOI] [PubMed] [Google Scholar]
- 24.Fang L., Wang C., Li S., Rabbani H., Chen X., Liu Z., “Attention to lesion: Lesion-aware convolutional neural network for retinal optical coherence tomography image classification,” IEEE Trans. Med. Imaging 38(8), 1959–1970 (2019). 10.1109/TMI.2019.2898414 [DOI] [PubMed] [Google Scholar]
- 25.Rasti R., Rabbani H., Mehridehnavi A., Hajizadeh F., “Macular OCT classification using a multi-scale convolutional neural network ensemble,” IEEE T. Med. Imaging 37(4), 1024–1034 (2018). 10.1109/TMI.2017.2780115 [DOI] [PubMed] [Google Scholar]
- 26.Wang J., Perez L., “The effectiveness of data augmentation in image classification using deep learning,” ArXiv171204621 Cs (2019).
- 27.Qian X., Hua X. S., Chen P., Ke L., “PLBP: An effective local binary patterns texture descriptor with pyramid representation,” Pattern Recogn. 44(10-11), 2502–2515 (2011). 10.1016/j.patcog.2011.03.029 [DOI] [Google Scholar]
- 28.Burlina P. M., Joshi N., Pekala M., Pacheco K. D., Freund D. E., Bressler N. M., “Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks,” JAMA Ophthalmol. 135(11), 1170–1176 (2017). 10.1001/jamaophthalmol.2017.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y., Xu J., Wu X., Wang F., Chen W., “A visual analytical approach for transfer learning in classification,” Inf. Sci. 390, 54–69 (2017). 10.1016/j.ins.2016.03.021 [DOI] [Google Scholar]
- 30.Castelvecchi D, “Can we open the black box of AI?” Nature 538(7623), 20–23 (2016). 10.1038/538020a [DOI] [PubMed] [Google Scholar]
- 31.Zeiler M. D., Fergus R., “Visualizing and understanding convolutional networks,” in 2014 13th European Conference on Computer Vision (ECCV) (2014), pp. 818–833. [Google Scholar]
- 32.Schlegl T., Waldstein S. M., Bogunovic H., Endstraßer F., Sadeghipour A., Philip A. M., Podkowinski D., Gerendas B. S., Langs G., Schmidt-Erfurth U., “Fully automated detection and quantification of macular fluid in OCT using deep learning,” Ophthalmology 125(4), 549–558 (2018). 10.1016/j.ophtha.2017.10.031 [DOI] [PubMed] [Google Scholar]
- 33.Schmidt-Erfurth U., Klimscha S., Waldstein S. M., Bogunović H., “A view of the current and future role of optical coherence tomography in the management of age-related macular degeneration,” Eye 31(1), 26–44 (2017). 10.1038/eye.2016.227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee C. S., Tyring A. J., Deruyter N. P., Wu Y., Rokem A., Lee A. Y., “Deep-learning based, automated segmentation of macular edema in optical coherence tomography,” Biomed. Opt. Express 8(7), 3440–3448 (2017). 10.1364/BOE.8.003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy A. G., Conjeti S., Karri S. P. K., Sheet D., Katouzian A., Wachinger C., Navab N., “ReLayNet: Retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomed. Opt. Express 8(8), 3627–3642 (2017). 10.1364/BOE.8.003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhammad H., Fuchs T. J., Cuir N. D., Moraes C. G. D., Blumberg D. M., Liebmann J. M., Ritch R., Hood D. C., “Hybrid deep learning on single wide-field optical coherence tomography scans accurately classifies glaucoma suspects,” J. Glaucoma. 26(12), 1086–1094 (2017). 10.1097/IJG.0000000000000765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu X., Lee K., Zhang L., Sonka M., Abràmoff M. D., “Stratified sampling voxel classification for segmentation of intraretinal and subretinal fluid in longitudinal clinical OCT data,” IEEE Trans. Med. Imaging 34(7), 1616–1623 (2015). 10.1109/TMI.2015.2408632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li A., Cheng J., Wong D. W. K., Liu J., “Integrating holistic and local deep features for glaucoma classification,” in 2016 38th Annual International Conference of the IEEE-Engineering-in-Medicine-and-Biology-Society (2016), pp. 1328–1331. [DOI] [PubMed] [Google Scholar]
- 39.Yang M., Zhang L., Shiu S. C. K., Zhang D., “Robust kernel representation with statistical local features for face recognition,” IEEE Trans. Neur. Net. Lear. 24(6), 900–912 (2013). 10.1109/TNNLS.2013.2245340 [DOI] [PubMed] [Google Scholar]
- 40.He K., Zhang X., Ren S., Sun J., “Deep residual learning for image recognition,” in 2016 IEEE Conference on Computer Vision and Pattern Recognition (2016), pp. 770–778. [Google Scholar]
- 41.Lecun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521(7553), 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 42.Rahimy E., “Deep learning applications in ophthalmology,” Curr. Opin. Ophthalmol. 29(3), 254–260 (2018). 10.1097/ICU.0000000000000470 [DOI] [PubMed] [Google Scholar]
- 43.Schmidt-Erfurth U., Sadeghipour A., Gerendas B. S., Waldstein S. M., Bogunović H., “Artificial intelligence in retina,” Prog. Retinal Eye Res. 67, 1–29 (2018). 10.1016/j.preteyeres.2018.07.004 [DOI] [PubMed] [Google Scholar]
- 44.Christopher M., Belghith A., Bowd C., Proudfoot J. A., Goldbaum M. H., Weinreb R. N., Girkin C. A., Liebmann J. M., Zangwill L. M., “Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs,” Sci. Rep. 8(1), 16685–13 (2018). 10.1038/s41598-018-35044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Son J., Shin J. Y., Kim H. D., Jung K. H., Park K. H., Park S. J., “Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images,” Ophthalmology, 1–10 (2019). [DOI] [PubMed]
- 46.Chalakkal R. J., Abdulla W. H., Thulaseedharan S. S., “Quality and content analysis of fundus images using deep learning,” Comput. Biol. Med. 108, 317–331 (2019). 10.1016/j.compbiomed.2019.03.019 [DOI] [PubMed] [Google Scholar]