Abstract
Plants regulate responses towards herbivory through fine-tuning of defence-related hormone production, expression of defence genes, and production of secondary metabolites. Jasmonic acid (JA) plays a key role in plant–herbivorous arthropod interactions. To understand how pepper (Capsicum annuum) responds to herbivory, leaf transcriptomes and metabolomes of two genotypes different in their susceptibility to spider mites were studied. Mites induced both JA and salicylic acid (SA) signalling. However, mite infestation and exogenous JA resulted in distinct transcriptome profiles. Compared with JA, mites induced fewer differentially expressed genes involved in metabolic processes (except for genes involved in the phenylpropanoid pathway) and lipid metabolic processes. Furthermore, pathogen-related defence responses including WRKY transcription factors were more strongly induced upon mite infestation, probably as a result of induced SA signalling. Untargeted analysis of secondary metabolites confirmed that JA treatment induced larger changes in metabolism than spider mite infestation, resulting in higher terpenoid and flavonoid production. The more resistant genotype exhibited a larger increase in endogenous JA and volatile and non-volatile secondary metabolites upon infestation, which could explain its stronger defence. Reasoning that in JA–SA antagonizing crosstalk, SA defences are prioritized over JA defences, we hypothesize that lack of SA-mediated repression of JA-induced defences could result in gain of resistance towards spider mites in pepper.
Keywords: Capsicum annuum, JA/SA crosstalk, plant–arthropod interactions, specialized metabolites, transcriptional changes, two-spotted spider mites
Lack of SA-mediated repression of JA-induced specialized metabolites results in gain of resistance towards spider mites in pepper
Introduction
Plants have evolved broad phenotypic plasticity to defend themselves against herbivores and pathogens. Constitutively present leaf surface waxes, morphological structures including trichomes, and various secondary metabolites form a first barrier discouraging herbivore attack (Furstenberg-Hagg et al., 2013). Should these barriers not be sufficient, plants can produce defensive proteins and secondary metabolites in response to feeding to affect the herbivore’s growth and reproduction directly. In addition to these induced direct defences, arthropod herbivory also results in release of volatiles that attract natural enemies of the herbivores, protecting the plant indirectly (Kappers et al., 2005; War et al., 2012; Gols, 2014) Ubiquitous or species-specific plant secondary metabolites belonging to different biochemical classes including phenolics, flavonoids, terpenoids, and green leaf volatiles (GLVs) play a key role in these direct and indirect defences (Snoeren et al., 2010; Kappers et al., 2011; Cheynier et al., 2013; Scala et al., 2013; Tholl, 2015).
Plants can distinguish herbivore presence from mechanical damage by recognizing herbivore oral secretions or oviposition fluids (Little et al., 2007; Bandoly et al., 2015). These elicitors, together with the physical damage caused by the herbivore, trigger electrical signals and change Ca2+ homeostasis subsequently, followed by downstream defence responses regulated through hormone signalling pathways of which jasmonic acid (JA), ethylene (ET), and salicylic acid (SA) are the major players involved (Bari and Jones, 2009). Both the individual hormones and the crosstalk between them play an essential role in fine-tuning defence responses to specific attackers (Kawazu et al., 2012; Proietti et al., 2018).
Jasmonates (JAs) mediate multiple aspects of plant development including root and trichome formation, flower development, and leaf senescence, as well as plant responses to various biotic and abiotic stresses (Wasternack and Hause, 2013). Biosynthesis of JA starts with oxygenation of α-linolenic acid released from chloroplast membranes via the octadecanoid pathway (Turner et al., 2002). The isoleucine-conjugated form of JA (JA-Ile) accumulates in response to wounding or herbivory and is considered to be the active form (Staswick and Tiryaki, 2004). In unstimulated conditions, basic helix–loop–helix (bHLH) transcription factor genes including MYC2, MYC3, and MYC4 are repressed, while upon stress MYC2 binds to JA-responsive elements (G-boxes) in the promoters of JA-regulated genes (Pauwels et al., 2010; Fernandez-Calvo et al., 2011). In Arabidopsis, MYC2 positively regulates expression of wound-responsive genes such as VEGETATIVE STORAGE PROTEIN (VSP) and LIPOXYGENASE (LOX) (Lorenzo et al., 2004). MYC2 enhances the production of flavonoids in Medicago truncatula (Adolfsson et al., 2017) and is required for JA-mediated tolerance to the generalist herbivore Helicoverpa armigera (Dombrecht et al., 2007). JA has been identified as the hormone responsible for the accumulation of several different classes of bioactive secondary metabolites (Memelink et al., 2001).
SA is synthesized from phenylalanine via phenylalanine ammonia lyase (PAL) and from chorismate through isochorismate synthase (ICS) action. SA is generally known to accumulate upon pathogen infection but also upon herbivory by phloem stylet-feeding whitefly (Zarate et al., 2007) and aphids (Kloth et al., 2016). The volatile conversion product of SA, methyl-SA, can be detected in the headspace of lima bean, Arabidopsis, tomato, and cucumber within hours after the onset of herbivory (Ament et al., 2004; De Boer and Dicke, 2004; Kappers et al., 2005, 2011). Methyl-SA alone or in combination with other volatile compounds is known to be attractive to predators of herbivores, including mites predaceous to spider mites (De Boer and Dicke, 2004). NON-EXPRESSOR OF PR GENES1 (NPR1), a master transcriptional co-regulator in the SA signalling pathway, is regulated by the cellular redox state (Fu and Dong, 2013) and activates SA-dependent gene transcription by binding to transcription factors of the TGACG sequence-specific binding protein (TGA) family (Wang et al., 2006).
SA acts antagonistically with JA signalling (Bari and Jones, 2009; Erb et al., 2012; Furstenberg-Hagg et al., 2013). Increasing SA accumulation in tomato and Arabidopsis resulted in inhibition of JA-induced responses (Doherty et al., 1988). This supports the antagonistic relationship between JA and SA pathways, but also suggests that SA-dependent defences are being prioritized over JA-related defences (Spoel et al., 2003). Bypassing JA biosynthesis by exogenous application of methyl-JA in a JA biosynthesis-deficient mutant aos/dde2 did not affect SA-mediated suppression of JA-responsive genes PDF1.2 and VSP in Arabidopsis (Leon-Reyes et al., 2010). In Arabidopsis, WRKY transcription factors were found to be essential for mediating crosstalk between JA and SA (Spoel et al., 2003). Overexpression of the NPR1-independent transcription factor gene WRKY70 resulted in constitutive expression of SA-induced pathogenesis-related genes and increased resistance to virulent pathogens, while antisense suppression of WRKY70 activated JA-responsive/COI-independent genes (Li et al., 2004). Several other WRKY transcription factors are also involved in downstream transcriptional responses to SA (Eulgem and Somssich, 2007; Kloth et al., 2016).
Capsicum annuum, including sweet and hot peppers, belongs to the nightshade family Solanaceae, and is a worldwide grown and economically important vegetable crop (Carrizo García et al., 2016; http://www.fao.org). Tetranychus urticae (two-spotted spider mite, TSSM) is a generalist herbivorous arthropod reported to infest >150 crop species, especially within the Solanaceae family (Fasulo and Denmark, 2016), including pepper. TSSMs feed through their stylet, penetrating leaf tissue via stomatal openings or by the intercellular space in between epidermal pavement cells, thus maintaining the integrity of the leaf epidermis (Bensoussan et al., 2016). Intriguingly, closure of plant stomata is one of the first plant physiological responses to TSSM feeding, leading to reduced photosynthetic rates and crop yields (De Freitas Bueno et al., 2009), along with down-regulation of photosynthetic gene expression (Mercke et al., 2004). Overall transcriptome responses to TSSM feeding have been studied in, for example, Arabidopsis (Zhurov et al., 2014), tomato (Martel et al., 2015), and grape (Díaz-Riquelme et al., 2016), and suggest JA to be the dominant player in plant defence responses. As TSSMs quickly develop resistance towards chemical acaricides (Van Leeuwen et al., 2010), the use of predaceous arthropods as biological control agents has become of increasing interest. For example, the predatory mites Phytoseiulus persimilis feed on both TSSM adults and their eggs, and use the odour emitted by infested plants as a cue to locate their prey (van den Boom et al., 2004; Kappers et al., 2005, 2011).
To obtain a comprehensive insight into the defence responses of pepper against TSSM and how stress-related hormones, especially JA, play a role in TSSM-induced defences, we performed comparative transcriptome and metabolome analysis upon TSSM infestation and JA treatment in two C. annuum genotypes that differ in TSSM susceptibility.
Materials and methods
Plants, arthropods, and experimental set-up
Two-spotted spider mites (Tetranychus urticae) were propagated on lima bean plants (Phaseolus lunatus) for many generations. Seeds of Capsicum annuum varieties ‘Vania’ (bell pepper inbred line originally from INRA, Genotype 8, G8) and ‘Ta Pien Chiao’ [hot pepper, land race originating from China, obtained via CGN (Wageningen University), Genotype 29, G29] were germinated at 23 °C and seedlings were cultivated in potting soil (Lentse potgrond, Katwijk, The Netherlands) in a greenhouse [16 h day (23 °C)/8 h night (18 °C), 50–60% relative humidity]. Plants were watered daily and received half-strength Hoagland nutrient solution once a week. Five-week week-old vegetative plants were treated with JA (100 µM, 0.001% Tween-80) for either 6 h or 24 h prior to harvest, or infested with ~300 adult TSSMs for 3 d, or were left untreated. TSSMs were collected by washing lima bean leaves in water. Adults of different ages were checked for aliveness and transferred to a Petri dish using a fine brush before transferring to the pepper leaves. JA was applied by spraying leaves until drops fell off the leaves. For each treatment, three independent biological replicates were generated. For each individual sample, leaves of two different plants were combined. RNA isolations, and metabolite and phytohormone extractions were performed using the same sample. To evaluate susceptibility of both pepper genotypes to TSSMs, additional plants were placed in between TSSM-infested lima bean plants for 3 weeks and the number of adults, nymphs, and eggs, and chlorotic spots on the leaves, were counted using a binocular microscope. To evaluate predator preferences, female P. persimilis mites were tested towards the odours of TSSM-infested pepper plants using a Y-shaped olfactometer as previously described (Kappers et al., 2005). For analysis of WRKY gene expression, plants were sprayed with SA (0.1 µM, 0.001% Tween-80) similar to as described for JA, or infested with 100 TSSMs for 3 d.
RNA isolation, library preparation, sequencing, and validation
Total RNA was extracted from leaf tissue using TriPure (Roche, Mannheim, Germany), cleaned with the RNeasy Plant Mini Kit (Qiagen, USA), and the DNA was digested using RNase-free DNase (Qiagen, USA). RNA integrity was evaluated by 1.0% agarose gel electrophoresis. Total RNA for RNA sequencing (RNA-Seq) was quantified and, for each condition, two samples were sent for cDNA library construction and sequencing (Bioscience, WUR, Wageningen, The Netherlands). Transcriptome libraries were constructed using the TruSeq™ RNA sample Prep Kit (Illumina, CA, USA) and sequenced using the Illumina HiSeq™2500 (Illumina, CA, USA) platform which produced 9–20 million bp paired-end reads per sample. Trimmomatic (Bolger et al., 2014) was used to remove adaptor sequences, empty reads, short reads (<25 bp), reads with an N-ratio >10%, and low quality sequences. For quantitative reverse transcription–PCR (RT–qPCR), 1 µg of DNA-free RNA was reverse transcribed using iScript Reverse Transcriptase (BioRad, USA). Gene-specific primers were designed based on sequences obtained by BLAST search in the pepper genome database. RT–qPCR analyses were performed in biological triplicates and technical duplicates, using iQ SYBR Green Supermix (BioRad, USA) and the following PCR program: 3 min at 95 °C; 40 cycles of 15 s at 95 °C, and 30 s at 58.0 °C. Relative expression values were obtained using the 2–∆∆Ct method with ACTIN and RPL2 as reference genes.
Quality control, mapping, and functional annotation of sequences
FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for quality control and to generate clean reads in FASTQ format. Clean reads were mapped to the Pepper Zunla-1 reference genome (http://peppersequence.genomics.cn/page/species/index.jsp). Transcript assembly, quantification, normalization, and differential expression analysis were performed using the CLC Genomics Server 7.0.3 (Mortazavi et al., 2008) with default settings for RNA-Seq mapping and analysis, using quantile normalization. Gene sequences were downloaded from the Pepper Genome Database (release 2.0, Pepper Institute, Zunyi Academy of Agricultural Science). Assembled sequences were putatively annotated using NCBI-blast-2.2 in the case of >90% identity and an E-value of <0.00001.
Gene expression and enrichment analysis
RNA-Seq reads were mapped to assembled sequences to calculate read counts for each unigene. Transcript levels of each unigene were calculated and normalized as reads per kilobase of transcript per million mapped reads (RPKM). Differently expressed genes (DEGs) between different experimental conditions were filtered using a Benjamini and Hochberg (1995) false discovery rate (FDR) of 0.05 and a threshold of log2-transformed fold changes (treatment/control) >|1.5|. Capsicum genes were blasted against the Arabidopsis genome using the CLC Workbench (QIAGEN Bioinformatics). DEGs were mapped to Gene Ontology (GO) terms in the general GO database (Du et al., 2010). Gene numbers were calculated for every GO term and enrichments analysed by agriGO (http://bioinfo.cau.edu.cn/agriGO/analysis.php) using an FDR of 0.05 to find significant enrichments.
Stress-related phytohormones
Endogenous stress-related hormones, namely SA, JA, its biosynthetic precursor cis-12-oxo-phytodienoic acid (OPDA), and the biologically active metabolite JA-Ile, were extracted according to Floková et al. (2014) with modifications as described in Supplementary Protocol S1 at JXB online and analysed using ultra-performance liquid chromatography (UPLC)-MS/MS analysis on a Acquity UPLC® System (Waters, USA) coupled to a triple quadrupole mass spectrometer Xevo™ TQ-S (Waters, UK). For detailed description of analytical conditions, see Supplementary Protocol S1.
Volatile metabolites
Headspace of TSSM-infested JA-treated and non-treated plants was collected on Tenax absorbent using dynamic headspace sampling in a climate-controlled cabinet for 2 h and analysed on a Thermal Desorber TD100-xr (Markes, UK) connected to a GC-quadrupole time of flight (QToF; Agilent Technologies, USA). Experimental conditions and GC-QToF operating conditions can be found in Supplementary Protocol S1. Chromatograms were analysed for the presence of plant-derived compounds using MassHunter Unknown Analysis deconvolution software (Agilent Techologies, USA), in combination with NIST98 and Adams (2017) spectral libraries. Data files generated from the GC-MS platform were processed with MZmine V2.23 (mzmine.github.io/) for baseline correction, mass spectra extraction, and mass signal alignment.
Endogenous semi-polar metabolites
For untargeted analysis of semi-polar metabolites, 20 mg of leaf tissue was extracted with 2 ml of 75% MeOH (0.125% formic acid) according to De Vos et al. (2007). Samples were analysed using UPLC (Waters Alliance HPLC2695) coupled to a QToF mass spectrometer (Synapt G25 QToFMS Ultima, Waters, US). LC-MS operating conditions can be found in Supplementary Protocol S1. Data files generated from the LC-MS platform were processed with Metalign (Lommen, 2009) for baseline correction, mass spectra extraction, and mass signal alignment. MSClust (Tikunov et al., 2012) was used for data reduction by unsupervised clustering and extracting putative metabolites from mass spectra from ion-wise chromatographic alignment data. Selected endogenous metabolites were putatively annotated based on their molecular weight using the knapsack database (http://kanaya.naist.jp/knapsack_jsp/top.html) taking possible adduct formation into account.
Multivariate analysis
Multidimensional analyses of log2-transformed data of DEGs and metabolites were performed using MetaboAnalyst 3.0 (Xia et al., 2016). Hierarchical clustering analysis (HCA) distances were calculated using Pearson correlation and summarized using UPGMA. Both unsupervised principal component analysis (PCA) and supervised partial least squares-discriminant analysis (PLS-DA) algorithms were used to identify clustering among samples. Cluster IDs that define class separation on PLS-DA were obtained to identify genes and metabolites. Individual metabolites and plant hormones were analysed for significant changes upon infestation using Student t-tests.
Results
Spider mite performance on two Capsicum genotypes
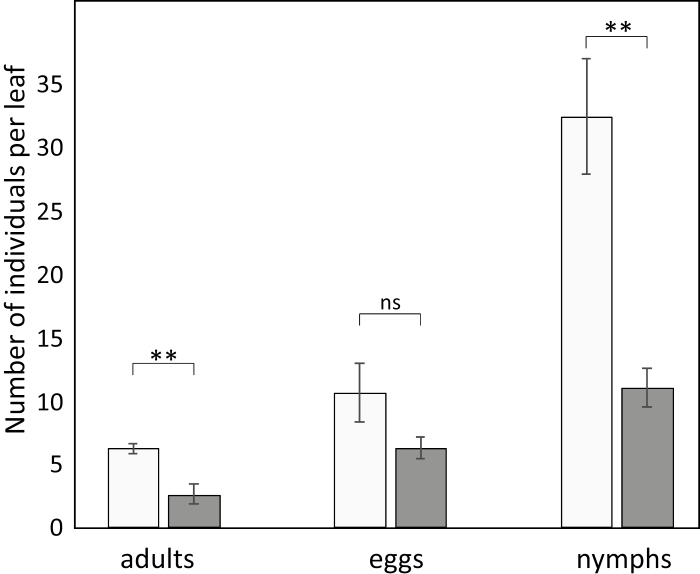
Selection of genotypes used in this study was based on previously found differences in relative resistance to TSSMs as well as in relative attractiveness towards predatory mites (unpublished). In the current study, the inbred line ‘Vania’ (G8) was confirmed to be more susceptible to TSSMs compared with the land race ‘Ta Pien Chiao’ (G29) as more offspring were found on G8 3 d after introducing female adults on whole plants (Fig. 1). In particular, more eggs developed into young nymphs on leaves of G8 compared with G29. Damage inflicted by TSSMs was assessed by observing chlorotic spots on leaves after 3 weeks of infestation, and this occurred to a greater extent in G8 than in G29 (6.9±0.3 and 3.2±0.4 spots cm–2, respectively).
Fig. 1.
Spider mite population developing on Capsicum. Individuals including adults and offspring (eggs and nymphs) found on Capsicum annuum inbred line ‘Vania’ (G8, light grey bars) and land race ‘Ta Pien Chiao’ (G29, dark grey bars), 4 d after introduction of adult mites. Data are means (±SE) of five replicate plants and were tested for significance using a two-tailed Student t-test. ns, not significant, **P<0.01.
Induced responses to spider mite feeding in Capsicum
Global transcriptome changes
To characterize the mechanism of TSSM-induced defence responses in Capsicum and disentangle the role that plant hormones in general and JA in particular play in this process, TSSM-induced transcriptome responses were compared with those induced by JA. As visible TSSM damage appears first after 4–5 d on the susceptible genotype, early mite-induced responses were captured after 3 d of infestation. To cover the relatively transient response induced by JA, two time points (6 h and 24 h) of JA induction were analysed.
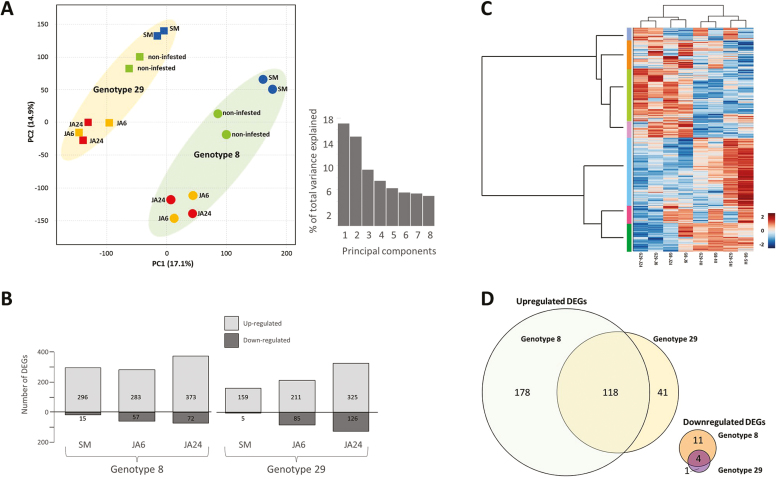
Unsupervised PCA of all detected transcripts (log2 values, normalized to RPKM), showed that the first two PCs explain 32% of the total variation (Fig. 2A). PCA shows that genotypes different in their susceptibility towards TSSM infestation have different transcriptome profiles regardless of whether they are infested or not. Upon TSSM feeding, transcriptional profiles change in the same direction in both genotypes, while they changes in opposite directions, in both genotypes, upon application of JA.
Fig. 2.
Transcriptional changes upon spider-mite infestation or JA application. (A) Principal component analysis (score plot) of all transcripts (RPKM values) detected in leaves. Data points represent the different samples. SM, leaves infested with TSSMs for 3 d; JA6, leaves treated with jasmonic acid for 6 h; JA24, leaves treated with jasmonic acid for 24 h. The first three PCs explain 41.4% of total variation. (B) Number of differentially expressed genes (DEGs) that are up- and down-regulated in each of the experimental conditions. (C) Clustering analysis and heat map of expression measures of DEGs detected in each of the experimental conditions. Colour coding represents the range of log2 fold induction (red) or repression (blue). (D) Number of TSSM-induced DEGs characteristic for or overlapping in both genotypes.
To reduce the complexity of the data set, 1955 DEGs between treated and non-treated conditions were selected. In all experimental conditions and in both genotypes, the number of up-regulated DEGs was higher than that of down-regulated genes (Fig. 2B). DEGs partly overlap between TSSM-infested and JA-treated plants, but are mostly unique for both treatments (Table S1; Figs S1, S2, available at the Dryad Digital Repository, https://doi:10.5061/dryad.n34h180). RT–qPCR analysis for a selection of genes validates our RNA-seq data as reproducible (Table S2 at Dryad). HCA classified DEGs into seven main groups of transcripts with similar expression profiles over different genotypes and treatments (Fig. 2C). GO enrichment for each of these groups showed that JA predominantly induced genes associated with metabolic processes, response to jasmonic acid, response to fungus, osmotic stress, salt stress, water deprivation, wounding, oxidative stress, and response to cadmium ion, while TSSM infestation resulted in over-representation of DEGs in processes associated with responses to fungus, wounding, response to jasmonic acid, and response to salicylic acid, as well as protein phosphorylation (Table S3 at Dryad).
Less than one-fifth of TSSM-induced DEGs were in common with those induced by JA (13.5% in G8, 19.5% in G29), of which genes involved in lipid metabolic process and stress-related processes were over-represented based on GO enrichment (Table S3 at Dryad). Genes repressed by TSSMs as well as JA include those with homology to a tomato abscisic acid (ABA) and environmental stress-inducible protein-encoding gene TAS14 (Parra et al., 1996) and a JA-dependent disease-resistant zinc finger CCCH domain-containing protein-encoding gene C3H12 (Deng et al., 2012).
Defence-related plant hormones
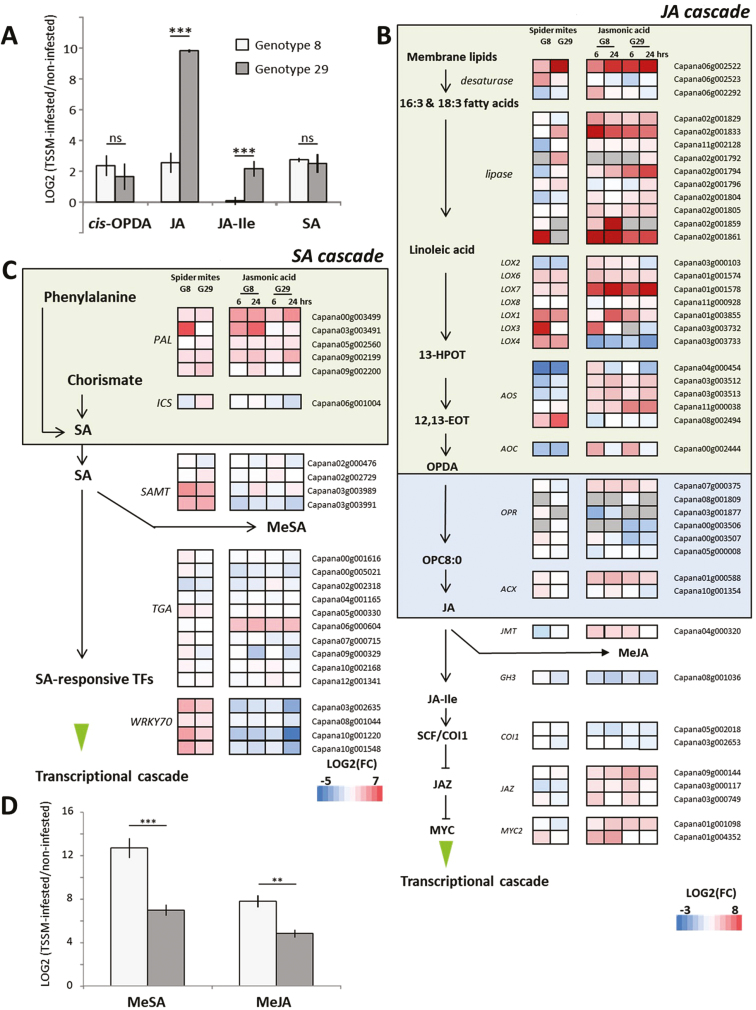
TSSM infestation significantly increased cis-OPDA, JA, and SA in both genotypes, and JA-Ile in G29 (Fig. 3A), suggesting that TSSMs induced the production of both JA and SA metabolites in Capsicum. In contrast, a slight but consistent decrease in ABA was found as a result of TSSM infestation (Table S4 at Dryad). In non-infested plants, basal concentrations of JA and SA were 242-fold (P=0.023) and 1.64-fold (P=0.007) higher in G8 than in G29, while cis-OPDA was 2-fold lower in G8 (P=0.006) and JA-Ile, and ABA did not differ. Accumulation of JA and JA-Ile in response to TSSMs was higher in G29 [log2(infested:non-infested) 9.82 and 2.16, respectively] compared with those in G8 [log2(infested:non-infested) 2.57 and 0.08]. Accumulation of SA in response to TSSMs was similar in both genotypes.
Fig. 3.
Spider mite-induced responses in JA and SA pathways. (A) Log2 fold changes in stress-related hormones upon TSSM infestation. (B) Heat map of log2 fold changes of genes involved in the jasmonic acid (JA) biosynthesis and signalling cascade. (C) Heat map of log2 fold changes of genes involved in salicylic acid (SA) biosynthesis and signalling cascade. In schemes of cascades, compounds are shown in bold and enzymes in italics. cis-OPDA, cis-(+)-12-oxo-phytodienoic acid; JA-Ile, JA-isoleucine conjugate; LOX, lipoxygenase; AOS, allene oxide synthase; AOC, allene oxide cyclase; OPR, oxidoreductase; ACX, acyl coenzyme-A oxidase; GH3, GH3 family proteins; COI1, coronatine-insensitive protein 1; JAZ, Jasmonate ZIM domain; MYC2, bHLH transcription factor; PAL, phenylalanine ammonia lyase; ICS, isochorismate synthase, SAMT, salicylate-O-methyl transferase; TGA, TGACG sequence-specific binding proteins; WRKY70, (T)TGAC(C/T) sequence-specific binding proteins. Colour coding represents the range of log2 fold changes. (D) Log2 fold changes of methyl salicylate (MeSA) and methyl jasmonate (MeJA), volatile metabolites of SA and JA, respectively, as a result of TSSM infestation.
To zoom in on how TSSM influences expression of genes involved in JA and SA biosynthesis and signalling cascades, we compared Capsicum transcripts with their tomato homologues involved in these processes using the lay-out as presented by Martel et al. (2015). TSSM infestation resulted in differential expression of multiple genes putatively involved in the biosynthesis of JA and SA (Fig. 3B, C). As the specific genes involved in these pathways are mostly not yet characterized for Capsicum, we depicted the most likely gene candidates. Genes involved in the formation of α-linoleic acid, an early intermediate in JA biosynthesis, include desaturases and lipases that play a role in the breakdown of membrane lipids. Several of these genes were differentially regulated in both genotypes upon TSSM infestation. Multiple 13-OH LOX-encoding genes, involved in the conversion of linoleic acid into 13(S)-hydroperoxylinolenic acid (13-HPOT), were up-regulated upon TSSM infestation. In contrast, most of the ALLENE OXIDE SYNTHASE (AOS) and ALLENE OXIDE CYCLASE gene homologues show repression of transcription, with the exception of Capana08G002494 that is up-regulated upon TSSM feeding, indicating that this AOS might be one that is associated with induced JA biosynthesis in Capsicum.
Interestingly, application of JA resulted in up-regulation of multiple genes in the biosynthetic pathway upstream of JA, including gene candidates encoding desaturases and lipases. Also 13-OH LOX genes including LOX6 and LOX7 (Sarde et al., 2018), multiple AOS homologues, 12-OXOPHYTODIENOATE REDUCTASE, ACYL COENZYME-A OXIDASE, and JASMONIC ACID CARBOXYL METHYLTRANSFERASE gene candidates were up-regulated upon JA treatment (Fig. 3B). Transcripts of genes related to JA signalling including JASMONATE ZIM DOMAIN (JAZ) and MYC2 homologues were induced by JA, while in response to TSSMs only minor changes in transcription of these genes were detected. In addition, multiple Capsicum homologues to known JA/ET-responsive genes including PATHOGENESIS-RELATED 4 (PR4) and CHIB (Table S3 at Dryad) indicate that TSSMs induced the JA signalling pathway in Capsicum.
Both TSSM- and JA-induced genes were related to the conversion of phenylalanine to SA, while transcript changes in a homologue of ICS were only minor and different in both genotypes.
TSSM infestation induced genes associated with SA signalling, including genes with homology to WRKY40 and WRKY70, while JA treatment repressed their expression. Furthermore, TSSM infestation, but not JA, resulted in induction of SALICYLATE O-METHYLTRANSFERASE candidates, responsible for the conversion of SA into methyl-SA. SA-inducible genes, including GLUTAREDOXIN (Ndamukong et al., 2007), the SA positive regulator RECEPTOR LECTIN KINASE (Luo et al., 2017), PR4, and several fungal defence-related genes which are important components of insect and pathogen defences especially in the Solanaceae family (Kim et al., 2009), were also induced by TSSMs. Emission of the volatile methyl esters of SA and JA strongly increased upon TSSM infestation and was higher in G8 than in G29 (Fig. 3D).
Volatile and non-volatile specialized metabolism
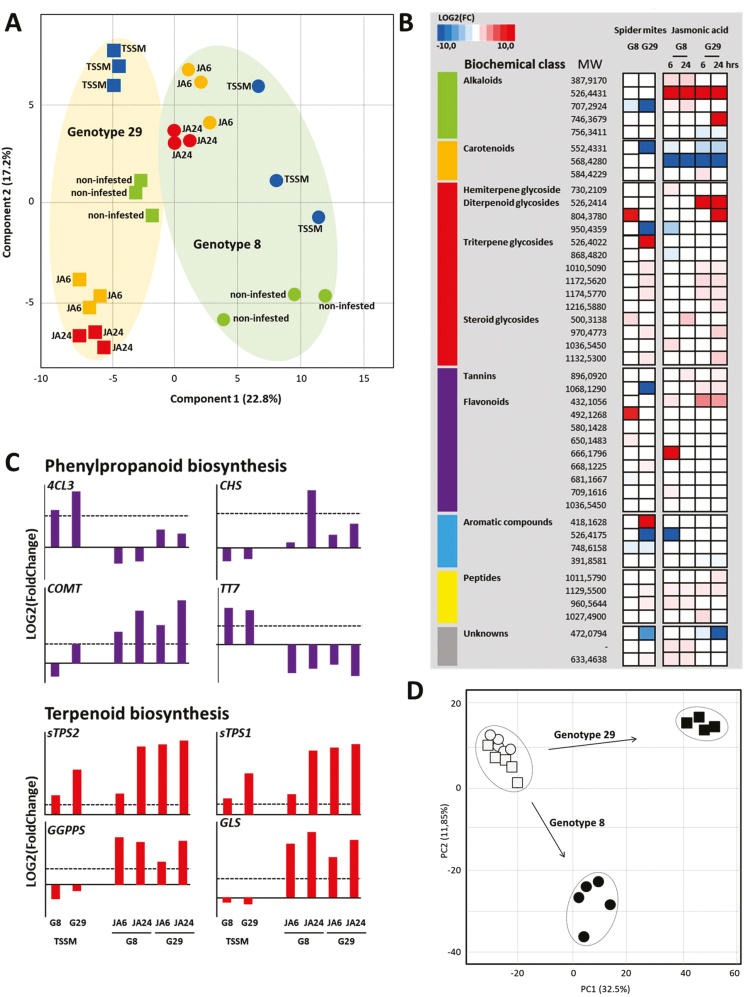
PLS-DA representation of non-volatile metabolites explains 40% of the data set variation in the first two components (Fig. 4A). Cross-validation indicates the first three components relevant for classification of the variation, illustrative for different directions of response to JA and TSSMs in both genotypes. After 3 d of TSSM infestation, prior to visible damage, 14 metabolites were significantly up-regulated in G29 when compared with non-infested plants, while seven were suppressed (Fig. 4B). The number of metabolite features altered by TSSMs was smaller in G8 (four induced, three repressed). Most of the metabolites induced upon TSSM feeding are glycosylated terpenoids, including several di- and triterpene glucosides. Furthermore, a flavonoid-O-glucoside strongly increased upon TSSM infestation in G8, specifically. Only two metabolites, a putative alkaloid and an aromatic compound, were repressed in both genotypes.
Fig. 4.
Changes in specialized metabolism upon spider mite infestation or JA application. (A) Partial least squares discriminant analysis (score plot) of endogenous semi-polar metabolites detected in leaves. Data points represent the different samples. TSSM, leaves infested with spider mites for 3 d; JA6, leaves treated with jasmonic acid for 6 h; JA24, leaves treated with jasmonic acid for 24 h. The first two components explain 40% of the total variation. (B) Metabolic features (accurate masses and putative biochemical class) that were found to be significantly induced or repressed in each of the experimental conditions. Colour coding indicates log2(FC) of induction (red) or repression (blue). (C) Log2(FC) of selected genes involved in biosynthesis of specialized metabolites. The dotted line indicates log2(FC)=1. 4CL3, 4-coumarate-CoA ligase; CHS, chalcone synthase; COMT, caffeic acid 3-O-methyltransferase; TT7, cytochrome P450 flavonoid biosynthesis related; sTPS, sesquiterpene synthase; GGPPS, geranylgeranyl diphosphate synthase; GLS, geranyl linalool synthase. For gene identifiers, see Supplementary Table S9 at Dryad. (D) Principal component analysis (score plot) of volatile metabolites emitted by non-infested and TSSM-infested Capsicum plants. Open circles indicate non-infested G8, open squares indicate non-infested G29, and filled symbols indicate TSSM-infested samples. Arrows indicate the direction of response within the first and second PC for both genotypes.
JA induced larger changes in more metabolites than did infestation with TSSMs, including terpenoids, flavonoids, and alkaloids (Fig. 4B; Table S5 at Dryad). In addition, JA significantly repressed the abundance of the carotenoids β-cryptoxanthin (mol. wt 552.4331 Da) and zeaxanthin (mol. wt 568.4280 Da) in both genotypes, while TSSMs only repressed β-cryptoxanthin in G29 and had no effect on carotenoids in G8. Xanthophylls are accessory pigments for photosynthesis and assist chlorophylls in light harvesting (Xu et al., 2015), and their lower abundance is consistent with the JA-repressed transcripts of photosynthesis-related genes found in the transcriptome analysis of both genotypes (Table S6 at Dryad).
With regard to volatile metabolites, TSSMs induced changes in the emitted blend of both genotypes. The first two components of the PCA explained 44.3% of the total variation and, interestingly, the direction of the response differs between the two genotypes (Fig. 4D). Within the top 20 most distinctive features contributing to the altered volatile blend, at least 12 of them are terpenoids, including the monoterpenes linalool and (E)-β-ocimene, six sesquiterpenes, and the homoterpenes (3E)-4,8-dimethyl-1,3,7-nonatriene and (E,E)-4,8,12-trimethyltrideca-1,3,7,11-tetraene (TMTT) (Table S7 at Dryad). Most of these compounds are volatiles detected in the blend of multiple herbivore-infested plant species. (E)-2,4-Hexadiene was the most distinct induced compound present in the volatile blend of TSSM-infested G8 plants, while other GLVs were more strongly induced in TSSM-infested G29. Furthermore, the induced volatile blend of G29 plants predominantly consisted of sesquiterpenes while methyl-SA was more strongly induced in G8 compared with G29 (Fig. 3D). Except for methyl-SA, benzenoid compounds are the most distinctive group of compounds whose emission was repressed in both genotypes. As cinnamoyl-CoA and benzoyl-CoA are involved in the biosynthesis of both SA and benzenoids in, for example, petunia flowers (Klempien et al., 2012), this suggests that upon TSSM infestation common precursors are converted to SA and methyl-SA instead of benzenoids.
While JA induced multiple genes involved in primary metabolic processes including biosynthesis and metabolism of isopentenyl diphosphate, fatty acids, glutamine, and aromatic amino acids, TSSM infestation did not have any effect on transcription of these genes. Also genes related to photosynthesis were repressed by JA, but not by TSSMs (Table S6 at Dryad). JA induced genes involved in the biosynthesis of several secondary metabolites, including terpenoids, phenylpropanoids, and flavonoids (Fig. 4C; Table S3 at Dryad). TSSM infestation for 3 d did not result in visible chlorotic spots, and only a limited number of metabolic processes showed up-regulated gene expression, including lipid and fatty acid metabolism, and, in G8 only, phenylpropanoid metabolism (Table S3 at Dryad). Nevertheless, a number of JA-induced TERPENE SYNTHASE genes were induced by early TSSM infestation, including multiple putative sesquiterpene synthase genes (Fig. 4C). In contrast, a geranylgeranyl diphosphate synthase, providing the precursor for diterpenes and TMTT, was found to be repressed upon TSSM infestation, while it was induced by JA (Fig. 4D). Also, GERANYL LINALOOL SYNTHASE was found to be induced upon JA treatment, and, based on the expression profiles of all samples, this gene strongly correlated (Pearson correlation >0.90) with 34 genes, including a number of CYTOCHROME P450 MONOOXYGENASES (P450) genes (Table S8 at Dryad). Further research will elucidate which of these P450 genes are responsible for the formation of volatile TMTT and hydroxylated geranyl linalool, which is the precursor for non-volatile diterpene glycosides.
Next to terpenoid-related metabolism, genes involved in the biosynthesis of flavonoids, including those with homology to CAFFEIC ACID 3-O-METHYLTRANSFERASE, whose product catalyses multistep methylations in the lignin and flavonoid biosynthetic pathway (Do et al., 2007), and CHALCONE SYNTHASE whose product catalyses the condensation of 4-coumaroyl-CoA and malonyl-CoA to form the flavonoid naringenin (Coburn et al., 2015), were found to be induced upon JA treatment, while transcriptional differences were minute in TSSM-infested leaves (Fig. 4C). In contrast, transcripts of 4CL3 and TT7 were induced upon TSSM infestation, while they were repressed by JA.
Across the different treatments, the changes in the metabolome are more or less consistent with the changes in the transcriptome. Together they suggest that JA induces extensive changes in specialized metabolism while early TSSM infestation predominantly induces genes involved in the lipid metabolic process and the phenylpropanoid pathway.
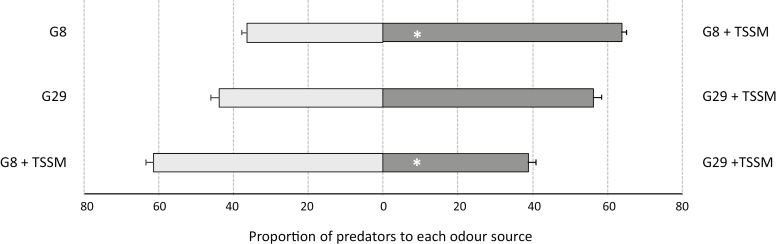
Both genotypes exhibited enhanced attractiveness to predatory mites upon TSSM infestation, but the odour blend of G8 was more attractive than that of G29 (Fig. 5). G29 had higher induced emission of multiple GLVs and terpenoids, including (E)-β-ocimene, (E,E)-α-farnesene, TMTT, and (E)-3-hexenol, which are compounds suggested to be involved in the attraction of natural enemies in many plant species (Schnee et al., 2006; Zhang et al., 2009; Kappers et al., 2011). Intriguingly, only a few volatile metabolites were induced to a higher level in the relatively susceptible genotype G8, including (E)-2,4-hexadiene and methyl-SA. Methyl-SA has been extensively reported to be attractive to predatory mites in many plant species (De Boer and Dicke, 2004; Ishiwari et al., 2007; Shimoda, 2010; Kappers et al., 2011). Although the attractiveness to predatory mites is most likely to be the result of the blend of volatile metabolites emitted, our data seem to confirm that methyl-SA is an important compound determining predatory mite preferences, also in pepper.
Fig. 5.
Attraction of predatory mites. Percentage of Phytoseiulus persimilis adult females that were attracted to the odours of TSSM-infested C. annuum plants in a two-choice olfactometer. Experiments were repeated four times for comparisons between TSSM-infested and non-infested plants in each genotype, and eight times in the comparison between TSSM-infested plants of both genotypes. In each experiment, 20 individual adult predatory mites were tested (χ 2 test, *P<0.05).
Constitutive defences of both genotypes are comparable
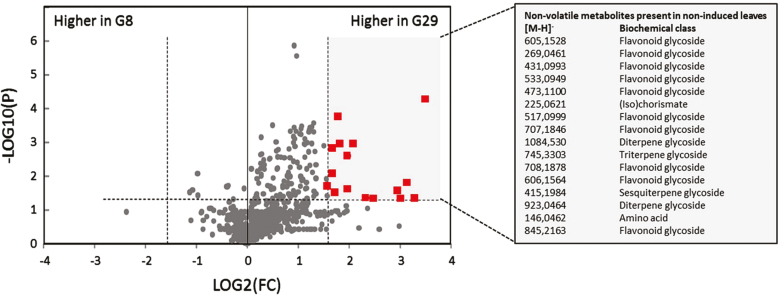
The final level of defence towards herbivores is determined by the magnitude of induced defences upon infestation as well as the basal defence level. We therefore compared transcriptomes of both genotypes under non-infested/treated conditions. In the relatively resistant G29 plants, no GO term was found significantly (FDR <0.05) enriched in genes with higher expression levels compared with those in G8. In contrast, the GO terms response to biotic stimulus and response to stress were enriched in genes with higher expression in G8. Apparently these constitutively expressed genes do not play an important role in defence as G8 is more susceptible. In contrast, when comparing metabolite profiles of non-induced plants, no metabolite features were found to be more than 3-fold [log2(1.58)] higher in G8 plants, while in G29 a total of 16 metabolite features were significantly (P<0.05) more abundant in non-infested plants compared with G8 (Fig. 6). The majority of these metabolites are flavonoids and there are also two diterpene glycosides.
Fig. 6.
Constitutive defences. Volcano plot indicating endogenous semi-polar metabolites in non-induced plants. Indicated as red squares are those compounds that significantly [–log10(P)>1.30] differ between both genotypes with a log2(FC)>2.3. Data are based on three repetitions for each genotype.
Gain of herbivore resistance by lack of SA-induced suppression of JA defences?
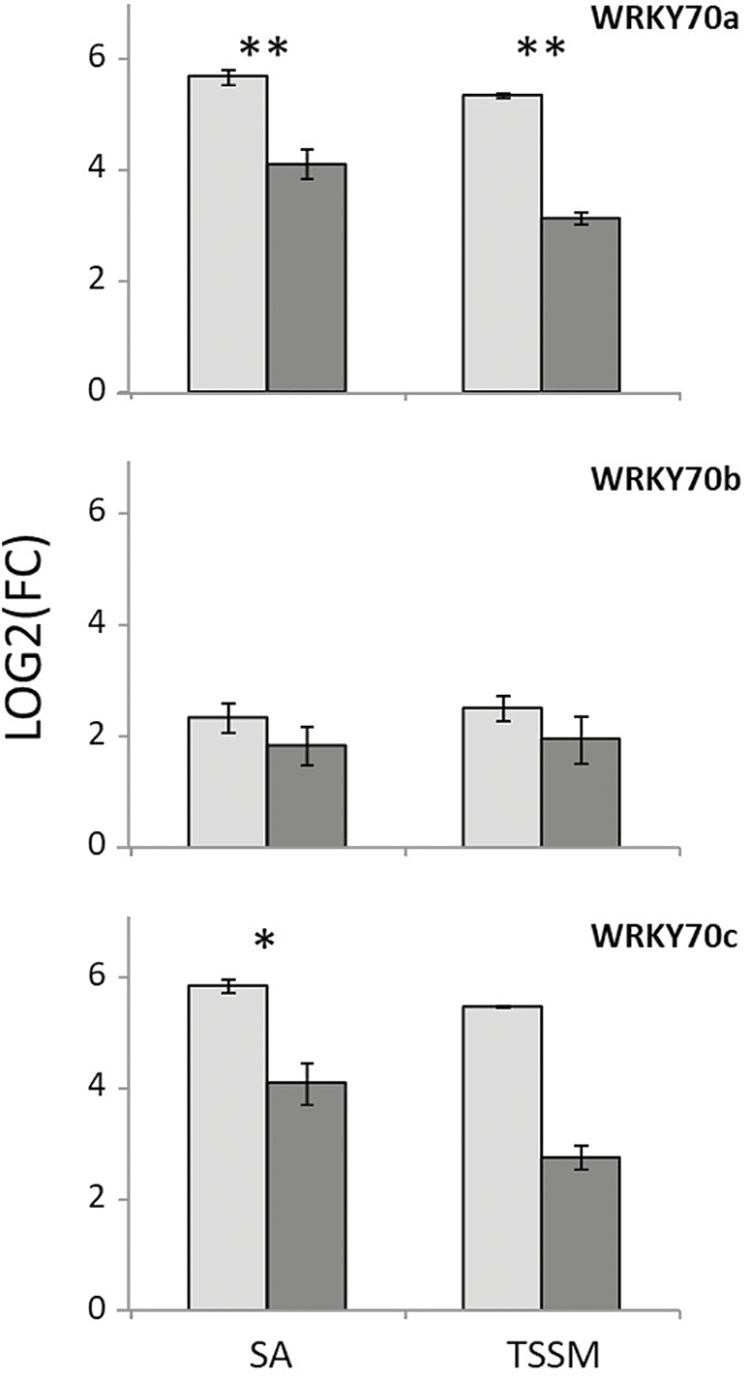
Upon TSSM feeding, G8 displayed about twice more transcriptional changes than G29 (Fig. 2D). Interestingly, a relatively large portion of DEGs was specific for each genotype, with 60% of up-regulated DEGs specifically induced in G8 while 25% of up-regulated DEGs found in G29 were not detected in G8. To compare transcriptional changes quantitatively, we compared transcriptomes of TSSM-infested leaves between both genotypes directly. Upon TSSM infestation, more genes were found enriched in response to SA in G8, including genes with homology to the SA-responsive transcription factor gene WRKY70 and several other SA-inducible genes (Table S3 at Dryad) while G29 showed stronger enrichment in secondary metabolic processes including the phenylpropanoid biosynthetic pathway. With the hypothesis that TSSMs induced SA signalling, which in turn repressed JA signalling, we examined JA-induced genes that were not induced or even repressed by TSSMs. In G29, genes enriched in amino acid metabolic process and wound response were detected, while in G8, genes enriched in lipid metabolic process and various secondary metabolic processes including isoprenoid biosynthetic pathway, phenylpropanoid biosynthetic pathway, lignin metabolic process, terpenoid metabolic process, amino acid metabolic process, and wound responses were found. In G8, more genes responsive to JA were not induced or even repressed upon TSSM infestation than in G29, which could support the hypothesis that there is stronger suppression of JA responses in G8. Indeed, TSSM-induced accumulation of methyl-SA was larger in G8 (Fig. 3), suggesting a higher induction of SA-related defences. WRKY70 transcription factor genes are known to have a regulatory role in the crosstalk between SA- and JA-induced defences, and Capsicum WRKY70 homologues were more strongly induced by TSSMs in G8 (Fig. 3C). When plants of both genotypes were equally induced by SA, or infested with an equal number of TSSMs, two out of three analysed WRKY70 genes were more strongly induced in G8 compared with G29 (Fig. 7), suggesting that the higher relative resistance of G29 to TSSMs could be partly due to a lack of SA-induced suppression of JA-related defences, possibly via WRKY70.
Fig. 7.
WRKY70 induced by spider mites. Induction of WRKY70 homologue genes upon spraying with SA or infestation with spider mites. Transcript levels are calculated as log2(FC), 2–ΔΔCt relative to control treatment and reference genes. Light grey bars indicate G8 and dark grey bars indicate G29. Data are means of three biological replicates ±SE.
Discussion
Many studies have shown that JA plays a prominent role in plant defence against herbivores (Bari and Jones, 2009) and induces production of metabolites including terpenoids, alkaloids, and proteins deterring herbivorous attackers directly or indirectly (Aerts et al., 1994; Van der Fits and Memelink, 2000; Ament et al., 2004; Chen et al., 2005). Here, we found that in C. annuum, spider mites induce distinctive transcriptional changes different from those in response to JA. JA treatment induced genes enriched in wound responses and JA biosynthesis genes. In multiple plant species it has been described that wound responses are mediated by COI1-dependent JA signalling (Wasternack et al., 2006; Wang et al., 2008) and that JA regulates JA biosynthesis via a positive feedback loop (Wasternack and Hause, 2013). However, COI1-independent induction of numerous genes and processes via OPDA have also been described in multiple plant species (Wasternack and Hause, 2016).
The expression of Capsicum genes related to different secondary metabolite biosynthesis pathways including those of terpenoids, flavonoids, and phenylpropanoids was induced by JA. In addition, multiple terpenoid, flavonoid, and alkaloid metabolites were found in higher abundance upon JA treatment. JA enhances flavonoid production probably via MYB transcription factors (An et al., 2015; Pireyre and Burow, 2015) in which MYC2 was shown to have a positive role (Dombrecht et al., 2007). Several terpenoids and alkaloids have been confirmed to be JA induced, and multiple biosynthetic genes and regulators in these pathways were identified in tomato (Ament et al., 2004), Artemisia annua (Yu et al., 2012), and Catharanthus roseus (Zhu et al., 2015). Although due to the complexity of different metabolic biosynthetic pathways and limited data sets of the current study, it is difficult to precisely connect metabolite production with gene expression, our data provide comprehensive evidence suggesting that JA induces multiple secondary metabolite biosynthetic pathways in pepper and is responsible for a burst of secondary metabolite production.
It is noteworthy that JA repressed genes functional in the tyrosine biosynthesis pathway. Tyrosine is generated from chorismate, an intermediate in the biosynthesis of phenylalanine, which is the starting point of the phenylpropanoid biosynthesis pathway (Vogt, 2010; Fraser and Chapple, 2011). In contrast to tyrosine biosynthesis, multiple transcripts related to the phenylpropanoid pathway were significantly induced by JA, suggesting that JA represses the production of tyrosine in favour of up-regulation of the phenylpropanoid pathway.
JA-induced secondary metabolites are important for Capsicum defence against spider mites
Based on the numbers of TSSMs that developed from eggs to nymphs and adults in the short time span of the bioassay, G8 is more susceptible than G29. Upon infestation, the flux in SA metabolism increased in G8, corresponding to the higher expression of SA-responsive genes found in this genotype. In contrast, JA production increased more strongly, and more endogenous metabolites increased upon TSSM feeding in G29. Transcriptome comparison between both genotypes in non-infested samples suggests that some stress-related genes are more highly expressed in G8 under unchallenged conditions, consistent with higher constitutive concentrations of SA and JA in G8. However, TSSM infestation leads to a stronger JA response in G29, resulting in higher expression of JA-responsive genes and the accumulation of various secondary metabolites, eventually resulting in a stronger induced defence correlating with the observed higher resistance for G29.
Spider mites induce both the SA and JA signalling pathways, but not all JA-induced metabolic processes
JA biosynthetic genes up-regulated byTSSM feeding caused the accumulation of cis-OPDA, JA, and JA-Ile, and up-regulated JA-responsive genes. Together, this confirms that TSSMs induced both JA biosynthesis and signalling in Capsicum. However, compared with large chewing herbivores such as caterpillars (Kawazu et al., 2012), TSSMs cause relatively less mechanical damage and, potentially via saliva-associated microorganisms, also induced the SA pathway, resulting in an increased SA flux and up-regulation of SA-responsive genes. This result is consistent with studies on TSSM-induced responses in tomato (Martel et al., 2015) and in grape (Díaz-Riquelme et al., 2016).
Our study shows that early TSSM infestation induces only a limited number of metabolite pathways compared with JA treatment. As a result, plants accumulated less secondary metabolites upon infestation than plants treated with JA, and distinctive metabolic profiles were found for JA-induced plants compared with TSSM-infested and control plants. However, both JA and TSSMs up-regulated the expression of genes encoding 4-coumarate-CoA ligase 2, in both genotypes. This enzyme is involved in the final step of the phenylpropanoid pathway, which precedes the biosynthesis of both SA (Chen et al., 2009) and flavonoids (Vogt, 2010). JA induced multiple genes in the flavonoid pathway, downstream of the phenylpropanoid pathway, and produced more flavonoid metabolites compared with non-treated plants. In contrast, TSSMs induced the SA pathway, but fewer flavonoids compared with JA treatment. This suggests that TSSMs induce the phenylpropanoid pathway only, and do not up-regulate the flavonoid pathway as JA does, resulting in the production of SA, but not flavonoids. In accordance with our results, overproduction of SA in tobacco plants by bacterial transgenes strongly inhibited the accumulation of the flavonoids quercetin, kaempferol, and rutin (Nugroho et al., 2002). However, exogenous SA application has been reported to boost the production of flavonoids in several plant species (Xu et al., 2009; Gondor et al., 2016), and SA induced by pathogens resulted in accumulation of flavonoid phytoalexins in a range of crops in the Brassicaceae, Fabaceae, Solanaceae, Vitaceae, and Poaceae (Ahuja et al., 2012). Altogether, we hypothesize that JA-induced flavonoid biosynthesis is restricted upon spider mite herbivory because the production of SA induced by the mites diverts a substantial part of the precursors available to SA biosynthesis.
Even though no GO terms related to terpenoid pathway metabolic processes were found enriched upon TSSM infestation, a number of terpene biosynthetic genes were induced, and multiple volatile and non-volatile terpenoids were found to be present in higher amounts after TSSM infestation. Our results indicate that the terpenoid biosynthetic pathway is mainly induced via JA signalling, while the SA signalling pathway activated by TSSM does not repress this biosynthetic pathway.
JA–SA crosstalk in spider mite-induced defence
In addition to multiple secondary metabolic pathway genes, we found many other genes responsive to exogenous JA but not induced by TSSMs even though JA signalling is induced by TSSM feeding. JA–SA antagonism has been demonstrated in many plant species (Thaler et al., 2012) and we propose that in Capsicum the SA pathway suppresses part of spider mite-induced JA responses. To verify this hypothesis and demonstrate a possible mechanism of antagonizing crosstalk, we identified transcription factors induced by JA and TSSMs, respectively, as well as genes whose expression strongly correlates with that of these transcription factors.
MYC2 is well known as the master regulator of JA signalling. Two homologues are identified in the C. annuum genome, Capana01g004352 and Capana01g001098. Since only Capana01g001098 shows strong correlations (Pearson correlation >0.85) with multiple DEGs, we predict that Capana01g001098 is the functional homologue of MYC2 in Capsicum. This Capsicum MYC2 homologue is induced by exogenous JA in both genotypes, though not statistically significantly (Tables S1, S3 at Dryad). Based on correlation analysis, 40 DEGs positively correlate with MYC2, and 24 and 25 of those 40 genes are induced by JA in G8 and G29, respectively, while none of them was induced by TSSMs in either of the genotypes. In Arabidopsis, protein levels of MYC2, MYC3, and MYC4 were strongly diminished by Pieris brassicae egg extract in an SA-dependent manner, suggesting that MYC transcription factors are targets of SA–JA antagonism (Schmiesing et al., 2016). Apart from the MYC2 branch of the JA signalling pathway, exogenous JA application also induced transcription factors from the JA/ET branch, including three ERF genes in G8, and ERF1 and MYB3 in G29. In total, 107 DEGs positively correlated with these JA-induced transcription factors in G8 and 51 of these DEGs were induced by JA while two were also induced by TSSMs. In G29, fewer DEGs (28) positively correlated with these JA-induced transcription factors, of which 25 were induced by JA and five also by TSSMs. Interestingly, TSSM feeding resulted in the up-regulation of an ERF-related gene (Capana12g000990) in both genotypes, annotated as TIFY10B which encodes the MYC repressor, JAZ, suggesting repression of the MYC branch in the JA signalling pathway by TSSMs. In our current work, we aim to elucidate the importance of this MYC branch in spider mite-induced defences.
As discussed, JA signalling was suppressed by spider mite-induced SA, but our data also show that exogenous JA application represses SA signalling. JA application represses the expression of WRKY70 homologues [Capana10g001548, log2(FC) –3.46 in G8; Capana10g001220, log2(FC) –60.16 in G29]. WRKY70 mediates the SA-dependent signalling pathway in multiple species (Pandey and Somssich, 2009). Overexpression of WRKY70 results in constitutive expression of SA-induced pathogenesis-related genes and enhances resistance to Erysiphe cichoracearum but represses JA responses. Conversely, antisense suppression of WRKY70 activates JA-responsive/COI1-dependent genes in Arabidopsis (Li et al., 2004, 2006).
In summary, this study gives a comprehensive overview of spider mite- and JA-induced responses in C. annuum by comparing transcriptome changes and corresponding metabolome changes, and provides insight into the antagonistic JA–SA crosstalk. JA induces extensive metabolic processes and represses photosynthesis, while spider mites induce SA biosynthesis and hence SA signalling as well as the JA biosynthesis pathway, resulting in the expression of SA- and some JA-responsive genes. Our data suggest that exogenous application of JA may suppress SA signalling by up-regulating the flavonoid biosynthetic pathway which may compete with SA biosynthesis and repress the SA-responsive regulator WRKY70. On the other hand, SA signalling induced upon TSSM infestation suppresses the MYC-regulated branch in the JA signalling pathway possibly by inducing expression of a MYC repressor. In our further studies, we aim to investigate the details of this mutual antagonizing JA–SA crosstalk in pepper and its consequences for specialized metabolism and direct and indirect defences towards cell-feeding chelicerate spider mites.
Supplementary data
Supplementary data are available at JXB online.
Protocol S1. Technical details on operating conditions of mass spectrometry platforms.
Data deposition
The following tables and figures are available at Dryad Data Repository: https://doi:10.5061/dryad.n34h180
Table S1. Differentially expressed genes (DEGs).
Table S2. RT–qPCR validation of selected gene transcripts.
Table S3. Gene Ontology enrichment of DEGs.
Table S4. Stress-related hormones.
Table S5. Non-volatile metabolites.
Table S6. Genes related to photosynthesis repressed by JA, but not by TSSMs.
Table S7. Volatile metabolites.
Table S8. Pearson correlation values between DEGs.
Table S9. Identifiers of genes discussed.
Fig. S1. GO enrichment of JA-induced genes not induced by TSSMs.
Fig. S2. Number of DEGs in both genotypes under TSSM infestation and early or late JA induction.
Acknowledgements
We thank Francel Verstappen, Bert Schippers, and Kristyna Flokova for technical assistance. Koppert BV, The Netherlands is acknowledged for supply of spider mites and predatory mites. This work is part of the Perspective programme Green Defences Against Pests financed by the Netherlands Organisation for Scientific Research (NWO, TTW-grant no. 13551) and partly by breeding companies ENZA seeds, RijkZwaan, and Syngenta through NWO. YZ acknowledges the China Scholarship Council (CSC) for a supporting grant. The authors declare no conflicts of interest
Author contributions
HB and IK conceived and designed the experiments; YZ performed the experiments; YZ and IK analysed the data; and all authors contributed to the writing of the manuscript. The authors declare no conflicts of interest
References
- Adams RP. 2017. Identification of essential oil components by gas chromatography/mass spectrometry, 5th edn. Gruver, TX: Texensis Pulishers. [Google Scholar]
- Adolfsson L, Nziengui H, Abreu IN, et al. 2017. Enhanced secondary- and hormone metabolism in leaves of arbuscular mycorrhizal Medicago truncatula. Plant Physiology 175, 392–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts RJ, Gisi D, Decarolis E, Deluca V, Baumann TW. 1994. Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids in Catharanthus and Cinchona seedlings. The Plant Journal 5, 635–643. [Google Scholar]
- Ahuja I, Kissen R, Bones AM. 2012. Phytoalexins in defense against pathogens. Trends in Plant Science 17, 73–90. [DOI] [PubMed] [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC. 2004. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiology 135, 2025–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An XH, Tian Y, Chen KQ, Liu XJ, Liu DD, Xie XB, Cheng CG, Cong PH, Hao YJ. 2015. MdMYB9 and MdMYB11 are involved in the regulation of the JA-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant & Cell Physiology 56, 650–662. [DOI] [PubMed] [Google Scholar]
- Bandoly M, Hilker M, Steppuhn A. 2015. Oviposition by Spodoptera exigua on Nicotiana attenuata primes induced plant defence against larval herbivory. The Plant Journal 83, 661–672. [DOI] [PubMed] [Google Scholar]
- Bari R, Jones JD. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Bensoussan N, Santamaria ME, Zhurov V, Diaz I, Grbić M, Grbić V. 2016. Plant–herbivore interaction: dissection of the cellular pattern of Tetranychus urticae feeding on the host plant. Frontiers in Plant Science 7, 1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrizo García C, Barfuss MH, Sehr EM, Barboza GE, Samuel R, Moscone EA, Ehrendorfer F. 2016. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Annals of Botany 118, 35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. 2005. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proceedings of the National Academy of Sciences, USA 102, 19237–19242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zheng Z, Huang J, Lai Z, Fan B. 2009. Biosynthesis of salicylic acid in plants. Plant Signaling & Behavior 4, 493–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheynier V, Comte G, Davies KM, Lattanzio V, Martens S. 2013. Plant phenolics: recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiology and Biochemistry: PPB 72, 1–20. [DOI] [PubMed] [Google Scholar]
- Coburn RA, Griffin RH, Smith SD. 2015. Genetic basis for a rare floral mutant in an Andean species of Solanaceae. American Journal of Botany 102, 264–272. [DOI] [PubMed] [Google Scholar]
- De Boer JG, Dicke M. 2004. The role of methyl salicylate in prey searching behavior of the predatory mite Phytoseiulus persimilis. Journal of Chemical Ecology 30, 255–271. [DOI] [PubMed] [Google Scholar]
- De Freitas Bueno A, De Freitas Bueno RCO, Nabity PD, Higley LG, Fernandes OA. 2009. Photosynthetic response of soybean to two-spotted spider mite (Acari: Tetranychydae) injury. Brazilian Archives of Biology and Technology 52, 825–834. [Google Scholar]
- Deng H, Liu H, Li X, Xiao J, Wang S. 2012. A CCCH-type zinc finger nucleic acid-binding protein quantitatively confers resistance against rice bacterial blight disease. Plant Physiology 158, 876–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos RC, Moco S, Lommen A, Keurentjes JJ, Bino RJ, Hall RD. 2007. Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nature Protocols 2, 778–791. [DOI] [PubMed] [Google Scholar]
- Díaz-Riquelme J, Zhurov V, Rioja C, et al. 2016. Comparative genome-wide transcriptome analysis of Vitis vinifera responses to adapted and non-adapted strains of two-spotted spider mite, Tetranyhus urticae. BMC Genomics 17, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thévenin J, Sibout R, Denoue D, Barrière Y, Lapierre C, Jouanin L. 2007. Both caffeoyl coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129. [DOI] [PubMed] [Google Scholar]
- Doherty HM, Selvendran RR, Bowles DJ. 1988. The wound response of tomato plants can be inhibited by aspirin and related hydroxy-benzoic acids. Physiological Molecular Plant Pathology 33, 377–384. [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb M, Meldau S, Howe GA. 2012. Role of phytohormones in insect-specific plant reactions. Trends in Plant Science 17, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Somssich IE. 2007. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology 10, 366–371. [DOI] [PubMed] [Google Scholar]
- Fasulo TR, Denmark HA. 2016. Two-spotted spider mite, Tetranychus Urticae Koch (Arachnida: Acari: Tetranychidae) https://edis.ifas.ufl.edu/pdffiles/IN/IN30700.pdf. Last accessed 22 July 2019.
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floková K, Tarkowská D, Miersch O, Strnad M, Wasternack C, Novák O. 2014. UHPLC-MS/MS based target profiling of stress-induced phytohormones. Phytochemistry 105, 147–157. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Chapple C. 2011. The phenylpropanoid pathway in Arabidopsis. The Arabidopsis Book 9, e0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Fürstenberg-Hägg J, Zagrobelny M, Bak S. 2013. Plant defense against insect herbivores. International Journal of Molecular Sciences 14, 10242–10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gols R. 2014. Direct and indirect chemical defences against insects in a multitrophic framework. Plant, Cell & Environment 37, 1741–1752. [DOI] [PubMed] [Google Scholar]
- Gondor OK, Janda T, Soós V, Pál M, Majláth I, Adak MK, Balázs E, Szalai G. 2016. Salicylic acid induction of flavonoid biosynthesis pathways in wheat varies by treatment. Frontiers in Plant Science 7, 1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiwari H, Suzuki T, Maeda T. 2007. Essential compounds in herbivore-induced plant volatiles that attract the predatory mite Neoseiulus womersleyi. Journal of Chemical Ecology 33, 1670–1681. [DOI] [PubMed] [Google Scholar]
- Kappers IF, Aharoni A, van Herpen TW, Luckerhoff LL, Dicke M, Bouwmeester HJ. 2005. Genetic engineering of terpenoid metabolism attracts bodyguards to Arabidopsis. Science 309, 2070–2072. [DOI] [PubMed] [Google Scholar]
- Kappers IF, Hoogerbrugge H, Bouwmeester HJ, Dicke M. 2011. Variation in herbivory-induced volatiles among cucumber (Cucumis sativus L.) varieties has consequences for the attraction of carnivorous natural enemies. Journal of Chemical Ecology 37, 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu K, Mochizuki A, Sato Y, Sugeno W, Murata M, Seo S, Mitsuhara L. 2012. Different expression profiles of jasmonic acid and salicylic acid inducible genes in the tomato plant against herbivores with various feeding modes. Arthropod-Plant Interactions 6, 221–230. [Google Scholar]
- Kim JY, Park SC, Hwang I, Cheong H, Nah JW, Hahm KS, Park Y. 2009. Protease inhibitors from plants with antimicrobial activity. International Journal of Molecular Sciences 10, 2860–2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempien A, Kaminaga Y, Qualley A, et al. 2012. Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. The Plant Cell 24, 2015–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloth KJ, Wiegers GL, Busscher-Lange J, van Haarst JC, Kruijer W, Bouwmeester HJ, Dicke M, Jongsma MA. 2016. AtWRKY22 promotes susceptibility to aphids and modulates salicylic acid and jasmonic acid signalling. Journal of Experimental Botany 67, 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon-Reyes A, Van der Does D, De Lange ES, Delker C, Wasternack C, Van Wees SC, Ritsema T, Pieterse CM. 2010. Salicylate-mediated suppression of jasmonate-responsive gene expression in Arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta 232, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Palva ET. 2004. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. The Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brader G, Kariola T, Palva ET. 2006. WRKY70 modulates the selection of signaling pathways in plant defense. The Plant Journal 46, 477–491. [DOI] [PubMed] [Google Scholar]
- Little D, Gouhier-Darimont C, Bruessow F, Reymond P. 2007. Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiology 143, 784–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommen A. 2009. MetAlign: interface-driven, versatile metabolomics tool for hyphenated full-scan mass spectrometry data preprocessing. Analytical Chemistry 81, 3079–3086. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Xu N, Huang J, Gao F, Zou H, Boudsocq M, Coaker G, Liu J. 2017. A lectin receptor-like kinase mediates pattern-triggered salicylic acid signaling. Plant Physiology 174, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Zhurov V, Navarro M, et al. 2015. Tomato whole genome transcriptional response to Tetranychus urticae identifies divergence of spider mite-induced responses between tomato and Arabidopsis. Molecular Plant-Microbe Interactions 28, 343–361. [DOI] [PubMed] [Google Scholar]
- Memelink J, Verpoorte R, Kijne JW. 2001. ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends in Plant Science 6, 212–219. [DOI] [PubMed] [Google Scholar]
- Mercke P, Kappers IF, Verstappen FW, Vorst O, Dicke M, Bouwmeester HJ. 2004. Combined transcript and metabolite analysis reveals genes involved in spider mite induced volatile formation in cucumber plants. Plant Physiology 135, 2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. 2007. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. The Plant Journal 50, 128–139. [DOI] [PubMed] [Google Scholar]
- Nugroho LH, Verberne MC, Verpoorte R. 2002. Activities of enzymes involved in the phenylpropanoid pathway in constitutively salicylic acid-producing tobacco plants. Plant Physiology Biochemistry 40, 755–760. [Google Scholar]
- Pandey SP, Somssich IE. 2009. The role of WRKY transcription factors in plant immunity. Plant Physiology 150, 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MM, del Pozo O, Luna R, Godoy JA, Pintor-Toro JA. 1996. Structure of the dehydrin tas14 gene of tomato and its developmental and environmental regulation in transgenic tobacco. Plant Molecular Biology 32, 453–460. [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pireyre M, Burow M. 2015. Regulation of MYB and bHLH transcription factors: a glance at the protein level. Molecular Plant 8, 378–388. [DOI] [PubMed] [Google Scholar]
- Proietti S, Caarls L, Coolen S, Van Pelt JA, Van Wees SCM, Pieterse CMJ. 2018. Genome-wide association study reveals novel players in defense hormone crosstalk in Arabidopsis. Plant, Cell & Environment 41, 2342–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarde SJ, Kumar A, Remme RN, Dicke M. 2018. Genome-wide identification, classification and expression of lipoxygenase gene family in pepper. Plant Molecular Biology 98, 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala A, Allmann S, Mirabella R, Haring MA, Schuurink RC. 2013. Green leaf volatiles: a plant’s multifunctional weapon against herbivores and pathogens. International Journal of Molecular Sciences 14, 17781–17811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiesing A, Emonet A, Gouhier-Darimont C, Reymond P. 2016. Arabidopsis MYC transcription factors are the target of hormonal salicylic acid/jasmonic acid cross talk in response to Pieris brassicae egg extract. Plant Physiology 170, 2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schnee C, Kollner TG, Held M, Turlings TCJ, Gershenzon J, Degenhardt J. 2006. The products of a single maize sesquiterpene synthase form a volatile defence signal that attracts natural enemies of maize herbivores. Proceedings of the National Academy of Sciences, USA 103, 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda T. 2010. A key volatile infochemical that elicits a strong olfactory response of the predatory mite Neoseiulus californicus, an important natural enemy of the two-spotted spider mite Tetranychus urticae. Experimental & Applied Acarology 50, 9–22. [DOI] [PubMed] [Google Scholar]
- Snoeren TA, Kappers IF, Broekgaarden C, Mumm R, Dicke M, Bouwmeester HJ. 2010. Natural variation in herbivore-induced volatiles in Arabidopsis thaliana. Journal of Experimental Botany 61, 3041–3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SM, et al. 2003. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. The Plant Cell 15, 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Tiryaki I. 2004. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell 16, 2117–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK. 2012. Evolution of jasmonate and salicylate signal crosstalk. Trends in Plant Science 17, 260–270. [DOI] [PubMed] [Google Scholar]
- Tholl D. 2015. Biosynthesis and biological functions of terpenoids in plants. Advances in Biochemical Engineering/Biotechnology 148, 63–106. [DOI] [PubMed] [Google Scholar]
- Tikunov YM, Laptenok S, Hall RD, Bovy A, de Vos RC. 2012. MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics 8, 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. 2002. The jasmonate signal pathway. The Plant Cell 14 Suppl, S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom CE, van Beek TA, Posthumus MA, de Groot A, Dicke M. 2004. Qualitative and quantitative variation among volatile profiles induced by Tetranychus urticae feeding on plants from various families. Journal of Chemical Ecology 30, 69–89. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. 2000. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. [DOI] [PubMed] [Google Scholar]
- Van Leeuwen T, Vontas J, Tsagkarakou A, Dermauw W, Tirry L. 2010. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: a review. Insect Biochemistry and Molecular Biology 40, 563–572. [DOI] [PubMed] [Google Scholar]
- Vogt T. 2010. Phenylpropanoid biosynthesis. Molecular Plant 3, 2–20. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. 2006. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cao G, Wang X, Miao J, Liu X, Chen Z, Qu LJ, Gu H. 2008. Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Reports 27, 125–135. [DOI] [PubMed] [Google Scholar]
- War AR, Paulraj MG, Ahmad T, Buhroo AA, Hussain B, Ignacimuthu S, Sharma HC. 2012. Mechanisms of plant defense against insect herbivores. Plant Signaling & Behavior 7, 1306–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Stenzel I, Hause B, Hause G, Kutter C, Maucher H, Neumerkel J, Feussner I, Miersch O. 2006. The wound response in tomato—role of jasmonic acid. Journal of Plant Physiology 163, 297–306. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2016. OPDA-Ile—a new JA-Ile-independent signal? Plant Signaling & Behavior 11, e1253646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wishart DS. 2016. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Current Protocols in Bioinformatics 55, 14.10.1–14.10.91. [DOI] [PubMed] [Google Scholar]
- Xu M, Dong J, Wang H, Huang L. 2009. Complementary action of jasmonic acid on salicylic acid in mediating fungal elicitor-induced flavonol glycoside accumulation of Ginkgo biloba cells. Plant, Cell & Environment 32, 960–967. [DOI] [PubMed] [Google Scholar]
- Xu P, Tian L, Kloz M, Croce R. 2015. Molecular insights into zeaxanthin-dependent quenching in higher plants. Scientific Reports 5, 13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY. 2012. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Molecular Plant 5, 353–365. [DOI] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. 2007. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiology 143, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PJ, Zheng SJ, Van Loon JJA, Boland W, David A, Mumm R, Dicke M. 2009. Whiteflies interfere with indirect plant defence against spider mites in lima bean. Proceedings of the National Academy of Sciences, USA 106, 21202–21207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bouwmeester HJ, Kappers IF. 2019. Data from: Combined transcriptome and metabolome analysis identifies defence responses in spider mite-infested pepper. Dryad Digital Repository. doi: 10.5061/dryad.n34h180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Wang M, Wen W, Yu R. 2015. Biosynthesis and regulation of terpenoid indole alkaloids in Catharanthus roseus. Pharmacognosy Reviews 9, 24–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurov V, Navarro M, Bruinsma KA, et al. 2014. Reciprocal responses in the interaction between Arabidopsis and the cell-content-feeding chelicerate herbivore spider mite. Plant Physiology 164, 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.