Abstract
To date, twelve protein lysine methyltransferases that modify translational elongation factors and ribosomal proteins (Efm1-7; Rkm 1-5) have been identified in the yeast Saccharomyces cerevisiae. Of these twelve, five (Efm1, Efm4-7) appear to be specific to elongation factor 1A (EF1A), the protein responsible for bringing aminoacyl-tRNAs to the ribosome. In S. cerevisiae the functional implications of lysine methylation in translation are mostly unknown. Here we assessed the physiological impact of disrupting EF1A methylation in a strain where four of the most conserved methylated lysine sites are mutated to arginine residues and in strains lacking either four or five of the Efm lysine methyltransferases specific to EF1A. We found that loss of EF1A methylation was not lethal but resulted in reduced growth rates, particularly under caffeine and rapamycin stress conditions, suggesting EF1A interacts with the TORC1 pathway, as well as altered sensitivities to ribosomal inhibitors. We also detected reduced cellular levels of the EF1A protein, which surprisingly was not reflected in its stability in vivo. We present evidence that these Efm methyltransferases appear to be largely devoted to the modification of EF1A, finding no evidence for the methylation of other substrates in the yeast cell. This work starts to illuminate why one protein can need five different methyltransferases for its functions and highlights the resilience of yeast to alterations in their posttranslational modifications.
Keywords: translation elongation factor, yeast, protein methylation, post-translational modification (PTM), stress response, translation
Graphical Abstract
INTRODUCTION
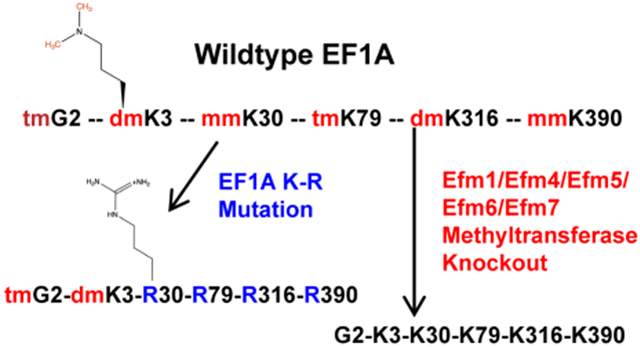
Methylation of proteins of the translational apparatus, including ribosomal proteins and elongation factors, has been well-characterized in recent years 1-6. One protein from Saccharomyces cerevisiae, elongation factor 1A (EF1A) stands out by the extensive methylation of its lysine residues. EF1A is primarily responsible for transporting the aminoacyl-tRNA to the ribosomal A site as a GTP complex and ensuring a correct codon-anticodon match 7. Additionally, EF1A has been shown to have a role in the assembly of the ribosomal subunits 8, the regulation of the actin cytoskeleton, and other cellular functions 9,10. Five distinct enzymes methylate EF1A at Lys 3 (Efm7), Lys-30 (Efm1), Lys-79 (Efm5), Lys-316 (Efm4), and Lys-390 (Efm6)2,3,11-15. Efm7 is also able to methylate the N-terminal amino group of Gly-2 13. It is presently unknown whether these methyltransferases are specific for EF1A or whether they also modify other cellular proteins. Methylation of EF1A is conserved between different species, with methylation at Lys-79 and Lys-316 being the most highly conserved.2,16.
Since the discovery of EF1A and its posttranslational modifications, the connection between EF1A function and its methylation has remained poorly characterized. To address the question of whether EF1A lysine methylation is necessary for EF1A’s functional roles in the cell, we used EF1A methyl-deficient strains and assayed function using multiple biochemical approaches. These approaches included measuring yeast growth under different stress conditions, ribosome sedimentation, and dual luciferase assays to assess translation fidelity.
Here we provide phenotypes associated with the disruption of EF1A methylation, including slow growth and sensitivity to translational inhibitors as well as to rapamycin and caffeine. This work demonstrates that methyl-deficient EF1A is still able to function in translation and ribosomal assembly but may disrupt the TORC1 pathway. Finally, we provide evidence that the five EF1A methyltransferases appear to be specific to EF1A and do not have additional major cellular targets, although we cannot rule out the methylation of minor species.
MATERIALS AND METHODS
Yeast strains and growth media.
All yeast strains were grown in 10 g/L yeast extract, 20 g/L peptone, and 20 g/L dextrose (YPD, Fisher) at 30 °C. For spot test analyses, yeast strains were plated on 2% agar in YPD, or on such plates supplemented with hydrogen peroxide, NaCl, caffeine (Alfa Aesar, AA3921414), rapamycin (Alfa Aesar, AAJ62473MF), anisomycin (Millipore, 176880), cycloheximide (Sigma, C7698), tunicamycin (Sigma, T7765) and puromycin (Sigma-Aldrich, P8833) as described in the figure legends. Solid growth media was also made as YPG with 10 g/L yeast extract, 20 g/L peptone, 3% glycerol and 20 g/L agar, or as lactate media with 3 g/L yeast extract, 0.5 g/L dextrose, 0.5 g/L CaCl2, 0.5 g/L NaCl, 0.6 g/L MgCl2, 1 g/L NH4Cl, 1 g/L KH2PO4, 8 g/L NaOH, 22 mL of 90% DL-lactic acid per L and 20 g/L agar.
Strains used in this study are listed in Table 1 below. The efm1456Δ and efm14567Δ deletions strains were based on the efm1Δ strain obtained from Dharmacon online yeast knockout collection. Each successive deletion was created through homologous recombination following the protocol as described 17. Each primer contained either 40 base pairs upstream or downstream of the corresponding ORFs to be deleted. For the knockout using the KIURA3 cassette, we used the KIURA3 found in the CORE cassette as a template. The mutants were confirmed through PCR using primers upstream and downstream of the corresponding gene.
Table 1:
Yeast Strains Used in This Study
| Strain | Genotype |
|---|---|
| BY4742 | MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 |
| efm1Δefm4Δefm5Δefm6Δ | BY4742 background, yhl039wΔ::hphMX, yil064wΔ::HIS3, ygr001cΔ::kanMX, ynl024cΔ:: URA3 |
| efm1Δefm4Δefm5Δefm6Δefm7Δ | BY4742 background, yhl039wΔ::hphMX, yil064wΔ::HIS3, ygr001cΔ::kanMX, ynl024cΔ::URA3, ylr285wΔ::LYS2 |
| EF1A K(30, 79, 316, 390)R | tef1Δ::kanMX, tef2Δ::hphMX, TEF1 K(30, 79, 316, 390)R/pUG23 |
For introducing arginine substitutions at lysine codons in TEF1, the endogenous yeast TEF1 gene was cloned into pUG23 (CEN/ARS HIS3 vector) under its native promoter and terminator using a standard cloning protocol. The point mutations were introduced via site-directed mutagenesis using QuikChange Lightning mutagenesis (Agilent #210518, 210515). Sanger Sequencing of the TEF1 open reading frame (ORF) was used to confirm the point mutations. Starting with wildtype yeast cells, the TEF1 ORF was first deleted with a kanMX cassette, and then the plasmid harboring the quadruple TEF1 mutant was transformed into cells and selected for under growth in -HIS. TEF2 was then deleted with the hphMX cassette, and the absence of both TEF1 and TEF2 was confirmed by PCR.
Lysis Method 1.
Yeast cells grown in YPD (7 OD600 nm) were washed 3 times with 1 ml of water and then resuspended in 0.2 mL of lysis buffer (0.2% SDS, 0.7 mM phenylmethylsulfonylfluoride (PMSF)). 0.2 g of baked glass beads (Biospec Products, 11079105) was added and the cells were lysed with 7 cycles of 1 min on vortex and 1 min on ice. Lysates were separated from beads using a gel loading tip and then clarified by centrifugation at 12,000 × g for 15 min.
Lysis Method 2.
Yeast cells grown in YPD (7 OD600 nm) were washed once with 1 mL ice cold water, spun at 4000 × g for 4 min and then washed again with 1 ml of ice cold water supplemented with 100 μg/ml PMSF. Cells were lysed by the method of Yaffe et al. 18 with the following modifications. Washed cells were incubated for 10 min in 150 μL of ice cold 1.85 M NaOH containing 2% 2-mercaptoethanol. After 10 min, ice cold 50% (wt/vol) trichloroacetic acid was added and the mixture incubated on ice for another 10 min. The mixture was centrifuged for 2 min and the resulting pellet washed with 1 mL of cold acetone and centrifuged again. The pellet was dried using vacuum centrifugation for 2 min. The pellet was then resuspended in 200 μL of sample buffer prepared from 500 μL of 0.2 M Tris-HCl pH 6.8, 6% SDS, 30% glycerol, 500 μL water, 12.5 μL 2-mercaptoethanol, 25 μL of 1 M Tris base, and 100 μg of PMSF and heated for 3 min at 95 °C. After the determination of protein concentration by Lowry analysis after trichloroacetic acid precipitation 19, a small amount of solid bromophenol blue was added and samples analyzed by SDS-PAGE.
SDS-PAGE.
Cell lysates were fractionated on 4-12% Bis-Tris precast polyacrylamide gel (GenScript) with 1X MOPS buffer (6.06 g/L Tris base, 10.46 g/L MOPS, 1 g/L SDS and 0.3 g/L EDTA, GenScript) for 1 h at 140 V. An unstained protein marker ladder used to determine protein size. The gel was Coomassie stained (50% methanol, 10% acetic acid, 40% water, 0.2% Brilliant Blue R-250 (w/v)) for 1 h and destained in 10% acetic acid and 15% methanol until bands became visible.
EF1A Purification.
The method of purification described below was adapted from Francisco et al. 20. A 50 mL overnight culture grown in YPD from wildtype or mutant strain was used to inoculate 2 flasks of 4 L of YPD and cells were grown to an OD600 nm of ~ 2.5. The cells were centrifuged at 664 × g in pre-weighed centrifuge bottles and the weight of the pellet recorded. Cells were stored at −80 °C until lysis could be performed. The pellet was resuspended in 2 mL/g of pellet in ice cold lysis buffer (60 mM Tris-Cl pH 7.5, 50 mM NH4Cl, 5 mM MgCl2, 0.1 mM EDTA pH 8, 10% glycerol, 1 mM dithiothreitol (DTT) and 0.2 mM PMSF) and lysed by passing through an emulsifier (EmulsiFlex-C3) four times at greater than 25,000 pounds per square inch pressure. Cell debris was removed by centrifugation at 11,300 × g for 30 min at 4 °C and then the supernatant clarified at 76,300 × g for 1.5 h at 4 °C. The supernatant was added to diethylaminoethyl cellulose resin (DE52, Whatman) that was pre-equilibrated with buffer 1 (20 mM Tris-Cl pH 7.5, 0.1 mM EDTA pH 8, 25% glycerol, 1 mM DTT and 0.2 mM PMSF) and 100 mM KCl for 1 h with light stirring at 4 °C.
Unbound EF1A was recovered by transferring to a 50 mL conical tube and centrifugation at 2,000 × g for 3 min. The supernatant was then incubated with 25 mL of sulphopropyl-Sepharose (fast flow, Sigma) also equilibrated with buffer 1 containing 100 mM KCl for 1 h with light stirring at 4 °C. Unbound material was removed by centrifugation as before and then EF1A eluted by incubating the resin with 25 mL of buffer 1 containing 500 mM KCl for 1 h with light stirring at 4 °C. Eluted proteins were then recovered by centrifugation at 2,000 × g for 3 min and dialyzed overnight in 3 L of buffer 1 with no salt. Lastly, the dialyzed protein was applied to 15 mL of carboxymethyl cellulose resin (CM52, Whatman) equilibrated with buffer 1 containing 50 mM KCl packed into a column and allowed to elute by gravity flow with a step-wise salt gradient 100 mM KCl, 150 mM KCl, 200 mM KCl, 300 mM KCl, 350 mM KCl and 500 mM KCl). 1.5 ml fractions were collected and analyzed by SDS-PAGE to determine where EF1A eluted. Fractions containing pure EF1A were pooled and dialyzed into buffer 1 containing 100 mM KCl overnight at 4°C for storage at −80 °C.
Immunoprecipitation.
Seven OD units of yeast cells grown to an OD600nm of ~0.7 was grown in S-adenosyl-[methyl-3H]methionine, using the method described 21. Next the labeled cells were washed with water, resuspended in 1 mL binding buffer (20 mM Tris, 100 mM KCl, 10% glycerol, 1% Triton X-100, 200 μg/mL PMSF) and lysed with 0.2 g of baked glass beads using 7 rounds of 30 s vortexing followed by 30 s on ice. The radiolabeled lysates were collected and clarified at 5,000 × g for 5 min. Ten microliters were set aside as the input material. Protein A beads were prepared in binding buffer with three washes at 700 × g for 2 min and kept on ice until needed. To start the immunoprecipitation, the labeled lysates (500 μg protein by Lowry assay) were incubated with 5 μL of anti-EF1A antibody (Kerafast, ED7001) for 3.5 h and then with protein A beads for 2 h. Following centrifugation as above, the protein-antibody-protein A bead complex was heated at 100 °C in 50 μL of 5X SDS- buffer (250mM Tris-Cl pH 6.8, 10% SDS, 30% glycerol, 0.5 M DTT, 0.02% bromophenol blue) for 8 min to release protein. Forty microliters of each sample and 5 μL of each input sample were analyzed by SDS-PAGE as described above. The destained gel was incubated in water overnight, then treated with En3hance (Perkin Elmer) for 1 h followed by a 30 min water wash. The dried gel was then exposed to film at −80 °C.
Protein Stability Assay.
Yeast cells were inoculated the night before in YPD media at 30 °C to give an OD600 nm of about 0.7 the following morning. The inhibitor chase was performed as described by Buchanan et al. 22 with the changes described below. Samples were collected at various time points and were spun down and frozen at −20 °C until lysis. Puromycin or cycloheximide was used to perform the chase. Lysis was performed using method 2 described above and the lysates fractionated in duplicate using SDS-PAGE (described above). Protein sizes were determined using a Bio-Rad broad range unstained molecular weight ladder and equal amounts of protein (by Lowry assay after precipitation with trichloroacetic acid) were loaded for each strain tested. One gel was stained and destained as above. A second gel was transferred to PVDF membrane for western blot analysis with 7 μL of Amersham full range ECL rainbow ladder as described below.
Immunoblot Analysis.
Proteins from lysates separated by SDS-PAGE were transferred to PVDF membrane (Hybond-P) at 30 V for 1 h. The membrane was then blocked overnight at 4 °C in 5% dried nonfat milk in Tris-buffered saline with 0.1% Tween 20 (v/v, TBST) or 0.5% BSA (w/v)/0.02% (w/v) SDS in phosphate-buffered saline with 0.1% Tween 20 (v/v, PBST). After blocking, the membranes were washed in 1X TBST or 1X PBST and incubated with primary antibodies (1:10000 rabbit anti-EF1A, Kerafast, ED7001) diluted into 1% dried nonfat milk in 1X TBST or 0.5% BSA/ 0.20% SDS in PBST, as indicated, for 1.5 h at 4 °C. After washing with the respective buffers, the membrane was incubated with anti-rabbit IgG-HRP (1:6666; Cell Signaling, 7074) secondary antibody in 1% dried nonfat milk or LICOR anti-goat fluorescent antibody in 0.5% BSA/ 0.02% SDS in PBST for 1 h at room temperature. ECL was used to visualize bands probed with HRP secondary antibody (Amersham Biosciences ECL Prime Western blotting, GE Healthcare, RPN2232) and LICOR Odyssey imager for the fluorescent probe. After probing, membranes were stained with Ponceau S or Coomassie to determine transfer efficiency.
Dual Luciferase (DLR) Assay.
For amino acid misincorporation, the CTY775/luc CAAAFF K529N plasmid was used and for programmed frameshift, the pJD376 (L-A) termed PRF −1 and pJD377 (Ty1) PRF +1 plasmid was used. These plasmids were transformed into the wildtype and mutant strains using the lithium acetate-ssDNA-PEG method 23. Transformed strains were grown in SD –Ura (minimal synthetic defined medium lacking uracil; 0.07% (w/v) CSM-Ura powder, 0.17% (w/v) yeast nitrogen base without amino acids or ammonium sulfate, 0.5% (w/v) ammonium sulfate, and 2% (w/v) dextrose) to an OD600 nm of 0.5-0.8. Next 0.5 OD600 nm units were harvested by centrifugation at 5,000 × g and stored on ice until ready for use. The DLR reagents, from Promega, were thawed to room temperature and diluted according to the assay manual. Harvested cells were individually lysed with 0.5 mL of passive lysis buffer, and then 6 μL transferred to a white (Greiner bio-one, 82050-736) 96 well plate. 30 μL of LARII solution was added and immediately read using SpectraMax M5 microplate reader, giving firefly luminescence; then 30 μL of Stop and Glo buffer immediately added to that same well and read to give Renilla luminescence. SpectraMax parameters were set as: read type - endpoint; read mode - luminescence with 1500 ms integration time; wavelength – all; automix – off; autocal – on; setting time – off; autoread –off.
RESULTS
Generation of yeast strains deficient in multiple EF1A methyltransferases or with arginine substitutions of EF1A methyl-accepting lysine residues.
To assess the functional role of the methylation of elongation factor 1A (EF1A) N-terminal glycine residue and lysine residues 3, 30, 79, 316 and 390, two approaches were taken. First, we constructed yeast strains lacking the five methyltransferases responsible for methylation at all of these sites (efm14567Δ) or the four methyltransferases that methylate lysine residues 30, 79, 316, and 390 (efm1456Δ) through marker-based gene deletions. Secondly, we mutated a plasmid-borne TEF1 gene encoding one copy of EF1A to replace lysine codons at positions 30, 79, 316, and 390 with arginine codons (Tef1 K(30,79,316,390)R) and then deleted both endogenous genes (TEF1 and TEF2) encoding EF1A as described in the "Experimental Procedures" section. The N-terminal modifications are still present in this strain (trimethyl Gly-2 and dimethyl Lys-3). The successful construction of the efm14567Δ mutant strain indicates that the loss of all five methyltransferase genes does not result in lethality.
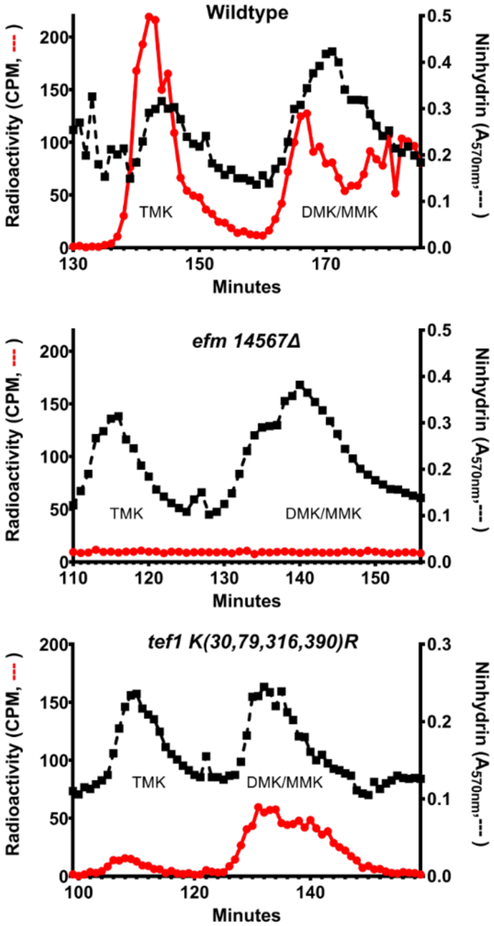
We then analyzed the extent of lysine methylation in wildtype, efm14567Δ, and TEF1 K(30,79,316,390)R strains labeled in vivo with S-adenosyl-[methyl-3H]methionine11. We performed acid hydrolysis on the 50 kDa polypeptides separated by SDS-PAGE that contain EF1A and analyzed the radiolabeled methylated lysine derivatives by high-resolution cation exchange chromatography. We were able to clearly resolve a peak of the 3H-trimethylated species (TMK) and a poorly-resolved peak that included both the 3H-dimethylated and 3H-monomethylated derivatives (DMK and MMK) (Fig. 1). In wildtype hydrolysates, all three lysine 3H-methylated species were detected whereas in the efm14567Δ strain, no radioactivity was detected at the positions of TMK, DMK, and MMK, confirming biochemically the loss of the Efm1, Efm4, Efm5, Efm6, and Efm7 methyltransferases. On the other hand, we observed reduced TMK and DMK/MMK methylation of tef1 K(30,79,316,390)R EF1A (Fig. 1). Although we expected some 3H-MMK and 3H-DMK from the methylation at Lys-2, we were surprised to see the formation of a small amount of 3H-TMK. These results suggest that alternative lysine residues may become available for methylation when lysines 30, 79, 316, and 390 are converted to arginine residues.
Figure 1: Loss of methylated lysine residues in EF1A from a strain lacking five Efm methyltransferases and a strain with lysine to arginine substitutions at positions 30, 79, 316, and 390 in EF1A.
EF1A purified from yeast cells that were labelled with S-adenosyl-[methyl-3H]methionine, acid hydrolyzed, and the methylated amino acid derivatives separated by high resolution cation exchange chromatography using the method described 21 with the modifications shown below. Wildtype and tef1 K(30,79,316,390)R hydrolysates were fractionated, mixed with standards of 2 μmol of ε-trimethyllysine (TMK) and 1.4 μmol ε-dimethyllysine (DMK) while efm14567Δ was fractionated with the same amount of TMK and DMK with the addition of 0.6 μmol of ε-monomethyllysine (MMK). The column was eluted with a sodium citrate buffer (0.3 M Na+) at pH 3.8. Radioactivity (red circles and line) was measured in 975 μl of the fractions eluting in the positions of the methylated lysine standards that were determined by ninhydrin assay in 25 μl aliquots (black squares and line; performed at 68 °C for 15 min). Data from the middle panel is from one experiment; the data in the upper and lower panels are from one experiment of two replicates.
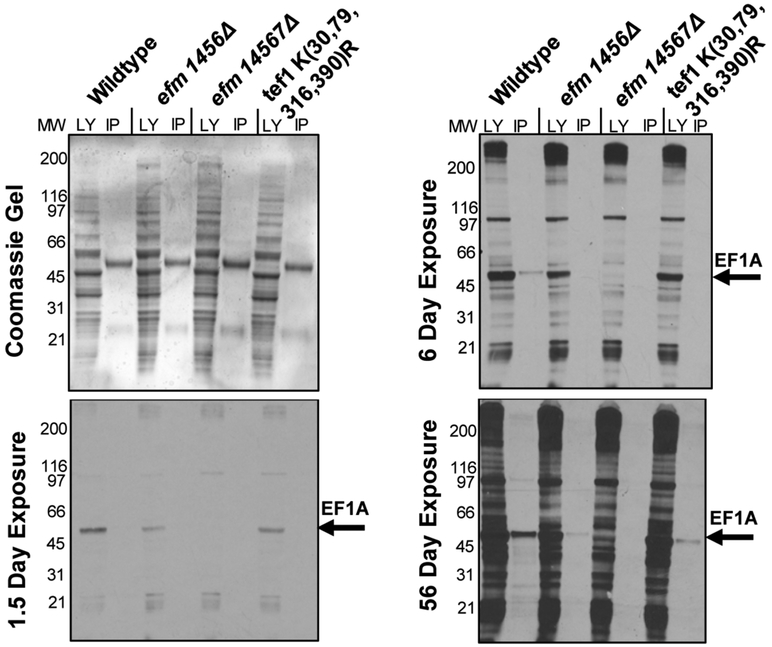
To confirm the reduction or absence of methylation of EF1A in the mutant strains, we labeled intact yeast cells with S-adenosyl-[methyl-3H] methionine and then analyzed 3H-methylated polypeptides by SDS-PAGE before and after immunoprecipitation with antibodies to EFlA. Even in long exposures, no radioactivity was detected at the 50 kDa position of EF1A in the efm14567Δ strain lacking all of the EF1A methyltransferases, and reduced methylation was observed in the efm1456Δ strain at shorter exposures (Fig. 2). As shown for the amino acid analysis experiment described above, we found significant 3H-methylation in the 50 kDa immunoprecipitated EF1A in the K(30,79,316,390)R strain, again suggesting that alternate methylation sites may be used when these four lysine residues were unavailable (Fig. 2).
Figure 2: Immunoprecipitation of EF1A from methylation-deficient cells shows specificity of elongation factor methyltransferases.
Yeast cells from wildtype and mutant strains that were labeled with S-adenosyl-[methyl-3H]methionine, and immunoprecipitated with an anti-EF1A polyclonal antibody as described in “Experimental Procedures”. The top left panel is a Coomassie-stained polyacrylamide gel, which serves as a protein loading control. The remaining panels show the detection of radioactive material in each sample at different time intervals. The longer exposure reveals that EF1A methylation is decreased in the methyltransferase knockout mutants. The LY lane shows the total lysate before the immunoprecipitation while the IP lanes show what was pulled down with the EF1A-antibody. The figure shown is a representative from one out of two separate experiments.
To probe if the EF1A methyltransferases had alternative methylation substrates, we also analyzed the entire spectrum of methylated polypeptides in lysates of the intact cells labeled with S-adenosyl-[methyl-3H] methionine (Fig. 2, Supplemental Fig. S1). Here we looked closely for evidence of methylated polypeptides on SDS-PAGE that were reduced or not found in any of the three mutant strains on the fluorograph. The Coomassie-stained gel was a control for protein loading and to show the electrophoretic mobility of EF1A. The heavy and light chains of the EF1A antibody (~60 kDa and ~25 kDa respectively) can be seen on the Coomassie-stained gel bracketing the ~50 kDa position of EF1A (Supplemental Fig. S1). In both the experiment shown in Fig. 2 and the replicate experiment shown in Fig. S1 we observed a complete loss of methylation in the efm14567Δ strain, confirming that the major methylated species at this polypeptide size was EF1A. However, we were unable to detect any reduction in the methylation of any other polypeptide band seen in the fluorographs (Fig. 2, Supplemental Fig. S1). These results suggest that none of the five EF1A methyltransferases catalyze the modification of non-EF1A polypeptides although we would not be able to detect the loss of minor methylated species.
Methylation deficient cells exhibit a slow growth phenotype and alter growth in response to cellular stress.
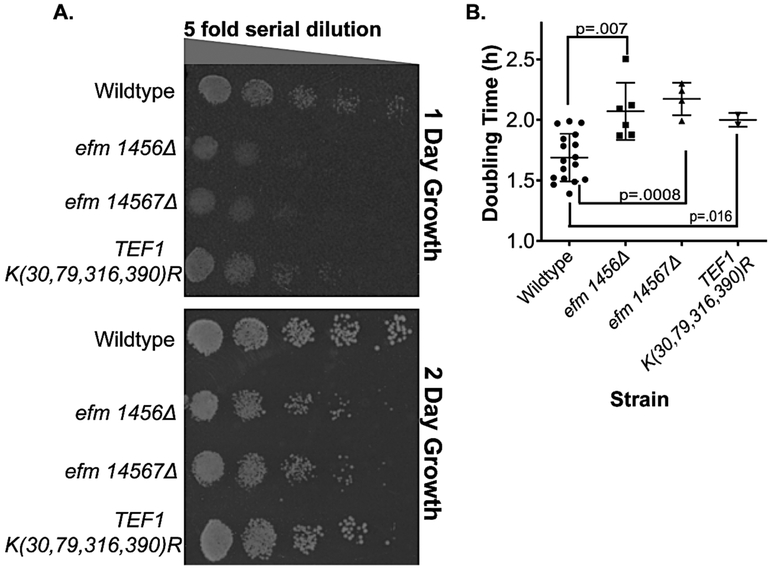
We then assessed differences in the growth of the EF1A methylation-deficient strains. In Fig. 3A, we show yeast growth on plates containing yeast extract, peptone and dextrose (YPD). Serially diluted strains were spotted and allowed to grow for 1 day (early growth) and 2 days (later growth). At both stages, colonies of the efm1456Δ strain as well as the efm14567Δ strain were much smaller than the wildtype colonies. These defects were confirmed and quantitated by observing slower growth in liquid YPD media as well. We found an increase in doubling times from about 1.7 h for the wildtype cells to 2.1 h for the efm1456Δ strain and 2.2 h for the efm14567Δ strain (Fig. 3B).
Figure 3: Loss of Efm methyltransferases results in slow growth in solid and liquid YPD growth media while EF1A with four lysine to arginine mutations shows slow growth in only liquid media.
A) Yeast cells from wildtype and mutant strains grown at 30 °C in YPD to an OD600 nm of about 0.5 and 3 μl of a cell suspension starting at 0.1 OD600 nm were then serially diluted and plated on YPD agar plates at 30 °C. Colonies were photographed for a representative experiment after 1 day or 2 days. In replicate experiments, we found that colonies for the efm1456Δ mutant were significantly smaller than wildtype colonies in 16 out of 23 experiments; in the other 7 cases colonies were roughly the same size. Colonies for the efm14567Δ mutant were significantly smaller than wildtype colonies in 19 out of 21 replicate experiments; in the other 2 cases colonies were roughly the same size. In 23 replicate experiments the colony sizes for the TEF1 K(30,79,316,390)R mutants were indistinguishable from the wildtype. B) Doubling times for growth in liquid YPD media at 30 °C were calculated from the linear portion of exponential growth measured by OD600 nm over a 12 h time frame. Each point is a biological replicate. Error bars indicate standard deviation values and Student t-test p values (unpaired, two tails) are shown.
When similar experiments were performed for the TEF1 K(30,79,316,390)R strain, somewhat reduced colony sizes were observed after 1 day of plate growth but not after 2 days (Fig. 3A). In liquid medium, we found a significantly increased doubling time of 2.1 h compared to 1.7 h for the wildtype (Fig. 3B). Thus, it is clear that the loss of either four or five of the EF1A methyltransferase genes, or the replacement of four of the methylated lysine residues on EF1A, results in significant decreases in the rate of growth.
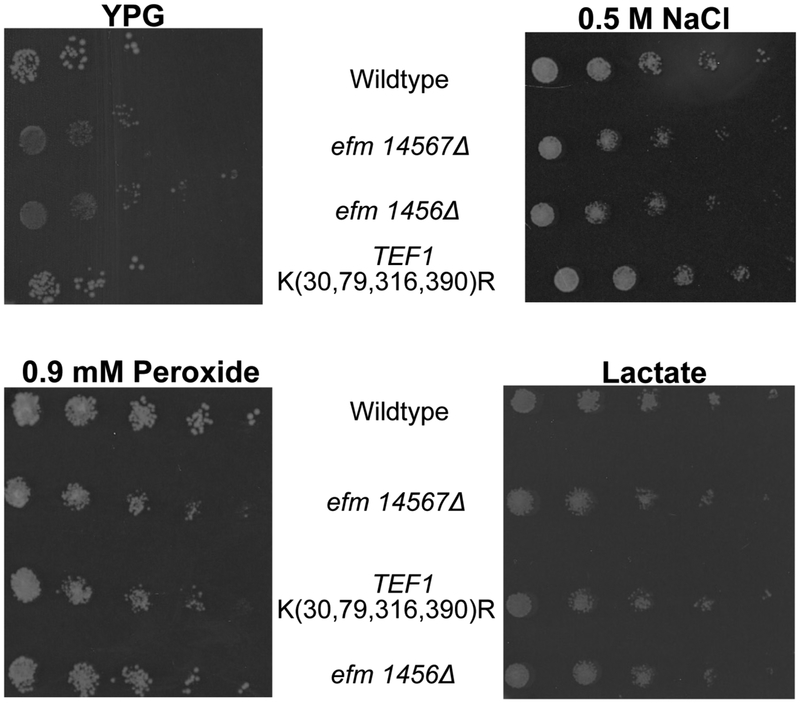
We then tested the growth of the mutant strains under respiratory, osmotic and oxidative stress conditions. When cells were grown on agar plates containing glycerol (YPG) as the carbon source or YPD plates containing 0.5 M NaCl or 0.9 mM hydrogen peroxide, the colonies of both the efm1456Δ and the efm14567Δ strains were markedly smaller than the wildtype strain (Fig. 4). We found that colonies of the TEF1 K(30,79,316,390)R strain on the plates were somewhat smaller than wildtype colonies under osmotic and oxidative stress conditions (Fig. 4). The TEF1 K(30,79,316,390)R colonies on YPG plates did not have any difference in size compared to wildtype. When cells were grown on lactate plates we observed no difference in the colony size of the mutants compared to wildtype (Fig 4). These results demonstrate the EF1A methylation deficient cells are less able to adapt to at least some stress conditions. However, it is unclear why these deficient cells are able to grow equally as well as wildtype cells with non-fermentable carbon sources. It is possible that reduced rates of translation in non-fermentative conditions allow the EF1A methylation deficient cells to grow at the same reduced rate as wildtype cells when EF1A function is not rate-limiting for growth.
Figure 4: Loss of Efm methyltransferases causes sensitivity under different cellular stress conditions.
Representative images showing yeast cells that were grown in YPD, serially diluted and then spotted on YPD agar containing 0.5 M NaCl, or 0.9 mM peroxide, or YPG, or lactate media at 30 °C as described in the Figure 3 legend. Colonies were imaged after 2 days. In YPG, colonies for the efm1456Δ and efm14567Δ mutant were significantly smaller than wildtype colonies in 2 out of 3 replicate experiments whereas the TEF1 K(30,79,316,390)R mutant always grew relatively the same as wildtype in those replicates. Under oxidative stress, colonies for the efm1456Δ and efm14567Δ mutant were significantly smaller than wildtype colonies in 3 replicate experiments whereas the TEF1 K(30,79,316,390)R mutant always grew relatively the same as wildtype in three replicates. In the presence of sodium chloride, mutant colonies were smaller compared to wildtype in four replicate experiments. No difference in colony size was observed in lactate media for six replicates.
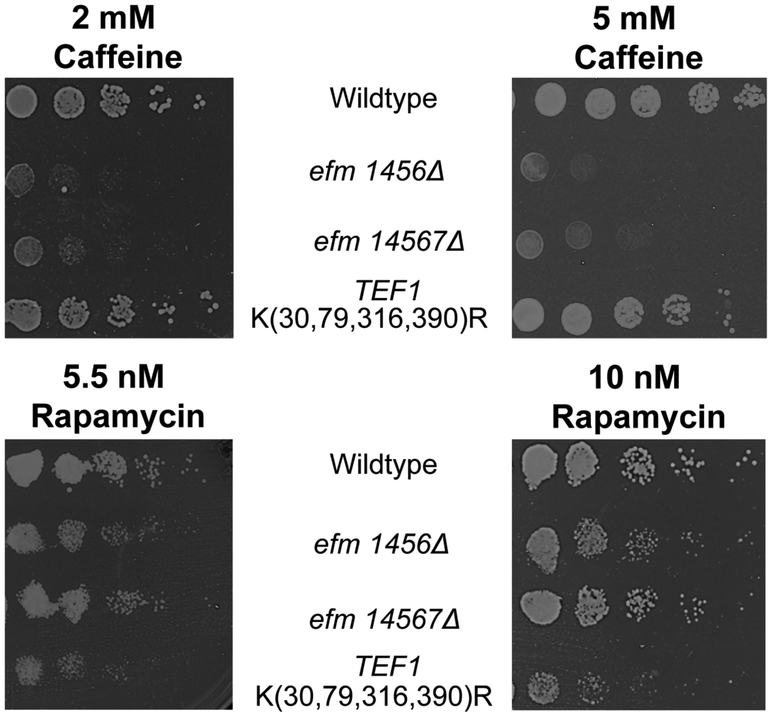
Lastly we assessed growth when the yeast cells were grown on YPD media containing caffeine or rapamycin (Fig. 5). The efm1456Δ and the efm14567Δ colonies were somewhat smaller compared to wildtype under rapamycin growth whereas they were significantly smaller compared to wildtype for caffeine growth (Fig. 5). The colonies of the TEF1 K(30,79,316,390)R strain on the caffeine plates grew similarly to wildtype (Fig. 5). Interestingly, we also observed smaller colonies for the TEF1 K(30,79,316,390)R strain in both rapamycin conditions tested (Fig. 5). Both rapamycin and caffeine affect protein synthesis and cellular growth through the TORC1 pathway 24,25. Since growth under rapamycin stress was altered in the efm1456Δ and TEF1 K(30,79,316,390)R strains, it suggests that there may be some interaction between methylated EF1A and the TORC1 pathway that is disrupted when EF1A is unmethylated. Alternatively, some or all of these methyltransferases may have additional methyl-accepting substrates (other than EF1A) in the TORC1 pathway.
Figure 5: Methylation-deficient EF1A growth inhibited by caffeine and rapamycin.
Representative images showing yeast cells that were grown in YPD, serially diluted and then spotted on YPD agar containing 2 mM or 5 mM caffeine and 5.5 nM or 10 nM rapamycin (diluted from a 50 mg/ml stock solution in ethanol) at 30 °C as described in the Figure 3 legend. Colonies were imaged after 2-4 days. In 2 mM caffeine, colonies for the efm1456Δ and efm14567Δ mutant were significantly smaller than wildtype colonies in all 4 replicate experiments whereas the TEF1 K(30,79,316,390)R mutant always grew relatively the same as wildtype in those replicates. At 5 mM caffeine, colonies for the efm1456Δ and efm14567Δ mutant were significantly smaller than wildtype colonies in all 5 replicate experiments whereas the TEF1 K(30,79,316,390)R mutant always grew relatively the same as wildtype in all replicates. In the presence of 5.5 nM and 10 nM rapamycin, all mutant colonies were smaller compared to wildtype in two replicate experiments each.
EF1A methyltransferase deficient cells have altered sensitivity to translation inhibitors.
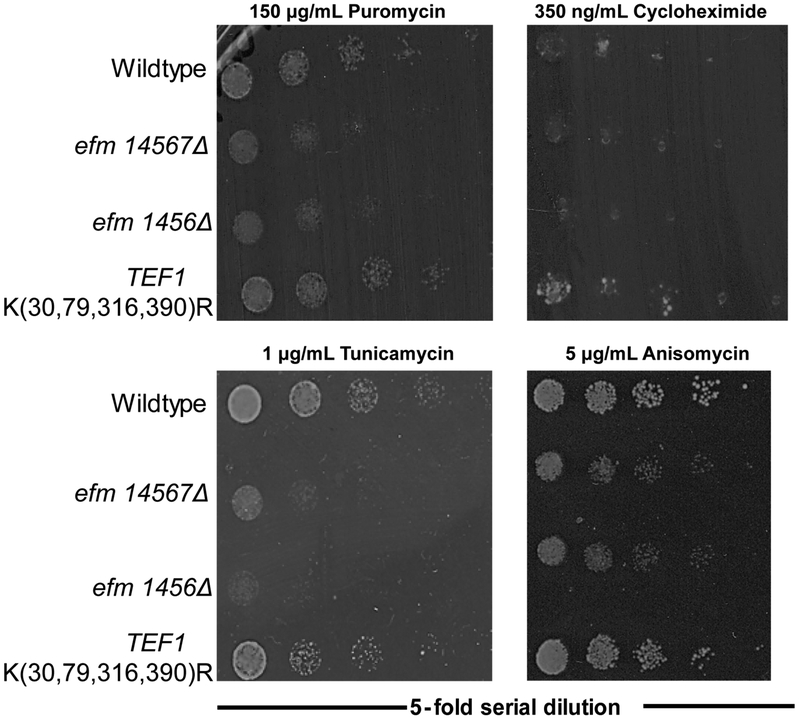
A major cellular role of EF1A is bringing aminoacyl-tRNAs to the ribosomal A decoding site. To address whether this role was dependent or affected by its methylation we first treated yeast cells with different translational inhibitors and assessed growth on YPD plates (Fig. 6). With puromycin, a drug that causes premature polypeptide chain release from the ribosome 26,27, tunicamycin, a drug that activates the unfolded protein response and inhibits translation 28,29, and anisomycin, a drug that interferes with the ribosomal acceptor site 30, we observed much smaller colonies of the efm1456Δ and the efm14567Δ strains compared to the wildtype strain. No decrease in cell size was seen with any of these inhibitors for the TEF1 K(30,79,316,390)R strain (Fig. 6). Finally, we detected no decrease in colony size with cycloheximide, a drug that blocks translation elongation 31, in any of the EF1A methylation deficient strains. These results indicate that changes in ribosomal architecture mediated by these inhibitors can affect translation more when EF1A is unmethylated, although the mechanisms for these effects are unknown.
Figure 6: Loss of Efm methyltransferases and mutation of four lysine residues to arginine residues in EF1A results in differential responses to translational inhibitors.
Representative image of yeast cells grown in YPD and then serially diluted onto agar plates as described in the Figure 3 legend but supplemented with either puromycin, cycloheximide, tunicamycin and anisomycin. In puromycin, 8 out of 10 replicates for efm14567Δ, and 10 out of 12 replicates for efm1456Δ strain had smaller colonies compared to wildtype colonies. Colonies for the TEF1 K(30,79,316,390)R strain always had similar sized colonies compared to wildtype colonies in 12 replicates. Growth on cycloheximide displayed no difference in colony size compared to wildtype colonies for the efm14567Δ mutant (four replicate experiments), efm1456Δ mutant (eight replicate experiments), and the TEF1 K(30,79,316,390)R mutant (eight replicate experiments). Tunicamycin colony sizes were always smaller than wildtype for efm1456Δ (six replicate experiments) and efm14567Δ (two replicated experiments) but remained unchanged for TEF1 K(30,79,316,390)R mutant. (six replicate experiments). On anisomycin plates, there were smaller colonies 4 out of 6 replicates for efm14567Δ and 5 out of 6 replicates for the efm1456Δ mutant compared to wildtype colonies. The TEF1 K(30,79,316,390)R strain had similar sized colonies compared to wildtype colonies with the exception of 2 out of 6 replicates where the colony sizes were bigger.
Stability of EF1A in methylation deficient cells.
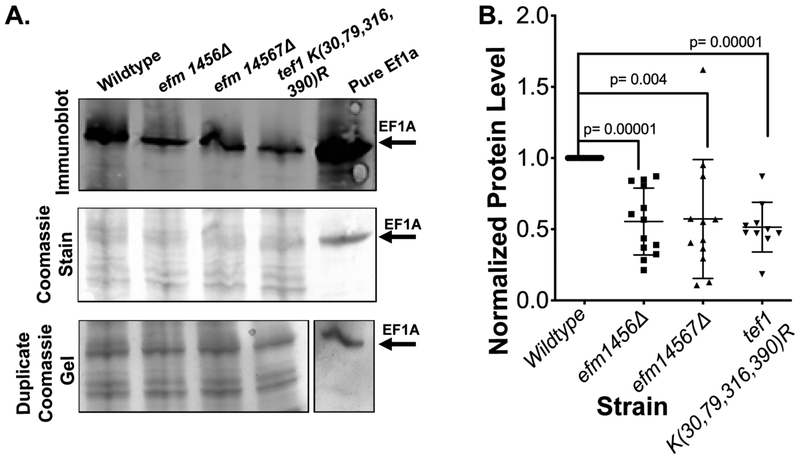
We then asked if the phenotypes seen might result from changes in the level of the EF1A protein itself. We thus measured EF1A by immunoblotting whole cell lysates of wildtype and methylation deficient strains with a polyclonal antibody specific to the entire yeast EF1A protein (Fig. 7A). Quantitation of the immunoblot signal demonstrated that the deficient strains contained about half of the EF1A present in wildtype strains, although there was considerable variability (Fig. 7B). This may explain the slowed growth rates and responses to translation inhibitors observed previously. Under these experimental conditions it is also possible that the reduction of EF1A level in the TEF1 K(30,79,316,390)R strain could be due to its plasmid expressing only one copy of the EF1A gene.
Figure 7: Loss of Efm methyltransferases and mutation of four lysine residues to arginine residues in EF1A affects protein abundance levels.
Seven OD600nm units of yeast cells were harvested after being grown in YPD media at 30 °C, then lysed using “method 2”, fractionated using SDS-PAGE and immunoblotted for antibody detection of EF1A with the LICOR secondary antibody as described in "Experimental Procedures". A) A representative experiment showing a Coomassie-stained PVDF membrane, the LICOR detected immunoblot showing EF1A protein levels, a duplicate Coomassie-stained gel of the lysates. B) EF1A protein expression levels determined from the comparison of peak areas of immunoblots probed for EF1A in yeast lysates. For each strain, each point represents a biological replicate. The densitometric signals for EF1A in the mutant strains were normalized to that of the wildtype strain in each experiment (making all wildtype values = 1) and were quantified using image J. Student t-test p values (unpaired, two tails) are shown.
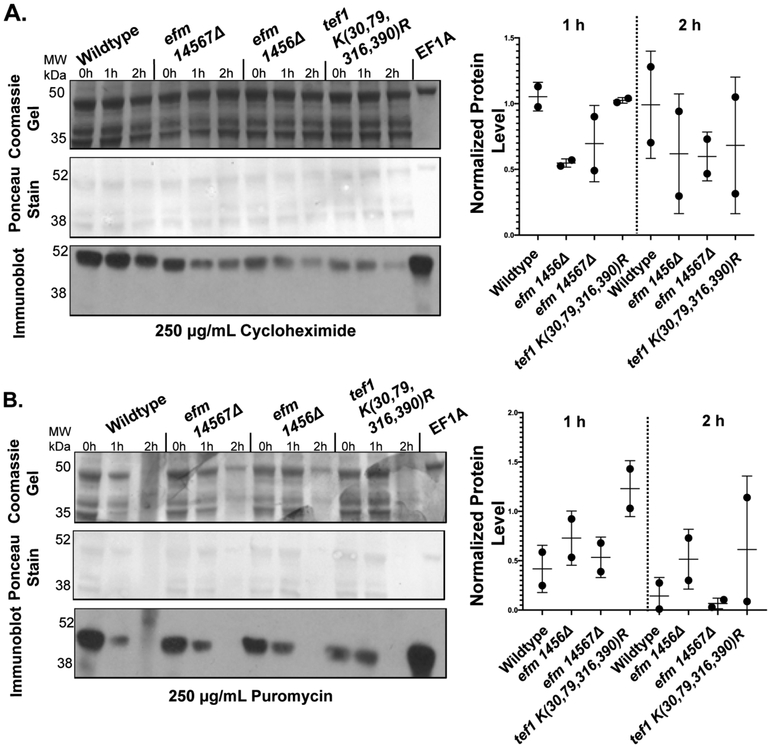
We considered the possibility that the absence of lysine methylation may enhance one or more ubiquitin-dependent proteolytic pathways. EF1A has been known to interact with ubiquitinated proteins to assist in ubiquitin-mediated degradation 9. We thus examined the stability of EF1A in intact cells grown in YPD after the addition of puromycin and cycloheximide to prevent new protein synthesis. In Fig. 8, we show the levels of EF1A by immunoblotting over a 2 h time course. In the puromycin chase experiment, we found that EF1A levels fell rapidly but in a similar fashion in both the wildtype and mutant strains and that there was a similar loss of total protein as well as indicated by the Coomassie and Ponceau staining. For the cycloheximide chase experiment, we also found little change in the relative loss of EF1A over 2 h in the wild type and mutant strains although the mutant strains had less EF1A at the zero time point and the total protein remained fairly constant. The similarity in the degradation rates of EF1A in the wild type and mutant cells was quantitated by densitometry in replicate cycloheximide and puromycin chase experiments (Fig. 8). From these data, we concluded that there was no large difference in the degradation of EF1A in the wildtype and mutant strains.
Figure 8: EF1A protein levels remain equally stable in the presence of cycloheximide and puromycin in wildtype cells and in cells deficient in EF1A methylation.
Yeast cells were grown to an OD of about 0.7 at 600 nm in YPD media at 30 °C. Cycloheximide (A) or puromycin (B) was then added individually to 3 mL of cells containing 7.5 OD600nm to a final concentration of 250 μg/ml. One mL aliquots of the 7.5 OD600nm cells were collected at the indicated times per strain and drug condition, lysed (using method 2) and then the proteins were fractionated by SDS-PAGE as described in the "Experimental Procedures" section. A representative gel and immunoblot is shown for both conditions. The top panel shows a Coomassie-stained gel. The middle panel is a Ponceau S-stained PVDF membrane from a duplicate gel. An immunoblot using antibodies to EF1A is shown in the lower panel. This experiment was performed twice for cycloheximide and twice for puromycin. The relative EF1A protein levels in each wildtype or mutant strain at the 1 h or 2 h time points compared to the EF1A level of the same strain at the zero time point were quantified using Image J densitometry and is shown to the right of its respective drug condition.
Ribosome assembly is unaffected by loss of EF1A methylation.
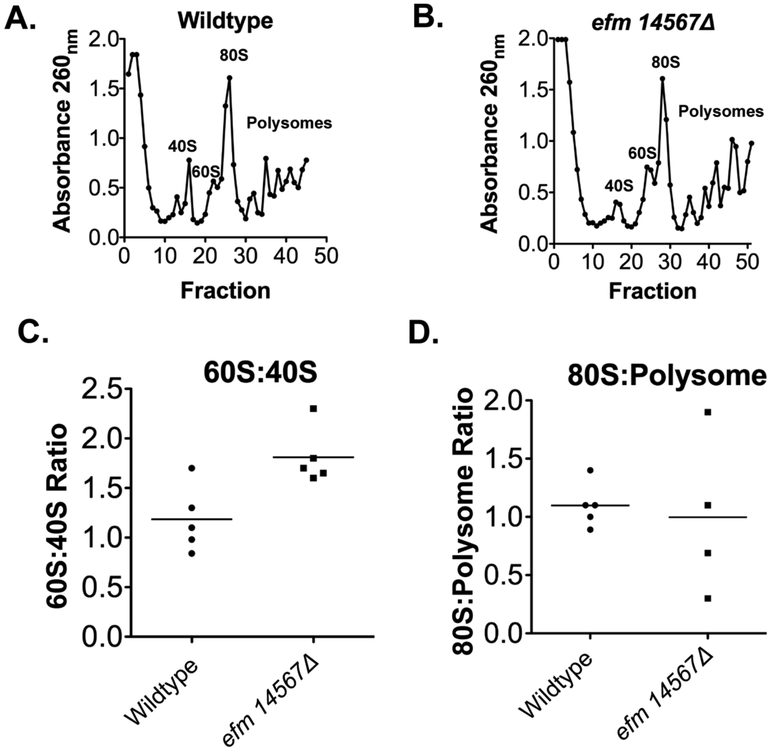
Although EF1A is primarily responsible for the transport of aminoacylated tRNA to the ribosomal A site it has been shown that it can also directly affect the assembly of the ribosomal subunits 8. We then asked if methylation of EF1A influenced levels of ribosomal subunits, polyribosomes, or ribosomes. Fig. 9A and B show a representative experiment of the separation of ribosomal subunits in the presence of cycloheximide for wildtype and efm14567Δ strains, respectively. Cycloheximide is used to stall translation to capture actively translating ribosomes on a transcript in order to analyze the differences in the amount of small ribosome subunit (40S), large ribosome subunit (60S), single fully formed active ribosomes (80S) and polysomes (more than one active ribosome on transcript) found. We were able to clearly resolve the 40S, 60S, 80S peaks and the polysomes peaks. We found that the efm14567Δ cells had a reduction in 40S subunits since the ration of 60S:40s: was about 50% higher than that of wildtype cells. However, this change was not statistically significant as determined by Student’s t-test ( Fig. 9C, p = 0.16). No significant differences were also seen when the 80S:polysome ratios were quantified (Fig. 9D, p = 0.79). Under these conditions, it appears that methylation of EF1A is not necessary for the assembly of ribosome subunits, although we cannot rule out small effects.
Figure 9: Deletion of EF1A methyltransferases Efm1, 4, 5, 6, and 7 does not affect ribosome assembly.
Ribosomes were prepared from yeast cells grown to an OD600nm of ~0.7 as described 32 and analyzed with the modifications described below. The top panels (A and B) show the fractionation of ribosomes by sucrose gradient centrifugation of 7 A260nm units. In each case, 100 μL fractions were collected and the A260 nm value of each fraction plotted. The absorbance of each of the peaks was summed to quantify the ratio of the 80S:polysome ribosomal subunits (panel C) and the ratio of the 60S:40S subunits (Panel D) with the mean value indicated by the horizontal line.
Protein synthesis fidelity is unaffected in methylation-deficient cells.
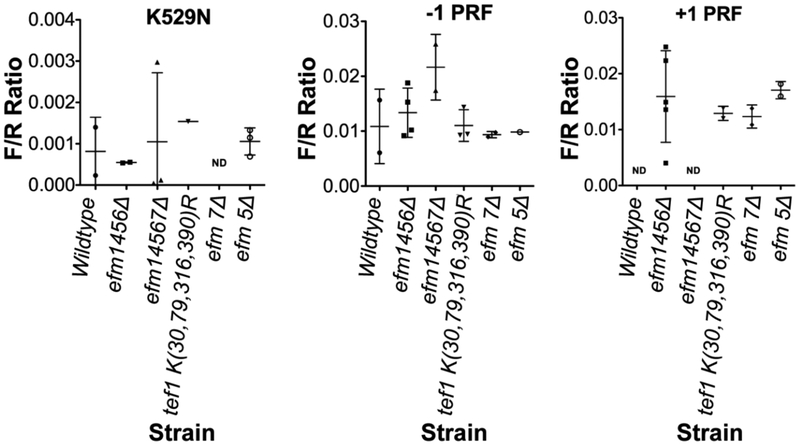
Lastly we examined the translation fidelity of the methylation-deficient strains using the dual luciferase reporter system (DLR) 33-35, examining both amino acid misincorporation and programmed frameshift errors. In these experiments, plasmids expressing fusion proteins of an N-terminal Renilla luciferase and a C-terminal firefly luciferase allow the expression of the firefly luciferase only when translational errors are made. For the frameshift plasmids a viral programmed frameshift is placed in the linker region and when it is bypassed firefly luminescence is detected35. Alternatively, the amino acid misincorporation plasmid has a mutation in the firefly gene itself that changes lysine 529 to an asparagine residue that results in the loss of luciferase activity36. As shown in Fig. 10, we found no differences in the misincorporation or frameshift rate with the efm1456Δ, efm14567Δ, or the TEF1 K(30,79,316,390)R strains. These results suggest that the loss of methylation does not result in the loss of translational fidelity, at least in this system under normal growth conditions.
Figure 10: Loss of Efm methyltransferases and mutation of four lysine residues to arginine residues in EF1A does not affect translation fidelity.
Yeast cells were prepared as described in "Experimental Procedures". Ratios of firefly and Renilla luciferase luminescence values are shown with each point representing a biological replicate. There was no statistical difference in the ratios with any of the strains shown here. ND - not done. The efm5 and efm7 mutants were used here to show there was no effect in single deletion strains as well.
DISCUSSION
EF1A is extensively post-translationally modified across all organisms. It can be ubiquitinated at lysine residues 37, phosphorylated at serine and threonine residues 38,39, acetylated 40, methyl esterified at its C-terminal lysine residue 41, methylated at multiple lysine residues and an N-terminal glycine residue 2,3,11,12,14,15,42 and glutaminylated at a glutamic acid residue 43. However, the functional relevance of these EF1A modifications are largely unknown. In this study we characterized two types of EF1A methylation-deficient yeast strains to elucidate the roles that lysine methylation of EF1A may have on its functions.
Extensive lysine methylation of EF1A is seen in a variety of eukaryotic species including humans 2, rabbits 16 , chickens 44, brine shrimp 16, corn 16, Arabidopsis 45, and the zygomycotan fungi Mucor racemosus 46 in addition to the ascomycotan yeast S. cerevisiae. However, lysine methylation of the corresponding EF-Tu protein in prokaryotes is not as extensive - Escherichia coli and Pseudomonas aeruginosa both only have one site of lysine methylation - dimethylation at Lys-57 and trimethylation at Lys-5 respectively 47-50. Sequence analysis using BLASTp revealed no clear orthologs of yeast Efm1, 4, 5, 6, or 7 in the prokaryotic species or in M. racemosus. However, a FungiDB search revealed orthologs of Efm 1, 4, 6, and 7 in Mucor circinelloides. On the other hand, there are clear orthologs for Efm4 and Efm5 in humans 13,51.
Thus far, the functional relevance of EF1A methylation has been studied in S. cerevisiae 52 M. racemosus 53, E. coli 54,55, P. aeruginosa 48,56, chicken 44 and humans 42,57,58. A similar point mutant strain was used in the S. cerevisiae study but was unavailable so we constructed our own. In that study, it was found that this strain was viable and had no in vitro difference in poly(U)-directed polyphenylalanine synthesis and GTP binding 52. In E. coli, the methylation at Lys-57 was shown to affect the aa-tRNA-induced GTP hydrolysis in vitro 55. Unmethylated EF1A did not affect EF1A’s ability to bind GTP or the aa-tRNA in M. racemosus 53 or affect translation fidelity in P. aeruginosa 56. Significantly, the extensive methylation characteristic to lysine residues on EF1A (about 8 methyl groups ) in M. racemosus was not found in the protein isolated from the spores of this organism. Additionally, E. coli EF-Tu was more methylated when cells were grown without nitrogen, phosphate, or carbon present 54. These changes suggest some regulation of the prokaryotic methyltransferases under growth conditions.
This is the first study showing that the five known methyltransferases responsible for methylating EF1A in S. cerevisiae do not appear to have any major additional substrates. Recently, evidence for the in vitro methylation of an EF1A-derived peptide containing Lys-253 in S. cerevisiae by Efm1 was presented 59. This lysine residue is found in a similar sequence motif as the Efm1 Lys-30 site. It is possible that methylation at Lys 253 could be contributing to the mono-methylation peak observed in the cation exchange chromatography of the TEF1 K(30,79,316,390)R mutant. However, whether this site is definitively methylated in vivo is not known.
We tested the ability of our S. cerevisiae strains to adapt to changing environments. As described above, the methylation of both M. racemosus and E. coli is dependent upon the stage of growth and nutritional status 53,60. We did observe increased sensitivity of our yeast methyltransferase mutant strains compared to the wildtype strain when grown with glycerol as a carbon source, or under oxidative and osmotic stress conditions. However, growth is also impaired in the mutant strains compared to wildtype in YPD media. Therefore, these stress-induced phenotypes we are seeing may not be a specific result of respiratory growth or environmental stress. We should point out that subtle growth differences may be masked in these assays. For example, the TEF1 K(30,79,316,390)R mutant did not appear to have reduced growth on solid media but a slight reduction in growth was observed in liquid media. On the other hand, all of the EF1A methylation-deficient mutant strains had reduced growth in the presence of rapamycin and caffeine. These effects on growth may to be due to stress-induced phenotypes of unmethylated EF1A in the TORC1 pathway24. It is also possible that one or more of the Efm methyltransferases 1,4,5,6, and 7 can methylate non-EF1A substrates that affect TORC1 signaling. Further examination of the individual methyltransferase knockout strains may be useful in distinguishing these possibilities.
From our examination of EF1A protein levels, we found that the mutant strains had significantly less EF1A present. This protein expression phenotype appears to be an additive effect of methyltransferase loss since single knockout methyltransferase mutations in yeast did not have alterations of EF1A protein levels (data not shown). In the prokaryote P. aeruginosa, loss of the single EftM methyltransferase does not result in the marked reduction of Ef-Tu 56. In yeast, it is unclear how the loss of EF1A methylation affects its protein abundance since we showed that the rate of degradation in the presence of cycloheximide or puromycin is unaltered in our methylation-deficient strains. On the other hand, the overexpression of EF1A also does not affect global translation efficiency 61. Thus it appears translation fidelity is independent of the amount of EF1A present.
Interestingly, although less EF1A protein is present in methylation-deficient strains, the translational function of EF1A remains apparently unimpaired. When amino acid incorporation, and programmed frameshift were measured by the dual luciferase translational fidelity assay system and ribosome assembly assessed using polysome analysis, the methylation deficient strains performed similarly to the wildtype strain. There may be compensatory mechanisms in our mutant strains that allow translational functions with reduced EF1A levels.
CONCLUSIONS
We have shown that the five protein lysine methyltransferases that modify elongation factor 1A in S. cerevisiae (Efm1, Efm4, Efm5, Efm6, and Efm7) are not essential to the viability of yeast. However, their loss results in slow growth and a particular sensitivity to caffeine and rapamycin, inhibitors of the Tor1 protein kinase component of the TORC1 signaling complex. Further work will be required to establish the mechanism(s) of these effects. The loss of these methyltransferases did not affect the fidelity of translation or the assembly of ribosomal subunits. We present evidence that EF1A is the major if not the sole substrate for these five methyltransferases. It appears that the fine tuning of EF1A function by modification at five distinct sites by five distinct methyltransferase enzymes optimizes cell physiology.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Maria Dzialo and Kyle Travaglini for their contributions to the initial stages of this work. We would like to thank Clarke lab members for suggestions on this work and David Bedwell and Ming Du from the University of Alabama and Dr. Jonathan Dinman at the University of Maryland for graciously provided the DLR plasmids. J.T.W. was the recipient of a NSF-LSAMP Bridge to Doctorate Fellowship; J.G., K.R.R., and C.W. were supported by the Ruth L Kirschstein National Research Service Award GM007185. This work was supported by grants to S.G.C. from the National Science Foundation (MCB-1714569) and the UCLA Academic Senate Faculty Research Program and a grant to G.C. from the National Institutes of Health (R35 GM130370).
Footnotes
Supporting Information
Immunoprecipitation of EF1A from methylation-deficient cells (Figure S1).
REFERENCES
- (1).Kako K, Kim J-D, and Fukamizu A (2019) Emerging impacts of biological methylation on genetic information. J. Biochem 165, 9–18. [DOI] [PubMed] [Google Scholar]
- (2).Hamey JJ, and Wilkins MR (2018) Methylation of elongation factor 1A: Where, who, and why? Trends Biochem. Sci 43, 211–223. [DOI] [PubMed] [Google Scholar]
- (3).Falnes PØ, Jakobsson ME, Davydova E, Ho A, and Małecki J (2016) Protein lysine methylation by seven-β-strand methyltransferases. Biochem. J 473, 1995–2009. [DOI] [PubMed] [Google Scholar]
- (4).Clarke SG (2018) The ribosome: A hot spot for the identification of new types of protein methyltransferases. J. Biol. Chem 293, 10438–10446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Egorova KS, Olenkina OM, and Olenina LV (2010) Lysine methylation of nonhistone proteins is a way to regulate their stability and function. Biochemistry (Mosc) 75, 535–548. [DOI] [PubMed] [Google Scholar]
- (6).Polevoda B, and Sherman F (2007) Methylation of proteins involved in translation. Mol. Microbiol 65, 590–606. [DOI] [PubMed] [Google Scholar]
- (7).Loveland AB, Demo G, Grigorieff N, and Korostelev AA (2017) Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature 546, 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Herrera F, Correia H, Triana L, and Fraile G (1991) Association of ribosomal subunits. A new functional role for yeast EF-1alpha in protein biosynthesis. Eur. J. Biochem 200, 321–327. [DOI] [PubMed] [Google Scholar]
- (9).Mateyak MK, and Kinzy TG (2010) eEF1A: thinking outside the ribosome. J. Biol. Chem 285, 21209–21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sasikumar AN, Perez WB, and Kinzy TG The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip. Rev. RNA 3, 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Dzialo MC, Travaglini KJ, Shen S, Loo JA, and Clarke SG (2014) A new type of protein lysine methyltransferase trimethylates Lys-79 of elongation factor 1A. Biochem. Biophys. Res. Commun 455, 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zhang L, Hamey JJ, Hart-Smith G, Erce MA, and Wilkins MR (2014) Elongation factor methyltransferase 3-A novel eukaryotic lysine methyltransferase. Biochem. Biophys. Res. Commun 451, 229–234. [DOI] [PubMed] [Google Scholar]
- (13).Hamey JJ, Winter DL, Yagoub D, Overall CM, Hart-Smith G, and Wilkins MR (2016) Novel N-terminal and lysine methyltransferases that target translation elongation factor 1A in yeast and human. Mol. Cell. Proteomics 15, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hart-Smith G, Chia SZ, Low JKK, McKay MJ, Molloy MP, and Wilkins MR (2014) Stoichiometry of Saccharomyces cerevisiae lysine methylation: Insights into non-histone protein lysine methyltransferase activity. J. Proteome Res 13, 1744–1756. [DOI] [PubMed] [Google Scholar]
- (15).Jakobsson ME, Davydova E, Małecki J, Moen A, and Falnes PØ (2015) Saccharomyces cerevisiae eukaryotic elongation factor 1A (eEF1A) Is methylated at lys-390 by a METTL21-like methyltransferase. PLoS One 10, e0131426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cavallius J, Zoll W, Chakraburtty K, and Merrick WC (1993) Characterization of yeast EF-1α: Non-conservation of post-translational modifications. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol 1163, 75–80. [DOI] [PubMed] [Google Scholar]
- (17).Longtine MS, Mckenzie III A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- (18).Yaffe MP, and Schatz G (1984) Two nuclear mutations that block mitochondrial protein import in yeast. Proc. Natl. Acad. Sci. U. S. A 81, 4819–4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lowry OH, Rosebrough NJ, Farr AL and Randall RJ (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem 193, 265–275. [PubMed] [Google Scholar]
- (20).Carvalho JF, Carbalho ME, and Merrick WC (1984) Purification of various forms of elongation factor 1 from rabbit reticulocytes. Arch. Biochem. Biophys 234, 591–602. [DOI] [PubMed] [Google Scholar]
- (21).Dzialo MC, Travaglini KJ, Shen S, Roy K, Chanfreau GF, Loo JA, and Clarke SG (2014) Translational roles of elongation factor 2 protein lysine methylation. J. Biol. Chem 289, 30511–30524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Buchanan BW, Lloyd ME, Engle SM, and Rubenstein EM (2016) Cycloheximide chase analysis of protein degradation in Saccharomyces cerevisiae. J. Vis. Exp 18, No. (110):53975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Daniel Gietz R, and Woods RA (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- (24).Zheng Y, and Jiang Y (2015) mTOR Inhibitors at a glance. Mol. Cell. Pharmacol 7, 15–20. [PMC free article] [PubMed] [Google Scholar]
- (25).Kuranda K, Leberre V, Sokol S, Palamarczyk G, and François J (2006) Investigating the caffeine effects in the yeast Saccharomyces cerevisiae brings new insights into the connection between TOR, PKC and Ras/cAMP signalling pathways. Mol. Microbiol 61, 1147–1166. [DOI] [PubMed] [Google Scholar]
- (26).Carrasco L, Barbacid M, and Vazquez D (1973) The trichodermin group of antibiotics, inhibitors of peptide bond formation by eukaryotic ribosomes. Biochim. Biophys. Acta - Nucleic Acids Protein Synth 312, 368–376. [DOI] [PubMed] [Google Scholar]
- (27).Cundliffe E, Cannon M, and Davies J (1974) Mechanism of inhibition of eukaryotic protein synthesis by trichothecene fungal toxins. Proc. Natl. Acad. Sci. U. S. A 71, 30–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Brewer JW, Hendershot LM, Sherr CJ, and Diehl JA (1999) Mammalian unfolded protein response inhibits cyclin D1 translation and cell-cycle progression. Proc. Natl. Acad. Sci. U. S. A 96, 8505–8510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chan S-W, and Egan PA (2005) Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 19, 1510–1512. [DOI] [PubMed] [Google Scholar]
- (30).Kirillov S, Porse BT, Vester B, Woolley P, and Garrett RA (1997) Movement of the 3′-end of tRNA through the peptidyl transferase centre and its inhibition by antibiotics. FEBS Lett. 406, 223–233. [DOI] [PubMed] [Google Scholar]
- (31).Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, and Liu JO (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol 6, 209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Al-Hadid Q, Roy K, Munroe W, Dzialo MC, Chanfreau GF, and Clarke SG (2014) Histidine methylation of yeast ribosomal protein Rpl3p is required for proper 60S subunit assembly. Mol. Cell. Biol 34, 2903–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).McNabb DS, Reed R, and Marciniak RA (2005) Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot. Cell 4, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, and Atkins JF (1998) A dual-luciferase reporter system for studying recoding signals. RNA 4, 479–486. [PMC free article] [PubMed] [Google Scholar]
- (35).Harger JW, and Dinman JD (2003) An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9, 1019–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Branchini BR, Murtiashaw MH, Magyar RA, and Anderson SM (2000) The role of lysine 529, a conserved residue of the acyl-adenylate-forming enzyme superfamily, in firefly luciferase. Biochemistry 39, 5433–5440. [DOI] [PubMed] [Google Scholar]
- (37).Starita LM, Lo RS, Eng JK, von Haller PD, and Fields S (2012) Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics 12, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Lin KW, Yakymovych I, Jia M, Yakymovych M, and Souchelnytskyi S (2010) Phosphorylation of eEF1A1 at Ser300 by TβR-I Results in Inhibition of mRNA Translation. Curr. Biol 20, 1615–1625. [DOI] [PubMed] [Google Scholar]
- (39).Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JEP, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, and Hunt DF (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U. S. A 104, 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Wu X, Oh M-H, Schwarz EM, Larue CT, Sivaguru M, Imai BS, Yau PM, Ort DR, and Huber SC (2011) Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 155, 1769–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Zobel-Thropp P, Yang MC, Machado L, and Clarke S (2000) A novel post-translational modification of yeast elongation factor 1A. Methylesterification at the C terminus. J. Biol. Chem 275, 37150–37158. [DOI] [PubMed] [Google Scholar]
- (42).Małecki J, Aileni VK, Ho AYY, Schwarz J, Moen A, Sørensen V, Nilges BS, Jakobsson ME, Leidel SA, and Falnes PØ (2017) The novel lysine specific methyltransferase METTL21B affects mRNA translation through inducible and dynamic methylation of Lys-165 in human eukaryotic elongation factor 1 alpha (eEF1A). Nucleic Acids Res. 45, 4370–4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Jank T, Belyi Y, Wirth C, Rospert S, Hu Z, Dengjel J, Tzivelekidis T, Andersen GR, Hunte C, Schlosser A, and Aktories K (2017) Protein glutaminylation is a yeast-specific posttranslational modification of elongation factor 1A. J. Biol. Chem 292, 16014–16023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Vermillion KL, Lidberg KA, and Gammill LS (2014) Cytoplasmic protein methylation is essential for neural crest migration. J Cell Biol 204, 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Alban C, Tardif M, Mininno M, Brugière S, Gilgen A, Ma S, Mazzoleni M, Gigarel O, Martin-Laffon J, Ferro M, and Ravanel S (2014) Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS One 9, e95512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Hiatt WR, Garcia R, Merrick WC, and Sypherd PS (1982) Methylation of elongation factor 1 alpha from the fungus Mucor. Proc. Natl. Acad. Sci. U. S. A 79, 3433–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Owings JP, Kuiper EG, Prezioso SM, Meisner J, Varga JJ, Zelinskaya N, Dammer EB, Duong DM, Seyfried NT, Alberti S, Conn GL, and Goldberg JB (2015) Pseudomonas aeruginosa EftM is a thermoregulated methyltransferase. J. Biol. Chem 291, 3280–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Barbier M, Owings JP, Martínez-Ramos I, Damron FH, Gomila R, Blázquez J, Goldberg JB, and Albertí S (2013) Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. MBio 4, e00207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Kraal B, Lippmann C, and Kleanthous C (1999) Translational regulation by modifications of the elongation factor Tu. Folia Microbiol. (Praha). 44, 131–141. [DOI] [PubMed] [Google Scholar]
- (50).Toledo H, and Jerez CA (1985) In vitro methylation of the elongation factor EF-Tu from Escherichia coli. FEBS Lett. 193, 17–21. [DOI] [PubMed] [Google Scholar]
- (51).Shimazu T, Barjau J, Sohtome Y, Sodeoka M, and Shinkai Y (2014) Selenium-based S-adenosylmethionine analog reveals the mammalian seven-beta-strand methyltransferase METTL10 to be an EF1A1 lysine methyltransferase. PLoS One 9, e105394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Cavallius J, Popkie AP, and Merrick WC (1997) Site-directed mutants of post-translationally modified sites of yeast eEF1A using a shuttle vector containing a chromogenic switch. Biochim. Biophys. Acta - Gene Struct. Expr 1350, 345–358. [DOI] [PubMed] [Google Scholar]
- (53).Sherman M, and Sypherd PS (1989) Role of lysine methylation in the activities of elongation factor 1α. Arch. Biochem. Biophys 275, 371–378. [DOI] [PubMed] [Google Scholar]
- (54).Young CC, Alvarez JD, and Bernlohr RW (1990) Nutrient-dependent methylation of a membrane-associated protein of Escherichia coli. J. Bacteriol 172, 5147–5153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Noort JM, Kraal B, Sinjorgo KMC, Persoon NLM, Johanns ESD, and Bosch L (1986) Methylation in vivo of elongation factor EF-Tu at lysine-56 decreases the rate of tRNA-dependent GTP hydrolysis. Eur. J. Biochem 160, 557–561. [DOI] [PubMed] [Google Scholar]
- (56).Prezioso SM, Duong DM, Kuiper EG, Deng Q, Albertí S, Conn GL, and Goldberg JB (2019) Trimethylation of elongation factor-tu by the dual thermoregulated methyltransferase EftM does not impact its canonical function in translation. Sci. Rep 9, 3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Liu S, Hausmann S, Carlson SM, Fuentes ME, Francis JW, Pillai R, Lofgren SM, Hulea L, Tandoc K, Lu J, Li A, Nguyen ND, Caporicci M, Kim MP, Maitra A, Wang H, Wistuba II, Porco JA, Bassik MC, Elias JE, Song J, Topisirovic I, Van Rechem C, Mazur PK, and Gozani O (2019) METTL13 methylation of eEF1A increases translational output to promote tumorigenesis. Cell 176, 491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Jakobsson ME, Małecki J, Nilges BS, Moen A, Leidel SA, and Falnes PØ (2017) Methylation of human eukaryotic elongation factor alpha (eEF1A) by a member of a novel protein lysine methyltransferase family modulates mRNA translation. Nucleic Acids Res. 45, 8239–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Hamey JJ, Separovich RJ, and Wilkins MR (2018) MT-MAMS: protein methyltransferase motif analysis by mass spectrometry. J. Proteome Res 17, 3485–3491. [DOI] [PubMed] [Google Scholar]
- (60).Young CC, and Bernlohr RW (1991) Elongation factor Tu is methylated in response to nutrient deprivation in Escherichia coli. J. Bacteriol 173, 3096–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Munshi R, Kandl KA, Carr-Schmid A, Whitacre JL, Adams AE, and Kinzy TG (2001) Overexpression of translation elongation factor 1A affects the organization and function of the actin cytoskeleton in yeast. Genetics 157, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.