Abstract
Background:
Identifying kidney allograft recipients who are predisposed to acute rejection (AR) could allow for optimization of clinical treatment to avoid rejection and prolong graft survival. It has been hypothesized that part of this predisposition is caused by the inheritance of specific genetic variants. There are many publications reporting a statistically significant association between a genetic variant, usually in the form of a single nucleotide polymorphism (SNP), and AR. However, there are additional publications reporting a lack of this association when a different cohort of recipients is analyzed for the same SNP.
Methods:
In this report we attempted to validate 75 common genetic variants, which have been previously reported to be associated with AR, using a large kidney allograft recipient cohort of 2,390 European-Americans and 482 African Americans.
Results:
Of those variants tested, only one variant, rs2910164, which alters expression of the microRNA MIR146A, was found to exhibit a significant association within the African American cohort. Suggestive variants were found in the genes CTLA and TLR4.
Discussion:
Our results show that most variants previously reported to be associated with AR were not validated in our cohort. This shows the importance of validation when reporting associations with complex clinical outcomes such as AR. Additional work will need to be done to understand the role of MIR146A in the risk of AR in kidney allograft recipients.
Keywords: Acute rejection, single nucleotide polymorphisms, SNPs, kidney, transplant, graft dysfunction
Introduction
Kidney allograft transplantation is the treatment of choice for end-stage kidney disease. Unfortunately, graft function decreases with the occurrence of chronic rejection (interstitial fibrosis/tubular atrophy; IF/TA). Acute rejection (AR) is a major risk factor for IF/TA and associated graft loss in kidney allograft recipients (1–3), particularly when renal function does not return to baseline. Clinical care of kidney allograft recipients could be greatly improved if individuals at risk for AR could be identified before transplantation, allowing for better individualized clinical care. AR is a complex event with several different presentations including early and late AR, antibody-mediated rejection (ABMR) and T cell-mediated rejection (TCMR). Classification of AR is continually being updated based on new and emerging techniques in histopathology, based in part on the integration of new genetic biomarkers (4, 5). It has been hypothesized that some individuals have increased risk for AR due to the inheritance of specific genetic variants (6). To understand the impact of genetic variation on AR, numerous studies in the last several decades have been undertaken to identify genetic variants associated with AR (7–70). Table 1 shows 75 genetic variants, as single nucleotide polymorphisms (SNPs), previously reported to be associated with AR. Unfortunately, there are also many reports showing failed attempts to validate some of these variants (53, 71, 72).
Table 1.
Candidate SNPs associated with acute rejection reported in the literature.
| SNP rs# | Proxy† | Gene | Chrm | Position†† | Nucleotide Change | Protein Change | Ref |
|---|---|---|---|---|---|---|---|
| rs2032582 | rs4148738 | ABCB1 | 7 | 87531302 | c.2677T>G | p.Ser893Ala | 7 |
| rs2269475 | AIF | 6 | 31616154 | c.43C>T | p.Arg15Trp | 8 | |
| rs5186 | AT1R | 3 | 148742201 | c.*86A>C | 3’ UTR | 9 | |
| rs10765602 | rs55637918 | DEUP1 (CCDC67) | 11 | 93314999 | g.93048165G>T | 5’ of gene | 10 |
| rs1024611 | CCL2 | 17 | 34252769 | c.-2582A>G | 5’ of gene | 11 | |
| rs2107538 | CCL5 | 17 | 35880776 | c.-471G>A | 5’ of gene | 12 | |
| rs1799864 | CCR2 | 3 | 46357717 | c.190G>A | p.Val64Ile | 13, 14 | |
| rs1799987 | CCR5 | 3 | 46370444 | c.-301+246A>G | Intronic | 13, 14, 15 | |
| rs3116496 | rs10490574 | CD28 | 2 | 203729789 | c.243+17T>C | Intronic | 16, 17 |
| rs1129055 | CD86 | 3 | 122119472 | c.592G>A | p.Ala198Thr | 17 | |
| rs733618 | CTLA4 | 2 | 203866221 | c.-1722T>C | 5’ of gene | 18 | |
| rs5742909 | CTLA4 | 2 | 203867624 | c.-319C>T | 5’ of gene | 19, 20 | |
| rs231775 | CTLA4 | 2 | 203867991 | c.49A>G | p.Thr17Ala | 21, 22 23, 24 | |
| rs3087243 | CTLA4 | 2 | 203874196 | c.1421G>A | 3’ of gene | 24 | |
| rs4073 | CXCL8 | 4 | 73740307 | c.-352A>T | 5’ of gene | 25 | |
| rs2515641 | CYP2E1 | 10 | 133537858 | c.1263C>G | p.Phe421 = | 26 | |
| rs776746 | CYP3A5 | 7 | 99672916 | c.219–237G/A | Intronic | 27 | |
| rs1042032 | rs4149257 | EPHX2 | 8 | 27544557 | c.35A>G | 3’ UTR | 28 |
| rs1799963 | F2 | 11 | 46739505 | c.97G>A | 3’ UTR | 29 | |
| rs6025 | F5 | 1 | 169549811 | c.1601G>A | p.Arg534Gln | 29, 30 | |
| rs7851696 | FCN2 | 9 | 134887245 | c.772G>C | p.Ala258Ser | 31 | |
| rs1801274 | FCGR2A | 1 | 161509955 | c.500A>G | p.His166Arg | 32 | |
| rs9296068 | HLA-DOA | 6 | 33020918 | g.4470122T>G | 5’ of gene | 33 | |
| rs1063320 | HLA-G | 6 | 29830972 | c.*233C>G | 3’ UTR | 34 | |
| rs5498 | ICAM1 | 19 | 10285007 | c.1405A>G | p.Lys469Glu | 35 | |
| rs1143634 | IL1B | 2 | 112832813 | c.315C>T | p.Phe105= | 36 | |
| rs2069762 | IL2 | 4 | 122456825 | c.-385T>G | 5’ of gene | 37 | |
| rs228942 | IL2RB | 22 | 37128579 | c.1173C>A | p.Asp391Glu | 38 | |
| rs228953 | rs2284033 | IL2RB | 22 | 37135396 | c.750C>T | p.Gly250= | 39 |
| rs181781 | rs657075 | IL3 | 5 | 132059422 | c.-1285G>A | 5’ of gene | 39 |
| rs2073506 | rs61527852 | IL3 | 5 | 132059045 | c.-1662C>A | 5’ of gene | 39 |
| rs40401 | rs31480 | IL3 | 5 | 132060785 | c.79C>T | p.Pro27Ser | 39 |
| rs2243250 | IL4 | 5 | 132673462 | c.-589C>T | 5’ of gene | 40 | |
| rs1801275 | IL4R | 16 | 27363079 | c.1727A>G | p.Gln576Arg | 41 | |
| rs1800796 | rs1524107 | IL6 | 7 | 22726627 | c.-636G>C | 5’ of gene | 42 |
| rs1800795 | IL6 | 7 | 22727026 | c.-237C>G | 5’ of gene | 43 | |
| rs1800896 | IL10 | 1 | 206773552 | c.-1117A>G | 5’ of gene | 44, 45 | |
| rs1800871 | IL10 | 1 | 206773289 | c.-854T>C | 5’ of gene | 44 | |
| rs1800872 | IL10 | 1 | 206773062 | c.-627A>C | 5’ of gene | 7, 44 | |
| rs763780 | IL17F | 6 | 52236941 | c.482A>G | p.His161Arg | 46 | |
| rs187238 | IL18 | 11 | 112164265 | c.-368G>C | 5’ of gene | 47 | |
| rs2278293 | IMPDH1 | 7 | 128400698 | c.579+119G>A | Intronic | 48 | |
| rs2278294 | IMPDH1 | 7 | 128400645 | c.580–106G>A | Intronic | 48 | |
| rs11706052 | IMPDH2 | 3 | 49026677 | c.819+10T>C | Intronic | 7 | |
| rs2430561 | INFG | 12 | 68158742 | c.115–483A>T | Intronic | 49 | |
| rs3757385 | rs3807307 | IRF5 | 7 | 128937250 | c.-811T>G | 5’ of gene | 50 |
| rs5918 | ITGB3 | 17 | 47283364 | c.176T>C | p.Leu59Pro | 51 | |
| rs7096206 | MBL2 | 10 | 52771925 | c.-290C>G | 5’ of gene | 52 | |
| rs5030737 | MBL2 | 10 | 52771482 | c.154C> | p.Arg52Cys | 52 | |
| rs180045 | MBL2 | 10 | 52771475 | c.161G>A | p.Gly54Asp | 52 | |
| rs1800451 | MBL2 | 10 | 52771466 | c.170G>A | p.Gly57Glu | 52 | |
| rs1801133 | MTHFR | 1 | 11796321 | c.788C>T | p.Ala222Val | 29, 53 | |
| rs2426295 | NFATC2 | 20 | 51398762 | c.2663–32T>A | Intronic | 54 | |
| rs28362491 | NFKB1 | 4 | 102500998 | c.-798_-795delATTG | 5’ of gene | 55 | |
| rs696 | rs8904 | NFKBIA | 14 | 35401887 | c.*126G>A | 3’ UTR | 55 |
| rs2227982 | PDCD1 | 2 | 241851281 | c.644C>T | p.Ala215Val | 56 | |
| rs689466 | rs12734919 | PTGS2 | 1 | 186681619 | c.-1329A>G | 5’ of gene | 8 |
| rs2476601 | PTPN22 | 1 | 113834946 | c.1858C>T | p.Arg620Trp | 57 | |
| rs7976329 | rs7137890 | PTPRO | 12 | 15449705 | c.76–34269T>C | Intronic | 10 |
| rs7574865 | STAT4 | 2 | 191099907 | c.274–23582A>C | Intronic | 58 | |
| rs1800470 | TGFB | 19 | 41353016 | c.29C>T | p.Pro10Leu | 44, 59, 60 | |
| rs1800471 | TGFB | 19 | 41352971 | c.74G>C | p.Arg25Pro | 44, 49, 59 | |
| rs3775291 | TLR3 | 4 | 186082920 | c.1234C>G | p.Leu412Phe | 61 | |
| rs4986790 | TLR4 | 9 | 117713024 | c.896A>G | p.Asp299Gly | 62 | |
| rs10759932 | TLR4 | 9 | 117702866 | c.-1847T>C | 5’ to gene | 63 | |
| rs1800629 | TNF | 6 | 31575254 | c.-488G>A | 5’ to gene | 6, 36, 44, 45, 59, 64, 65 | |
| rs1625895 | TP53 | 17 | 7674797 | c.672+62A>G | Intronic | 66 | |
| rs17868320 | UGT1A9 | 2 | 233669782 | c.-2152C>T | 5’ to gene | 67 | |
| rs6714486 | UGT1A9 | 2 | 233671659 | c.-276T>A | 5’ to gene | 67 | |
| rs7439366 | UGT2B7 | 4 | 69098620 | c.802T>C | p.Tyr268His | 68 | |
| rs699947 | VEGFA | 6 | 43768652 | c.-2055A>C | 5’ to gene | 69 | |
| rs1570360 | rs3025007 | VEGFA | 6 | 43770093 | c.-614A>G | 5’ to gene | 69 |
| rs2910164 | rs2961920 | MIR146A | 5 | 160485411 | n.60C>G | 70 | |
| rs11614913 | rs4759316 | MIR196A2 | 12 | 53991815 | n.78C>T | 70 | |
| rs3746444 | rs3746436 | MIR499A | 20 | 34990448 | n.73A>G | 70 |
- SNP was used as a proxy when a variant was not present in the genotyping chip
- Assembly CRCh38.p7 used for nucleotide position
In this report, we attempted to validate 75 variants previously reported in the literature to be associated with AR using DNA from combing two multicenter cohorts of kidney allograft recipients enrolled in genome-wide association studies (GWAS). The sample size of these two cohorts combined is larger than most of the previous studies and the candidate-SNP approach instead of a genome-wide analysis can maximize our power to validate previous findings.
Materials and Methods
The design of the Deterioration of Kidney Allograft Function (DeKAF) Genomics and the Genomics of Transplantation (GEN-03) cohorts along with each participant’s characteristics has been previously reported (73–75). For this analysis, the DeKAF Genomics and the GEN-03 studies were combined and the kidney transplant recipients with GWAS data were identified and divided into two sub-cohorts consisting of 2,390 European-Americans (EA) and 482 African Americans (AA) kidney allograft recipients and tested separately. Though self-reported race was available in the clinical information, subjects were separated into EA and AA sub-cohorts based on ancestry principal components. Subjects were enrolled at time of transplant and signed informed consents were approved by the Institutional Review Boards of the enrolling centers. This study is registered at www.clinicaltrials.gov ( and ).
Clinical information was obtained from the respective medical records (74, 75). Induction therapy was administered as per transplant center preference but mainly consisted of rabbit anti-thymocyte globulin (rATG), basiliximab or Campath-1H. Immunologically high-risk patients were more likely to receive rATG, such as those with donor specific antibody, pregnancies, or repeat transplants. AR was defined as time to first T-cell, antibody mediated, or mixed T-cell and antibody mediated rejection post-transplant as determined by the enrolling center and treating physician. Rejection was biopsy confirmed in 96% of the cases. The median time to first 12 month AR was 53 days and the median time to first all-time AR was 105.5 days. Both first 12 month AR and all time AR were used in the analysis.
Papers evaluating genetic variants associated with AR were identified through Pubmed. Variants which were shown to have a statistically significant association (p-value <0.05) with AR in solid organ transplantation patients were included in this study (6). The variants which were chosen from the literature for validation in this report are shown in Table 1. All but 6 variants were reported in studies using cohorts of kidney allograft recipients. Those six SNPs not identified in kidney recipients were reported in liver allograft recipients (rs9296068 in HLA-DOA; rs1063320 in HLA-G; rs1800796 in IL6; rs3757385 in IRF5; rs2476601 in PTPN22; rs3775291 in TLR3). Genotype information for this study was extracted from our previous study using a custom genome-wide Affymetrix Axiom Transplant Array chip created specifically for analysis of allograft recipients (71, 76). The 75 variants analyzed are located in 58 genes. For those variants which were not part of the GWAS chip, a proxy SNP was selected which was present on the chip and where genotypes were available. In all cases, the r2 between the reported variant and the proxy SNP was 1.0 as determined by the SNP Annotation and Proxy Search program (SNAP) (77). All selected SNPs were tested for Hardy-Weinberg Equilibrium (HWE). SNPs with a HWE test p-value <0.001 or sample missing rate > 1% were replaced by imputation using the IMPUTE2 program (78). The imputation quality (info) score were above 0.95 for all but two SNPs (rs2426295 and rs2430561, info ~ 0.7 and 0.8, respectively).
Differences in baseline characteristics of recipients without AR vs. with AR were tested using t-tests for continuous variables and chi-sq tests for categorical variables.
Cox proportional-hazard models were used to test the association between each literature identified SNP and time to first AR per person in our cohort. SNPs were coded using an additive genetic model, i.e., the number of copies of a reference allele. The at-risk time period began on the day of transplant and lasted until the earliest event of AR, death, graft failure, last date of follow up, or common close out date. For the outcome of AR in the first 12 months post-transplant, an additional censoring date of one year post-transplant was added. When testing a single SNP association, we stratified by transplant center, and adjusted for variables determined using model selection. We performed backwards model selection with a retention p-value of 0.10 on the outcome of all time AR, separately for AA and EA cohorts, using all of the variables listed in Table 2. For the EA cohort, the retained variables were: gender, primary cause of ESRD, need for dialysis in the first 14 days post-transplant, T- or B-cell crossmatch positive, plasmapheresis prior to transplant, greater than zero HLA mismatches, type of antibody induction, calcineurin inhibitor type at transplant, age at transplant, and donor age. For the AA cohort, the retained variables were: greater than zero % panel reactive antibodies, T- or B-cell crossmatch positive, plasmapheresis prior to transplant, smoking status, calcineurin inhibitor type at transplant, and SPK. Significance for an association between a SNP and AR was set at p<6.6×10−4 (Bonferroni correction with 75 independent tests). Analyses were conducted using SAS v9.4 (The SAS Institute, Cary, NC, USA, http://www.sas.com).
Table 2.
Characteristics and comparisons of European American and African Americans kidney transplant recipients with and without acute rejection; % (no. of recipients)
| European Americans | African Americans | |||||||
|---|---|---|---|---|---|---|---|---|
| Characteristic | All | No AR | AR | P-value | All | No AR | AR | P-value |
| Total | 2390 | 82.4% (1969) | 17.6% (421) | 482 | 85.3% (411) | 14.7% (71) | ||
| Ethnicity; % (no. of recipients): | ||||||||
| Not Hispanic/Latino | 99.4% (2318) | 99.4% (1909) | 99.5% (409) | 0.831 | 99.5% (446) | 99.5% (381) | 100% (65) | 0.5593 |
| Hispanic/Latino | 0.6% (13) | 0.6% (11) | 0.5% (2) | 0.5% (2) | 0.5% (2) | |||
| Gender; % (no. of recipients): | ||||||||
| Female | 37.2% (890) | 38.0% (749) | 33.5% (141) | 0.079 | 37.1% (179) | 36.7% (151) | 39.4% (28) | 0.664 |
| Male | 62.8% (1500) | 62.0% (1220) | 66.5% (280) | 62.9% (303) | 63.3% (260) | 60.6% (43) | ||
| Mean age at transplant in years; years (SD): | ||||||||
| 50.39 (14.7) | 50.88 (14.3) | 48.11 (15.9) | 0.0004 | 46.94 (12.2) | 47.17 (12.2) | 45.56 (12.0) | 0.301 | |
| Primary Cause of End Stage Kidney Disease; % (no. of recipients): | ||||||||
| Diabetes | 27.2% (650) | 27.3% (537) | 26.8% (113) | 0.024 | 23.7% (114) | 24.6% (101) | 18.3% (13) | 0.218 |
| Glomerular disease | 25.2% (601) | 24.4% (481) | 28.5% (120) | 19.1% (92) | 18.0% (74) | 25.4% (18) | ||
| Hypertension | 6.2% (149) | 6.9% (135) | 3.3% (14) | 38.2% (184) | 38.7% (159) | 35.2% (25) | ||
| Other | 21.5% (513) | 21.3% (420) | 22.1% (93) | 11.2% (54) | 11.0% (45) | 12.7% (9) | ||
| Polycystic kidney dis. | 15.8% (378) | 16.3% (320) | 13.8% (58) | 5.2% (25) | 4.6% (19) | 8.5% (6) | ||
| Unknown | 4.1% (99) | 3.9% (76) | 5.5% (23) | 2.7% (13) | 3.2% (13) | |||
| Donor Status; % (no. of recipients): | ||||||||
| Deceased | 32.3% (773) | 33.5% (659) | 27.1% (114) | 0.011 | 67.8% (327) | 68.9% (283) | 62.0% (44) | 0.251 |
| Living | 67.7% (1617) | 66.5% (1310) | 72.9% (307) | 32.2% (155) | 82.6% (128) | 38.0% (27) | ||
| Mean donor age in years; years (SD): | ||||||||
| 41.99 (13.53) | 41.58 (13.74) | 43.89 (12.34) | 0.0014 | 36.93 (13.93) | 37.01 (14.13) | 36.52 (12.86) | 0.788 | |
| Donor Gender; % (no. of recipients): | ||||||||
| Missing | .(1) | .(1) | 0.316 | .(5) | .(4) | .(1) | 0.072 | |
| Female | 53.4% (1275) | 52.9% (1041) | 55.6% (234) | 44.4% (212) | 42.8% (174) | 54.3% (38) | ||
| Male | 46.6% (1114) | 47.1% (927) | 44.4% (187) | 55.6%6 (265) | 57.2% (233) | 45.7% (32) | ||
| Cold Ischemia Time; % (no. of recipients): | ||||||||
| Missing | .(98) | .(87) | .(11) | 0.125 | .(62) | .(58) | .(4) | 0.185 |
| <= 24 h | 96.5% (2213) | 96.3% (1812) | 97.8% (401) | 80.7% (339) | 79.6% (281) | 86.6% (58) | ||
| >24 h | 3.5% (79) | 3.7% (70) | 2.2%(9) | 19.3% (81) | 20.4% (72) | 13.4% (9) | ||
| Prior Kidney Transplant; % (no. of recipients): | ||||||||
| No Prior Transplants | 83.9% (2006) | 84.8% (1669) | 80.0% (337) | 0.017 | 89.0% (429) | 89.3% (367) | 87.3% (62) | 0.624 |
| Prior Transplant | 16.1% (384) | 15.2% (300) | 20.0% (84) | 11.0% (53) | 10.7% (44) | 12.7% (9) | ||
| Need for dialysis in the first 14 days post-transplant; % (no. of recipients): | ||||||||
| No Dialysis | 92.8% (2217) | 93.1% (1834) | 91.0% (383) | 0.119 | 83.2% (401) | 84.9% (349) | 73.2% (52) | 0.015 |
| Dialysis | 7.2% (173) | 6.9% (135) | 9.0% (38) | 16.8% (81) | 15.1% (62) | 26.3% (19) | ||
| Panel Reactive Antibodies; % (no. of recipients): | ||||||||
| Missing | .(5) | .(5) | 0.244 | |||||
| Zero % | 46.7% (1115) | 47.3% (929) | 44.2% (186) | 56.2% (271) | 59.1% (243) | 39.4% (28) | 0.002 | |
| Greater than zero | 53.3% (1270) | 52.7% (1035) | 55.8% (235) | 43.8% (211) | 40.9% (168) | 60.6% (43) | ||
| T or B Cell Crossmatch; % (no. of recipients): | ||||||||
| Missing | .(37) | .(29) | .(8) | 0.0027 | .(2) | .(2) | 0.0013 | |
| Negative | 93.8% (2207) | 94.5% (1833) | 90.6% (374) | 94.2% (452) | 95.6% (391) | 85.9% (61) | ||
| Positive | 6.2% (146) | 5.5% (107) | 9.4% (39) | 5.8% (28) | 4.4% (18) | 14.1% (10) | ||
| Plasmapheresis Prior to Transplant; % (no. of recipients): | ||||||||
| Missing | .(129) | .(110) | .(19) | <.0001 | .(10) | .(9) | .(1) | 0.0013 |
| No Plasmapheresis | 97.2% (2198) | 98.0% (1822) | 93.5% (376) | 97.2% (459) | 98.3% (395) | 91.4% (64) | ||
| Plasmapheresis | 2.8% (63) | 2.0% (37) | 6.5% (26) | 2.8% (13) | 1.7% (7) | 8.6% (6) | ||
| HLA mismatches; % (no. of recipients): | ||||||||
| Missing | .(18) | .(17) | .(1) | <.0001 | .(1) | .(1) | 0.582 | |
| Greater than zero | 87.3% (2070) | 85.7% (1673) | 94.5% (397) | 94.4% (454) | 94.2% (386) | 95.7% (68) | ||
| Zero | 12.7% (302) | 14.3% (279) | 5.5% (23) | 5.6% (27) | 5.8% (24) | 4.3% (3) | ||
| Type of Antibody Induction; % (no. of recipients): | ||||||||
| Combination | 2.6% (63) | 2.1% (42) | 5.0% (21) | <.0001 | 2.5% (12) | 2.7% (11) | 1.4% (1) | 0.0046 |
| Monoclonal | 36.0% (861) | 37.6% (740) | 28.7% (121) | 42.7% (206) | 45.0% (185) | 29.6% (21) | ||
| None | 2.2% (52) | 2.4% (47) | 1.2% (5) | 1.7% (8) | 1.7% (7) | 1.4% (1) | ||
| Polyclonal | 59.3% (1416) | 58.0% (1142) | 65.1% (274) | 53.5% (258) | 51.1% (210) | 67.6% (48) | ||
| Smoking status; % (no. of recipients): | ||||||||
| Missing | .(90) | (80) | .(10) | 0.678 | .(9) | .(8) | .(1) | 0.179 |
| Current | 7.9% (181) | 8.1% (152) | 7.1% (29) | 12.3% (58) | 11.2% (45) | 18.6% (13) | ||
| Past | 36.4% (837) | 36.5% (691) | 35.5% (146) | 24.7% (117) | 25.5% (103) | 20.0% (14) | ||
| Never | 55.7% (1282) | 55.4% (1046) | 57.4% (236) | 63.0% (298) | 63.3% (255) | 61.4% (43) | ||
| Preemptive Transplan; % (no. of recipients)t: | ||||||||
| Not Preemptive | 62.4% (1491) | 62.3% (1227) | 62.7% (264) | 0.880 | 92.1% (444) | 92.5% (380) | 90.1% (64) | 0.504 |
| Preemptive | 37.6% (899) | 37.7% (742) | 37.3% (157) | 7.9% (38) | 7.5% (31) | 9.9% (7) | ||
| Steroid Use at Day 14 Post-Transplant; % (no. of recipients): | ||||||||
| On Steroids | 60.6% (1448) | 61.5% (1211) | 56.3% (237) | 0.047 | 58.9% (284) | 58.2% (239) | 63.4% (45) | 0.408 |
| Off Steroids | 39.4% (942) | 38.5% (758) | 43.7% (184) | 41.1% (198) | 41.8% (172) | 36.6% (26) | ||
| Calcineurin Inhibitor Type at Transplant; % (no. of recipients): | ||||||||
| Both | 0.1% (2) | 0.1% (1) | 0.2% (1) | <.0001 | 0.2% (1) | 0.2% (1) | 0.0002 | |
| Cyclosporine | 23.2% (555) | 21.8% (429) | 29.9% (126) | 11.2% (54) | 8.8% (36) | 25.4% (18) | ||
| None | 2.0% (47) | 1.6% (31) | 3.8% (16) | 3.1% (15) | 2.7% (11) | 5.6% (4) | ||
| Tacrolimus | 74.7% (1786) | 76.6% (1508) | 66.0% (278) | 85.5% (412) | 88.3% (363) | 69.0% (49) | ||
| Simultaneous Pancreas Kidney Transplant (SPK); % (no. of recipients): | ||||||||
| non-SPK | 93.9% (2245) | 94.1% (1854) | 92.9% (391) | 0.316 | 96.5% (465) | 97.1% (399) | 93.0% (66) | 0.082 |
| SPK | 6.1% (145) | 5.9% (115) | 7.1% (30) | 3.5% (17) | 2.9% (12) | 7.0% (5) | ||
| Prior Non-kidney Transplants; % (no. of recipients): | ||||||||
| No Prior Transplants | 87.5% (2091) | 88.1% (1735) | 84.6% (356) | 0.045 | 96.1% (463) | 96.6% (397) | 93.0% (66) | 0.146 |
| Prior Transplant | 12.5% (299) | 11.9% (234) | 15.4% (65) | 3.9% (19) | 3.4% (14) | 7.0% (5) | ||
| Cytomegalovirus Recipient/Donor Status; % (no. of recipients): | ||||||||
| Missing | .(76) | .(67) | .(9) | 0.199 | .(12) | .(8) | .(4) | 0.950 |
| Recipient(-)/Donor(-) | 28.0% (647) | 27.5% (523) | 30.1%(124) | 8.1% (38) | 7.9% (32) | 9.0% (6) | ||
| Recipient (+) | 51.8% (1199) | 52.7% (1002) | 47.8% (197) | 78.9% (371) | 79.2% (319) | 77.6% (52) | ||
| Recipient(-)/Donor(+) | 20.2% (468) | 19.8% (377) | 22.1% (91) | 13.0% (61) | 12.9% (52) | 13.4% (9) | ||
Single SNP Cox proportional hazard models were used to the analysis of variants associated with time to death-censored chronic graft failure (DCGF). Backward selection with a retention p-value of 0.10 was performed separately for European-American and African-American cohorts. In the European-American cohort (DCGF events = 273), models were adjusted for gender, primary cause of ESRD, need for dialysis, cross T- or B-cell match, plasmapheresis prior to transplant, HLA mismatches, type of antibody induction, CNI at baseline, age and donor age. In the African-American cohort (DCGF events = 105), models were adjusted for PRA positive, cross T- or B-cell match, plasmapheresis prior to transplant, smoking status, CNI at baseline and SPK. Both cohorts were stratified by transplant center.
Results
Characteristics of the two cohorts are shown in Table 2. Significant differences (p<0.002) between recipients with and without AR for the EA cohort are, mean age at enrollment in years (p=0.0004), mean donor age in years (p=0.0014), plasmapheresis prior to transplant (p<0.0001), HLA mismatches (p<0.0001), type of antibody induction (p<0.0001) and calcineurin inhibitor type (p<0.001). Significant differences between recipients with and without AR for the AA cohort are, panel reactive antibodies (p=0.002), T or B cell crossmatch (0.0013), plasmapheresis prior to transplant (p<0.0013), and calcineurin inhibitor type at time of transplant (p<0.0002).
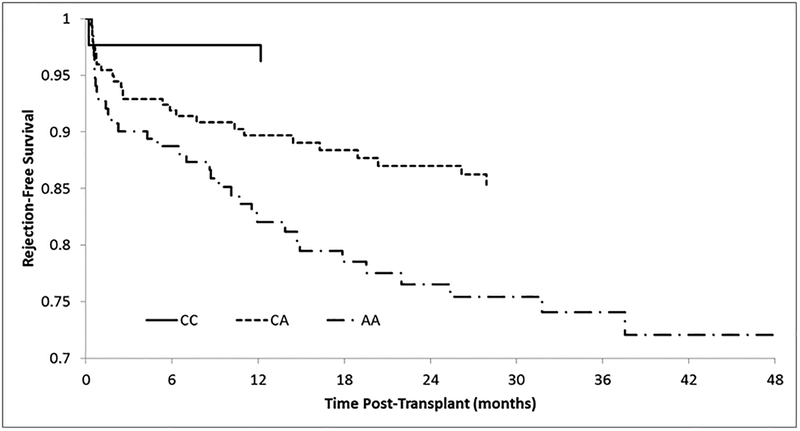
The results of the association analysis for the SNPs tested are found in Table 3. The only significant SNP was rs2961920 (p=1.1×10−4), a proxy for rs2910164, which is located in the MIR146A gene. This SNP was only significant in the AA cohort for all-time AR. The variant was also marginally significant (p=1.9×10−3) for AR within 12 months. The hazard ratio (95% CI) for this variant for AR within 12 months was 2.28 (1.42–3.89) and for AR all time was 2.43 (1.50–3.48). A Kaplan-Meier analysis for AR by MIR146A genotype is shown in Figure 1. At 28 months, 85% of recipients heterozygous for the risk allele (C/A) were AR free whereas individuals homozygous for the risk allele (A/A), at 28 months, only 75% were AR free. This variant was not significant in the EA cohort (p=0.59; 12 months AR and p=0.45; all time AR).
Table 3.
Frequencies of tested alleles and P-values of SNPs for 12 month and all time acute rejection for both European-American and African-American cohorts adjusted for clinical factors.
| European-American Recipients | African-American Recipients | ||||||
|---|---|---|---|---|---|---|---|
| SNP rs# | Gene | TAF | 12 month P-value | All Time P-value | TAF | 12 month P-value | All Time P-value |
| rs4148738 | ABCB1 | 0.558 | 0.65 | 0.98 | 0.756 | 0.19 | 0.051 |
| rs2269475 | AIF | 0.146 | 0.67 | 0.89 | 0.103 | 0.63 | 0.65 |
| rs5186 | AT1R | 0.301 | 0.14 | 0.15 | 0.054 | 0.25 | 0.41 |
| rs55637918 | DEUP1 (CCDC67) | 0.100 | 0.12 | 0.51 | 0.303 | 0.36 | 0.075 |
| rs1024611 | CCL2 | 0.272 | 0.35 | 0.23 | 0.188 | 0.97 | 0.51 |
| rs2107538 | CCL5 | 0.166 | 0.49 | 0.44 | 0.429 | 0.21 | 0.20 |
| rs1799864 | CCR2 | 0.098 | 0.18 | 0.31 | 0.173 | 0.74 | 0.94 |
| rs1799987 | CCR5 | 0.444 | 0.42 | 0.53 | 0.585 | 0.77 | 0.89 |
| rs10490574 | CD28 | 0.184 | 0.048 | 0.051 | 0.056 | 0.55 | 0.32 |
| rs1129055 | CD86 | 0.274 | 0.62 | 0.56 | 0.172 | 0.084 | 0.096 |
| rs733618 | CTLA4 | 0.081 | 0.17 | 0.26 | 0.141 | 0.53 | 0.14 |
| rs5742909 | CTLA4 | 0.095 | 0.0049 | 0.012 | 0.016 | 0.94 | 0.58 |
| rs231775 | CTLA4 | 0.391 | 0.034 | 0.018 | 0.399 | 0.27 | 0.13 |
| rs3087243 | CTLA4 | 0.427 | 0.96 | 0.85 | 0.200 | 0.53 | 0.63 |
| rs4073 | CXCL8 | 0.525 | 0.23 | 0.15 | 0.224 | 0.69 | 0.99 |
| rs2515641 | CYP2E1 | 0.892 | 0.66 | 0.80 | 0.395 | 0.82 | 0.68 |
| rs776746 | CYP3A5 | 0.068 | 0.21 | 0.050 | 0.694 | 0.67 | 0.56 |
| rs4149257 | EPHX2 | 0.254 | 0.54 | 0.41 | 0.787 | 0.44 | 0.19 |
| rs1799963 | F2 | 0.017 | 0.45 | 0.74 | 0.004 | 0.99 | 0.99 |
| rs6025 | F5 | 0.970 | 0.077 | 0.083 | 0.997 | 0.082 | 0.37 |
| rs7851696 | FCN2 | 0.114 | 0.23 | 0.37 | 0.220 | 0.19 | 0.55 |
| rs1801274 | FCGR2A | 0.507 | 0.26 | 0.12 | 0.554 | 0.37 | 0.030 |
| rs9296068 | HLA-DOA | 0.331 | 0.18 | 0.094 | 0.539 | 0.94 | 0.22 |
| rs1063320 | HLA-G | 0.490 | 0.015 | 0.050 | 0.602 | 0.053 | 0.47 |
| rs5498 | ICAM1 | 0.422 | 0.90 | 0.84 | 0.189 | 0.92 | 0.44 |
| rs1143634 | IL1B | 0.236 | 0.29 | 0.11 | 0.140 | 0.60 | 0.099 |
| rs2069762 | IL2 | 0.301 | 0.76 | 0.90 | 0.093 | 0.68 | 0.90 |
| rs228942 | IL2RB | 0.176 | 0.25 | 0.42 | 0.095 | 0.22 | 0.19 |
| rs2284033 | IL2RB | 0.425 | 0.65 | 0.94 | 0.430 | 0.76 | 0.44 |
| rs657075 | IL3 | 0.102 | 0.41 | 0.41 | 0.026 | 0.36 | 0.27 |
| rs61527852 | IL3 | 0.090 | 0.25 | 0.18 | 0.126 | 0.066 | 0.027 |
| rs31480 | IL3 | 0.221 | 0.71 | 0.82 | 0.145 | 0.77 | 0.36 |
| rs2243250 | IL4 | 0.143 | 0.98 | 0.78 | 0.657 | 0.92 | 0.84 |
| rs1801275 | IL4R | 0.214 | 0.24 | 0.51 | 0.678 | 0.58 | 0.90 |
| rs1524107 | IL6 | 0.050 | 0.048 | 0.20 | 0.085 | 0.45 | 0.37 |
| rs1800795 | IL6 | 0.574 | 0.15 | 0.26 | 0.925 | 0.67 | 0.96 |
| rs1800896 | IL10 | 0.490 | 0.75 | 0.64 | 0.356 | 0.19 | 0.090 |
| rs1800871 | IL10 | 0.764 | 0.75 | 0.83 | 0.610 | 0.91 | 0.85 |
| rs1800872 | IL10 | 0.764 | 0.75 | 0.83 | 0.610 | 0.91 | 0.85 |
| rs763780 | IL17F | 0.048 | 0.21 | 0.38 | 0.074 | 0.59 | 0.81 |
| rs187238 | IL18 | 0.270 | 0.11 | 0.094 | 0.215 | 0.67 | 0.83 |
| rs2278293 | IMPDH1 | 0.454 | 0.63 | 0.86 | 0.481 | 0.66 | 0.98 |
| rs2278294 | IMPDH1 | 0.350 | 0.88 | 0.67 | 0.425 | 0.84 | 0.52 |
| rs11706052 | IMPDH2 | 0.104 | 0.72 | 0.74 | 0.014 | 0.72 | 0.22 |
| rs2430561 | INFG | 0.238 | 0.50 | 0.59 | 0.167 | 0.95 | 0.76 |
| rs3807307 | IRF5 | 0.476 | 0.68 | 0.95 | 0.293 | 0.041 | 0.052 |
| rs5918 | ITGB3 | 0.149 | 0.59 | 0.46 | 0.108 | 0.078 | 0.15 |
| rs7096206 | MBL2 | 0.779 | 0.20 | 0.51 | 0.844 | 0.66 | 0.58 |
| rs5030737 | MBL2 | 0.073 | 0.92 | 0.77 | 0.006 | 0.23 | 0.41 |
| rs1800450 | MBL2 | 0.134 | 0.67 | 0.75 | 0.034 | 0.75 | 0.50 |
| rs1800451 | MBL2 | 0.015 | 0.89 | 0.84 | 0.231 | 0.28 | 0.45 |
| rs1801133 | MTHFR | 0.329 | 0.94 | 0.76 | 0.107 | 0.84 | 0.45 |
| rs2426295 | NFATC2 | 0.073 | 0.22 | 0.82 | 0.063 | 0.36 | 0.38 |
| rs28362491 | NFKB1 | 0.383 | 0.29 | 0.71 | 0.513 | 0.22 | 0.34 |
| rs8904 | NFKBIA | 0.380 | 0.87 | 0.42 | 0.594 | 0.93 | 0.75 |
| rs2227982 | PDCD1 | 0.009 | 0.24 | 0.52 | 0.01 | 0.99 | 0.69 |
| rs12734919 | PTGS2 | 0.178 | 1.00 | 0.68 | 0.035 | 0.49 | 0.31 |
| rs2476601 | PTPN22 | 0.880 | 0.95 | 0.84 | 0.985 | 0.52 | 0.77 |
| rs7137890 | PTPRO | 0.343 | 0.18 | 0.34 | 0.174 | 0.79 | 0.92 |
| rs7574865 | STAT4 | 0.773 | 0.90 | 0.84 | 0.845 | 0.94 | 0.68 |
| rs1800470 | TGFB | 0.620 | 0.37 | 0.93 | 0.546 | 0.87 | 0.69 |
| rs1800471 | TGFB | 0.077 | 0.47 | 0.42 | 0.067 | 0.89 | 0.96 |
| rs3775291 | TLR3 | 0.292 | 0.70 | 0.76 | 0.074 | 0.35 | 0.27 |
| rs4986790 | TLR4 | 0.052 | 0.30 | 0.19 | 0.070 | 0.95 | 0.78 |
| rs10759932 | TLR4 | 0.139 | 0.30 | 0.78 | 0.238 | 0.0089 | 0.0046 |
| rs1800629 | TNF | 0.195 | 0.75 | 0.90 | 0.110 | 0.86 | 0.55 |
| rs1625895 | TP53 | 0.882 | 0.97 | 0.98 | 0.726 | 0.72 | 0.49 |
| rs17868320 | UGT1A9 | 0.060 | 0.46 | 0.50 | 0.025 | 0.79 | 0.54 |
| rs6714486 | UGT1A9 | 0.061 | 0.41 | 0.44 | 0.197 | 0.96 | 0.36 |
| rs7439366 | UGT2B7 | 0.462 | 0.48 | 0.19 | 0.706 | 0.068 | 0.17 |
| rs699947 | VEGFA | 0.507 | 0.23 | 0.25 | 0.791 | 0.29 | 0.32 |
| rs3025007 | VEGFA | 0.456 | 0.83 | 0.82 | 0.334 | 0.61 | 0.62 |
| rs2961920 | MIR146A | 0.768 | 0.59 | 0.45 | 0.576 | 0.0019 | 0.00011 |
| rs4759316 | MIR196A2 | 0.558 | 0.84 | 0.68 | 0.604 | 0.96 | 0.95 |
| rs3746436 | MIR499A | 0.189 | 0.48 | 0.69 | 0.165 | 0.43 | 0.38 |
TAF – Tested allele frequency
Figure 1. A Kaplan-Meier analysis of proxy SNP rs2910164 in MIR146A for acute rejection.
A Kaplan-Meier analysis for rejection-free kidney recipients based on the genotypes of MIR146A. The solid line represents the CC genotype, dashed line represents the CA genotype and the dash-dot-dash line represents the AA genotype. The A allele was found to be associated with a greater risk of acute rejection.
There were two suggestive variants. In the EA cohort, SNP rs5742909 (p=0.0049; 12 months AR and p=0.012; all time AR) within the CTLA4 gene and in the AA cohort, SNP rs10759932 (p=0.0089; 12 months AR and p=0.0046; all time AR) within the TLR4 gene. All other tested variants were not significant (p > 0.02).
All variants were also tested against time to death-censored graft function (DCGF) using a Cox proportional hazard model (Table 1S). There were no significant associations with any of the variants, but suggestive associations were found in the AA cohort for SNP rs61527852 in the IL3 gene (p=0.0087; all time AR) and for SNP rs2961920 in the MIR146A locus in both the EA cohort (p=0.0046; all time AR) and the AA cohort but less significant (p=0.049; all time AR)
Discussion
Since the beginning of kidney allograft transplantation, there has been considerable reduction in occurrence of AR and improved treatments, resulting in an almost 95% graft survival rates for the first year after transplantation. Unfortunately, there remains an insidious rate of late graft dysfunction and loss and one of the major clinical problems in transplantation. The risk of graft loss has been shown to be increased in the event of AR (1). Being able to reduce AR events would improve graft survival. It has been hypothesized that some recipients are genetically predisposed to increased risk for AR (6, 79). Those genetic variants with the greatest impact on AR are within the human leukocyte antigens (HLA) related loci within the major histocompatibility complex (MHC) (80). HLA alleles are strong predictors of AR and matching HLA alleles between the recipient and donor organ greatly decreases the risk for AR. Though the HLA loci plays an important role in AR, other genes which impact the immune system also have genetic variation and these alleles may also impact AR risk. To this end, many variants within these genes have been reported for their association with AR.
In this analysis, 75 SNPs, previously reported to be associated with the risk of AR, were tested in our EA and AA cohorts of kidney allograft recipients. Only one of these SNPs was found to be significant; A SNP within the microRNA 146a in the AA cohort. In a previous report it was found that rs2910164 in the MIR146A gene was associated with lowest overall survival among 350 North Indian renal allograft recipients and a three-fold higher risk for AR (70). The hazard ratio was similar to the previous report on this variant, 2.43 vs. 2.63. In the previous report, the recipients were from north India and in our analysis the recipients with the significant p-value were within the AA cohort. The EA cohort was not significant for this variant. We speculate that the lack of significance in the EA cohort may be the result of not having additional variants in other genes which were present in the north Indian and AA populations and act synergistically with rs2910164.
MIR146A has a number of targets, including mRNAs from genes involved in immune regulation including regulatory T-cells (81). An in silico analysis identified several target genes including an interleukin-1 receptor-associated kinase (IRAK1) gene, and TNF-receptor associated factor (TRAF-6) gene (70). MIR146A is thought to help modulate the immune system by suppressing inflammatory responses, in part through the NF-κB signaling pathway. The variant allele has been shown to reduce the expression of this microRNA, possibly resulting in an enhanced inflammatory response to the allograft, increasing the risk of AR (82, 83). This variant has also been reported to be associated with type 2 diabetes with increased fasting glucose and HbA1C levels and cardiovascular disease risk factors such as increased diastolic blood pressure and triglycerides (84).
The three variants which exhibited suggestive evidence for an association with AR included SNP rs5742909 within the cytotoxic T-lymphocyte associated protein 4 (CTLA4) gene and SNP rs10759932 within the toll like receptor 4 (TLR4) gene. The CTLA4 gene plays an inhibitory role in T-cell signaling and the TLR4 gene product is a lipopolysaccharide receptor and plays a fundamental role in pathogen recognition and activation of innate immunity (85, 86). We also tested all variants for their association with DCGF, but none were significant.
There are many reasons for the high number of variants that did not replicate in this study (87, 88). For the most part, most of the published studies are underpowered. An additional source of error in replication may be differences between populations and clinical care. Also, six of these variants were identified in liver recipients and may not be important in kidney allograft recipients. In this analysis, we used recipients from a single multicenter study in which identical clinical variables were collected for all individuals in the cohort. Many of the published studies analyzed only single univariate associations and did not adjust for clinical characteristics which can improve the power of statistical testing. Additionally, the follow-up period to AR is often too short to see an impact of the variant on AR risk and no consideration is given to linkage disequilibrium. Additional reasons have been stated in a report which attempted to identify donor specific variants associated with long- and short-term outcomes using a GWAS in renal allograft recipients (89). In this study a genome-wide association study was done on 2,094 renal transplant-pairs, but no variants outside of the HLA region were found to be statistically significant in a 5,866 replication cohort. The authors suggested that both phenotype heterogeneity and the lack of statistical power due to limited sample size is a possible cause of no statistically significant variants being identified. The AR phenotype is most likely both clinically and genetically heterogeneous making identification of associated variants unlikely unless larger populations are used.
Other variables which may impact AR and/or graft loss include subclinical rejection and immunosuppressant adherence. In both cases this information was not available from the published papers and we did not collect this data in our cohort so the inclusion of these variables in our analysis was not possible, though both of these have been shown to be important in rejection risk and health of the allograft (90, 91).
The positive association of the MIR146A with AR provides a novel pathway to study and may provide additional genes and their variants as candidates for recipient risk for AR and possible therapeutic targets to reduce this risk.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to thank the research subjects for their participation in this study. We acknowledge the dedication and hard work of our coordinators at each of the DeKAF Genomics and GEN03 clinical sites: University of Alberta, Nicoleta Bobocea, Tina Wong, Adrian Geambasu and Alyssa Sader; University of Manitoba, Myrna Ross and Kathy Peters; University of Minnesota, Mandi DeGrote, Monica Myers and Danielle Berglund; Hennepin Healthcare, Lisa Berndt; Mayo Clinic, Tom DeLeeuw; University of Iowa, Wendy Wallace and Tammy Lowe; University of Alabama, Jacquelin Vaughn, Valencia Stephens and Tena Hilario. We also acknowledge the dedicated work of our research scientists Marcia Brott and Amutha Muthusamy. This study was supported by NIH/NIAID grants 5U19-AI070119, 5U01-AI058013 and K01 AI130409.
ABBREVIATIONS PAGE
- IF/TA
interstitial fibrosis/tubular atrophy
- AR
acute rejection
- ABMR
antibody-mediated rejection
- TCMR
T cell-mediated rejection
- SNPs
single nucleotide polymorphisms
- GWAS
genome-wide association studies
- DeKAF
Deterioration of Kidney Allograft Function
- EA
European-Americans
- AA
African Americans
- rATG
rabbit anti-thymocyte
- SNAP
SNP Annotation and Proxy Search program
- HLA
human leukocyte antigens
- MHC
major histocompatibility complex
Footnotes
Clinical Trial Notation: and .
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Matas AJ, Gillingham KJ, Payne WD, Najarian JS. The impact of an acute rejection episode on long-term renal allograft survival (t1/2). Transplantation 1994;57:857–859. [DOI] [PubMed] [Google Scholar]
- 2.Paraskevas S, Kandaswamy R, Humar A, et al. Risk factors for rising creatinine in renal allografts with 1 and 3 yr survival. Clin Transplant 2006;20:667–672. [DOI] [PubMed] [Google Scholar]
- 3.Lentine KL, Gheorghian A, Axelrod D, Kalsekar A, L’italien G, Schnitzler MA. The implications of acute rejection for allograft survival in contemporary U.S. kidney transplantation. Transplantation. 2012;94:369–376. [DOI] [PubMed] [Google Scholar]
- 4.Solez K, Colvin RB, Racusen LC, et al. Banff 07 classification of renal allograft pathology: updates and future directions. Am J Transplant. 2008;8:753–760. [DOI] [PubMed] [Google Scholar]
- 5.Loupy A, Haas M, Solez K, et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am J Transplant. 2017;17:28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorr CR, Oetting WS, Jacobson PA, Israni AK. Genetics of acute rejection after kidney transplantation. Transpl Int. 2018;31:263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grinyo J, Vanrenterghem Y, Nashan B, et al. Association of four DNA polymorphisms with acute rejection after kidney transplantation. Transpl Int. 2008;21:879–891. [DOI] [PubMed] [Google Scholar]
- 8.Vu D, Tellez-Corrales E, Shah T, Hutchinson I, Min DI. Influence of Cyclooxygenase-2 (COX-2) gene promoter-1195 and allograft inflammatory factor-1 (AIF-1) polymorphisms on allograft outcome in Hispanic kidney transplant recipients. Hum Immunol. 2013;74:1386–1391. [DOI] [PubMed] [Google Scholar]
- 9.Zhang G, Wang H, Wang F, et al. Gene polymorphisms of the renin-angiotensin-aldosterone system and angiotensin II type 1-receptor activating antibodies in renal rejection. Tohoku J Exp Med. 2007;213:203–214. [DOI] [PubMed] [Google Scholar]
- 10.Ghisdal L, Baron C, Lebranchu Y, et al. Genome-Wide Association Study of Acute Renal Graft Rejection. Am J Transplant. 2017;17:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang SW, Park SJ, Kim YW, et al. Association of MCP-1 and CCR2 polymorphisms with the risk of late acute rejection after renal transplantation in Korean patients. Int J Immunogenet. 2008;35:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kruger B, Boger CA, Obed A, et al. RANTES/CCL5 polymorphisms as a risk factor for recurrent acute rejection. Clin Transplant. 2007;21:385–390. [DOI] [PubMed] [Google Scholar]
- 13.Abdi R, Tran TB, Sahagun-Ruiz A, et al. Chemokine receptor polymorphism and risk of acute rejection in human renal transplantation. J Am Soc Nephrol. 2002;13:754–758. [DOI] [PubMed] [Google Scholar]
- 14.Yigit B, Bozkurt N, Berber I, Titiz I, Isbir T. Analysis of CC chemokine receptor 5 and 2 polymorphisms and renal transplant survival. Cell Biochem Funct. 2007;25:423–426. [DOI] [PubMed] [Google Scholar]
- 15.Cha RH, Yang SH, Kim HS, et al. Genetic interactions between the donor and the recipient for susceptibility to acute rejection in kidney transplantation: polymorphisms of CCR5. Nephrol Dial Transplant. 2009;24:2919–2925. [DOI] [PubMed] [Google Scholar]
- 16.Pawlik A, Dabrowska-Zamojcin E, Dziedziejko V, Safranow K, Domanski L. Association between IVS3 +17T/C CD28 gene polymorphism and the acute kidney allograft rejection. Transpl Immunol. 2014;30:84–87. [DOI] [PubMed] [Google Scholar]
- 17.Han FF, Fan H, Wang ZH, et al. Association between co-stimulatory molecule gene polymorphism and acute rejection of allograft. Transpl Immunol. 2014;31:81–86. [DOI] [PubMed] [Google Scholar]
- 18.Gao JW, Guo YF, Fan Y, et al. Polymorphisms in cytotoxic T lymphocyte associated antigen-4 influence the rate of acute rejection after renal transplantation in 167 Chinese recipients. Transpl Immunol. 2012;26:207–211. [DOI] [PubMed] [Google Scholar]
- 19.Ruhi C, Sallakci N, Yegin O, Suleymanlar G, Ersoy FF. The influence of CTLA-4 single nucleotide polymorphisms on acute kidney allograft rejection in Turkish patients. Clin Transplant. 2015;29:612–618. [DOI] [PubMed] [Google Scholar]
- 20.Duan Z, Zhang Y, Pan F, et al. Association between CTLA4 gene polymorphisms and acute rejection of kidney transplantation: a meta-analysis. J Nephrol. 2012;25:996–1002. [DOI] [PubMed] [Google Scholar]
- 21.Misra MK, Kapoor R, Pandey SK, Sharma RK, Agrawal S. Association of CTLA-4 gene polymorphism with end-stage renal disease and renal allograft outcome. J Interferon Cytokine Res. 2014;34:148–161. [DOI] [PubMed] [Google Scholar]
- 22.Gendzekhadze K, Rivas-Vetencourt P, Montano RF. Risk of adverse post-transplant events after kidney allograft transplantation as predicted by CTLA-4 +49 and TNF-alpha −308 single nucleotide polymorphisms: a preliminary study. Transpl Immunol. 2006;16:194–199. [DOI] [PubMed] [Google Scholar]
- 23.Gao JW, Zhou ZH, Guo SC, Guo YF, Guo F. A deeper understanding of the association between CTLA4 +49A/G and acute rejection in renal transplantation: an updated meta-analysis. Ren Fail. 2015;37:165–174. [DOI] [PubMed] [Google Scholar]
- 24.Canossi A, Aureli A, Delreno F, et al. Influence of cytotoxic T-lymphocyte antigen-4 polymorphisms on acute rejection onset of cadaveric renal transplants. Transplant Proc. 2013;45:2645–2649. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, Kesarwani P, Ahirwar DK, Kapoor R, Mittal RD. Interleukin 8 −251T>A and Interferon gamma +874A>T polymorphism: potential predictors of allograft outcome in renal transplant recipients from north India. Transpl Immunol. 2009;21:13–17. [DOI] [PubMed] [Google Scholar]
- 26.Kim SK, Park HJ, Seok H, et al. Association studies of cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1) gene polymorphisms with acute rejection in kidney transplantation recipients. Clin Transplant. 2014;28:707–712. [DOI] [PubMed] [Google Scholar]
- 27.Quteineh L, Verstuyft C, Furlan V, et al. Influence of CYP3A5 genetic polymorphism on tacrolimus daily dose requirements and acute rejection in renal graft recipients. Basic Clin Pharmacol Toxicol. 2008;103:546–552. [DOI] [PubMed] [Google Scholar]
- 28.Gervasini G, Garcia-Cerrada M, et al. A 3’-UTR Polymorphism in Soluble Epoxide Hydrolase Gene Is Associated with Acute Rejection in Renal Transplant Recipients. PLoS One. 2015;10:e0133563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heidenreich S, Junker R, Wolters H, et al. Outcome of kidney transplantation in patients with inherited thrombophilia: data of a prospective study. J Am Soc Nephrol. 2003;14:234–239. [DOI] [PubMed] [Google Scholar]
- 30.Hocher B, Slowinski T, Hauser I, et al. Association of factor V Leiden mutation with delayed graft function, acute rejection episodes and long-term graft dysfunction in kidney transplant recipients. Thromb Haemost. 2002;87:194–198. [PubMed] [Google Scholar]
- 31.Dabrowska-Zamojcin E, Czerewaty M, Malinowski D, et al. Ficolin-2 Gene rs7851696 Polymorphism is Associated with Delayed Graft Function and Acute Rejection in Kidney Allograft Recipients. Arch Immunol Ther Exp (Warsz). 2018;66:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yuan FF, Watson N, Sullivan JS, et al. Association of Fc gamma receptor IIA polymorphisms with acute renal-allograft rejection. Transplantation. 2004;78:766–769. [DOI] [PubMed] [Google Scholar]
- 33.Ningappa M, Ashokkumar C, Higgs BW, et al. Enhanced B Cell Alloantigen Presentation and Its Epigenetic Dysregulation in Liver Transplant Rejection. Am J Transplant. 2016;16:497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thude H, Janssen M, Sterneck M, Nashan B, Koch M. 14-bp ins/del polymorphism and +3142C>G SNP of the HLA-G gene have a significant impact on acute rejection after liver transplantation. Hum Immunol. 2016;77:1159–1165. [DOI] [PubMed] [Google Scholar]
- 35.Tajik N, Salari F, Ghods AJ, Hajilooi M, Radjabzadeh MF, Mousavi T. Association between recipient ICAM-1 K469 allele and renal allograft acute rejection. Int J Immunogenet. 2008;35:9–13. [DOI] [PubMed] [Google Scholar]
- 36.Manchanda PK, Mittal RD. Analysis of cytokine gene polymorphisms in recipient’s matched with living donors on acute rejection after renal transplantation. Mol Cell Biochem. 2008;311:57–65. [DOI] [PubMed] [Google Scholar]
- 37.Morgun A, Shulzhenko N, Rampim GF, et al. Interleukin-2 gene polymorphism is associated with renal but not cardiac transplant outcome. Transplant Proc. 2003;35:1344–1345. [DOI] [PubMed] [Google Scholar]
- 38.Park SJ, Yoon YC, Kang SW, et al. Impact of IL2 and IL2RB genetic polymorphisms in kidney transplantation. Transplant Proc. 2011;43:2383–1287. [DOI] [PubMed] [Google Scholar]
- 39.Lee DY, Song SB, Moon JY, et al. Association between interleukin-3 gene polymorphism and acute rejection after kidney transplantation. Transplant Proc. 2010;42:4501–4504. [DOI] [PubMed] [Google Scholar]
- 40.Poole KL, Gibbs PJ, Evans PR, Sadek SA, Howell WM. Influence of patient and donor cytokine genotypes on renal allograft rejection: evidence from a single centre study. Transpl Immunol. 2001;8:259–265. [DOI] [PubMed] [Google Scholar]
- 41.Lee HJ, Kim TH, Kang SW, et al. Association Interleukin-4 and Interleukin-4 Receptor Gene Polymorphism and Acute Rejection and Graft Dysfunction After Kidney Transplantation. Transplant Proc. 2016;48:813–819. [DOI] [PubMed] [Google Scholar]
- 42.Yao J, Feng XW, Yu XB, et al. Recipient IL-6–572C/G genotype is associated with reduced incidence of acute rejection following liver transplantation. J Int Med Res. 2013;41:356–364. [DOI] [PubMed] [Google Scholar]
- 43.Marshall SE, McLaren AJ, McKinney EF, et al. Donor cytokine genotype influences the development of acute rejection after renal transplantation. Transplantation. 2001;71:469–476. [DOI] [PubMed] [Google Scholar]
- 44.Alakulppi NS, Kyllonen LE, Jantti VT, et al. Cytokine gene polymorphisms and risks of acute rejection and delayed graft function after kidney transplantation. Transplantation. 2004;78:1422–1428. [DOI] [PubMed] [Google Scholar]
- 45.Sankaran D, Asderakis A, Ashraf S, et al. Cytokine gene polymorphisms predict acute graft rejection following renal transplantation. Kidney Int. 1999;56:281–288. [DOI] [PubMed] [Google Scholar]
- 46.Haouami Y, Sfar I, Dhaouadi T, et al. Impact of Interleukin-17F Gene Polymorphisms in Outcome of Kidney Transplantation in Tunisian Recipients. Transplant Proc. 2018;50:110–114. [DOI] [PubMed] [Google Scholar]
- 47.Kim CD, Ryu HM, Choi JY, et al. Association of G-137C IL-18 promoter polymorphism with acute allograft rejection in renal transplant recipients. Transplantation. 2008;86:1610–1614. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Yang JW, Zeevi A, et al. IMPDH1 gene polymorphisms and association with acute rejection in renal transplant patients. Clin Pharmacol Ther. 2008;83:711–717. [DOI] [PubMed] [Google Scholar]
- 49.Tinckam K, Rush D, Hutchinson I, et al. The relative importance of cytokine gene polymorphisms in the development of early and late acute rejection and six-month renal allograft pathology. Transplantation. 2005;79:836–841. [DOI] [PubMed] [Google Scholar]
- 50.Yu X, Wei B, Dai Y, et al. Genetic polymorphism of interferon regulatory factor 5 (IRF5) correlates with allograft acute rejection of liver transplantation. PLoS One. 2014;9:e94426. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Salido E, Martin B, Barrios Y, et al. The PlA2 polymorphism of the platelet glycoprotein IIIA gene as a risk factor for acute renal allograft rejection. J Am Soc Nephrol. 1999;10:2599–2605. [DOI] [PubMed] [Google Scholar]
- 52.Golshayan D, Wojtowicz A, Bibert S, et al. Polymorphisms in the lectin pathway of complement activation influence the incidence of acute rejection and graft outcome after kidney transplantation. Kidney Int. 2016;89:927–938. [DOI] [PubMed] [Google Scholar]
- 53.Oetting WS, Zhu Y, Brott MJ, Matas AJ, Cordner GK, Pan W. Validation of genetic variants associated with early acute rejection in kidney allograft transplantation. Clin Transplant. 2012;26:418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Z, Yang H, Si S, et al. Polymorphisms of nucleotide factor of activated T cells cytoplasmic 2 and 4 and the risk of acute rejection following kidney transplantation. World J Urol. 2018;36:111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Misra MK, Mishra A, Pandey SK, Kapoor R, Sharma RK, Agrawal S. Association of functional genetic variants of transcription factor Forkhead Box P3 and Nuclear Factor-kappaB with end-stage renal disease and renal allograft outcome. Gene. 2016;581:57–65. [DOI] [PubMed] [Google Scholar]
- 56.Zolfaghari L, Solgi G, Nafar M, et al. Association of programmed cell death 1 and programmed cell death 1 ligand gene polymorphisms with delayed graft function and acute rejection in kidney allograft recipients. Iran J Kidney Dis. 2015;9:138–145. [PubMed] [Google Scholar]
- 57.Dullin R, Koch M, Sterneck M, Nashan B, Thude H. Association between a gain-of-function variant of PTPN22 and rejection in liver transplantation. Transplantation. 2015;99:431–437. [DOI] [PubMed] [Google Scholar]
- 58.Yang H, Zhou Q, Chen ZM, Chen WQ, Wang MM, Chen JH. Polymorphisms in STAT4 increase the risk of acute renal allograft rejection in the Chinese population. Transpl Immunol. 2011;24:216–219. [DOI] [PubMed] [Google Scholar]
- 59.Park JY, Park MH, Park H, Ha J, Kim SJ, Ahn C. TNF-alpha and TGF-beta1 gene polymorphisms and renal allograft rejection in Koreans. Tissue Antigens. 2004;64:660–666. [DOI] [PubMed] [Google Scholar]
- 60.Zhang XX, Bian RJ, Wang J, Zhang QY. Relationship between cytokine gene polymorphisms and acute rejection following liver transplantation. Genet Mol Res. 2016;15:gmr.15027599. [DOI] [PubMed] [Google Scholar]
- 61.Citores MJ, Banos I, Noblejas A, Rosado S, Castejon R, Cuervas-Mons V. Toll-like receptor 3 L412F polymorphism may protect against acute graft rejection in adult patients undergoing liver transplantation for hepatitis C-related cirrhosis. Transplant Proc. 2011;43:2224–2226. [DOI] [PubMed] [Google Scholar]
- 62.Ducloux D, Deschamps M, Yannaraki M, et al. Relevance of Toll-like receptor-4 polymorphisms in renal transplantation. Kidney Int. 2005;67:2454–2461. [DOI] [PubMed] [Google Scholar]
- 63.Hwang YH, Ro H, Choi I, et al. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation. 2009;88:699–705. [DOI] [PubMed] [Google Scholar]
- 64.Pawlik A, Domanski L, Rozanski J, et al. IL-2 and TNF-alpha promoter polymorphisms in patients with acute kidney graft rejection. Transplant Proc. 2005;37:2041–2043. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Fructuoso AI, Perez-Flores I, Valero R, et al. The Polymorphism −308G/A of Tumor Necrosis Factor-alpha Gene Modulates the Effect of Immunosuppressive Treatment in First Kidney Transplant Subjects Who Suffer an Acute Rejection. J Immunol Res. 2016;2016:2197595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Azarpira N, Kazemi K, Darai M. Influence of p53 (rs1625895) polymorphism in kidney transplant recipients. Saudi J Kidney Dis Transpl. 2014;25:1160–1165. [DOI] [PubMed] [Google Scholar]
- 67.Pazik J, Oldak M, Dabrowski M, et al. Association of UDP-glucuronosyltransferase 1A9 (UGT1A9) gene polymorphism with kidney allograft function. Ann Transplant. 2011;16:69–73. [DOI] [PubMed] [Google Scholar]
- 68.Pazik J, Oldak M, Lewandowski Z, et al. Uridine diphosphate glucuronosyltransferase 2B7 variant p.His268Tyr as a predictor of kidney allograft early acute rejection. Transplant Proc. 2013;45:1516–1519. [DOI] [PubMed] [Google Scholar]
- 69.Shahbazi M, Fryer AA, Pravica V, et al. Vascular endothelial growth factor gene polymorphisms are associated with acute renal allograft rejection. J Am Soc Nephrol. 2002;13:260–264. [DOI] [PubMed] [Google Scholar]
- 70.Misra MK, Pandey SK, Kapoor R, Sharma RK, Agrawal S. Genetic variants of MicroRNA-related genes in susceptibility and prognosis of end-stage renal disease and renal allograft outcome among north Indians. Pharmacogenet Genomics. 2014;24:442–450. [DOI] [PubMed] [Google Scholar]
- 71.Oetting WS, Schladt DP, Leduc RE, et al. Validation of single nucleotide polymorphisms associated with acute rejection in kidney transplant recipients using a large multi-center cohort. Transpl Int 2011;24:1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.de Mare-Bredemeijer EL, Mancham S, Utomo WK, et al. Genetic polymorphisms in innate immunity receptors do not predict the risk of bacterial and fungal infections and acute rejection after liver transplantation. Transpl Infect Dis 2013;15:120–133. [DOI] [PubMed] [Google Scholar]
- 73.Oetting WS, Schladt DP, Guan W, et al. Genomewide Association Study of Tacrolimus Concentrations in African American Kidney Transplant Recipients Identifies Multiple CYP3A5 Alleles. Am. J. Transplant 2016;16:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Israni A, Leduc R, Holmes J, et al. Single-nucleotide polymorphisms, acute rejection, and severity of tubulitis in kidney transplantation, accounting for center-to-center variation. Transplantation. 2010;90:1401–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pulk RA, Schladt DS, Oetting WS, et al. Multigene predictors of tacrolimus exposure in kidney transplant recipients. Pharmacogenomics 2015;16:841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li YR, van Setten J, Verma SS, et al. Concept and design of a genome-wide association genotyping array tailored for transplantation-specific studies. Genome Medicine. 2015;7:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson AD, Handsaker RE, Pulit S, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap Bioinformatics. 2008;24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genetics 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldfarb‐Rumyantzev AS, Naiman N. Genetic prediction of renal transplant outcome. Curr Opin Nephrol Hypertens 2008;17:573–579. [DOI] [PubMed] [Google Scholar]
- 80.Sheldon S, Hasleton PS, Yonan NA, et al. Rejection in heart transplantation strongly correlates with HLA‐DR antigen mismatch. Transplantation 1994;58:719–722. [PubMed] [Google Scholar]
- 81.Paterson MR, Kriegel AJ. MiR-146a/b: a family with shared seeds and different roots. Physiol Genomics. 2017;49:243–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2008;105:7269–7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Alipoor B, Ghaedi H, Meshkani R, Omrani MD, Sharifi Z, Golmohammadi T. The rs2910164 variant is associated with reduced miR-146a expression but not cytokine levels in patients with type 2 diabetes. J Endocrinol Invest. 2018;41:557–566. [DOI] [PubMed] [Google Scholar]
- 84.Alipoor B, Meshkani R, Ghaedi H, Sharifi Z, Panahi G, Golmohammadi T. Association of miR-146a rs2910164 and miR-149 rs2292832 variants with susceptibility to type 2 diabetes. Clin. Lab 2016;62:1553–1561. [DOI] [PubMed] [Google Scholar]
- 85.Adams AB, Ford ML and Larsen CP: Costimulation blockade in autoimmunity and transplantation: The CD 28 pathway. J Immunol. 2016;197:2045–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Funami K, Matsumoto M, Oshiumi H, Inagaki F, Seya T. Functional interfaces between TICAM-2/TRAM and TICAM-1/TRIF in TLR4 signaling. Biochem Soc Trans. 2017;45:929–935. [DOI] [PubMed] [Google Scholar]
- 87.Pallet N, Thervet E. The genetics of kidney transplantation. Hum Genet. 2012;131:317–323. [DOI] [PubMed] [Google Scholar]
- 88.Oetting WS, Wu B, Schladt DP, et al. Attempted validation of 44 reported SNPs associated with tacrolimus troughs in a cohort of kidney allograft recipients. Pharmacogenomics. 2018;19:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hernandez-Fuentes MP, Franklin C, Rebollo-Mesa I, et al. Long- and short-term outcomes in renal allografts with deceased donors: A large recipient and donor genome-wide association study. Am J Transplant. 2018;18:1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Doyle IC, Maldonado AQ, Heldenbrand S, Tichy EM, Trofe-Clark J. Nonadherence to therapy after adult solid organ transplantation: A focus on risks and mitigation strategies. Am J Health Syst Pharm. 2016;73:909–920. [DOI] [PubMed] [Google Scholar]
- 91.Loupy A, Vernerey D, Tinel C, et al. Subclinical rejection phenotypes at 1 year post‐transplant and outcome of kidney allografts. J Am Soc Nephrol. 2015;26:1721–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.