Abstract
In the clinical practice of pathology, trichrome stains are commonly used to highlight collagen and to help evaluate fibrosis. Such stains do delineate collagen deposits but are not molecularly specific and can suffer from staining inconsistencies. Moreover, performing histochemical stain evaluation requires the preparation of additional sections beyond the original hematoxylin- and eosin-stained slides, as well as additional staining steps, which together add cost, time, and workflow complications. We have developed a new microscopy approach, termed DUET (DUal-mode Emission and Transmission) that can be used to extract signals that would typically require special stains or advanced optical methods. Our preliminary analysis demonstrates the potential of using the resulting signals to generate virtual histochemical images that resemble trichrome-stained slides and can support clinical evaluation. We demonstrate advantages of this approach over images acquired from conventional trichrome-stained slides and compare them with images created using second harmonic generation microscopy.
1. Introduction
Molecules from the collagen family comprise the main component of connective tissue and are the most abundant protein class in mammals [1]. In addition to their obvious presence in tissues such as ligament, bone, skin and cartilage, various types of collagen are also widely present in the extracellular matrix, blood vessels, and basement membranes of essentially all organs, playing important roles in physiology and response to injury.
A common sequela of ongoing tissue injury involves the development of fibrosis, namely an excess accumulation of extracellular matrix primarily composed of collagen. This process can be a normal reactive and reparative response, especially in situations where injury is sustained over long periods of time. With disease progression, more and more of the normal tissue architecture can be replaced with fibrotic scar, leading to eventual loss of organ function. In many disease settings, the overall extent of fibrosis reflects the level of irreversible end-organ injury and is the best recognized indicator for disease stage and prognosis. This is especially true for medical diseases of the kidney and liver, as well as transplant pathology in these organs. In such settings, histologic evaluation is essential for determining the extent and distribution of fibrosis and is considered standard of care. Recently, it has been increasingly recognized that abnormal collagen composition and deposition is also intimately involved in the interaction between cancers and their adjacent stroma, and subsequent tumor behavior can be shaped through mechanisms thought to involve tumor cell adhesion, proliferation, migration and invasion [2]. These interactions can have profound impact on tumor progression and prognosis via imputed biochemical and mechanical mechanisms [3–7].
While dense fibrosis is fairly straightforward to recognize using standard hematoxylin- and eosin- (H&E-) stained histologic slides, more sensitive methods are often required for optimal detection of fibrosis [8]. In the clinical setting, trichrome is the most common staining method, but comes with a number of drawbacks. As with other histochemical stains, trichrome can exhibit significant stain variation between institutions and laboratories. Even the same laboratory may have inconsistent staining, with results varying from day to day. Also, trichrome is not specific to collagen and will stain other material to exhibit a similar blue hue. Conversely, some studies show that trichrome stains can also underestimate collagen content [9–11].
Other histochemical stains such as picrosirius red are technically superior to trichrome for collagen detection [6,7], offering finer spatial resolution of the collagen fibers. However, such stains have not found widespread adoption in the clinical setting, possibly due to established practice, pathologist preference or the requirement for polarization optics for optimal performance. Additional histochemical stains are used routinely in clinical situations to highlight other collagen-containing structures. For instance, periodic acid-Schiff (PAS) and Jones methenamine silver stains are routinely used in kidney biopsy evaluation to increase the contrast of glomerular and tubular basement membrane as well as the mesangium within glomeruli. In all these settings, using histochemical stains in addition to H&E-stained sections require the preparation of additional sections, availability of the necessary (and unexpired) staining reagents, and either automated equipment or trained technicians to perform the procedures, all of which adds cost and time, and affects laboratory workflow.
Alternative approaches based on optical contrast, such as second-harmonic generation (SHG) microscopy or polarization techniques to highlight collagen have also been explored [12,13]. SHG is an expensive approach that requires ultrashort pulsed lasers, high-numerical aperture objectives and precision scanning, and is limited to generating signals only from the non-centrosymmetric collagens such as types I, and II [14]. The intensity of the detected collagen signals can also be highly orientation-dependent. Raman microscopy/spectroscopy is another method that can be used with or without labels to investigate the molecular makeup of tissue samples, though this method comes with relatively slow acquisition times and depends on expensive, complicated optics [15–17]. Fluorescence lifetime imaging has been also used to image regular H&E slide in fluorescence mode to create contrast between different macromolecules on H&E slides [18,19]. This method has been shown to identify and highlight components based on their fluorescence lifetime values. However, it is again expensive and complicated, requiring pulsed lasers and modulated sensors or photon counting devices to acquire images. Multispectral images of slides taken in brightfield mode can be used to extract collagenous or other components as well [20–27], but this method requires longer acquisition times, specialized acquisition optics and large data sets. Finally, autofluorescence of unstained slides has been used as input to neural networks for highlighting regions of imputed collagen deposition, but this method requires extensive training and validation [28].
Given the drawbacks of histochemical stains and or alternative optical or computational approaches, it would be ideal if standard H&E-stained slides could be used for quantitative, high-resolution visualization of extracellular matrix components. It was noted as early as 1969 that the eosin in H&E-stained slides was strongly fluorescent [29,30]. Various reports have since highlighted some of the extra information provided by fluorescence-based imaging of H&E-stains, including: visualizing immunoglobulins in kidney [31]; visualization of the septa in spleen frozen sections [32]; and the evaluation of elastin in arteries with confocal microscopy [33]. Utilizing H&E slides would be particularly advantageous because they are almost universally prepared for clinical and as well as research use, and typically accompany other histochemical or immunohistochemical stains.
Our innovation involves exploitation of previously underappreciated color (spectral) differences in H&E fluorescence. The method described here, Dual-mode Emission Transmission microscopy (DUET), highlights collagen distribution on already prepared H&E slides based on color content in fluorescence mode, without relying on complicated optics or additional histochemical or immunohistochemical staining. DUET combines brightfield and fluorescence images, along with a spectral phasors approach, to highlight collagen distribution. Due to its reliance simply on H&E-stained slides, its low cost, simplicity, rapid imaging speed, and non-destructive nature, DUET may be suitable for translation into clinical settings. It can also be extended to provide additional value when used to image frozen sections that are currently not typically evaluated with special stains because of time constraints. Finally, the additional information provided by DUET, over and above what is available from simple brightfield imaging, may enhance artificial-intelligence (AI) approaches that use image data as input.
2. Methods and materials
2.1. Histology sample preparation
Thin sections (4 µm) cut from formalin-fixed paraffin-embedded tissues, mounted on glass slides and stained with H&E were obtained from archival files of the Department of Pathology (UC Davis and Johns Hopkins University). The samples were provided under IRB biobank protocol and deidentified prior to imaging. The project did not include any interaction or intervention with human subjects.
2.2. Imaging instrumentation and control software
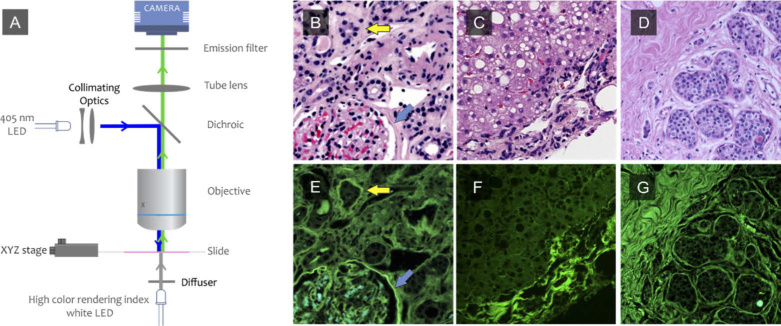
A DUET whole-slide scanning system comprises a spectrally broad, white LED array (CCLU026-1201C1 Super High-CRI Model, Citizen) integrated with a diffuser located below the slide to generate even transillumination across the whole slide at once. Epifluorescence excitation provided by a 405-nm LED (LZ1-00UB00-LED Engin) is directed to the sample via a broadband dichroic (Di03-R405-t1-25 × 36, Semrock) and focused on the sample using a 10X objective (Nikon, Plan Apo, 0.45 NA). A long-pass emission filter (FF01-430/LP-25, Semrock) is present in front of the sensor to reject scattered and reflected light when imaging in fluorescence mode. It remains in place for brightfield imaging and does not adversely affect brightfield color rendition. The slide is positioned on a movable stage with a travel range of 25 mm by 50 mm, and a Z positioner is also included to provide automated focusing capabilities (Zaber Technologies). For the most part, the camera used to capture the images was a 9-megapixel scientific-grade CCD color camera (Ximea, MD091CU-SY) connected via a 200-mm tube lens (Thorlabs ILT 200). The specimen was scanned field by field, and at each capture position, brightfield and fluorescence images were taken in quick succession in near-perfect pixel registration. To assess the impact of camera choice on image quality and downstream analysis, two other sensors were tested: a 12-megapixel color CMOS sensor (Ximea, MC124CG-SY) and a greyscale CCD camera (Ximea, MD028MU-SY) with a rotating filter wheel equipped with red, green and blue filters (Astronomiks). For spectral imaging, the Ximea camera was replaced with a Nuance multispectral tunable-filter-based camera (PerkinElmer) and spectral image stacks were captured from 420 nm to 720 nm at 15-nm intervals. Image acquisition, alternation between light sources, stage movement and focusing was performed using control software developed in the .NET environment.
2.3. SHG imaging
In order to compare DUET images with optically visualized collagen signals, a multiphoton microscope was used to simultaneously record second-harmonic generation (SHG) and two-photon fluorescence (2PF) images [34]. Briefly, a femtosecond laser (Coherent Chameleon) coupled into a laser scanning confocal microscope (Olympus), tuned to 940 nm was focused onto the sample with a 40X, 0.9 NA objective. The laser power after the objective was 15 mW. SHG signals were collected in the forward direction with a 0.55 NA condenser lens and acquired by a photomultiplier tube with a 470 ± 20 nm bandpass filter. Two-photon-excited fluorescence (510-590 nm) images were simultaneously detected in the epi-direction. The scanning speed was 10 µs/pixel.
2.4. Phasor analysis
Extraction of image components from DUET data was initially performed using a spectral phasor approach for segmentation of 3-channel and multichannel spectral data. Phasor methods in microscopy and imaging involve a graphical representation of multichannel data and were originally used to analyze lifetime images [35]. The concept has been extended into the spectral domain by author FF and has been implemented to analyze spectral images of cells, in-vivo human skin, Raman signals, MRI images, histology samples and remote sensing data [36–40]. Phasor analysis was performed using an ImageJ plugin (www.Spechron.com); the mathematical concept is elaborated elsewhere [37,41]. Briefly, the sine and cosine transformation of the multi-channel spectra (which includes RGB color data as well) are computed at first harmonics and normalized to total intensity:
| (1) |
Where a is the collected intensity at channel l and L is the total number of channels. By plotting G vs. S on a 2D plot, the phasor histogram is created by linking each pixel on the original image with its corresponding phasor point. This provides a straightforward means for visualizing clusters of pixels with similar spectral properties. Mapping the pixels from those phasor clusters back to the original image allows for spatially resolved segmentation or unmixing, as well as visual assessment of signal distribution.
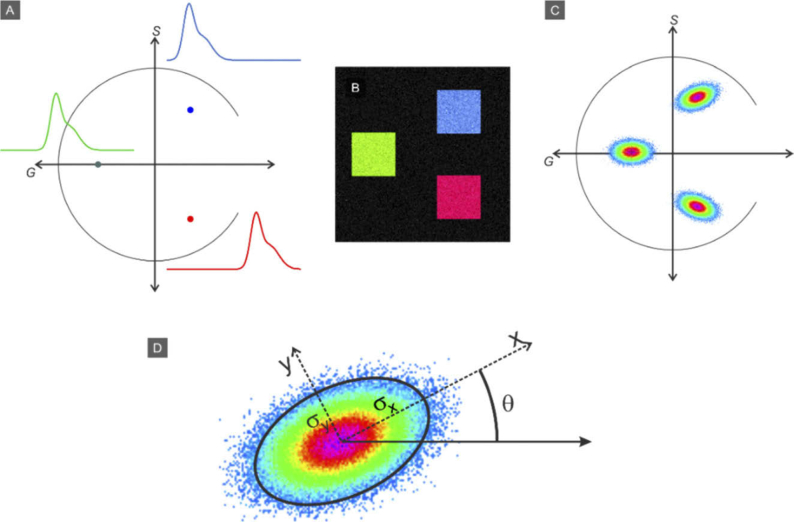
Figure 2 illustrates the concept. Panel A: Three example spectra are converted into phasor points representing their spectral content and are located in three positions on the phasor plot. Panel B shows a synthetic image composed of three components indicated by blue, green and red squares, with Poisson noise added. The phasor transformation of this image is shown in Panel C, which contains 3 corresponding point clouds. A reciprocal transformation from the phasor cloud back to image coordinates enables a straightforward segmentation approach. After creating the phasor plot from the fluorescence RGB images, we use this method to identify and extract the collagen signal.
Fig. 2.
Concept of the spectral phasor approach. A) Transformation of three example spectra into phasor points within the unit circle. B) Synthetic image composed of three (noisy) components. C) The phasor transformation of the image in panel B. D) Fitting a rotating two-dimensional Gaussian function to a phasor plot lobe and the fit parameters.
A rotating two-dimensional Gaussian function is defined to segment the region of interest on the phasor plot data and to perform the reciprocal transformation.
| (2) |
Where is a scaling factor and:
Figure 2(D) shows the parameters schematically. and are the RMS with of the Gaussian function on the rotated x and y axis. After initial evaluation of the phasor plot to identify the collagen lobe, the image processing procedure involves user selecting any region on the collagen lobe and then the rotating Gaussian function is found by fitting f from Eq. (2) to phasor plot data using a nonlinear least-squares method. The average intensity of corresponding pixels from the image to phasor points are multiplied by the resulted Gaussian function, to provide grayscale representation after the initial binary segmentation results.
2.5. Creating overlaid images
In order to highlight the collagen signal on the brightfield image, extracted signals can be pseudo-colored and layered on top. Assuming the illumination intensity of at each pixel, the collected signal for a given optical density is computed by Beer-Lambert Law:
| (3) |
In order to add the collagen content to the brightfield image, first we need to extract the optical density value from the brightfield image:
| (4) |
The overlaid image is computed by adding the collagen signal to the original image (expressed in optical density units) before converting the result into a simulated transmission image using the Beer-Lambert law:
| (5) |
The indices are selected in a way that the desired overlay color is achieved. For 8-bit color images,
3. Results
3.1. Fluorescence imaging of H&E slides
Pixel-registered brightfield and fluorescence images of H&E-stained slides from kidney, liver and breast cancer were acquired using DUET (Fig. 3). The fluorescence images demonstrate a predominantly green appearance, reflecting mostly emissions from the eosin stain excited at 405 nm and collected through a 420 nm long-pass emission filter. Although similar structures are visible in both modes, there are some components that can be appreciated only in fluorescence, for example, the basement membrane structures identified by arrows in panels B and E. Similar improvements in collagen contrast can be seen in the fluorescence images from liver and breast (panels C,D,F and G). However, the images features shown in this figure are visualized only due to increased brightness for collagen and basement membrane components. Such intensity differences are unlikely by themselves to enable robust signal extraction across a variety of specimens. For that it would be important to show that there are color (spectral) properties that can be relied on as well. Evidence for this is elaborated below.
Fig. 3.
DUET dual-mode imaging. An H&E slide is illuminated from below using a high color rendering index white LED and a color image is acquired; the white LEDs are turned off, and a 405-nm LED then is used in standard epifluorescence mode to excite fluorescence signals that are then collected using the same light path and camera. The two image modes are naturally pixel-registered. The process is repeated at each position of a whole-slide scanning procedure, using an XY stage and 10X lens. Brightfield image of human (B) kidney (C) liver and (D) breast from H&E slide. E-F: fluorescence images of human kidney, liver and breast from the same regions shown on B, C and D.
3.2. Multispectral analysis
The existence of extra information on the fluorescence image motivated the use of using multispectral analysis to explore whether there might be also spectral features in H&E-slide fluorescence. Accordingly, multispectral emission data were collected using a liquid-crystal tunable-filter-equipped camera (Nuance, PerkinElmer). The resulting spectra, averaged from a small number of adjacent pixels selected within regions corresponding to different structures, are shown in Fig. 4(C). The spectral curves obtained from collagen-, basement membrane- and tubular cytoplasm-containing regions (Fig. 4(C)) proved to be remarkably similar, consistent with why the potential utility of H&E-fluorescence for distinguishing such components may have previously been underappreciated. Red cells, not surprisingly, displayed more distinct spectral properties than the other components. Figure 4(D) shows the same spectra in log scale for increased clarity in the green to blue range.
Fig. 4.
Multispectral fluorescence imaging and phasor plot analysis of an H&E-stained slide of human kidney. A) Brightfield image; B) fluorescence color image synthesized from a 420-720-nm spectral data stack; C) spectra from representative regions containing collagen, basement membrane, tubules, and red blood cells; D) same spectra plotted in log scale for better visibility. E) Phasor plot of spectral data from the complete image with phasor clusters representing specific components identified as follows: Col (collagen); AF (autofluorescence); BM (basement membrane); PT (proximal tubules); and RBC (red blood cells). F-I) phasor-segmented images corresponding to the outlined clusters.
However, when these data were subjected to spectral phasor analysis (Fig. 4(E)) and plotted, clear clusters became apparent. These phasor clusters were then mapped back to the originating image space in grayscale mode, thereby highlighting specific constituents such as collagen, basement membrane, red blood cells, and renal proximal tubular cytoplasm (Fig. 4, (F-I)). The distal tubules fluorescence is usually weaker than the other compartments and their fluorescence spectrum is very close to autofluorescence spectrum and are not clustered here. The fact that nuclei are not visible in the fluorescence images (because hematoxylin is not a fluorescent dye) is an advantage, since their absence simplifies downstream analysis.
3.3. RGB 3-color analysis
Such multispectral data revealed the potential information content intrinsic in H&E-fluorescence images. However, it would be preferable if such information could be captured using simpler, snapshot RGB imaging systems that could be implemented at an order-of-magnitude lower price than multispectral approaches, and could capture images more quickly as well. Phasor-based analyses of RGB color-camera-captured fluorescence images are shown in Fig. 5. Even though these fluorescence images were acquired using simple RGB sensors, there was still sufficient spectral content, as revealed in the phasor plots, to reliably extract collagen signals from the bulk eosin-based tissue fluorescence, as shown the righthand-most two columns.
Fig. 5.
RGB images of H&E-stained slide fluorescence. Top row: kidney; middle row: ovarian cancer; bottom row: breast cancer. Phasor plots are shown in the middle column, with unmixed collagen signals shown as monochrome images and as blue overlays on top of the original brightfield images. Compared to the multispectral phasor plot shown in Fig. 4, these plots have fewer clusters, and it is not possible to separate collagen from basement membrane signals. The shapes of the phasor clusters differ from each other, reflecting the fact the slides were stained in different laboratories, and that 3 different color cameras (CCD, CMOS and 3-band filter-wheel-equipped monochrome) were used to capture these images.
However, unlike the relatively complex spectral phasor clouds demonstrated in the multispectral image shown in Fig. 4, the phasor data from the RGB images, not unexpectedly, displayed fewer clusters. The two major visible clusters highlighted collagen versus everything else, but it was not possible to pull out an additional basement-membrane cloud that had been discernible via the multispectral approach shown in Fig. 4. The RGB phasor clouds also varied in appearance from sample to sample. This variation could be attributable to differences in specimen type, staining laboratory with differences in dye components and procedures, and/or color camera choice. The kidney was imaged with a 12-megapixel CMOS sensor; the cervical cancer slide was captured using a greyscale CCD camera with a rotating filter wheel equipped with Astronomik color filters; and the breast specimen was captured using a 9-megapixel color CCD camera (all sensors from Ximea). Nevertheless, despite variations in the shapes and orientation of the phasor clouds, it was always possible to highlight the collagen, which appeared as an individual cluster on each plot. Clustering software tools are under development that should allow such assignments to be made automatically.
3.4. Comparison between slides from different institutions
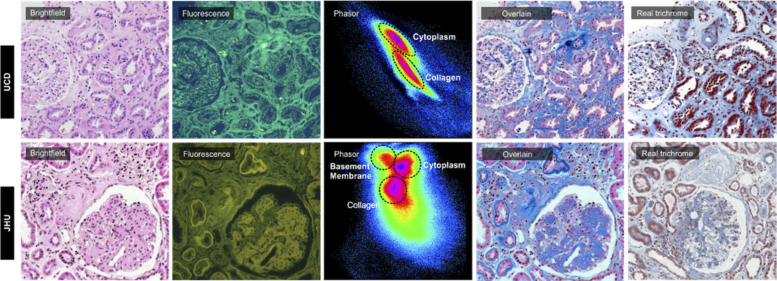
Comparisons between H&E-stained slides prepared at UC Davis and Johns Hopkins, and imaged using the same color camera, further indicate the variability that can be encountered from institution to institution. As shown in Fig. 6, while the brightfield images are similar, the fluorescence ones are quite distinct, with the JHU version having much less signal in the blue range (as indicated by the greenish-yellow tint). Although the fluorescence images are different in color, the resulted bulk collagen overlay images look very similar from two institutions and correspond well with the true trichrome stains prepared at each location (rightmost column). Intriguingly, despite the smaller overall spectral range in the JHU image, there proved to be more spectral content, as is evident in the respective phasor plots, and it was possible to extract a basement-membrane signal not visible in the UC Davis RGB phasor plot.
Fig. 6.
Brightfield and fluorescence images from the slides processed at UC Davis and Johns Hopkins University. Phasor plots demonstrate extractable components in spite of large color discrepancy between the fluorescence images from two institutions.
3.5. Comparison with independent detection methods
DUET signals attributed to connective tissue components were compared to independent collagen-detection methods: Mallory’s trichrome (histochemical), second-harmonic generation (SHG) microscopy (optical), and collagen III immunostaining.
SHG signals were acquired from the same slide imaged via DUET, so the results could be compared directly; the other methods were applied to serial sections, so while pixel-wise comparisons were not possible, general distribution patterns could be compared. Figure 7 illustrates a comparison between trichrome, SHG and DUET in a liver specimen manifesting diagnostic features consistent with NASH (non-alcoholic steatohepatitis). In this example, DUET highlights not only the relatively dense collagen signal around the central vein, but also fine “chicken-wire” collagen fibers surrounding some of the hepatocytes (a diagnostic feature of NASH) as seen in the region outlined in the yellow box on Panel F. That said, it is important to confirm that these signals truly arise from collagen deposits; the SHG image, taken from the same slide, does in fact indicate that these fibers can be visualized with both DUET and SHG—with minor differences reflecting the fact that the contrast mechanisms for the two methods are so distinct—demonstrating that the DUET signal can faithfully highlight the presence of delicate collagenous deposits. Such delicate features are hard to appreciate in the trichrome image (C), and as shown in panel G, are not visible at all on the H&E brightfield image.
Fig. 7.
A) Brightfield and B) fluorescence image of human liver biopsy from H&E slide. C) Serial section from the same region stained with trichrome. D) SHG signal and E) collagen image from DUET imaged from H&E slide. F) Overlaid image of collagen distribution from DUET on brightfield image. Zoomed in version of G) brightfield, H) overlaid, I) SHG and J) collagen extracted from DUET setup.
DUET is not only sensitive to fine structures, but also visualizes collagen deposits not actually detectable with SHG. H&E slides of human kidney were imaged using DUET and SHG, and a serial section was stained with trichrome (Fig. 8). After DUET analysis, the collagen channel (Fig. 8(D)) was overlaid onto the brightfield image to generate a virtual-trichrome appearance (Fig. 8(F)), which can be directly compared to the actual trichrome histochemical stain shown in Fig. 8(C). However, if the DUET signal is compared to the SHG image, it is evident that the interstitial collagen distribution detected by both methods is similar, while the glomerular signals can be appreciated only in the DUET version, since SHG does not detect any intraglomerular deposits. As indicated by [14] the main collagen class within typical, non-fibrotic glomeruli is type IV. This collagen species is not birefringent and consequently does not generate an SHG signal using commonly used femtosecond pulses. Moreover, DUET is sensitive to collagen type III, found in the inter-tubular regions, whereas SHG does not generate imageable contrast [14]. Antibody-based studies (Fig. 9) are also widely used—mostly in research–because, if properly performed, they provide information on the spatial distribution of specific molecular components. For example, collagen III is consistently detected in the renal interstitium, appearing as fine strands between normal tubules, as well as in broader areas or fibrosis, and in the presence of intimal fibrosis of larger arteries [42]. Our preliminary results with immunostaining of human kidney for collagen III and imaging the serial section reveal very similar distribution patterns between collagen III IHC and co-registered DUET signals.
Fig. 8.
A) Brightfield and B) fluorescence image of kidney biopsy from an H&E slide. C) Image of a serial section stained with trichrome. D) SHG image of the same region. Unlike DUET, SHG is does not highlight collagen structures (mostly collagen IV) within the glomerulus. E) Collagen distribution from the DUET-analyzed fluorescence image. F) Overlaid image of collagen distribution from DUET onto the corresponding brightfield image.
Fig. 9.
Serial section images of kidney stained with H&E (A), DUET overlaid (B), IHC staining for collagen type III (C). The blue pseudocolor in panel B correlates well with COLIII IHC.
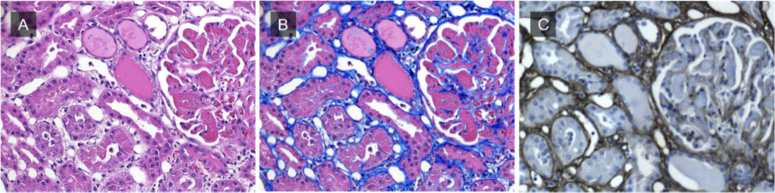
While good sensitivity is desirable, it should not come at the cost of diminished specificity. In fact, it appears that DUET may be in fact be more specific than trichrome, as shown in Fig. 10, which presents a comparison of DUET and trichrome staining of a collagenous region in a human liver biopsy. The H&E brightfield image is shown in panel A, with panel B displaying the corresponding DUET-extracted collagen signal overlaid onto the same brightfield image. Panel C shows the appearance of a trichrome-stained serial section.
Fig. 10.
A) Brightfield and B) DUET overlay image. C) Image of a serial section stained with trichrome. The yellow arrow points to the location of a nerve and the green arrow indicates the smooth muscle of arterial wall. These structures do not contain collagen, and do not show as positive in the DUET overlay image; however, they do appear blue (positive for collagen) in the trichrome-stained slide.
While there is good correlation between the virtual and real trichrome image on the overall collagen-containing regions surrounding the blood vessels, a few significant differences are evident on closer examination. The yellow and green arrows (Panels B and C) indicate the location of a nerve bundle and arteriole respectively. Both of these structures demonstrate false-positive blue coloration in the trichrome-stained specimen (C) while DUET correctly avoids attributing to them a collagen signal (B). This suggests that DUET can be more specific in detection of collagen-containing structures than the commonly used trichrome approach.
4. Conclusion and discussion
We have demonstrated that a simple, novel approach using dual-mode imaging of standard H&E-stained clinical histology specimens, combining brightfield and fluorescence, can reveal high-quality, spatially resolved collagen and basement-membrane signals without requiring special stains, expensive optics, or the preparation of extra slides. Being able to extract quantifiable, intrinsically digital collagen signals quickly and simply may be useful in both clinical and research settings.
In terms of health and disease, tissue collagen deposition is recognized as central to processes linked both to homeostasis as well as many underlying pathologies. As such, its evaluation is a critical part of biological research and clinical diagnostics. Parenthetically, it is instructive to consider the central role that collagen and triple-helical molecules generally has played in the earliest biological evolution of multicellular animals [43]. Type IV collagen in particular has specifically been implicated as the first molecule involved in this evolutionary process [44].
To study these essential molecules and their histological appearance, the usual methods involve conventional collagen stains such as trichrome, picrosirius red, or antibody-based approaches. However, these are time-consuming, potentially nonspecific, and can involve toxic reagents; the requisite additional sections can also consume potentially scarce specimen material. We show here that DUET was able to generate collagen signals that appear to be of higher quality than can be achieved using the commonly used trichrome stain, as follows:
-
1)
DUET images can have higher specificity than trichrome that can generate non-collagen signals in structures such as nerves or vessels;
-
2)
DUET can generate overall better images than trichrome, which can obscure nuclear and cytoplasmic detail. In contrast, the digital collagen signal is overlaid as a separate and “toggle-able” layer on top of the H&E image, which of course retains its original histologic appearance and quality (best appreciated in Fig. 1).
Fig. 1.
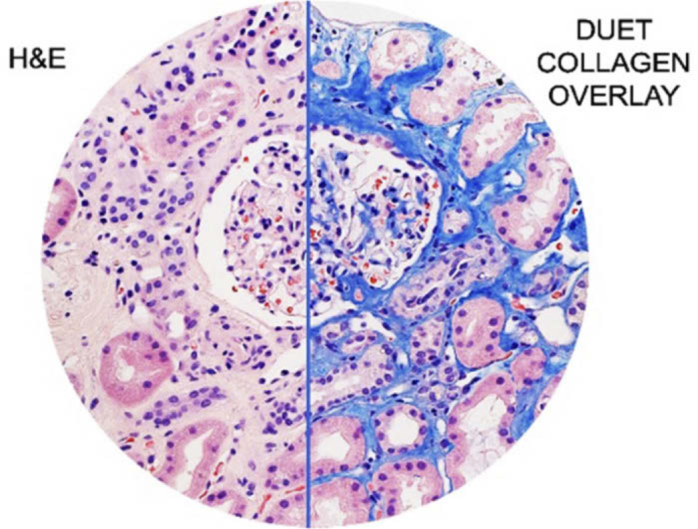
(Visual abstract) DUET imaging can identify collagen faithfully on H&E-stained slides and generate images that can improve upon conventional histochemical stains for better diagnostics and patient care.
As discussed above, there are other non-destructive techniques for visualizing collagens directly on unstained or H&E-stained slides; a widely used approach in research settings involves second-harmonic generation (SHG) microscopy, which can provide background-free and fairly specific collagen signals. In this report, SHG, as an orthogonal optical technique, was useful for validating DUET’s collagen detection performance. In fact, DUET, even though requiring much simpler and less expensive optics than SHG, proved to be almost as sensitive to collagen overall, but was also able to detect species, such as types III and IV, that are not readily detectable using SHG, since the contrast mechanism used in DUET interactions with eosin rather than intrinsic properties of (some) collagen molecules. In addition, unlike SHG, which depends on non-linear optical properties that mandating the use of high-NA (high-magnification) objectives, DUET can operate at any magnification and is able to scan whole slides within a few minutes.
Previous work using spectral phasor approaches demonstrated its value in a number of setting [38]. Here spectral phasors were deployed to extract collagen (and other) signals from the fluorescence channel. However, while collagen phasor clouds could be readily visualized, their position within the phasor plot did vary if differences in H&E-staining methods or camera performance were present. Such variability currently mandates some degree of manual supervision. However, this step can be automated in the future by virtue of cluster analysis techniques based on, for example, K-means [45] or hierarchical clustering [46] tools to automatically analyze phasor plots and separate collagen from the other tissue constituents and also to employ the morphological information to inspect the segmented image to see if it is collagen or not.
We found that it possible to separate overall collagen signals from other cellular constituents using just a 3-color RGB camera. However, when fluorescence images were collected across 30 spectral channels using a multispectral camera, the resulting spectral phasor plots were, not surprisingly, more complex and informative, and it proved possible to extract basement membranes as a separate signal. This demonstrates that the necessary information is indeed present within the fluorescence data—however, it would be more convenient if it the desired components did not need full spectral data for their detection.
Finally, the collagen signals are intrinsically digital and thus straightforward to introduce into image analysis tools of various levels of sophistication. Future research will focus on developing and testing semi- and fully automated tools for analysis and quantitation of the tissue-component distributions, designed, at least initially, to mimic current manual fibrosis scoring methods for renal and hepatic disease assessment.
In conclusion, we describe DUET, a new and simple approach for collagen (and potentially other macromolecule) signal detection directly from H&E-stained slides. The methodology, originally explored in kidney and liver pathologies, should be extensible to other organ systems, including cancer-related diseases. Since only H&E-stained slides are needed for DUET imaging, major archival-based research is feasible across many disease categories in which special stains may not have been originally or routinely performed. Archived slides also allow the use of long (retrospective) follow-up, so clinical outcomes can be readily linked to collagen distribution phenotypes. Another avenue can include true prospective clinical studies that would use DUET data, possibly coupled, possibly using AI analytics, with patient outcomes, to actively guide patient management. We think that this work promises to have significant positive impact on fibrosis disease research, clinical practice, and ultimately on clinical care.
Acknowledgments
We would like to express our thanks to UC Davis Health Department of Pathology and Laboratory Medicine and UC Davis Comprehensive Cancer Center to support this work.
Funding
National Cancer Institute10.13039/100000054 (R33 CA202881-01).
Disclosures
FF: HistoliX (I,P) RL: HistoliX (I,P)
References
- 1.Shoulders M. D., Raines R. T., “Collagen structure and stability,” Annu. Rev. Biochem. 78(1), 929–958 (2009). 10.1146/annurev.biochem.77.032207.120833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frantz C., Stewart K. M., Weaver V. M., “The extracellular matrix at a glance,” J. Cell Sci. 123(24), 4195–4200 (2010). 10.1242/jcs.023820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokarz D., Cisek R., Joseph A., Golaraei A., Mirsanaye K., Krouglov S., Asa S.L., Wilson B.C., Barzda V., “Characterization of Pancreatic Cancer Tissue using Multiphoton Excitation Fluorescence and Polarization-Sensitive Harmonic Generation Microscopy,” Front. Oncol. 9, 272 (2019). 10.3389/fonc.2019.00272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamauchi M., Barker T. H., Gibbons D. L., Kurie J. M., “The fibrotic tumor stroma,” J. Clin. Invest. 128(1), 16–25 (2018). 10.1172/JCI93554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z.-S.J., Yang D.-H., Protein Kinase Inhibitors as Sensitizing Agents for Chemotherapy (Academic Press, 2018).
- 6.Natal R. A., Vassallo J., Paiva G. R., Pelegati V. B., Barbosa G. O., Mendonça G. R., Bondarik C., Derchain S. F., Carvalho H. F., Lima C. S., “Collagen analysis by second-harmonic generation microscopy predicts outcome of luminal breast cancer,” Tumor Biol. 40(4), 1010428318770953 (2018). 10.1177/1010428318770953 [DOI] [PubMed] [Google Scholar]
- 7.Drifka C. R., Loeffler A. G., Mathewson K., Keikhosravi A., Eickhoff J. C., Liu Y., Weber S. M., Kao W. J., Eliceiri K. W., “Highly aligned stromal collagen is a negative prognostic factor following pancreatic ductal adenocarcinoma resection,” Oncotarget 7, 76197 (2016). 10.18632/oncotarget.12772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tretheway D., Jain A., LaPoint R., Sharma R., Orloff M., Milot P., Bozorgzadeh A., Ryan C., “Should trichrome stain be used on all post–liver transplant biopsies with hepatitis C virus infection to estimate the fibrosis score?” Liver Transpl. 14, 695–700 (2008). 10.1002/lt.21422 [DOI] [PubMed] [Google Scholar]
- 9.Cason J. E., “A rapid one-step Mallory-Heidenhain stain for connective tissue,” Stain Technol. 25(4), 225–226 (1950). 10.3109/10520295009110996 [DOI] [PubMed] [Google Scholar]
- 10.Lillie R., Miller G., “Histochemical acylation of hydroxyl and amino groups. Effect on the periodic acid Schiff reaction, anionic and cationic dye and Van Gieson collagen stains,” J. Histochem. Cytochem. 12(11), 821–841 (1964). 10.1177/12.11.821 [DOI] [PubMed] [Google Scholar]
- 11.Whittaker P., Kloner R., Boughner D., Pickering J., “Quantitative assessment of myocardial collagen with picrosirius red staining and circularly polarized light,” Basic Res. Cardiol. 89(5), 397–410 (1994). 10.1007/BF00788278 [DOI] [PubMed] [Google Scholar]
- 12.Drifka C. R., Loeffler A. G., Mathewson K., Mehta G., Keikhosravi A., Liu Y., Lemancik S., Ricke W. A., Weber S. M., Kao W. J., “Comparison of picrosirius red staining with second harmonic generation imaging for the quantification of clinically relevant collagen fiber features in histopathology samples,” J. Histochem. Cytochem. 64(9), 519–529 (2016). 10.1369/0022155416659249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shribak M., “Polychromatic polarization microscope: bringing colors to a colorless world,” Sci. Rep. 5(1), 17340 (2015). 10.1038/srep17340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Nadiarynkh O., Plotnikov S., Campagnola P. J., “Second harmonic generation microscopy for quantitative analysis of collagen fibrillar structure,” Nat. Protoc. 7(4), 654–669 (2012). 10.1038/nprot.2012.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andronia L., Mireșan V., Coroian A., Pop I., Răducu C., Rotaru A., Cocan D., Pânzaru S.C., Domșa I., Coroian C.O., “Raman Spectroscopy of the Hematoxylin-Eosin Stained Tissue,” ProEnvironment/ProMediu 8(24), 590–600 (2015). [Google Scholar]
- 16.Rodner E., Bocklitz T., von Eggeling F., Ernst G., Chernavskaia O., Popp J., Denzler J., Guntinas-Lichius O., “Fully convolutional networks in multimodal nonlinear microscopy images for automated detection of head and neck carcinoma: Pilot study,” Head & Neck 41(1), 25489 (2018). 10.1002/hed.25489 [DOI] [PubMed] [Google Scholar]
- 17.Leoncini E., Ricciardi W., Cadoni G., Arzani D., Petrelli L., Paludetti G., Brennan P., Luce D., Stucker I., Matsuo K., “Adult height and head and neck cancer: a pooled analysis within the INHANCE Consortium,” Eur. J. Epidemiol. 29(1), 35–48 (2014). 10.1007/s10654-013-9863-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu J., Fu C. Y., Ng B. K., Gulam Razul S., Lim S. K., “Quantitative diagnosis of cervical neoplasia using fluorescence lifetime imaging on haematoxylin and eosin stained tissue sections,” J. Biophotonics 7(7), 483–491 (2014). 10.1002/jbio.201200202 [DOI] [PubMed] [Google Scholar]
- 19.Luo T., Lu Y., Liu S., Lin D., Qu J., “Enhanced Visualization of Hematoxylin and Eosin Stained Pathological Characteristics by Phasor Approach,” Anal. Chem. 89(17), 9224–9231 (2017). 10.1021/acs.analchem.7b01999 [DOI] [PubMed] [Google Scholar]
- 20.Abeysekera S., Ooi M.P.-L., Kuang Y.C., Tan C.P., Hassan S.S., Detecting spongiosis in stained histopathological specimen using multispectral imaging and machine learning, 2014 IEEE Sensors Applications Symposium (SAS), IEEE, 2014, pp. 195–200. [Google Scholar]
- 21.Bautista P.A., Yagi Y., “Localization of Eosinophilic Esophagitis from H&E stained images using multispectral imaging,” Diagn. Pathol. 6(S1), S2 (2011). 10.1186/1746-1596-6-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto N., Murakami Y., Bautista P. A., Yamaguchi M., Obi T., Ohyama N., Uto K., Kosugi Y., “Multispectral image enhancement for effective visualization,” Opt. Express 19(10), 9315–9329 (2011). 10.1364/OE.19.009315 [DOI] [PubMed] [Google Scholar]
- 23.Bautista P. A., Abe T., Yamaguchi M., Yagi Y., Ohyama N., “Digital staining for multispectral images of pathological tissue specimens based on combined classification of spectral transmittance,” Computerized Medical Imaging and Graphics 29(8), 649–657 (2005). 10.1016/j.compmedimag.2005.09.003 [DOI] [PubMed] [Google Scholar]
- 24.Levenson R. M., Fornari A., Loda M., “Multispectral imaging and pathology: seeing and doing more,” Expert Opin. Med. Diagn. 2(9), 1067–1081 (2008). 10.1517/17530059.2.9.1067 [DOI] [PubMed] [Google Scholar]
- 25.Bautista P. A., Yagi Y., “Digital simulation of staining in histopathology multispectral images: enhancement and linear transformation of spectral transmittance,” J. Biomed. Opt. 17(5), 056013 (2012). 10.1117/1.JBO.17.5.056013 [DOI] [PubMed] [Google Scholar]
- 26.Bautista P.A., Yagi Y., “Localization of eosinophilic esophagitis from H&E stained images using multispectral imaging,” Diagn. Pathol. 6(S1), S2 (2011). 10.1186/1746-1596-6-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bautista P.A., Abe T., Yamaguchi M., Ohyama N., Yagi Y., Multispectral image enhancement for H&E stained pathological tissue specimens, Medical Imaging 2008: Visualization, Image-Guided Procedures, and Modeling, International Society for Optics and Photonics, 2008, pp. 691836. [Google Scholar]
- 28.Rivenson Y., Wang H., Wei Z., de Haan K., Zhang Y., Wu Y., Günaydın H., Zuckerman J. E., Chong T., Sisk A. E., “Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning,” Nat. Biomed. Eng. 3(6), 466–477 (2019). 10.1038/s41551-019-0362-y [DOI] [PubMed] [Google Scholar]
- 29.Goldstein D., “The fluorescence of elastic fibres stained with eosin and excited by visible light,” Histochem. J. 1(3), 187–198 (1969). 10.1007/BF01081407 [DOI] [PubMed] [Google Scholar]
- 30.Lev R., Stoward P. J., “On the use of eosin as a fluorescent dye to demonstrate mucous cells and other structures in tissue sections,” Histochemie 20(4), 363–377 (1969). 10.1007/BF00263753 [DOI] [PubMed] [Google Scholar]
- 31.McMahon J. T., Myles J. L., Tubbs R. R., “Demonstration of Immune Complex Deposits Using Fluorescence Microscopy of Hematoxylin and Eosin–Stained Sections of Hollande's Fixed Renal Biopsies,” Mod. Pathol. 15(9), 988–997 (2002). 10.1097/01.MP.0000027202.51385.85 [DOI] [PubMed] [Google Scholar]
- 32.Jakubovský J., Guller L., Černá M., Balážová K., Polák Š, Jakubovská V., Babál P., “Fluorescence of hematoxylin and eosin-stained histological sections of the human spleen,” Acta Histochem. 104(4), 353–356 (2002). 10.1078/0065-1281-00684 [DOI] [PubMed] [Google Scholar]
- 33.de Carvalho H. F., Taboga S. R., “Fluorescence and confocal laser scanning microscopy imaging of elastic fibers in hematoxylin-eosin stained sections,” Histochem. Cell Biol. 106(6), 587–592 (1996). 10.1007/BF02473274 [DOI] [PubMed] [Google Scholar]
- 34.Awasthi S., Izu L. T., Mao Z., Jian Z., Landas T., Lerner A., Shimkunas R., Woldeyesus R., Bossuyt J., Wood B. M., “Multimodal SHG-2PF imaging of microdomain Ca2+-contraction coupling in live cardiac myocytes,” Circ. Res. 118(2), e19–e28 (2016). 10.1161/CIRCRESAHA.115.307919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Digman M. A., Caiolfa V. R., Zamai M., Gratton E., “The phasor approach to fluorescence lifetime imaging analysis,” Biophys. J. 94(2), L14–L16 (2008). 10.1529/biophysj.107.120154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fereidouni F., Bader A. N., Colonna A., Gerritsen H. C., “Phasor analysis of multiphoton spectral images distinguishes autofluorescence components of in vivo human skin,” J. Biophotonics 7, 589–596 (2014). 10.1002/jbio.201200244 [DOI] [PubMed] [Google Scholar]
- 37.Fereidouni F., Bader A. N., Gerritsen H. C., “Spectral phasor analysis allows rapid and reliable unmixing of fluorescence microscopy spectral images,” Opt. Express 20(12), 12729–12741 (2012). 10.1364/OE.20.012729 [DOI] [PubMed] [Google Scholar]
- 38.Fereidouni F., Griffin C., Todd A., Levenson R., “Multispectral analysis tools can increase utility of RGB color images in histology,” J. Opt. 20(4), 044007 (2018). 10.1088/2040-8986/aab0e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu D., Xie X. S., “Reliable cell segmentation based on spectral phasor analysis of hyperspectral stimulated Raman scattering imaging data,” Anal. Chem. 86(9), 4115–4119 (2014). 10.1021/ac500014b [DOI] [PubMed] [Google Scholar]
- 40.Vergeldt F. J., Prusova A., Fereidouni F., Van Amerongen H., Van As H., Scheenen T. W., Bader A. N., “Multi-component quantitative magnetic resonance imaging by phasor representation,” Sci. Rep. 7(1), 861 (2017). 10.1038/s41598-017-00864-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fereidouni F., Reitsma K., Gerritsen H. C., “High speed multispectral fluorescence lifetime imaging,” Opt. Express 21(10), 11769–11782 (2013). 10.1364/OE.21.011769 [DOI] [PubMed] [Google Scholar]
- 42.Farris A. B., Adams C. D., Brousaides N., Della Pelle P. A., Collins A. B., Moradi E., Smith R. N., Grimm P. C., Colvin R. B., “Morphometric and visual evaluation of fibrosis in renal biopsies,” J. Am. Soc. Nephrol. 22(1), 176–186 (2011). 10.1681/ASN.2009091005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fidler A. L., Boudko S. P., Rokas A., Hudson B. G., “The triple helix of collagens–an ancient protein structure that enabled animal multicellularity and tissue evolution,” J. Cell. Sci. 131(7), jcs203950 (2018). 10.1242/jcs.203950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fidler A. L., Darris C. E., Chetyrkin S. V., Pedchenko V. K., Boudko S. P., Brown K. L., Jerome W. G., Hudson J. K., Rokas A., Hudson B. G., “Collagen IV and basement membrane at the evolutionary dawn of metazoan tissues,” elife 6, e24176 (2017). 10.7554/eLife.24176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Likas A., Vlassis N., Verbeek J. J., “The global k-means clustering algorithm,” Pattern Recognition 36(2), 451–461 (2003). 10.1016/S0031-3203(02)00060-2 [DOI] [Google Scholar]
- 46.Deininger S.-O., Ebert M. P., Futterer A., Gerhard M., Rocken C., “MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers,” J. Proteome Res. 7(12), 5230–5236 (2008). 10.1021/pr8005777 [DOI] [PubMed] [Google Scholar]