Abstract
The identification and quantification of actionable mutations are of critical importance for effective genotype-directed therapies, prognosis and drug response monitoring in patients with non-small-cell lung cancer (NSCLC). Although tumor tissue biopsy remains the gold standard for diagnosis of NSCLC, the analysis of circulating tumor DNA (ctDNA) in plasma, known as liquid biopsy, has recently emerged as an alternative and noninvasive approach for exploring tumor genetic constitution. In this study, we developed a protocol for liquid biopsy using ultra-deep massively parallel sequencing (MPS) with unique molecular identifier tagging and evaluated its performance for the identification and quantification of tumor-derived mutations from plasma of patients with advanced NSCLC. Paired plasma and tumor tissue samples were used to evaluate mutation profiles detected by ultra-deep MPS, which showed 87.5% concordance. Cross-platform comparison with droplet digital PCR demonstrated comparable detection performance (91.4% concordance, Cohen’s kappa coefficient of 0.85 with 95% CI = 0.72–0.97) and great reliability in quantification of mutation allele frequency (Intraclass correlation coefficient of 0.96 with 95% CI = 0.90–0.98). Our results highlight the potential application of liquid biopsy using ultra-deep MPS as a routine assay in clinical practice for both detection and quantification of actionable mutation landscape in NSCLC patients.
Introduction
Cancer of the lung is the leading type of cancer, responsible for the highest number of new cases and the largest number of deaths worldwide [1]. Non-small cell lung cancer (NSCLC) is the most common subtype, accounting for approximately 85% of all cases [2]. The majority of NSCLC patients display advanced disease when diagnosed and thus have poor prognosis [2, 3]. Treatment options for NSCLC patients are based on the stage of the cancer but high recurrence rate of 30–70% is expected after surgical resection [4]. In patients with advanced stage or tumor recurrence, the mutation profiles of cancer tissue are vital to guide targeted therapy and monitor the tumor recurrence, thereby improving the survival rate of advanced NSCLC patients [4, 5].
Acquired genetic alterations in the EGFR, KRAS, NRAS, BRAF, ROS1 and ALK oncogenes are the most common mutations in NSCLC and certain mutations are associated with drug sensitivity or resistance [6, 7]. Advanced NSCLC patients harbouring activating EGFR mutations including deletion in exon 19 (del19) or a point mutation L858R in exon 21 (L858R) exhibited longer progressive-free survival after receiving treatment with gefitinib, a tyrosine kinase inhibitor (TKI) [8–10]. However, patients treated with the first and second generation TKI drugs such as afatinib and gefitinib often develop a TKI resistant mutation T790M in EGFR exon 20 after a median period of 12 months [11, 12]. In such cases, a third generation TKI drug, osimertinib, has been shown to be effective against cells with the T790M mutation [13]. Apart from mutations in EGFR, a significant proportion of NSCLC patients harbour somatic mutations in other oncogenes, downstream effector molecules of the EGFR pathway, including KRAS (15–25%), BRAF (1–3%) and NRAS (1%) [14, 15]. It has been reported that carriers of NRAS and BRAF mutations display distinct clinicopathologic features and that BRAF mutation testing has recently been recommended for NSCLC patients by American Society of Clinical Oncology (ASCO) [14, 16, 17]. Patients with KRAS mutations were shown to develop resistance to the current EGFR targeted therapies, supporting the use of KRAS mutations as negative prediction biomarkers [18]. However, its clinical significance has been challenged by recent meta-analysis studies reporting inconsistent results amongst different patient cohorts [19–21]. Nevertheless, these studies highlighted that comprehensive mutation analysis of cancer driver genes is essential to provide NSCLC patients with the optimal treatment regimen.
Tumor tissue biopsy is regarded as the gold standard for tumor genetic profiling in current clinical practice [22]. However, since this is an invasive procedure, it is not always feasible to carry out the biopsy to assess patients’ responses following initial treatment, particularly in those who are in advanced stages or do not have sufficient tumor tissues [23]. Liquid biopsy has recently been shown to better reflect the whole genetic complexity of tumor tissues and enables real-time monitoring of treatment-associated resistance [24, 25]. This approach involves detecting genetic alterations in circulating tumor DNAs (ctDNA), which are 160–200 bp DNA fragments released into the blood circulation by tumor cells undergoing cell death [24]. However, the low abundance of ctDNA as well as low variant allele frequency (VAF) of somatic mutations in human plasma necessitates the use of a highly sensitive analytical technique for genetic assessment in liquid biopsy [26]. Several methods have been developed to detect low VAF mutations in plasma, including targeted methods such as amplification refractory mutation system (ARMS) and droplet digital PCR (ddPCR) or non-targeted genome wide massively parallel sequencing (MPS) [27–30]. However, both ARMS and MPS are not sensitive enough to detect low VAF mutations in plasma samples, discouraging its application in liquid biopsy [29, 31, 32]. In contrast, ddPCR has been shown to achieve high sensitivity and accuracy for both identification and quantification of mutations in ctDNA, enabling the evaluation of the intra-tumor progression of drug sensitive or resistant mutant clones [27, 33]. However, this technology relies on prior knowledge of tumor genetic constitution and only allows analyzing a limited number of mutations per reaction.
Recent advances in MPS technology such as unique molecular barcoding has made substantial improvements on its sensitivity and accuracy [34–37]. Unlike ddPCR, MPS is capable of exploring the complete mutation landscape of multiple driver genes simultaneously [38]. This provides particular advantage for longitudinal monitoring of tumor progression and recurrence following initial treatments, and could lead to the discovery of novel mutations that might be of clinical significance [38]. With enhanced sensitivity and accuracy, we believe ultra-deep MPS with unique molecular identifier tagging represents a promising method applicable for liquid biopsy.
In the present study, we adopted ultra-deep MPS for liquid biopsy and evaluated its clinical use for both detection and quantification of plasma circulating tumor DNA in advanced NSCLC patients. The performance of ultra-deep MPS was also compared against that of ddPCR to demonstrate a comparable performance with added benefit of detecting more mutations in more target genes.
Methods
Patient recruitment
A total of 58 patients diagnosed with NSCLC from Pham Ngoc Thach hospital, Thu Duc district hospital, Ho Chi Minh City and National cancer hospital Vietnam were recruited to this study, 40 of which provided paired samples of tissue biopsies and plasma, while the remaining 18 provided only plasma samples (Fig 1). Written informed consents were obtained from all patients. Comprehensive details of patients’ clinical factors were summarised in S1 Table and listed in S2 Table. This study was approved by The Ethic Committee of University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam (Ethic number: 027/DHYD-HD). Case No. 50, 51, 52 and 53 were confirmed to experience TKI treatment.
Fig 1. Schematic diagram of the sample handling procedure.
Clinical sample collection
Prior to tissue biopsy, 10 mL of peripheral blood was drawn in K2-EDTA tubes (BD Vacutainer, USA), stored at room temperature for maximum of 8 hours before 2 rounds of centrifugation (2,000 x g for 10 min then 16,000 x g for 10 min) to separate plasma from blood cells. The plasma (4–6 mL) was then collected, aliquoted (2 mL per aliquot) and stored at -80°C until cell free DNA extraction. Tissue biopsies were collected, formalin-fixed and paraffin-embedded (FFPE) and then the tumor-rich areas of the FFPE tissues that contain at least 50% of tumor cells identified by a hematoxylin and eosin staining were micro-dissected.
DNA isolation
Cell free DNA was extracted from an aliquot of 2 mL of plasma using the MagMAX Cell-Free DNA Isolation kit (Thermo Fisher, USA) following the manufacturer’s instructions. Tumor tissue-derived DNA was extracted from FFPE samples using QIAamp DNA FFPE Tissue Kit (Qiagen, USA) following the manufacturer’s instructions. Both cell free DNA from plasma and genomic DNA from FFPE (2μl of sample) were then quantified using the QuantiFluor dsDNA system (Promega, USA) and Quantus Fluorometer (Promega, USA).
Ultra-deep massively parallel sequencing with unique molecular identifier tagging
For cell free DNA (cfDNA), library with unique molecular identifier tagging were prepared from 2 ng of cfDNA using the Accel-NGS 2S Plus DNA library kit (Swift Biosciences, USA) following the manufacturer’s instructions. Library concentrations were quantified with a QuantiFluor dsDNA system (Promega, USA). Equal amounts of libraries were pooled together and hybridized with xGen Lockdown probes for four targeted genes EGFR, KRAS, NRAS, BRAF (IDT DNA, USA). Sequencing was performed using NextSeq 500/550 High output kits v2 (150 cycles) on Illumina NextSeq 550 system (Illumina, USA) with the coverage of 10.000X.
For genomic DNA from FFPE, libraries were prepared from 2 ng of cfDNA using the NEBNext Ultra II FS DNA library prep kit (New England Biolabs, USA) following the manufacturer’s instructions. Similar to ctDNA libraries, FFPE libraries were pooled before hybridization with the xGen Lockdown probes and sequencing in the Illumina NextSeq 550 system. Both cfDNA and FFPE samples exhibited 5–10% on-target reads.
Variant calling using Mutect2
For ctDNA, each sample was barcoded with a single 8-bp index in the P7 primer and each DNA fragment were tagged with a unique identifier consisting of a random 9-bp sequence within the P5 primer. Pair-end (PE) reads and the correspondent unique identifier sequences were generated using bcl2fastq package (Illumina). The reads were aligned to human genome (hg38) using BWA package and then grouped by the unique identifier in order to determine a consensus sequence for each fragment, eliminating sequencing and PCR errors that account for less than 50% of reads per fragment. The consensus reads were used for final variant calling using Mutect2. A custom pipeline with call to BWA, Picard, Samtools and Fulcrum genomic analysis packages were built to perform the above-mentioned analysis steps.
For genomic DNA from FFPE samples, each sample was barcoded with dual indexes in the P7 and P5 primer. The PE reads were generated by bcl2fastq package (Illumina) and aligned to human genome (hg38) using BWA package. Duplicate reads were marked using MarkDuplicates from Picard tools (Broad Institute). Somatic variants were called using Mutect2 package (Broad Institute). A custom pipeline with call to BWA, Picard, and Samtools packages were built to perform the above-mentioned analysis steps.
ddPCR method
A four-step ddPCR procedure was performed using reagents and equipment from Bio-Rad (unless otherwise stated) following the manufacturer’s instruction [39]. Briefly, the PCR mix was first prepared by mixing 1 × ddPCR Supermix for Probes, primers and probes (IDT DNA) and DNA template (0.8 or 1.6 ng). Next, 20 μl of the PCR mix was transferred into the Droplet Generator DG8TM Cartridge followed by 70 μl of the Droplet Generation Oil before placing in a QX100TM Droplet Generator to generate droplets. Subsequently, the droplets were transferred to a 96-well plate before placing in a thermal cycler (C1000 Touch, Bio-Rad) for PCR amplification. The PCR thermal program was performed as follows: 95°C for 10 min, then 40 successive cycles of amplification (94°C for 30 sec; 55°C for 60 sec) and 98°C for 10 min. Lastly, the droplet reading was acquired by the QX 200 Droplet reader and analyzed using the QuantaSoft Software. Positive and negative droplets were assigned based on the fluorescence threshold that was set as previously described by Deprez et al. [40].
To detect T790M and L858R mutations in exon 20 and 21 of the EGFR gene, one reaction of ddPCR was used with two sets of primers and probes as follows: T790M primer F-GCCTGCTGGGCATCTG; T790M primer R-TCTTTGTGTTCCCGGACATAGTC; T790M mutation probe FAM- ATGAGCTGCTGATGAG-ZEN/3'IBFQ; L858R primer F-GCAGCATGTCAAGATCACAGATT; L858R primer R-CCTCCTTCTGCATGGTATTCTTTCT; L858R mutation probe HEX-AGTTTGGCCGCCCAA- ZEN/3'IBFQ. For detection of 15 deletion sites in exon 19 (del19) of the EGFR gene, a commercially available ddPCR reaction (Bio-rad) was used (ddPCR™ EGFR Exon 19 Deletions Screening Kit #12002392).
Determination of limit of detection
To determine the limit of detection (LOD) for our assays, we first performed fragmentation of reference wild type (WT) and mutant DNA (Tru-Q1 and Tru-Q2, Horizon) to create 100–200 bp fragments corresponding to the general length of plasma cell-free DNA. Subsequently, these mutant DNA fragments were spiked into fragmented WT DNA to obtain a series of standard samples with a desired variant allele frequency (VAF) range. The LOD value was defined as the lowest VAF that can be reliably detected by ddPCR or Ultra deep MPS. The LOD values of ddPCR and ultra-deep MPS assay for detecting major driver mutations in plasma were 0.5% and 1%, equivalent to 5 and 10 mutant copies per 1,000 copies of DNA input, respectively.
Statistical analysis
All statistical tests and visualisation plots were performed using R, the ggplot2 and ggpubr packages. Cohen’s Kappa coefficient and its confidence intervals using the psych package were employed to assess the reliability of mutation detection by ddPCR and MPS. Pearson’s correlation coefficient and Bland-Altman’s plot were performed to examine the correlations and agreement, respectively, between VAF results obtained by the two methods. To assess the reliability of VAF quantification, Intraclass correlation coefficient (ICC) estimates and their 95% confident intervals were calculated using irr package based on single rater type, consistency definition, and a 2-way random-effects model.
Results
High concordance between mutations detected by paired liquid and tissue biopsy
In this study, we developed a liquid biopsy protocol based on ultra-deep Illumina sequencing with unique molecular identifier tagging for detecting mutations in four genes EGFR, KRAS, NRAS and BRAF for patients with advanced NSCLC. To evaluate the mutations detected by liquid biopsy, we examined the concordance between mutations detected from plasma samples and from tissue samples in the cohort of 40 patients who provided paired plasma-tissue samples (Table 1). Within this cohort, liquid biopsy detected 9 types of mutations in two genes EGFR and KRAS, while no mutation was detected in either BRAF or NRAS gene (Table 1). Deletions in exon 19 of EGFR (del19) were the most common, found in 5 plasma samples (Table 1, Case No. 4–8), followed by L858R in EGFR in 3 samples (Table 1, Case No. 1–3), then insertion in EGFR exon 20 in 2 samples (Table 1, Case No. 14 and 35). The remaining mutations were found in one sample each, including H773A, S768C, in EGFR (Table 1, Case No. 15 and 21) and G12V, G12C and G12D in KRAS (Table 1, Case No. 30, 32 and 37). One plasma sample was found to harbour double mutations of L858R and T790M, a TKI resistant mutation, in EGFR (Table 1, Case No. 11). Compared to the mutations detected in tissue samples, the assays showed high concordance rate of 87.5% (35/40), including 15 patients with matching mutation profiles and 20 patients with no mutation detected (Table 1). Among the five discordant cases, four were positive for mutations in EGFR in tissue but negative in plasma (Table 1, Case No. 10,12, 13 and 24), while one was negative in tissue but positive for EGFR del19 in plasma (Table 1, Case No. 5).
Table 1. Mutation results of 40 plasma and matched tumor tissue samples detected by ultra-deep MPS.
| Case No. | Sample ID | NGS Results | |||
|---|---|---|---|---|---|
| Plasma | Tumor tissues | ||||
| Mutation | VAF (%) | Mutation | VAF (%) | ||
| 1 | LBL015 | EGFR L858R | 89 | EGFR L858R | 65 |
| 2 | LBL017 | EGFR L858R | 2 | EGFR L858R | 90 |
| 3 | L10055 | EGFR L858R | 1 | EGFR L858R | 55 |
| 4 | L10019 | EGFR del19 | 17 | EGFR del19 | 11 |
| 5 | L10021 | EGFR del19 | 6 | (-) | |
| 6 | L10036 | EGFR del19 | 50 | EGFR del19 | 44 |
| 7 | L10072 | EGFR del19 | 8 | EGFR del19 | 50 |
| 8 | L10076 | EGFR del19 | 8 | EGFR del19 | 34 |
| 9 | LBL021 | (-) | (-) | ||
| 10 | LBL033 | (-) | EGFR del19 | 20 | |
| 11 | L10022 | EGFR L858R | 1 | EGFR L858R | 37 |
| EGFR T790M | 5 | EGFR T790M | 43 | ||
| 12 | LBL026 | (-) | EGFR del19 | 23 | |
| 13 | LBL030 | (-) | EGFR del19 | 10 | |
| 14 | LBL001 | EGFR ins20 | 1.5 | EGFR ins20 | 25 |
| 15 | LBL002 | EGFR H773A | 15 | EGFR H773A | 1 |
| 16 | LBL003 | (-) | (-) | ||
| 17 | LBL004 | (-) | (-) | ||
| 18 | LBL005 | (-) | (-) | ||
| 19 | LBL006 | (-) | (-) | ||
| 20 | LBL007 | (-) | (-) | ||
| 21 | LBL008 | EGFR S768C | 1.5 | EGFR S768C | 45 |
| 22 | LBL009 | (-) | (-) | ||
| 23 | LBL012 | (-) | (-) | ||
| 24 | LBL013 | (-) | EGFR ins20 | 20 | |
| 25 | LBL014 | (-) | (-) | ||
| 26 | LBL016 | (-) | (-) | ||
| 27 | LBL020 | (-) | (-) | ||
| 28 | LBL022 | (-) | (-) | ||
| 29 | LBL023 | (-) | (-) | ||
| 30 | LBL024 | KRAS G12V | 5 | KRAS G12V | 1 |
| 31 | LBL025 | (-) | (-) | ||
| 32 | LBL027 | KRAS G12C | 3.5 | KRAS G12C | 23 |
| 33 | LBL028 | (-) | (-) | ||
| 34 | LBL029 | (-) | (-) | ||
| 35 | LBL031 | EGFR ins20 | 2.9 | EGFR ins20 | 25 |
| 36 | LBL034 | (-) | (-) | ||
| 37 | LBL036 | KRAS G12D | 1 | KRAS G12D | 1 |
| 38 | LBL037 | (-) | (-) | ||
| 39 | LBL040 | (-) | (-) | ||
| 40 | LBL041 | (-) | (-) | ||
(-): negative for tested mutation
Comparable performance between ultra-deep MPS and droplet digital PCR (ddPCR) for EGFR mutation detection in plasma samples
Droplet digital PCR (ddPCR) has been reported to achieve high sensitivity and specificity for the detection of low frequency mutations such as those in ctDNA from plasma, with a limit of detection of less than 0.001% (1 copy of mutant DNA per 100,000 copies of wild-type DNA background) [26]. Using a commercially available ddPCR (Bio-rad) assay as a reference standard, we conducted a cross-platform comparison with ultra-deep MPS for the detection of the three most common actionable EGFR mutations (del19, L858R and T790M) in 58 plasma samples comprising the 40 previously tested samples and 18 additional samples (Table 2). The concordant rate between the two methods were 91.4% (53/58 samples), whereby for 19 cases, both methods agreed upon the mutation identified and 35 more cases were also congruent, by not having any of the 3 target mutations (Table 3).
Table 2. Mutational profile and variant allele frequency (VAF) determined by ultra-deep MPS and ddPCR in 58 plasma samples.
| Case No. | Sample ID | Ultra-deep MPS Results | ddPCR Results |
||
|---|---|---|---|---|---|
| Mutation | VAF (%) | Mutation | VAF (%) |
||
| 1 | LBL015 | EGFR L858R | 89 | EGFR L858R | 86.0 |
| 2 | LBL017 | EGFR L858R | 2 | EGFR L858R | 2.8 |
| 3 | L10055 | EGFR L858R | 1 | EGFR L858R | 4.6 |
| 4 | L10019 | EGFR del19 | 17 | EGFR del19 | 28.6 |
| 5 | L10021 | EGFR del19 | 6 | EGFR del19 | 7.0 |
| 6 | L10036 | EGFR del19 | 50 | EGFR del19 | 68.4 |
| 7 | L10072 | EGFR del19 | 8 | EGFR del19 | 12.8 |
| 8 | L10076 | EGFR del19 | 8 | EGFR del19 | 9.5 |
| 9 | LBL021 | (-) | EGFR del19 | 0.7 | |
| 10 | LBL033 | (-) | EGFR del19 | 0.5 | |
| 11 | L10022 | EGFR L858R | 1 | EGFR L858R | 0.75 |
| EGFR T790M | 5 | EGFR T790M | 0.75 | ||
| 12 | LBL026 | (-) | (-) | ||
| 13 | LBL030 | (-) | (-) | ||
| 14 | LBL001 | EGFR ins20 | 1.5 | NA | |
| 15 | LBL002 | EGFR H773A | 15 | NA | |
| 16 | LBL003 | (-) | (-) | ||
| 17 | LBL004 | (-) | (-) | ||
| 18 | LBL005 | (-) | (-) | ||
| 19 | LBL006 | (-) | (-) | ||
| 20 | LBL007 | (-) | (-) | ||
| 21 | LBL008 | EGFR S768C | 1.5 | NA | |
| 22 | LBL009 | (-) | (-) | ||
| 23 | LBL012 | (-) | (-) | ||
| 24 | LBL013 | (-) | (-) | ||
| 25 | LBL014 | (-) | (-) | ||
| 26 | LBL016 | (-) | (-) | ||
| 27 | LBL020 | (-) | (-) | ||
| 28 | LBL022 | (-) | (-) | ||
| 29 | LBL023 | (-) | (-) | ||
| 30 | LBL024 | KRAS G12V | 5 | NA | |
| 31 | LBL025 | (-) | (-) | ||
| 32 | LBL027 | KRAS G12C | 3.5 | NA | |
| 33 | LBL028 | (-) | (-) | ||
| 34 | LBL029 | (-) | (-) | ||
| 35 | LBL031 | EGFR ins20 | 2.9 | NA | |
| 36 | LBL034 | (-) | (-) | ||
| 37 | LBL036 | KRAS G12D | 1 | NA | |
| 38 | LBL037 | (-) | (-) | ||
| 39 | LBL040 | (-) | (-) | ||
| 40 | LBL041 | (-) | (-) | ||
| 41 | LBL019 | EGFR L858R | 0.8 * | EGFR L858R | 1.2 |
| 42 | LBL032 | (-) | EGFR L858R | 0.9 | |
| 43 | LBL035 | EGFR L858R | 0.7 * | EGFR L858R | 2.8 |
| 44 | L10002 | EGFR L858R | 3 | EGFR L858R | 5.8 |
| 45 | L10045 | EGFR L858R | 20 | EGFR L858R | 15.9 |
| 46 | L10077 | EGFR L858R | 8 | EGFR L858R | 6.1 |
| 47 | LBL038 | (-) | EGFR del19 | 3.4 | |
| 48 | LBL010 | EGFR del19 | 5 | EGFR del19 | 12.1 |
| 49 | L10043 | EGFR del19 | 23 | EGFR del19 | 36.0 |
| 50 | L10005 | EGFR L858R | 4 | EGFR T790M | 1.5 |
| EGFR L858R | 3.9 | ||||
| 51 | L10046 | EGFR L858R | 24 | EGFR T790M | 18 |
| EGFR T790M | 56 | EGFR L858R | 61.7 | ||
| 52 | L10007 | EGFR T790M | 16 | EGFR T790M | 12.9 |
| EGFR del19 | 20 | EGFR del19 | 34 | ||
| 53 | L10074 | EGFR T790M | 15 | EGFR T790M | 15 |
| EGFR del19 | 31 | EGFR del19 | 37.7 | ||
| 54 | LBL011 | (-) | (-) | ||
| 55 | LBL018 | (-) | (-) | ||
| 56 | LBL039 | (-) | (-) | ||
| 57 | LBL042 | (-) | (-) | ||
| 58 | LBL043 | (-) | (-) | ||
(-): negative for tested mutations; NA: mutations not analysed by ddPCR;
(*): VAF below the limiting detection of Ultra-deep MPS
Table 3. Evaluation of performance of ddPCR and MPS for mutation detection in 58 plasma samples.
| NGS vs ddPCR | NGS | Performance results | |||
|---|---|---|---|---|---|
| ddPCR | Mutation | Wild type | Total | ||
| Mutation | 19 | 5 | 24 | Sensitivity | 79.2% |
| Wild type | 0 | 34 | 34 | Specificity | 100.0% |
| Total | 19 | 39 | 58 | Concordance | 91.4% |
Among the 5 discordant cases, 4 were negative by ultra-deep MPS while positive for del19 (Table 2, Case No. 9, 10 and 47) or L858R (Table 2, Case No. 42) by ddPCR; and 1 were a double mutation (L858R & T790M) by ddPCR but a single L858R mutation by ultra-deep MPS (Table 2, case No. 50). There were 7 cases where ultra-deep MPS detected mutations other than the three types targeted by ddPCR, illustrating an advantage of MPS over ddPCR (Table 2, case No. 14, 15, 21, 30, 32, 35 and 37).
If we considered ddPCR as a reference method and counted the 7 samples with mutations outside of ddPCR detectable mutations as wild type, the sensitivity and specificity of the ultra-deep MPS assay for EGFR mutation detection in plasma samples were 79.2% (19/24, 95% CI = 57.8%-92.9%) and 100% (34/34), respectively, with an accuracy of 91.4% (53/58) (Table 3). The Cohen’s kappa coefficient was 0.85 (95% CI = 0.72–0.97), suggesting good agreement between the two methods. Taken together, these results demonstrated that liquid biopsy using ultra-deep MPS achieved good agreement with ddPCR for the detection of mutations from ctDNA in plasma samples.
Quantitative measurement of mutation allelic frequency by ultra-deep sequencing and ddPCR
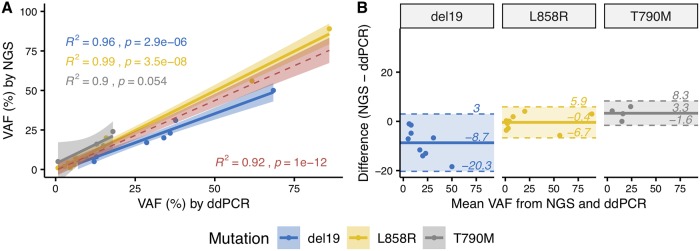
Besides high mutation detection sensitivity in ctDNA, ddPCR also shows the capability of absolute mutation quantification, allowing better disease prognosis and therapy response monitoring [26, 41]. To evaluate the quantitative measurement of VAF by ultra-deep sequencing, we compared the VAF for the three EGFR mutations (del19, L858R and T790M) with those reported by ddPCR. VAFs reported by the two methods exhibited a strong overall Pearson’s linear correlation (R2 = 0.92, P <0.0001) (Fig 2A). More specifically, VAFs of L858R mutation showed the best correlation (R2 = 0.99, P <0.0001), followed by VAFs of del19 (R2 = 0.96, P <0.0001), then by VAFs of T790M (R2 = 0.90, P = 0.05) (Fig 2A). Intraclass correlation coefficient (ICC) for the two methods was estimated at 0.96 (95% CI = 0.90–0.98), indicates excellent reliability. Bland-Altman analysis revealed relatively high level of agreement between two methods, of which del19 mutations showed the largest range of limits of agreement (LOA) from -20.3% to 3%, followed by L858R from -6.7% to 5.9% and T790M from -1.6 to 8.3% (Fig 2B). Thus, the liquid biopsy based on ultra-deep sequencing exhibited comparable quantitative measurement of VAF to that of ddPCR.
Fig 2. Comparing mutation allele frequency quantified from plasma by ultra-deep MPS and ddPCR.
(A) Linear regression and Pearson’s correlation coefficients of VAFs in plasma samples as determined by ddPCR and ultra-deep MPS with unique molecular identifier tagging. VAFs of del19, L858R and T790M mutations in EGFR were analysed separately (blue, yellow and grey, respectively) and combined (red) to show that MPS achieved significant correlation with ddPCR. (B) Bland-Altman plots demonstrating the agreement between ultra-deep MPS and ddPCR in quantifying VAFs of the three mutation types in EGFR from plasma samples.
Discussion
The American Society of Clinical Oncology (ASCO) has stated that the identification of somatic driver mutations is essential for designing optimal treatment regimens for NSCLC patients [17]. The technical and clinical limitations of traditional tissue biopsy necessitate the development of liquid biopsy, a procedure of detecting and quantifying tumor-derived mutations from ctDNA found in plasma samples of cancer patients [23, 24]. The choice of analytical platform for liquid biopsy requires proper evaluation, taking into account sensitivity, repeatability, discoverability and feasibility in clinical settings [42, 43].
In this study, we aimed to demonstrate that ultra-deep MPS with unique molecular identifier tagging is suitable for liquid biopsy to detect and quantify mutations in ctDNA of NSCLC patients. First, we used paired plasma and tumor tissue samples to examine whether liquid biopsy using ultra-deep MPS could detect the mutation profiles found in tumor tissues. Despite of the small sample size of this cohort (n = 40), the mutation profiles identified support previous findings that the majority of adenocarcinomas associated mutations occur in EGFR exon 19 and 21 and that KRAS and EGFR mutations are mutually exclusive [44–46]. Although NRAS and BRAF mutations have been found in NSCLC patients, none of the cases in this cohort was identified to carry such mutations. At high concordance rate of 87.5% between liquid and tissue biopsies, our results indicated that ultra-deep MPS could be useful for exploring the mutational landscape of NSCLC in clinical practice. There were four cases where mutations in EGFR (3 del19 and 1 ins20) were found in tissue but not in paired plasma samples, probably due to the low abundance of ctDNA in plasma [47]. Indeed, assaying these three plasma samples (del19 in tissue) by ddPCR showed that two were also negative and 1 with low VAF of 0.5% (Table 1, Case No. 10, 12 and 13). In contrast, there was one case where EGFR del19 mutation was detected in plasma but not in its paired tissue. This could be explained by the intratumoral genetic heterogeneity with the presence multiple cancer clones [47]. To address these issues, the current ASCO guidelines recommend that positive testing results in plasma would allow drawing definitive conclusion about the presence of mutation and that wild-type testing results in liquid biopsy be retested using tissue biopsy [42].
Second, by using ddPCR targeting three clinically actionable mutations in EGFR (del19, L858R and T790M) as the reference method, we conducted a cross-platform comparison of the performance of ultra-deep MPS in detecting these three mutations in 58 plasma samples. Ultra-deep MPS exhibited excellent concordance with ddPCR (91.4%), including 4/5 cases of double mutations (del19&T790M and L858R&T790M) (Table 2). The presence of T790M mutation in these patients was consistent with their previous treatment with first generation TKIs, suggesting that they might benefit from a third generation TKI therapy [41]. Ultra-deep MPS achieved sensitivity and specificity of 79.2% (19/24, 95% CI = 57.8%-92.9%) and 100% (34/34), respectively (Table 2). Of note, there were 5 cases positive by ddPCR but negative by ultra-deep MPS; three of which had VAF values lower than the LOD of the ultra-deep MPS assay (1%) while the other two also had low VAF (1.5% and 3.4%). Among those five cases, three did not have matched tissues to confirm ddPCR results (case No. 42, 47 and 50); one case was confirmed to have the mutation (del19) identified by ddPCR in matched tissue (case No. 10) and one case did not show any detectable mutation in matched tissue (case No. 9). Our data was consistent with previous studies reporting sensitivity value ranging from 70 to 80% for mutation detection in plasma of advanced NSCLC patients [48–50]. The Cohen’s kappa coefficient was 0.85 (95% CI = 0.72–0.97), further confirmed that ultra-deep MPS is comparable to ddPCR for the detection of 3 actionable mutations in EGFR. In additions, ultra-deep MPS showed extra advantage of ddPCR, capable of detecting more mutations than the limited set in ddPCR assays (Table 2).
Third, we investigated the ability of ultra-deep MPS with unique identifier tagging to quantify VAF in plasma samples. It has been reported that the relative abundance of activating and resistant mutations in EGFR is associated with patient survival rate and that the dynamic and quantitative analysis of EGFR mutations could guide personalized interventions [51]. Here, we demonstrated that ultra-deep MPS achieved accurate measurement of VAF values, showing great agreement with ddPCR (ICC = 0.96 with 95% CI = 0.90–0.98). However, the levels of agreement varied among the three mutations. Bland-Altman analysis (Fig 2B) showed that the LOA range is broadest for del19 mutation and ultra-deep MPS was more likely to give lower VAF estimates for del19 compared to those by ddPCR.
There were some limitations in our study. We could not calculate the costs of running ultra-deep MPS versus ddPCR in clinical settings. However, the reagent cost of a 4-gene panel using ultra-deep MPS was approximately that of ddPCR assays to detect two genetic alterations. Not all mutations detected by ultra-deep MPS were validated by ddPCR due to the limited number of assayed mutations in ddPCR. Although ALK and ROS1 are clinically actionable genes in NSCLC, we did not include them in our ultra-deep MPS analysis because the genetic alterations frequently occur in these genes are rearrangement. Future work is required to solve the challenge of detecting gene rearrangements from ctDNA.
In conclusions, we have demonstrated that, in the context of liquid biopsy, our ultra-deep MPS with unique molecular identifier tagging achieved comparable performance to ddPCR for both the detection and quantification of clinically actionable mutations on plasma ctDNA. Altogether, our results highlight the potential application of liquid biopsy using modified MPS as a routine assay in clinical practice for both detection and quantification of actionable mutation landscape in NSCLC patients.
Supporting information
(XLSX)
(XLSX)
Data Availability
Sequencing data from individuals are only available to interested researchers upon individual requests due to ethical restrictions imposed by The Ethic Committee of University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam. To access the data, please contact Medical Genetic Institute (MGI): Medical Genetic Institute (MGI); Address: 186-188 Nguyen Duy Duong, District 10, Ho Chi Minh city, Vietnam; Email: mgi@suckhoeditruyen.vn; Phone: +84 28 6660 8050.
Funding Statement
This research is funded by The Vietnam National Foundation for Science and Technology Development (NAFOSTED, https://nafosted.gov.vn/) under grant number 108.01-2017.306 (to HNN) and Ho Chi Minh city Department of Science and Technology under grant number 257/2017/HD-SKHCN (to HNN) and Gene Solutions (GS-003). We confirm that the funder Gene Solutions provided support in the form of salaries for authors LS Tran, HAT Pham, VU Tran, TT Tran, BT Vo, HTT Dao, NH Nguyen, HN Do, M-D Phan and H Giang but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 2.Sher T, Dy GK, Adjei AA. Small cell lung cancer. Mayo Clinic proceedings 2008;83:355–67. 10.4065/83.3.355 [DOI] [PubMed] [Google Scholar]
- 3.Birring SS, Peake MD. Symptoms and the early diagnosis of lung cancer. Thorax 2005;60:268–9. 10.1136/thx.2004.032698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang H, Huang J, Wang B, Liu Z, He J, Liang W. The role of liquid biopsy in predicting post-operative recurrence of non-small cell lung cancer. Journal of thoracic disease 2018;10:S838–s45. 10.21037/jtd.2018.04.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Absenger G, Terzic J, Bezan A. ASCO update: lung cancer. Memo 2017;10:224–7. 10.1007/s12254-017-0373-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tafe LJ, Pierce KJ, Peterson JD, de Abreu F, Memoli VA, Black CC, et al. Clinical Genotyping of Non-Small Cell Lung Cancers Using Targeted Next-Generation Sequencing: Utility of Identifying Rare and Co-mutations in Oncogenic Driver Genes. Neoplasia (New York, N.Y.) 2016;18:577–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindeman NI, Cagle PT, Beasley MB, Chitale DA, Dacic S, Giaccone G, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 2013;8:823–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine 2010;362:2380–8. 10.1056/NEJMoa0909530 [DOI] [PubMed] [Google Scholar]
- 9.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine 2009;361:947–57. 10.1056/NEJMoa0810699 [DOI] [PubMed] [Google Scholar]
- 10.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, McCormack R, Webster A, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. British journal of cancer 2014;110:55–62. 10.1038/bjc.2013.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2013;31:3327–34. [DOI] [PubMed] [Google Scholar]
- 12.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clinical cancer research: an official journal of the American Association for Cancer Research 2013;19:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. The New England journal of medicine 2018;378:113–25. 10.1056/NEJMoa1713137 [DOI] [PubMed] [Google Scholar]
- 14.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer research 2002;62:6997–7000. [PubMed] [Google Scholar]
- 15.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069–75. 10.1038/nature07423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinno T, Tsuta K, Shiraishi K, Mizukami T, Suzuki M, Yoshida A, et al. Clinicopathological features of nonsmall cell lung carcinomas with BRAF mutations. Annals of oncology: official journal of the European Society for Medical Oncology 2014;25:138–42. [DOI] [PubMed] [Google Scholar]
- 17.Kalemkerian GP, Narula N, Kennedy EB, Biermann WA, Donington J, Leighl NB, et al. Molecular Testing Guideline for the Selection of Patients With Lung Cancer for Treatment With Targeted Tyrosine Kinase Inhibitors: American Society of Clinical Oncology Endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology Clinical Practice Guideline Update. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2018;36:911–9. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine 2005;2:e17 10.1371/journal.pmed.0020017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loriot Y, Mordant P, Deutsch E, Olaussen KA, Soria JC. Are RAS mutations predictive markers of resistance to standard chemotherapy? Nature reviews. Clinical oncology 2009;6:528–34. 10.1038/nrclinonc.2009.106 [DOI] [PubMed] [Google Scholar]
- 20.Roberts PJ, Stinchcombe TE. KRAS mutation: should we test for it, and does it matter? Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2013;31:1112–21. [DOI] [PubMed] [Google Scholar]
- 21.Pesek M, Benesova L, Belsanova B, Mukensnabl P, Bruha F, Minarik M. Dominance of EGFR and insignificant KRAS mutations in prediction of tyrosine-kinase therapy for NSCLC patients stratified by tumor subtype and smoking status. Anticancer research 2009;29:2767–73. [PubMed] [Google Scholar]
- 22.Melosky B. Current Treatment Algorithms for Patients with Metastatic Non-Small Cell, Non-Squamous Lung Cancer. Frontiers in oncology 2017;7:38 10.3389/fonc.2017.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim SS, Choi HJ, Kim JJ, Kim MS, Lee IS, Byun B, et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Scientific reports 2018;8:543 10.1038/s41598-017-18642-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nature reviews. Clinical oncology 2013;10:472–84. 10.1038/nrclinonc.2013.110 [DOI] [PubMed] [Google Scholar]
- 25.NN S, JA P, H M, A Z, D Y, J T, et al.—Plasma DNA-based molecular diagnosis, prognostication, and monitoring of patients. D—101705370 T—ppublish. [DOI] [PMC free article] [PubMed]
- 26.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Analytical chemistry 2011;83:8604–10. 10.1021/ac202028g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu J, Zang W, Liu B, Li L, Huang L, Li S, et al. Evaluation of digital PCR for detecting low-level EGFR mutations in advanced lung adenocarcinoma patients: a cross-platform comparison study. Oncotarget 2017;8:67810–20. 10.18632/oncotarget.18866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Guo Z, Liu Y, Zheng X, Yang G, Zheng G. A novel ARMS-based assay for the quantification of EGFR mutations in patients with lung adenocarcinoma. Oncology letters 2018;15:2905–12. 10.3892/ol.2017.7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyawali B, West HJ. Plasma vs Tissue Next-Generation Sequencing in Non-Small Cell Lung Cancer-Either, Both, or Neither? JAMA oncology 2018. [DOI] [PubMed] [Google Scholar]
- 30.Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA oncology 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng WN, Gu WQ, Zhao N, Pan YM, Luo W, Zhang H, et al. Comparison of the SuperARMS and Droplet Digital PCR for Detecting EGFR Mutation in ctDNA From NSCLC Patients. Translational oncology 2018;11:542–5. 10.1016/j.tranon.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma L, Zhang F, Xu Q, Tang X, Mao M. 2016;- 11.
- 33.Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Scientific reports 2017;7:2409 10.1038/s41598-017-02217-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Science translational medicine 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacConaill LE, Burns RT, Nag A, Coleman HA, Slevin MK, Giorda K, et al. Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC genomics 2018;19:30 10.1186/s12864-017-4428-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman AM, Bratman SV, To J, Wynne JF, Eclov NC, Modlin LA, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature medicine 2014;20:548–54. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.JG S, MSD M, LP L, KT P, LM S, GR O, et al.—Sensitivity of next-generation sequencing assays detecting oncogenic fusions in. D—8800805–96–9. [DOI] [PubMed]
- 38.Zhang H, Liu R, Yan C, Liu L, Tong Z, Jiang W, et al. Advantage of Next-Generation Sequencing in Dynamic Monitoring of Circulating Tumor DNA over Droplet Digital PCR in Cetuximab Treated Colorectal Cancer Patients. Translational oncology 2019;12:426–31. 10.1016/j.tranon.2018.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bio-Rad Laboratories I. Rare Mutation Detection Best Practices Guidelines. [Google Scholar]
- 40.L D, P C, AM K, S M, R BH, S T, et al.—Validation of a digital PCR method for quantification of DNA copy number. D—101667177–29–39. [DOI] [PMC free article] [PubMed]
- 41.Li JY, Ho JC, Wong KH. T790M mutant copy number quantified via ddPCR predicts outcome after osimertinib treatment in lung cancer. Oncotarget 2018;9:27929–39. 10.18632/oncotarget.25332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meador C, Hu Y, Yang JC-H, Mok T, Laus G, Hovey T, et al. Refining the sensitivity of plasma cell-free DNA (cfDNA) genotyping by controlling for plasma tumor content. Journal of Clinical Oncology 36:9071-. [Google Scholar]
- 43.TA C, JH C, M K, JD H, N C, DS L, et al.—Analytical Validation of a Hybrid Capture-Based Next-Generation Sequencing. D—100893612–686–702.
- 44.Kohsaka S, Petronczki M, Solca F, Maemondo M. Tumor clonality and resistance mechanisms in EGFR mutation-positive non-small-cell lung cancer: implications for therapeutic sequencing. Future oncology (London, England) 2019;15:637–52. [DOI] [PubMed] [Google Scholar]
- 45.Suda K, Tomizawa K, Mitsudomi T. Biological and clinical significance of KRAS mutations in lung cancer: an oncogenic driver that contrasts with EGFR mutation. Cancer metastasis reviews 2010;29:49–60. 10.1007/s10555-010-9209-4 [DOI] [PubMed] [Google Scholar]
- 46.Gainor JF, Varghese AM, Ou SH, Kabraji S, Awad MM, Katayama R, et al. ALK rearrangements are mutually exclusive with mutations in EGFR or KRAS: an analysis of 1,683 patients with non-small cell lung cancer. Clinical cancer research: an official journal of the American Association for Cancer Research 2013;19:4273–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bai H, Mao L, Wang HS, Zhao J, Yang L, An TT, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 2009;27:2653–9. [DOI] [PubMed] [Google Scholar]
- 48.Couraud S, Vaca-Paniagua F, Villar S, Oliver J, Schuster T, Blanche H, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: a proof-of-concept study from BioCAST/IFCT-1002. Clinical cancer research: an official journal of the American Association for Cancer Research 2014;20:4613–24. [DOI] [PubMed] [Google Scholar]
- 49.Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutant lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clinical cancer research: an official journal of the American Association for Cancer Research 2014;20:1698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plagnol V, Woodhouse S, Howarth K, Lensing S, Smith M, Epstein M, et al. Analytical validation of a next generation sequencing liquid biopsy assay for high sensitivity broad molecular profiling. PloS one 2018;13:e0193802 10.1371/journal.pone.0193802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X, Zhuo M, Ye X, Bai H, Wang Z, Sun Y, et al. Quantification of mutant alleles in circulating tumor DNA can predict survival in lung cancer. Oncotarget 2016;7:20810–24. 10.18632/oncotarget.8021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
Sequencing data from individuals are only available to interested researchers upon individual requests due to ethical restrictions imposed by The Ethic Committee of University of Medicine and Pharmacy at Ho Chi Minh City, Vietnam. To access the data, please contact Medical Genetic Institute (MGI): Medical Genetic Institute (MGI); Address: 186-188 Nguyen Duy Duong, District 10, Ho Chi Minh city, Vietnam; Email: mgi@suckhoeditruyen.vn; Phone: +84 28 6660 8050.