Abstract
Programmed cell death, or apoptosis, is an essential process in development and homeostasis, and disruptions in associated pathways are responsible for a wide variety of diseases such as cancer, developmental abnormalities, and Alzheimer’s disease. On the other hand, cell death, in many cases, is the desired outcome of therapeutic treatments targeting diseases such as cancer. Recently, metabolic imaging based on two-photon fluorescence microscopy has been developed and shown to be highly sensitive to certain cell death processes, most notably apoptosis, thus having the potential as an advanced label-free screening tool. However, the typically low acquisition rates of this imaging technique have resulted in a limited throughput approach, allowing only a small population of cells to be tracked at well-separated time points. To address this limitation, a high-speed two-photon fluorescence lifetime imaging microscopy (2P-FLIM) platform capable of video-rate imaging is applied to study and further characterize the metabolic dynamics associated with cell death. Building upon previous work demonstrating the capabilities of this system, this microscope is utilized to study rapid metabolic changes during cell death induction, such as dose-dependency of metabolic response, response in invasive vs. noninvasive cancer cells, and response in an apoptosis-resistant cell line, which is further shown to undergo autophagy in response to toxic stimuli. Results from these experiments show that the early apoptosis-related metabolic dynamics are strongly correlated with important cellular parameters including responsiveness to apoptosis-inducing stimuli. The high speed and sensitivity of the presented imaging approach enables new investigations into this highly dynamic and complex process.
1. Introduction
Programmed cell death is a vital process responsible for homeostasis in the human body [1]. Seemingly minor aberrations in cell death pathways can lead to a variety of human diseases, such as Alzheimer’s disease, in the case of increased levels of cell death [2], and cancer, in the case of decreased levels [3]. Apoptosis, a well-regulated form of programmed cell death, is known to be the preferred pathway of cell death under normal conditions. In apoptosis, cells undergo compartmentalized self-destruction where containment of intracellular material in membrane-enclosed blebs allows for efficient removal by the immune system. Additionally, necrotic and autophagic cell death represent important alternative cell death pathways. Necrosis is an acute form of cell death, causing cells to expand and lyse, leaking intracellular materials into the extracellular space, which leads to inflammation in vivo [4]. Autophagic cell death represents a process by which autophagy, or the degradation of dysfunctional intracellular components, occurs with or causes cell death [5]. One of the distinguishing biochemical features of most programmed cell death processes such as apoptosis is the increased rate of ATP production due to the increased demand for energy necessary to dismantle the cell while avoiding inflammatory processes [6]. Indeed, a previous study has shown that by depleting intracellular ATP pools, cells will tend to undergo necrosis rather than apoptosis [7]. It was further shown here that the cellular generation of ATP is also critical to execute apoptotic cell death [7]. These observations point to the central role of metabolism in determining cell fate and the particular cell death pathway that an individual cell may undergo [8].
Due to its subcellular resolution capabilities, optical microscopy methods have shown great promise in identifying, tracking, and studying cell death processes in living cells [9,10]. In particular, several nonlinear optical microscopy techniques have been developed to directly probe the metabolic state of cells with high-sensitivity based on endogenous contrast from metabolic co-enzymes [11]. Two-photon excited fluorescence lifetime imaging microscopy (2P-FLIM) of reduced nicotinamide adenine dinucleotide (phosphate) (NAD(P)H) specifically has shown promise in assessing relative rates of aerobic and anaerobic metabolism. It has been demonstrated that free NADH in the cell cytoplasm, which mainly reflects the levels of glycolysis, has a significantly shorter fluorescence lifetime than that of NADH that binds to complex I in the mitochondria during oxidative phosphorylation [12]. Thus, these two contributions can be separated through fluorescence lifetime detection, allowing for the determination of changes in relative rates of glycolysis and oxidative phosphorylation with high sensitivity [13]. This method has proven exceptionally useful, for example, in the characterization of cancer at the cellular level, providing direct in vivo evidence of the Warburg effect whereby cancer cells can be identified by their increased glycolytic levels compared to normal, healthy cells [14]. More recently, the use of these methods to assess therapeutic endpoints has been proposed and shown to be highly predictive of patient outcomes [15]. Generally, however, one primary limitation with these optical metabolic imaging techniques is the low image acquisition rates, allowing only a small number of cells to be imaged at relatively few time points, limiting high throughput analysis. While higher-throughput 2P-FLIM approaches have been developed [16–20], few have focused on the application of label-free metabolic imaging for tracking dynamic cellular processes. This limitation primarily arises due to the reliance of many currently utilized 2P-FLIM platforms on a technique known as time-correlated single-photon counting (TCSPC). This photon counting-based method measures the arrival times of individually detected fluorescence photons in reference to the excitation laser pulse and builds up a histogram of arrival times with very high temporal resolution and accuracy [21]. However, in order to obtain high quality data and images, long pixel dwell times are often needed to build temporal histograms of sufficient signal-to-noise ratio (SNR). Recently, high-speed imaging has been demonstrated using an analog detection scheme for NAD(P)H lifetime imaging, allowing visualization of intracellular metabolic dynamics in response to an apoptotic stimulus, demonstrating metabolic changes that occur on the order of seconds [16].
While metabolic imaging of cell death has been performed both in vitro [22–24] and in vivo [25], the early cellular metabolic dynamics in response to apoptotic stimuli remain to be studied. Specifically, previous studies utilizing high temporal resolution TCSPC-based FLIM systems have shown a dramatic increase in the NAD(P)H mean lifetime occurring rapidly after treatment with staurosporine (STS), a potent inducer of apoptosis. Previous studies directly correlated these lifetime observations with metabolic indicators and classical markers of apoptosis such as mitochondrial membrane potential, cleaved caspase-3, and ATP levels to further study the mechanism responsible for this NAD(P)H lifetime signature [26]. However, due to the slow acquisition rates of the TCSPC instruments used in these studies, the fast dynamics associated with apoptosis induction could not be fully captured. Investigating these early dynamics is important to better understand the intricate relationship between metabolism and cell death, as well as to assess the potential of metabolic imaging as a high-throughput therapeutic screening tool. Building upon our previous work developing and characterizing a high-speed 2P-FLIM imaging platform for metabolic imaging [16], this technique is applied here to provide a deeper investigation of metabolic dynamics in response to apoptotic stimuli under a wide range of conditions in order to investigate important effects such as the dose-dependent dynamics, the response of malignant vs. nonmalignant cells, and the metabolic response of apoptosis-resistant cells. Furthermore, high-throughput imaging of large fields-of-view, typically impractical using TCSPC-based techniques, is demonstrated for monitoring the heterogeneous response to cell death-inducing stimuli. This study explores the unique capabilities of this high-speed metabolic imaging platform for investigating the complex role of metabolism in cell death.
2. Methods
2.1. High-speed two-photon fluorescence lifetime imaging microscopy
A high-speed 2P-FLIM platform was utilized for this study, which has been described previously in detail [16]. Briefly, a titanium:sapphire laser (Spectra-Physics Mai Tai HP) centered at 750 nm was used as a laser excitation source for the 2P-FLIM imaging system. The beam was scanned across the focal plane of the microscope using an 8 kHz resonant scanner (EOPC SC-30) for the fast axis and a galvanometer scanning mirror (Cambridge Technology) for the slow-axis. The emitted fluorescence was collected in the epi (backward) direction through a 1.05 NA water immersion objective (Olympus XLPLN-25X-WMP2—1.05 NA; 25× magnification) and detected with a compact analog photomultiplier tube (PMT) (Hamamatsu H10721-20). The signal photocurrent generated by the PMT was amplified by a high-gain transimpedance amplifier (Hamamatsu C5594) and sampled through a high-speed digitizer (AlazarTech ATS 9360) at 1.6 gigasamples per second.
2.2. Analysis of high-speed 2P-FLIM data
Lifetime processing and analysis of high-speed 2P-FLIM data was based on an exponential fitting procedure. Each individual FLIM micrograph was first preprocessed into a three-dimensional array encoding the spatial (x, y) and temporal (t) dimensions. Following this, the fluorescence decay was analyzed at each pixel through least-squares fitting with a single exponential model, as has been previously demonstrated in several similar approaches [14–17]. While a bi-exponential model is typically appropriate for assessing the fluorescence dynamics of NAD(P)H due to the free and protein-bound contributions, the wide instrument response function as well as the limited temporal sampling limits the sensitivity of the 2P-FLIM system to these subtle decay dynamics. However, even with a single exponential decay model, this high-speed imaging approach has been shown to provide sensitive contrast between different metabolic processes [16]. To reduce the effect of background and suppress the lifetime contribution from nuclear regions during image analysis, the cytoplasm from each cell was extracted and used for quantification of fluorescence dynamics, as was performed previously [16]. Segmentation was performed using the software package CellProfiler [27] for this procedure.
2.3. Video-rate imaging
Video-rate 2P-FLIM imaging was performed by rapidly acquiring datasets at 20 frames per second (fps), streaming this data directly to the high-speed solid-state drive (SSD) in the acquisition PC. Datasets were acquired at 256 × 256 pixel image size with a temporal sampling rate of 0.625 ns. A dwell time of approximately 0.25 µs was utilized.
2.4. Large field-of-view imaging
To obtain a large field-of-view (FOV) image of the specimen under investigation, a mosaicing procedure was performed using a motorized translation stage, averaging 20 frames recorded at 20 fps at each stage position, and stitching together the frames during post-processing. For all datasets, 10-by-10 mosaics were acquired over an area of approximately 1.8 mm x 1.8 mm in order to analyze large cellular populations, potentially on the order of 10,000 cells, using the high-speed 2P-FLIM system.
2.5. Cell culture
Cells lines used in this study were cultured using standard techniques. MCF-7 (ATCC HTB-22) cells were cultured in MEM alpha supplemented with 10% fetal bovine serum (FBS) and 1x antibiotics and passaged less than 10 times. MDA-MB-231 cells (ATCC HTB-26) were cultured in Leibovitz’s L15 medium supplemented with 10% FBS and 1x antibiotics. MCF 10-A cells were cultured in basal media (Lifeline MammaryLife Basal Medium) and passaged less than 10 times. The apoptosis-resistant cell line (BAX-BAK double knock out (DKO) SV40 mouse embryonic fibroblasts; ATCC CRL-2913) and its non-resistant counterpart (SV40 mouse embryonic fibroblast wild type (WT); ATCC CRL-2907) were cultured in IMDM supplemented with 10% FBS, 1x non-essential amino acids, and 1x antibiotics. All cells were cultured at 37˚C with 5% CO2, except for MDA-MB-231 cells which were cultured at 37˚C without CO2. For imaging, cells were plated in glass bottom imaging dishes (MatTek P35GC-0-14-C) with phenol red-free media formulations of the base media described above. All imaging was performed in ambient conditions at room temperature due to the short duration of the experiments. Previous studies of the NAD(P)H lifetime dynamics in cultured cells have shown that the NAD(P)H lifetime dynamics associated with STS treatment are minimally affected when performing similar experiments at room temperature compared to physiological conditions [22]. Authentication of all cell lines was performed by the manufacturer and confirmed through microscopic examination of cell morphology when possible.
2.6. Cell death induction
Cell death was induced by adding staurosporine (STS), a potent and fast-acting apoptosis inducing compound [28], to the cell culture with either manual pipetting or a syringe pump. For video-rate imaging during introduction of STS, the media in each dish was first removed via vacuum suction. Immediately following, a 120-second video acquisition sequence was initiated, and the syringe pump was activated to introduce the proper cell culture media supplemented with the desired concentration of STS to the imaging dish.
2.7. Alamar Blue cell viability assay
For validation of imaging results, Alamar Blue, a metabolic assay typically utilized for cell death quantification, was used [29]. This assay has been demonstrated previously to be sensitive to changes in mitochondrial metabolism, and capable of validating our 2P-FLIM imaging results [30]. Twenty-four hours following seeding of MCF-7 cells at a density of 250k cells/mL into 35 mm diameter plates, the Alamar Blue (ThermoFisher) assay was performed according to manufacturer instructions. Media was removed from the cell plates and 2 mL of a solution of 90% (vol) media and 10% (vol) Alamar Blue was applied. Cells were incubated with the Alamar Blue and media solution for 1.5 hours. After incubation, the Alamar Blue solution was removed from the cell plates and replaced with standard media. Fifteen minutes following treatment and imaging, the Alamar Blue protocol was repeated, again applying 2 mL of a solution of 90% (vol) media and 10% (vol) Alamar Blue to the cells and incubating for 1.5 hours, before removing the solution. The Alamar Blue and media solution was read in triplicate using a plate reader (BioTek) to measure absorbance at 570 and 600 nm, additionally utilizing a media-only blank and an Alamar Blue/media blank. The relative metabolic rate was subsequently calculated using absorbance values. The amount of reduced Alamar Blue was calculated at each timepoint by taking:
| (1) |
where A570 and A600 are the blank-subtracted absorbance values at the respective wavelengths. R0 is a correction factor based on the ratio of absorbance values at 570 nm and 600 nm of a 10% Alamar Blue and media solution that has not been exposed to cells. The reduced Alamar Blue absorbs at 570 nm and oxidized at 600 nm. The amount of reduced Alamar Blue was then normalized to the initial value.
3. Results and discussion
3.1. Video-rate and longitudinal imaging of STS-induced apoptosis
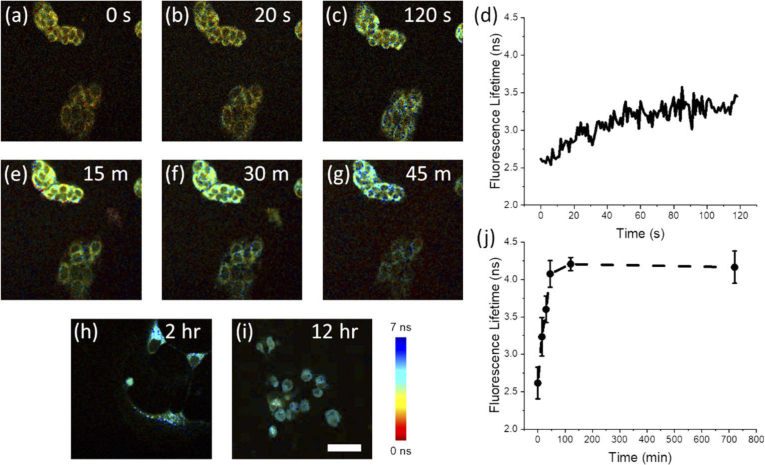
To first demonstrate the capabilities of this high-throughput imaging platform, MCF-7 breast cancer cells were imaged during the addition of STS to the culture dish. Figure 1(a)-(c) shows snapshots from video acquisitions over two minutes of cells treated with 1 µM STS. As shown previously [16], this high-speed microscope can capture the rapid increase in NAD(P)H fluorescence lifetime within cells in direct response to this apoptotic stimulus. Figure 1(d) shows these longitudinal dynamics of the NAD(P)H lifetime following STS treatment averaged across many cells in the FOV (N = 11 cells) exhibiting the characteristic rapid increase in lifetime consistent across all cells observed here. In order to correlate these short-term dynamics with the long-term metabolic effects of apoptosis induction with STS, longitudinal imaging of the same cells was performed at 15 minutes, 30 minutes, 45 minutes, 2 hours, and 12 hours (Fig. 1(e)-(i) respectively). Analysis of cellular NAD(P)H lifetime levels shows a sharp increase following addition of 1 µM STS that reaches a peak value after 2 hours and persists at this level at 12 hours (Fig. 1(j)).
Fig. 1.
Longitudinal 2P-FLIM imaging of apoptosis induction in MCF-7 cells treated with 1 µM STS. (a-c) Short-term imaging of the same FOV (a) before treatment, and (b) 20 seconds and (c) 120 seconds following treatment. (d) Longitudinal trace of the lifetime averaged across cells in the FOV (N = 11 cells) from (a-c). (e-g) Imaging of the same FOV as (a-c) at (e) 15 minutes, (f) 30 minutes, and (g) 45 minutes post-treatment. Further imaging at (h) 2 hours and (i) 12 hours was performed in order to track long-term changes in NAD(P)H lifetime. (j) Long-term dynamics of apoptosis induction up to 12 hours. Scalebar is 50 µm.
These results demonstrate the relationship between the short-term and previously studied long-term metabolic dynamics [22,26] associated with apoptosis induction in MCF-7 cells. The measured lifetime increased rapidly within seconds of STS treatment. The results measured at later timepoints are consistent with previous studies utilizing TCSPC-based 2P-FLIM systems observing a rapid, significant increase in NAD(P)H mean lifetime [22,26]. Interestingly, this initial phase exhibits the greatest rate of change in fluorescence lifetime over the course of the experiment, suggesting that STS treatment evokes a sudden increase in aerobic metabolism that continues to increase until a steady-state plateau is reached.
3.2. High-throughput screening of cell death
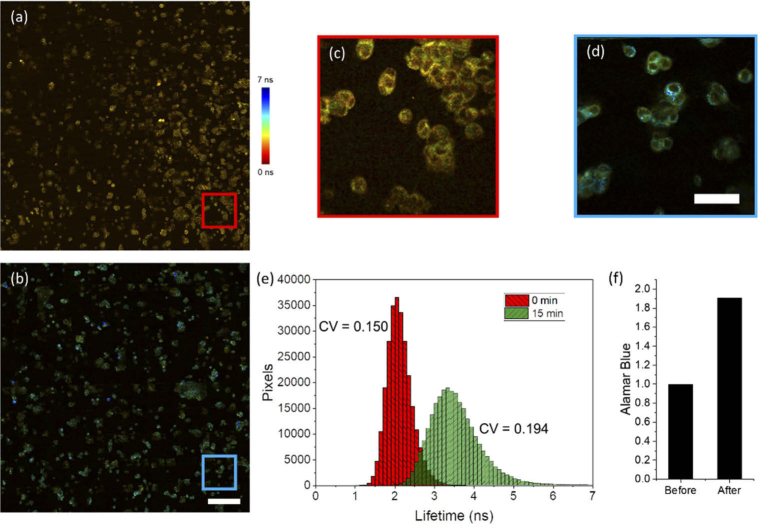
Next, in order to demonstrate the capabilities of this high-throughput imaging system for analyzing large cellular populations, large FOV imaging of the cellular metabolic response to STS treatment was performed (Fig. 2). Large FOV (∼2 × 2 mm) images of in vitro MCF-7 cells were acquired before (Fig. 2(a) and 2(c)) and 15 minutes after (Fig. 2(b) and 2(d)) treatment with 1 µM of STS, each requiring approximately 2 minutes total per acquisition. This is a significantly shorter time compared to previous studies using TCSPC, which required several hours for acquisition of a comparable FOV. Using this acquisition strategy, both metabolism and morphology of large and heterogeneous cell populations can be screened in a relatively short period of time.
Fig. 2.
Wide FOV 2P-FLIM imaging of STS-induced apoptosis induction in MCF-7 cells. (a, b) Wide FOV images across an approximately 1.8 mm x 1.8 mm FOV (a) before and (b) 15 minutes following 1 µM STS treatment. (c, d) Zoomed views from the mosaics in (a) and (b) respectively. (e) Histogram of NAD(P)H lifetime values from pixels in cellular regions before (red) and 15 minutes following the treatment (green). The histogram shows an increase in the mean NAD(P)H lifetime value across analyzed cells as well as a broadened distribution following treatment. (f) Results from Alamar Blue analysis of these cell cultures before treatment and immediately after treatment, demonstrating a large relative increase in oxidative phosphorylation in treated cells shortly after treatment. Scalebar is 200 µm in (b) and 50 µm in (d). CV – coefficient of variation.
Figure 2(e) shows histograms of lifetime values corresponding to pixels from cellular regions of these images. Results here show the dramatic increase in NAD(P)H lifetime, as was observed previously. A noticeable increase in the coefficient of variation (CV), from 0.150 before treatment to 0.194 at 15 minutes post-treatment, of the NAD(P)H lifetime distribution following STS treatment that may be associated with the heterogeneous response of cells to the treatment is also observed. In addition, Alamar Blue staining demonstrates a large increase following treatment (Fig. 2(f)), providing further evidence that STS evokes a strong increase in levels of cellular metabolism. Combining powerful high-throughput screening tools with this high-speed imaging approach could yield a highly sensitive workflow process that adds functional cellular imaging results into a high-content cell and therapeutic response screening platform.
3.3. Dose response of NAD(P)H lifetime dynamics
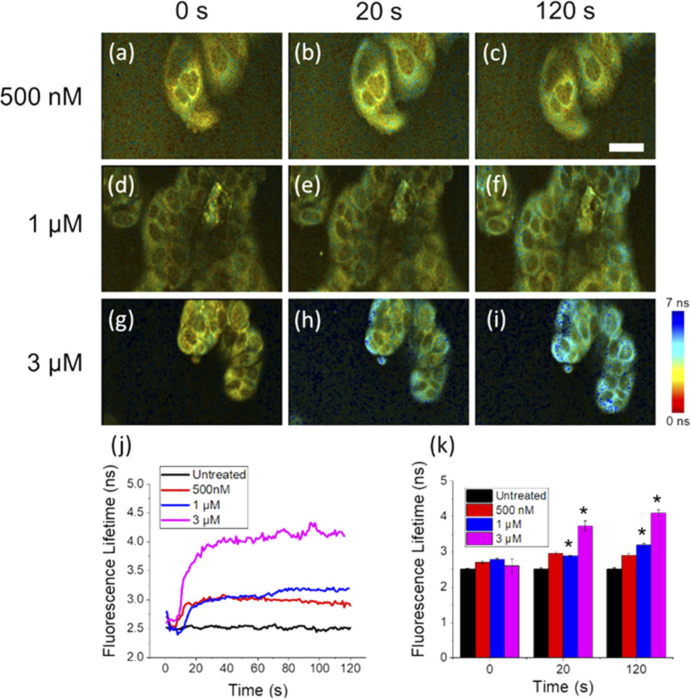
Previous NAD(P)H 2P-FLIM studies of STS-induced apoptosis reported a significant difference between the longitudinal NAD(P)H lifetime dynamics of HeLa cells treated with low (50 nM) and high (1 µM) doses of STS, with larger STS concentrations showing more rapid and dramatic increases in mean lifetime [26]. In order to assess and better characterize the dose-dependent response of STS-induced apoptosis in MCF-7 cells, STS treatments of 500 nM (Fig. 3(a)-(c)), 1 µM (Fig. 3(d)-(f)), and 3 µM (Fig. 3(g)-(i)) were introduced to plated MCF-7 cells while imaging with the high-speed 2P-FLIM microscope (Visualization 1 (2.2MB, mp4) ). NAD(P)H lifetime dynamics from individual cells are shown in Fig. 3(k). From these plots, a rapid increase in fluorescence lifetime can be observed immediately after the addition of STS, for all doses. Furthermore, this effect appears to be clearly associated with the STS concentration used. At higher concentrations, the characteristic NAD(P)H lifetime dynamics occur more rapidly with a dramatically increased lifetime compared to lower concentrations, indicating a faster increase in cellular oxidative metabolism rates (Fig. 3(j)). Analysis shows a statistically significant increase in the NAD(P)H lifetime observed in MCF-7 cells treated with 1 µM and 3 µM STS, compared to untreated cells at 20 seconds and 120 seconds (Fig. 3(k)). In addition, a statistically significant increase in NAD(P)H lifetime was observed in the 3 µM STS-treated group compared to the 1 µM STS-treated group. These results suggest that increasing the dose of STS applied to MCF-7 cells in vitro induces a larger, more rapid increase in aerobic metabolism that can be observed nearly immediately after treatment, supported by previous studies making similar observations at much later time points [26]. The observed differences in metabolic rate may be attributed to the effect of the STS dose on the rate of cell-death related events in the cell. Previous studies investigating the dose-dependent effects of STS-induced apoptosis in HeLa cells have also observed a significantly slower rate of caspase-3 activation, and a delay in the formation of mitochondrial membrane pores, often considered as a “point-of-no-return” [31], in cells treated with lower STS doses that appear correlated with these NAD(P)H dynamics [26]. Accordingly, metabolic rates, including the initial transient dynamics observed here, may be directly related to the rate of activation of apoptosis-specific processes in response to the applied stimulus. Thus, this high-speed 2P-FLIM imaging platform constitutes a sensitive technique to probe the metabolic processes and rates associated with apoptosis induction.
Fig. 3.
Dose-dependent dynamics of NAD(P)H lifetime in MCF-7 cells treated with STS. (a-i) Snapshots of MCF-7 cells at (a, d, g) 0 seconds, (b, e, h) 20 seconds, and (c, f, i) 120 seconds following treatment with (a-c) 500 nM, (d-f), 1 µM, and (g-i) 3 µM STS. (j) Short-term dynamics plotted as the average lifetime across multiple cells in the FOV show a stronger initial increase with larger STS doses that persists at 2 minutes following treatment (see Visualization 1 (2.2MB, mp4) ). (k) Cells treated with 1 µM and 3 µM STS show a statistically significantly increased NAD(P)H lifetime at 20 seconds and 120 seconds compared to the untreated group. Scale bar is 25 µm. *p < 0.01.
3.4. Apoptosis-related metabolic dynamics in cancer and non-tumorigenic cell lines
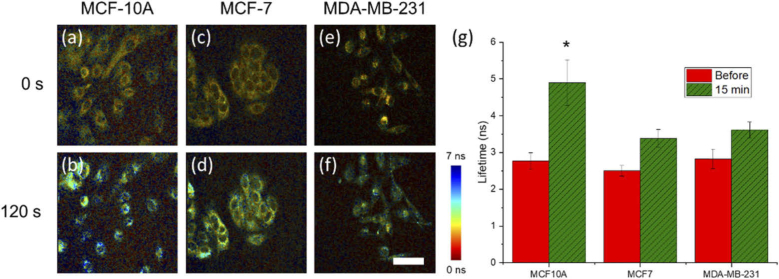
To determine the role that the baseline metabolic state of a cell has on the early-stage initiation of metabolic changes following treatment with STS, non-tumorigenic (MCF10-A, Fig. 4(a)-(b)), noninvasive cancer (MCF-7, Fig. 4(c)-(d)), and invasive cancer (MDA-MB-231, Fig. 4(e)-(f)) cell lines were imaged in response to 1 µM STS treatment. Images at 0 and 120 seconds post-treatment show previously observed NAD(P)H lifetime increases in all cell lines. However, the non-tumorigenic MCF-10A cell line showed a statistically significant increase in NAD(P)H lifetime response over similar baseline levels across all cell lines after 120 seconds, compared to the two cancer cell lines (Fig. 4(g)). This implies that this non-tumorigenic cell line may have greater aerobic respiration capabilities to respond to the STS stimulus earlier, compared to the two cancer cell lines which have been characterized previously as having altered metabolic profiles, in the case of MDA-MB-231 cells [32], or may be missing key pro-apoptotic proteins such as caspase-3, in the case of MCF-7 cells [33].
Fig. 4.
Comparison of NAD(P)H lifetime response to 1 µM STS in malignant and non-malignant cancer cells. 2P-FLIM images of (a, b) MCF10-A, (c, d) MCF-7, and (e, f) MDA-MB-231 at (a, c, e) 0 seconds and (b, d, f) 120 seconds following treatment with 1 µM STS. (g) Statistical analysis of imaged cells shows a significant increase in the NAD(P)H lifetime increase over the course of imaging in MCF10-A cells compared to both MCF-7 and MDA-MB-231 cells. Scalebar is 50 µm. * p < 0.01.
These observations have important consequences for future high-throughput screening applications. Most importantly, this unique imaging technique provides an informative approach to potentially probe the metabolic capacity and response of different cellular conditions to an apoptotic stimulus. These results are further supported by previous studies demonstrating that the IC50 of MCF-10A cells is at least an order-of-magnitude smaller than IC50 of both MCF-7 and MDA-MB-231 cells [34]. This suggests further that there may be a correlation between cellular sensitivity to apoptosis-inducing compounds and the initiation dynamics observed by our high-speed 2P-FLIM imaging results that may be further validated in the future through comparison to traditional biochemical assays.
3.5. Metabolic dynamics of apoptosis-resistant cells
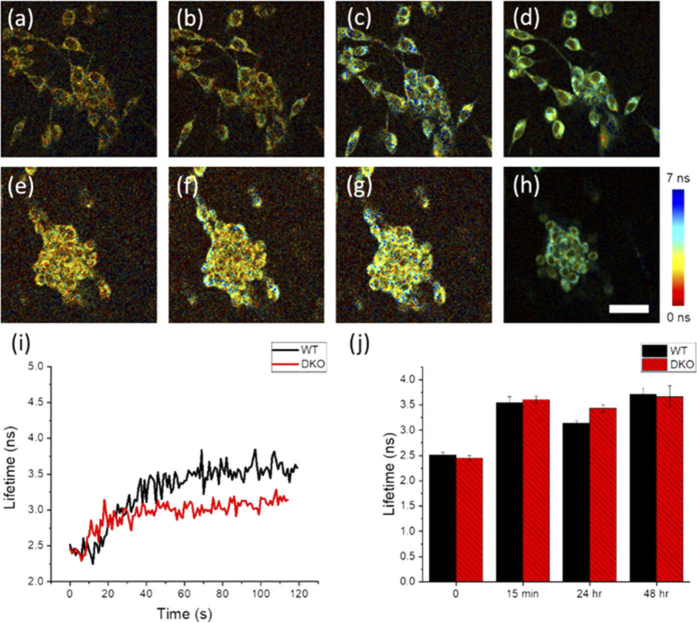
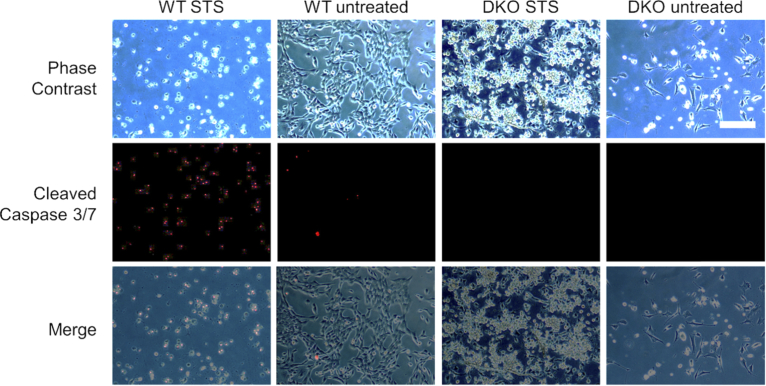
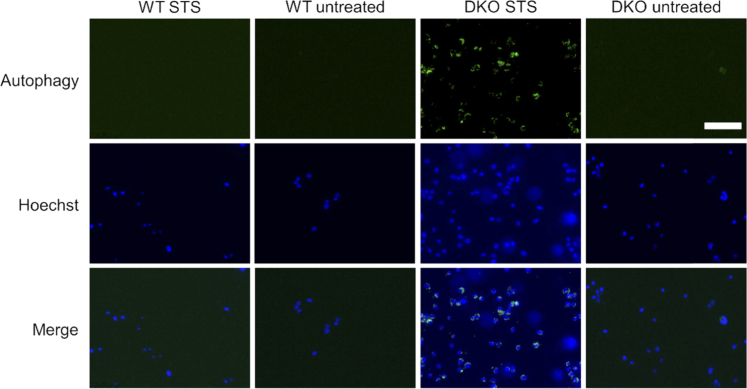
Finally, in order to study conditions of inhibited cell death induction, the metabolic dynamics of a genetically modified apoptosis-resistant cell line (BAX-BAK double knock out (DKO) SV40 mouse embryonic fibroblasts; ATCC CRL-2913) were compared to their non-resistant counterpart (SV40 mouse embryonic fibroblast wild type (WT); ATCC CRL-2907) following treatment with 1 µM STS. These cells are missing BAX and BAK, two key pro-apoptotic molecules necessary for mitochondrial-mediated apoptosis [35]. High-speed 2P-FLIM imaging results are shown in Fig. 5 and Visualization 2 (4.3MB, mp4) . WT (Fig. 5(a)-(d)) and DKO (Fig. 5(e)-(h)) cells were treated with 1 µM STS and snapshots of the NAD(P)H lifetime are shown at 0 seconds (Fig. 5(a) and 5(e)), 20 seconds (Fig. 5(b) and 5(f)), 120 seconds (Fig. 5(c) and 5(g)), and 15 minutes (Fig. 5(d) and 5(h)). Both WT and DKO cells exhibit the previously observed rapid lifetime increase following STS treatment. However, The WT cells had a statistically significant increase in lifetime compared to DKO cells at 2 minutes following treatment (Fig. 5(i)). Imaging was again performed at 24 hours and 48 hours following treatment (Fig. 5(j)). Interestingly, the NAD(P)H lifetimes at these later time points were very similar, suggesting that the metabolic dynamics between the resistant and non-resistant cells differ only in the initial stages of apoptosis. Furthermore, apoptotic cell death was confirmed in the WT cells 24 hours after treatment (Fig. 6), but nearly no cell death was observed in the apoptosis-resistant cell line at 24 or 48 hours (Figs. 6 and 7). It has been previously reported that these DKO cells, while resistant to apoptosis, will undergo autophagy, a natural regulatory mechanism of cells to remove unwanted or dysfunctional organelles, in order to survive the apoptotic stimulus [36]. In our study, autophagy was confirmed using an autophagy flux stain (Enzo Life Sciences CYTO-ID Autophagy detection kit) 48 hours post-treatment (Fig. 7). Results suggest that while many metabolic trends at the later stages of STS treatment in resistant and non-resistant cells may be similar, the subtle differences in the early dynamics may elucidate some of the dysfunctional apoptotic mechanisms in DKO cells.
Fig. 5.
Analysis of NAD(P)H lifetime dynamics in apoptosis-resistant cell line (BAX/BAK DKO) compared to the apoptosis non-resistant cell line (WT). (a-h) Snapshots of (a-d) WT and (e-h) BAX/BAK DKO cells at (a, e) 0 seconds, (b, f) 20 seconds, (c, g) 120 seconds, and (d, h) 15 minutes following 1 µM STS treatment (see Visualization 2 (4.3MB, mp4) ). (i) Short-term tracking of NAD(P)H lifetime dynamics reveals a statistically significant increase in the NAD(P)H lifetime of WT cells compared to DKO cells at approximately 120 seconds following treatment (p < 0.01). (j) NAD(P)H lifetime at later times (up to 48 hours) shows similar NAD(P)H lifetime values for WT and DKO cells. Scalebar is 50 µm.
Fig. 6.
Cleaved-caspase-3/7 analysis of WT and DKO cells untreated and treated with 1 µM STS after 24 hours. WT cells treated with STS show a strong increase in cleaved caspase-3/7 marker staining implying increased rates of apoptosis, while DKO cells show negligible cleaved caspase-3/7 marker signal. Scalebar is 100 µm.
Fig. 7.
Autophagy flux staining at 48 hours in STS treated WT and DKO cells. STS treated DKO cells show an increase in autophagic flux compared to WT and untreated DKO cells 48 hours following treatment with 1 µM STS. Scalebar is 100 µm.
Interestingly, BAX activation and translocation from the cytoplasm to the mitochondria in the non-resistant cells is known to occur within 30 minutes of STS treatment [37], inducing a loss of mitochondrial transmembrane potential and cell death. Observed alterations in early metabolic dynamics observed in DKO cells may be associated with the absence of BAX and its mitochondrial interactions that occur following STS treatment in WT cells. The role of metabolism in cell survival and the dependence of DKO cells on autophagy merits further study. While clear morphological changes can be observed in autophagic, STS-treated DKO cells at 24 hours following treatment (Fig. 6), the NAD(P)H lifetime is quite similar between treated DKO and WT cells at these time points. This suggests that the increased oxidative metabolism elicited by the introduction of STS may drive apoptosis in the case of WT cells, and autophagy in the case of DKO cells, but that the preferred pathway is likely determined by intracellular factors including cell death-associated proteins such as BAX and BAK.
4. Conclusions
In this study, the dynamic and heterogeneous metabolic effects of STS-induced cell death were investigated through the development and application of a high-speed 2P-FLIM imaging platform. In the study of longitudinal short-term dynamics of metabolic effects across a large population of cells, the role of metabolism in apoptosis was probed in cells with different metabolic profiles as well as in apoptosis-resistant cells. Results show the strong link between early metabolic dynamics and these cellular parameters. Specifically, a strong dose-dependent response is observed when STS is used to induce apoptosis in MCF-7 cells. Additionally, the relationship between metabolic dynamics and cellular sensitivity to apoptosis was studied in different cancer cell lines as well as in an apoptosis-resistant cell line. Results from these experiments showed that cell lines that are more responsive to STS-induced apoptosis generally exhibit a more dramatic initial increase in mean NAD(P)H lifetime upon treatment. While the results presented here are consistent with the longitudinal long-term dynamics presented previously [22,26], it is an important area of future work to directly compare results from high lifetime resolution TCSPC-based microscopes to the high-speed TP-FLIM results presented here in order to further validate these results and better quantify the specific tradeoffs inherent between these two imaging approaches. Additionally, the role of noise, background signal, and the limited temporal resolution of the high-speed 2P-FLIM instrument on the extracted lifetime values should be further studied to better understand the error sources of this fast system. As current sustained imaging durations are currently limited to only 2-3 minutes, future technical improvements should also be made to allow long-term high-speed imaging to continuously image this dynamic process over longer time scales. Further study combining this approach with standard high-throughput metabolic and cell health assays such as an NAD/NADH assay should be performed in order to better understand the precise mechanism responsible for the observed metabolic dynamics. In addition, further high-speed 2P-FLIM imaging of the cellular effects of clinically relevant cell-death inducing agents should be undertaken in order to assess the utility of this technique as a clinical screening tool, as has been suggested by several optical metabolic imaging studies [38,15]. With further investigation, the heterogeneous metabolic response to a variety of apoptotic stimuli can be studied in a high-throughput fashion, enabled through further development and application of this promising imaging approach.
Acknowledgments
We thank Darold Spillman for assistance with logistical and information technology support. Additional information can be found at http://biophotonics.illinois.edu.
Funding
National Institutes of Health10.13039/100000002 (T32 EB019944); National Institute of Biomedical Imaging and Bioengineering10.13039/100000070 (R01 EB023232); National Cancer Institute10.13039/100000054 (R01 CA213149); Air Force Office of Scientific Research10.13039/100000181 (FA9550-17-1-0387); National Science Foundation10.13039/100000001 (CBET 18-41539, DGE-1144245).
Disclosures
The authors declare no conflicts of interest.
References
- 1.Elmore S., “Apoptosis: a review of programmed cell death,” Toxicol. Pathol. 35(4), 495–516 (2007). 10.1080/01926230701320337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obulesu M., Lakshmi M. J., “Apoptosis in Alzheimer’s disease: an understanding of the physiology, pathology and therapeutic avenues,” Neurochem. Res. 39(12), 2301–2312 (2014). 10.1007/s11064-014-1454-4 [DOI] [PubMed] [Google Scholar]
- 3.Gerl R., Vaux D. L., “Apoptosis in the development and treatment of cancer,” Carcinogenesis 26(2), 263–270 (2004). 10.1093/carcin/bgh283 [DOI] [PubMed] [Google Scholar]
- 4.Rock K. L., Kono H., “The inflammatory response to cell death,” Annu. Rev. Pathol.: Mech. Dis. 3(1), 99–126 (2008). 10.1146/annurev.pathmechdis.3.121806.151456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levine B., Kroemer G., “Autophagy in the pathogenesis of disease,” Cell 132(1), 27–42 (2008). 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsumi T., Shiraishi J., Keira N., Akashi K., Mano A., Yamanaka S., Matoba S., Fushiki S., Fliss H., Nakagawa M., “Intracellular ATP is required for mitochondrial apoptotic pathways in isolated hypoxic rat cardiac myocytes,” Cardiovasc. Res. 59(2), 428–440 (2003). 10.1016/S0008-6363(03)00391-2 [DOI] [PubMed] [Google Scholar]
- 7.Leist M., Single B., Castoldi A. F., Kühnle S., Nicotera P., “Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis,” J. Exp. Med. 185(8), 1481–1486 (1997). 10.1084/jem.185.8.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green D. R., Galluzzi L., Kroemer G., “Metabolic control of cell death,” Science 345(6203), 1250256 (2014). 10.1126/science.1250256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kupcho K., Shultz J., Hurst R., Hartnett J., Zhou W., Machleidt T., Grailer J., Worzella T., Riss T., Lazar D., Cali J. J., Niles A., “A real-time, bioluminescent annexin V assay for the assessment of apoptosis,” Apoptosis 24(1-2), 184–197 (2019). 10.1007/s10495-018-1502-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okada M., Smith N. I., Palonpon A. F., Endo H., Kawata S., Sodeoka M., Fujita K., “Label-free Raman observation of cytochrome c dynamics during apoptosis,” Proc. Natl. Acad. Sci. U. S. A. 109(1), 28–32 (2012). 10.1073/pnas.1107524108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z., Pouli D., Alonzo C. A., Varone A., Karaliota S., Quinn K. P., Münger K., Karalis K. P., Georgakoudi I., “Mapping metabolic changes by noninvasive, multiparametric, high-resolution imaging using endogenous contrast,” Sci. Adv. 4(3), eaap9302 (2018). 10.1126/sciadv.aap9302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuttleworth C. W., “Use of NAD(P)H and flavoprotein autofluorescence transients to probe neuron and astrocyte responses to synaptic activation,” Neurochem. Int. 56(3), 379–386 (2010). 10.1016/j.neuint.2009.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lakowicz J. R., Szmacinski H., Nowaczyk K., Johnson M. L., “Fluorescence lifetime imaging of free and protein-bound NADH,” Proc. Natl. Acad. Sci. U. S. A. 89(4), 1271–1275 (1992). 10.1073/pnas.89.4.1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skala M. C., Riching K. M., Gendron-Fitzpatrick A., Eickhoff J., Eliceiri K. W., White J. G., Ramanujam N., “In vivo multiphoton microscopy of NADH and FAD redox states, fluorescence lifetimes, and cellular morphology in precancerous epithelia,” Proc. Natl. Acad. Sci. U. S. A. 104(49), 19494–19499 (2007). 10.1073/pnas.0708425104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walsh A. J., Cook R. S., Sanders M. E., Aurisicchio L., Ciliberto G., Arteaga C. L., Skala M. C., “Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer,” Cancer Res. 74(18), 5184–5194 (2014). 10.1158/0008-5472.CAN-14-0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bower A. J., Li J., Chaney E. J., Marjanovic M., Spillman D. R., Boppart S. A., “High-speed imaging of transient metabolic dynamics using two-photon fluorescence lifetime imaging microscopy,” Optica 5(10), 1290–1296 (2018). 10.1364/OPTICA.5.001290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dow X. Y., Sullivan S. Z., Muir R. D., Simpson G. J., “Video-rate two-photon excited fluorescence lifetime imaging system with interleaved digitization,” Opt. Lett. 40(14), 3296–3299 (2015). 10.1364/OL.40.003296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eibl M., Karpf S., Weng D., Hakert H., Pfeiffer T., Kolb J. P., Huber R., “Single pulse two photon fluorescence lifetime imaging (SP-FLIM) with MHz pixel rate,” Biomed. Opt. Express 8(7), 3132–3142 (2017). 10.1364/BOE.8.003132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giacomelli M. G., Sheikine Y., Vardeh H., Connolly J. L., Fujimoto J. G., “Rapid imaging of surgical breast excisions using direct temporal sampling two photon fluorescent lifetime imaging,” Biomed. Opt. Express 6(11), 4317–4325 (2015). 10.1364/BOE.6.004317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu J., Kang U., Kim J., Kim H., Kang J. H., Kim H., Sohn D. K., Jeong J. H., Yoo H., Gweon B., “Real-time visualization of two-photon fluorescence lifetime imaging microscopy using a wavelength-tunable femtosecond pulsed laser,” Biomed. Opt. Express 9(7), 3449–3463 (2018). 10.1364/BOE.9.003449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Becker W., Bergmann A., Hink M. A., König K., Benndorf K., Biskup C., “Fluorescence lifetime imaging by time-correlated single-photon counting,” Microsc. Res. Tech. 63(1), 58–66 (2004). 10.1002/jemt.10421 [DOI] [PubMed] [Google Scholar]
- 22.Wang H.-W., Ghukasyan V., Chen C.-T., Wei Y.-H., Guo H.-W., Yu J.-S., Kao F.-J., “Differentiation of apoptosis from necrosis by dynamic changes of reduced nicotinamide adenine dinucleotide fluorescence lifetime in live cells,” J. Biomed. Opt. 13(5), 054011 (2008). 10.1117/1.2975831 [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y., Marjanovic M., Chaney E. J., Graf B. W., Mahmassani Z., Boppart M. D., Boppart S. A., “Longitudinal label-free tracking of cell death dynamics in living engineered human skin tissue with a multimodal microscope,” Biomed. Opt. Express 5(10), 3699–3716 (2014). 10.1364/BOE.5.003699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghukasyan V. V., Kao F.-J., “Monitoring cellular metabolism with fluorescence lifetime of reduced nicotinamide adenine dinucleotide,” J. Phys. Chem. C 113(27), 11532–11540 (2009). 10.1021/jp810931u [DOI] [Google Scholar]
- 25.Bower A. J., Marjanovic M., Zhao Y., Li J., Chaney E. J., Boppart S. A., “Label-free in vivo cellular-level detection and imaging of apoptosis,” J. Biophotonics 10(1), 143–150 (2017). 10.1002/jbio.201600003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu J.-S., Guo H.-W., Wang H.-W., Wang C.-H., Wei Y.-H., “Increase of reduced nicotinamide adenine dinucleotide fluorescence lifetime precedes mitochondrial dysfunction in staurosporine-induced apoptosis of HeLa cells,” J. Biomed. Opt. 16(3), 036008 (2011). 10.1117/1.3560513 [DOI] [PubMed] [Google Scholar]
- 27.Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I. H., Friman O., Guertin D. A., Chang J. H., Lindquist R. A., Moffat J., Golland P., Sabatini D. M., “CellProfiler: image analysis software for identifying and quantifying cell phenotypes,” Genome Biol. 7(10), R100 (2006). 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palchaudhuri R., Lambrecht M. J., Botham R. C., Partlow K. C., van Ham T. J., Putt K. S., Nguyen L. T., Kim S.-H., Peterson R. T., Fan T. M., Hergenrother P. J., “A small molecule that induces intrinsic pathway apoptosis with unparalleled speed,” Cell Rep. 13(9), 2027–2036 (2015). 10.1016/j.celrep.2015.10.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Back S. A., Khan R., Gan X., Rosenberg P. A., Volpe J. J., “A new Alamar Blue viability assay to rapidly quantify oligodendrocyte death,” J. Neurosci. Methods 91(1-2), 47–54 (1999). 10.1016/S0165-0270(99)00062-X [DOI] [PubMed] [Google Scholar]
- 30.Rampersad S. N., “Multiple applications of Alamar Blue as an indicator of metabolic function and cellular health in cell viability bioassays,” Sensors 12(9), 12347–12360 (2012). 10.3390/s120912347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galluzzi L., Zamzami N., de La Motte Rouge T., Lemaire C., Brenner C., Kroemer G., “Methods for the assessment of mitochondrial membrane permeabilization in apoptosis,” Apoptosis 12(5), 803–813 (2007). 10.1007/s10495-007-0720-1 [DOI] [PubMed] [Google Scholar]
- 32.Gatenby R. A., Gillies R. J., “Why do cancers have high aerobic glycolysis?” Nat. Rev. Cancer 4(11), 891–899 (2004). 10.1038/nrc1478 [DOI] [PubMed] [Google Scholar]
- 33.Yang X.-H., Sladek T. L., Liu X., Butler B. R., Froelich C. J., Thor A. D., “Reconstitution of caspase-3 sensitizes MCF-7 breast cancer cells to doxorubicin- and etoposide-induced apoptosis,” Cancer Res. 61, 348–354 (2001). [PubMed] [Google Scholar]
- 34.Cyr L., Langler R., Lavigne C., “Cell cycle arrest and apoptosis responses of human breast epithelial cells to the synthetic organosulfur compound p-methoxyphenyl p-toluenesulfonate,” Anticancer Res. 28, 2753–2763 (2008). [PubMed] [Google Scholar]
- 35.Wei M. C., Zong W.-X., Cheng E. H.-Y., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J., “Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death,” Science 292(5517), 727–730 (2001). 10.1126/science.1059108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu S., Kanaseki T., Mizushima N., Mizuta T., Arakawa-Kobayashi S., Thompson C. B., Tsujimoto Y., “Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes,” Nat. Cell Biol. 6(12), 1221–1228 (2004). 10.1038/ncb1192 [DOI] [PubMed] [Google Scholar]
- 37.Kashkar H., Krönke M., Jürgensmeier J. M., “Defective BAX activation in Hodgkin B-cell lines confers resistance to staurosporine-induced apoptosis,” Cell Death Differ. 9(7), 750–757 (2002). 10.1038/sj.cdd.4401024 [DOI] [PubMed] [Google Scholar]
- 38.Alam S. R., Wallrabe H., Svindrych Z., Chaudhary A. K., Christopher K. G., Chandra D., Periasamy A., “Investigation of mitochondrial metabolic response to doxorubicin in prostate cancer cells: An NADH, FAD and Tryptophan FLIM assay,” Sci. Rep. 7(1), 10451 (2017). 10.1038/s41598-017-10856-3 [DOI] [PMC free article] [PubMed] [Google Scholar]