ABSTRACT
Shiga toxin-producing Escherichia coli (STEC) colonize the gastrointestinal tract of animals; however, STEC may also cause severe diarrheal diseases. Food-producing animals have been acting as reservoirs and disseminators of multidrug-resistant (MDR) bacteria and antimicrobial resistance genes (ARGs); however, there are few studies characterizing molecularly bacterial isolates from sheep. Therefore, this study aimed to characterize E. coli isolates obtained from feces of sheep in a Brazilian farmhouse. A total of 14 MDR E. coli isolates were obtained from 100 feces samples, six of which were classified as non-O157 STEC (stx1, stx2 and ehxA). MDR E. coli isolates presented different ARGs [blaCTX-M-Gp9, blaCMY, blaSHV, qnrS, oqxB, aac(6ʹ)-Ib, tet(A), tet(B), tet(C), sul1, sul2, and cmlA] and plasmids (IncI1, IncFrepB, IncFIB, IncFIA, IncHI1, IncK, and ColE-like). In addition, mutations in the quinolone-resistance determining region of GyrA (Ser83Leu; Asp87Asn) and ParC (Glu84Asp) were detected. PFGE showed a high genetic diversity (30.9 to 83.9%) and thirteen STs were detected (ST25, ST48, ST155, ST162, ST642, ST1247, ST1518, ST1725, ST2107, ST2522, ST3270, ST5036, and ST7100). Subtyping of the fimH gene showed seven fimH-type (25, 32, 38, 41, 54, 61, and 366). The results found in the present study showed high genetic diversity among MDR ARGs-producing E. coli obtained from a farmhouse. This study reports for the first time, the presence of MDR STEC and non-STEC belonging to ST25, ST162, ST642, ST1247, ST1518, ST1725, ST2107, ST3270, ST5036, and ST7100 in sheep, and contributes to the surveillance studies associated with One Health concept.
KEYWORDS: Escherichia coli, multidrug-resistant, antimicrobial resistance, Diarrheagenic virulence genes, fimH, MLST, sheep, One Health
Introduction
Diarrheagenic Escherichia coli are responsible for diarrheal diseases in animals and humans, which are classified into well-defined pathotypes. Among them, Shiga toxin-producing Escherichia coli (STEC) are defined as zoonotic pathogens that colonize the gastrointestinal tract of animals (e.g. sheep and bovine); however, STEC may also cause severe diarrheal diseases [1,2]. Diarrheal diseases are classified as a public health problem, which affect the developing countries and industrialized countries, causing high rates of morbidity and mortality, as well as high health care costs [3].
Resistance to antimicrobials in bacteria is a global public health problem and multidrug-resistant (MDR) bacteria, including E. coli, have been spreading to different sources, which is worrying. The One Health concept has been applied worldwide due to the global challenge of bacterial resistance to antimicrobials [4]. The animals have been acting as reservoirs and disseminators (e.g. for humans and the environment) of MDR bacteria carrying several antimicrobial resistance genes (ARGs), including the extended-spectrum β-lactamases (ESBL) [5–8].
Many studies related to bacterial resistance to antimicrobials are performed in food-producing animals (i.e. chickens, cattle and pigs); however, there are few studies characterizing molecularly bacterial isolates from sheep. Therefore, the present study aimed to characterize E. coli isolates obtained from feces of sheep in a Brazilian farmhouse regarding resistance to antimicrobials, ARGs, plasmids, diarrheagenic virulence genes and serotypes, as well as sequence types, phylogenetic groups and fimH-type.
Materials and methods
Obtaining and identification of isolates
A total of 100 fecal samples were obtained in a farmhouse (4 ha) from Jardinópolis City, São Paulo State, Brazil. The feces samples were collected using sterile recipients and transported to the laboratory on the same day. The fecal samples (1 g) were added in sterile saline solution (5 mL) (0.9% NaCl) and, subsequently, seeded on MacConkey Agar (Oxoid, UK) and incubated at 37 ºC for 24 hours. A glucose-fermenting colony from each sample was selected and stocked at −80ºC in Brain Heart Infusion broth (Oxoid, UK) plus 15 % glycerol.
Molecular identification
The GenElute™ Bacterial Genomic DNA Kit (Sigma-Aldrich, USA) was used for the extraction of genomic DNA. The sequencing of the 16S rDNA gene was performed for the identification of the isolates using the BigDye™ Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific, USA) [9].
Detection of diarrheagenic virulence genes and E. coli serotyping
Detection of diarrheagenic virulence genes (ipaH, stx1, stx2, ehxA, aaiC, aatA, eae, bfpA, aggR, elt, est, aap, aggR, and AA probe) was performed by PCR [10–14]. Serotyping was performed by agglutination assays in 96-well Microtiter™ microplates (Thermo Fisher Scientific, USA), using rabbit antisera against 1 to 187 somatic (O) and 53 flagellar (H) antigens (SERUNAM, registered trademark in Mexico, 323,158/2015) [15,16].
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by disk diffusion [17]. Thirty-two antimicrobials were tested, including aminoglycosides (streptomycin, gentamicin, tobramycin, amikacin), β-lactams (ampicillin, amoxicillin-clavulanate, ampicillin-sulbactam, piperacillin-tazobactam, cefoxitin, cefazolin, cefuroxime, cefaclor, cefepime, cefotaxime, ceftriaxone, ceftazidime, ertapenem, meropenem, imipenem, aztreonam), (fluoro) quinolones (nalidixic acid, ciprofloxacin, levofloxacin, ofloxacin, norfloxacin, lomefloxacin), tetracyclines (minocycline, doxycycline, tetracycline), nitrofurans (nitrofurantoin), sulfonamides (trimethoprim-sulfamethoxazole), and phenicols (chloramphenicol). The isolates were classified as multidrug-resistant when presented non-susceptibility to ≥ 1 antimicrobial in ≥ 3 antimicrobial categories [18].
Detection of ARGs and plasmid replicon typing
The ARGs were detected by PCR in non-susceptible (resistant or intermediate) isolates for (fluoro) quinolones (oqxAB, qepA, qnrA, qnrB, qnrS), tetracyclines [tet(A) to (E), tet(G), tet(J), tet(L), tet(M), tet(O), tet(P), tet(Q), tet(S), and tet(X)], β-lactams [blaCTX-M (groups 1, 2, 8 and 9), blaCMY, blaVEB, blaPER, blaOXA-1-like, blaSHV], phenicols (floR, cmlA), sulfonamides (sul1, sul2, sul3), and aminoglycosides [aph(3ʹ)-Ia, aph(3′)-VI, aac(6ʹ)-Ib, aac(6′)-Ih, aac(3′)-Ia, aac(3′)-IIa, ant(2”)-Ia] [19–28]. The amplicons were sequenced for confirmation.
Detection of mutations in the quinolone-resistance determining region (QRDR) of GyrA (encoded by gyrA gene) and ParC (encoded by parC gene) was also performed [29,30]. Plasmids were detected by PCR-based replicon typing for twenty plasmid families (IncFrepB, IncFIB, IncFIA, IncFIC, IncU, IncR, IncHI1, IncHI2, IncK, IncY, IncI1, IncL/M, IncW, IncP, IncN, IncA/C, IncT, IncX, ColE-like) [31,32].
Pulsed-field gel electrophoresis (PFGE)
Genetic relatedness of E. coli isolates was performed by PFGE using 50U of XbaI restriction enzyme (Thermo Fisher Scientific, USA). Salmonella Braenderup H9812 was used as a molecular mass standard. The electrophoresis was performed on the CHEF-DR III system (Bio-Rad, USA) at 14 °C (Voltage: 6 V; Initial switch time: 6.76 seconds; Final switch time: 35.38 seconds; Included angle: 120°; Run time: 19 hours). A similarity dendrogram was constructed on the BioNumerics v. 7.6 (Applied Math, Belgium) using unweighted pair group method with arithmetic mean (UPGMA) and DICE similarity coefficient (optimization: 1.5%; band position tolerance: 1.5%). Discrimination index (DI) was evaluated using the Simpson’s diversity [33].
Phylogenetic groups, multilocus sequence typing (MLST) and fimH subtyping
The phylo-typing method was used to determine phylogenetic groups (A, B1, B2, C, D, E, and F) using five target genes (chuA, yjaA, TspE4.C2, arpA, and trpA) [34]. MLST analysis was performed as described by Achtman scheme (housekeeping genes [adk, gyrB, icd, fumC, purA, mdh, and recA]) available in Escherichia MLST website (https://pubmlst.org/escherichia/) and subtyping of fimH gene was performed using the FimTyper 1.0 [35,36].
Results
Isolates, diarrheagenic virulence genes and serotypes
In this study, 14 MDR E. coli isolates were obtained from feces of sheep from a Brazilian farmhouse. These isolates were identified by sequencing of the 16S rDNA (GenBank access no. MN147823-MN147836). Three diarrheagenic virulence genes (stx1, stx2 and ehxA) related to STEC pathotype were detected in six isolates (SJA6, SJA11, SJA29, SJA31, SJA80, and SJA92). The other diarrheagenic virulence genes (ipaH, aaiC, aatA, eae, bfpA, aggR, elt, est, aap, aggR, and AA probe) were not detected. Thirteen serotypes were assigned in E. coli isolates, including O153:H12 (2), O8:H21 (1), O127:H43 (1), O173:HNM (1), O100:HNM (1), O147:H19 (1), O154:H9 (1), O54:H21 (1), ONT:H10 (1), O102:H21 (1), O8:H19 (1), O78:HNM (1) (Figure 1).
Figure 1.
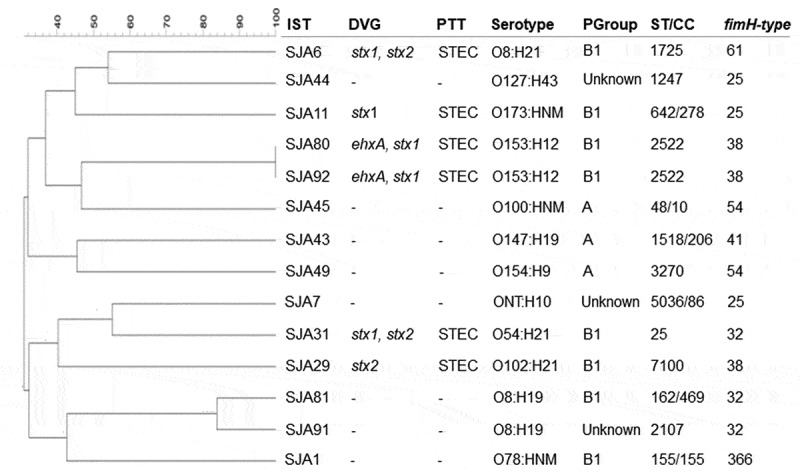
Dendrogram based on PFGE XbaI fingerprints representing the genetic relatedness among the MDR E. coli isolates obtained from sheep. IST, isolate; DVG, diarrheagenic virulence genes; PTT, pathotype; STEC, Shiga toxin-producing E. coli; PGroup, phylogenetic group; ST, sequence type; CC, clonal complex.
Antimicrobial resistance profile
All isolates were classified as MDR and presented several ARGs for β-lactams, aminoglycosides, tetracyclines, (fluoro) quinolones, phenicols, and sulfonamides (Table 1). All isolates were resistant to ampicillin, tetracycline and doxycycline, 12 (85.7%) to cefazolin, cefuroxime, cefaclor and trimethoprim-sulfamethoxazole, 5 (35.7%) to streptomycin, gentamicin and tobramycin, three (21.4%) to ampicillin-sulbactam, nalidixic acid, ciprofloxacin, levofloxacin, norfloxacin, lomefloxacin and ofloxacin, and two (12.3%) to chloramphenicol (Table 1).
Table 1.
STEC and non-STEC isolates according to antimicrobial resistance profile, ARGs and plasmids found.
| Isolate | Antimicrobial resistance profilea | ARGsb | Plasmid incompatibility (Inc) groups | |
|---|---|---|---|---|
| STEC | SJA6 | AMP, STP, GEN, TOB, TET, DOX, SXT, CLO | tet(A), tet(B), sul2, cmlA | FIA, FIB, ColE-like |
| SJA11 | AMP, CFZ, CRX, CFC, STP, GEN, TOB, TET, DOX | tet(A), tet(B), aac(6ʹ)-Ib | I1, ColE-like | |
| SJA29 | AMP, ASB, CFZ, CRX, CFC, STP, GEN, TOB, TET, DOX, NAL, CIP, LVX, NOR, LMX, OFX, SXT | blaCTX-M-Gp9, blaCMY, tet(A), tet(B), aac(6ʹ)-Ib, sul2 | FrepB, FIA, I1 | |
| SJA31 | AMP, CFZ, CRX, CFC, TET, DOX | tet(B), tet(C) | FrepB, FIA, I1 | |
| SJA80 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | blaCTX-M-Gp9, tet(B), sul2 | I1 | |
| SJA92 | AMP, CFZ, CRX, CFC, STP, GEN, TOB, TET, DOX, NAL, CIP, LVX, NOR, LMX, OFX, SXT | blaSHV, tet(B), qnrS, oqxB, sul1 | HI1 | |
| Non-STEC | SJA1 | AMP, TET, DOX, SXT, CLO | tet(A), tet(B), sul1, cmlA | FIB |
| SJA7 | AMP, ASB, CFZ, CRX, CFC, TET, DOX, SXT | blaCTX-M-Gp9, tet(A), tet(B), sul1 | FIB, I1, ColE-like | |
| SJA43 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | blaCTX-M-Gp9, tet(A), tet(B), sul2 | FrepB, I1 | |
| SJA44 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | blaCTX-M-Gp9, tet(A), tet(B), sul1 | I1, ColE-like | |
| SJA45 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | blaCTX-M-Gp9, blaSHV, tet(A), tet(B), sul1 | I1, K, ColE-like | |
| SJA49 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | tet(A), tet(B), sul1 | FrepB, I1 | |
| SJA81 | AMP, CFZ, CRX, CFC, TET, DOX, SXT | tet(A), tet(B), sul2 | HI1, I1 | |
| SJA91 | AMP, ASB, CFZ, CRX, CFC, STP, GEN, TOB, TET, DOX, NAL, CIP, LVX, NOR, LMX, OFX, SXT | blaCTX-M-Gp9, blaSHV, qnrS, tet(A), tet(B), sul2 | FIB, HI1, I1 |
a AMP, ampicillin; ASB, ampicillin-sulbactam; CFZ, cefazolin; CRX, cefuroxime; CFC, cefaclor; STP, streptomycin; GEN, gentamicin; TOB, tobramycin; TET, tetracycline; DOX, doxycycline, SXT, trimethoprim-sulfamethoxazole; NAL, nalidixic acid; CIP, ciprofloxacin; LVX, levofloxacin; NOR, norfloxacin; LMX, lomefloxacin; OFX, ofloxacin; CLO, chloramphenicol.
b ARGs, antimicrobial resistance genes.
ARGs and plasmids
Twelve different ARGs were detected in the MDR E. coli isolates and all presented at least two ARGs investigated. All isolates presented tet(B), followed by tet(A) (11), blaCTX-M-Gp9 (7), sul1 (6), sul2 (6), blaSHV (3), qnrS (2), cmlA (2), aac(6ʹ)-Ib (2), blaCMY (1), tet(C) (1), and oqxB (1) (Table 1). Among the fluoroquinolone-resistant E. coli isolates (SJA29, SJA91 and SJA92), only SJA29 showed mutations in QRDR of GyrA (Ser83Leu; Asp87Asn) and ParC (Glu84Asp) (GenBank accession no. MN148169-MN148182). Among the plasmid families, the IncI1 (11) was the most prevalent, followed by ColE-like (5), IncFrepB (4), IncFIB (4), IncFIA (3), IncHI1 (3), and IncK (1) (Table 1).
Epidemiological analysis
PFGE showed a high genetic diversity (30.9 to 83.9%) among the MDR E. coli isolates and thirteen sequence types (STs) belonging to six clonal complexes (CCs) were detected (ST25, ST48/CC10, ST155/CC155, ST162/CC469, ST642/CC278, ST1247, ST1518/CC206, ST1725, ST2107, ST2522, ST3270, ST5036/CC86, and ST7100). Subtyping of the fimH gene showed seven fimH-type (fimH25, 32, 38, 41, 54, 61 and 366) (Figure 1) (GenBank access no. MN148183-MN148196). Three phylogenetic groups [B1 (8), A (3) and Unknown (3)] were detected. Interestingly, the isolates SJA80 and SJA92 presented 100% of genetic similarity and the same diarrheagenic virulence genes, pathotype, serotype, ST, phylogenetic group and fimH-type; however, they presented different resistance profile as well as ARGs and plasmids (Table 1; Figure 1).
Discussion
STEC produces Shiga toxins [Stx1 (stx1) and Stx2 (stx2)], which may be associated with the enterohemolysin (ehxA), a virulence marker. STEC can cause diarrheal disease and outbreaks by STEC have been reported and are principally related to the consumption of contaminated products [1,2,37,38]. Infections by non-O157 STEC belonging to several serogroups (O8, O78, O100, O127, O153, and O154) have been increasing significantly over time in the United States, mainly in children (1 to 4 years) [39,40].
Resistance to β-lactams (ampicillin and cephalosporins), tetracyclines and sulfonamides have been increasingly detected in E. coli isolates obtained from different spheres. This MDR profile is associated with the presence of β-lactamases (CTX-M-like, SHV and CMY), acquired efflux pumps (TetABC) and changed dihydropteroate synthase (Sul) [41,42]. For other antimicrobials, such as aminoglycosides, phenicols and (fluoro) quinolones, the resistance is correlated to the presence of aminoglycoside-modifying enzymes (APH, ANT and AAC), efflux pumps (ClmA and FloR), and mutation in QRDR and/or plasmid-mediated quinolone resistance genes (PMQR), respectively [42–44].
Detection of antimicrobial-resistant E. coli, including MDR non-O157 STEC, obtained from animals (e.g. sheep) have been reported worldwide, including in Brazil [45–49]. Many studies characterize only the antimicrobial resistance profile of non-O157 STEC obtained from animals; however, there are few studies reporting different ARGs related to this phenotype. Srinivasa et al. [50], Ferdous et al. [51] and Bai et al. [52] reported non-O157 STEC carrying ARGs for tetracyclines (tet genes), (fluoro) quinolones (qnrS and oqxA) and sulfonamides (sul1 and sul2).
CTX-M-like β-lactamases have extended-spectrum against β-lactams antimicrobials and have been increasingly detected worldwide, including non-O157 STEC from animals [51,53–56]. The ARGs detected in the present study are commonly reported in plasmids [e.g. IncI1, IncF (FrepB, FIA and FIB), HI1, K, and ColE-like], which carrying principally encoding genes for β-lactamases, PMQR and efflux pumps [56–59]. IncI1 plasmids were the most detected in this study and have been reported carrying several ARGs, including tet(A), tet(B), blaCTX-M-like, sul1 and sul2 in E. coli isolates obtained from humans and animals [8,60,61].
The association of molecular typing and subtyping methods (i.e. phylogenetic group, MLST and fimH-type) has been used for epidemiological studies related to antimicrobial resistance and virulence in E. coli isolates. From this association, is possible to differentiate within the same ST/CC [34,36,62]. According to Enterobase Database (https://enterobase.warwick.ac.uk/), the ST48/CC10, ST155/CC155 and ST2522 have already been detected in sheep; however, all the other STs detected in the present study have been reported carrying ARGs in different sources (e.g. human, animal, food and the environment).
Curiously, E. coli isolates assigned as B1-ST2522-fimH38, B1-ST642-fimH25 and B1-ST155-fimH366 were previously reported in food, human, animals (i.e. food-producing animal, companion animal and wild animal), and in the environment (i.e. soil and water). Besides, ESBL-producing E. coli belonging to CC10 and CC155 are commonly reported causing infections in humans [63,64]. Therefore, to the best of our knowledge, this is the first report in the world of MDR STEC and non-STEC belonging to ST25, ST162/CC469, ST642/CC278, ST1247, ST1518/CC206, ST1725, ST2107, ST3270, ST5036/CC86, and ST7100 in sheep.
In conclusion, the results found in the present study showed high genetic diversity among MDR ARGs-producing E. coli, including non-O157 STEC, obtained from a farmhouse. These results contribute to the surveillance studies associated with One Health concept and call attention to the monitoring of MDR E. coli in animals, mainly in sheep.
Funding Statement
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant number 2018/19539-0].
Acknowledgments
The authors thank the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) [grant no. 88882.180855/2018-01 and code 001] and the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [grant no. 2018/01890-3] for fellowships.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Ferens WA, Hovde CJ.. Escherichia coli O157: H7:animal reservoir and sources of human infection. Foodborne Pathog Dis. 2011;8:465–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kumar A, Taneja N, Kumar Y, et al. Detection of shiga toxin variants among shiga toxin-forming Escherichia coli isolates from animal stool, meat and human stool samples in India. J Appl Microbiol. 2012;113:1208–1216. [DOI] [PubMed] [Google Scholar]
- [3].Farthing M, Salam MA, Lindberg G, et al. Acute diarrhea in adults and children: a global perspective. J Clin Gastroenterol. 2013;47:12–20. [DOI] [PubMed] [Google Scholar]
- [4].Bidaisee S, Macpherson CN.. Zoonoses and one health: a review of the literature. J Parasitol Res. 2014;2014:874345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Salyers A, Shoemaker NB. Reservoirs of antibiotic resistance genes. Anim Biotechnol. 2006;17:137–146. [DOI] [PubMed] [Google Scholar]
- [6].Geser N, Stephan R, Hächler H. Occurrence and characteristics of extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae in food producing animals, minced meat and raw milk. BMC Vet Res. 2012;7:8–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Toner E, Adalja A, Gronvall GK, et al. Antimicrobial resistance is a global health emergency. Health Secur. 2015;13:153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gozi KS, Froes JR, Deus ALPT, et al. Dissemination of multidrug-resistant commensal Escherichia coli in feedlot lambs in Southeastern Brazil. Front Microbiol. 2019;10:1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Weisburg WG, Barns SM, Pelletier BA, et al. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schmidt H, Knop C, Franke S, et al. Development of PCR for screening of enteroaggregative Escherichia coli. J Clin Microbiol. 1995;33:701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Aranda K, Fabbricotti SH, Fagundes-Neto U, et al. Single multiplex assay to identify simultaneously enteropathogenic, enteroaggregative, enterotoxigenic, enteroinvasive and shiga toxin producing Escherichia coli strains in Brazilian children. FEMS Microbiol Lett. 2007;267:145–150. [DOI] [PubMed] [Google Scholar]
- [13].Lima IF, Boisen N, Quetz Jda S, et al. Prevalence of enteroaggregative Escherichia coli and its virulence-related genes in a case-control study among children from north-eastern Brazil. J Med Microbiol. 2013;62:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cerna JF, Nataro JP, Estrada-Garcia T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol. 2003;41:2138–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Orskov F, Orskov I. Serotyping of Escherichia coli. Meth Microbiol. 1984;14:43–112. [Google Scholar]
- [16].Scheutz F, Cheasty T, Woodward D, et al. Designation of O174 and O175 to temporary O groups OX3 and OX7, and six new E. coli O groups that include Verocytotoxin-producing E. coli (VTEC): O176, O177, O178, O179, O180 and O181. APMIS. 2004;112:569–584. [DOI] [PubMed] [Google Scholar]
- [17].CLSI Performance standards for antimicrobial susceptibility testing: twenty-seventh informational supplement. CLSI document M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- [18].Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–281. [DOI] [PubMed] [Google Scholar]
- [19].Cattoir V, Poirel L, Rotimi V, et al. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60:394–397. [DOI] [PubMed] [Google Scholar]
- [20].Karczmarczyk M, Abbott Y, Walsh C, et al. Characterization of multidrug-resistant Escherichia coli isolates from animals presenting at a university veterinary hospital. Appl Environ Microbiol. 2011;77:7104–7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen X, Zhang W, Pan W, et al. Prevalence of qnr, aac(6ʹ)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. 2012;56:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ng LK, Martin I, Alfa M, et al. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. 2001;15:209–215. [DOI] [PubMed] [Google Scholar]
- [23].Dallenne C, Costa A, Decré D, et al. Development of a set of multiplex PCR assays for the detection of genes enconding importante beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65:490–495. [DOI] [PubMed] [Google Scholar]
- [24].Keyes K, Hudson C, Maurer JJ, et al. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob Agents Chemother. 2000;44:421–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gordon L, Cloeckaert A, Doublet B, et al. Complete sequence of the floR-carrying multiresistance plasmid pAB5S9 from freshwater Aeromonas bestiarum. J Antimicrob Chemother. 2008;62:65–71. [DOI] [PubMed] [Google Scholar]
- [26].Kerrn MB, Klemmensen T, Frimodt-Mǿller N, et al. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. J Antimicrob Chemother. 2002;50:513–516. [DOI] [PubMed] [Google Scholar]
- [27].Perreten V, Boerlin P. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob Agents Chemother. 2003;47:1169–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Noppe-Leclercq I, Wallet F, Haentjens S, et al. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for Acinetobacter baumannii. Res Microbiol. 1999;150:317–322. [DOI] [PubMed] [Google Scholar]
- [29].Jeong HS, Kim JA, Shin JH, et al. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb Drug Resist. 2011;17:551–557. [DOI] [PubMed] [Google Scholar]
- [30].Jia W, Wang J, Xu H, et al. Resistance of Stenotrophomonas maltophilia to fluoroquinolones: prevalence in a university hospital and possible mechanisms. Int J Environ Res Public Health. 2015;12:5177–5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Carattoli A, Bertini A, Villa L, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. [DOI] [PubMed] [Google Scholar]
- [32].García-Fernández A, Fortini D, Veldman K, et al. Characterization of plasmids harbouring qnrS1, qnrB2 and qnrB19 genes in Salmonella. J Antimicrob Chemother. 2009;63:274–281. [DOI] [PubMed] [Google Scholar]
- [33].Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Clermont O, Christenson JK, Denamur E, et al. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. [DOI] [PubMed] [Google Scholar]
- [35].Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: bIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Roer L, Tchesnokova V, Allesøe R, et al. Development of a web tool for Escherichia coli subtyping based on fimH Alleles. J Clin Microbiol. 2017;55:2538–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Folster JP, Pecic G, Taylor E, et al. Characterization of isolates from an outbreak of multidrug-resistant, Shiga toxin-producing Escherichia coli O145 in the United States. Antimicrob Agents Chemother. 2011;55:5955–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Etcheverría AI, Padola NL. Shiga toxin-producing Escherichia coli: factors involved in virulence and cattle colonization. Virulence. 2013;4:366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Tseng M, Sha Q, Rudrik JT, et al. Increasing incidence of non-O157 Shiga toxin-producing Escherichia coli (STEC) in Michigan and association with clinical illness. Epidemiol Infect. 2016;144:1394–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Centers for Disease Control and Prevention (CDC) National STEC surveillance annual report, 2015. Atlanta, Georgia: US Department of Health and Human Services, CDC; 2017. [Google Scholar]
- [41].Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54:969–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Roberts MC, Schwarz S. Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J Environ Qual. 2016;45:576–592. [DOI] [PubMed] [Google Scholar]
- [43].Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41:S120–S126. [DOI] [PubMed] [Google Scholar]
- [44].Krause KM, Serio AW, Kane TR, et al. Aminoglycosides: an overview. Cold Spring Harb Perspect Med. 2016;6:a027029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Cergole-Novella MC, Pignatari AC, Castanheira M, et al. Molecular typing of antimicrobial-resistant Shiga-toxin-producing Escherichia coli strains (STEC) in Brazil. Res Microbiol. 2011;162:117–123. [DOI] [PubMed] [Google Scholar]
- [46].Ghanbarpour R, Kiani M. Characterization of non-O157 shiga toxin-producing Escherichia coli isolates from healthy fat-tailed sheep in southeastern of Iran. Trop Anim Health Prod. 2013;45:641–648. [DOI] [PubMed] [Google Scholar]
- [47].Amézquita-López BA, Quiñones B, Soto-Beltrán M, et al. Antimicrobial resistance profiles of Shiga toxin-producing Escherichia coli O157 and non-O157 recovered from domestic farm animals in rural communities in Northwestern Mexico. Antimicrob Resist Infect Control. 2016;5:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kusumoto M, Hikoda Y, Fujii Y, et al. Emergence of a multidrug-resistant shiga toxin-producing enterotoxigenic Escherichia coli lineage in diseased swine in Japan. J Clin Microbiol. 2016;54:1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mukherjee S, Mosci RE, Anderson CM, et al. Antimicrobial drug-resistant shiga toxin-producing Escherichia coli infections, Michigan, USA. Emerg Infect Dis. 2017;23:1609–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Srinivasa TR, Gill JPS, Kumar GVVPS, et al. Multi drug resistance patterns of Shiga toxin – producing Escherichia coli (STEC) and non – STEC isolates from meats, RTE meat foods, drinking water and human diarrhoeic samples of Punjab, India. Arch Clin Microbiol. 2011;2:3. [Google Scholar]
- [51].Ferdous M, Friedrich AW, Grundmann H, et al. Molecular characterization and phylogeny of Shiga toxin-producing Escherichia coli isolates obtained from two Dutch regions using whole genome sequencing. Clin Microbiol Infect. 2016;22:642.e1–9. [DOI] [PubMed] [Google Scholar]
- [52].Bai L, Hurley D, Li J, et al. Characterisation of multidrug-resistant Shiga toxin-producing Escherichia coli cultured from pigs in China: co-occurrence of extended-spectrum β-lactamase- and mcr-1-encoding genes on plasmids. Int J Antimicrob Agents. 2016;48:445–458. [DOI] [PubMed] [Google Scholar]
- [53].Valat C, Haenni M, Saras E, et al. CTX-M-15 extended-spectrum β-lactamase in a shiga toxin-producing Escherichia coli isolate of serotype O111:H8. Appl Environ Microbiol. 2012;78:8–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mellmann A, Harmsen D, Cummings CA, et al. Prospective genomic characterization of the German enterohemorrhagic Escherichia coli O104: h4outbreak by rapid next generation sequencing technology. PLoS One. 2011;6:e22751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cantón R, González-Alba JM, Galán JC. CTX-M Enzymes: origin and Diffusion. Front Microbiol. 2012;3:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ewers C, Stamm I, Stolle I, et al. Detection of shiga toxin- and extended-spectrum β-lactamase-producing Escherichia coli O145: nMand Ont: nMfrom calves with diarrhoea. J Antimicrob Chemother. 2014;69:2005–2007. [DOI] [PubMed] [Google Scholar]
- [57].Pallecchi L, Riccobono E, Sennati S, et al. Characterization of small ColE-like plasmids mediating widespread dissemination of the qnrB19 gene in commensal enterobacteria. Antimicrob Agents Chemother. 2010;54:678–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. [DOI] [PubMed] [Google Scholar]
- [59].Yang QE, Sun J, Li L, et al. IncF plasmid diversity in multi-drug resistant Escherichia coli strains from animals in China. Front Microbiol. 2015;6:964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Kao CY, Chen JW, Liu TL, et al. Comparative genomics of Escherichia coli sequence type 219 clones from the same patient: evolution of the IncI1 blaCMY-carrying plasmid in vivo. Front Microbiol. 2018;9:1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Abraham S, Kirkwood RN, Laird T, et al. Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. Isme J. 2018;12:2352–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rogers BA, Ingram PR, Runnegar N, et al. Sequence type 131 fimH30 and fimH41 subclones amongst Escherichia coli isolates in Australia and New Zealand. Int J Antimicrob Agents. 2015;45:351–358. [DOI] [PubMed] [Google Scholar]
- [63].Toval F, Köhler CD, Vogel U, et al. Characterization of Escherichia coli isolates from hospital inpatients or outpatients with urinary tract infection. J Clin Microbiol. 2014;52:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Gauthier L, Dortet L, Cotellon G, et al. Diversity of carbapenemase-producing Escherichia coli isolates in France in 2012–2013. Antimicrob Agents Chemother. 2018;62:e00266–18. [DOI] [PMC free article] [PubMed] [Google Scholar]


