Abstract
Large scale simultaneous recording of fast patterns of neural activity remains challenging. Volumetric imaging modalities such as scanning-beam light-sheet microscopy (LSM) and wide-field light-field microscopy (WFLFM) fall short of the goal due to their complex calibration procedure, low spatial resolution, or high-photobleaching. Here, we demonstrate a hybrid light-sheet light-field microscopy (LSLFM) modality that yields high spatial resolution with simplified alignment of the imaging plane and the excitation plane. This new modality combines the selective excitation of light-sheet illumination with volumetric light-field imaging. This modality overcomes the current limitations of the scanning-beam LSM and WFLFM implementations. Compared with LSM, LSLFM captures volumetric data at a frame rate 50× lower than the rate of LSM and requires no dynamic calibration. Compared with WFLFM, LSLFM produces moderate improvements in spatial resolutions, 10 times improvement in the contrast when imaging fluorescent beads, and 3.2× the signal-to-noise ratio in the detection of neural activity when imaging live zebrafish expressing a genetically encoded calcium sensor.
1. Introduction
Volumetric fluorescence imaging is a powerful tool to uncover the patterns of neural activity that underlie brain function. The ability to simultaneously access hundreds to thousands of neurons enables sophisticated studies of the collective activity from individual or multiple neural populations [1,2]. Extracting such population-scale activity at cellular resolution is thus a key focus of optical technology development; such development seeks to increase both the scale and the resolution of neural recordings [3,4].
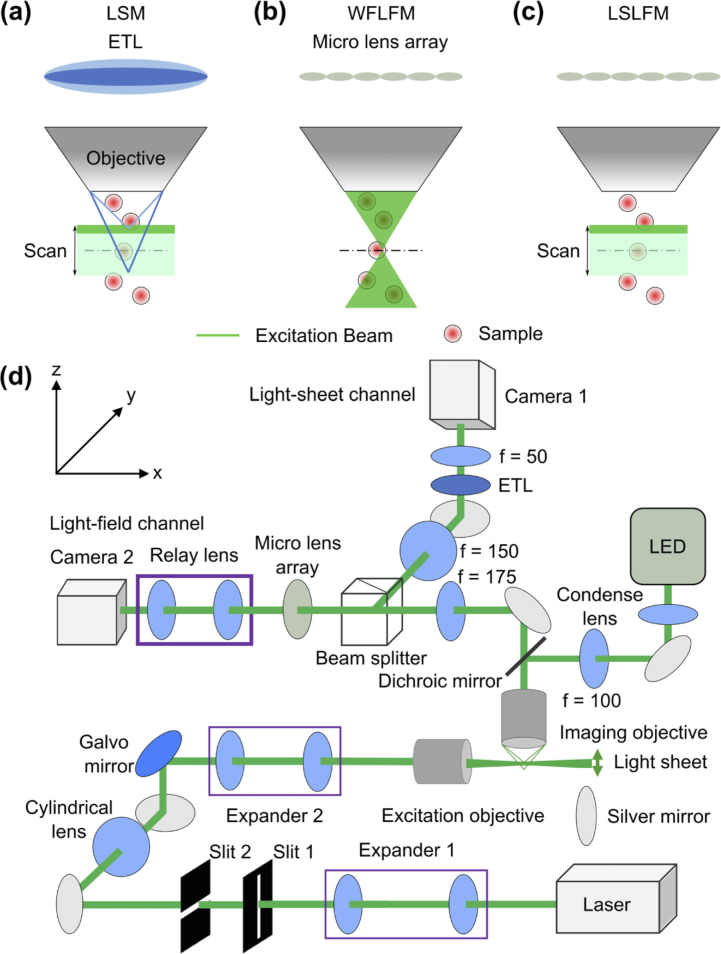
Two existing forms of fluorescence microscopy currently attempt to extract population-scale activity from model organisms – light-sheet microscopy (LSM) and light-field microscopy. The two modalities attain high resolution, large-scale imaging with two different principles. LSM attains high resolution by illuminating the sample with a micrometers-thin sheet [5]. The thinness of the sheet provides the high axial resolution needed to separate neurons at different depths of the sample. LSM attains large scale recordings by imaging the two-dimensional (2D) sheets over hundreds of micrometers in the planar direction of the sheet; the microscope then extends from sheet imaging to volumetric imaging by scanning the sheet in the remaining third dimension. Recent developments have attempted to increase the temporal resolution of this microscopy modality by increasing the scan speed. Traditional light-sheet microscopes mechanically shifted the sample relative to the microscope to acquire the structure of the fruit fly or zebrafish brain [6,7]. More recent work bypassed the mechanical speed limitations in physically moving the sample or microscope by only mechanically scanning the excitation beam [8–12]. One form of volumetric LSM simultaneously matched the light-sheet excitation plane controlled by a galvo with the imaging plane controlled by an electrically tunable lens (ETL) (Fig. 1(a)) [8]. The Swept confocally-aligned planar excitation (SCAPE) light-sheet configuration further increased the imaging speed by sweeping an oblique light-sheet through the sample and simultaneously de-scanning the excitation sheet to a fixed imaging plane [9]. This LSM configuration could potentially achieve kilohertz imaging rates of multiple layers, but taxed the speed of the existing camera electronics.
Fig. 1.
Conceptual schematics show that the LSLFM simplified the detection (vs. LSM) and limited the illumination to the region of interest for volumetric imaging (vs. WFLFM). (a) LSM used a scanning light-sheet excitation matched to an imaging plane modulated by an ETL. Dark green: excitation laser beam. Light green: Scanning volume of light-sheet. Dark blue and light blue lines represent the imaging paths focused at two positions controlled by the ETL that matched the excitation planes. (b) WFLFM used wide-field illumination and light-field imaging. (c) LSLFM used a scanning light-sheet for excitation and a micro-lens array for light-field imaging. (d) Schematic drawing shows the imaging setup.
A second form of microscopy called light-field microscopy attained large-scale volumetric data by capturing the 2D spatial and the 2D angular information of the incident light-field in one snapshot. The 2D angular information in turn relayed depth information for volumetric reconstruction [13]. The light-field microscope captured the light-field by adding a microlens array between the tube lens and the camera of a traditional wide-field microscope. Wide-field light-field microscopy (WFLFM) (Fig. 1(b)) employed wide-field epifluorescent illumination, which in turn produced high background and photobleaching. Although deconvolution processing can computationally remove the background, the process was time-consuming [14–16]. Recent work has also attempted to reduce the background within light-field imaging through selective volume illumination. One type of selective volume illumination employed light-sheet illumination patterned by a digital micromirror device (DMD): an incident beam illuminated a large area of a DMD and the light reflected by the ‘ON’ state pixels produced the excitation light-sheet [17]. By turning different pixels to the ‘ON’ state, the DMD scanned the light-sheet to different sample depths. However, this modality inefficiently employed the excitation light because only a fraction of the incident laser light contributed to the light-sheet. Another type of selective volume light-field imaging drew inspiration from multi-view light-sheet microscopy: two orthogonal imaging paths imaged two views of the sample, while an excitation objective in the third orthogonal direction illuminated the sample [18]. This modality achieved isotropic light-field imaging, but required complex reconstruction to fuse the two views into one 3D image. Lastly, researchers have also generated a galvo-scanned light-sheet using a cylindrical lens, and thus incorporated all of the energy from the incident laser into the resulting light-sheet [19]. Both the multiview light-field imaging and galvo-scanned light-field imaging compared their light-field modalities to light-sheet microscopy using sequentially acquired datasets, and thus did not quantify simultaneously acquired light-sheet imaging and light-field imaging data.
In this work, we combine the capability of light-field imaging to simultaneously capture volumetric images and the capability of light-sheet excitation to optically section the sample volume together into a hybrid light-sheet light-field microscopy (LSLFM) system (Fig. 1(c)). LSLFM excites a limited axial volume of the sample with a scanned light-sheet and images the sample with light-field microscopy. This hybrid system can potentially overcome the limitations of WFLFM and LSM. Compared to WFLFM, our imaging modality limits illumination to a small axial range of the sample. Such illumination would reduce background contributions from sources far out of plane and would potentially improve axial and lateral resolution. Compared to LSM, LSLFM reduces the required camera frame rate. LSLFM would image at the volumetric imaging rate instead of at multiples of the volumetric imaging rate corresponding to the number of layers within each volume captured with LSM. LSLFM also bypasses the challenging synchronization between the light-sheet excitation plane and the light-sheet imaging plane. The proposed LSLFM overcomes the limitations of the aforementioned selective illumination light-field imaging modalities. In comparison with the light-field imaging using DMD generated light-sheet illumination, LSLFM makes more use of the incident beam by using a cylindrical lens for light-sheet generation and a galvo for scanning. Our configuration of the LSLFM also enables simultaneous acquisition of both light-sheet and light-field channels, thereby providing direct comparison between the light-sheet and light-field imaging modalities lacking in the existing reports of multiview light-field imaging or galvo-scanned light-field imaging.
In executing our hybrid scheme, we found that the LSLFM improved contrast and resolution compared to WFLFM when imaging calibration samples and live larval zebrafish. Zebrafish is a fitting model organism for optical fluorescence imaging of neural activity: zebrafish have nearly transparent bodies and there exist established tools for integrating protein sensor sequences into its genome [20]. Our scheme demonstrates large-scale and simultaneous recording of neural activity from hundreds of neurons in the zebrafish brain reported by the green-fluorescent calmodulin protein (GCaMP6s) sensor [21].
2. Methods
2.1. Optical setup
To compare WFLFM, LSM, and LSFLM, we designed a system that integrated the three different modalities on one platform (Fig. 1(d)). For illumination, the system included one wide-field excitation path for WFLFM illumination and one light-sheet excitation path for LSM and LSLFM illumination. The wide-field illumination path used a light-emitting diode (LED; Thorlabs, 505L3-505 nm) as the excitation source. The source illuminated the sample in the Kohler illumination configuration through a 20×/1.0 NA water-immersion imaging objective (Olympus, XLUMPlanFL). The light-sheet excitation path used a 505 nm laser (Coherent, OBIS) as the excitation source. This path first magnified the laser beam by a factor of 8 through the first relay lens pair and then cropped the beam with one slit each in the x and z directions, respectively. A round cylindrical achromatic-doublet (Thorlabs, ACY254-050-A) subsequently focused one dimension of the beam. A second relay lens pair further magnified the beam by a factor of 1.6 and focused the beam at the back aperture of a 10×/0.3 NA objective (Nikon, Plan Fluor) to produce the light-sheet. The rotation of a 1D galvo scanner (Thorlabs, GVS001) between the intermediate plane of the first and second relay pairs shifted the light-sheet along the axial direction of the imaging path and enabled volumetric imaging.
The imaging system consisted of one light-sheet imaging channel and one light-field imaging channel. These two channels originated from the same 20×/1.0 NA imaging objective and then passed through a 175 mm tube lens. The common path then bifurcated at a 50:50 beam splitter into the light-sheet and light-field imaging channels. In the light-field channel, a microlens array (RPC Photonics, MLA-S100-f10) placed at the conjugate imaging plane generated the light-field. The microlens array had a f-number of f/10, matching the f-number in the used portion of the tube lens (f/10.2). A five-axis kinematic mount (Thorlabs, K5X1) provided fine alignment of the microlens array to the optical axis. A 1:1 relay macro lens (Tamron, 70-300 mm) then imaged the focal plane of the microlens array onto a 4.2-megapixel scientific complementary metal-oxide-semiconductor (sCMOS) camera (Hamamatsu, Flash4v3). The light-sheet channel employed a second relay lens pair to de-magnify the intermediate image by a factor of three onto a second Hamamatsu camera. An ETL (Optotune, EL-10-30-Ci) placed between this relay pair shifted the detection focal plane for volumetric imaging. To maintain the full NA of the imaging objective, the aperture of the ETL () has to be larger than
| (1) |
where is the focal length of the first relay lens (a 150 mm lens in our setup), is the back focal aperture of the imaging objective (18 mm), and is the focal length of the tube lens (a 175 mm lens in our setup). Based on the above equation, an ETL aperture of 15 mm would avoid clipping the emission path. However, commercially available ETLs with sufficiently fast response to image volumetrically at 10 Hz have apertures limited to 10 mm, which slightly clipped the fluorescence emission.
2.2. Dynamic calibration of the light sheet
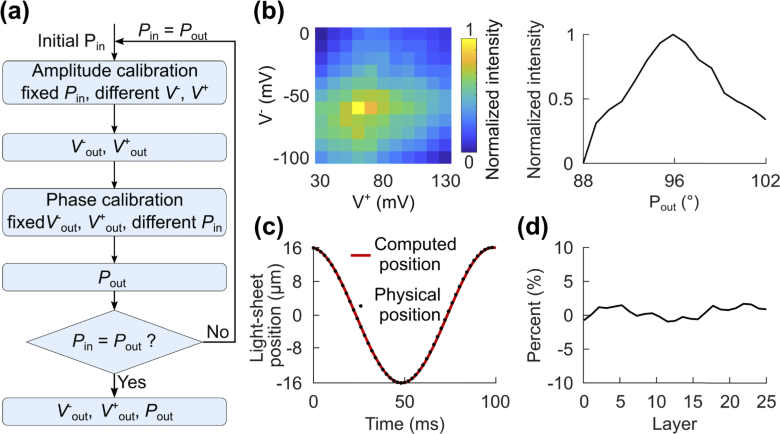
The first step toward engineering the hybrid LSLFM is the synchronization between the light-field acquisitions and the individual passes of the LS excitation. This procedure also synchronizes the galvo position with the ETL curvature to enable traditional LSM imaging that will serve as the high-resolution control for our new imaging modality. We controlled the light-sheet excitation plane position by modulating a galvo mirror; we controlled the imaging plane position by modulating an ETL. We borrowed from existing dynamic calibration procedures [8] (Fig. 2(a)) to match the amplitudes and phases of the sinusoidal control signals to the galvo mirror and ETL. Proper alignment of these two properties maximized the imaging intensity throughout one full cycle of the sinusoid, as all features would be in focus during such a scan. We imaged a volume of 350×300×32 µm3 with the imaging depth centered on the principal focal plane of the imaging objective.
Fig. 2.
Dynamic calibration synchronized the displacement of the light-sheet with the focus shift of ETL. (a) The flow chart outlines the dynamic calibration procedures. Calibration of amplitude and phase identified the best peak voltage (V+out), trough voltage (V-out), and phase (Pout) for the sine wave applied to the galvo mirror. (b) Left: To calibrate the amplitude, we applied different peak and trough voltages in combination, and maximized the total pixel intensity over one cycle of the light-sheet scanning. Right: Once we obtained the optimal voltage input combination (V+out = 60 mV and V-out = −60 mV), we found the phase (Pout = 95°) that likewise maximized the pixel intensity. (c) The computed light-sheet position over one cycle of scanning matched the physical position determined by sample translation. (d) The intensity difference between the images taken in one half cycle using either the scanning-beam or the sample translational LSM was minimal.
We executed this calibration by iterating over two steps. First, to calibrate the amplitude, we applied a range of peak voltages (V+) and trough voltages (V-) to the galvo, while holding the input phase (Pin) constant. For each set of V+ and V-, we acquired volumetric stacks over three sinusoidal cycles of 50 frames each, identified the maximum pixel intensity for each frame, and computed the average of these maximum pixel intensities as the measure of synchronization. The maximum value of this measure identified the best set of V+out and V-out for a given Pin (Fig. 2(b); left). Second, to calibrate the phase, we applied a range of phases to the galvo while maintaining the amplitude of the signal at the V+out and V-out obtained from the previous amplitude calibration. For each set of phase, we again acquired images over three sinusoidal cycles and computed the average of the maximum pixel values. This measure identified the best phase value (Pout) for a given V+out and V-out (Fig. 2(b); right). We continued to iterate through these two main steps until Pout matched Pin. We initialized the search at V+ = 10 mV, V- = −10 mV, and Pin = 90°. We found that V+out = 60 mV, V-out = −60 mV and Pout = 95° maximized the average of the maximum pixel intensities, and thus best synchronized the traditional light-sheet excitation and emission pathways. For calcium activity imaging, we calibrated the galvo and ETL for volumetric imaging at 10 Hz, but our calibration method is applicable for any other imaging speed. We calibrated the amplitude and phase independently to reduce the data size of each video for easier processing as we iterated between calibrating for phase or amplitude: each video contained calibration results for combinations of either 1 phase with 11 amplitudes or 15 phases with 1 amplitude. A more comprehensive search could potentially acquire all 165 (11×15) possible combinations of phase and amplitude in one movie and find the best set of parameters in one calibration search.
Using the best set of V+out, V-out and phase, we calculated the light-sheet position () over one cycle of the scanning (Fig. 2(c), red line) through Eq. (2):
| (2) |
where is the focal distance of the scan lens (63 mm), is the focal distance of the tube lens of the excitation path (100 mm), is the focal distance of the excitation objective (20 mm), and V is the input voltage.
To validate the effectiveness of the calibrated scanning-beam LSM, we fixed the light-sheet at the principal focal plane of the imaging objective and moved the sample across the light-sheet. We then matched the 25 imaging layers of this stack with the same 25 layers produced by moving the excitation sheet with the galvo mirror (Fig. 2(c)). The 25 imaged layers covered a depth of 32 µm. The depth of each imaged layer matched the physical position (black dots in Fig. 2(c)) in the first half of the scanning cycle. The acquired data showed high agreement between the intensities collected with the two LSM procedures, revealing a less than 5% offset in the difference in pixel intensities between the two sets of volumetric data (Fig. 2(d)).
2.3. Data acquisition
We acquired data for LSM and LSLFM simultaneously as both modalities employed the same light-sheet for excitation. For both fluorescent beads imaging and zebrafish imaging, we imaged a volume of 350×300×32 µm3, equal to the volume used during the dynamic calibration. The acquisition frame rates were 500 Hz and 10 Hz for the LSM camera and the LSLFM camera, respectively. We synchronized the two acquisitions by using the rising edge of the digital output trigger from the ETL to initiate the scanning of the galvo and the frame capture of the cameras. For both LSLFM and WFLFM, we imaged the light-field channel at 10 Hz; we performed these two light-field imaging experiments independently using either lateral laser excitation or epifluorescent LED illumination, respectively.
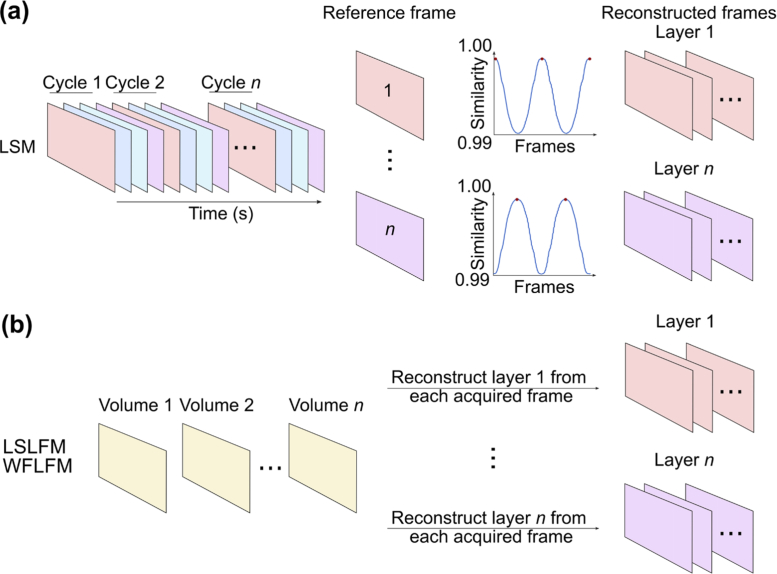
2.4. Image reconstruction and contrast calculation
LSM reconstruction computed the time series for the imaged volume by first setting each of the 25 imaging planes within the first acquired volume as the reference planes. At each subsequent time point, we matched each of the 25 imaged planes of the volume to one of the reference planes by finding the reference plane that maximized similarity to each imaging plane (Fig. 3(a)) [22]. This sequence of matched imaging planes formed the time series for that imaging plane.
Fig. 3.
Schematic shows the reconstruction procedures of (a) light-sheet and (b) light-field imaging. In LSM reconstruction, we set a series of reference frames from data within the first scanning cycle and computed the similarity of all other frames to this set of reference frames. We then combined the frames showing the highest similarity in each cycle (labeled with red dots) sequentially over time, forming the reconstructed sequence of frames for each layer within our imaging depth. Light-field imaging acquired only one frame for each imaging volume. We first reconstructed each depth in the volume individually, and then combined these reconstructed images sequentially. Similarly, we repeated the same reconstruction procedure for all the imaging depths.
Light-field image reconstruction reshaped the two-dimensional light-field image into a three-dimensional volume. We first obtained the lenslet parameters from the light-field image using the guides within the Light Field Display software (Version 2010), an open-source, cross-platform, GPU-accelerated software package for real-time viewing of microscope light fields provided by the Stanford Computer Graphics Laboratory [23]. We then employed these parameters within the shift-and-add algorithm to refocus the volumetric image stack [24,25]. We adjusted the shift parameter to match the computational refocused plane to each plane from the LSM volumetric imaging (Fig. 3(b)). To compare all light-field imaging modalities under the same conditions, we did not deconvolve our light-field data.
In bead imaging experiments, we used the Weber contrast to quantify contrast for the reconstructed image obtained with different modalities. The Weber contrast was the ratio of the difference in intensity between the bead and background to the intensity of the background [26].
2.5. Fish preparation and imaging
We imaged albino zebrafish expressing GCaMP6s under the huc promoter using the Tol2kit system at 5-7 days post fertilization [20]. Before imaging, we embedded the fish within low melting point agarose gel (1.5% by weight in egg water) and then drew the fish and agarose inside a glass capillary with an inner diameter of 1.0 mm. After the agarose gel solidified, we extruded two-thirds of the larvae body from the capillary. We then mounted the capillary horizontally inside a 3D printed water tank filled with egg water. The water tank’s side consisted of thin transparent glass plates that allowed optical access for the light-sheet illumination. For all imaging experiments, we used an illumination intensity of 0.5 mW/mm2. We imaged all specimens at room temperature.
During experiments that explored the auditory response of the zebrafish, we delivered auditory stimulus through a speaker (Logitech S120) controlled by a Raspberry Pi. We synchronized the start and stop of the entire auditory stimulus waveform with the image acquisition by activating an LED located in the detection light path for 0.2 s. The signal indicated the start of the entire auditory stimulus pattern, which included an initial 20 s period of silence and then three 1 s tones with 6 s gaps between the tones. The tones had a frequency of 300 Hz, near the peak of auditory sensitivity for zebrafish [27–29].
2.6. Neuron detection
To locate putative neurons from the imaging movies, we first registered the reconstructed videos with a subpixel image registration algorithm [30]. We then spatially filtered the imaged planes with a Gaussian spatial filter with low and high spatial cutoff frequencies at 33 cycles/mm and 200 cycles/mm, respectively. Lastly, we extracted active neurons and calcium transients with the CaImAn neuron analysis method [31], and displayed them with a color depth-encoded map.
2.7. Ethics statement
The Duke Institutional Animal Care and Use Committee (IACUC) approved all animal experiments.
3. Results
3.1. Fluorescent bead imaging
To experimentally validate the LSLFM’s potential for high-resolution imaging, we imaged red fluorescent beads embedded in agarose with WFLFM, LSLFM, and LSM. We restricted the LSLFM scanning volume to a depth of 32 µm centered on the principal focal plane of the imaging objective. Because we expected LSM imaging to produce focused beads at all depths and achieve superior lateral resolution compared to the light-field modalities, we did not strive to achieve diffraction-limited resolution. We instead imaged large objects (500 nm beads) and undersampled the image with ∼3× demagnification when quantifying the spot size of our imaging modalities. These design choices enabled simultaneous 10 Hz volumetric imaging with both LSM and light-field imaging. Whereas this volumetric imaging speed corresponded to a 10 Hz camera frame rate when using WFLFM and LSLFM, it corresponded to a 500 Hz camera frame rate when using LSM to image 50 layers per imaging volume.
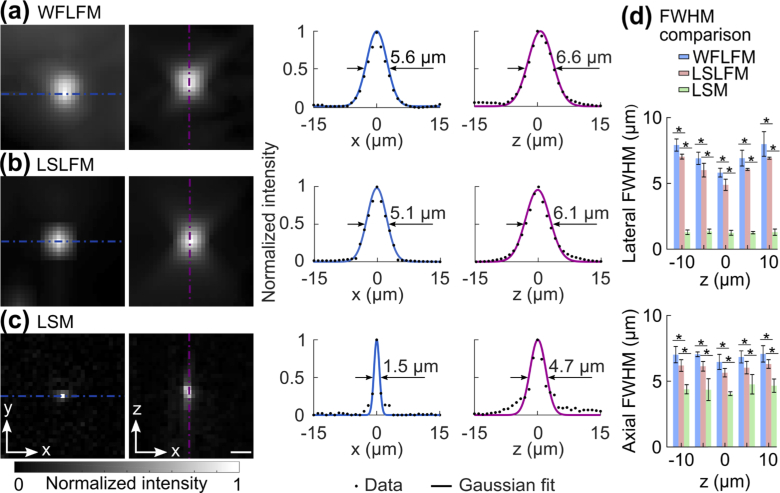
We first quantitatively compared the resolution of the three imaging modalities at −10 µm, −5 µm, 0 µm, 5 µm, and 10 µm relative to the principal focal plane. We fit the intensity profiles of representative beads to Gaussian profiles and computed the full-widths at half-maximum (FWHMs) of the fits (Fig. 4(a)). At the depth of 0 µm, WFLFM produced a FWHM of 5.6 µm and 6.6 µm in the lateral and axial directions, respectively. The light-sheet excitation of LSLFM limited the excitation to a limited axial volume, and significantly suppressed the background; LSLFM produced a spot size of 5.1 µm in the lateral direction and 6.1 µm in the axial direction (Fig. 4(b)). At the principal focal plane, LSM produced FWHMs of 1.5 µm in the lateral direction and 4.7 µm in the axial direction (Fig. 4(c)). We compared the lateral and axial spot sizes of the three imaging modalities over all five depths (Fig. 4(d)). The lateral and axial spot sizes of WFLFM and LSLFM grew as the distance from the principal focal plane increased from 0 µm to 10 µm. At each depth, the spot size of the LSLFM was smaller than that of the WFLFM. The LSM produced the smallest spot sizes at all depths, and produced nearly a constant resolution in both the lateral and axial directions over all depths. This is expected, as LSM images focused beads at all depths. The low resolution of the light-field imaging modalities originates from the trade-off between lateral resolution and angular resolution: light-field imaging samples information from a set of lateral locations equal to the pitch of the microlens array in the sample plane, which improves axial information but blurs lateral information.
Fig. 4.
Imaging of sub-diffraction beads demonstrated that LSLFM imaged with finer resolution than WFLFM. Images of individual 0.5 µm beads and the intensity profiles along the dashed lines obtained with (a) WFLFM, (b) LSLFM, and (c) LSM. Fits to Gaussian profiles determined the FWHM. Scale bar: 5 µm. (d) Comparison of FWHM between the three tested imaging modalities (mean ± std.). The lateral and axial FWHMs for LSLFM were smaller than those of the WFLFM, *p < 0.05 (n = 6, two-sided Wilcoxon rank-sum test).
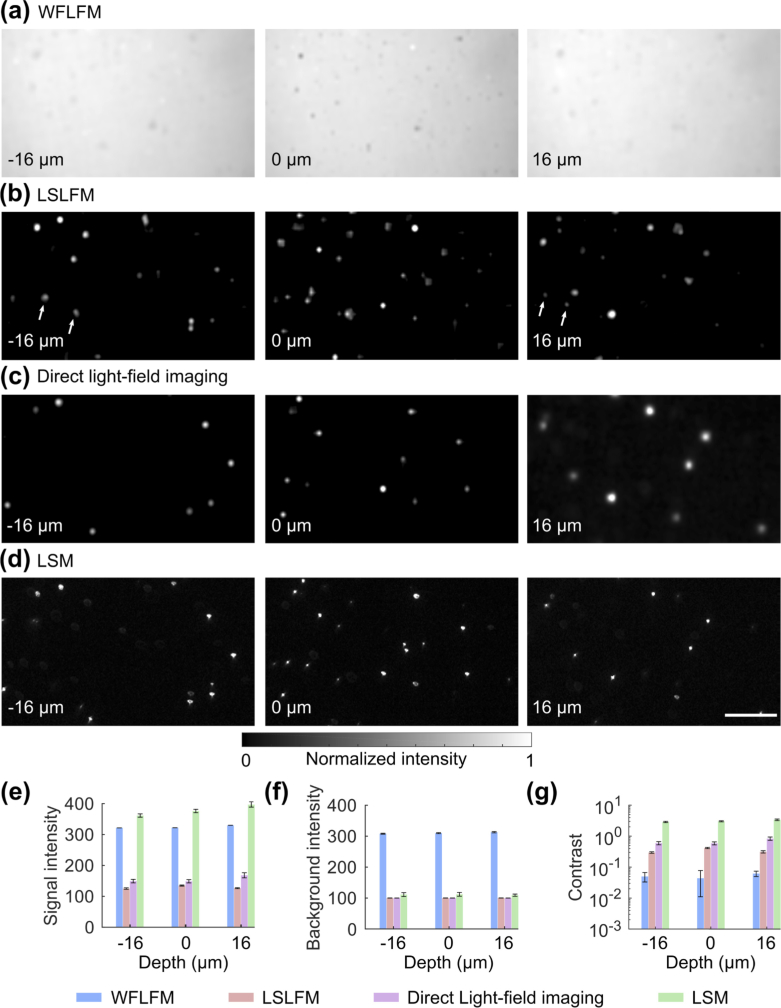
After quantifying the resolution, we quantified the contrast of the imaging modalities. We imaged the same bead and examined the resulting planes at depths of −16 µm, 0 µm and 16 µm (Fig. 5(a) and Fig. 5(b)). As a control, we also imaged the light-field directly (Fig. 5(c) when excited by the same light-sheet used in LSM at the same frame rate as the LSM (Fig. 5(d)). To quantify the contrast, we computed a bead’s averaged absolute intensity as the signal (Fig. 5(e)), and the averaged absolute intensity of a bead’s surroundings as the background (Fig. 5(f)). The ratio between the signal and background was the Weber contrast (Fig. 5(g)). The background intensity of WFLFM was higher than LSLFM, direct light-field imaging, and LSM (Fig. 5(e)). WFLFM produced signal intensity comparable to background intensity, resulting in low contrast. In comparison, LSLFM produced higher signal intensity than background intensity, thereby generating higher contrast than WFLFM. LSM’s signal intensity was much higher than its background intensity; LSM provided the highest contrast as expected.
Fig. 5.
LSLFM produced higher contrast and lower background than WFLFM when imaging sub-diffraction beads. Bead images acquired at different depths within a 32 µm deep volume using (a) WFLSM, (b) LSLFM, (c) direct light-field imaging and (d) scanning-beam LSM. Scale bar: 50 µm. WFLFM did not clearly reveal the individual beads due to the high background. In contrast, LSLFM clearly showed the individual beads over different depths, similar to the results of direct-light-field imaging and LSM. The white arrows in panel (b) indicate beads that appeared in multiple planes due to the blurring of the axial point spread function in the LSLFM image. The same beads were not seen in the images of direct-light-field imaging and LSM. Quantitative analysis of the beads’ (e) absolute signal intensity, (f) background intensity, and (g) Weber contrast for the four imaging modalities (n = 5, mean ± std.).
WFLFM broadly excited fluorescence above and below the imaging volume and thus generated significant amounts of excess out-of-focus illumination. This background severely blurred the image and obscured the beads (Fig. 5(a)), producing an image contrast of 0.03 (Fig. 5(e)-(g)). In comparison, LSLFM took advantage of light-sheet excitation and significantly reduced the illumination outside of the imaging volume. This microscopy modality’s images more clearly resolved individual beads over the entire depth of the imaging volume (Fig. 5(b)). The contrast of the beads over the imaged volume was 0.3, ∼10× higher than that of the WFLFM. The larger point-spread-function (PSF) of the LSLFM blurred beads across reconstructed planes; the same beads appeared over multiple layers (labeled with white arrows). Direct light-field imaging limited the excitation to only one imaging plane; beads appeared only in selected layers when imaging in this way (Fig. 5(c)). Despite the more selective illumination, images of beads produced by direct light-field imaging only produced slightly higher contrast than the beads imaged by LSLFM. All imaged beads matched beads imaged with LSM (Fig. 5(d)), which produced a contrast around three times the contrast of LSLFM (Fig. 5(g)). This increased contrast matched theoretical predictions that combined the effect of the 1:3 magnification ratio between the light-sheet imaging channel and the light-field imaging channel and the effect of the ETL’s small aperture within the light-sheet imaging channel: The 1:3 magnification ratio improved the contrast by a factor of 9, while the small aperture of the ETL in the LSM imaging channel reduced the contrast by a factor of ∼3.
3.2. Zebrafish imaging
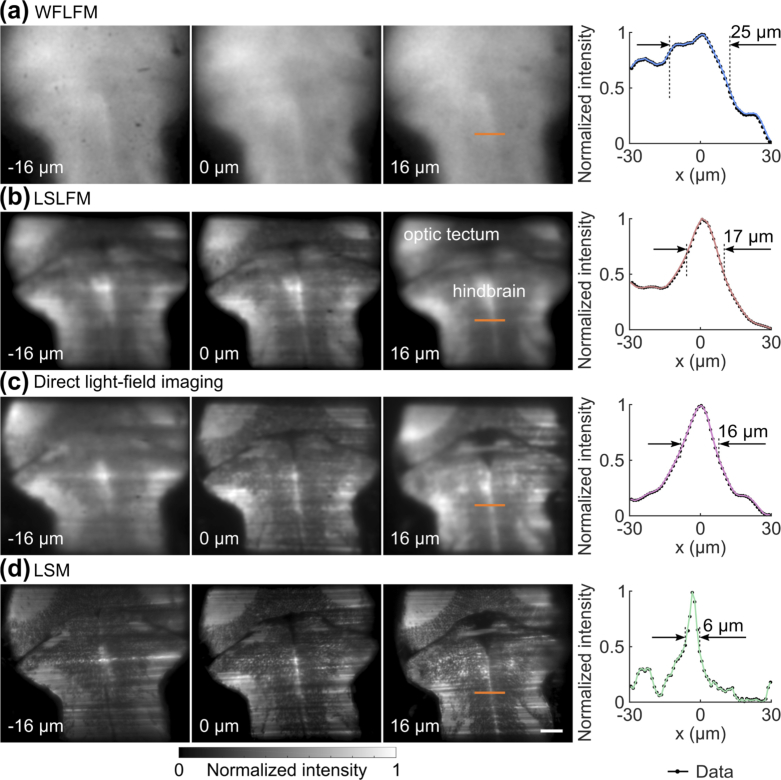
We next verified the feasibility of the LSLFM to image live animal models through in vivo imaging of larval zebrafish expressing GCaMP6s. Matching our bead experiments, we performed volumetric zebrafish imaging with WFLFM, LSLFM, direct light-field imaging, and LSM (Fig. 6). For all experiments, we imaged a volume of 350×300×32 µm3. Again, WFLFM illuminated significant portions of the out-of-focus volume, leading to significant blurring of structures within the zebrafish brain at all depths (Fig. 6(a)). In contrast, LSLFM resolved the sub-structure of the zebrafish brain more clearly at all depths (Fig. 6(b)) and demarcated the optic tectum and the hindbrain. LSLFM also generated comparable results as direct light-field imaging (Fig. 6(c)). LSM revealed the structure and individual neurons across multiple depths (Fig. 6(d)). We extracted the intensity of a pronounced fluorescent feature near the midline of the brain (labeled with orange lines). We then quantified the feature size as the distance between the half-feature heights: we chose the local minimum as the background, used the profiles to find the locations of the half feature heights on either side of the peak. We then calculated the distance between these two locations as the feature size (Fig. 6, right panels). The feature from the LSM image had a width of 6 µm, but the same feature from WFLFM had a larger width of 25 µm. The image from LSLFM had improved sharpness; the feature when imaged by LSLFM had a width of 17 µm, comparable to the size of the feature when imaged by direct light-field imaging (16 µm).
Fig. 6.
LSLFM imaging of the zebrafish brain expressing GCaMP6s in vivo produced more detail than WFLFM imaging. Images of the zebrafish brain acquired with (a) WFLFM, (b) LSLFM, (c) direct light-field imaging, and (d) LSM at depths of −16 µm, 0 µm, and 16 µm. LSLFM revealed the structure of sub-regions within the larval zebrafish brain, comparable to direct light-field imaging. WFLFM failed to resolve such structure due to the high background. Right: We quantified the feature size as the distance between the half feature heights. Scale bar: 50 µm.
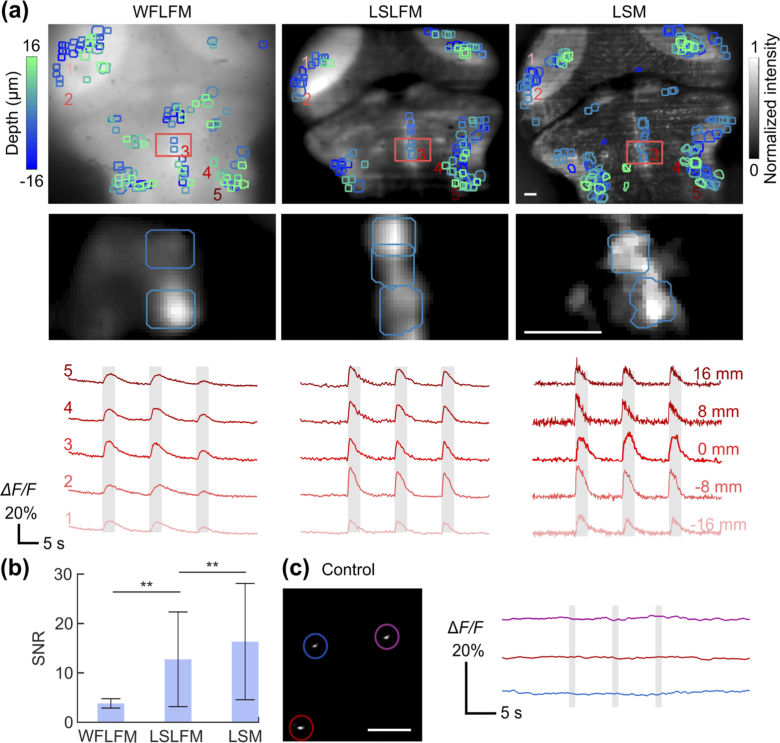
Finally, we validated the performance of LSLFM by recording the brain activity of GCaMP6s-expressing larval zebrafish. We imaged the same volume of zebrafish brain with WFLFM, LSLFM, and LSM during the delivery of auditory stimulus. Within the imaging volume, WFLFM revealed 115 ± 10 (mean ± std., n = 4 fish) active neurons; these neurons produced ΔF/F traces with a signal-to-noise ratio (SNR, defined as the ratio of peak ΔF/F to the baseline noise) of 4.0 ± 1.0 (mean ± std., n = 460 neurons) (Fig. 7(a)-b). LSLFM revealed 147 ± 15 active neurons per fish (mean ± std., n = 4 fish), significantly more than the number revealed by WFLFM (n = 4; p < 0.05, two-sided Wilcoxon rank-sum test). LSLFM recorded these neurons with an average SNR of 12.9 ± 9.6 (mean ± std., n = 558 neurons), significantly higher than the average SNR of neurons recorded by WFLFM (p < 0.01, two-sided Wilcoxon rank-sum test) (Fig. 7(b)). Neurons detected within LSM imaging volumes produced an SNR of 16.5 ± 11.8 (mean ± std., n = 648 neurons), higher than that of LSLFM (p < 0.01, two-sided Wilcoxon rank-sum test). LSM also identified more active neurons (165 ± 11; mean ± std., n = 4 fish) than LSLFM (p < 0.05, two-sided Wilcoxon rank-sum test). As a control, we also imaged beads with LSM during auditory stimulus (Fig. 7c). The stability of the fluorescence traces demonstrated that most noise arose from fish motion as opposed to mechanical instability.
Fig. 7.
LSLFM imaging of the zebrafish brain during auditory stimulus improves the quality of recorded calcium transients compared to WFLFM. (a) Active neurons and the fluorescence traces acquired with WFLFM (left), LSLFM (middle) and LSM (right). Top row: Image of depth-encoded labeling of active neurons (colored masks) on top of brain structures (grayscale); Middle row: an enlarged view of the regions labeled with red boxes in the top images. The intensity corresponds to the normalized difference between the mean image of the frames with calcium transients and the mean image of the frames without neuron transients. Bottom row: Fluorescence traces from the same five representative neurons at different depths (−16 µm, −8 µm, 0 µm, 8 µm and 16 µm). Gray bars indicate auditory stimulus. (b) SNR comparison between neurons imaged with WFLFM, LSLFM, and LSM (mean ± std., n = 460, 558, and 648 neurons for WFLFM, LSLFM and LSM, respectively. ** p < 0.01, two-sided Wilcoxon rank-sum test). (c) Fluorescent beads imaged with LSM under the identical condition as the fish experiments. All scale bars are 20 µm.
4. Discussion
Cellular resolution volumetric imaging is important for simultaneous recording of individual neurons within large neural populations. Here, we demonstrated a hybrid light-sheet light-field imaging modality for high-resolution large-scale imaging of dynamic neural activity. LSLFM improved the quality of images compared to WFLFM while bypassing the technical challenges such as the complex calibration for LSM. We demonstrated the feasibility of LSLFM through imaging of fluorescent beads and live zebrafish expressing GCaMP6s. When imaging static fluorescent beads, LSLFM outperformed WFLFM in imaging contrast by approximately10 times and reduced the spot size. When imaging live zebrafish, LSLFM produced SNR approximately 3.2 times the SNR of WFLFM when detecting fluorescence calcium transients from active neurons. LSLFM also identified more active neurons than WFLFM. LSLFM offered the same volumetric imaging speed as LSM at 2% of the imaging frame rate of LSM. Simultaneous LSLFM and LSM measurements of beads or live zebrafish found that the two modalities reported the same structure and dynamics with good agreement.
LSLFM could potentially target specific temporal imaging regimes that challenge LSM. LSM requires fast imaging frame rates to isolate each imaging plane individually. In our experiment, LSM imaged at 50 times the frame rate of LSLFM’s frame rate. These frame rate differences presented differences in data sizes: LSM datasets were 6.45 GB in size while LSLFM datasets were 1.64 GB in size. The rapid scan rate of LSM also requires high camera frame rates and fast computer control electronics. These requirements for the camera will potentially preclude LSM from future scaling: volumetric imaging of protein voltage sensors that report action potentials on the millisecond timescale [32,33] will necessitate camera frame rates in the thousands of frames per second. Such frame rates are orders of magnitude faster than the frame rates of existing sCMOS cameras that have full-frame readout at 100 Hz. LSLFM will likely bypass the limitations of camera speeds more readily; the camera frame rate of this modality will only be the volumetric imaging rate, and thus only multi-fold increases in camera speeds would be needed to record the response of fast fluorescent sensors.
Our LSLFM design is modular, and thus is capable of improved imaging metrics if updated with existing technologies. For example, the quality of light-sheet will affect the image quality of LSLFM. Our LSLFM used a static light-sheet generated by a cylindrical lens to illuminate the entire imaging plane. One limitation of a static light-sheet is the appearance of stripe artifacts caused by non-uniform illumination arising from the refraction, scattering, and absorption of the coherent excitation light at the lateral surface of the fish or interfaces within the fish. Light-sheet formed by sweeping a Gaussian, Bessel, or Airy beam over the lateral direction would greatly reduce such artifacts [6,34,35]. Alternatively, light-sheet generated by diffuse beam could also alleviate stripe artifacts [27]. Likewise, using superior individual optical elements within the microscope, such as higher NA objectives or microlens arrays with higher lens densities, would increase the resolution of our design.
Volumetric neuroimaging regularly produces large data sets and thus requires appropriate data processing and analysis for high-resolution neuron segmentation and activity extraction. LSLFM could potentially reduce the computational burden of analyzing large-scale imaging datasets through its resolving power. In our direct comparison, LSLFM images had substantially higher contrast than WFLFM images because LSLFM reduced the illumination of out-of-focus background. Thus, LSLFM can resolve fine features and neural calcium transients with higher fidelity than WFLFM without employing deconvolution [14]. Bypassing the computationally-intensive deconvolution step could potentially speed up the extraction of neural activity from imaging data. Traditional machine learning [36] and deep-learning algorithms [37] have demonstrated robust performance in spike detection and neuron segmentation on multiple forms of calcium imaging data. Such algorithms will take advantage of LSLFM’s high resolving power to improve the speed of data processing and accuracy of calcium transient detection as well. Coupled with advanced image processing methods, LSLFM should improve the analysis and interpretation of large-scale neural recordings.
Funding
Beckman Young Investigator Award; National Institutes of Health10.13039/100000002 Brain Iniative (1UF1NS107678); National Science Foundation10.13039/100000001 Career Award (1847540); Brain Research Foundation10.13039/100000882 Seed Grant; National Science Foundation10.13039/100000001 Brain Iniative (NCS-FO 1533598); Vallee Young Investigator Award.
Disclosures
The authors declare no conflicts of interest.
References
- 1.Lemon W. C., Pulver S. R., Höckendorf B., McDole K., Branson K., Freeman J., Keller P. J., “Whole-central nervous system functional imaging in larval Drosophila,” Nat. Commun. 6(1), 7924 (2015). 10.1038/ncomms8924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quirin S., Vladimirov N., Yang C.-T., Peterka D. S., Yuste R., Ahrens M. B., “Calcium imaging of neural circuits with extended depth-of-field light-sheet microscopy,” Opt. Lett. 41(5), 855–858 (2016). 10.1364/OL.41.000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang W., Yuste R., “In vivo imaging of neural activity,” Nat. Methods 14(4), 349–359 (2017). 10.1038/nmeth.4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koroshetz W., Gordon J., Adams A., Beckel-Mitchener A., Churchill J., Farber G., Freund M., Gnadt J., Hsu N. S., Langhals N., “The state of the NIH Brain initiative,” J. Neurosci. 38(29), 6427–6438 (2018). 10.1523/JNEUROSCI.3174-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Power R. M., Huisken J., “A guide to light-sheet fluorescence microscopy for multiscale imaging,” Nat. Methods 14(4), 360–373 (2017). 10.1038/nmeth.4224 [DOI] [PubMed] [Google Scholar]
- 6.Krzic U., Gunther S., Saunders T. E., Streichan S. J., Hufnagel L., “Multiview light-sheet microscope for rapid in toto imaging,” Nat. Methods 9(7), 730–733 (2012). 10.1038/nmeth.2064 [DOI] [PubMed] [Google Scholar]
- 7.Ahrens M. B., Orger M. B., Robson D. N., Li J. M., Keller P. J., “Whole-brain functional imaging at cellular resolution using light-sheet microscopy,” Nat. Methods 10(5), 413–420 (2013). 10.1038/nmeth.2434 [DOI] [PubMed] [Google Scholar]
- 8.Fahrbach F. O., Voigt F. F., Schmid B., Helmchen F., Huisken J., “Rapid 3D light-sheet microscopy with a tunable lens,” Opt. Express 21(18), 21010–21026 (2013). 10.1364/OE.21.021010 [DOI] [PubMed] [Google Scholar]
- 9.Bouchard M. B., Voleti V., Mendes C. S., Lacefield C., Grueber W. B., Mann R. S., Bruno R. M., Hillman E. M., “Swept confocally-aligned planar excitation (SCAPE) microscopy for high-speed volumetric imaging of behaving organisms,” Nat. Photonics 9(2), 113–119 (2015). 10.1038/nphoton.2014.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryan D. P., Gould E. A., Seedorf G. J., Masihzadeh O., Abman S. H., Vijayaraghavan S., Macklin W. B., Restrepo D., Shepherd D. P., “Automatic and adaptive heterogeneous refractive index compensation for light-sheet microscopy,” Nat. Commun. 8(1), 612 (2017). 10.1038/s41467-017-00514-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustavsson A.-K., Petrov P. N., Lee M. Y., Shechtman Y., Moerner W., “3D single-molecule super-resolution microscopy with a tilted light sheet,” Nat. Commun. 9(1), 123 (2018). 10.1038/s41467-017-02563-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meddens M. B., Liu S., Finnegan P. S., Edwards T. L., James C. D., Lidke K. A., “Single objective light-sheet microscopy for high-speed whole-cell 3D super-resolution,” Biomed. Opt. Express 7(6), 2219–2236 (2016). 10.1364/BOE.7.002219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levoy M., Ng R., Adams A., Footer M., Horowitz M., “Light field microscopy,” ACM Trans. Graph 25(3), 924–934 (2006). 10.1145/1141911.1141976 [DOI] [Google Scholar]
- 14.Broxton M., Grosenick L., Yang S., Cohen N., Andalman A., Deisseroth K., Levoy M., “Wave optics theory and 3-D deconvolution for the light field microscope,” Opt. Express 21(21), 25418–25439 (2013). 10.1364/OE.21.025418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prevedel R., Yoon Y.-G., Hoffmann M., Pak N., Wetzstein G., Kato S., Schrödel T., Raskar R., Zimmer M., Boyden E. S., “Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy,” Nat. Methods 11(7), 727–730 (2014). 10.1038/nmeth.2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skocek O., Nöbauer T., Weilguny L., Traub F. M., Xia C. N., Molodtsov M. I., Grama A., Yamagata M., Aharoni D., Cox D. D., “High-speed volumetric imaging of neuronal activity in freely moving rodents,” Nat. Methods 15(6), 429–432 (2018). 10.1038/s41592-018-0008-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wolff J. M., Castro D., Arbeláez P., Forero-Shelton M., “Light-sheet enhanced resolution of light field microscopy for rapid imaging of large volumes,” in Three-Dimensional and Multidimensional Microscopy: Image Acquisition and Processing XXV, (International Society for Optics and Photonics, 2018), 104991U. [Google Scholar]
- 18.Wagner N., Norlin N., Gierten J., de Medeiros G., Balázs B., Wittbrodt J., Hufnagel L., Prevedel R., “Instantaneous isotropic volumetric imaging of fast biological processes,” Nat. Methods 16(6), 497–500 (2019). 10.1038/s41592-019-0393-z [DOI] [PubMed] [Google Scholar]
- 19.Truong T. V., Holland D. B., Madaan S., Andreev A., Troll J. V., Koo D. E., Keomanee-Dizon K., McFall-Ngai M., Fraser S. E., “Selective volume illumination microscopy offers synchronous volumetric imaging with high contrast,” bioRxiv, 403303 (2018).
- 20.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C. B., “The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs,” Dev. Dyn. 236(11), 3088–3099 (2007). 10.1002/dvdy.21343 [DOI] [PubMed] [Google Scholar]
- 21.Chen T.-W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V., “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499(7458), 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Bovik A. C., Sheikh H. R., Simoncelli E. P., “Image quality assessment: from error visibility to structural similarity,” IEEE Trans. on Image Process. 13(4), 600–612 (2004). 10.1109/TIP.2003.819861 [DOI] [PubMed] [Google Scholar]
- 23.Ng R., Levoy M., Brédif M., Duval G., Horowitz M., Hanrahan P., “Light field photography with a hand-held plenoptic camera,” Computer Science Technical Report CSTR 2, 1–11 (2005). [Google Scholar]
- 24.Wilburn B., Joshi N., Vaish V., Talvala E.-V., Antunez E., Barth A., Adams A., Horowitz M., Levoy M., “High performance imaging using large camera arrays,” in ACM Trans. Graph, (ACM, 2005), 765–776. [Google Scholar]
- 25.Tian L., Wang J., Waller L., “3D differential phase-contrast microscopy with computational illumination using an LED array,” Opt. Lett. 39(5), 1326–1329 (2014). 10.1364/OL.39.001326 [DOI] [PubMed] [Google Scholar]
- 26.Weber E. H., EH Weber on the tactile senses (Psychology Press, 1996). [Google Scholar]
- 27.Taylor M. A., Vanwalleghem G. C., Favre-Bulle I. A., Scott E. K., “Diffuse light-sheet microscopy for stripe-free calcium imaging of neural populations,” J. Biophotonics 11(12), e201800088 (2018). 10.1002/jbio.201800088 [DOI] [PubMed] [Google Scholar]
- 28.Kunst M., Laurell E., Mokayes N., Kramer A., Kubo F., Fernandes A. M., Förster D., Dal Maschio M., Baier H., “A cellular-resolution atlas of the larval zebrafish brain,” Available at SSRN 3257346 (2018). [DOI] [PubMed]
- 29.Bhandiwad A. A., Zeddies D. G., Raible D. W., Rubel E. W., Sisneros J. A., “Auditory sensitivity of larval zebrafish (Danio rerio) measured using a behavioral prepulse inhibition assay,” J. Exp. Biol. 216(18), 3504–3513 (2013). 10.1242/jeb.087635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guizar-Sicairos M., Thurman S. T., Fienup J. R., “Efficient subpixel image registration algorithms,” Opt. Lett. 33(2), 156–158 (2008). 10.1364/OL.33.000156 [DOI] [PubMed] [Google Scholar]
- 31.Giovannucci A., Friedrich J., Gunn P., Kalfon J., Brown B. L., Koay S. A., Taxidis J., Najafi F., Gauthier J. L., Zhou P., “CaImAn an open source tool for scalable calcium imaging data analysis,” eLife 8, e38173 (2019). 10.7554/eLife.38173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gong Y., Huang C., Li J. Z., Grewe B. F., Zhang Y., Eismann S., Schnitzer M. J., “High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor,” Science 350(6266), 1361–1366 (2015). 10.1126/science.aab0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong Y., Wagner M. J., Li J. Z., Schnitzer M. J., “Imaging neural spiking in brain tissue using FRET-opsin protein voltage sensors,” Nat. Commun. 5(1), 3674 (2014). 10.1038/ncomms4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vettenburg T., Dalgarno H. I., Nylk J., Coll-Lladó C., Ferrier D. E., Čižmár T., Gunn-Moore F. J., Dholakia K., “Light-sheet microscopy using an Airy beam,” Nat. Methods 11(5), 541–544 (2014). 10.1038/nmeth.2922 [DOI] [PubMed] [Google Scholar]
- 35.Chen B.-C., Legant W. R., Wang K., Shao L., Milkie D. E., Davidson M. W., Janetopoulos C., Wu X. S., Hammer J. A., Liu Z., “Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution,” Science 346(6208), 1257998 (2014). 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theis L., Berens P., Froudarakis E., Reimer J., Rosón M. R., Baden T., Euler T., Tolias A. S., Bethge M., “Benchmarking spike rate inference in population calcium imaging,” Neuron 90(3), 471–482 (2016). 10.1016/j.neuron.2016.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soltanian-Zadeh S., Sahingur K., Blau S., Gong Y., Farsiu S., “Fast and robust active neuron segmentation in two-photon calcium imaging using spatiotemporal deep learning,” Proceedings of the National Academy of Sciences, 201812995 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]