Abstract
This article comments on:
Chia T, Chirico M, King R et al. 2019. A carbohydrate-binding protein, B-granule content 1 influences starch granule-size distribution in a dose dependent manner in polyploid wheat. Journal of Experimental Botany 70, 105–115.
Keywords: A- and B-granules, compound starch, starch granules, protein targeting to starch, wheat
Starch is both an important plant metabolite and a feedstock in many industrial processes. Most plants accumulate granules with a unimodal size distribution; however some tissues, such as wheat endosperm, demonstrate a bimodal distribution containing a mixture of large A-granules and small B-granules. A quantitative trait locus (QTL) in wheat has been identified that greatly influences B-granule formation, and Chia et al. (2019) isolated a single gene (B-GRANULE CONTENT 1, BGC1) from within that QTL responsible for the phenotype. It is similar in sequence to other genes that have recently been shown to be involved in starch metabolism.
Starch is a glucose polymer that accumulates as granules within plastids and is composed of two structurally distinct polymers, amylose and amylopectin. Its importance in plant biology comes both for its role as a carbon store where it influences plant growth and yield (Lloyd and Kossmann, 2019), as well as for its use in many industrial processes (Zeeman et al. 2010). Research over the past thirty years has uncovered a plethora of genes encoding enzymes that synthesize the starch polymers including multiple isoforms of starch synthase, starch branching enzyme and starch debranching enzymes. More recent work has moved on to the identification of proteins that affect starch granule formation but do not necessarily form the polymer directly.
Many roles for new genes affecting starch metabolism
A new class of proteins involved in starch metabolism was identified five years ago in rice (Peng et al. 2014) where a mutant gene named floury endosperm 6 (flo6) was shown to encode a polypeptide comprising both carbohydrate binding and coiled coil domains. The mutation led to the accumulation of starch granules with altered shape in rice endosperm. Three genes related to FLO6 have been identified in Arabidopsis: PROTEIN TARGETING TO STARCH (PTST or PTST1), PTST2 and PTST3, and OsFLO6 is orthologous to AtPTST2. There appear to be similar genes encoding PTST-like proteins in many plants including green algae, but grasses have lost PTST3 (Seung et al. 2015, 2017).
Due the presence of coiled coil domains it was assumed that these polypeptides would interact with other proteins and there is good evidence that they guide catalytically active enzymes to the surface of the starch granule (Peng et al. 2014, Seung et al. 2015). PTST1-like proteins have been shown to interact with the amylose-synthesizing granule bound starch synthase (Seung et al. 2015; Bull et al. 2018). On the other hand, PTST2-like proteins interact with several polypeptides (Peng et al. 2014; Seung et al. 2017) and are thought to be involved in starch granule initiation as ptst2 mutants accumulate fewer (but larger) starch granules (Seung et al. 2017). Some of the proteins they interact with have been shown to be directly involved in granule initiation (Roldán et al. 2007; Szydlowski et al. 2009, Seung et al. 2018, Vandromme et al. 2019). No interacting partners have yet been identified for PTST3, but, like PTST2, it is involved in starch granule initiation in Arabidopsis leaves (Seung et al. 2017). A summary of the phenotype of mutations in PTST-like genes from various species alongside known interacting protein partners is provided in Box 1.
Box 1. The diverse roles of PTST like proteins in starch synthesis.
PTST1-like proteins have been studied in Arabidopsis leaves (Seung et al. 2015), cassava roots (Bull et al. 2018) as well as, barley and rice endosperm (Wang et al. 2019; Zhong et al. 2019). In Arabidopsis, rice and barley, these proteins have been shown to interact with the amylose synthesizing GRANULE BOUND STARCH SYNTHASE (GBSS). Mutations of this protein in Arabidopsis (Seung et al. 2015), cassava (Bull et al. 2018) and rice (Wang et al. 2019) reduce amylose amounts in starch, while barley ptst1 mutants do not accumulate endosperm starch (Zhong et al. 2019).
PTST2 family proteins have been studied in rice, Arabidopsis, barley and now wheat. In rice the protein interacts with the starch debranching enzyme ISOAMYLASE 1 (ISA1) and a mutation affecting it (flo6) results in production of starch granules with altered shape in the endosperm (Peng et al. 2014). AtPTST2 is involved in starch granule initiation in Arabidopsis and has been shown to interact with three polypeptides, STARCH SYNTHASE 4 (SS4; Seung et al. 2017), MYOSIN-RESEMBLING CHLOROPLAST PROTEIN (MRC) and MAR BINDING FILAMENT LIKE PROTEIN1 (MFP1; Seung et al. 2018). MRC is also known as PROTEIN INVOLVED IN STARCH INITIATION (PII1) and has been shown to interact directly with SS4 (Vandromme et al. 2019). AtPTST2 is found partly bound to the thylakoid membrane, and MFP1 guides it to this position with the chloroplast (Seung et al. 2018). No interacting partners have been identified for the barley and wheat proteins, but mutations (named franubet (fra) in barley and b-granule content 1 (bgc1) in wheat) affecting them lead to altered starch granule shape (Saito et al. 2018, Chia et al. 2019).
AtPTST3 is known to affect starch granule initiation in Arabidopsis (Seung et al. 2017), but it is unclear how as no PTST3-interacting proteins have yet been identified. Some species, such as the grasses, have lost the gene encoding PTST3 (Seung et al. 2017).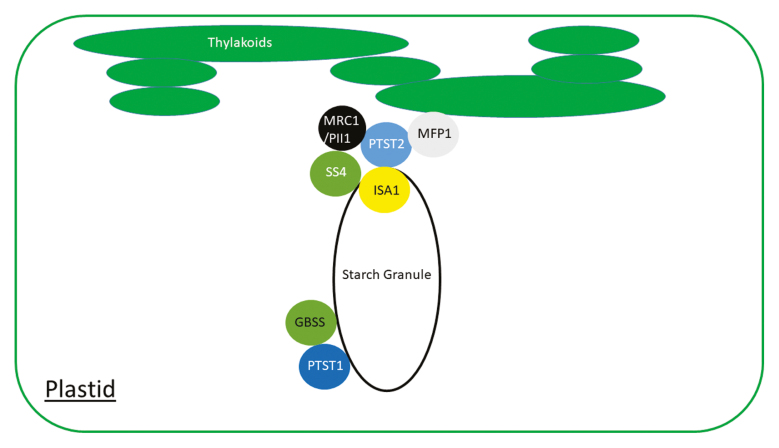
A wheat member of the PTST2 family influences starch granule formation
The focus of a paper by Chia and co-workers (2019) has been the examination of a protein that influences the formation of different sized starch granules in wheat endosperm. Triticeae species are unusual as they accumulate two distinct starch granule size classes within their seeds, known as A- and B-granules. The formation of both types is temporally and spatially distinct, with one large A-granule being initiated per plastid early in endosperm development, followed later by the initiation of several smaller B-granules.
Some wild Triticeae show differing proportions of A and B granules within their endosperm and this has been used to identify a gene involved in their formation. By studying the progeny of a cross between a B-granule-free species (Aegilops peregrina Hack.) with a synthetic tetraploid, a QTL affecting B-granule formation (named B-GRANULE CONTENT1 (BGC1)) was identified on chromosome 4S (Howard et al. 2011). This led to the hypothesis that other Triticeae, including hexaploid bread wheat (Triticum aestivum), would contain syntenous genetic elements affecting granule size on chromosome 4. This was tested in bread wheat by combining large deletions within chromosomes 4A and 4D that span the QTL, and it was found that double deletion mutants lacked B-granules (Chia et al. 2017).
The current paper uses an elegant series of experiments to identify a single gene that is responsible for the variation in B-granule content. The BGC1 gene encodes a PTST-like protein that is most similar to members of the PTST2 family. To examine the role of this gene further, the authors identified TILLING mutants in the tetraploid species T. durum where BGC1 genes were interrupted by either nonsense or missense mutations. Mutant lines with a nonsense mutation in one genome and missense in the other accumulated fewer B-granules.
As mentioned in Box 1, the barley fra mutant affects a PTST2-like protein (Saito et al. 2018). Although closely related to bread wheat, the phenotype observed in the fra mutant is different to that in bgc1 plants as it affects granule shape. Chia et al. (2019) studied the fra mutant in more detail and showed both that B-starch granules are not eliminated and that starch accumulates as semi-compound granules where one granule is composed of several sub-granules. As the bread wheat line lacking B-granules was constructed with deletions within chromosomes 4A and 4D, it was hypothesized that the difference may be due to the presence of a BGC1 gene on chromosome 4B. To examine this, a bread wheat mutant was created where BGC1 genes in all three (A, B and D) genomes were either deleted, or mutated through introduction of a stop codon. Starch from this mutant appeared more similar to that found in the fra mutant as compound and semi-compound granules formed alongside reduced numbers of B-granules. This shows that the effect of BGC1 in bread wheat is dependent on gene dose, with mutations in one genome having little or no effect, within two genomes leading to elimination of B-granules, and within all three to the formation of granules greatly altered in shape. Given these phenotypes, the authors propose that BGC1 limits starch granule formation in plastids during early endosperm development, allowing the formation of A-granules that arise from single initiations, but then stimulates the formation of B-granules later in seed development.
A to B and beyond
This work is significant as it helps in understanding starch granule formation in a major group of plants that are important both for human calorie intake as well as for the production of industrially relevant starches. It would be useful to know how the physicochemical properties of starches from the various wheat lines change with gene dose as this would provide insight to potential industrial applications. From a biological perspective it will be important to understand how BGC1 leads to the observed effect on starch. Given the known role of other PTST proteins, it is likely to involve an interaction with one or more other proteins, and one candidate is the starch debranching enzyme, isoamylase. This is both because the ortholog in rice, FLO6, has been shown to interact with this enzyme (Peng et al. 2014; Box 1) and because barley mutants lacking isoamylase have been demonstrated to accumulate compound starch granules (Burton et al. 2002). However, PTST2-like proteins from Arabidopsis are known to interact with other proteins involved in granule initiation (Box 1; Seung et al. 2017, 2018) which may also interact. More generally, an effort to identify all proteins that interact with PTST proteins across several species is needed to fully understand their role(s) in starch metabolism. It would be especially interesting to examine this in non-vascular plants to elucidate if the roles of these proteins have changed over evolutionary time.
References
- Bull SE, Seung D, Chanez C, et al. 2018. Accelerated ex situ breeding of GBSS- and PTST1-edited cassava for modified starch. Science Advances 4, eaat6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton R, Jenner H, Carrangis L, et al. 2002. Starch granule initiation and growth are altered in barley mutants that lack isoamylase activity. Plant Journal 31, 97–112. [DOI] [PubMed] [Google Scholar]
- Chia T, Adamski NM, Saccomanno B, Greenland A, Nash A, Uauy C, Trafford K. 2017. Transfer of a starch phenotype from wild wheat to bread wheat by deletion of a locus controlling B-type starch granule content. Journal of Experimental Botany 68, 5497–5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia T, Chirico M, King R, et al. 2020. A carbohydrate-binding protein, B-granule content 1 influences starch granule-size distribution in a dose dependent manner in polyploid wheat. Journal of Experimental Botany 71, 105–115. [DOI] [PubMed] [Google Scholar]
- Howard T, Rejab NA, Griffiths S, Leigh F, Leverington-Waite M, Simmonds J, Uauy C, Trafford K. 2011. Identification of a major QTL controlling the content of B-type starch granules in Aegilops. Journal of Experimental Botany 62, 2217–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Kossmann J. 2019. Starch trek: The search for yield. Frontiers in Plant Science 9, 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng C, Wang Y, Liu F, et al. 2014. FLOURY ENDOSPERM6 encodes a CBM48 domain-containing protein involved in compound granule formation and starch synthesis in rice endosperm. Plant Journal 77, 917–930. [DOI] [PubMed] [Google Scholar]
- Roldán I, Wattebled F, Mercedes Lucas M, Delvallé D, Planchot V, Jiménez S, Pérez R, Ball S, D’Hulst C, Mérida Á. 2007. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant Journal 49, 492–504. [DOI] [PubMed] [Google Scholar]
- Saito M, Tanaka T, Sato K, Vrinten P, Nakamura T. 2018. A single nucleotide polymorphism in the “Fra” gene results in fractured starch granules in barley. Theoretical and Applied Genetics 131, 353–364. [DOI] [PubMed] [Google Scholar]
- Seung D, Soyk S, Coiro M, Miller BA, Eicke S, Zeeman SC. 2015. PROTEIN TARGETING TO STARCH is required for localising GRANULE BOUND STARCH SYNTHASE to starch granules and for normal amylose synthesis on Arabidopsis. PLoS Biology 13 e1002080. doi: 10.1371/journal.pbio.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Boudet J, Monroe J, Schreier TB, Davids LC, Abt M, Lu K-J, Zanella M, Zeeman SC. 2017. Homologs of PROTEIN TARGETING TO STARCH control starch granule initiation in Arabidopsis leaves. Plant Cell 29, 1657–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seung D, Schreier TB, Bürgy L. Eicke S, Zeeman SC. 2018. Two plastidial coiled-coil proteins are essential for normal starch granule initiation in Arabidopsis. Plant Cell 30, 1523–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szydlowski N, Ragel P, Raynaud S, et al. 2009. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 21, 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandromme C, Spriet C, Dauvillée D, Courseaux A, Putaux P, Wychowski A, Krzewinski F, Facon M, D’Hulst C, Wattebled F. 2019. PII1: A protein involved in starch initiation that determines granule number and size in Arabidopsis chloroplast. New Phytologist 221, 256–270. [DOI] [PubMed] [Google Scholar]
- Wang W, Wei X, Jiao G, et al. 2019. GBSS-BINDING PROTEIN, encoding a CBM48 domain-containing protein, affects rice quality and yield. Journal of Integrative Plant Biology. doi: 10.1111/jipb.12866. [DOI] [PubMed] [Google Scholar]
- Zeeman SC, Kossmann J, Smith AM. 2010. Starch: Its metabolism, evolution and biotechnological modification in plants. Annual Review of Plant Biology. 61, 15.1–15.26. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Blennow A, Kofoed-Enevoldsen O, Jiang D, Hebelstrup KH. 2019. Protein targeting to Starch 1 is essential for starchy endosperm development in barley. Journal of Experimental Botany 70, 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]


