Abstract
In recent years, tissue-resident memory T cells (TRM) have emerged as essential components of immunological memory. Following antigenic challenge, TRM remain in nonlymphoid tissues and defend against re-exposure. Although accumulating evidence suggests important roles for TRM in mediating protective immunity, fundamental aspects of the population biology of TRM remain poorly understood. Here we discuss how results from different systems shed light on the ecological dynamics of TRM in mice and humans. We highlight the importance of dissecting processes contributing to TRM maintenance, and how these might vary across phenotypically and spatially heterogeneous subsets. We also discuss how the diversity of TRM communities within specific tissues may evolve under competition and in response to antigenic perturbation. Throughout, we illustrate how mathematical models can clarify inferences obtained from experimental data, and help elucidate the homeostatic mechanisms underpinning the ecology of TRM populations.
Introduction
Following primary infection and resolution of the associated cellular immune response, memory T cells are generated to protect against secondary exposures. It is well established that memory T cells comprise heterogeneous subsets. While central memory T cells (TCM) and effector memory T cells (TEM) circulate between the blood and secondary lymphoid organs (SLOs) or peripheral tissues, respectively, tissue-resident memory T cells (TRM) are found in diverse tissue sites and provide crucial defenses against previously encountered pathogens (1–13). TRM are widely dispersed throughout the body and have been associated with protection against viral and bacterial infections, anti-tumor immunity, and the pathology of allergic and autoimmune diseases (3, 7, 14–21).
TRM are phenotypically and transcriptionally distinct from TEM and TCM, and the factors involved in their differentiation and maintenance have been studied extensively (reviewed in refs. (22–28)). As shown in parabiosis experiments involving the conjoining of two mice, TRM persist within a wide variety of nonlymphoid tissues (NLT) (17, 29, 30). TRM have also been identified in draining SLOs following local infection in the female reproductive tract (FRT) and skin (31). Evidence that TRM persist in the presence of agents which deplete circulating peripheral T cells (e.g. FTY720 or anti-T cell antibodies) further indicates that TRM are non-circulating and largely maintained independently of circulating populations (14, 32, 33). While TRM are phenotypically heterogeneous, and there are no perfect markers of tissue residency, the most commonly associated marker in mice is CD69. Although CD69 is often attributed to recent antigenic stimulation, it may also function to retain TRM within NLT (22), and parabiosis experiments have confirmed its utility in distinguishing tissue-resident from circulating cells (11, 17, 25). Another frequently used marker is CD103, which is predominantly associated with CD8+ TRM (34), although CD103+ CD4+ TRM are found in skin and FRT (3, 34–37). Other markers, such as CD11a and CXCR3, are associated with the TRM phenotype in certain tissues (11, 32, 34, 38–40).
Despite this extensive characterization, many fundamental aspects of TRM biology remain poorly understood. For example, what determines the longevity of memory encoded by TRM? How does the presence of TRM impact the recruitment of new antigen-specific T cells upon exposure to related or unrelated pathogens? Is there competition between pre-existing and newly generated TRM, and if so what factors mediate this competition? To address these questions requires formulating a quantitative ecology of tissue-resident memory, so that we may understand how continual exposure to environmental and infectious antigens impacts the distribution, diversity and persistence of TRM at different sites across the body.
By pairing quantitative models with experimental observations one can test different hypotheses regarding cellular turnover and interactions, estimate quantities that may not be directly measured, and generate quantitative, testable predictions. Notably, mathematical models have elucidated mechanisms underlying the generation and resolution of effector T cell responses, and the maintenance of naive and circulating memory T cell populations (reviewed in ref. (41); see also, for example, refs. (42–52)). Further afield, there is a rich literature employing models to understand the ecological processes sustaining communities of plants, animals, and infectious agents (see, for example, refs. (53–56)). In this article we collate information regarding the ecological dynamics of TRM, including current quantitative estimates of growth and loss rates in different tissues. We first consider studies performed in mice, as these comprise the bulk of the work to date, and then discuss our understanding of TRM ecology in humans. Throughout this review we illustrate how mathematical tools can be harnessed to refine and enhance current experimental insights, and highlight open questions and areas for future work.
Identifying ontogenic pathways
The ontogeny of TRM across different tissues has not been fully characterized. Although, in general, it seems that TRM are enriched for relatively long-lived, quiescent cells in an early-differentiated state (57, 58), a definitive differentiation pathway remains elusive. One barrier to consensus in this area is that T cell responses are heterogeneous, and move through high-dimensional phenotypic trajectories, of which any given experiment only views a projection. However, by allowing us to frame mechanistic descriptions of the dynamics of proliferation, loss and differentiation, mathematical models can be used to explore competing hypotheses regarding patterns of T cell differentiation (45, 47, 59–61).
In addition to identifying precursor populations, quantifying the rate of TRM generation from these populations, and whether this rate changes over both short timescales (such as during the contraction phase of an immune response) and throughout life is important for understanding how TRM populations are established and maintained. Recent work suggests that host age modulates the rate of TRM generation, as activated T cells in infants show reduced capacity to differentiate into lung TRM compared to cells in adults (62). Such hypotheses can be explored using models that test the impact of host and/or cell age on population kinetics (49, 52, 63). For instance, a comparison of models describing different mechanisms of naive T cell homeostasis identified increasing cell survival with post-thymic age as the most likely determinant of population stability throughout life (49). Such methods could be used to dissect the roles of host and cell age in modulating TRM formation.
Discriminating between models of T cell responses in vivo, and of TRM ontogeny in particular, requires frequent sampling of the phenotypic profile of T cells during the acute, resolution and memory phases, both in the tissue and draining lymph nodes. Such data are typically cross-sectional, but even data from single cell fate-mapping experiments leave uncertainty regarding lineage relationships (45, 59, 64, 65). Nevertheless, if candidate mechanisms or pathways can be expressed as mathematical models, there exist statistical methods to compare the support for each (see, for example, ref. (66)). Such methods assess the ability of different models to describe observations while penalizing those which are too complex and tend to ‘over-fit’ – that is, those models which are overly tailored to the particulars of a given (inevitably noisy) dataset, and do not generalize well to data from other experiments. The most parsimonious models can thus be identified from a set of candidates, and used to infer biological parameters and underlying mechanisms. Additional data may then be used to test the predictions of models, and further assess their biological relevance.
Quantifying and dissecting contributions to TRM longevity
Perhaps the key motivation for understanding mechanisms of TRM development and maintenance is to optimize immunization protocols and maximize the longevity of tissue-resident immunity. Important questions include: does the longevity of TRM clones depend on antigen-specificity and/or tissue location? Does longevity map directly to the phenotypic heterogeneity of TRM? And to what extent does competition between TRM, either at the level of the whole population or among antigen-specific populations, influence longevity? To tackle these questions quantitatively, we first note that the longevity of a TRM population is determined by the balance between self-renewal, loss, and recruitment from circulating precursor populations (Box 1). In particular, if there is proliferative self-renewal of TRM, the lifetime of individual TRM cells may be shorter than the lifetime of the population as a whole. In what follows, then, we use the term ‘longevity’ to define the persistence of a population, combining the net effects of influx, division, and loss.
Box 1: Modeling TRM homeostasis.
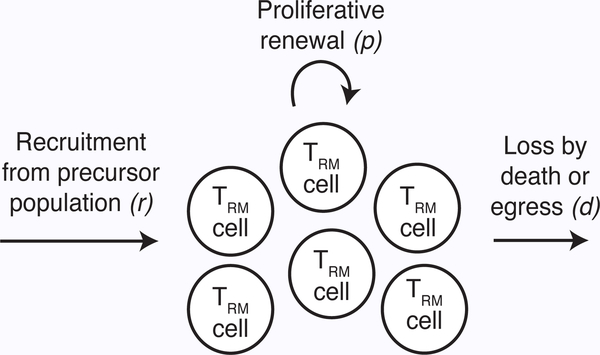
Consider a single homogeneous population of TRM, T. A simple model of homeostasis assumes that newly recruited TRM cells enter the population from a precursor subset at a constant rate, r. They then divide in situ at rate p and are lost at some rate d, which reflects both cell death and egress from the tissue (Fig. 1). The average residence time of an individual cell in the tissue is 1/d. These processes can be expressed mathematically as
| (1) |
where d − p is the net loss rate from the population, which is the balance of loss and self-renewal through division.
If there is no recruitment from precursor populations, then r = 0 and Eq. 1 can be solved to give
| (2) |
where T(0) is the initial number of TRM. Thus, assuming cells are lost at a faster rate than they divide (i.e. d > p), the population will decline exponentially at net loss rate (d − p). This quantity, rather than the residence time of any particular cell, is most relevant for quantifying the duration of immunity (i.e. the persistence of a T cell population that undergoes both loss and self-renewal). It follows that the average time for this population to halve in size is ln(2)/(d − p). One can use this net loss rate to estimate, for example, the time taken for the population size to fall below a defined threshold of protection.
Finally, if r 6= 0, then some cell loss will be masked by recruitment of new TRM. Models that fail to account for this recruitment will underestimate the net loss rate, (d − p), and thus overestimate the corresponding population halflife. If r is constant and d > p, the population will eventually reach an equilibrium, T*, where influx and renewal are balanced by loss. This steady-state population size can be found be setting dT/dt = 0, giving T* = r/(d − p).
Figure 1 –
Schematic of TRM homeostasis model. Cells are recruited at a constant rate, r, self-renew through division at rate p, and are lost at rate d, leading to exponentiallydistributed residence times within the tissue. Extensions of this model include different residence time distributions, or decomposing the decay rate into distinct terms for cell death and tissue egress (109).
Measuring longevity is hampered by the censoring of experimental information – estimates can be dependent on the duration of sampling. Nevertheless, broad patterns have emerged that indicate potential tissue-specificity in TRM longevity: populations in the lungs and liver persist for weeks to months (7, 11, 32, 67–70), whereas those in tissues such as the brain and skin may persist for months to years (Table 1) (4–6, 8, 12, 17, 35). While the longevity of TRM may also be phenotype-specific, as it is in circulating memory subsets (41), little is known in this regard. Despite evidence that CD8+ memory cell populations persist longer than CD4+ memory cells in tissues such as the skin and FRT (5, 35, 71, 72), there have been no direct comparisons of the longevity of CD4+ and CD8+ TRM. Similarly, although some evidence suggests regulatory CD4+ T cells, another distinct subset of resident memory, can persist for over 50 days in the skin and lungs, their population dynamics are, in general, more poorly defined than those of conventional CD4+ and CD8+ TRM (73, 74).
Table 1 – Experimental estimates of CD8+ TRM population parameters in different murine tissues and infection settings.
Time detectable represents the maximum observed duration of TRM persistence; tissues are approximately arranged in order of increasing estimates.
| Tissue | System | Time detectable |
Population half-life* |
Self renewal |
References |
|---|---|---|---|---|---|
| Lungs | Influenza | < 210d | 5–7d | No BrdU uptake detected | (7, 69, 79) |
| Liver | Malaria | > 100d | 28–36d | (11, 70) | |
| LCMV | > 120d | (17) | |||
| FRT | HSV | > 80d | (5, 35) | ||
| LCMV | > 120d | (17) | |||
| Intestine | YPTB | > 120d | (16) | ||
| LCMV | > 120d | (17) | |||
| Brain | VACV | > 120d | <1% BrdU+ in 1 wk | (76) | |
| LCMV | > 240d | 9% Ki67+ | (12) | ||
| Skin | VACV | > 160d | (6) | ||
| LCMV | > 200d | (69) | |||
| HSV | > 540d | <5% BrdU+ in 1wk | (3, 4, 58, 83) | ||
FRT = female reproductive tract; HSV = herpes simplex virus; LCMV = Lymphocytic choriomeningitis; VACV = vaccinia virus; YPTB = Yersinia pseudotuberculosis.
Half-life estimates do not account for any ongoing recruitment from circulating subsets.
Although experimental studies have provided valuable insights into longevity in different tissues, the majority measure TRM abundance over time and cannot untangle the contributions of recruitment, self-renewal, and loss. For instance, the rapid loss of influenza-specific TRM in the lung has been attributed to all three components of population longevity: high levels of apoptosis-driven loss, low levels of proliferation, and decreasing recruitment of circulating precursors over time (67, 69, 75). However, direct quantitative comparisons of the relative roles of each are currently lacking. Ultimately, to develop a mechanistic understanding of TRM homeostasis, we need to quantify the relative contributions of proliferation, recruitment, and loss. These factors are discussed in detail below.
The extent of TRM recruitment at steady state (in the absence of infection) varies across NLT, cell phenotypes, and possibly infections. Recently-generated CD4+ TRM in the lung, and CD8+ TRM in the brain and skin, remain stable in numbers in the absence of input from the circulation (32, 35, 76). In contrast, CD8+ TRM in the lung are at least partly replenished by circulating memory precursors (69, 77), and there may also be low level seeding in the FRT (17). Given the sensitivity of the above analyses to sampling times, and the potential for changes in the rate of recruitment from precursors over time (69), further work is needed to measure the long-term contribution of recruitment to TRM maintenance.
Although TRM populations expand efficiently in response to antigenic restimulation (12, 58, 78), the extent to which they self-renew at steady state is unclear. BrdU-labeling experiments have indicated that low-level proliferation occurs in the lung, skin, and brain, with 0–5% BrdU uptake over seven days (3, 69, 76). However, it has also been shown that a more substantial fraction (around 9%) of established TRM in the brain express Ki67, a marker of recent division (12). Evidence that these Ki67hi cells are established TRM, rather than recently-divided immigrants, implies a more important role for self-renewal in certain tissues (12, 76).
Finally, loss of TRM can result from cell death (turnover) or tissue egress. In some systems, such as the lungs, TRM express lower levels of the anti-apoptotic protein Bcl-2, and increased levels of the pro-apoptotic caspase 3/7, than their circulating counterparts, suggesting impaired cell survival (69). However, this impairment is not apparent for TRM in the skin, illustrating that loss rates likely vary across different tissues and cell phenotypes.
Combining these experimental observations with quantitative models will enable (i) characterization of homeostatic dynamics as cells die, divide, and/or differentiate; and (ii) rates of recruitment, self-renewal, and loss to be inferred (41, 46, 48). Although some quantitative methods have been employed to infer population half-lives in the liver and lungs (Table 1; see refs. (11, 70, 79)), these may be overestimates as cell recruitment was not taken into account (Box 1). To discriminate self-renewal of existing TRM from the recruitment of new cells, one can use fate-mapping systems, or adoptive transfer of congenically labeled T cells prior to secondary challenge. As an example, the former has been employed to track the differentiation of congenically labeled cells that develop normally in the thymus and are released into a lymphoreplete periphery (47). In combination with BrdU-labeling, models were used to quantify heterogeneity in the dynamics of effector and memory CD4+ T cells, and the rate of naive cell recruitment into these populations. Applying similar approaches to TRM could disentangle the processes of recruitment and renewal, and identify heterogeneity in population dynamics. In particular, heterogeneity is suggestive of TRM subpopulations with distinct homeostatic niches (Box 2), and the combination of modeling and fate-mapping may reveal whether these coexist independently or whether there is interconversion between them. These methods also allow projection of TRM population dynamics beyond typical experimental timecourses, to develop a more complete picture of TRM homeostasis and persistence (Box 3).
Box 2. Heterogeneity in TRM populations.
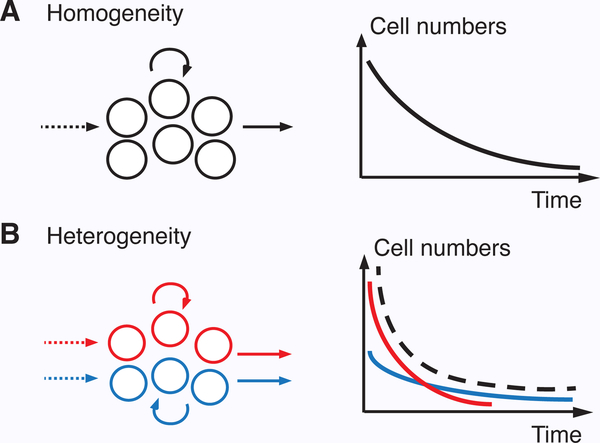
The homogeneous model of TRM homeostasis (Eq. 1) assumes the rates of input, loss, and division are constant over time, and across all cells in the population. However, these assumptions may not always hold. For instance, input rates may wane over time, as has been found in the lung (69), and phenotypic heterogeneity may lead to different rates of division and/or loss, as has been found in other T cell subsets (43, 47). Such mechanisms can lead to heterogeneous population dynamics. For example, differences in net loss rates among phenotypic subsets can manifest as multiple phases of exponential decline, rather than the single phase of a homogeneous population (Fig. 2) (46). By modifying Eq. 1 to reflect these scenarios, heterogeneity in TRM ontogeny and homeostasis can be identified and compared (although the ability to detect such heterogeneity requires sufficient data, such as detailed timecourses from fate-mapping and DNA labeling) (47, 110, 111).
Figure 2 – Impact of heterogeneity on population dynamics.
(A) Assuming precursor input is negligible, homogeneity in the net loss rate across the population leads to a single exponential decay function (Eq. 2). (B) Conversely, if there is heterogeneity in net loss among cell subsets (e.g. due to different death rates), then the observed decay (dashed line) reflects the sum of the decay kinetics of each subset (red and blue lines).
Box 3: Mathematical analysis of published data.
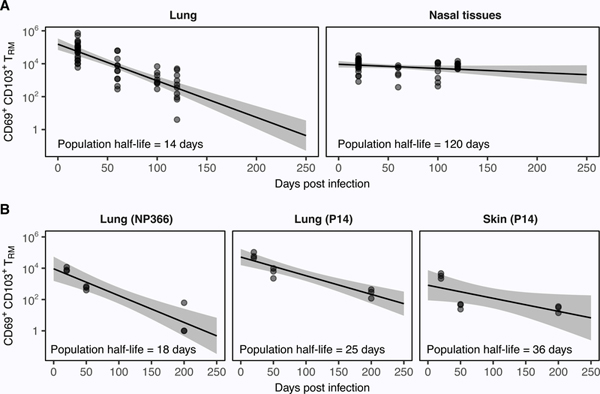
To illustrate the insights that can be gained from mathematical modeling, we collated data from two independent studies of CD8+ TRM persistence in mice. One describes the number of OT-I TRM in the lungs and nasal tissues (NT) from 20–120 days following infection with the recombinant X31-OVA strain of influenza virus (68). The other follows the number of P14 TRM in the skin and lungs 20–200 days after infection with either Vaccinia-GP33 (skin) or PR8-GP33 (lungs) (69). The latter study also follows the endogenous TRM response to the NP366 protein following infection with the PR8 strain of influenza.
Due to sparse information on the extent of TRM recruitment in each tissue, we make the simplifying assumption that r = 0; i.e. influx is negligible over the experimental time frame. Although this assumption may be applicable to the NT and skin, it is unlikely to hold strongly in the lung (35, 68, 69). We also assume cells within each population are homogeneous with respect to their rates of division (p) and loss (d). TRM decline can then be modeled using Eq. 2 to estimate the half-life of the population (Fig. 3). If in situ proliferation is minimal, then p ≈ 0 and the net loss rate of the population (d − p) is equivalent to the loss rate of individual cells (d); this may be the case for lung and NT TRM (68, 69). More generally, however, p ≠ 0 (12), and distinguishing the contributions of p and d to the net loss rate requires further data regarding TRM self-renewal (e.g. Ki67 expression), death, or egress. Note also that the net loss rate will be underestimated if there is significant TRM recruitment (Box 1).
Importantly, mathematical models can be used to project population dynamics beyond experimental time frames. For example, our model predicts that OT-I (Fig. 3A) and NP366 (Fig. 3B) TRM cells will become undetectable in 230 and 210 days, respectively. These estimates are in agreement with the observation that lung TRM are undetectable seven months (approximately 210 days) after infection (7).
Figure 3 – Mathematical analysis of published TRM data.
Eq. 2 fitted to data from refs. (68) (A) and (69) (B). Solid lines are the model predictions, points are the data, and grey shaded regions are prediction envelopes obtained by sampling parameters from their 95% confidence intervals. Least-squares optimization was performed on the log-scale and residuals were normally-distributed (using the Shapiro-Wilk test), except those for the NT. The NT fits are thus included for approximate comparison only. Note that we use the above approach to illustrate how a simple model can be applied to experimental data. In a more thorough analysis, these predictions would then be compared with those from alternative models to identify the most parsimonious description of the data. Code for the above analysis can be found at https://github.com/SineadMorris/TRMecology.
TRM competition and community composition
Community composition refers to the diversity and spatial distribution of cells coexisting within a tissue. This composition may be altered if TRM compete with other cells (including other TRM subsets) within their local niche. In the following section we consider competition and its subsequent impact on cell diversity within NLT, first by focusing on competition arising during homeostasis, or following a single inflammatory response, and then by considering how populations change across multiple exposures.
First, it is important to understand the extent to which TRM compete with one another for homeostatic resources. In particular, competition implies the existence of a ‘carrying capacity’ that may limit the amount of TRM a tissue can support. Such a restriction on population size may be important in the skin following HSV challenge, where TRM numbers eventually plateau with increasing numbers of transferred effector cells, despite a continued increase in numbers in the spleen (58). Although saturation in TRM formation must be carefully distinguished from saturation of other upstream immune responses (80), identifying potential carrying capacities is important as the density of antigen-specific TRM within a tissue can influence the efficacy of protection (11, 58). Models can provide further insights into these systems by exploring the density of TRM required to provide effective protection and predicting when populations will drop below this threshold (79, 81).
One mechanism that may drive cell competition is cytokine-mediated signaling. For example, TRM maintenance and homeostatic division in the skin, salivary glands, liver, and kidney may be IL-15 dependent, with IL-15 deficiency leading to significant reductions in cell numbers (34, 70, 82–84). However, these data do not address whether IL-15 is limiting at steady state and thus are not direct evidence for cytokine-mediated competition under normal conditions. Another mechanism through which TRM may compete is antigen availability. If TRM formation is antigen-dependent, immunodominance hierarchies can arise amongst the newly generated population: more abundant epitopes stimulate greater numbers of the corresponding clone. Such hierarchies have been observed in the lungs, skin, and FRT (3, 8, 68, 85, 86). The clonal composition of TRM at a tissue site may be further shaped by influx from memory precursors (17, 69) and subsequent antigen exposure (78).
One caveat to the above discussion is that most studies of TRM maintenance use specific pathogen-free (SPF) or germ-free mice, which are typically isolated from exposure to environmental antigens. As such, the density and diversity of TRM in tissues may be relatively low. In contrast, greater insights may be gained from so-called ‘dirty mice’ which are exposed to more diverse environmental antigens than those in traditional SPF facilities, and in which TRM are typically found at greater densities (31). The populated tissues of these mice may more closely reflect normal conditions, and thus provide a better system to study competition between cells.
With respect to sequential exposures, multiple antigenic stimuli may change the diversity of the TRM community as newly recruited and pre-existing cells compete for resources within a particular niche (87). In the skin, proliferation of pre-existing TRM following HSV rechallenge is accompanied by increased apoptosis, suggesting prior population expansion may be controlled during recall responses (58). Despite this, a stable population of original cells can coexist with new recruits following either homologous or heterologous infection (58). In the FRT, TRM numbers increase following consecutive homologous challenge, primarily due to in situ proliferation of pre-existing cells (78). Finally in the liver, pre-existing and newly established TRM coexist following consecutive transfers of three heterologous populations (70). Together these results suggest heterogeneous TRM populations can persist in a number of NLT following multiple exposure events. However, the extent to which this applies over longer timescales, across other sites, and against more biologically relevant exposure histories (e.g. SPF vs dirty mice) has yet to be determined.
An additional challenge in understanding community composition is to identify the specific rules of replacement – is TRM homeostasis democratic, in the sense that all specificities or phenotypes compete equally for space and/or homeostatic resources in the absence of antigen? Do pre-existing TRM tend to exclude new immigrants, or do newer cells have a competitive advantage? Does competition take place on a global level (independent of antigen specificity), or are there antigen-specific niches, created either by persistent antigen, or cell congregation at previous sites of infection? These questions can be addressed by further measuring the persistence of epitope-specific TRM after multiple exposures to related and heterologous pathogens (70). Such insights will be of critical importance in understanding how host immunity changes in response to sequential infections.
The ecology of tissue-resident memory in humans
TRM in humans are less well studied, primarily due to difficulties in sampling NLT. Although in vivo challenge experiments have been carried out with healthy volunteers (10, 88), such studies are generally rare. Nonetheless, recent insights have been gained from a variety of valuable sources, including tissues sampled from organ donors and patients undergoing surgical procedures (86, 89–94). These snapshots in time reveal the diversity of TRM populations that accumulate throughout life in response to multiple pathogen exposures. In addition, they complement studies of germ-free and SPF mice that do not consider dynamics in the context of pre-existing immune landscapes.
In general, the phenotypes of human TRM are relatively well-characterized. Despite differences in the transcription factors regulating their differentiation (95–99), human TRM share core transcriptional and phenotypic profiles with their murine counterparts, including high expression of CD69 and enrichment for CD103 in CD8+ populations (86, 90, 96, 100–104). Additional surface markers, including CD49a, CXCR6, and CD101, are also enriched at multiple sites (96, 105). Another notable finding from human studies is that TRM are more widely distributed than in laboratory mice: TRM have been identified in human lymphoid tissues (e.g. spleen, lymph nodes, and tonsils) in addition to NLT, and are the most abundant T cell subset at these sites (90, 96). This greater dissemination may reflect increased exposure to environmental antigens (103), and is consistent with recent findings that TRM accumulate in the draining lymph nodes of mice following repeated antigenic stimulation (31). Overall, these studies highlight the importance of considering environment and host ecology when developing a conceptual framework of TRM dynamics.
Determining the dynamics of TRM maintenance in human tissues is particularly challenging given the difficulty of tracking cell populations over time. Nevertheless, cross-sectional analyses of mucosal and lymphoid tissues has revealed that TRM are maintained at remarkably stable frequencies from childhood well into old age, at levels that are tissue-specific (106, 107). Important insights have also been gained from longitudinal studies. Firstly, TRM clones specific for the allergen diphenylcyclopropenone (DPCP) persist in healthy human skin for at least four months (10). Secondly, donor TRM persist in lung and intestine transplant recipients for over one year following transplantation (93, 94). Collectively, these findings suggest stable maintenance of human TRM, although quantitative estimates from other tissues and antigens will be required before broad conclusions can be drawn.
The relative contributions of recruitment, self-renewal, and loss to overall population maintenance are also not well known. However, the gradual formation of recipient TRM following lung transplantation suggests continual recruitment can occur (94). In addition, cross-sectional data from donor tissues indicate homeostatic proliferation of resident CD8+ and CD4+ cells is detectible, but low, across mucosal sites (91). In particular, few resident skin and intestine cells express Ki67 (93, 108), and expression levels in the lung are less than 8% (96). Although these observations suggest relatively low average rates of homeostatic self-renewal, recent partitioning of TRM based on their ability to efflux fluorescent dye has revealed underlying differences in Ki67 expression: cells capable of efflux (efflux+) show reduced rates of proliferation compared to efflux− cells (106). Moreover, the efflux+ subset exhibited increased CD127 expression and enhanced IL-7 signaling following ex vivo stimulation, suggesting greater survival capacity. Together, these findings suggest important heterogeneity in cell quiescence and survival that modulates TRM longevity at the population level.
Finally, with respect to competition and community composition, humans are exposed to a greater number and diversity of environmental antigens than germ-free or SPF mice. Thus, one important question is how the accumulation and composition of different TRM communities is impacted by these more frequent and multifaceted challenges. Recent sampling of donor tissue suggests a diverse range of influenza-specific TRM clonotypes can be maintained in the adult human lung (86). A high degree of TCR diversity has also been observed in healthy human skin, and is largely maintained in vitro during IL-2 and IL-15 induced expansion (89). These findings hint that human NLT can support an array of coexisting TRM communities. Mapping the dynamics of such communities, in the face of differential individual exposure history and genetic background, will be a key challenge for the future. In the absence of longitudinal sampling, one approach may be to use quantitative imaging of human tissue sections, including markers of proliferation and apoptosis, to establish the spatial organization of TRM in different organs and how self-renewal and turnover reflect tissue location and local TRM densities.
Concluding remarks
TRM are a crucial component of the adaptive immune system and provide frontline defenses against invading pathogens. Understanding the mechanisms underlying population persistence and community composition is therefore crucial to enhancing immune memory and improving vaccine-mediated protection. Although recent experimental studies have provided many insights into the molecular factors governing cell differentiation and maintenance, a broader picture of TRM ecology is still lacking. Mathematical modeling can help bridge this gap by providing a quantitative framework for exploring multiple different aspects of TRM ecology. In particular, integrating models with data will allow us to disentangle the contributions of recruitment, proliferation, and loss to TRM longevity, and project population dynamics beyond current experimental limits. Success in this endeavor will require longitudinal data from NLT and SLOs – a particularly challenging prospect for human studies.
References
- 1.Sallusto F, Lenig D, Forster R, Lipp M, and Lanzavecchia A. 1999. Two subsets of memory Tlymphocytes with distinct homing potentials. Nature 401: 708–712. [DOI] [PubMed] [Google Scholar]
- 2.Masopust D, Vezys V, Marzo AL, and Lefran Lçois. 2001. Preferential localization of effectormemory cells in nonlymphoid tissue. Science 291: 2413–2417. [DOI] [PubMed] [Google Scholar]
- 3.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, and Carbone FR. 2009. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10: 524–530. [DOI] [PubMed] [Google Scholar]
- 4.Ariotti S, Beltman JB, Chodaczek G, Hoekstra ME, van Beek AE, Gomez-Eerland R, Ritsma L, van Rheenen J, Maree AFM, Zal T, et al. 2012. Tissue-resident memory CD8+ T cells continuously patrol skin epithelia to quickly recognize local antigen. Proc. Natl. Acad. Sci. 109: 19739–19744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shin H and Iwasaki A. 2012. A vaccine strategy that protects against genital herpes by establishinglocal memory T cells. Nature 491: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, and Kupper TS. 2012. Skin infectiongenerates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature 483: 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu T, Hu Y, Lee Y-T, Bouchard KR, Benechet A, Khanna K, and Cauley LS. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95: 215–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iijima N and Iwasaki A. 2014. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 346: 93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheridan BS, Pham QM, Lee YT, Cauley LS, Puddington L, and Lefran¸ois L. 2014. Oralinfection drives a distinct population of intestinal resident memory CD8+ T cells with enhanced protective function. Immunity 40: 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, Robins H, Krueger JG,Clark RA, and Kupper TS. 2015. Common clonal origin of central and resident memory T cells following skin immunization. Nat. Med. 21: 647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V,Cozijnsen A, Collins N, et al. 2016. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45: 889–902. [DOI] [PubMed] [Google Scholar]
- 12.Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, Drexler I, Pinschewer D, Korn T, and Merkler D. 2016. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J. Exp. Med. 213: 1571–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zens KD, Chen JK, and Farber DL. 2016. Vaccine-generated lung tissue-resident memory Tcells provide heterosubtypic protection to influenza infection. JCI Insight 1: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA,Barber DL, Kawamura KS, et al. 2010. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207: 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. 2012. Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J. Immunol. 188: 4866–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergsbaken T and Bevan MJ. 2015. Proinflammatory microenvironments within the intestineregulate the differentiation of tissue-resident CD8+ T cells responding to infection. Nat. Immunol. 16: 406–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, and Masopust D. 2015. Quantifying memory CD8 T cells reveals regionalization of immunosurveillance. Cell 161: 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molodtsov A and Turk MJ. 2018. Tissue resident CD8 memory T cell responses in cancer andautoimmunity. Front. Immunol. 9: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muruganandah V, Sathkumara HD, Navarro S, and Kupz A. 2018. A systematic review: Therole of resident memory T cells in infectious diseases and their relevance for vaccine development. Front. Immunol. 9: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, et al. 2018. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24: 986–993. [DOI] [PubMed] [Google Scholar]
- 21.Park SL, Buzzai A, Rautela J, Hor JL, Hochheiser K, Effern M, McBain N, Wagner T, Edwards J, McConville R, et al. 2019. Tissue-resident memory CD8+ T cells promote melanoma-immune equilibrium in skin. Nature 565: 366–371. [DOI] [PubMed] [Google Scholar]
- 22.Park CO and Kupper TS. 2015. The emerging role of resident memory T cells in protectiveimmunity and inflammatory disease. Nat. Med. 21: 688–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosato PC, Beura LK, and Masopust D. 2017. Tissue resident memory T cells and viralimmunity. Curr. Opin. Virol. 22: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milner JJ and Goldrath AW. 2018. Transcriptional programming of tissue-resident memoryCD8+ T cells. Curr. Opin. Immunol. 51: 162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takamura S 2018. Niches for the long-term maintenance of tissue-resident memory T cells. Front. Immunol. 9: 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topham DJ and Reilly EC. 2018. Tissue-resident memory CD8+T cells: From phenotype tofunction. Front. Immunol. 9: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Wu P, Shen Y, Jiang X, and Xu F. 2018. CD8+ resident memory T cells and viralinfection. Front. Immunol. 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szabo PA, Miron M, and Farber DL. 2019. Location, location, location: Tissue resident memory T cells in mice and humans. Sci. Immunol. 4: eaas9673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masopust D and Schenkel JM. 2013. The integration of T cell migration, differentiation andfunction. Nat. Rev. Immunol. 13: 309–320. [DOI] [PubMed] [Google Scholar]
- 30.Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, et al. 2016. Interleukin-2-dependent allergen-specific tissue-resident memory cells drive asthma. Immunity 44: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ,Schenkel JM, Mitchell JS, Vezys V, Fife BT, et al. 2018. T Cells in nonlymphoid tissues give rise to lymph-node-resident memory T cells. Immunity 48: 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner DL, Bickham KL, Thome JJ, Kim CY, D’Ovidio F, Wherry EJ, and Farber DL.2014. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal Immunol. 7: 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, Carbone FR, and Gebhardt T. 2015. Cutting Edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J. Immunol. 194: 2059–2063. [DOI] [PubMed] [Google Scholar]
- 34.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon M-L, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. 2013. The developmental pathway for CD103+CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14: 1294–1301. [DOI] [PubMed] [Google Scholar]
- 35.Gebhardt T, Whitney PG, Zaid A, MacKay LK, Brooks AG, Heath WR, Carbone FR,and Mueller SN. 2011. Different patterns of peripheral migration by memory CD4+and CD8+T cells. Nature 477: 216–219. [DOI] [PubMed] [Google Scholar]
- 36.Bromley SK, Yan S, Tomura M, Kanagawa O, and Luster AD. 2013. Recirculating memoryT cells are a unique subset of CD4+ T cells with a distinct phenotype and migratory pattern. J. Immunol. 190: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iijima N and Iwasaki A. 2015. Tissue instruction for migration and retention of TRM cells. Trends Immunol. 36: 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner DL and Farber DL. 2014. Mucosal resident memory CD4 T cells in protection andimmunopathology. Front. Immunol. 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai S, Kauffman KD, Schenkel JM, McBerry CC, Mayer-Barber KD, Masopust D, andD. L. Barber. 2014. Cutting Edge: Control of mycobacterium tuberculosis infection by a subset of lung parenchyma-homing CD4 T cells. J. Immunol. 192: 2965–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, andS. M. Kaech. 2014. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41: 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Boer RJ and Perelson AS. 2013. Quantifying T lymphocyte turnover. J. Theor. Biol. 327: 45–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vezys V, Yates A, Casey KA, Lanier G, Ahmed R, Antia R, and Masopust D. 2009. MemoryCD8 T-cell compartment grows in size with immunological experience. Nature 457: 196–199. [DOI] [PubMed] [Google Scholar]
- 43.Mandl JN, Liou R, Klauschen F, Vrisekoop N, Monteiro JP, Yates AJ, Huang AY, and Germain RN 2012. Quantification of lymph node transit times reveals differences in antigen surveillance strategies of naive CD4+ and CD8+ T cells. Proc. Natl. Acad. Sci. 109: 18036–18041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao H, Jin X, Perelson AS, and Wu H. 2012. Evaluation of multitype mathematical modelsfor CFSE-labeling experiment data. Bull. Math. Biol. 74: 300–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buchholz VR, Flossdorf M, Hensel I, Kretschmer L, Weissbrich B, Gräf P, Verschoor A, Schiemann M, ofer TH, and Busch DH. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science 340: 630–635. [DOI] [PubMed] [Google Scholar]
- 46.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature 552: 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gossel G, Hogan T, Cownden D, Seddon B, and Yates AJ. 2017. Memory CD4 T cell subsetsare kinetically heterogeneous and replenished from naive T cells at high levels. Elife 6: e23013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.del Amo PC, Beneytez JL, Boelen L, Ahmed R, Miners KL, Zhang Y, Roger L, Jones RE, Marraco SA, Speiser DE, et al. 2018. Human TSCM cell dynamics in vivo are compatible with long-lived immunological memory and stemness. PLoS Biol. 16: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rane S, Hogan T, Seddon B, and Yates AJ. 2018. Age is not just a number: Naive T cellsincrease their ability to persist in the circulation over time. PLoS Biol. 16: e2003949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seddon B and Yates AJ. 2018. The natural history of naive T cells from birth to maturity.Immunol. Rev. 285: 218–232. [DOI] [PubMed] [Google Scholar]
- 51.Pandit A and de Boer RJ. 2019. Stochastic inheritance of division and death times determinesthe size and phenotype of CD8+ T cell families. Front. Immunol. 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reynaldi A, Smith NL, Schlub TE, Tabilas C, Venturi V, Rudd BD, and Davenport MP.2019. Fate mapping reveals the age structure of the peripheral T cell compartment. Proc. Natl. Acad. Sci. 116: 3974–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levin SA, Hallam TG, and Gross LJ. 1989. Applied Mathematical Ecology. Springer-Verlag,Berlin Heidelberg. [Google Scholar]
- 54.Anderson RM and May RM. 1991. Infectious Diseases of Humans: Dynamics and Control.Oxford University Press, Oxford, UK: ISBN 0198545991. [Google Scholar]
- 55.Hilborn R and Mangel M. 1997. The Ecological Detective. Princeton University Press, Princeton,USA: ISBN 9780691034973. [Google Scholar]
- 56.Otto SP and Day T. 2007. A Biologist’s Guide to Mathematical Modeling in Ecology and Evolution. Princeton University Press, Princeton, USA: ISBN 9780691123448. [Google Scholar]
- 57.Chang JT, Wherry EJ, and Goldrath AW. 2014. Molecular regulation of effector and memoryT cell differentiation. Nat. Immunol. 15: 1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL,Russell TA, Gebhardt T, et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses article. Nat. Immunol. 19: 183–191. [DOI] [PubMed] [Google Scholar]
- 59.Arsenio J, Kakaradov B, Metz PJ, Kim SH, Yeo GW, and Chang JT. 2014. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell geneexpression analyses. Nat. Immunol. 15: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pandit A and De Boer RJ. 2019. Stochastic Inheritance of Division and Death Times Determinesthe Size and Phenotype of CD8+ T Cell Families. Front Immunol 10: 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hogan T, Nowicka M, Cownden D, Pearson C, Yates AJ, and Seddon B. 2019. Differentialimpact of self and environmental antigens on the ontogeny and maintenance of CD4+ T cell memory. bioRxiv BIORXIV/2019/632281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zens KD, Chen JK, Guyer RS, Wu FL, Cvetkovski F, Miron M, and Farber DL.2017. Reduced generation of lung tissue-resident memory T cells during infancy. J. Exp. Med. 214: 2915–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hogan T, Gossel G, Yates AJ, and Seddon B. 2015. Temporal fate mapping reveals age-linkedheterogeneity in naive T lymphocytes in mice. Proc. Natl. Acad. Sci. 112: E6917–E6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerlach C, Rohr JC, Perié L, van Rooij N, van Heijst JWJ, Velds A, Urbanus J, Naik SH, Jacobs H, Beltman JB, et al. 2013. Heterogeneous differentiation patterns of individual CD8+ T cells. Science 890: 635–640. [DOI] [PubMed] [Google Scholar]
- 65.Flossdorf M, Rossler J, Buchholz VR, Busch DH, and Hofer T. 2015. CD8+ T cell diversification by asymmetric cell division. Nat. Immunol. 16: 891–893. [DOI] [PubMed] [Google Scholar]
- 66.Burnham KP and Anderson DR. 2002. Model Seleciton and Multimodel Inference: A Practical Information-Theoretic Approach. Springer-Verlag, New York, USA, second edition ISBN 0387953647. [Google Scholar]
- 67.Shen C-H, Ge Q, Talay O, Eisen HN, Garcia-Sastre A, and Chen J. 2008. Loss of IL-7R andIL-15R expression is associated with disappearance of memory T cells in respiratory tract following influenza infection. J. Immunol. 180: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pizzolla A, Nguyen THO, Smith JM, Brooks AG, Kedzierska K, Heath WR, Reading PC, and Wakim LM. 2017. Resident memory CD8+ T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci. Immunol. 2: eaam6970. [DOI] [PubMed] [Google Scholar]
- 69.Slütter B, Van Braeckel-Budimir N, Abboud G, Varga SM, Salek-Ardakani S, and Harty JT.2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2: eaag2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holz LE, Prier JE, Freestone D, Bertolino P, Fernandez-Ruiz D, and Heath WR. 2018. CD8+ T cell activation leads to constitutive formation of liver tissue-resident memory T cells that seed a large and flexible niche in the liver. Cell Rep. 25: 68–79. [DOI] [PubMed] [Google Scholar]
- 71.Beura LK, Fares-Frederickson NJ, Steinert EM, Scott MC, Thompson EA, Fraser KA, Schenkel JM, Vezys V, and Masopust D. 2019. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216: 1214–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carbone FR and Gebhardt T. 2019. Should I stay or should I go–Reconciling clashing perspectiveson CD4+ tissue-resident memory T cells. Sci. Immunol. 4: eaax5595. [DOI] [PubMed] [Google Scholar]
- 73.Sanchez AM, Zhu J, Huang X, and Yang Y. 2012. The development and function of memoryregulatory T cells after acute viral infections. J. Immunol. 189: 2805–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenblum MD, Way SS, and Abbas AK. 2016. Regulatory T cell memory. Nat. Rev. Immunol. 16: 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reagin KL and Klonowski KD. 2018. Incomplete memories: The natural suppression of tissue-resident memory CD8 T cells in the lung. Front. Immunol. 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wakim LM, Woodward-Davis A, and Bevan MJ. 2010. Memory T cells persisting within thebrain after local infection show functional adaptations to their tissue of residence. Proc. Natl. Acad. Sci. 107: 17872–17879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takamura S and Kohlmeier JE. 2019. Establishment and maintenance of conventional and circulation-driven lung-resident memory CD8+ T cells following respiratory virus infections. Front. Immunol. 10: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S,Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. 2018. Intravital mucosal imaging of CD8+resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory article. Nat. Immunol. 19: 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zarnitsyna VI, Handel A, McMaster SR, Hayward SL, Kohlmeier JE, and Antia R.2016. Mathematical model reveals the role of memory CD8 T cell populations in recall responses to influenza. Front. Immunol. 7: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Badovinac VP, Haring JS, and Harty JT. 2007. Initial T cell receptor transgenic cell precursorfrequency dictates critical aspects of the CD8+ T cell response to infection. Immunity 26: 827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kadolsky UD and Yates AJ. 2015. How is the effectiveness of immune surveillance impactedby the spatial distribution of spreading infections? Philos. Trans. R. Soc. London B Biol. Sci. 370: e20140289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verbist KC, Field MB, and Klonowski KD. 2011. Cutting Edge: IL-15-independent maintenance of mucosally generated memory CD8 T cells. J. Immunol. 186: 6667–6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, Braun A, Masson F, Kallies A, Belz GT, et al. 2015. T-box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity 43: 1101–1111. [DOI] [PubMed] [Google Scholar]
- 84.Schenkel JM, Fraser KA, Casey KA, Beura LK, Pauken KE, Vezys V, and Masopust D.2016. IL-15-independent maintenance of tissue-resident and boosted effector memory CD8 T cells. J. Immunol. 196: 3920–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Muschaweckh A, Buchholz VR, Fellenzer A, Hessel C, König P-A, Tao S, Tao R, Heikenwälder M, Busch DH, Korn T, et al. 2016. Antigen-dependent competition shapes the local repertoire of tissue-resident memory CD8+ T cells. J. Exp. Med. 2013: 3075–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pizzolla A, Nguyen TH, Sant S, Jaffar J, Loudovaris T, Mannering SI, Thomas PG, Westall GP, Kedzierska K, and Wakim LM. 2018. Influenza-specific lung-resident memory t cells are proliferative and polyfunctional and maintain diverse TCR profiles. J. Clin. Invest. 128: 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Souquette A and Thomas PG. 2018. Past life and future effects-how heterologous infections alterimmunity to influenza viruses. Front. Immunol. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, Almond M, Wong EH, Sykes A, Maybeno M, et al. 2015. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat. Commun. 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clark RA, Chong BF, Mirchandani N, Yamanaka K-I, Murphy GF, Dowgiert RK, and Kupper TS. 2006. A novel method for the isolation of skin resident T cells from normal and diseased human skin. J. Invest. Dermatol. 126: 1059–1070. [DOI] [PubMed] [Google Scholar]
- 90.Sathaliyawala T, Kubota M, Yudanin N, Turner DL, Camp P, Thome JJ, Bickham KL,Lerner H, Goldstein M, Sykes M, et al. 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity 38: 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thome JJ, Yudanin N, Ohmura Y, Kubota M, Grinshpun B, Sathaliyawala T, Kato T, Lerner H, Shen Y, and Farber DL. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell 159: 814–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, and Kupper TS. 2011. Residentmemory T cells (TRM) are abundant in human lung: Diversity, function, and antigen specificity. PLoS One 6: e16245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Casado RB, Landsverk OJB, Chauhan SK, Richter L, Phung D, Greiff V, Risnes LF,Yao Y, Neumann RS, Yaqub S, et al. 2019. Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med. jem.20190414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, Carpenter D, Marboe C, Benvenuto L, Shah L, et al. 2019. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci. Immunol. 4: eaav5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mackay LK, Minnich M, Kragten NAM, Liao Y, Freestone D, Braun A, Wynne-jones E,Behr FM, and Stark R. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352: 459–464. [DOI] [PubMed] [Google Scholar]
- 96.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho SH, Lerner H, et al. 2017. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 20: 2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJ, Burton AR, Stegmann KA, Schurich A, Swadling L, et al. 2017. IL-2 high tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J. Exp. Med. 214: 1567–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oja AE, Piet B, Helbig C, Stark R, Van Der Zwan D, Blaauwgeers H, Remmerswaal EB, Amsen D, Jonkers RE, Moerland PD, et al. 2018. Trigger-happy resident memory CD4+ T cells inhabit the human lungs. Mucosal Immunol. 11: 654–667. [DOI] [PubMed] [Google Scholar]
- 99.Smolders J, Heutinck KM, Fransen NL, Remmerswaal EBM, Hombrink P, ten Berge IJM, van Lier RAW, Huitinga I, and Hamann J. 2018. Tissue-resident memory T cells populate the human brain. Nat. Commun. 9: 4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, Elco CP, Huang V,Matos TR, Kupper TS, et al. 2015. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci. Transl. Med. 7: 279ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, Brasser G, Jongejan A, Jonkers RE, Nota B, et al. 2016. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17: 1467–1478. [DOI] [PubMed] [Google Scholar]
- 102.Wong MT, Ong DEH, Lim FSH, Teng KWW, McGovern N, Narayanan S, Ho WQ,Cerny D, Tan HKK, Anicete R, et al. 2016. A high-dimensional atlas of human T cell diversity reveals tissue-specific trafficking and cytokine signatures. Immunity 45: 442–456. [DOI] [PubMed] [Google Scholar]
- 103.Kumar BV, Connors TJ, and Farber DL. 2018. Human T cell development, localization, andfunction throughout life. Immunity 48: 202–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schreiner D and King CG. 2018. CD4+ memory T cells at home in the tissue: Mechanisms forhealth and disease. Front. Immunol. 9: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, Gibbs A, Detlofsson E, Introini A, Forkel M, et al. 2017. CD49a expression defines tissue-resident CD8+T cells poised for cytotoxic function in human skin. Immunity 46: 287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumar BV, Kratchmarov R, Miron M, Carpenter DJ, Senda T, Lerner H, Friedman A, Reiner SL, and Farber DL. 2018. Functional heterogeneity of human tissue-resident memory T cells based on dye efflux capacities. JCI Insight 3: e123568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Senda T, Dogra P, Granot T, Furuhashi K, Snyder ME, Carpenter DJ, Szabo PA, Thapa P, Miron M, and Farber DL. 2019. Microanatomical dissection of human intestinal T-cell immunity reveals site-specific changes in gut-associated lymphoid tissues over life. Mucosal Immunol. 12: 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McCully ML, Ladell K, Andrews R, Jones RE, Miners KL, Roger L, Baird DM, Cameron MJ, Jessop ZM, Whitaker IS, et al. 2018. CCR8 expression defines tissue-resident memory T cells in human skin. J. Immunol. 200: 1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ganusov VV and Auerbach J. 2014. Mathematical modeling reveals kinetics of lymphocyterecirculation in the whole organism. PLoS Comput. Biol. 10: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ganusov VV, Borghans JAM, and de Boer RJ. 2010. Explicit kinetic heterogeneity: Mathematical models for interpretation of deuterium labeling of heterogeneous cell populations. PLoS Comput. Biol. 6: e1000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.de Boer RJ, Perelson AS, and Ribeiro RM. 2012. Modelling deuterium labelling of lymphocyteswith temporal and/or kinetic heterogeneity. J. R. Soc. Interface 9: 2191–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]