Abstract
The microbiota composition of the offspring of women with gestational diabetes mellitus (GDM), a common pregnancy complication, is still little known. We investigated whether the GDM offspring gut microbiota composition is associated with the maternal nutritional habits, metabolic variables or pregnancy outcomes. Furthermore, we compared the GDM offspring microbiota to the microbiota of normoglycemic-mother offspring. Fecal samples of 29 GDM infants were collected during the first week of life and assessed by 16S amplicon-based sequencing. The offspring’s microbiota showed significantly lower α-diversity than the corresponding mothers. Earlier maternal nutritional habits were more strongly associated with the offspring microbiota (maternal oligosaccharide positively with infant Ruminococcus, maternal saturated fat intake inversely with infant Rikenellaceae and Ruminococcus) than last-trimester maternal habits. Principal coordinate analysis showed a separation of the infant microbiota according to the type of feeding (breastfeeding vs formula-feeding), displaying in breast-fed infants a higher abundance of Bifidobacterium. A few Bacteroides and Blautia oligotypes were shared by the GDM mothers and their offspring, suggesting a maternal microbial imprinting. Finally, GDM infants showed higher relative abundance of pro-inflammatory taxa than infants from healthy women. In conclusion, many maternal conditions impact on the microbiota composition of GDM offspring whose microbiota showed increased abundance of pro-inflammatory taxa.
Introduction
It is well known that the maternal environment affects the offspring health. The newborn gut microbiota is strongly influenced by maternal health and pregnancy conditions and participates in the development programming of the newborns [1–3]. Early disruption of the infant microbiota has been associated with many inflammatory, immune-mediated, allergic, and dysmetabolic diseases in later life [1,4–5]. Children’s obesity, non-alcoholic hepatic liver diseases, aberrant cardiac growth are, among others, the conditions that have been associated with maternal/newborn dysbiosis [1,6–7]. Nevertheless, uncertainty still exists about the microbiota offspring colonization, and both the modality and the timing of microbial exposure for the fetus/newborn are controversial [8–11].
Gestational diabetes mellitus (GDM), the most frequent complication of pregnancy, has been associated with increased risk for the offspring of developing dysmetabolic diseases, such as obesity and diabetes mellitus [12]. Many prenatal and postnatal factors could determine an increased metabolic risk, such as epigenetic changes induced by in-utero exposure to maternal hyperglycemia, impaired secretion of fetal hormones (insulin and leptin), or fetal “mal-programming” of hypothalamic neuropeptidergic neurons, leading to hyperphagia [13–14]. GDM was found to be associated with specific changes in the gut microbiota composition [15–19]. The altered microbiome may be a bystander of the underlying metabolic dysregulation that underpins the pathogenesis of gestational hyperglycemia, as well as the consequence of the increased adiposity frequently co-existing in GDM patients [20–21]. However, the finding that a different microbial pattern precedes the onset of GDM leads to the hypothesis that microbiota alterations might have a role in the pathogenesis of GDM [22–23]. Even more important is the reporting that specific taxa associated with GDM can be transmitted to the offspring and differentiate their gut microbiota from that of the offspring of normoglycemic women [16,24–25].
The possibility of early modulation of the offspring gut microbiota by acting on specific maternal factors and/or characteristics is of potential great interest. Nevertheless, only few data are available about the associations between maternal characteristics and newborn microbiota pattern. Maternal fasting glucose concentrations were correlated positively with the relative abundance of phylum Actinobacteria and genus Acinetobacter, and negatively with the phylum Bacteroidetes and the genus Prevotella [26]. We hypothesized that maternal dietary habits and metabolic characteristics may impact on the offspring gut microbiota.
Aims of the present observational study were therefore evaluating whether maternal nutritional habits and/or metabolic variables and pregnancy outcomes of GDM patients are associated with the gut microbiota composition of their offspring. Furthermore, the gut microbiota composition of these infants was compared to the gut microbiota of the offspring of healthy normoglycemic women to evaluate possible differences between the two cohorts of newborns.
Materials and methods
Patients recruitment
All pregnant women routinely performed an oral glucose tolerance test at 24–28 gestational weeks at the “Città della Salute e della Scienza” Hospital of Turin. The first 50 consecutive subjects who met both the criteria for GDM according to international guidelines (fasting plasma glucose ≥92 mg/dL and/or 1h post-test glycemia ≥180 mg/dL and/or 2h post-test glycemia ≥153 mg/dL) and the inclusion criteria (see below) were enrolled [17]. Out of them, 41 women participated in an observational study aiming to evaluate the changes in gut microbiota composition across pregnancy [17]. The fecal samples of their offspring were collected in the first week of life. Indeed, as the mode of collection and/or storage of the offspring stool samples by 12 mothers resulted inappropriate, data of 29 infants (70.7%) could be analyzed only (S1 Table). Inclusion criteria were: GDM diagnosed by a 75g oral glucose tolerance test (OGTT) between 24–28 weeks gestational age and European origin with both parents born in Europe. Women with twin pregnancy, any pathological conditions before or during pregnancy (known diabetes mellitus, hypertension, cardiovascular, pulmonary, autoimmune, joint, liver or kidney diseases, thyroid dysfunction, cancer, any other disease/condition), no compliance to the study protocol, on prebiotics/probiotics, antibiotics or any drug during pregnancy were excluded. All the GDM patients received dietary counselling and the recommendation of performing 30-min daily moderate exercise (i.e. brisk walking) and were instructed to self-monitor finger-prick capillary blood glucose at least 4 times per day. Participants completed a 3-day food record (2 weekdays and 1 weekend day) and the Minnesota-Leisure-Time-Physical Activity Questionnaire at enrolment and at the study end [17]. Detailed information on how to record the food and drink consumed by using common household measures was provided to all participants. Two dieticians checked all questionnaires for completeness, internal coherence and plausibility [17].
Patients were considered to be compliant to the given dietary recommendations in the presence of all the following criteria: at least 20 g/day fiber consumption (or increasing fiber intake more than 50% than enrolment), sugar reduction to less than 10% of total energy and abolition of alcohol intake. Only 14 out of 41 (34.1%) of the patients were compliant to dietary recommendations. Finally, all mothers were suggested to breastfeed their children. However, only 10 of them followed this suggestion. All the others declared to have used formulas without added probiotic/prebiotic compounds.
In order to compare our offspring to infants from normoglycemic women, we have used the recently released 16S rRNA gene sequence from 19 Italian infants collected in the first week of life [27] (NCBI SRA, under BioProject ID PRJNA378341). Infants from healthy women showed the following characteristic: (i) born from vaginal delivery, (ii) breastfed, (iii) no use of antibiotic/probiotic during pregnancy [27]. We used as comparison the results of the 4th day of life.
Ethical aspects
The present study conforms to the principles outlined in the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of the “Città della Salute e della Scienza” hospital of Turin (approval 707/2016). All patients provided written informed consent prior to participation in the study protocol.
Data and samples collections
Anthropometric values, 3-days food record questionnaires, fasting blood samples (for the determination of metabolic variables and C-reactive protein levels) and stool samples of the mothers were collected both at the time of GDM diagnosis, at 24–28 weeks of gestational age, and at 38 weeks, or before delivery, in the case of preterm delivery [17]. Fecal samples of the offspring were collected between the 3rd and the 5th day of life, after meconium expulsion. Data relative to the type of delivery, birth weight, gestational age and the type of feeding were extracted from medical records.
Insulin therapy was added to diet when fasting blood glucose levels were consistently ≥90 mg/dL, 1-hour levels consistently ≥130 mg/dL, or 2-hour levels ≥120 mg/dL, according to guidelines [28].
Mothers were instructed to collect the stool samples and all materials were provided in a convenient, refrigerated, specimen collection kit (VWR, Milan, Italy) [17]. The fecal samples were transferred to the sterile sampling containers using a polypropylene spoon (3 spoons of about 10 g) and immediately stored at 4 °C. The specimens were transported to the laboratory within 12 hours of collection at a refrigerated temperature. Containers were immediately stored at −80 °C for DNA extraction. No storage medium was used [17].
In the case of infants from normoglycemic mothers, fecal samples have been reported to be collected from diapers using standard sterile collection tubes [27].
Fecal DNA extraction and sequencing
Total DNA from the feces collected was extracted using the RNeasy Power Microbiome KIT (Qiagen, Milan, Italy) following the manufacturer’s instructions. One microliter of RNase (Illumina Inc. San Diego. CA) was added to digest RNA in the DNA samples, with an incubation of 1 h at 37 °C. DNA was quantified using the QUBIT dsDNA Assay kit (Life Technologies, Milan, Italy) and standardized at 5 ng/μL. DNA was used as a template in a PCR reaction in order to amplify the V3-V4 region of the 16S rRNA gene [29]. Library preparation and sequencing (2X250bp) was performed as already reported [17].
Bioinformatics analyses
Paired-end reads were first assembled using FLASH software and quality filtering using QIIME 1.9.0 software [30] and the pipeline already reported [17]. OTUs picked at 97% of similarity were rarefied to the lowest number of sequences per sample. The OTU table obtained through QIIME displays the higher taxonomy resolution that was reached; when the taxonomy assignment was not able to reach the genus, family name was displayed.
In order to identify sub-OTUs populations in our offspring cohort, reads assigned to genera Blautia, Bacteroides and Bifidobacterium were extracted and entropy analysis and oligotyping were carried out [31] since these were the only OTUs showing higher level of entropy able to obtain sub-OTUs. After the first round of oligotyping, high entropy positions were chosen (-C option): 8, 12, 113, 223, 224, 247, 432, 433 for Blautia; position 8, 12, 40, 74, 75, 116, 117, 124, 129, 130, 390, 397, 452 and 453 were chosen for Bacteroides; while position 8, 12, 14, 81, 114, 115, 158, 159, 160, 328, 329, 389, 437, 438, 439, 440, 441, 442, 443 and 444 were chosen for Bifidobacterium. In order to reduce the noise, each oligotype should appear in at least 10 samples, occur in more than 1.0% of the reads for at least ten samples, represent a minimum of 750 reads in all samples combined, and have a most abundant unique sequence with a minimum abundance of 50. A cladogram of representative sequences was generated using the ANVIOs software [32].
Statistical analyses
Gut microbiota α-diversity was calculated by the diversity function of the vegan package [33] in R environment (http://www.r-project.org). ADONIS and ANOSIM statistical test were used to detect significant differences in the overall microbial community by using the unweighted UniFrac distance matrices or OTU table. Offspring variables were compared with maternal variables by paired-sample t-test, or Wilcoxon matched pairs test, as appropriate. Differences in infant gut microbiota between mothers who were compliant or not to the dietary recommendations, or between the GDM offspring and healthy-women offspring were calculated by t-Student test or Mann-Whitney test. Pairwise Spearman’s non-parametric correlations were used to study the relationships between the infant relative abundance of microbial taxa and maternal dietary and metabolic variables. The degree of agreement (concordance) among maternal oligotypes at enrolment, maternal oligotypes at the study-end and infant oligotypes was assessed by the Friedman ANOVA & Kendall’s test.
Multiple regression analyses were performed to evaluate the associations between infant microbial taxa abundance (dependent variable) and maternal nutrient intakes or laboratory variables, after adjusting for gestational weight change, breastfeeding and Cesarean section (Statistica, ver. 7.0; StatSoft Inc., Tulsa, OK, USA).
The post-hoc power estimated on partial R2 according to the multivariate linear regression model adjusted for maternal weight change, breastfeeding and Cesarean section was 0.80 with α = 0.05. A P value of 0.002 or lower was considered as statistically significant.
Results
Maternal and infant gut microbiota composition
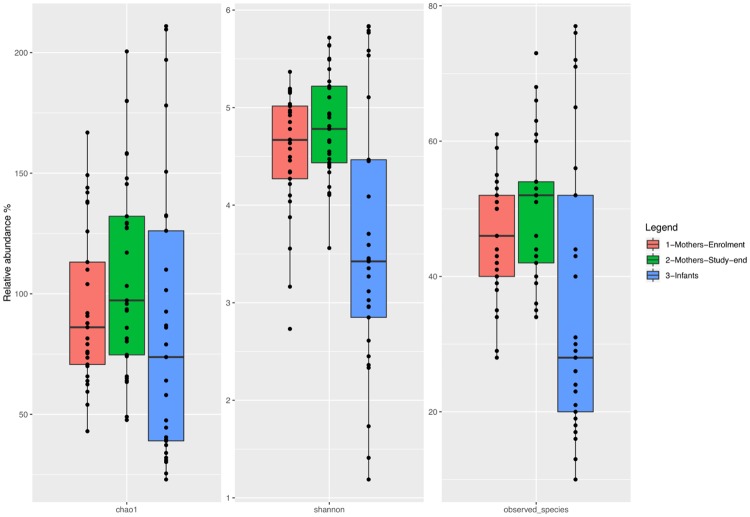
The microbial richness (alpha diversity index) was lower in infant stools than in the corresponding maternal samples, and fewer taxa were present in the offspring (Fig 1). An increase in the rarefaction measure across pregnancy was evident in GDM patients. The number of observed species was significantly different between mother samples (both at enrolment and at the study-end) and the corresponding offspring samples (for the Chao-1 diversity index, P for paired data <0.001). The decrease in the complexity and the number of taxa observed in offspring samples was not a consequence of the lower number of infant sequences because the analysis was performed by using a rarefied down-sampling operational taxonomic unit (OTU) table.
Fig 1. α-diversity measures of fecal microbiota.
Boxplots describe α-diversity measures of fecal microbiota of GDM patients at enrolment (red bars), of GDM patients at the study end (green bars) and of their offspring (blue bars). Individual points and brackets represent the richness estimate and the theoretical standard error range, respectively.
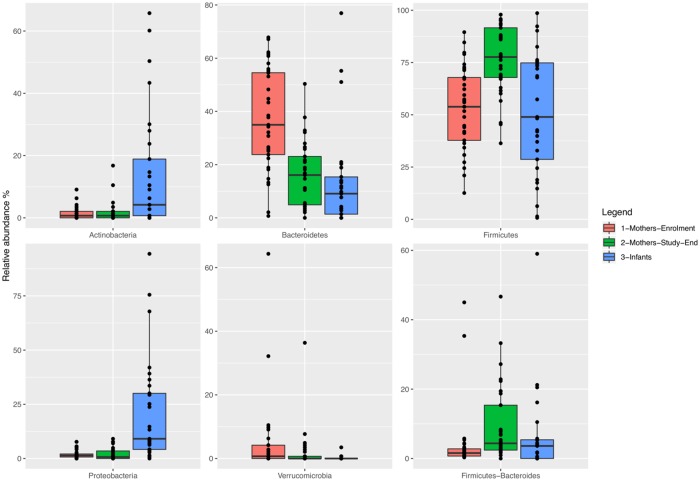
Patients with GDM showed a predominance of Firmicutes (increasing during the gestational period) and Bacteroidetes (whose relative abundance decreased), while the infant gut microbiota was characterized by an increased relative abundance of Actinobacteria and Proteobacteria (Fig 2).
Fig 2. Phylum-level abundance profiles.
Boxplots describe the relative abundance of phyla in GDM patients at enrolment (red bars), in GDM patients at the study end (green bars) and in the infants (blue bars).
Hierarchical clustering analysis at genus level (S1 Fig) showed a separation between the infant microbiota and the mother microbiota (anosim P<0.01). Infants displayed a higher abundance of Bifidobacterium, Streptococcus, Escherichia, Staphylococcus and Enterococcaceae while mothers had a more complex microbiota composition (S1 Fig).
Infant gut microbiota and maternal dietary habits and laboratory variables
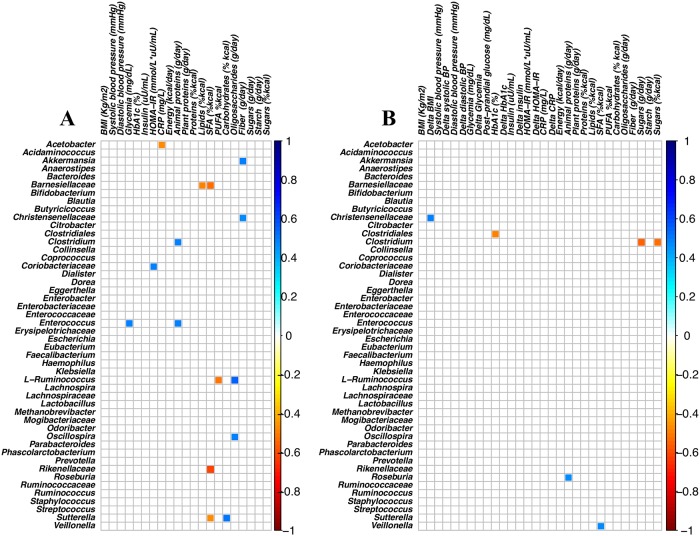
No clear separation of the infant gut microbiota was observed (anosim P > 0.05) according to the maternal compliance to dietary recommendations (S2 Fig). The correlation plot between maternal nutrient intakes and infant gut microbiota showed a higher number of significant associations with maternal variables at enrolment than at the study-end (Fig 3).
Fig 3. Spearman’s rank correlation of OTUs dietary information and blood variables.
Spearman’s rank correlation matrix of OTUs with > 0.2% abundance in at least 10 fecal samples, dietary information and blood variables. The colors of the scale bar denote the nature of the correlation, with 1 indicating a perfectly positive correlation (dark blue) and -1 indicating a perfectly negative correlation (dark red) between the two datasets. Only correlations with P-values <0.002 are shown. Data at enrolment (plot A) or at study-end (Plot B).
Maternal oligosaccharides derived mainly from dairy products, cereals, fruit and vegetables, while saturated fatty acids (SFA) derived mainly from meat and cheese. In a multiple regression model, after adjusting for weight change, breastfeeding and Cesarean section, associations between infant Ruminococcus with maternal oligosaccharide (positive association) and with maternal intake of SFA (inverse association) were found, even if not reaching the defined cutoff of p-values. The inverse relationship between Rikenellaceae and maternal SFA intake remained statistically significant (Table 1). In the multivariate model, no significant association between infant gut microbiota and maternal dietary habits at the study-end was found.
Table 1. Statistically significant associations between infant microbiota composition and maternal variables by Spearman’s correlations for continuous variables (left) and multiple regression analyses (right).
| Rho | Beta | 95% CI | P | |
|---|---|---|---|---|
| Dietary habits at enrolment | ||||
| Oligosaccharides | ||||
| Ruminococcus | 0.55 | 0.09 | 0.04 0.14 | 0.005 |
| Saturated fatty acids (%kcal) | ||||
| Rikenellaceae | -0.61 | -0.24 | -0.37–0.11 | 0.001 |
| Ruminococcus | -0.44 | -0.76 | -1.21–0.31 | 0.004 |
| Maternal laboratory variables | ||||
| Delta HbA1c | ||||
| Clostridiales | -0.49 | -0.32 | -0.48–0.16 | <0.001 |
| Breastfeeding* | ||||
| Bifidobacterium | - | 22.9 | 10.1 35.7 | 0.0017 |
Model adjusted for maternal weight change, breastfeeding, and Cesarean section;
*Model adjusted for maternal weight change and Cesarean section
The direct association between maternal fasting glucose at enrolment and Enterococcus (Rho = 0.49) was no longer confirmed in the multivariate model, while the inverse association between Clostridiales and maternal changes across pregnancy (delta) in glycated hemoglobin (HbA1c) levels was statistically significant (Table 1). No significant association between maternal anthropometric or metabolic variables and the gestational changes in these variables and infant gut microbiota composition was found. Two out of 29 women were treated with insulin; none received metformin. After adjusting for insulin treatment, the results of the multiple regression analyses did not change.
Infant gut microbiota and delivery outcomes and breastfeeding
No significant associations between infant gut microbiota and delivery (Cesarean section vs vaginal delivery) was found.
Ten out of the 29 (34.5%) newborns were breastfed by their mothers, while 19 (65.5%) were formula-fed. We observed a non-significant reduction in microbial richness in breastfed infants when compared to infants fed with artificial milk.
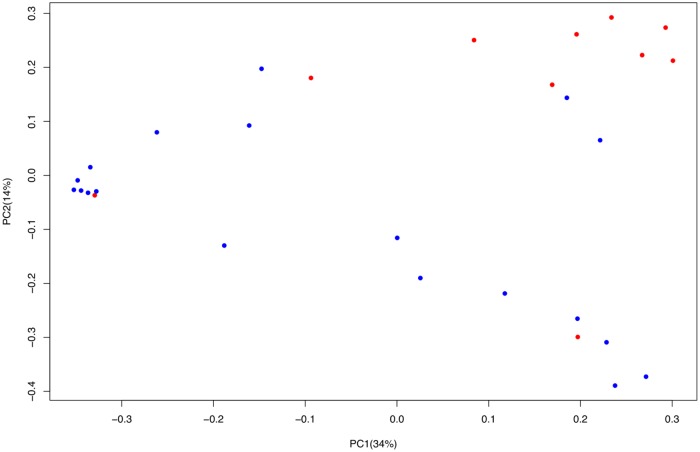
The β–diversity calculation based on unweighted UniFrac distance matrices assessed by Principal coordinate analysis (PCoA) showed a separation of the infant microbiota only as a function of the type of feeding (breast vs formula) as confirmed by ANOSIM and ADONIS statistical test (P <0.001) (Fig 4).
Fig 4. Principal coordinates analysis (PCoA) of unweighted UniFrac distances matrix for 16S rRNA gene sequence data.
Infant artificially-fed (blue dots) or breast-fed (red dots).
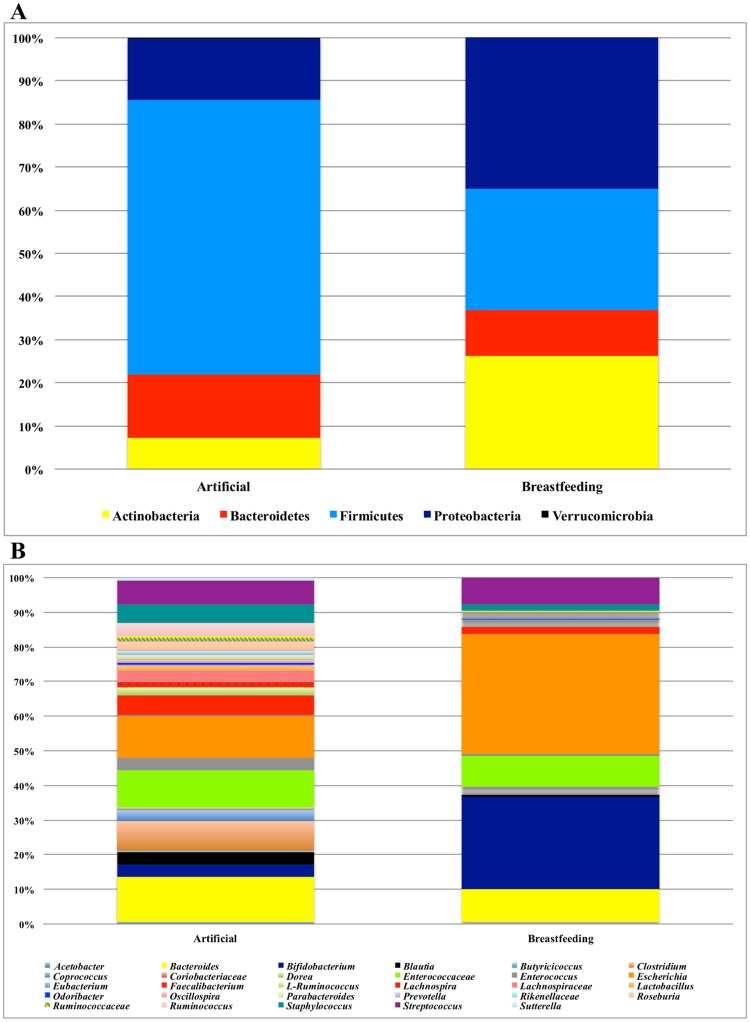
At phylum level, we observed that breastfed infants showed a higher abundance of Actinobacteria and Proteobacteria, while in formula-fed infants we observed a higher proportion of Firmicutes phyla (Fig 5A). At genus level, breastfed infants displayed an increased abundance of Escherichia and Bifidobacterium, while formula-fed infants had a varied microbiota mainly composed of Bacteroides, Clostridium, Enterococcaceae, Escherichia, Faecalibacterium, Staphylococcus and Streptococcus (Fig 5B). In multiple regression analyses, a significant association between breastfeeding and the relative abundance of Bifidobacterium in the infant gut microbiota was found (β = 22.9; 95%CI = 10.1–35.7; P = 0.0017).
Fig 5. Relative phylum-level abundance profiles.
Phylum-level abundance profiles of breastfed infants and artificially fed infants (Panel A); relative genus-level abundance of breastfed infants and artificially fed infants (Panel B).
Gut microbiota signature at sub-genus level
In order to explore the possible mother-to-infant gut microbiota transmission at sub-OTUs level, we carried out oligotyping on sequences of Blautia, Bacteroides and Bifidobacterium since these were the only genera showing a Shannon entropy index sufficient to identify all nucleotide positions that would resolve the oligotypes. We considered the dominant oligotypes shared by mothers and their offspring only to explore possible mother-to-child oligotype transmission. We then selected the oligotypes which were present in at least 5 mother-to-child pairs. We found 18 oligotypes for Bacteroides, 30 for Blautia, and 81 for Bifidobacterium. Only for Bifidobacterium, oligotypes of the offspring were taken into account, since resolving Bifidobacterium in mothers was not possible. A high inter-individual diversity in the Bifidobacterium oligotypes was observed in the offspring samples (S3 Fig). We observed that the relative abundance of the Blautia oligotypes increased during the pregnancy and were lower in the infants (S4A Fig). A high degree of mother-offspring concordance was found for B11 (P = 0.008) and B42 (P<0.001); both were identified as Blautia wexlerae by best BLASTn match. A reduction in the relative abundance of Bacteroides oligotypes was observed across pregnancy while few oligotypes were observed to be dominant in the offspring samples (S4B Fig).
Comparison between gut microbiota from GDM offspring and healthy women offspring
The gut microbiota of our offspring was compared with those of 19 breastfed infants from healthy normoglycemic women. A higher α-diversity was evident in the latter (for the Chao-1 diversity index, P = 0.001). However, when comparing to those infants the 10 breastfed infants from GDM women, no significant difference in α-diversity was found. At phylum level, we observed higher abundance of Actinobacteria and Bacteroidetes in the offspring from GDM women (S5A Fig) (P<0.001). At genus level, higher abundance of Staphylococcus, Ralstonia, Lactobacillus and some members of Enterobacteriaceae were observed in the offspring from healthy women (S5B Fig).
In the comparison between the two groups of breastfed infants, the offspring of GDM women showed a significantly higher relative abundance of Escherichia and Parabacteroides (P<0.001).
Discussion
The microbiota of infants from GDM women showed a low complexity, a high inter-individual variability, and was influenced by early maternal nutrition and breastfeeding. Intriguingly, in comparison with the offspring of healthy women, our infants showed a higher relative abundance of pro-inflammatory taxa, in particular Escherichia and Parabacteroides. A few mother-to-child oligotype transmissions were found.
The offspring gut microbiota composition substantially differs from the microbiota composition of their GDM mothers, being the former characterized by limited species-level complexity (i.e. a lower α-diversity) and a greater inter-individual variability of genus abundance (i.e. a higher β-diversity), as already reported [22,34–39]. The microbiota complexity indeed progressively increases with the infant growth, with the gradual development of a microbial community resembling the adult microbiota at the age of 3 years with a lower inter-individual variability [40–41]. Actinobacteria and Proteobacteria dominated the gut microbiota of our infants from GDM women, similarly to the infants of healthy mothers and in accordance with previous reports [36–37,42–43]. The relative abundance of these phyla has been reported to be higher in the meconium of GDM newborns compared to offspring from healthy controls [26]. Bifidobacterium, Streptococcus, Escherichia, Staphylococcus and members of the Enterococcaceae family displayed a higher relative abundance in the feces of our GDM infants. Accordingly, these bacteria have been detected in the feces or meconium of full-term newborns in several studies [24,34,35–37,39,44–48].
A distinct microbial composition was found in our breastfed GDM offspring when comparing them with breastfed neonates from healthy controls, consistent with literature [16,24–25]. In particular, a significantly higher relative abundance of Escherichia and Parabacteroides was found in our GDM infants. These taxa can be considered as pro-inflammatory and have been found to be higher in the meconium of babies from T2DM mothers [24]. Furthermore, Parabacteroides have been reported to be enriched in GDM patients when compared to healthy subjects [18]. In our previous study [17], we have observed an increased, though not statistically significant, relative abundance of Parabacteroides across pregnancy from enrolment to study end. Similarly, Escherichia is more abundant in pregnant overweight women, particularly in GDM patients [49–50] Diabetes has been considered as an inflammatory disease and, not surprisingly, the offspring microbiota may be influenced by this pregnancy pathological condition; a direct transmission from mother to child of some pro-inflammatory taxa like Parabacteroides or Escherichia might be hypothesized, even if the contribution of not analyzed environmental conditions could not be ruled out.
No differences in the Firmicutes-to-Bacteroidetes ratio, indicating an increased risk for developing obesity [51], was found neither between the breast-fed GDM infants when compared to the formula-fed GDM infants, nor between breast-fed GDM infants vs breast-fed infants form normoglycemic women.
The microbiota in the first days of life of our GDM offspring might be one of the contributing factors to the 2- to 8-fold increased future risk of dysmetabolic diseases of those offspring when compared to offspring of healthy women [52]. We observed that the infant gut microbiota composition is influenced by nutritional maternal habits. The offspring relative abundance of Ruminococcus was directly associated with the maternal intake of oligosaccharides and inversely with the maternal intake of SFA, as previously reported in infants from healthy women [53]. The Ruminococcus genus produces both butyrate and a bacteriocin, ruminococcin A, able to inhibit the growth of the potentially harmful Clostridium species, therefore potentially playing a beneficial role for the newborns [54]. Furthermore, maternal SFA intake was inversely associated with the relative abundance of Rikenellaceae, a butyrate-producers family [55] that has been related to favorable metabolic outcomes and a healthy gut [56–57]. These associations are remarkable and could explain the previously reported adverse impact of SFA on maternal [58] and neonatal health [59–60] even if did not reach the established significance threshold, after multiple adjustments.
Nutritional recommendations were given to our women between 24–28 weeks of gestational age, at the time of the oral glucose challenge test and GDM diagnosis [17]. Only 34.1% of our participants resulted compliant to the nutritional advice; indeed, dietary adherence and nutritional habits at the end of pregnancy did not influence the offspring microbiota. Contrarily to a few previous human studies showing that the infant gut microbiota was associated with the last-trimester maternal diet [61], we found stronger associations between infant gut bacteria composition and early maternal nutrition. Even if the mechanisms by which the maternal diet affects the offspring microbiota remain unclear, the nutritional habits of late pregnancy might have a lower impact, owing to the lower maternal-fetal time of contact and seeding possibility in a period when fetal growth is already advanced. This hypothesis needs to be confirmed by further studies. Our results indeed highlight the importance of a proper maternal nutrition, low in SFA and with a high content of fiber and prebiotic oligosaccharides, starting from early pregnancy and probably even before gestation, in order to favorably modulate the offspring microbiota.
Many other factors have been reported to influence and shape the neonatal microbiota during intrauterine life [62–63], among which those that are more frequently implicated are maternal glycemic status [24], weight gain during pregnancy [64], pre-pregnancy BMI [64–65], and antibiotic use [66]. We failed to demonstrate any associations between maternal anthropometric indexes with the offspring microbiota composition, indeed the insulin resistance and hyperglycemia of our participants, all suffering from GDM, may have obscured these relationships. Furthermore, other studies did not show relationships between maternal BMI or clinical characteristics and the infant microbiota composition [24–25].
We found an inverse association between Clostridiales and HbA1c changes across pregnancy. The Clostridiales order includes several genera and in particular Roseburia and Faecalibacterium prausnitzii, both butyrate-producing bacteria with beneficial functions, which have been found to be reduced in GDM women [15,18,22].
The microbiota of our GDM offspring did not meaningfully vary according to the way of delivery. Contrasting data are available about this topic, since a different microbial pattern [43,67–69] or no differences [8,24,36] have been reported between C-section-delivered infants and vaginally-delivered infants. It could be hypothesized that the exposure to an adverse environment in our GDM offspring could have had a major role on their gut microbiota, partly overshadowing the effects of the exposure during birth. Furthermore, the influence of delivery mode on the infant microbiota rapidly decline after delivery [40,70] and is quickly overridden by the role of lactation [39,71–72].
A clear separation of infant microbiota according to the type of feeding (breast milk vs formula) was evident, as previously reported [5,73]. In particular, our formula-fed infants showed increased microbial complexity and levels of strict anaerobes and facultative anaerobes, and higher abundance of Firmicutes, such as Clostridium, Streptococcus and Staphylococcus, in line with literature [2,70,74–75]. On the other hand, our breastfed infants showed a less complex microbiota with higher abundance of Proteobacteria and Actinobacteria, being the latter mainly derived from the Bifidobacterium genus that resulted to be strongly associated with breastfeeding, accordingly with most studies [2,75–78]. The higher abundance of Bifidobacterium in breast-fed infants can be explained by the presence of human milk oligosaccharides (HMOs), sugar polymers with prebiotic effects that promote the growth of specific microbial communities, including Bifidobacterium spp. with beneficial protective effects on the infant health [79–81]. A higher, though not significantly different abundance of Proteobacteria was evident in our breast-fed infants when compared to formula-fed infants. Literature is discordant on this topic, since either a higher abundance of Proteobacteria, in particular of Enterobacteriaceae, has been reported in formula-fed infants [82] or the opposite was observed, in particular a decrease in the relative abundance of Proteobacteria in non-breast-fed vs breast-fed infants [83]. Differences in breastmilk microbiota and HMOs content, maternal diet and type of formula milk used across populations might be responsible for these heterogeneous results.
A possible vertical mother-to-child transmission of maternal gut bacteria has already been reported, even if, to date, certainty about the way of intrauterine microbial acquisition is lacking [37,84–85]. Besides breastfeeding and vaginal microbiota, also placenta and amniotic fluid have been reported to be a vehicle for this transmission [22,86]. By using the oligotype pipeline, the transmission of different sub-OTUs from our mothers to their infants was observed in line with data of a recent metagenomic study confirming that mother’s dominant strains were transmitted to their children [42]. In particular, we have found a few Bacteroides and Blautia oligotypes significantly shared by the GDM mothers and their offspring, suggesting a maternal microbial imprinting. On the other hand, the infant Bifidobacterium oligotypes of the newborns were not present in the feces of the GDM women, and this directs towards a post-natal acquisition. Indeed, the infant Bifidobacteria mostly derive from breast-feeding, being either isolated from human milk and vertically transmitted [86–87] or induced by the milk prebiotic HMO [88]. The Bifidobacterium oligotypes were highly different among infants but only one third of them was breastfed and the human milk has a heterogeneous composition and contains variable amounts of glycans and bifidogenic oligosaccharides [72,89].
In conclusion, the gut microbiota changes might be one of the conditions determining the increased metabolic risk of the GDM offspring with both pre and postnatal effects, since in utero exposure to maternal nutrition and dysmetabolic conditions, and post-natal breastfeeding can both modulate the offspring microbial community.
Several limitations of the present study should be recognized. The sample size was low, the standard errors were wide, and we could analyze the fecal samples of only 70.7% of the offspring of the initially enrolled women. Therefore, we might not have detected modest differences in bacterial composition. Many associations that we found to be not statistically significant, could have become so if we had analyzed a larger sample. However, the post-hoc power was 0.80 with α = 0.05; furthermore, we have set-up a lower p-value cut-off value (p<0.002) as statistically significant. In doing so, we have considered only the strongest associations; however, we cannot exclude the possibility that a type II error could have occurred.
Infant fecal samples were collected between the 3rd and the 5th days of life. We cannot exclude that the difference of a few days could have influenced the results due to the high instability of the infant microbiota.
Conclusions
Early maternal eating habits and breastfeeding may have a significant influence on the fetal gut microbiota composition of infants from GDM women. These results are worthy to be replicated in larger studies owing to their potential practical implications on the health of future generations.
Supporting information
Heatplot showing OTUs of GDM patients at enrolment (red bars), study end (green bars) and their offspring (blue bars). Rows and columns are clustered by means of Ward linkage hierarchical clustering. The intensity of the colors represents the degree of correlation between the samples and OTUs as measured by Spearman’s correlations.
(TIF)
Samples are color-coded according to compliance to dietary recommendations (red) or non-compliance to dietary recommendations (blue).
(TIF)
Inner black bars indicate the presence of an oligotype in a given sample.
(TIF)
Plot showing the sequence distribution in GDM patients at enrolment (red bars), study end (green bars) and their offspring (blue bars). Inner bars indicate the presence of an oligotype in a given sample. Outer circle, if colored, denotes oligotype abundance with high degree of mother-offspring concordance.
(TIF)
Plot shows the relative phyla (panel A) and genus (panel B) abundance in the offspring from healthy and GDM women.
(TIF)
(DOCX)
Data Availability
All the sequencing data were deposited at the Sequence Read Archive of the National Center for Biotechnology Information (SRA accession number: SRP189631 and SRP135886)
Funding Statement
This study was supported by a grant from the Ministry of Education, University and Research of Italy (ex-60% 2017). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Milani C, Duranti S, Bottacini F, Casey E, Turroni F, Mahony J, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol Mol Biol Rev. 2017; 81: e00036–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chong CYL, Bloomfield FH, O’Sullivan JM. Factors affecting gastrointestinal microbiome development in neonates. Nutrients. 2018; 10: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchanan KL, Bohórquez DV. You are what you (first) eat. Front Hum Neurosci. 2018; 12: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gohir W, Ratcliffe EM, Sloboda DM. Of the bugs that shape us: Maternal obesity, the gut microbiome, and long-term disease risk. Pediatr Res. 2015; 77: 196–204. 10.1038/pr.2014.169 [DOI] [PubMed] [Google Scholar]
- 5.Ficara M, Pietrella E, Spada C, Della Casa Muttini E, Lucaccioni L, Iughetti L, et al. Changes of intestinal microbiota in early life. J Matern Neonatal Med. 2018; 10: 1–8. [DOI] [PubMed] [Google Scholar]
- 6.Soderborg TK, Clark SE, Mulligan CE, Janssen RC, Babcock L, Ir D, et al. The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun. 2018; 9: 4462 10.1038/s41467-018-06929-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guzzardi MA, Ait Ali L, D’Aurizio R, Rizzo F, Saggese P, Sanguinetti E, Weisz A, et al. Fetal cardiac growth is associated with in utero gut colonization. Nutr Metab Cardiovasc Dis. 2019; 29: 170–176. 10.1016/j.numecd.2018.10.005 [DOI] [PubMed] [Google Scholar]
- 8.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017; 23: 314–326. 10.1038/nm.4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehbinder EM, Lødrup Carlsen KC, Staff AC, Angell IL, Landrø L, Hilde K, et al. Is amniotic fluid of women with uncomplicated term pregnancies free of bacteria? Am J Obstet Gynecol. 2018; 219: 289.e1–289.e12. [DOI] [PubMed] [Google Scholar]
- 10.Lim E, Rodriguez C, Holtz LR. Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome. 2018; 6: 87 10.1186/s40168-018-0475-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassandra W. Baby’s first bacteria. Nature. 2018; 553: 264–266.29345664 [Google Scholar]
- 12.Chandler-Laney PC, Bush NC, Granger WM, Rouse DJ, Mancuso MS, Gower BA. Overweight status and intrauterine exposure to gestational diabetes are associated with children’s metabolic health. Pediatr Obes. 2012; 7: 44–52. 10.1111/j.2047-6310.2011.00009.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franzago M, Fraticelli F, Stuppia L, Vitacolonna E. Nutrigenetics, epigenetics and gestational diabetes: consequences in mother and child. Epigenetics. 2019; 14: 215–235. 10.1080/15592294.2019.1582277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dabelea D. The Predisposition to Obesity and Diabetes in Offspring of Diabetic Mothers. Diabetes Care. 2007; 30: S169–S174. 10.2337/dc07-s211 [DOI] [PubMed] [Google Scholar]
- 15.Crusell MKW, Hansen TH, Nielsen T, Allin KH, Rühlemann MC, Damm P, et al. Gestational diabetes is associated with change in the gut microbiota composition in third trimester of pregnancy and postpartum. Microbiome. 2018; 6: 89 10.1186/s40168-018-0472-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Zheng J, Shi W, Du N, Xu X, Zhang Y, et al. Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut. 2018; 67: 1614–1625. 10.1136/gutjnl-2018-315988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferrocino I, Ponzo V, Gambino R, Zarovska A, Leone F, Monzeglio C et al. Changes in the gut microbiota composition during pregnancy in patients with gestational diabetes mellitus (GDM). Sci Rep. 2018; 8: 12216 10.1038/s41598-018-30735-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuang YS, Lu JH, Li SH, Li JH, Yuan MY, He JR, et al. Connections between the human gut microbiome and gestational diabetes mellitus. Gigascience. 2017; 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cortez RV, Taddei CR, Sparvoli LG, Ângelo AGS, Padilha M, Mattar R, et al. Microbiome and its relation to gestational diabetes. Endocrine. 2018, 10.1007/s12020-018-1813-z [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 20.Saad MJ, Santos A, Prada PO. Linking Gut Microbiota and Inflammation to Obesity and Insulin Resistance. Physiology. 2016; 31: 283–293. 10.1152/physiol.00041.2015 [DOI] [PubMed] [Google Scholar]
- 21.Zhou L, Xiao X. The role of gut microbiota in the effects of maternal obesity during pregnancy on offspring metabolism. Biosci Rep. 2018; 38: BSR20171234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koren O, Goodrich JK, Cullender TC, Spor AA, Laitinen K, Backhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2013; 150: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mokkala K, Houttu N, Vahlberg T, Munukka E, Rönnemaa T, Laitinen K. Gut microbiota aberrations precede diagnosis of gestational diabetes mellitus. Acta Diabetol. 2017; 54: 1147–1149. 10.1007/s00592-017-1056-0 [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, et al. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One. 2013; 8: e78257 10.1371/journal.pone.0078257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasan S, Aho V, Pereira P, Paulin L, Koivusalo SB, Auvinen P, et al. Gut microbiome in gestational diabetes: a cross-sectional study of mothers and offspring 5 years postpartum. Acta Obstet Gynecol Scand. 2018; 97: 38–46. 10.1111/aogs.13252 [DOI] [PubMed] [Google Scholar]
- 26.Su M, Nie Y, Shao R, Duan S, Jiang Y. Diversified gut microbiota in newborns of mothers with gestational diabetes mellitus. PLoS one. 2018; 13: e0205695 10.1371/journal.pone.0205695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, et al. The bacterial ecosystem of mother’s milk and infant’s mouth and gut. Front Microbiol. 2017; 8: 1214 10.3389/fmicb.2017.01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Associazione Medici Diabetologi (AMD); Società Italiana di Diabetologia (SID). Standard Italiani per la Cura del Diabete 2018 (Italian). https://aemmedi.it/wp-content/uploads/2009/06/AMD-Standard-unico1.pdf
- 29.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013; 41: e1 10.1093/nar/gks808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7: 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013; 4: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015; 3: e1319 10.7717/peerj.1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon P. VEGAN, a package of R functions for community ecology. J Veg Sci. 2003; 14: 927–930. [Google Scholar]
- 34.Chernikova DA, Koestler DC, Hoen AG, Housman ML, Hibberd PL, Moore JH, et al. Fetal exposures and perinatal influences on the stool microbiota of premature infants. J Matern Fetal Neonatal Med. 2016; 29: 99–105. 10.3109/14767058.2014.987748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016; 6: 23129 10.1038/srep23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Chierico F, Vernocchi P, Petrucca A, Paci P, Fuentes S, Praticò G, et al. Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One. 2015; 10: e0137347 10.1371/journal.pone.0137347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gosalbes MJ, Llop S, Vallès Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy. 2013; 43: 198–211. 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- 38.Madan JC, Salari RC, Saxena D, Davidson L, O’Toole GA, Moore JH, et al. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed. 2012; 97: F456–62. 10.1136/fetalneonatal-2011-301373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moles L.; Gómez M.; Heilig H.; Bustos G.; Fuentes S.; de Vos W.; Fernández L.; Rodríguez J.M.; Jiménez E. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 2013, 8, e66986 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bäckhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015; 17: 690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 41.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012; 486: 222–227. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yassour M.; Jason E.; Hogstrom L.J.; Arthur T.D.; Tripathi S.; Siljander H.; Selvenius J.; Oikarinen S.; Hyöty H.; Virtanen S.M.; et al. Strain-level analysis of mother-to-child bacterial transmission during the first few months of life. Cell Host Microbe 2018, 24, 146–154.e4. 10.1016/j.chom.2018.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, et al. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One. 2014; 9: e90784 10.1371/journal.pone.0090784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hornef M, Penders J. Does a prenatal bacterial microbiota exist? Mucosal Immunol. 2017; 10: 598–601. 10.1038/mi.2016.141 [DOI] [PubMed] [Google Scholar]
- 45.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut. 2007; 56: 661–667. 10.1136/gut.2006.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 2010; 156: 20–25. 10.1016/j.jpeds.2009.06.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiménez E, Marín ML, Martín R, Odriozola JM, Olivares M, Xaus J, et al. Is meconium from healthy newborns actually sterile? Res Microbiol. 2008; 159: 187–193. 10.1016/j.resmic.2007.12.007 [DOI] [PubMed] [Google Scholar]
- 48.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, et al. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One. 2015; 10: e0133320 10.1371/journal.pone.0133320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010; 104: 83–92. 10.1017/S0007114510000176 [DOI] [PubMed] [Google Scholar]
- 50.Cortez RV, Taddei CR, Sparvoli LG, Ângelo AGS, Padilha M, Mattar R, Daher S. Microbiome and its relation to gestational diabetes. Endocrine. 2019; 64: 254–264. 10.1007/s12020-018-1813-z [DOI] [PubMed] [Google Scholar]
- 51.Damm P, Houshmand-Oeregaard A, Kelstrup L, Lauenborg J, Mathiesen ER, Clausen TD. Gestational diabetes mellitus and long-term consequences for mother and offspring: a view from Denmark. Diabetologia. 2016; 59: 1396–1399. 10.1007/s00125-016-3985-5 [DOI] [PubMed] [Google Scholar]
- 52.Damm P. Future risk of diabetes in mother and child after gestational diabetes mellitus. Int J Gynecol Obstet. 2009; 104: S25–S26. [DOI] [PubMed] [Google Scholar]
- 53.Mandal S, Godfrey KM, McDonald D, Treuren WV, Bjørnholt JV, Midtvedt T, et al. Fat and vitamin intakes during pregnancy have stronger relations with a pro-inflammatory maternal microbiota than does carbohydrate intake. Microbiome. 2016; 4: 55 10.1186/s40168-016-0200-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coppa GV, Gabrielli O, Zampini L, Galeazzi T, Ficcadenti A, Padella L, et al. Oligosaccharides in 4 different milk groups, Bifidobacteria, and Ruminococcus obeum. J Pediatr Gastroenterol Nutr 2011; 53: 80–87. 10.1097/MPG.0b013e3182073103 [DOI] [PubMed] [Google Scholar]
- 55.Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, et al. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol. 2014; 64: 2986–2991. 10.1099/ijs.0.063917-0 [DOI] [PubMed] [Google Scholar]
- 56.Lippert K, Kedenko L, Antonielli L, Kedenko I, Gemeier C, Leitner M, et al. Gut microbiota dysbiosis associated with glucose metabolism disorders and the metabolic syndrome in older adults. Benef Microbes. 2017; 8: 545–556. 10.3920/BM2016.0184 [DOI] [PubMed] [Google Scholar]
- 57.Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, et al. Connection between BMI-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab. 2018; 103: 1491–1501. 10.1210/jc.2017-02114 [DOI] [PubMed] [Google Scholar]
- 58.Bo S, Menato G, Lezo A, Signorile A, Bardelli C, De Michieli F, et al. Dietary fat and gestational hyperglycaemia. Diabetologia. 2001; 44: 972–978. 10.1007/s001250100590 [DOI] [PubMed] [Google Scholar]
- 59.Horan MK, Donnelly JM, McGowan CA, Gibney ER, McAuliffe FM. The association between maternal nutrition and lifestyle during pregnancy and 2-year-old offspring adiposity: analysis from the ROLO study. Zeitschrift Fur Gesundheitswissenschaften. 2016; 24: 427–436. 10.1007/s10389-016-0740-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murrin C, Shrivastava A, Kelleher CC. Maternal macronutrient intake during pregnancy and 5 years postpartum and associations with child weight status aged five. Eur J Clin Nutr. 2013; 67: 670–679. 10.1038/ejcn.2013.76 [DOI] [PubMed] [Google Scholar]
- 61.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, et al. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med. 2016; 8: 77 10.1186/s13073-016-0330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker RW, Clemente JC, Peter I, Loos RJF. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes. 2017; 12: 3–17. 10.1111/ijpo.12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mulligan CM, Friedman JE. Maternal modifiers of the infant gut microbiota: metabolic consequences. J Endocrinol. 2017; 235: R1–R12. 10.1530/JOE-17-0303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother ‘s weight on infant ‘s microbiota acquisition, composition, and activity during early infancy : a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010; 92: 1023–1030. 10.3945/ajcn.2010.29877 [DOI] [PubMed] [Google Scholar]
- 65.Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014; 9: e113026 10.1371/journal.pone.0113026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell SL, Gold MJ, Reynolds LA, Willing BP, Dimitriu P, Thorson L, et al. Perinatal antibiotic-induced shifts in gut microbiota have differential effects on inflammatory lung diseases. J Allergy Clin Immunol. 2015; 135: 100–109.e5. 10.1016/j.jaci.2014.06.027 [DOI] [PubMed] [Google Scholar]
- 67.Singh SB, Madan J, Coker M, Hoen A, Baker ER, Karagas MR, et al. Does birth mode modify associations of maternal pre-pregnancy BMI and gestational weight gain with the infant gut microbiome? Int J Obes. 2019, 10.1038/s41366-018-0273-0 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U. S. A. 2010; 107: 11971–11975. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rutayisire E, Huang K, Liu Y, Tao F. The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants’ life: a systematic review. BMC Gastroenterol. 2016; 16: 86 10.1186/s12876-016-0498-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I, et al. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015; 60: 825–833. 10.1097/MPG.0000000000000752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiménez E, Fernández L, Marín ML, Martín R, Odriozola JM, Nueno-Palop C, et al. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol. 2005; 51: 270–274. 10.1007/s00284-005-0020-3 [DOI] [PubMed] [Google Scholar]
- 72.Korpela K, Salonen A, Hickman B, Kunz C, Sprenger N, Kukkonen K, et al. Fucosylated oligosaccharides in mother’s milk alleviate the effects of caesarean birth on infant gut microbiota. Sci Rep. 2018; 8: 13757 10.1038/s41598-018-32037-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pannaraj PS, Li F, Cerini C, Bender JM, Yang S, Rollie A, et al. Association between breast milk bacterial communities and establishment and development of the infant gut microbiome. JAMA Pediatr. 2017; 171: 647–654. 10.1001/jamapediatrics.2017.0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuyama M, Gomez-Arango LF, Fukuma NM, Morrison M, Davies PSW, Hill RJ. Breastfeeding: a key modulator of gut microbiota characteristics in late infancy. J Dev Orig Health Dis. 2019; 10: 206–213. 10.1017/S2040174418000624 [DOI] [PubMed] [Google Scholar]
- 75.Lee SA, Lim JY, Kim BS, Cho SJ, Kim NY, Kim O, et al. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract 2015; 9: 242–248. 10.4162/nrp.2015.9.3.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grönlund MM, Gueimonde M, Laitinen K, Kociubinski G, Grönroos T, Salminen S, et al. Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the Bifidobacterium microbiota in infants at risk of allergic disease. Clin Exp Allergy. 2007; 37: 1764–1772. 10.1111/j.1365-2222.2007.02849.x [DOI] [PubMed] [Google Scholar]
- 77.Garrido D, Barile D, Mills DA. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv Nutr. 2012; 3: 415S–21S. 10.3945/an.111.001586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fallani M, Young D, Scott J, Norin E, Amarri S, Adam R, et al. Intestinal microbiota of 6-week-old infants across europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J Pediatr Gastroenterol Nutr. 2010; 51: 77–84. 10.1097/MPG.0b013e3181d1b11e [DOI] [PubMed] [Google Scholar]
- 79.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, et al. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PLoS One. 2013; 8: e78331 10.1371/journal.pone.0078331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barile D, Rastall RA. Human milk and related oligosaccharides as prebiotics. Curr Opin Biotechnol. 2013; 24: 214–219. 10.1016/j.copbio.2013.01.008 [DOI] [PubMed] [Google Scholar]
- 81.American Academy Of Pediatrics. Breastfeeding and the use of human milk. Pediatrics. 2012; 129: e827–41. 10.1542/peds.2011-3552 [DOI] [PubMed] [Google Scholar]
- 82.Lee SA, Lim JY, Kim BS, Cho SJ, Kim NY, Kim OB, Kim Y. Comparison of the gut microbiota profile in breast-fed and formula-fed Korean infants using pyrosequencing. Nutr Res Pract. 2015; 9: 242–248. 10.4162/nrp.2015.9.3.242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ho NT, Li F, Lee-Sarwar KA, Tun HM, Brown BP, Pannaraj PS, et al. Meta-analysis of effects of exclusive breastfeeding on infant gut microbiota across populations. Nat Commun. 2018; 9: 4169 10.1038/s41467-018-06473-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ferretti P, Pasolli E, Tett A, Asnicar F, Gorfer V, Fedi S, et al. Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe. 2018; 24: 133–145.e5. 10.1016/j.chom.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asnicar F, Manara S, Zolfo M, Truong DT, Scholz M, Armanini F, et al. Studying vertical microbiome transmission from mothers to infants by strain-level metagenomic profiling. mSystems. 2017; 2: e00164–16. 10.1128/mSystems.00164-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turroni F, Foroni E, Serafini F, Viappiani A, Montanini B, Bottacini F, et al. Ability of Bifidobacterium breve to grow on different types of milk: exploring the metabolism of milk through genome analysis. Appl Environ Microbiol. 2011; 77: 7408–7417. 10.1128/AEM.05336-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Murphy K, Curley D, O’callaghan TF, O’shea CA, Dempsey EM, O’toole PW, et al. The composition of human milk and infant faecal microbiota over the first three months of life: A pilot study. Sci Rep. 2017; 7: 40597 10.1038/srep40597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010; 58: 5334–5340. 10.1021/jf9044205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome. 2015; 3: 13 10.1186/s40168-015-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Heatplot showing OTUs of GDM patients at enrolment (red bars), study end (green bars) and their offspring (blue bars). Rows and columns are clustered by means of Ward linkage hierarchical clustering. The intensity of the colors represents the degree of correlation between the samples and OTUs as measured by Spearman’s correlations.
(TIF)
Samples are color-coded according to compliance to dietary recommendations (red) or non-compliance to dietary recommendations (blue).
(TIF)
Inner black bars indicate the presence of an oligotype in a given sample.
(TIF)
Plot showing the sequence distribution in GDM patients at enrolment (red bars), study end (green bars) and their offspring (blue bars). Inner bars indicate the presence of an oligotype in a given sample. Outer circle, if colored, denotes oligotype abundance with high degree of mother-offspring concordance.
(TIF)
Plot shows the relative phyla (panel A) and genus (panel B) abundance in the offspring from healthy and GDM women.
(TIF)
(DOCX)
Data Availability Statement
All the sequencing data were deposited at the Sequence Read Archive of the National Center for Biotechnology Information (SRA accession number: SRP189631 and SRP135886)