ABSTRACT
Autism spectrum disorder (ASD) is associated with several oropharyngeal abnormalities, including dysbiosis in the oral microbiota. Since the oral cavity is the start of the gastrointestinal tract, this strengthens and extends the notion of a microbial gut-brain axis in ASD and even raises the question whether a microbial oral-brain axis exists. It is clear that oral bacteria can find their way to the brain through a number of pathways following routine dental procedures. A connection between the oral microbiota and a number of other brain disorders has been reported. As the evidence so far for an association between the oral microbiota and ASDs rests on a few reports only, further studies in this field are necessary. The current review discusses a possible relationship between oral bacteria and the biologic and symptomologic aspects of ASD, focusing on the clinical implications for diagnostic and therapeutic development.
KEYWORDS: Autism, oral microbiota, microbial oral-brain axis, dissemination of oral bacteria, neurological disorders
Autism spectrum disorder (ASD), which appears in the first years of life, is associated with abnormalities such as buccal sensory sensitivity, taste and texture aversions, speech apraxia and changes in salivary ribonucleic acid expression [1–9]. It has been estimated that one in 59 American children is affected by ASD, and there has been a marked increase in its incidence and prevalence over the last decades [10]. There are a number of co-occurring pathologies in ASD (Table 1). Early clinical interventions can improve symptom trajectory, but do not completely abrogate ASD symptoms, and pharmacologic interventions are limited. One novel avenue for diagnostic and therapeutic research is the emerging association between ASD and oral bacteria communities [1,11]. This review will discuss the possible relationship between oral bacteria and the biologic and symptomologic aspects of ASD, focusing particularly on the clinical implications for diagnostic and therapeutic development.
Table 1.
Co-occurring diseases in ASD (from ref [29])
| Brain-related comorbidities |
|---|
| Altered metabolite profile in urine and blood |
| Fragile X syndrome, Rett syndrome and tuberous sclerosis |
| Mitochondrial dysfunction |
| Gut-related co-morbidities |
| Gastrointestinal symptoms |
| Increased permeability of the intestinal epithelial barrier |
| Decreased expression of brush-border disaccharides in the intestinal epithelium |
| Other co-morbidities |
| Altered expression of tight junction protein in the BBL |
| Increased amounts of activated microglia cells |
Dental problems in ASD
Children with autism can have multiple medical and behavioral problems that make adequate oral hygiene and dental treatment difficult to perform. In a study of 61 children with ASD, aged 6–16 years (45 males and 16 females), higher caries prevalence, poor oral hygiene and extensive unmet needs for dental treatment compared to controls without autism were reported [12]. This could promote dissipation of oral bacteria to the circulation and potentially the brain [13], initiated by widespread dental plaque-induced diseases such as caries and gingivitis/periodontitis [14–16].
Studies on oral bacteria in ASD
Qiao et al. [11] used high throughput sequencing to compare the oral microbiota in children with ASD to healthy controls (Table 2). Approximately 1 ml of non-stimulated, naturally outflowed saliva was first collected. Then, supragingival plaques were obtained separately from caries-free molars in four quadrants (upper right, upper left, lower right and lower left) per subject. The 111 samples were divided into four groups: 1) salivary samples from healthy controls (HS; n = 27); 2) dental samples from healthy controls (HP; n = 26); 3) salivary samples from ASD patients (AS; n = 32); 4) dental samples from ASD patients (AP; n = 26). The transcriptional activity of the salivary and dental microbiota in ASD patients differed markedly from that of healthy children. In children with ASD, a lower bacterial diversity was demonstrated than in controls, consistent with findings from the gut [17,18]. This finding was particularly pronounced in dental plaque samples. The genera Haemophilus in saliva and Streptococcus in dental plaque were significantly more abundant in ASD whereas Prevotella, Selenomonas, Actinomyces, Porphyromonas and Fusobacterium were reduced. A depletion of the Prevotellaceae family co-occurrence network was also detected in plaque from ASD patients. In saliva, no phylotypes were highly correlated with decayed, missing, filled teeth or surfaces (DMFT/S). In dental plaque, however, six phylotypes including Streptococcus, Actinomyces and Capnocytophaga were positively associated with DMFT/S. Accordingly, presence of dental caries was more related to the microbiota of dental plaque than to that of saliva. Aggregatibacter segnis (OTU220) was positively associated with bleeding on probing, gingival index and periodontitis. The bacterial patterns observed in individuals with ASD suggested a possible role for microorganisms in this disorder, but did not establish a causal relationship. The results also suggested that aversion of ASD patients to dental hygiene interventions might be one mechanism for oral dysbiosis.
Table 2.
Clinical trials performed on the oral microbiota in children with autism spectrum disorder (ASD)
| Authors/ref | Age (yrs) | Method | Groups | Results and Conclusions |
|---|---|---|---|---|
| Hicks et al. [1] | 2–6 |
|
|
|
| Qiao et al. [11] | 7–14 |
|
|
|
ASD = autism spectrum disorder; DD = developmental delay; TD = typically developing
In a second study [1], changes in the salivary microbiome of children 2–6 years old were identified across three developmental profiles: ASD (n = 180), non-ASD with development delay (DD; n = 60) and typically developing (TD; n = 106) children (Table 2). Actively transcribing taxa were quantified and tested for differences between the groups and within ASD endophenotypes. Between the developmental groups, 12 bacterial taxa differed. Of particular note, 28 taxa were distinctly active among ASD patients with gastrointestinal (GI) disturbance. By group classification, five microbial ratios distinguished ASD from TD children (79.5% accuracy), three separated ASD from DD (76.5% accuracy) and three identified ASD children with GI disturbance from ASD peers without GI comorbidities (85.7% accuracy). There were significant differences in microbial transcription of energy metabolism and lysine degradation pathways across the ASD, TD and DD groups. The results indicated that GI microbial disruption in ASD likely extends to the oropharynx. Given the largely unidirectional transit of bacteria from the oropharynx to the lower GI tract, this implies that oral dysbiosis may actually serve as a primary source for a portion of the fecal dysbiosis reported in numerous ASD studies [19–21].
Oral microbiota affecting the intestine
Studies in animals and humans have demonstrated that oral bacteria can be transferred to the gut, changing its microbial composition and perhaps even host immune responses [22–24]. Oral bacteria and stool bacteria overlapped in almost half (45%) of the subjects in the Human Microbiome Project [25]. The ectopic transfer of oral bacteria has also been reported in patients with systemic diseases, such as inflammatory bowel disease [26]. Co-occurring GI problems are common in children with ASD [27]. GI symptoms were four times more prevalent in children with ASD than in children with typical development [28]. The GI symptoms seen in individuals with autism can include constipation, diarrhea, bloating, abdominal pain, reflux, vomiting, gaseousness and foul-smelling stools (for a review see [29]). Such symptoms may be related to the lower bacterial diversity reported in children with ASD [17].
Ectopic transfer of oral bacteria can occur in patients with localized ‘chronic’ periodontitis. Porphyromonas gingivalis, which is proposed as a keystone bacterium in this disease [30,31], causes dysbiosis in the periodontal microbiota. This may lead to microbial dysregulation in the gut since each day 108–1010 of P. gingivalis can be swallowed [32,33]. Changes in the gut microbiota composition could induce permeability of the gut barrier and immune activation leading to systemic inflammation. Ectopic colonization of oral bacteria in the intestines has been found to drive T-helper (TH)-1 cell induction and inflammation [22]. In one case study, Klebsiella spp. isolated from the saliva of a patient with inflammatory bowel disease were marked inducers of TH-1 cells. Ongoing colonization by oral bacteria was suggested to perpetuate gut microbiota dysbiosis and chronic inflammation. In this setting, the oral cavity can serve as a reservoir for potential intestinal pathobionts that aggravate intestinal disease. Wang et al. [27] and Ashwood [34] have also reported abnormalities in intestinal immunity in children with ASD.
Dysbiosis of the intestinal microbiota
Dysbiosis of the intestinal microbiota is an emerging etiological factor proposed for ASD [17,29,35–40]. The GI microbiome is thought to influence host behavior and neurodevelopment through the ‘microbial-gut-brain axis’ [41,42]. Imbalance in the intestinal microbiota or its metabolites may affect several complex behaviors (such as emotional and anxiety-like behaviors), and influence brain development or modulate cognition [11,43–45]. A microbiota-gut-brain axis is based on a bidirectional physiologic connection where information between the host microbiome, gut and brain are exchanged [46]. This likely involves cross talk between the central nervous system and microbes within the GI tract through direct neural activation, immune modulation, and hormonal, peptidergic and epigenetic signaling [47–50]. Below, we consider how each of these factors may be translated to an ‘oral-brain axis’.
Oral microbiota and the brain
How oral microbiota may reach the brain
There are several plausible pathways for bacteria in the mouth to reach the brain and directly influence neuro-immune activity and inflammation [51] (Figure 1). Even routine dental procedures can cause bacteremia [52], and a portion of these microbes may traverse the blood–brain barrier (BBB). Altered transcript expression has been described in microglia of ASD individuals, and disrupted microglia function could impair BBB integrity [53]. Increased permeability of the BBB has been described in children with ASD [54]. This could expose the brain to bacterial metabolites, thereby triggering an inflammatory response and altering metabolic activity within the central nervous system [29]. Prolonged disruption of energy metabolism within neurons, oligodendrocytes and glia could lead to structural changes in the cortex, hippocampus, amygdala or cerebellum, which have all been documented in ASD individuals [29].
Figure 1.
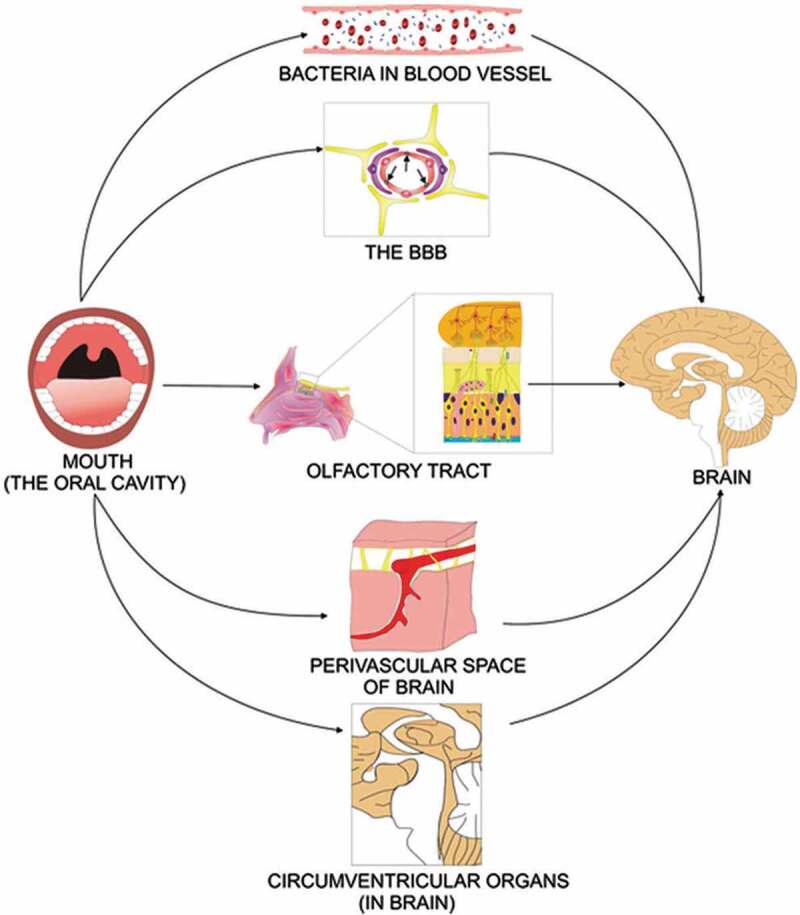
Direct and indirect mechanisms of infecting the brain. In the direct mechanism, the oral cavity infects the olfactory tract, and the olfactory nerve transfer the bacteria to the brain. In other mechanisms, bacteria inside the mouth infect the blood and find their way via blood, blood–brain barrier (BBB), perivascular spaces and circumventricular organs to the brain (figure is based on concepts presented in ref [51] and collected from ref [55])
How oral microbiota may affect the brain: inflammation
Central nervous system inflammation has been a prominent feature in studies of both animal models and post-mortem brains from individuals with ASD. For example, a study by Morgan and colleagues described the up-regulation of microglia in the ASD brain [56]. Cytokines and chemokines are also elevated in the cerebrospinal fluid of ASD patients [57,58]. Moreover, genes associated with immune and inflammatory responses are activated in the ASD cortex [59]. There appears to be a general dysregulation of the immune system towards a pro-inflammatory phenotype in ASD individuals [58,60]. Such inflammation in the developing brain may lead to synapse malfunction [61]. A significant reduction of both synaptic transmission and excitability has been observed when hypoxia and inflammation occur in combination, whereas re-oxygenation leads to neuronal hyper-excitability [62]. Malfunctioning synapses may cause the release of vasopressin, which has been shown to affect social behavior [61]. Interestingly, induction of inflammation early in gestation may promote an ASD-like phenotype through increased synaptic excitation [58] (Figure 2). In this process, early life exposure to inflammation might prime microglial cells to become hyper-responsive to subsequent insults [63]. Notably, chronic application of periodontal pathogens in mice have resulted in the development of neuropathological changes consistent with Alzheimer’s disease (a condition in which cortical inflammation is a decisive factor) [64]. Oral bacteria reaching the brain could reduce the anti-oxidative capacity and lead to reduction in the ability of mitochondria to produce energy in ASD individuals [65]. Gram-negative, putative periodontal pathogens, are rich in lipopolysaccharide (LPS) which has pro-inflammatory activity. Leakage of LPS through the BBB in ASD individuals could lead to inflammation in the central nervous system. Furthermore, increased levels of LPS in individuals with autism have been found to correlate with high levels of IL-6, a pro-inflammatory cytokine [66].
Figure 2.
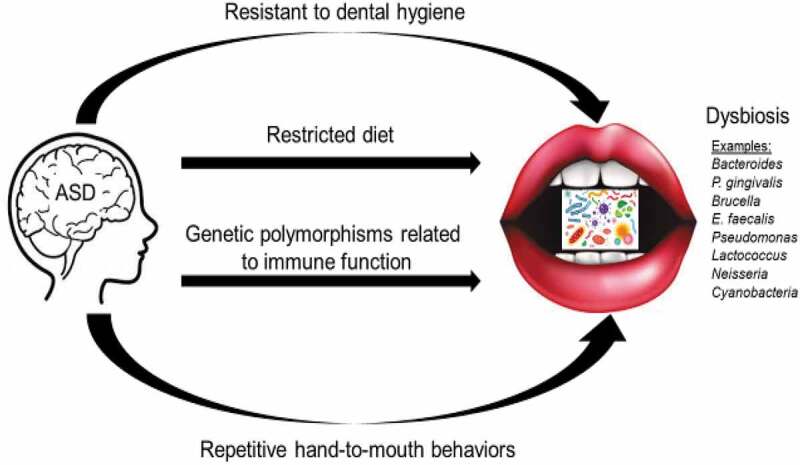
ASD phenotype can lead to oral dysbiosis
How oral microbiota may affect the brain: metabolic alterations
Microbial communities have a significant impact on metabolism within the human GI tract [67,68]. Thus, oral dysbiosis in ASD could lead to disruptions in the metabolome – a putative mechanism for ASD pathogenesis [69–71]. There are indications that increases in acetate and propionate, as well as decreases in butyrate (short-chain fatty acids of bacterial origin), can be involved in the development of ASD together with indoles [29] (Figure 3). There are also increased levels of 3-(3-hydroxyphenyl)-3-hydroxypropionic acid, 3-hydroxyphenylacetic acid and 3-hydroxyhippuric acid in children with ASD, which together indicate potential perturbations in the phenylalanine metabolism [72]. These metabolites are related to the abundance of Clostridium spp. and associated with aggravated restricted and repetitive behaviors in children. The high abundance of intestinal Clostridium detected in ASD may reflect a pathogenic role for these particular organisms [73]. Whether metabolic changes result from the oral microbiota composition in children with ASD remains to be determined. However, a study of oral microbe transcription across 346 children (including 180 with ASD) identified ASD-specific changes in pathways involving lysine degradation – a precursor to the neurotransmitter glutamate, that has been implicated in ASD pathogenesis [1]. By using saliva samples from this same cohort, the authors also described ASD-specific alterations in human microRNA expression that were associated with microbial activity, and implicated in cell growth and metabolism pathways [1]. Such findings provide a framework for human–microbial interaction at the biochemical level that may have functional consequences for host behavior.
Figure 3.
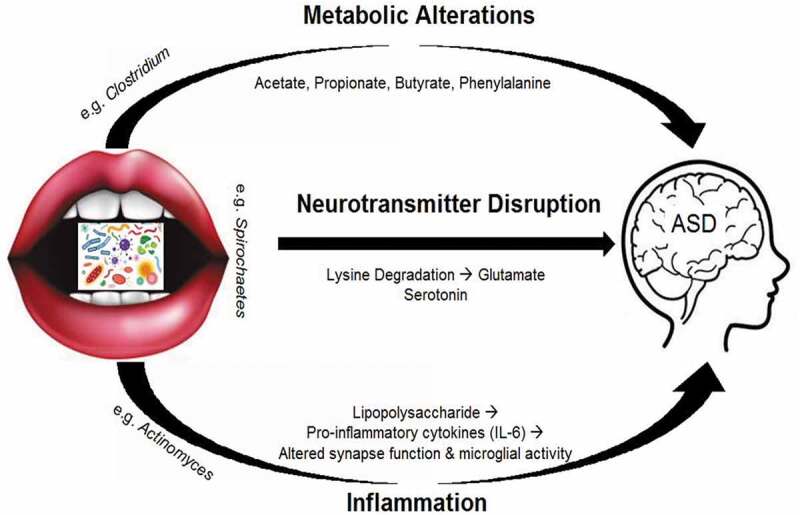
Proposed mechanisms for development of ASD
Cause–effect relationship between ASD and microbes?
Although numerous studies have identified microbial disruptions in patients with ASD and linked those disruptions to symptoms and behavior, we still do not fully understand the mechanism by which microbial communities are dysregulated in individuals with ASD. Furthermore, it is unclear if the microbial patterns described in individuals with ASD cause ASD symptoms, or result from behaviors common to the ASD phenotype.
Phenotype
Microbial dysbiosis may be influenced by the ASD phenotype. This could occur through resistance to dental hygiene, lack of a varied diet, and placing objects into the mouth as sensory seeking behavior. Discontinuation of oral hygiene in 29 orally healthy individuals for 4, 7 and 10 days, and assessment 14 days after resumption of oral hygiene, was associated with a significant increase in relative abundance of potential cariogenic Leptotrichia species and a decrease in Streptococcus species [74]. This study demonstrated the importance of regular oral hygiene on the maintenance of oral homeostasis. Furthermore, dental caries can be caused by ecological imbalance of commensal microbiota (mainly due to lack of a varied diet, such as frequent carbohydrate consumption) [75]. Placing foreign objects (e.g. toys, dirt, etc.) in the mouth is yet another source of dysbiosis, because these objects can be contaminated with microorganisms from unwashed hands in contact with other human body fluids [76].
Discerning the importance of ASD phenotype as a modulator of the oral and intestinal microbiota will likely require parallel studies in both humans and animal models. One potential strategy to help elucidate the cause/effect dilemma involving microbial disruption and ASD, is to establish ASD in a gnotobiotic animal and examine the potential of major members of the oral and intestinal microbiota (for example, oral P. gingivalis and Klebsiella spp. [22,77], and intestinal Clostridium spp. [17]) to induce ASD symptoms. Furthermore, promising pilot studies on microbiota transfer therapy (i.e. fecal transfer) should be extended to include double-blinded placebo controlled trials with well-defined a-priori hypotheses for functional outcome measures. Studies carefully examining the interaction between probiotic therapies (e.g. Bifidobacterium) and antibiotic therapies (e.g. vancomycin, minocycline) may also provide useful information about whether microbial modulation can alter ASD behaviors [78,79].
Environmental and genetic factors
Although most cases of ASD are idiopathic [17], both environmental and genetic factors are likely important for ASD development [80,81]. Exposure to environmental risk factors or genetic risk transmission can affect the maternal microbiome [21]. Offspring acquires a large portion of their microbiome from mothers during the birth process. Whether birth occurs via the vaginal canal or by cesarean section significantly affects the infant’s microbiome [82–84]. Thus, delivery mode might play a role in certain neurodevelopmental disorders. To date, studies examining the relationship between ASD and cesarean sections have demonstrated mixed results [85–87]. Changes in the microbiome due to stress might also be transferred to offspring during birth, initiating microbial dysbiosis that lasts into adulthood [88–90]. It has been reported that early life exposures to plastics and other chemicals can affect the infant microbiota [21]. Disentangling the relationship between these exposures, microbiome profiles and developmental trajectories is a difficult task that will require careful, comprehensive data collection, and powerful statistical models that can account for the interplay of many different environmental factors.
Clinical implications for microbial dysbiosis in ASD
Biomarkers
Many children with ASD exhibit hyperserotonemia [91], augmented oxidative stress [92] and increased expression of neuro-inflammatory markers [93–95]. Such disturbances may be related to disruptions in glutamate [96] and brain-derived neurotrophic factor [97]. In a study using mass spectrometry, West et al. [98] identified several blood plasma metabolites that could be of value in diagnosing ASD in 4 to 6 years old children. Amino acid metabotypes have been proposed as biomarkers for diagnostic subtypes of ASD [99]. Metabolites detected in blood and urine such as short-chain fatty acids, indoles and LPSs of bacterial origin might have diagnostic utility [29]. However, this biologic approach has not demonstrated an ability to differentiate children with ASD from peers with non-ASD developmental delay – a comparison that forms the crux of the ASD diagnostic dilemma. At the present time, the diagnosis of ASD remains dependent on clinical evaluation of behavioral symptoms, with no laboratory or objective biologic tests [100]. There is, however, growing evidence that oral microbes may be useful as a diagnostic aid in ASD [1,11]. This is an extension of a larger body of evidence relating ASD to the gut microbiome. Recently, salivary poly-omic RNA measurement was described as a novel approach to accurately identify children with ASD [101]. This objective, quantitative algorithm accurately discriminated children with ASD from peers with either developmental delay, or typical development. It could one day be used as a rapid, biologic aid for ASD diagnosis. This would constitute an important advancement, given the evidence that early diagnosis and intervention lead to the improvement of developmental trajectories for children with ASD.
Therapeutics
In animal studies, the microbiome has been shown to modulate social behavior through dysbiosis, while microbiome restoration may ameliorate ASD symptoms [102,103]. Hsaio et al. [36] demonstrated that microbial shifts within the gut of a maternal immune activation (MIA) mouse model that is known to display features of ASD, changed metabolites in the serum and that these caused autism-like behaviors. Notably, administration of a beneficial bacterium, Bacteroides fragilis, reversed the observed physiological, neurological and immunological anomalies. Wang et al. [104] reported that oral probiotics prevented ASD-like behaviors in offspring induced by maternal immune activation. Bifidobacterium (e.g. B. longum, B. breve and B. infantis) and Lactobacillus (e.g. L. helveticus and L. rhamnosus) are commonly employed probiotics in human patients. These probiotics have demonstrated promising effects on behaviors such as anxiety, depression, ASD, obsessive-compulsive disorder, and memory (including spatial and non-spatial memory) [105]. In a recent review by Ng et al. [106] it was concluded that prebiotics played a limited role in alleviating the GI and behavioral symptoms in children with ASD, but when combined with an exclusion diet (gluten and casein-free) could potentially impact sociability. Significant support for a microbial-gut-brain axis in ASD arises from studies demonstrating that microbiota transfer therapy changes the gut ecosystem and improves gastrointestinal and autism symptoms in children [107]. When microbiota transfer therapy was combined with antibiotics, bowel cleanse, and a stomach-acid suppressant, 18 individuals with ASD demonstrated significant improvements in GI symptoms, autism-related symptoms, and gut microbiota [108]. Follow-up 2 years after treatment found that most improvements in GI symptoms were maintained, and some ASD-related symptoms also remained improved. Notably, there was a significant increase in bacterial diversity and relative abundances of bifidobacteria and Prevotella. Well-designed, randomized, placebo-controlled clinical trials are needed to assess the effectiveness of probiotics and microbial transfer therapies in the treatment of ASD. Choice of appropriate strains, dose, and timing of treatment are all important factors to consider [109].
Conclusions
Microbial studies of ASD have focused largely on fecal samples [45]. It is worth noting that the oral microbiota and a possible microbial oral-brain axis have been disregarded in this context. The mouth is an extension of the digestive tract and has an abundant microbiome that includes more than 700 identified bacterial species (http://www.homd.org). Oral bacteria can enter the circulation and cause bacteremia following routine procedures such as chewing, flossing, brushing and dental cleaning [52]. Oral microbiota may contribute to several neurological diseases, including Alzheimer’s disease [51,77,110–113], epileptic seizures [114], multiple sclerosis [115], migraines [116], and Parkinson’s disease [117–119]. Whether a microbial oral-brain axis exists in ASD has yet to be definitively demonstrated. However, the relationship of oral bacteria with neurological function makes the existence of such an axis highly plausible.
Disclosure statement
SDH serves as a paid consultant and scientific advisory board member for Quadrant Biosciences Inc. Quadrant has licensed intellectual property from the Penn State College of Medicine involving saliva ribonucleic acid profiles as a diagnostic aid for autism. SDH is named as a co-inventor on this patent.
References
- [1].Hicks SD, Uhlig R, Afshari P, et al. Oral microbiome activity in children with autism spectrum disorder. Autism Res. 2018;11(9):1286–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Whiteley P, Rodgers J, Shattock P.. Feeding patterns in autism. Autism. 2000;4(2):207–211. [Google Scholar]
- [3].Desnos Y, Maris S, Segond H.. Sensory integration devices as a new modality of therapeutic care for autistic people with mental retardation. J Visual Impairness Blindness. 2009;101(1):32–43. [Google Scholar]
- [4].Sharp WG, Jaquess DL. Bite size and texture assessments to prescribe treatment for severe food selectivity in autism. Beha Interventions. 2009;24(3):157–170. [Google Scholar]
- [5].Cermak SA, Curtin C, Bandini LG. Food selectivity and sensory sensitivity in children with autism spectrum disorders. J Am Diet Assoc. 2010;110(2):238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shriberg LD, Paul R, Black LM, et al. The hypothesis of apraxia of speech in children with autism spectrum disorder. J Autism Dev Disord. 2011;41(4):405–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kuhl PK, Coffey‐Corina S, Padden D, et al. Links between social and linguistic processing of speech in preschool children with autism: behavioral and electrophysiological measures. Dev Sci. 2005;8(1):F1–2. [DOI] [PubMed] [Google Scholar]
- [8].Fakhruddin KS, El Batawi HY. Effectiveness of audiovisual distraction in behavior modification during dental caries assessment and sealant placement in children with autism spectrum disorder. Dent Res J (Isfahan). 2017;14(3):177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mayes SD, Zickgraf H. Atypical eating behaviors in children and adolescents with autism, ADHD, other disorders, and typical development. Res Autism Spectr Disord. 2019;1(64):76–83. [Google Scholar]
- [10].Kurzius-Spencer M, Zahorodny W, Rosenberg CR, et al. Prevalence of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, USA, 2014. Morb Mortal Wkly Rep. 2018;67(6):1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Qiao Y, Wu M, Feng Y, et al. Alterations of oral microbiota distinguish children with autism spectrum disorders from healthy controls. Sci Rep. 2018;8(1):1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jaber MA. Dental caries experience, oral health status and treatment needs of dental patients with autism. J Appl Oral Sci. 2011;19(3):212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hedström SÅ, Nord CE, Ursing B. Chronic meningitis in patients with dental infections. Scand J Infect Dis. 1980;12(2):117–121. [DOI] [PubMed] [Google Scholar]
- [14].Li X, Kolltveit KM, Tronstad L, et al. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13(4):547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gurenlian JR. Inflammation: the relationship between oral health and systemic disease. Dent Assist. 2009;78(2): 8–10, 12–4, 38–40; quiz 41–43. [PubMed] [Google Scholar]
- [16].Scannapieco FA. The oral microbiome: its role in health and in oral and systemic infections. Clin Microbiol Newsl. 2013;35(20):163–169. [Google Scholar]
- [17].Kang DW, Park JG, Ilhan ZE, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8(7):e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Finegold SM, Summanen PH, Downes J, et al. Detection of Clostridium perfringens toxin genes in the gut microbiota of autistic children. Anaerobe. 2017;45:133–137. [DOI] [PubMed] [Google Scholar]
- [19].Krajmalnik-Brown R, Lozupone C, Kang DW, et al. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb Ecol Health Dis. 2015;26. DOI: 10.3402/mehd.v26.26914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li Q, Han Y, Dy AB, et al. The gut microbiota and autism spectrum disorders. Front Cell Neurosci. 2017;11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vuong HE, Hsiao EY. Emerging roles for the gut microbiome in autism spectrum disorder. Biol Psychiatry. 2017;81(5):411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Atarashi K, Suda W, Luo C, et al. Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science. 2017;358(6361):359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kato T, Yamazaki K, Nakajima M, et al. Oral administration of Porphyromonas gingivalis alters the gut microbiome and serum metabolome. mSphere. 2018;3(5):pii: e00460–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. 2019;11(1):1586422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Segata N, Haake SK, Mannon P, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang M, Zhou J, He F, et al. Alteration of gut microbiota-associated epitopes in children with autism spectrum disorders. Brain Behav Immun. 2019;75:192–199. [DOI] [PubMed] [Google Scholar]
- [28].Marler S, Ferguson BJ, Lee EB, et al. Association of rigid-compulsive behavior with functional constipation in autism spectrum disorder. J Autism Dev Disord. 2017;47(6):1673–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Srikantha P, Mohajeri MH. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int J Mol Sci. 2019;20(9):pii: E2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91(9):816–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].von Troil-lindén B, Torkko H, Alaluusua S, et al. Salivary levels of suspected periodontal pathogens in relation to periodontal status and treatment. J Dent Res. 1995;74(11):1789–1795. [DOI] [PubMed] [Google Scholar]
- [33].Saygun I, Nizam N, Keskiner I, et al. Salivary infectious agents and periodontal disease status. J Periodontal Res. 2011;46(2):235–239. [DOI] [PubMed] [Google Scholar]
- [34].Ashwood P. Differential T cell levels of tumor necrosis factor receptor-II in children with autism. Front Psychiatry. 2018;9:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Finegold SM, Dowd SE, Gontcharova V, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16(4):444–453. [DOI] [PubMed] [Google Scholar]
- [36].Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Adams JB, Johansen LJ, Powell LD, et al. Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Williams BL, Hornig M, Parekh T, et al. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3(1):pii: e00261–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Macfabe D. Autism: metabolism, mitochondria, and the microbiome. Glob Adv Health Med. 2013;2(6):52–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Sone JS, Zheng LJ, Rowehl LM, et al. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the simons simplex collection. PLoS One. 2015;10(10):e0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cryan JF, O’Mahony SM. The microbiome-gut-brain axis: from bowel to behavior. Neurogastroenterol Motil. 2011;23(3):187–192. [DOI] [PubMed] [Google Scholar]
- [42].Vuong HE, Yano JM, Fung TC, et al. The microbiome and host behavior. Annu Rev Neurosci. 2017;40:21–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735–742. [DOI] [PubMed] [Google Scholar]
- [44].Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. [DOI] [PubMed] [Google Scholar]
- [45].Rosenfeld CS. Microbiome disturbances and autism spectrum disorders. Drug Metab Dispos. 2015;43(10):1557–1571. [DOI] [PubMed] [Google Scholar]
- [46].Santocchi E, Guiducci L, Fulceri F, et al. Gut to brain interaction in autism spectrum disorders: a randomized controlled trial on the role of probiotics on clinical, biochemical and neurophysiological parameters. BMC Psychiatry. 2016;16:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schroeder BO, Bäckhed F. Signals from the gut microbiota to distant organs in physiology and disease. Nat Med. 2016;22(10):1079–1089. [DOI] [PubMed] [Google Scholar]
- [48].Kelly JR, Minuto C, Cryan JF, et al. Cross talk: the microbiota and neurodevelopmental disorders. Front Neurosci. 2017;11:490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cenit MC, Nuevo IC, Codoñer-Franch P, et al. Gut microbiota and attention deficit hyperactivity disorder: new perspectives for a challenging condition. Eur Child Adolesc Psychiatry. 2017;26(9):1081–1092. [DOI] [PubMed] [Google Scholar]
- [50].de Theije CG, Wopereis H, Ramadan M, et al. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:197–206. [DOI] [PubMed] [Google Scholar]
- [51].Olsen I, Singhrao SK. Can oral infection be a risk factor for Alzheimer’s disease? J Oral Microbiol. 2015;7:29143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Olsen I. Update on bacteraemia related to dental procedures. Transfus Apher Sci. 2008;39(2):173–178. [DOI] [PubMed] [Google Scholar]
- [53].Fiorentino M, Sapone A, Senger S, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol Autism. 2016;7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kealy J, Greene C, Campbell M. Blood-brain barrier regulation in psychiatric disorders. Neurosci Lett. 2018;133664. doi: 10.1016/j.neulet.2018.06.033. [DOI] [PubMed] [Google Scholar]
- [55].Ranjan R, Abhinay A, Mishra M. Can oral microbial infections be a risk factor for neurodegeneration? A review of the literature. Neurol India. 2018;66:344–351. [DOI] [PubMed] [Google Scholar]
- [56].Morgan JT, Chana G, Pardo CA, et al. Microglial activation and increased microglial density observed in the dorsolateral prefrontal cortex in autism. Biol Psychiatry. 2010;68:368–376. [DOI] [PubMed] [Google Scholar]
- [57].Vargas DL, Nascimbene C, Krishnan C, et al. Neurological activation and neuroinflammation in the brain of patients with autism. Ann Neurol. 2005;57:67–81. [DOI] [PubMed] [Google Scholar]
- [58].Mazarati AM, Lewis ML, Pittman QJ. Neurobehavioral comorbidities of epilepsy: role of inflammation. Epilepsia. 2017;58(Suppl 3):48–56. [DOI] [PubMed] [Google Scholar]
- [59].Voinegau I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Meltzer A, Van de Water J. The role of the immune system in autism spectrum disorder. Neuropsychopharmacology. 2017;42(1):284–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Madore C, Leyrolle Q, Lacabanne C, et al. Neuroinflammation in autism: plausible role of maternal inflammation, dietary omega 3, and microbiota. Neural Plast. 2016;2016:3597209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang YS, Son SJ, Choi JH, et al. Synaptic transmission and excitability during hypoxia with inflammation and reoxygenation in hippocampal CA1 neurons. Neuropharmacology. 2018;138:20–31. [DOI] [PubMed] [Google Scholar]
- [63].Bibo SD, Schwarz JM. The immune system and developmental programming of brain and behavior. Front Neuroendocrinol. 2012;33:267–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ilievski V, Zuchowska PK, Green SJ, et al. Chronic oral application of a periodontal pathogen results in brain inflammation, neurodegeneration and amyloid beta production in wild type mice. PLoS One. 2018;13(10):e0204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Castora FJ. Mitochondrial function and abnormalities implicated in the pathogenesis of ASD. Prog Neuropsychopharmacol Biol Psychiatry. 2019;92:83–108. [DOI] [PubMed] [Google Scholar]
- [66].Beurel E, Jope RS. Lipopolysaccharide-induced interleukin-6 production is controlled by glycogen synthase kinase-3 and STAT3 in the brain. J Neuroinflammation. 2009;6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017;35:8–15. [DOI] [PubMed] [Google Scholar]
- [68].Blacher E, Levy M, Tatirovsky E, et al. Microbiome-modulated metabolites at the interface of host immunity. J Immunol. 2017;198(2):572–580. [DOI] [PubMed] [Google Scholar]
- [69].Mussap M, Noto A, Fanos V. Metabolomics of autism spectrum disorders: early insights regarding mammalian-microbial cometabolites. Expert Rev Mol Diagn. 2016;16(8):869–881. [DOI] [PubMed] [Google Scholar]
- [70].Wang H, Liang S, Wang M, et al. Potential serum biomarkers from a metabolomics study of autism. J Psychiatry Neurosci. 2016;41(1):27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Cezar GG, Smith AM, inventors; Wisconsin Alumni Research Foundation, Stemina Biomarker Discovery Inc., assignee . Metabolic biomarkers of autism. USA patent application US 15/371, 692. 2017.
- [72].Li Q, Zhou JM. The microbiota-gut-brain axis and its potential therapeutic role in autism spectrum disorder. Neuroscience. 2016;324:131–139. [DOI] [PubMed] [Google Scholar]
- [73].Zeidán Chuliá F, Moreira JC. Clostridium bacteria and its impact in autism research: thinking “outside the box” of neuroscience. J Commun Disord Deaf Stud Hearing Aids. 2013;1:101. [Google Scholar]
- [74].Belstrøm D, Sembler-Møller ML, Grande MA, et al. Impact of oral hygiene discontinuation on supragingival and salivary microbiomes. JDR Clin Trans Res. 2018;3(1):57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Giacaman RA. Sugars and beyond. The role of sugars and the other nutrients and their potential impact on caries. Oral Dis. 2018;24(7):1185–1197. [DOI] [PubMed] [Google Scholar]
- [76].Little K, Cutcliffe S. The safe use of children’s toys within the healthcare setting. Nurs Times. 2006;102(38):34–37. [PubMed] [Google Scholar]
- [77].Dominy SS, Lynch C, Ermini F, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5(1):eaau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Soczynska JK, Mansur RB, Brietzke E, et al. Novel therapeutic targets in depression: minocycline as a candidate treatment. Behav Brain Res. 2012;235:302–317.22963995 [Google Scholar]
- [79].Borre YE, Moloney RD, Clarke G, et al. The impact of microbiota on brain and behavior: mechanisms & therapeutic potential. Adv Exp Med Biol. 2014;817:373–403. [DOI] [PubMed] [Google Scholar]
- [80].Sandin S, Lichtenstein P, Kuja-Halkola R, et al. The familial risk of autism. JAMA. 2014;311(17):1770–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Schaafsma SM, Gagnidze K, Reyes A, et al. Sex-specific gene-environment interactions underlying ASD-like behaviors. Proc Natl Acad Sci U S A. 2017;114(6):1383–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Biasucci G, Benenati B, Morelli L, et al. Cesarean delivery may affect the early biodiversity of intestinal bacteria. J Nutr. 2008;138:1796S–1800S. [DOI] [PubMed] [Google Scholar]
- [83].Biasucci G, Benenati B, Riboni S, et al. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev. 2010;86(Suppl 1):13–15. [DOI] [PubMed] [Google Scholar]
- [84].Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Curran EA, Dalman C, Kearney PM, et al. Association between obstetric mode of delivery and autism spectrum disorder: a population-based sibling design study. JAMA Psychiatry. 2015;72:935–942. [DOI] [PubMed] [Google Scholar]
- [86].Curran EA, O’Neill SM, Cryan JF, et al. Research review: birth by caesarean section and development of autism spectrum disorder and attention-deficit/hyperactivity disorder: a systematic review and meta-analysis. J Child Psychol Psychiatry. 2015;56:500–508. [DOI] [PubMed] [Google Scholar]
- [87].Curran EA, Cryan JF, Kenny LC, et al. Obstetrical mode of delivery and childhood behavior and psychological development in a British cohort. J Autism Dev Disord. 2016;46(2):603–614. [DOI] [PubMed] [Google Scholar]
- [88].Golubeva AV, Crampton S, Desbonnet L, et al. Prenatal stress-induced alterations in major physiological systems correlate with gut microbiota composition in adulthood. Psychoneuroendocrinology. 2015;60:58–74. [DOI] [PubMed] [Google Scholar]
- [89].Jasarevic E, Howerton CL, Howard CD, et al. Alterations in the vaginal microbiome by maternal stress are associated with metabolic reprogramming of the offspring gut and brain. Endocrinology. 2015;156:3265–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].De Palma G, Blennerhassett P, Lu J, et al. Microbiota and host determinants of behavioural phenotype in maternally separated mice. Nat Commun. 2015;6:7735. [DOI] [PubMed] [Google Scholar]
- [91].Chamberlain RS, Herman BH. A novel biochemical model linking dysfunctions in brain melatonin, propiomelanocortin peptides, and serotonin in autism. Biol Psychiatry. 1990;28(9):773–793. [DOI] [PubMed] [Google Scholar]
- [92].Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13(3):171–181. [DOI] [PubMed] [Google Scholar]
- [93].Corbett BA, Kantor AB, Schulman H, et al. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry. 2007;12(3):292–306. [DOI] [PubMed] [Google Scholar]
- [94].Goines P, Van de Water J. The immune system’s role in the biology of autism. Curr Opin Neurol. 2010;23(2):111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Onore C, Careaga M, Ashwood P. The role of immune dysfunction in the pathophysiology of autism. Brain Behav Immun. 2012;26(3):383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].El-Ansary A, Al-Ayadhi L. GABAergic/glutamatergic imbalance relative to excessive neuroinflammation in autism spectrum disorders. J Neuroinflammation. 2014;11:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Connolly AM, Chez M, Streif EM, et al. Brain-derived neurotrophic factor and autoantibodies to neural antigens in sera of children with autistic spectrum disorders, Landau-Kleffner syndrome, and epilepsy. Biol Psychiatry. 2006;59(4):354–363. [DOI] [PubMed] [Google Scholar]
- [98].West PR, Amaral DG, Bais P, et al. Metabolomics as a tool for discovery of biomarkers of autism spectrum disorder in the blood plasma of children. PLoS One. 2014;9(11):e112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Smith AM, King JJ, West PR, et al. Amino acid dysregulation metabotypes: potential biomarkers for diagnosis and individualized treatment for subtypes of autism spectrum disorder. Biol Psychiatry. 2019;85(4):345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Kulage KM, Smaldone AM, Cohn EG. How will DSM-5 affect autism diagnosis? A systematic literature review and meta-analysis. J Autism Dev Disord. 2014;44(8):1918–1932. [DOI] [PubMed] [Google Scholar]
- [101].Hicks SD, Rajan AT, Wagner KE, et al. Validation of a salivary RNA test for childhood autism spectrum disorder. Front Genet. 2018;9:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Buffington SA, Di Prisco GV, Auchtung TA, et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165(7):1762–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Kumar H, Sharma B. Minocycline ameliorates prenatal valproic acid induced autistic behaviour, biochemistry and blood brain barrier impairments in rats. Brain Res. 2016;1630:83–97. [DOI] [PubMed] [Google Scholar]
- [104].Wang X, Yang J, Zhang H, et al. Oral probiotic administration during pregnancy prevents autism-related behaviors in offspring induced by maternal immune activation via anti-inflammation in mice. Autism Res. 2019;12(4):576–588. [DOI] [PubMed] [Google Scholar]
- [105].Wang H, Lee IS, Braun C, et al. Effect of probiotics on central nervous system functions in animals and humans: a systematic review. J Neurogastroenterol Motil. 2016;22(4):589–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Ng QX, Loke W, Venkatanarayanan N, et al. A systematic review of the role of prebiotics and probiotics in autism spectrum disorders. Medicina (Kaunas). 2019;55(5):pii: E129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Kang DW, Adams JB, Gregory AC, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Kang DW, Adams JB, Coleman DM, et al. Long-term benefit of microbiota transfer therapy on autism symptoms and gut microbiota. Sci Rep. 2019;9(1):5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Fattorusso A, Di Genova L, Dell’Isola GB, et al. Autism spectrum disorders and the gut microbiota. Nutrients. 2019;11(3):pii: E521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Singhrao SK, Harding A, Simmons T, et al. Oral inflammation, tooth loss, risk factors, and association with progression of Alzheimer’s disease. J Alzheimers Dis. 2014;42(3):723–737. [DOI] [PubMed] [Google Scholar]
- [111].Shoemark DK, Allen SJ. The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer’s disease. J Alzheimers Dis. 2015;43(3):725–738. [DOI] [PubMed] [Google Scholar]
- [112].Olsen I, Singhrao SK. Poor oral health and its neurological consequences: mechanisms of Porphyromonas gingivalis involvement in cognitive dysfunction. Curr Oral Health Rep. 2019;6(2):120–129. [Google Scholar]
- [113].Singhrao SK, Olsen I. Assessing the role of Porphyromonas gingivalis in periodontitis to determine a causative relationship with Alzheimer’s disease. J Oral Microbiol. 2019;11(1):1563405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Costa AL, Yasuda CL, Shibasaki W, et al. The association between periodontal disease and seizure severity in refractory epilepsy patients. Seizure. 2014;23(3):227–230. [DOI] [PubMed] [Google Scholar]
- [115].Farrokhi V, Nemati R, Nichols FC, et al. Bacterial lipodipeptide, Lipid 654, is a microbiome-associated biomarker for multiple sclerosis. Clin Transl Immunology. 2013;2(11):e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gonzalez A, Hyde E, Sangwan N, et al. Migraines are correlated with higher levels of nitrate-, nitrite-, and nitric oxide-reducing oral microbes in the American gut project cohort. mSystems. 2016;1(5):pii: e00105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Pereira PAB, Aho VTE, Paulin L, et al. Oral and nasal microbiota in Parkinson’s disease. Parkinsonism Relat Disord. 2017;38:61–67. [DOI] [PubMed] [Google Scholar]
- [118].Chen CK, Wu YT, Chang YC. Periodontal inflammatory disease is associated with the risk of Parkinson’s disease: a population-based retrospective matched-cohort study. PeerJ. 2017;5:e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Chen CK, Huang JY, Wu YT, et al. Dental scaling decreases the risk of Parkinson’s disease: a nationwide population-based nested case-control study. Int J Environ Res Public Health. 2018;15(8):pii, E1587. [DOI] [PMC free article] [PubMed] [Google Scholar]


