Abstract
Apoptosis is a form of programmed cell death (PCD) that plays critical physiological roles in removing superfluous or dangerous cell populations that are unneeded or threatening to the health of the host organism. Although the molecular pathways leading to activation of the apoptotic program have been extensively studied and characterized starting in the 1970s, new evidence suggests that members of the gasdermin superfamily are novel pore-forming proteins that augment apoptosis by permeabilizing the mitochondria and participate in the final stages of the apoptotic program by inducing secondary necrosis/pyroptosis. These findings may explain outstanding questions in the field such as why certain gasdermin members sensitize cells to apoptosis, and why some apoptotic cells also show morphological features of necrosis. Furthermore, the interplay between the gasdermins and apoptosis may also explain why genetic and epigenetic alterations in these genes cause diseases and disorders like cancer and hearing loss. This review focuses on our current understanding of the function of several gasdermin superfamily members, their role in apoptosis, and how they may contribute to pathophysiological conditions.
Keywords: Gasdermins, Apoptosis, Pyroptosis, Secondary Necrosis, Caspases, Mitochondria, Cell Death
Introduction
Apoptosis is a genetically encoded form of programmed cell death (PCD) that is carried out by programmed molecular machinery and plays a variety of vitally important physiological roles in organismal development and homeostasis (Table 1). In humans, a coordinated series of complex cellular signaling events results in the transformation of a single-cell zygote to a fully mature adult composed of trillions of cells that make up very specialized organs. While mitosis results in the proliferation of cells that eventually composes these organs, it is actually apoptosis that fine-tunes their structure, resulting in the final form and function they eventually take. Apoptosis also plays important functions in eliminating transformed or pathogen-infected cells that are deleterious to the host. Apoptosis can be triggered intrinsically when cells sense their own irreparable cellular damage (e.g. DNA damage) and stress, or extrinsically through activation of death receptors on the plasma membrane by death-inducing cytokines released from cells as a result of infection with pathogens (Figure 1). Intrinsic apoptosis is triggered by signaling pathways controlled largely by members of the proapoptotic B Cell CLL/Lymphoma-2 (BCL-2) family which ultimately culminate on the BCL-2 effectors, BCL-2 Antagonist Killer 1 (BAK), and BCL-2-Associated X Protein (BAX). Activation of these proteins leads to their oligomerization on the outer mitochondrial membrane resulting in its permeabilization and the release of proapoptotic factors such as cytochrome c. The released cytochrome c activates the Apaf-1 apoptosome which in turn activates the executioner caspases 3 and 7 leading to cellular dismantling [1-5]. While BAK and BAX have been the most extensively characterized apoptotic mitochondrial pore-forming proteins, recent evidence demonstrates that members of the gasdermin superfamily also possess similar functions.
Table 1. Cell death definitions.
| Term | Definition |
| Programmed Cell Death (PCD) | Cell death that is regulated by genetically encoded molecules in response to physiological or pathophysiological stimuli |
| Apoptosis | An immunologically silent form of PCD driven by the executioner caspases that typically results in rapid engulfment by nearby phagocytes |
| Pyroptosis | An immunogenic form of PCD driven by the inflammatory caspase-mediated cleavage of GSDMD in response to pathogen- and danger- associated molecular patterns (PAMPs and DAMPs, respectively) |
| Secondary Necrosis | An immunogenic form of PCD driven by the caspase-3-mediated cleavage of GSDME that can sometimes follow the apoptotic program; secondary necrotic cells display very similar features to those undergoing pyroptosis |
| NETosis | An immunogenic form of PCD occurring specifically in neutrophils driven by the inflammatory caspase-mediated cleavage of GSDMD, resulting in the release of neutrophil extracellular traps (NETs) |
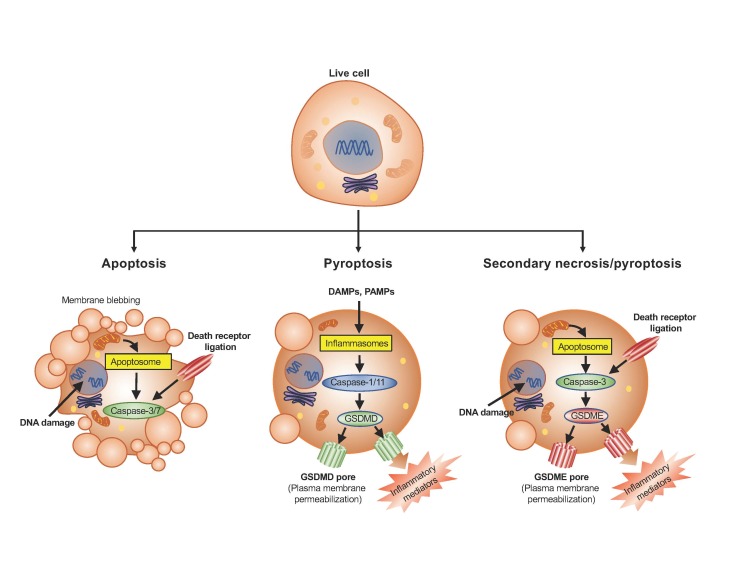
Figure 1.
Features of cells undergoing apoptosis, pyroptosis or secondary necrosis. Apoptosis is orchestrated by the executioner caspases 3 and 7 which are activated by the apoptosome or death receptor ligation. Cells undergoing apoptosis are characterized by their defining blebs around the plasma membrane, cellular shrinkage, fragmentation into apoptotic bodies, nuclear condensation, and mitochondrial outer membrane permeabilization. Pyroptosis is orchestrated by cleavage of GSDMD by inflammatory caspases 1 and 11 within the inflammasome complexes, whereas secondary necrosis is orchestrated by cleavage of GSDME by caspase-3 during apoptosis. Cells undergoing pyroptosis/secondary necrosis exhibit permeabilized plasma and organelle membranes, cellular swelling, and release of intracellular contents through the GSDMD or GSDME pores. These cells, unlike apoptotic cells, do not maintain their plasma membrane integrity.
The gasdermin superfamily is a group of proteins that has garnered much attention lately upon the discovery that these members possess the ability to form pores in the plasma membrane leading to a necrotic form of programmed cell death called pyroptosis (Table 1; Figure 1). This novel function was first discovered in gasdermin D (GSDMD) when it was shown that cleavage between the N- and C-terminal domains by the inflammatory caspases such as caspase-1 and caspase-11 liberates a cytotoxic N-terminus [6-8] that oligomerizes and permeabilizes the plasma membrane to drive pyroptosis [9-14]. Subsequently, it was identified that another member of the superfamily, GSDME, is also cleaved between its N- and C-terminal domains by caspase-3 during apoptosis which, similar to GSDMD, also liberates a cytotoxic N-terminus that permeabilizes the plasma membrane and may drive secondary necrosis (Table 1; Figure 1) [15,16]. Furthermore, this N-terminal pore-forming function is conserved in all superfamily members, except DFNB59, although the mechanism of activation of other members currently remains unknown [14]. Interestingly, recent evidence suggests that in addition to forming pores in the plasma membrane, this family also possesses the ability to form pores in mitochondrial membranes which is important in augmenting the apoptotic response and may serve to bridge activation of the pyroptotic pathway to the apoptotic pathway [17].
Apoptosis
Intrinsic Apoptotic Pathway
Mechanistically, apoptosis can be activated by one of two pathways: the intrinsic or the extrinsic apoptotic pathway. Activation of the intrinsic apoptotic pathway arises when cells sense intracellular or extracellular stresses such as DNA damage, viral or bacterial infection, growth factor withdrawal, glucocorticoids, ROS overload, or hypoxia. These stimuli begin a cascade of signaling by the pro-apoptotic BCL-2 family members whose expression and activation ultimately leads to disruption of mitochondrial function via mitochondrial outer membrane permeabilization (MOMP) [1,18-22].
All BCL-2 family members share one to four BCL-2 homology (BH) domains and are divided into three groups: the proapoptotic effectors, the antiapoptotic BCL-2-like members, and the proapoptotic BH3-only members [2]. The proapoptotic effectors include BAK, BAX, and BCL-2-related ovarian killer (BOK) which are unique in their ability to oligomerize and directly form pores in the outer mitochondrial membrane (OMM) leading to MOMP and the release of additional proapoptotic factors that potentiate apoptotic signaling [1-5]. While BAX cycles between the cytosol and the OMM and BAK permanently localizes to the OMM, both are inhibited from forming mitochondrial pores by the activity of the antiapoptotic BCL-2 family members [23-25]. These members include BCL-2, BCL-w, BCL-2-related gene, long isoform (BCL-xL), myeloid cell leukemia 1 (MCL-1), and BCL-2 related protein A1 (A1 or BFL-1) which reside predominantly in the OMM and function to antagonize the proapoptotic BCL-2 family members by directly binding to and sequestering them [26-29]. Binding of antiapoptotic BCL-2 family members to proapoptotic effectors prevents their ability to form oligomers in the OMM and can also promote their retrotranslocation away from the OMM to the cytosol [1,2,25]. Furthermore, antiapoptotic BCL-2 family members can block apoptosis by sequestering proapoptotic BH3-only members that directly activate BAK and BAX (Figure 2) [5,30,31].
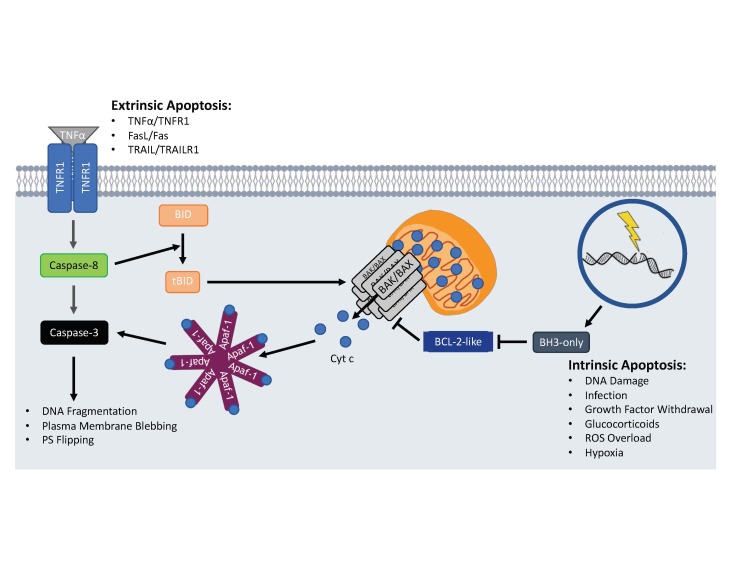
Figure 2.
Intrinsic and extrinsic apoptotic activation. Activation of the apoptotic program can occur intrinsically when cells sense internal damage and stress leading to activation of the proapoptotic BH3-only BCL-2 family members. The BH3-only proteins prevent anti-apoptotic BCL-2-like proteins from inhibiting Bak/Bax oligomerization on the OMM. Once oligomerized, proapoptotic factors like Cyt c escape into the cytosol and activate caspase-3 via the Apaf-1 apoptosome. Extrinsic apoptotic activation occurs when death ligands bind to cell surface death receptors leading to the recruitment and activation of pro-caspase-8. Active caspase-8 can then directly activate caspase-3 to begin dismantling the cell or cleave BID to activate the intrinsic apoptotic pathway.
To overcome the inhibitory effects of the antiapoptotic BCL-2 members, the proapoptotic BH3-only members which include BCL-2 antagonist of cell death (BAD), BCL-2 modifying factor (BMF), harakiri (HRK), p53-upregulated modulator of apoptosis (PUMA), BCL-2-interacting killer (BIK), NOXA, BCL-2-interacting mediator of cell death (BIM), and BCL-2-interacting domain death agonist (BID) are activated in response to cellular stresses or developmental cues. Activation occurs transcriptionally such as with the p53-mediated upregulation of PUMA or post-translationally such as with the caspase-8-medited proteolytic cleavage of BID [32-35]. Some proapoptotic BH3-only members (i.e. BID, BIM, PUMA, and NOXA) directly activate the intrinsic apoptotic pathway by binding to BAK and BAX leading to a conformational change that allows for their oligomerization in the OMM [36-38] while other members (i.e. BAD, BMF, HRK) can promote apoptosis indirectly by binding to and inhibiting the antiapoptotic BCL-2 family members from sequestering BAK and BAX [39-41].
Once BAK and BAX induce MOMP, proapoptotic factors including Cytochrome c (Cyt c), High Temperature Requirement Protein A2 (HtrA2/Omi), and second mitochondrial activator of caspases/direct IAP-binding protein with low pI (Smac/DIABLO) are released from the mitochondria into the cytosol where they continue the apoptotic signaling cascade [42-45]. HtrA2/Omi and Smac/DIABLO function by binding to and inactivating the inhibitors of apoptosis (IAPs) which normally bind to and inhibit activation of the caspases [43,46-49]. In addition, Cyt c binds to the cytosolic adaptor molecule apoptotic protease activating factor 1 (Apaf-1) leading to a conformational change that results in its oligomerization, formation of the apoptosome, and recruitment of the initiator caspase pro-caspase-9 [50,51]. Pro-caspase-9 autoproteolytically processes itself generating a fully active caspase-9 protease which then cleaves the effector caspases pro-caspase-3, -6, and -7 leading to their fully active proteolytic forms [51]. These caspases target hundreds of cellular substrates for proteolysis, leading to the characteristic morphological changes associated with cells undergoing apoptosis such as DNA fragmentation, plasma membrane blebbing, cell shrinkage, and the flipping of phosphatidylserine from the inner plasma membrane leaflet to the outer (Figures 1 and 2) [52-56].
Extrinsic Apoptotic Pathway
While the outcome of extrinsic and intrinsic apoptotic signaling is virtually identical, these pathways differ in their mode of activation. In contrast to the intrinsic apoptotic pathway, activation of the extrinsic apoptotic pathway occurs via the extracellular binding of ligands such as Fas ligand (FasL), tumor necrosis factor (TNF) α, and TNF-Related Apoptosis-Inducing Ligand (TRAIL) to the cell surface death receptors Fas (CD95 or APO-1), TNF receptor superfamily member 1A (TNFR1), and TRAIL receptor 1 (TRAILR1), respectively. Binding of these ligands to their receptors induces conformational changes in the intracellular trimerized receptor tails that allows for the recruitment of adaptor molecules like Fas associated death domain (FADD) protein to Fas and TRAILR1 or TNFR1 associated death domain (TRADD) protein to TNFR1 [57-63]. The initiator caspase pro-caspase-8 is then directly recruited to FADD via homotypic death effector domain (DED) interactions or indirectly recruited to TRADD via FADD-TRADD interactions leading to pro-caspase-8 dimerization and subsequent autoproteolytic activation [63-65]. Of note, TNFα-mediated apoptosis can be dependent or independent on the kinase activity of the serine/threonine kinase receptor-interacting protein kinase 1 (RIPK1) depending on RIPK1 phosphorylation status [66] . Once activated, caspase-8 cleaves and activates pro-caspase-3 which carries out the dismantling of the cellular machinery as outlined above [67,68].
Intriguingly, activation of the extrinsic apoptotic pathway can lead to activation of the intrinsic apoptotic pathway via the proapoptotic BH3-only member BID. BID is normally found inactive in the cytosol but is cleaved by caspase-8 generating an active C-terminal fragment called truncated BID (tBID) [32-34]. tBID translocates to the mitochondria where it binds to and directly activates oligomerization of BAX/BAK pores leading to MOMP and activation of the Apaf-1 apoptosome (Figure 2) [37,38]. It is interesting to note, however, that genetic knockout or knockdown of BID only partially reduces the magnitude and delays the kinetics of Cyt c release, caspase-3 activation, and cell death in mouse hepatocytes and human islet cells treated with TNFα or FasL, respectively, suggesting the possibility of other molecules that can bridge activation of the extrinsic apoptotic pathway to that of the intrinsic [69,70]. Indeed, it was recently demonstrated that similar to BID, GSDME is cleaved downstream of the extrinsic apoptotic pathway and that the active fragment, GSDME-N, independently induces the release of Cyt c from the mitochondria to promote caspase-3 activation [17].
The Gasdermin Superfamily
Humans encode six gasdermin superfamily members including GSDMA, GSDMB, GSDMC, GSDMD, GSDME/DFNA5, and DFNB59 while mice encode ten including three GSDMA orthologs (Gsdma1, Gsdma2, and Gsdma3), four GSDMC orthologs (Gsdmc1, Gsdmc2, Gsdmc3, and Gsdmc4), Gsdmd, Gsdme/Dfna5, and Dfnb59, but no GSDMB orthologs. All members share ~45% sequence homology and, except for DFNB59, adopt a two-domain architecture characterized by globular N- and C-terminal domains separated by a flexible linker region [7,14]. The N-terminal domains (GSDM-N) of all members except DFNB59 can oligomerize and form membrane-spanning pores in the plasma membrane resulting in the disruption of ionic gradients, osmotic cellular swelling, and cytolysis, ultimately resulting in necrotic cell death [9-14]. In healthy cells, however, the cytotoxic N-termini, are normally masked by the autoinhibitory C-termini which prevent the gasdermins from binding to negatively charged phospholipids, oligomerizing, and forming pores [14,71]. Interestingly, new evidence also demonstrates that several members including GSDMA-N, GSDMD-N, and GSDME-N can also form pores in the mitochondria leading to the release of proapoptotic molecules (Figures 3 and 4) [17].
GSDMD-N pores Induce Pyroptosis and NETosis
Pyroptosis is another form of PCD that plays important roles in pathogen clearance and activation of the adaptive immune system. In contrast to apoptotic cells, which maintain their plasma membrane integrity, cells undergoing pyroptosis are highly inflammatory due to permeabilization of their plasma membranes and the consequential release of pro-inflammatory signaling molecules (Figure 1). This form of PCD is initiated in cells of the innate immune system such as macrophages and dendritic cells upon sensing a variety of diverse pathogen- and danger-associated molecular patterns (PAMPs and DAMPs, respectively) such as lipopolysaccharide (LPS) and ATP. These signals lead to the formation of multiprotein complexes termed inflammasomes that recruit the inflammatory caspase pro-caspase-1 either directly or indirectly through the adaptor protein apoptosis-associated speck-like protein containing CARD (ASC) leading to its autoproteolytic activation (reviewed in [72]). Active caspase-1 is a cysteine protease that cleaves pro-interleukin (IL)-1β and pro-IL-18 to generate their active signaling forms, IL-1β and IL-18, respectively, which are released from cells undergoing pyroptosis to activate additional immune responses and inflammation [73,74]. In addition to canonical inflammasome-driven pyroptosis, a caspase-1-independent inflammasome called the noncanonical inflammasome can also activate pyroptosis. The noncanonical inflammasome is activated when intracellular LPS from gram-negative bacteria binds to the caspase activation and recruitment domain (CARD) of pro-caspase-11 of mice (or the human orthologs pro-caspase-4 or -5) leading to their oligomerization and autoactivation [75].
In 2015, using three different techniques (CRISPR/Cas-9 screening, ENU-forward mouse genetic screening, and high-sensitive quantitative mass spectrometry), three independent laboratories simultaneously demonstrated that GSDMD is a novel substrate of caspase-1, -4, and -5 (-11 in mice) that is cleaved between its N- and C-terminal domains downstream of canonical and noncanonical inflammasome activation and that the liberated N-terminus harbors intrinsic pyroptotic activity [6-8]. It wasn’t until a year later, however, that six independent groups then demonstrated mechanistically that GSDMD-N drives pyroptosis by oligomerizing and forming pores in the plasma membrane causing the cell swelling, lysis, and release of cytosolic contents like IL-1β, IL-18, and high mobility group box 1 (HMGB1) that characterize pyroptotic cell death (Figure 3) [9-14]. These studies also demonstrated that GSDMD-N binds specifically to phosphatidylinositol-4-phosphate [PI(4)P], phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], phosphatidylinositol-3,4,5-triphosphate [PI(3,4,5)P3], phosphatidylserine (PS), and cardiolipin (CL) phospholipids present in biomembranes, a feature that is also shared with GSDMA-N and GSDME-N [10,14,16,71]. As phosphatidylinositol phosphates and PS are enriched on the inner leaflet of the plasma membrane, GSDMD-N only attacks the plasma membrane intracellularly and not when present extracellularly [10]. Physiologically, GSDMD-N may play a beneficial role by directly killing bacteria by forming pores in their cardiolipin-enriched membranes and by initiating pyroptosis [10,14], however, excessive GSDMD-N-mediated pyroptosis plays a detrimental role in promoting lethal endotoxemia that occurs during LPS-induced septic shock [6].
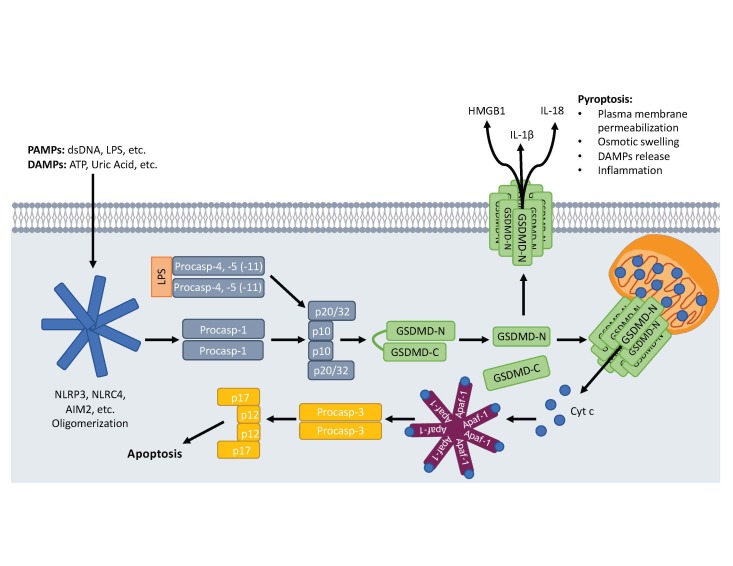
Figure 3.
GSDMD-mediated pyroptosis. Macrophages and dendritic cells sensing DAMPs and PAMPs will activate canonical inflammasome assemblies leading to the recruitment and activation of procaspase-1 (procasp-1). Intracellular LPS from gram-negative bacteria binds to and activates procaspases -4 or -5 (-11 in mice; procasp-4, -5, -11) forming the noncanonical inflammasome. Active inflammatory caspases cleave GSDMD liberating an ~30 kDa N-terminal fragment (GSDMD-N) that translocates and oligomerizes to form pores in the plasma membrane. These pores allow the release of proinflammatory molecules like HMGB1, IL-1β, and IL-18 and disrupt ionic gradients leading to cellular swelling and pyroptosis. GSDMD-N also permeabilizes the mitochondria, releasing proapoptotic factors like Cyt c leading to activation of caspase-3 via the Apaf-1 apoptosome.
In addition to its role in pyroptosis, GSDMD was recently shown to play a vital role in NETosis (Table 1). In response to microbial attack, neutrophils will release neutrophil extracellular traps (NETs) composed of DNA and antimicrobial proteins that trap and kill pathogens, although the mechanism of NET release has remained elusive. However, two groups demonstrated that activation of GSDMD is required not only for permeabilization of the plasma membrane that is required for NET extrusion, but also nuclear membrane permeabilization required for the release of DNA [76,77].
GSDME-N pores Switch Apoptosis to Secondary Necrosis
Apoptosis is a noninflammatory and immunologically silent form of PCD as cells maintain their plasma membrane integrity and do not release proinflammatory molecules. In addition, upon activation of the apoptotic pathway, dying cells display cell surface “eat-me” signals, such as PS and calreticulin, that are quickly recognized by neighboring scavenger cells allowing for rapid phagocytosis through a process called efferocytosis [78-80]. In some instances, however, the scavenging capacity of a particular system is inhibited or insufficient to remove apoptotic cells in a timely manner which causes these dying cells to progress to secondary necrosis (Table 1; Figure 1). In contrast to apoptosis, secondary necrotic cells lose their plasma membrane integrity, swell, and rupture leading to the release of proinflammatory, intracellular molecules including HMGB1 and activated caspase-3 [81,82]. Interestingly, secondary necrosis has been thought to be a result of a passive breakdown of the plasma membrane over time, however recent evidence suggests that GSDME likely induces secondary necrosis upon cleavage by caspase-3 during apoptosis. Indeed, activation of apoptosis by many stimuli including TNFα, ultraviolet irradiation, etoposide, encephalomyocarditis virus, and vesicular stomatitis virus infection activates the apoptotic pathway and caspase-3 leading to cleavage of GSDME between its N- and C-terminal domains (Figure 4) [15-17]. Like GSDMD, this cleavage liberates the necrotic N-terminus which over time permeabilizes the plasma membrane and leads to cellular swelling, cytolysis, and the release of proinflammatory molecules that characterizes secondary necrosis [15-17]. This function may contribute to the extensive intestinal tissue damage and inflammation that occurs in patients undergoing chemotherapy and may be a beneficial target for alleviating such side effects [16]. Although the role of GSDME in secondary necrosis downstream of caspase-3 activation is now well documented, GSDME deficiency may not prevent secondary necrosis in all cell types [83,84], suggesting that additional GSDME-independent and cell-type specific secondary necrosis mechanisms may play a dominant role in permeabilizing the plasma membrane downstream of caspase-3 activation.
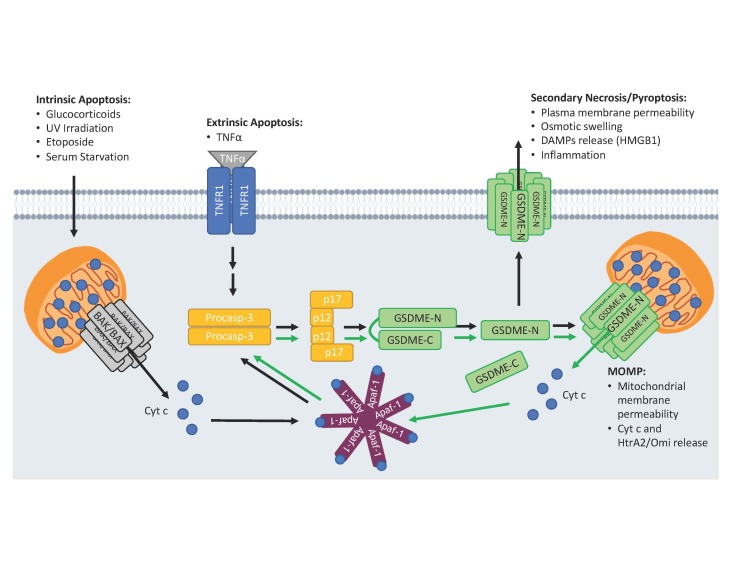
Figure 4.
GSDME-N permeabilizes the plasma and mitochondrial membranes. Activation of the intrinsic apoptotic pathway by glucocorticoids, UV irradiation, etoposide, or serum starvation or the extrinsic apoptotic pathway by TNFα leads to activation of caspase-3. Active caspase-3 cleaves GSDME generating a pore-forming (GSDME-N) ~30 kDa N-terminal fragment that translocates to and permeabilizes the mitochondria. Proapoptotic factors are released through GSDME-N mitochondrial pores and positively feedback on caspase-3 activation and GSDME cleavage (green arrows). After mitochondrial permeabilization, GSDME-N forms pores in the plasma membrane leading to secondary necrosis/pyroptosis, which allows the release of proinflammatory DAMP molecules like HMGB1.
Gasdermins Induce Apoptosis by Permeabilizing the Mitochondria
New evidence demonstrates that members of the gasdermin family also possess mitochondrial pore-forming functions. Indeed, downstream of intrinsic and extrinsic apoptotic activation, caspase-3 generated GSDME-N localizes to the mitochondria and is sufficient to form pores that release proapoptotic molecules like Cyt c and HtrA2/Omi [17]. This event creates a positive feedback loop that promotes caspase-3 activation and further GSDME cleavage and augments the apoptotic program (Figure 4) [17]. This function appears to be conserved in the superfamily as other members like GSDMA-N and GSDMD-N also permeabilize the mitochondria to release proapoptotic factors [17]. Consistent with this idea, previous reports have demonstrated that GSDMA-N, GSDMD-N, and GSDME-N all bind to CL and form pores in CL-enriched liposomes [10,14,16,71], a major phospholipid constituent of the inner mitochondrial membrane that translocates to the outer mitochondrial membrane during PCD [85-87]. These findings may also explain previous observations that these members can activate the apoptotic program, disrupt mitochondrial function, and suppress tumor growth.
Indeed, GSDMA has been shown to play a role in activating apoptosis in gastric pit cells. Overexpression of GSDMA in gastric cancer cell lines is sufficient to activate caspase-3/-7 as well as induce internucleosomal DNA cleavage, and physiologically, TGF-β-induced apoptosis in gastric pit cells is mediated by transcriptional upregulation of GSDMA via recruitment of the transcription factor LMO1 to the GSDMA promoter [88]. The N-terminus of GSDMA3 can translocate to the mitochondria via Hsp90 where it stimulates the production of mitochondrial ROS, dissipates mitochondrial membrane potential, and induces mitochondrial permeability transition (MPT), ultimately leading to cell death [89,90]. In addition, several mutant mouse lines harboring mutations in Gsdma3 display alopecia, hyperkeratosis, and skin inflammation due to a depletion of the bulge stem cells that give rise to hair follicles [91-94], and polymorphisms in GSDMA have been linked to the development of asthma [95,96], supporting its role as a pore-forming protein that promotes both apoptosis and inflammation.
During pyroptosis, GSDMD induces mitochondrial depolarization in a BAK/BAX-independent manner [97], and GSDMD-N has recently been shown to localize to the mitochondria where it causes mitochondrial ROS generation downstream of Shiga toxin/LPS-induced canonical and noncanonical inflammasome activation [98]. Furthermore, several lines of evidence suggest that activation of the pyroptotic pathway can also lead to activation of the apoptotic pathway. For example, the apoptotic caspase-7 is activated downstream of caspase-1 by a number of inflammasomes including the nod-like receptor family pyrin domain containing 3 (NLRP3) inflammasome to induce transcriptional upregulation of important pro-inflammatory cytokines [99], the nod-like receptor family CARD containing 4 (NLRC4) inflammasome in order to deliver bacteria to the lysosomal compartments for degradation [100], and the absent in melanoma 2 (AIM2) inflammasome [101]. In addition to canonical inflammasome-mediated activation of apoptotic caspases, the noncanonical inflammasome can also activate caspases-3/-7 and apoptosis independent of BID and caspase-1 in vitro and in vivo [102]. How these pathways are mechanistically linked has remained unknown, however a recent study demonstrates that GSDMD-N can form pores in the mitochondria and that macrophages deficient in GSDMD expression have a significant delay in caspase-3/-7 activation upon activation of the noncanonical inflammasome [17]. Therefore, GSDMD-N-mediated MOMP may explain how GSDMD disrupts mitochondrial function during pyroptosis and how these apoptotic caspases are activated downstream of inflammasome activation. It is also interesting to note, however, that caspase-3/-7 have also been shown to cleave GSDMD within its N-terminal domain, thus inactivating its pore-forming functions [15,103]. This mechanism suggests a potential negative feedback loop in GSDMD-N-mediated mitochondrial pore-formation, however future studies should determine the importance of this regulation.
Furthermore, overexpression of a gain-of-function mutant of GSDME that causes sensorineural hearing loss (GSDMEHL) activates apoptosis in HEK293T and yeast cells [17,104,105], and expression of WT GSDME sensitizes hepatocellular carcinoma, gastric, melanoma, thymoma, and acute lymphoblastic leukemia cancer cell lines to apoptosis [17,106-108]. In addition, GSDME-N and GSDMEHL colocalize with mitochondria and induce the production of ROS when expressed in HEK293T and yeast cells [17,105,109], and gene set enrichment analysis demonstrated that gene sets involved in apoptotic pathways are downregulated in GSDME-/- mice compared to GSDME+/+ mice [104] supporting the role of GSDME as a mitochondrial pore-forming protein that potentiates apoptotic signaling.
Gasdermins in Diseases and Disorders
Gasdermins in Cancer
Cancer results from the malignant transformation of cells that leads to their uncontrolled proliferation and metastasis where they spread and form tumors throughout the body that disrupt organ function and ultimately lead to death of the organism. Apoptosis is normally triggered by many of the changes that a healthy cell must undergo in order to become tumorigenic including DNA damage and genomic instability caused by rapid cell division, hypoxia, oncogene overexpression, and survival factor depletion. Overtime, however, some cells may continue accumulating mutations that desensitize them to activation of the apoptotic program and inhibit them from responding to these triggers, initiating cell death, and preventing cancer. In fact, evasion of PCD is recognized as one of the “Hallmarks of Cancer” that most, if not all, cells must adopt in order to transform and become malignant [110,111]. Mutations that drive cancer can over activate molecules that inhibit apoptosis (oncogenes) or inactivate molecules that trigger apoptosis (tumor suppressors).
Interestingly, many members of the gasdermin superfamily are thought to be tumor suppressors. Indeed, GSDMA expression is highly enriched in normal gastric and esophageal tissue but almost completely lost in a panel of 14 different gastric cancer cell lines and most primary esophageal squamous cell carcinomas [88,112,113], and overexpression of GSDMA in gastric cancer cell lines decreases colony formation [88]. GSDME also functions as a tumor suppressor as it is highly downregulated in breast, gastric, and colorectal cancers due to promoter hypermethylation [114-121], and its expression positively correlates with superior prognosis and 5-year survival rates in patients with esophageal squamous cell carcinoma [122] and negatively correlates with an increased risk of breast cancer metastasis [118]. Furthermore, expression of GSDME in melanoma, acute lymphoblastic leukemia, gastric, lung, liver, and colon cancer cell lines leads to decreased cell growth, survival, and colony formation [17,114,119,123,124]. One study also shows that deletion of GSDME from melanoma cells accelerates tumor growth and decreases survival outcomes in an in vivo allograft mouse model [17], however another study shows no significant difference in tumor growth when GSDME is deleted from lung cancer cells [125]. This discrepancy may be explained by particular tissue- and/or cancer-specific drivers that may desensitize lung cancer cells, but not melanoma cells, to the proapoptotic effects of GSDME on tumor growth.
GSDMD has also been studied in the context of cancer, however, its function as either a tumor suppressor or oncogene remains unclear. Support for its role as a tumor suppressor comes from findings demonstrating that GSDMD is downregulated in primary esophageal squamous cell carcinomas and gastric cancers although it is normally highly enriched in these tissues and that expression of GSDMD reduces colony formation and cell proliferation in gastric cancer cell lines and reduces their ability to form tumors in xenograft models [113,126]. However, GSDMD was also found to be oncogenic as it is upregulated in non-small cell lung cancer (NSCLC) tissue compared to adjacent, non-cancerous tissue and that knockdown of its expression in NSCLC cell lines restricts cell growth in vitro and in vivo [127]. These observations suggest that the proapoptotic and tumor suppressive functions of GSDMD may be cancer- and/or tissue-specific.
On the other hand, one superfamily member, GSDMB, may function as an oncogene as it is amplified and overexpressed in several gastric cancer cell lines and primary gastric and breast tumors [113,128,129]. Furthermore, its expression does not limit colony formation in the same gastric cancer cells that GSDMA and GSDMD do [113], and its expression in breast cancer promotes invasion and metastasis and decreases patient survival outcomes [130]. Interestingly, GSDMA is highly expressed in the nondividing, differentiated cells of the esophagus and stomach while GSDMB is expressed in the rapidly dividing stem cells of these same organs suggesting that GSDMB may play a role in promoting cell proliferation and/or migration, while GSDMA does not [113]. In addition, GSDMB-N does not bind CL like GSDMA-N, GSDMD-N, and GSDME-N suggesting that it does not interact with the mitochondria [131]. As all members of the gasdermin superfamily, except DFNB59, possess the ability to permeabilize the plasma membrane while there is only evidence that GSDMA, GSDMD, and GSDME possess mitochondrial pore-forming activity, it is possible that only the latter function determines whether or not these members are tumor suppressors.
GSDME Mutations and Hearing Loss
A variety of mutations in GSDME have been reported to cause autosomal dominant, progressive, sensorineural hearing loss (HL) with the age of onset being highly variable ranging from 0 to 50 years [132-138]. These mutations are found in introns 7 and 8 and include base pair substitutions and indels that disrupt splice sites and splice site selection sequences which ultimately lead to skipping of exon 8 during pre-mRNA splicing [135,137,139]. Splicing of exon 7 and 9 introduces a frameshift that translates amino acids 1-330 followed by an aberrant stretch of 41 amino acids and a premature stop codon. As the C-terminus of gasdermin members inhibit their pore-forming activity, this truncation represents a gain-of-function mutation by unmasking the toxic N-terminus. Supporting this notion, truncating mutations in an Iranian family in exon 5 that would lead to an inactive N-terminal protein product do not cause HL [140]. Furthermore, overexpression of GSDMEHL in HEK293T and yeast cells leads to apoptosis and necrosis [17,104,105], similar to GSDME-N, and histopathological analysis of inner ear tissue from patients with GSDME HL mutations revealed severe degeneration of the cochlea where WT GSDME is highly expressed [141], indicating that it is a gain-of-function mutation. However, as GSDME appears to have widespread tissue distribution, albeit at varying levels, it remains to be seen why only the cochlea is affected and patients harboring these mutations do not present any obvious additional abnormalities.
One possibility is that expression of the C-terminally truncated mutant results in a protein that is highly unstable and rapidly degraded by the proteasomal machinery. Indeed, Wang et al. show that overexpression of GSDMEHL in HeLa cells followed by treatment with cycloheximide resulted in nearly 100 percent degradation within 24 hours whereas little to no loss was observed in cells expressing WT GSDME or WT GSDME-N [16]. Furthermore, Val Laer et al. generated a mouse model mimicking the GSDME HL mutation by specifically deleting exon 8 and showed that although the truncated mRNA transcript is expressed in these mice, no mutant protein could be detected by immunoblotting [142]. These observations support the theory that the mutant protein likely has a short half-life and is degraded before it can cause extensive damage in other organs. This notion, however, still begs the question of how GSDMEHL specifically damages the cochlear hair cells to cause HL.
Hair cells are terminally differentiated sensory epithelium that transform physical noises into chemical signals and transmit them to the brain. These cells must last the lifetime of mammalian species as they are not regenerated to replace dead or damaged cells. Furthermore, outer hair cells are extremely sensitive to noise-induced trauma which can result in the accumulation of ROS, activation of apoptosis, and cell death [143]. It is therefore possible that hair cells express very low levels of GSDMEHL that are insufficient to activate apoptosis alone but instead further increase the sensitivity of these cells to noise-induced cell death by lowering the threshold for apoptosis activation. In this case, noise levels that would normally be insufficient to kill WT hair cells may be sufficient to kill hair cells expressing GSDMEHL, and accumulating exposure to these everyday noises would lead to a progressive increase in hair cell death, ultimately causing permanent HL. This theory would also explain the extreme variability in the age of onset and why mutations cause progressive HL, as the noise levels experienced by an individual on a daily basis can drastically differ.
Conclusions and Outlook
Recent advances in PCD research have greatly deepened our knowledge of gasdermin biology and provided novel avenues to pursue for therapeutic intervention for diseases and disorders like sepsis, cancer, and HL. While these past few years have taught us a lot, there are still many questions that remain open. GSDMD and GSDME are activated upon cleavage by the inflammatory caspases and caspase-3, respectively, but how are the other gasdermin family members activated? We know that all members except DFNB59 harbor pore-forming activity in their N-terminal domains, but how is this activity unleashed from the autoinhibitory C-terminus. Can other proteases such as granzymes also cleave members of the gasdermin family, and if so, under what conditions? In addition, we know that post-translational modifications (PTMs) like phosphorylation of the N-terminus of GSDME can inhibit oligomerization [17], however, can PTMs in the C-terminus of these molecules disrupt their autoinhibition and activate their pore-forming activity in the absence of proteolytic cleavage?
Furthermore, we now know that GSDMD plays important physiological roles in mediating pyroptosis and controlling pathogen replication, but the physiological role of GSDME remains unclear. GSDME-/- mice appear to develop normally ruling out the likelihood that GSDME plays important functions in augmenting the apoptotic pathways critical for embryonic development, however, it is activated in response to other stimuli such as viral infection suggesting that it may function similar to GSDMD in destroying cellular niches where microorganisms are hiding and replicating. As GSDME sensitizes cells to activation of the apoptotic program, it may significantly limit the spread of pathogens during infection, especially in cells that do not undergo pyroptosis. Future studies should determine how GSDME-mediated potentiation of apoptosis contributes to physiological or pathophysiological processes and if other superfamily members share similar or redundant functions.
Ultimately, knowledge of gasdermin biology may provide insight into many clinically useful applications in treating and preventing disease or alleviating unwanted side effects from toxic chemotherapeutics. Almost all members of the superfamily possess pro-death activities by either forming pores in the mitochondria and augmenting apoptosis and/or forming pores in the plasma membrane and inducing programmed necrosis. As numerous diseases and disorders are a result of dysregulated programmed cell death pathways, this family of proteins may reveal themselves as important, novel therapeutic targets. Indeed, inhibiting GSDME pore-formation may be useful in reducing intestinal damage and inflammation caused by chemotherapeutics or in preventing or delaying the onset of HL in patients with GSDME mutations. Conversely, cancer patients with tumors that have low GSDME expression may benefit from treatment with a methyltransferase inhibitor to decrease promoter methylation, restore GSDME expression, and sensitize tumor cells to apoptosis. Similarly, inhibition of GSDMD may be useful in preventing sepsis induced by gram negative bacterial infections. Future studies should also determine the clinical relevance of the gasdermin family members and identify specific inhibitors that prevent their pore-forming function.
Glossary
- PCD
programmed cell death
- BCL-2
B Cell CLL/Lymphoma-2
- BAK
BCL-2 Antagonist Killer 1
- BAX
BCL-2-Associated X Protein
- GSDMD
gasdermin D
References
- Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25(3):486–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion [Internet] Mol Cell. 2010. February;37(3):299–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamas-Din A, Kale J, Leber B, Andrews DW. Mechanisms of action of bcl-2 family proteins [Internet] Cold Spring Harb Perspect Biol. 2013. April;5(4):a008714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T, Follis AV, Kriwacki RW, Green DR. Many players in BCL-2 family affairs [Internet] Trends Biochem Sci. 2014. March;39(3):101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy [Internet] Nat Rev Mol Cell Biol. 2014. January;15(1):49–63. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Stowe IB, Lee BL, O’Rourke K, Anderson K, Warming S, et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling [Internet] Nature. 2015. October;526(7575):666–71. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death [Internet] Nature. 2015. October;526(7575):660–5. [DOI] [PubMed] [Google Scholar]
- He W, Wan H, Hu L, Chen P, Wang X, Huang Z, et al. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion [Internet] Cell Res. 2015. December;25(12):1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aglietti RA, Estevez A, Gupta A, Ramirez MG, Liu PS, Kayagaki N, Ciferri C, Dixit VM, Dueber EC. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc Natl Acad Sci U S A [Internet]. 2016. 07 12,;113(28):7858-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature [Internet]. 2016. 07 07,;535(7610):153-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo HM, Rathkey J, Boyd-Tressler A, Katsnelson MA, Abbott DW, Dubyak GR. Active caspase-1 induces plasma membrane pores that precede pyroptotic lysis and are blocked by lanthanides. J Immunol [Internet]. 2016. 08 15,;197(4):1353-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sborgi L, Rühl S, Mulvihill E, Pipercevic J, Heilig R, Stahlberg H, Farady CJ, Müller DJ, Broz P, Hiller S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J [Internet]. 2016. 08 15,;35(16):1766-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, He W, Hu L, Li J, Fang Y, Wang X, Xu X, Wang Z, Huang K, Han J. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res [Internet]. 2016. 09;26(9):1007-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Wang K, Liu W, She Y, Sun Q, Shi J, Sun H, Wang D, Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature [Internet]. 2016. 07 07,;535(7610):111-6. [DOI] [PubMed] [Google Scholar]
- Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun [Internet]. 2017. 01 03, [cited Apr 16, 2019];8:14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao W, Shi X, Ding J, Liu W, He H, Wang K, Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature [Internet]. 2017. 07 06,;547(7661):99-103. [DOI] [PubMed] [Google Scholar]
- Rogers C, Erkes DA, Nardone A, Aplin AE, Fernandes-Alnemri T, Alnemri ES. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat Commun [Internet]. 2019 04 11;10(1):1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold M, McPherson K, Reichardt H. Glucocorticoids in T cell apoptosis and function [Internet] Cell Mol Life Sci. 2006. January;63(1):60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez G, London L, Hockenbery D, Alexander M, McKearn JP, Korsmeyer SJ. Deregulated bcl-2 gene expression selectively prolongs survival of growth factor-deprived hemopoietic cell lines [Internet] J Immunol. 1990. May;144(9):3602–10. [PubMed] [Google Scholar]
- Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells [Internet] Mol Cell. 2017. May;66(3):306–19. [DOI] [PubMed] [Google Scholar]
- Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology [Internet] Nat Rev Cancer. 2016. January;16(1):20–33. [DOI] [PubMed] [Google Scholar]
- Sendoel A, Hengartner MO. Apoptotic cell death under hypoxia [Internet] Physiology (Bethesda). 2014. May;29(3):168–76. [DOI] [PubMed] [Google Scholar]
- Naghdi S, Várnai P, Hajnóczky G. Motifs of VDAC2 required for mitochondrial bak import and tBid-induced apoptosis [Internet] Proc Natl Acad Sci USA. 2015. October;112(41):5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Stojanovski D, Frazier AE, Kotevski A, Dewson G, Craigen WJ, et al. Inhibition of bak activation by VDAC2 is dependent on the bak transmembrane anchor [Internet] J Biol Chem. 2010. November;285(47):36876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich F, Banerjee S, Suzuki M, Cleland MM, Arnoult D, Wang C, et al. Bcl-x(L) retrotranslocates bax from the mitochondria into the cytosol [Internet] Cell. 2011. April;145(1):104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng EH, Wei MC, Weiler S, Flavell RA, Mak TW, Lindsten T, et al. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis [Internet] Mol Cell. 2001. September;8(3):705–11. [DOI] [PubMed] [Google Scholar]
- Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, bax, that accelerates programmed cell death [Internet] Cell. 1993. August;74(4):609–19. [DOI] [PubMed] [Google Scholar]
- Antonsson B, Conti F, Ciavatta A, Montessuit S, Lewis S, Martinou I, et al. Inhibition of bax channel-forming activity by bcl-2 [Internet] Science. 1997. July;277(5324):370–2. [DOI] [PubMed] [Google Scholar]
- Barclay LA, Wales TE, Garner TP, Wachter F, Lee S, Guerra RM, et al. Inhibition of pro-apoptotic BAX by a noncanonical interaction mechanism [Internet] Mol Cell. 2015. March;57(5):873–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, et al. A unified model of mammalian BCL-2 protein family interactions at the mitochondria [Internet] Mol Cell. 2011. November;44(4):517–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kanai M, Inoue-Yamauchi A, Tu H, Huang Y, Ren D, et al. An interconnected hierarchical model of cell death regulation by the BCL-2 family [Internet] Nat Cell Biol. 2015. October;17(10):1270–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross A, Yin XM, Wang K, Wei MC, Jockel J, Milliman C, et al. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/fas death [Internet] J Biol Chem. 1999. January;274(2):1156–63. [DOI] [PubMed] [Google Scholar]
- Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the fas pathway of apoptosis [Internet] Cell. 1998. August;94(4):491–501. [DOI] [PubMed] [Google Scholar]
- Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors [Internet] Cell. 1998. August;94(4):481–90. [DOI] [PubMed] [Google Scholar]
- Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53 [Internet] Mol Cell. 2001. March;7(3):683–94. [DOI] [PubMed] [Google Scholar]
- Kim H, Tu H, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, et al. -, Cheng EH-. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis [Internet] Mol Cell. 2009. November;36(3):487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, et al. Bid, bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane [Internet] Cell. 2002. November;111(3):331–42. [DOI] [PubMed] [Google Scholar]
- Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, et al. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c [Internet] Genes Dev. 2000. August;14(16):2060–71. [PMC free article] [PubMed] [Google Scholar]
- Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics [Internet] Cancer Cell. 2002. September;2(3):183–92. [DOI] [PubMed] [Google Scholar]
- Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, et al. BH3 domains of BH3-only proteins differentially regulate bax-mediated mitochondrial membrane permeabilization both directly and indirectly [Internet] Mol Cell. 2005. February;17(4):525–35. [DOI] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function [Internet] Mol Cell. 2005. February;17(3):393–403. [DOI] [PubMed] [Google Scholar]
- Adrain C, Creagh EM, Martin SJ. Apoptosis-associated release of Smac/DIABLO from mitochondria requires active caspases and is blocked by Bcl-2 EMBO J 2001. 6627 p DOI: 10.1093/emboj/20.23.6627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loo G, van Gurp M, Depuydt B, Srinivasula SM, Rodriguez I, Alnemri ES, et al. The serine protease omi/HtrA2 is released from mitochondria during apoptosis. omi interacts with caspase-inhibitor XIAP and induces enhanced caspase activity [Internet] Cell Death Differ. 2002. January;9(1):20–6. [DOI] [PubMed] [Google Scholar]
- Martins LM, Iaccarino I, Tenev T, Gschmeissner S, Totty NF, Lemoine NR, et al. The serine protease omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif [Internet] J Biol Chem. 2002. January;277(1):439–44. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c [Internet] Cell. 1996. July;86(1):147–57. [DOI] [PubMed] [Google Scholar]
- Cory S, Fenner F. Walter and Eliza Hall Institute of Medical Research. [Internet].; 2007. 345 p [Google Scholar]
- Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, et al. A conserved XIAP-interaction motif in caspase-9 and smac/DIABLO regulates caspase activity and apoptosis [Internet] Nature. 2001. March;410(6824):112–6. [DOI] [PubMed] [Google Scholar]
- Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition [Internet] Cell. 2000. July;102(1):33–42. [DOI] [PubMed] [Google Scholar]
- Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins [Internet] Cell. 2000. July;102(1):43–53. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of apaf-1/caspase-9 complex initiates an apoptotic protease cascade [Internet] Cell. 1997. November;91(4):479–89. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by apaf-1-mediated oligomerization [Internet] Mol Cell. 1998. June;1(7):949–57. [DOI] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level [Internet] Nat Rev Mol Cell Biol. 2008. March;9(3):231–41. [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Renvoizé C, Hamelin J, Riché N, Bertoglio J, Bréard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing [Internet] Nat Cell Biol. 2001. April;3(4):346–52. [DOI] [PubMed] [Google Scholar]
- Naito M, Nagashima K, Mashima T, Tsuruo T. Phosphatidylserine externalization is a downstream event of interleukin-1 beta-converting enzyme family protease activation during apoptosis [Internet] Blood. 1997. March;89(6):2060–6. [PubMed] [Google Scholar]
- Nagata S. DNA degradation in development and programmed cell death [Internet] Annu Rev Immunol. 2005;23:853–75. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Finucane DM, Amarante-Mendes GP, O’Brien GA, Green DR. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity [Internet] J Biol Chem. 1996. November;271(46):28753–6. [DOI] [PubMed] [Google Scholar]
- Ting AT, Bertrand MJM. More to life than NF-κB in TNFR1 signaling. Trends Immunol [Internet]. 2016. 08;37(8):535-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FK, Chun HJ, Zheng L, Siegel RM, Bui KL, Lenardo MJ. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling [Internet] Science. 2000. June;288(5475):2351–4. [DOI] [PubMed] [Google Scholar]
- Fu Q, Fu T, Cruz AC, Sengupta P, Thomas SK, Wang S, et al. Structural basis and functional role of intramembrane trimerization of the fas/CD95 death receptor [Internet] Mol Cell. 2016. February;61(4):602–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of fas and initiates apoptosis [Internet] Cell. 1995. May;81(4):505–12. [DOI] [PubMed] [Google Scholar]
- Tummers B, Green DR. Caspase-8: Regulating life and death. Immunol Rev [Internet]. 2017. 05;277(1):76-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H, Shu HB, Pan MG, Goeddel DV. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways [Internet] Cell. 1996. January;84(2):299–308. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes [Internet] Cell. 2003. July;114(2):181–90. [DOI] [PubMed] [Google Scholar]
- Kischkel FC, Lawrence DA, Chuntharapai A, Schow P, Kim KJ, Ashkenazi A. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5 [Internet] Immunity. 2000. June;12(6):611–20. [DOI] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization [Internet] Mol Cell. 1998. January;1(2):319–25. [DOI] [PubMed] [Google Scholar]
- Geng J, Ito Y, Shi L, Amin P, Chu J, Ouchida AT, Mookhtiar AK, Zhao H, Xu D, Shan B, Najafov A, Gao G, Akira S, Yuan J. Regulation of RIPK1 activation by TAK1-mediated phosphorylation dictates apoptosis and necroptosis. Nat Commun [Internet]. 2017. 08 25,;8(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudouin J, Liesche C, Aschenbrenner S, Hörner M, Eils R. Caspase-8 cleaves its substrates from the plasma membrane upon CD95-induced apoptosis [Internet] Cell Death Differ. 2013. April;20(4):599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennicke HR, Jürgensmeier JM, Shin H, Deveraux Q, Wolf BB, Yang X, et al. Pro-caspase-3 is a major physiologic target of caspase-8 [Internet] J Biol Chem. 1998. October;273(42):27084–90. [DOI] [PubMed] [Google Scholar]
- Chen X, Ding W, Ni H, Gao W, Shi Y, Gambotto AA, et al. Bid-independent mitochondrial activation in tumor necrosis factor alpha-induced apoptosis and liver injury [Internet] Mol Cell Biol. 2007. January;27(2):541–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar MV, Trivedi PM, Kay TW, Hawthorne WJ, O’Connell PJ, Jenkins AJ, et al. Human islet cells are killed by BID-independent mechanisms in response to FAS ligand [Internet] Apoptosis. 2016. April;21(4):379–89. [DOI] [PubMed] [Google Scholar]
- Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature [Internet]. 2018. 05;557(7703):62-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Kanneganti T. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol [Internet]. 2016. 06 20,;213(6):617-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantuzzi G, Dinarello CA. Interleukin-18 and interleukin-1 beta: two cytokine substrates for ICE (caspase-1) [Internet] J Clin Immunol. 1999. January;19(1):1–11. [DOI] [PubMed] [Google Scholar]
- Brough D, Rothwell NJ. Caspase-1-dependent processing of pro-interleukin-1beta is cytosolic and precedes cell death [Internet] J Cell Sci. 2007. March;120(Pt 5):772–81. [DOI] [PubMed] [Google Scholar]
- Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, et al. Inflammatory caspases are innate immune receptors for intracellular LPS [Internet] Nature. 2014. October;514(7521):187–92. [DOI] [PubMed] [Google Scholar]
- Chen KW, Monteleone M, Boucher D, Sollberger G, Ramnath D, Condon ND, von Pein JB, Broz P, Sweet MJ, Schroder K. Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci Immunol [Internet]. 2018. 08 24, [cited Jun 20, 2019];3(26) [DOI] [PubMed] [Google Scholar]
- Sollberger G, Choidas A, Burn GL, Habenberger P, Di Lucrezia R, Kordes S, Menninger S, Eickhoff J, Nussbaumer P, Klebl B, Krüger R, Herzig A, Zychlinsky A. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci Immunol [Internet]. 2018. 08 24, [cited Jun 20, 2019];3(26) [DOI] [PubMed] [Google Scholar]
- Silva MT. Secondary necrosis: the natural outcome of the complete apoptotic program [Internet] FEBS Lett. 2010. November;584(22):4491–9. [DOI] [PubMed] [Google Scholar]
- deCathelineau AM, Henson PM. The final step in programmed cell death: phagocytes carry apoptotic cells to the grave [Internet] Essays Biochem. 2003;39:105–17. [DOI] [PubMed] [Google Scholar]
- Gardai SJ, Bratton DL, Ogden CA, Henson PM. Recognition ligands on apoptotic cells: A perspective [Internet] J Leukoc Biol. 2006. May;79(5):896–903. [cited 2019 Sep 19]. [DOI] [PubMed] [Google Scholar]
- Silva MT, do Vale A. dos Santos, Nuno M. N. Secondary necrosis in multicellular animals: an outcome of apoptosis with pathogenic implications [Internet] Apoptosis. 2008. April;13(4):463–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Berghe T, Vanlangenakker N, Parthoens E, Deckers W, Devos M, Festjens N, et al. Necroptosis, necrosis and secondary necrosis converge on similar cellular disintegration features [Internet] Cell Death Differ. 2010. June;17(6):922–30. [DOI] [PubMed] [Google Scholar]
- Lee BL, Mirrashidi KM, Stowe IB, Kummerfeld SK, Watanabe C, Haley B, et al. ASC- and caspase-8-dependent apoptotic pathway diverges from the NLRC4 inflammasome in macrophages [Internet] Sci Rep. 2018. February;8(1):3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tixeira R, Shi B, Parkes MA, Hodge AL, Caruso S, Hulett MD, et al. Gasdermin E does not limit apoptotic cell disassembly by promoting early onset of secondary necrosis in jurkat T cells and THP-1 monocytes [Internet] Front Immunol. 2018. December;9:2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan VE, Chu CT, Tyurina YY, Cheikhi A, Bayir H. Cardiolipin asymmetry, oxidation and signaling [Internet] Chem Phys Lipids. 2014. April;179:64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug ZT, Gottlieb E. Cardiolipin acts as a mitochondrial signalling platform to launch apoptosis [Internet] Biochim Biophys Acta. 2009. October;1788(10):2022–31. [DOI] [PubMed] [Google Scholar]
- Garcia Fernandez M, Troiano L, Moretti L, Nasi M, Pinti M, Salvioli S, et al. Early changes in intramitochondrial cardiolipin distribution during apoptosis [Internet] Cell Growth Differ. 2002. September;13(9):449–55. [PubMed] [Google Scholar]
- Saeki N, Kim DH, Usui T, Aoyagi K, Tatsuta T, Aoki K, et al. GASDERMIN, suppressed frequently in gastric cancer, is a target of LMO1 in TGF-beta-dependent apoptotic signalling [Internet] Oncogene. 2007. October;26(45):6488–98. [DOI] [PubMed] [Google Scholar]
- Shi P, Tang A, Xian L, Hou S, Zou D, Lv Y, et al. Loss of conserved Gsdma3 self-regulation causes autophagy and cell death [Internet] Biochem J. 2015. June;468(2):325–36. [DOI] [PubMed] [Google Scholar]
- Lin P, Lin H, Kuo C, Yang L. N-terminal functional domain of gasdermin A3 regulates mitochondrial homeostasis via mitochondrial targeting [Internet] J Biomed Sci. 2015. June;22:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jiang X, Gu P, Chen W, Zeng X, Gao X. Gsdma3 mutation causes bulge stem cell depletion and alopecia mediated by skin inflammation [Internet] Am J Pathol. 2012. February;180(2):763–74. [DOI] [PubMed] [Google Scholar]
- Lunny DP, Weed E, Nolan PM, Marquardt A, Augustin M, Porter RM. Mutations in gasdermin 3 cause aberrant differentiation of the hair follicle and sebaceous gland [Internet] J Invest Dermatol. 2005. March;124(3):615–21. [DOI] [PubMed] [Google Scholar]
- Runkel F, Marquardt A, Stoeger C, Kochmann E, Simon D, Kohnke B, et al. The dominant alopecia phenotypes bareskin, rex-denuded, and reduced coat 2 are caused by mutations in gasdermin 3 [Internet] Genomics. 2004. November;84(5):824–35. [DOI] [PubMed] [Google Scholar]
- Lin H, Lin P, Wu S, Yang L. Inducible expression of gasdermin A3 in the epidermis causes epidermal hyperplasia and skin inflammation [Internet] Exp Dermatol. 2015. November;24(11):897–9. [DOI] [PubMed] [Google Scholar]
- Lluis A, Schedel M, Liu J, Illi S, Depner M, von Mutius E, Kabesch M, Schaub B. Asthma-associated polymorphisms in 17q21 influence cord blood ORMDL3 and GSDMA gene expression and IL-17 secretion. J Allergy Clin Immunol [Internet]. 2011. Jun;127(6):158,1594.e6. [DOI] [PubMed] [Google Scholar]
- Yu J, Kang M, Kim B, Kwon J, Song Y, Choi W, et al. Polymorphisms in GSDMA and GSDMB are associated with asthma susceptibility, atopy and BHR [Internet] Pediatr Pulmonol. 2011. July;46(7):701–8. [DOI] [PubMed] [Google Scholar]
- de Vasconcelos NM, Van Opdenbosch N, Van Gorp H, Parthoens E, Lamkanfi M. Single-cell analysis of pyroptosis dynamics reveals conserved GSDMD-mediated subcellular events that precede plasma membrane rupture [Internet] Cell Death Differ. 2019. January;26(1):146–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platnich JM, Chung H, Lau A, Sandall CF, Bondzi-Simpson A, Chen H, Komada T, Trotman-Grant AC, Brandelli JR, Chun J, Beck PL, Philpott DJ, Girardin SE, Ho M, Johnson RP, MacDonald JA, Armstrong GD, Muruve DA. Shiga toxin/lipopolysaccharide activates caspase-4 and gasdermin D to trigger mitochondrial reactive oxygen species upstream of the NLRP3 inflammasome. Cell Rep [Internet]. 2018. Nov 06,;25(6):152,1536.e7. [DOI] [PubMed] [Google Scholar]
- Erener S, Pétrilli V, Kassner I, Minotti R, Castillo R, Santoro R, et al. Inflammasome-activated caspase 7 cleaves PARP1 to enhance the expression of a subset of NF-κB target genes [Internet] Mol Cell. 2012. April;46(2):200–11. [DOI] [PubMed] [Google Scholar]
- Akhter A, Gavrilin MA, Frantz L, Washington S, Ditty C, Limoli D, et al. Caspase-7 activation by the Nlrc4/ipaf inflammasome restricts legionella pneumophila infection [Internet] PLoS Pathog. 2009. April;5(4):e1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko V, Vitak N, Vajjhala PR, Vince JE, Stacey KJ. Caspase-1 is an apical caspase leading to caspase-3 cleavage in the AIM2 inflammasome response, independent of caspase-8. J Mol Biol [Internet]. 2018. 01 19,;430(2):238-47. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Wang S, Kuida K, Yuan J. Distinct downstream pathways of caspase-11 in regulating apoptosis and cytokine maturation during septic shock response [Internet] Cell Death Differ. 2002. October;9(10):1115–25. [DOI] [PubMed] [Google Scholar]
- Taabazuing CY, Okondo MC, Bachovchin DA. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem Biol [Internet]. 2017. Apr 20, [cited Jun 20, 2019];24(4):50,514.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op de Beeck K, Van Camp G, Thys S, Cools N, Callebaut I, Vrijens K, et al. Van Tendeloo, Viggo F. I., Timmermans JP, Van Laer L. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein [Internet] Eur J Hum Genet. 2011. September;19(9):965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossom S, Op de Beeck K, Franssens V, Swinnen E, Schepers A, Ghillebert R, et al. The splicing mutant of the human tumor suppressor protein DFNA5 induces programmed cell death when expressed in the yeast saccharomyces cerevisiae [Internet] Front Oncol. 2012;2:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tang L, Shen D, Wang C, Yuan Q, Gao W, et al. The expression and regulation of DFNA5 in human hepatocellular carcinoma DFNA5 in hepatocellular carcinoma [Internet] Mol Biol Rep. 2013. December;40(12):6525–31. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yin B, Li D, Wang G, Han X, Sun X. GSDME mediates caspase-3-dependent pyroptosis in gastric cancer. Biochem Biophys Res Commun [Internet]. 2018. 01 01,;495(1):1418-25. [DOI] [PubMed] [Google Scholar]
- Lage H, Helmbach H, Grottke C, Dietel M, Schadendorf D. DFNA5 (ICERE-1) contributes to acquired etoposide resistance in melanoma cells [Internet] FEBS Lett. 2001. April;494(1-2):54–9. [DOI] [PubMed] [Google Scholar]
- Van Rossom S, Op de Beeck K, Hristovska V, Winderickx J, Van Camp G. The deafness gene DFNA5 induces programmed cell death through mitochondria and MAPK-related pathways [Internet] Front Cell Neurosci. 2015;9:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer [Internet] Cell. 2000. January;100(1):57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation [Internet] Cell. 2011. March;144(5):646–74. [DOI] [PubMed] [Google Scholar]
- Saeki N, Kuwahara Y, Sasaki H, Satoh H, Shiroishi T. Gasdermin (gsdm) localizing to mouse chromosome 11 is predominantly expressed in upper gastrointestinal tract but significantly suppressed in human gastric cancer cells [Internet] Mamm Genome. 2000;11(9):718–24. [DOI] [PubMed] [Google Scholar]
- Saeki N, Usui T, Aoyagi K, Kim DH, Sato M, Mabuchi T, et al. Distinctive expression and function of four GSDM family genes (GSDMA-D) in normal and malignant upper gastrointestinal epithelium [Internet] Genes Chromosomes Cancer. 2009. March;48(3):261–71. [DOI] [PubMed] [Google Scholar]
- Akino K, Toyota M, Suzuki H, Imai T, Maruyama R, Kusano M, et al. Identification of DFNA5 as a target of epigenetic inactivation in gastric cancer [Internet] Cancer Sci. 2007. January;98(1):88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoll G, Ma Y, Yang H, Kepp O, Zitvogel L, Kroemer G. Pro-necrotic molecules impact local immunosurveillance in human breast cancer [Internet] OncoImmunology. 2017;6(4):e1299302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DA, Weigel RJ. Characterization of a gene that is inversely correlated with estrogen receptor expression (ICERE-1) in breast carcinomas [Internet] Eur J Biochem. 1998. February;252(1):169–77. [DOI] [PubMed] [Google Scholar]
- Croes L, de Beeck KO, Pauwels P, Vanden Berghe W, Peeters M, Fransen E, et al. DFNA5 promoter methylation a marker for breast tumorigenesis [Internet] Oncotarget. 2017. May;8(19):31948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lebron C, Nagpal JK, Chae YK, Chang X, Huang Y, et al. Methylation of the DFNA5 increases risk of lymph node metastasis in human breast cancer [Internet] Biochem Biophys Res Commun. 2008. May;370(1):38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Chang X, Yamashita K, Nagpal JK, Baek JH, Wu G, et al. Aberrant promoter methylation and tumor suppressive activity of the DFNA5 gene in colorectal carcinoma [Internet] Oncogene. 2008. June;27(25):3624–34. [DOI] [PubMed] [Google Scholar]
- Yokomizo K, Harada Y, Kijima K, Shinmura K, Sakata M, Sakuraba K, et al. Methylation of the DFNA5 gene is frequently detected in colorectal cancer [Internet] Anticancer Res. 2012. April;32(4):1319–22. [PubMed] [Google Scholar]
- Croes L, Beyens M, Fransen E, Ibrahim J, Vanden Berghe W, Suls A, et al. Large-scale analysis of DFNA5 methylation reveals its potential as biomarker for breast cancer [Internet] Clin Epigenetics. 2018;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang Y, Yang D, Gong Y, Rao F, Liu R, et al. PLK1 kinase inhibitor enhances the chemosensitivity of cisplatin by inducing pyroptosis in oesophageal squamous cell carcinoma [Internet] EBioMedicine. 2019. March [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda Y, Futamura M, Kamino H, Nakamura Y, Kitamura N, Ohnishi S, et al. The potential role of DFNA5, a hearing impairment gene, in p53-mediated cellular response to DNA damage [Internet] J Hum Genet. 2006;51(8):652–64. [DOI] [PubMed] [Google Scholar]
- Wang C, Tang L, Shen D, Wang C, Yuan Q, Gao W, et al. The expression and regulation of DFNA5 in human hepatocellular carcinoma DFNA5 in hepatocellular carcinoma [Internet] Mol Biol Rep. 2013. December;40(12):6525–31. [DOI] [PubMed] [Google Scholar]
- Lu H, Zhang S, Wu J, Chen M, Cai M, Fu Y, et al. Molecular targeted therapies elicit concurrent apoptotic and GSDME-dependent pyroptotic tumor cell death [Internet] Clin Cancer Res. 2018. December;24(23):6066–77. [DOI] [PubMed] [Google Scholar]
- Wang WJ, Chen D, Jiang MZ, Xu B, Li XW, Chu Y, et al. Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins [Internet] J Dig Dis. 2018. February;19(2):74–83. [DOI] [PubMed] [Google Scholar]
- Gao J, Qiu X, Xi G, Liu H, Zhang F, Lv T, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/akt signaling and predicts a good prognosis in non small cell lung cancer [Internet] Oncol Rep. 2018. October;40(4):1971–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama H, Aoki A, Tanaka S, Maekawa H, Kato Y, Wada R, et al. Alu-derived cis-element regulates tumorigenesis-dependent gastric expression of GASDERMIN B (GSDMB) [Internet] Genes Genet Syst. 2010. February;85(1):75–83. [DOI] [PubMed] [Google Scholar]
- Hergueta-Redondo M, Sarrio D, Molina-Crespo Á, Vicario R, Bernadó-Morales C, Martínez L, et al. Gasdermin B expression predicts poor clinical outcome in HER2-positive breast cancer [Internet] Oncotarget. 2016. August;7(35):56295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hergueta-Redondo M, Sarrió D, Molina-Crespo Á, Megias D, Mota A, Rojo-Sebastian A, et al. Gasdermin-B promotes invasion and metastasis in breast cancer cells [Internet] PLoS One. 2014;9(3):e90099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao KL, Kulakova L, Herzberg O. Gene polymorphism linked to increased asthma and IBD risk alters gasdermin-B structure, a sulfatide and phosphoinositide binding protein. Proc Natl Acad Sci U S A [Internet]. 2017. 02 14,;114(7):E112-E1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Meng X, Zhang S, Zhao G, Hu L, Kong X. A 3-nucleotide deletion in the polypyrimidine tract of intron 7 of the DFNA5 gene causes nonsyndromic hearing impairment in a chinese family [Internet] Genomics. 2003. November;82(5):575–9. [DOI] [PubMed] [Google Scholar]
- Park H, Cho H, Baek J, Ben-Yosef T, Kwon T, Griffith AJ, et al. Evidence for a founder mutation causing DFNA5 hearing loss in east asians [Internet] J Hum Genet. 2010. January;55(1):59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio A, Noguchi Y, Sato T, Naruse TK, Kimura A, Takagi A, et al. DFNA5 mutation identified in japanese families with autosomal dominant hereditary hearing loss [Internet] Ann Hum Genet. 2014. March;78(2):83–91. [DOI] [PubMed] [Google Scholar]
- Chai Y, Chen D, Wang X, Wu H, Yang T. A novel splice site mutation in DFNA5 causes late-onset progressive non-syndromic hearing loss in a chinese family [Internet] Int J Pediatr Otorhinolaryngol. 2014. August;78(8):1265–8. [DOI] [PubMed] [Google Scholar]
- Bischoff AM. Luijendijk MWJ, Huygen PLM, van Duijnhoven G, De Leenheer, Els M. R., Oudesluijs GG, Van Laer L, Cremers FPM, Cremers, Cor W. R. J., Kremer H. A novel mutation identified in the DFNA5 gene in a dutch family: A clinical and genetic evaluation [Internet] Audiol Neurotol. 2004. Jan-Feb;9(1):34–46. [DOI] [PubMed] [Google Scholar]
- Cheng J, Han DY, Dai P, Sun HJ, Tao R, Sun Q, et al. A novel DFNA5 mutation, IVS8+4 A>G, in the splice donor site of intron 8 causes late-onset non-syndromic hearing loss in a chinese family [Internet] Clin Genet. 2007. November;72(5):471–7. [DOI] [PubMed] [Google Scholar]
- Wang H, Guan J, Guan L, Yang J, Wu K, Lin Q, Xiong W, Lan L, Zhao C, Xie L, Yu L, Bing D, Zhao L, Wang D, Wang Q. Further evidence for “gain-of-function” mechanism of DFNA5 related hearing loss. Sci Rep [Internet]. 2018-05-30;8(1):8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer L, Huizing EH, Verstreken M, van Zuijlen D, Wauters JG, Bossuyt PJ, et al. Nonsyndromic hearing impairment is associated with a mutation in DFNA5 [Internet] Nat Genet. 1998. October;20(2):194–7. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Meyer NC, Malekpour M, Riazalhosseini Y, Moghannibashi M, Kahrizi K, et al. A novel DFNA5 mutation does not cause hearing loss in an iranian family [Internet] J Hum Genet. 2007;52(6):549–52. [DOI] [PubMed] [Google Scholar]
- Nadol JB, Handzel O, Amr S. Histopathology of the human inner ear in a patient with sensorineural hearing loss caused by a variant in DFNA5 [Internet] Otol Neurotol. 2015. December;36(10):1616–21. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Pfister M, Thys S, Vrijens K, Mueller M, Umans L, et al. Mice lacking Dfna5 show a diverging number of cochlear fourth row outer hair cells [Internet] Neurobiol Dis. 2005. August;19(3):386–99. [DOI] [PubMed] [Google Scholar]
- Op de Beeck K, Schacht J, Van Camp G. Apoptosis in acquired and genetic hearing impairment: the programmed death of the hair cell [Internet] Hear Res. 2011. November;281(1-2):18–27. [DOI] [PMC free article] [PubMed] [Google Scholar]