Abstract
Coronary artery disease causes acute myocardial infarction and heart failure. Identifying coronary vascular progenitors and their developmental program could inspire novel regenerative treatments for cardiac diseases. The developmental origins of the coronary vessels have been shrouded in mystery and debated for several decades. Recent identification of progenitors for coronary vessels within the endocardium, epicardium and sinus venosus provides new insights into this question. In addition, significant progress has been achieved in elucidating the cellular and molecular programs that orchestrate coronary artery development. Establishing adequate vascular supply will be an essential component of cardiac regenerative strategies, and these findings raise exciting new strategies for therapeutic cardiac revascularization.
Keywords: Coronary origin, lineage tracing, coronary vascular endothelial cells, epicardium, endocardium, sinus venosus
To satiate its voracious appetite for oxygen and nutrients, the heart has a dedicated circulatory system, the coronary vasculature. Coronary artery disease causes myocardial infarction (MI) and heart failure, making it the leading cause of death worldwide.1–3 In MI, coronary artery occlusion causes death of as many as one billion cardiomyocytes.4 Lack of adequate numbers of functional cardiomyocytes coupled with chronic overload and subsequent dysfunction of remaining cardiomyocytes eventually results in heart failure and death.5, 6 Currently, angioplasty and coronary artery bypass surgery are the mainstays for coronary artery disease treatment.7 Although pro-angiogenic approaches have shown promise in animal models,8, 9 these have not yet been efficacious in clinical trials. Clearly, improved approaches for coronary revascularization and cardiomyocyte regeneration after myocardial infarction are urgently needed.9, 10 A better understanding of how coronary vessels are built during development will provide important insights that might be therapeutically deployed for cardiac regeneration.
Benjamin Franklin stated, “Originality is the art of concealing your sources”. Nature has done a good job of concealing the sources of the coronary arteries. Simply speaking, the coronary arteries consist of two layers of cells: the inner endothelial cell layer and outer smooth muscle cells. Overwhelming evidence indicates that most coronary smooth muscle cells originate from epicardium.11–14 However, the origin of coronary endothelium has ignited controversies over the past two decades and remains an enigma. This review focuses on the origin of coronary vascular endothelial cells (VECs) and on the developmental programs that regulate coronary angiogenesis in development and disease.
Genetic lineage tracing and its caveats
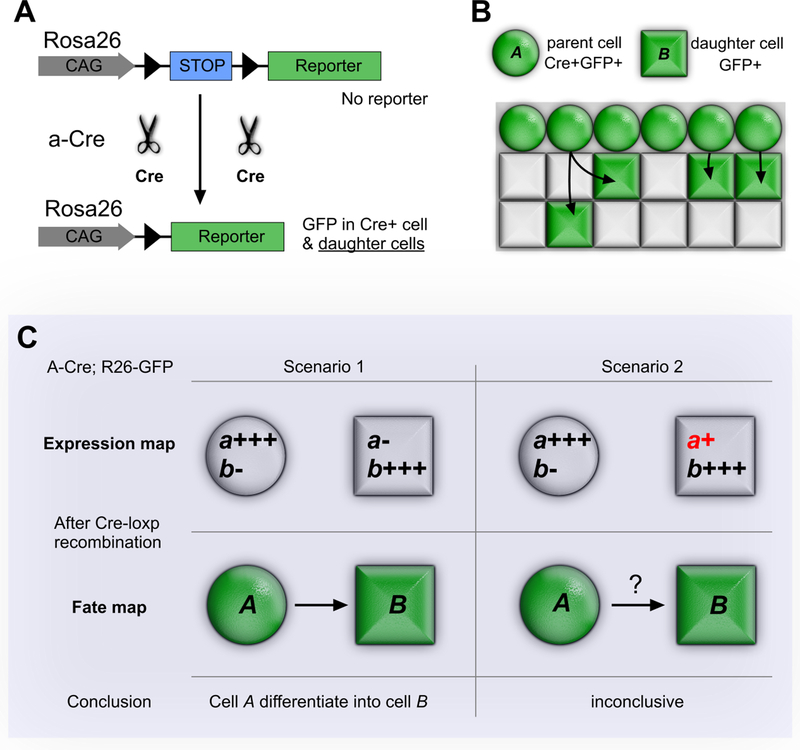
Most recent studies of coronary vessel development were performed in mouse, and state-of-the-art techniques have been used to trace the origin of coronary VECs. It is critical to first introduce genetic lineage tracing techniques and to discuss its caveats so that experiments can be appropriately analyzed and interpreted. Lineage tracing in mammalian systems largely relies on Cre-loxP recombination.15, 16 This technology is dependent upon two components: a Cre allele that selectively expresses in the progenitor cell of interest, and a Cre-activated reporter allele (Figure 1A). Selective expression of Cre recombinase in a progenitor cell of interest is achieved most commonly by identifying an endogenous marker gene selectively expressed in the progenitor cell and then placing Cre under control of the marker gene’s transcriptional regulatory sequences, e.g. by replacement of the marker gene’s open reading frame with Cre in the endogenous locus (knock-in). Typically, Cre-activated reporters are located in a widely permissive locus (e.g. Rosa26) and use a broadly expressed promoter (e.g. CAG) to drive expression of a reporter gene. In the absence of Cre, a loxP-flanked transcriptional stop cassette blocks reporter gene expression. Cre expression in the progenitor cell leads to excision of the floxed stop cassette and allows subsequent expression of the reporter gene (Figure 1A). Because this genetic label is heritable and permanent, the cell and all of its descendants are indelibly marked by reporter gene expression, regardless of whether or not their descendants actively express Cre.
Figure 1. Cre-loxP mediated genetic lineage tracing system.
A, Cre-loxP mediated recombination for lineage tracing is heritable and irreversible. In Cre expressing cells (eg. cell A), the loxP-flanked transcriptional stop cassette is removed, permitting reporter (e.g. GFP) expression in cell A and its descendants. Cre is driven by promoter a, which is strong in cell A but absent in cell B. Rosa26-loxp-stop-loxp-GFP (R26-GFP) is the Cre reporter line. B, Schematic figure showing cell A (Cre+) migrate and differentiate into cell B (Cre-). A and B cells both express lineage tracing marker GFP. C, Correct interpretation of lineage tracing data hinges on the data supporting negative Cre expression in Cell B or its non-A ancestors.
The powerful Cre-loxP system provides a history of a cell and its descendants by showing where they migrate and also the cell types into which they differentiate. For example, progenitor cell A expresses typical marker a strongly and specifically. Cre recombinase under promoter a would lead to lineage labeling of A cells. Cell B expresses neither marker a nor Cre recombinase from promoter a (Figure 1B). As lineage tracing is irreversible and inheritable, if B cells are labeled by a-Cre, then the straightforward interpretation is that A cells differentiate into B cells (Figure 1B). However, this interpretation should be made cautiously. If B cells express low level of a and therefore Cre, then one would not be able to interpret B cell marking by Cre as indicative of an A to B fate transition (Figure 1C). The interpretation of a Cre-loxP experiment therefore hinges on the “negative” expression of Cre in B cells, or indeed in any non-A cell type in the organism’s developmental history that could differentiate into B cells.
This type of expansive negative data can be difficult to acquire. Direct measurement of Cre expression is more desirable than measurement of the endogenous gene (because the Cre allele’s expression characteristics may differ from the endogenous gene). Since the expression of a gene is dynamic with time and strength, negative expression at one time point does not exclude its possible expression at another. This is an important limitation to the rigorous interpretation of lineage tracing data obtained with constitutively active Cre alleles. Thus, it is desirable to perform these experiments using inducible Cre alleles that are active only in the presence of an inducing agent, since then one can restrict the temporal window in which one needs detailed Cre expression data. On the other hand, inducible Cre alleles can be difficult to fully activate. As a result, labeling may be incomplete and so underestimate the extent of contribution of lineage A to lineage B. Complementary approaches that do not critically hinge on the expression or lack of expression of Cre in target cells (B cells) are essential to provide definitive evidence of A to B trans-differentiation.
A final factor that must be considered to interpret Cre-loxP data, and to compare between studies, is that Cre reporter genes differ significantly in their susceptibility to recombination.17, 18 This is true even when comparing between Cre reporters positioned in the same locus (e.g. Rosa26). While expression differences are graded, Cre recombination is all-or-none. As a result, the susceptibility of a Cre reporter to recombination imposes a “threshold” for Cre labeling, and slightly different thresholds can lead to very different Cre labeling maps (discussed in detail in ref. 18). A recombination-sensitive reporter would reveal a broader fate map that includes progenitors with lower level or transient Cre expression, while an insensitive reporter would reveal a more narrow fate map that corresponds with higher level or longer duration of Cre expression.19 Apparent descrepancies between the lineage tracing results reported by different labs can be linked to the use of Cre reporters with different sensitivities.18–20
We will incorporate these caveats in our interpretation of the different candidate origins for coronary vascular endothelium being most heavily investigated: (pro)epicardium, sinus venosus, and ventricular endocardium.
Coronary VECs from (Pro)epicardium
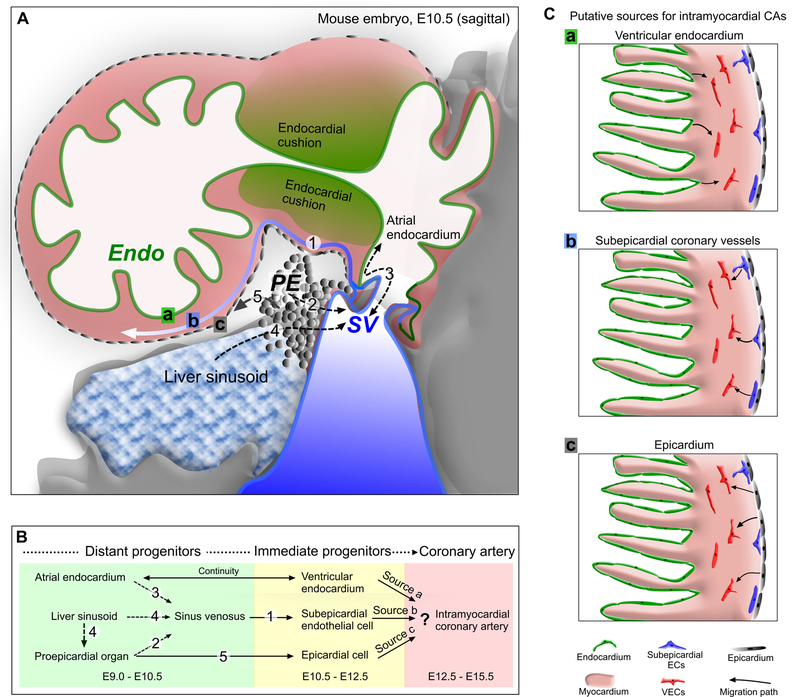
Proepicardium is a transient cauliflower-like protrusion originating from transversum septum and wedged below the atrioventricular junction at E9.5 (Figure 2A). Proepicardial cells migrate onto and spread over the surface of the heart, eventually covering the heart surface with an epithelial sheet, the epicardium.21–24 The epicardium plays an essential role in heart development and disease through two general mechanisms. First, it undergoes epithelial-to-mesenchymal transition (EMT) to form mesenchymal epicardium-derived cells (EPDCs), which subsequently differentiate into most of the non-cardiomyocyte cell types of the myocardial wall.22 Second, the myocardial-epicardial interface forms a critical signaling center that is necessary for both myocardial growth and coronary vessel development.22, 25, 26
Figure 2. Formation of the nascent coronary vessel plexus in the developing heart.
A and B, The three major sources of coronary vessels, the proepicardium (PE), sinus venosus (SV) and endocardium (Endo) are intimately associated with each other during heart development. The PE is a transient structure (grey) that wedged into the atrioventricular groove between liver sinusoids and SV, and eventually gives rise to the epicardium covering the heart. The SV (blue) is the venous inflow tract. Venous cells from SV sprout onto the heart and produce subepicardial coronary vessels. The endocardium (green) lines the heart lumen. Black dashed arrows denote movement from one compartment to another, potentially complicating lineage-tracing experiments. Numbers _in B correspond to those in A showing location of migration events. C, Three putative sources for intramyocardial coronary arteries (CAs) in the developing heart. Arrows indicate corresponding migration path.
For two decades, the (pro)epicardium was proposed to give rise to the coronary arteries based on three types of lineage tracing experiments in chick embryos: retroviral labeling, dye labeling, and quail-to-chick cell transplantation.11, 27, 28 In pioneering work, Mikawa and colleagues injected replication-deficient, lacZ expressing retrovirus into proepicardium. Later in heart development, lacZ-expressing cells were found in the endothelial, smooth muscle, and fibroblast lineages.27 These key experiments established the paradigm that coronary VECs and smooth muscle cells arise from proepicardium. This view was reinforced by transplantation of microdissected quail proepicardium into chicken embryos. Quail cells, detected by the quail-specific antibody QCPN, formed coronary VECs and smooth muscle cells.28
These seminal experiments had two key limitations. First, both faced potential contamination issues. For example, sinus venosus (SV) was co-transplanted with the proepicardium in the chimera assay,28 which may have been important in retrospect since the SV was recently found to be an important source of VECs in mouse embryos (see below).29 Second, neither study could quantitatively determine the fraction of coronary VECs that arise from pro-epicardium, an important issue if there are multiple different VEC sources. In addition, transplantation assays, although informative about the trans-differentiation potential of proepicardium into coronary endothelium, may not inform us on the actual differentiation of epicardial cells under physiological conditions. For example, mammary myoepithelial cells can adopt a multipotent fate and are able to regenerate a complete mammary gland upon transplantation. However, it still remains a controversy whether these myoepithelial cells really behave in this manner in vivo under physiological conditions.30, 31
The retroviral and transplantation assays described above suggest that at least some coronary VECs arise from proepicardium. The proepicardium is a transient structure that contains several morphologically distinct cell types. In addition to superficial mesothelial cells, the avian proepicardium also contains angioblast, endothelial and hematopoietic cells.32, 33 Poelmann et al. showed that endothelial cells from the SV expand into the proepicardium to reach the dorsal side of the atrioventricular sulcus, where most coronary vessels are first detected.34 Distinct populations of endothelial cells – those associated with SV or liver bud, and those not associated with either SV or liver bud – were also found in murine proepicardium.35 These results lead to the question of whether those coronary VECs found to arise from proepicardium develop from endothelial cells already present in proepicardium, or if they develop by transdifferentiation of the proepicardium’s non-endothelial cell types. Addressing this question requires use of cell-type specific labeling reagents, rather than spatial labeling that is the basis of retroviral or transplantation assay.
The origin of coronary VECs has been studied in the murine system using the Cre-loxP system, which achieves cell-type specific labeling. Cre alleles driven by regulatory elements of epicardial markers Wt1,14, 36–39 Gata5,40 Tbx18,41, Tcf21,42 Sema3d, or Scx,43 were used to study the fate of epicardium in the developing mammalian heart. These studies showed that at most a small fraction of coronary VECs arise from (pro)epicardium in mouse (Table 1), a surprising finding given the long-standing view, established by the avian studies, that coronary VECs develop from (pro)epicardium. Why were such divergent conclusions reached in avians compared to mice? In retrospect, the avian studies did not provide quantitative information on the fraction of coronary VECs that originate from (pro)epicardium. Second, the key cell population that generated coronary VECs in the regional labeling in the avian systems may not have the mesothelial populations labeled by these Cre alleles. For instance, proepicardial vascular cells may have yielded coronary VECs in the avian experiments. Third, Katz et al. demonstrated that proepicardial cells are heterogeneous and suggested that different proepicardial progenitors may contribute differently to specific lineages.43 However, the relevance of this hypothesis to coronary VECs awaits additional experimental data. Interestingly, in this case, the proepicardium itself may contribute to sinus venosus, which was also lineage labeled (Figure 2A, B).
Table 1.
Lineage tracing for intramyocardial coronary VECs in embryonic ventricle wall
| Mouse line | Targeting strategy | Constitutive / inducible | Origins / sources | % VECs labeled | Note | References |
|---|---|---|---|---|---|---|
| WT280Cre, YAC | Transgenic | Constitutive | Proepicardium | 14% | Labels most epicardium | (Wilm et al. 2005) |
| Wt1/IRES/GFP-Cre, BAC | Transgenic (RP23-266M16) | Constitutive | Proepicardium | Few | Labels most epicardium | (Wessels et al. 2012) |
| Wt1(RP23–8C14)-Cre | Transgenic (RP23–8C14) | Constitutive | Proepicardium | Few | Labels most epicardium | (Kolander et al. 2014) |
| Wt1-GFPCre | Knockin (ATG) | Constitutive | Proepicardium | Few | Labels most epicardium; caution regarding non-epicardial labeling under some conditions. |
(Zhou et al. 2008, 2012; Rudat and Kispert 2012). |
| Wt1-CreERT2 | Knockin (ATG) | Inducible | Proepicardium | Few | Labels most epicardium; caution regarding non-epicardial labeling under some conditions. |
(Zhou et al. 2008, 2012) |
| Tbx18-Cre | Knockin (5’ UTR) | Constitutive | Proepicardium | None | Labels most epicardium | (Cai et al. 2009) |
| Tcf21 MerCreMer | Knockin (ATG) | Inducible | Epicardium | None | Labels most epicardium | (Asha et al. 2011) |
| Tbx18-CreERT2 | Knockin (ATG) | Inducible | Epicardium | None | Labels most epicardium | (Moore-Morris et al. 2014) |
| VE-Cad CreERT2 | Transgenic | Inducible | Sinus venosus | Most | Clonal analysis | (Red-Horse et al. 2010) |
| Scx-GFPCre, BAC | Transgenic | Constitutive | Proepicardium | ND* (1) | Labels part of SV | (Katz et al. 2012) |
| Sema3D-Cre | Knockin (ATG) | Constitutive | Proepicardium | ND* (2) | Labels part of SV | (Katz et al. 2012) |
| Nfatc1-IRES-Cre | Knockin (3’ UTR) | Constitutive | Endocardium | 81% | Labels part of SV | (Wu et al. 2012) |
| Nfatc1-nrtTA, BAC | Transgenic | Inducible | Endocardium | Most | Clonal analysis | (Wu et al. 2012) |
| Nfatc1-CreERT2 | Knockin (ATG) | Inducible | Endocardium | Few | Labels most 2nd CVP | (Tian et al. 2014) |
| Apj-CreERT2 | Transgenic | Inducible | Sinus venosus | 40% LV | Labels part of SV | (Chen et al. 2014) |
| Apln-CreERT2 | Knockin (ATG) | Inducible | Subepicardial VECs | >90% | Labels subepicardial vessels | (Tian et al. 2013) |
Few means the minority, as there is no quantitative data available from published papers.
Most means the majority of clones show their relationship by clonal analysis
ATG means the Cre or CreERT2 cDNA cassette was knocked into the endogenous start codon.
3’UTR means the Cre or CreERT2 cDNA cassette is inserted within the 3’ untranslated region as IRES following endogenous gene.
ND, not determined; LV, left ventricle.
(1) 24.3% or (2) 6.9% of Cre-labeled cells were coronary VECs.
Another caveat in lineage tracing of epicardial cells into coronary VECs is that the Cre allele used should not be expressed in coronary VECs themselves (Figure 1C). The most widely used epicardial Cre lines are based on Wt1 regulatory elements, using different strategies including BAC and YAC transgenics and endogenous locus knockins.14, 36–39, 44 Although Wt1 is highly expressed in epicardium, it was also reported that Wt1 could be expressed in coronary vasculature in adult heart after myocardial infarction.45 Epicardial cells labeled by tamoxifen treatment of Wt1CreERT2 embryos at E10.5 contribute to few coronary endothelial cells at E18.5. However, tamoxifen treatment in late embryonic or early neonatal stages (e.g. E14.5-P4) labeled a substantial number of coronary endothelial cells at E18.5 and P7 respectively.19 Labeling of endothelial cells by E14.5 tamoxifen treatment likely reflected expression of Cre in situ in these coronary endothelial cells rather than their origin from epicardial cells.19 During development, epicardial cells mainly contribute to coronary smooth muscle cells and fibroblasts (Figure 3). Likewise, the inducible lineage tracing in adult heart after myocardial injury captures a small population of coronary VECs.46 However, these labeled coronary VECs were quite isolated in myocardium and far from reactivated epicardial layers in injured heart.46 As there was no spatial relationship between these labeled VECs and the epicardium even shortly after injury, labeling of these coronary VECs are likely due to Wt1-driven CreERT2 expression in some coronary VECs rather than EPDCs that had migrated into the myocardium.45, 46 These epicardial cells adopt smooth muscle and fibroblast fates in injured heart (Figure 3). Other studies based on a different Wt1-IRES-GFPCre line showed that a small population of coronary endothelial cells were labeled and they were interpreted to form de novo after myocardium infarction from re-activated epicardial cells.47 This interpretation should be made cautiously given the aforementioned caveats of Cre lineage tracing and the potential for Wt1 expression in occassional coronary endothelial cells.
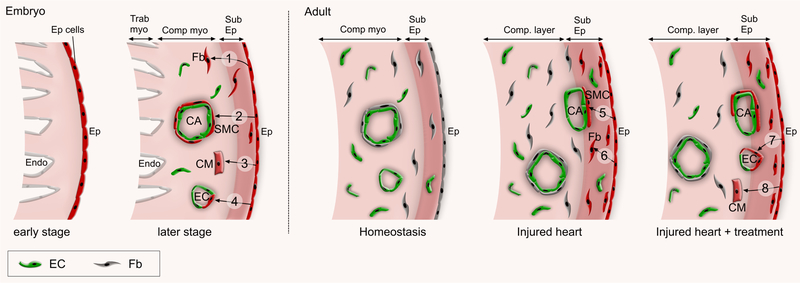
Figure 3. Epicardial contribution to developing and adult heart.
In early embryonic stage, epicardial cells form an epithelial sheet that covers the heart. At later embryonic stages, epicardial cells (Ep cells) form mesenchymal epicardium-derived cells (EPDCs) by EMT. EPDCs appear in the subepicardial layer (Sub Ep) and migrate into compact myocardium (Comp myo), where they differentiated into fibroblasts (Fb, 1), smooth muscle cells (SMC, 2), cardiomyocytes (CM, 3) and endothelial cells (EC, 4). In adult heart under normal homeostatic conditions, most epicardial cells remain quiescent (grey color). MI activates the embryonic program (red) and these epicardial cells differentiate into SMC (5) and Fb (6), but rarely if at all to CMs or ECs. Paracrine factors such as modRNA encoding Vegfa or thymosin β4 stimulates a subset of EPDCs to differentiate into EC (7) or CM (8) lineages, respectively in the post MI hearts.
Taken together, epicardium likely contributes to coronary vessels in the developing mouse heart, but quantitatively, its contribution is small (Table 1). The same is likely true in the avian system, although definitively establishing this would require use of assays in which the cell-type specific contribution of mesothelial cells to coronary VECs could be quantitatively estimated.
Crosstalk Between Epicardium and Myocardium
Although epicardial cells provide few coronary VECs, the epicardium is essential for coronary vessel formation during heart development. Epicardial cells secrete essential growth factors and cytokines that regulate cells of the nascent coronaries directly and that impact the underlying myocardium, which in return provides angiogenic factors that promote coronary vasculogenesis and angiogenesis.25, 48–50 Gene knockdown or knockout approaches have identified a number of epicardial or myocardial genes that are essential for coronary angiogenesis (Table 2). Vascular cell adhesion molecule 1 (Vcam1), erythropoietin receptor, connexin 43, and α4 integrin are required for formation, migration, or integrity of the epicardium.51–54 The transcriptional regulators Wt1 and Rxra are required within epicardial cells, where they influence the integrity of the epicardium as well as the formation of vascular cells by EMT.40, 55, 56 Wt1 and Rxra both regulate epicardial EMT through Wnt signaling pathways.40, 56 Wt1 also regulates expression of Ntrk2, a cytokine receptor necessary for normal coronary vessel development,57 and Raldh2, a key regulator of retinoic acid signaling confined to the epicardium was also regulated by Wt1.56 Tbx18, another epicardial transcription factor, is also required for proper development of coronary arteries.58
Table 2.
Genetically Engineered Mice With Coronary or Epicardial Phenotypes
| Gene | Expression | Model | Coronary plexus | Comp. Myo. | Additional Co/Epi Phenotypes | References | ||
|---|---|---|---|---|---|---|---|---|
| Ep | M | En | ||||||
| RXRa | X | X | MLC2vcre | Nl | Nl | no phenotype | (Chen et al., 1998) | |
| Gata5Cre | abnormal CA branching |
thin | abnormal Wnt/b-catenin signaling | (Merki et al., 2005) | ||||
| WT1 | X | KO | absent | thin | decreased SEMC, epicardium fails to envelope heart |
(Moore et al., 1999) | ||
| Ntrk2 | X | KO | impaired | Nl | normal epicardium | (Wagner et al., 2005) | ||
| BDNF | KO | normal, but EC apoptosis |
Nl | hemmorhage around subepicardial vessels | (Donovan et al., 2000) | |||
| EpoR | X | X | KO | impaired | thin | detached epicardium | (Wu et al., 1999) a4 | |
| integrin | X | KO | impaired | no epicardium; pericardial hemmorhage | (Yang et al., 1995) | |||
| VCAM1 | X | KO | premature death | thin | no epicardium; pericardial hemmorhage | (Kwee et al., 1995) | ||
| Tbx1 | X | KO | disorganized | NI | disordefective vascular plexus remodeling | (Wu et al. 2013) | ||
| PDGFrb | X | Gata5Cre | disrupted | thin | disrupted epicardial cell migration | (Mellgren et al. 2008) | ||
| Podoplanin | X | KO | abnormal | thin | impaired epicardial adhesion and spreading | (Mahtab et al. 2008) | ||
| ALK5 | X | X | X | Gata5Cre | defective | thin | aberrant capillary vessel formation | (Sridurongrit et al. 2008) |
| FGF9 | X | X | KO | impaired | thin | abnormal Hedgehog signaling | (Lavine et al., 2006) | |
| FGFR1+2 | X | MLC2vCre | impaired | thin | abnormal Hedgehog signaling | (Lavine et al., 2006) | ||
| Hedgehog signaling |
X | X | Smo, Mlc2v-Cre or Dermo1-Cre | Reduced | Nl | Rudeced coronary veins or intramyocardial arterial vessels |
(Lavine et al., 2008) | |
| Thymosin b4 | X | MLC2vCre-activated siRNA | Impaired | Nl | left ventricle noncompaction | (Smart et al., 2006) | ||
| Gata4 | X | X | X | TR-KO | premature death | thin | no proepicardium | (Watt et al., 2004) |
| Fog2-Gata4 loss | impaired | thin | epicardium intact, and decreased SEMC | (Crispino et al., 2001) | ||||
| Fog2 | X | X | X | KO | impaired | thin | epicardium intact, decreased SEMC | (Tevosian et al., 2000) |
| Vegfa | X | Tnnt2-Cre | imapired CAs | thin | epicardium intact, decreased intramyocardial CAs, no defect in coronary veins |
(Wu et al., 2012) | ||
| VegfR2 | X | Nfatc1-Cre | impaired CAs | thin | 88% decrease in intramyocardial CAs, 37% decrease in coronary veins |
(Wu et al., 2012) | ||
| Ang1 | X | aMHC-Cre | impaired coronary veins | thin | 74% decrease in intramyocardial CAs, 24% decrease in coronary veins |
(Arita et al., 2014) | ||
| BAF180 | X | X | X | KO | defective | NI | impaired EMT and epicardial maturation | (Huang et al., 2008) |
| BAF200 | X | X | X | KO | defective | thin | reduced intramyocardial coronary vessels | (He et al. 2014) |
TR-KO, tetraploid rescued knock out; Fog2-Gata4 loss, loss of interaction between Fog2 and Gata4; SEMC, subepicardial mesenchyme, Ep, epicardium; M, myocardium; En, endocardium; Comp. Myo., compact myocardium; Co/Epi, coronary/epicardium; NI, not indicated.
Within the myocardium, several major signaling pathways have been implicated in coronary vessel development. Here we will highlight three of the most studied pathways. First, normal formation of the coronary vascular plexus requires fibroblast growth factor (FGF) signaling. FGF9, secreted by the epicardium, stimulates FGFR1 and FGFR2c receptors on cardiomyocytes. The expression of FGF9 is regulated by retinoic acid signals from epicardium.59 Disruption of this FGF signal results in impaired myocardial secretion of angiopoietin-2 (Ang-2), vascular endothelial growth factor-A (VEGF-A), and VEGF-B60 VEGF was shown to be highest in the compact myocardium nearest the epicardium and subsequently become more evenly distributed transmurally, which coincides with sites of coronary vessel formation.61, 62 Additional fibroblast growth factors such as FGF1, 2, 7 secreted by either myocardium or epicardium, also positively regulate epicardial EMT.63 Sonic hedgehog is a critical mediator that links FGF signaling to VEGF-A expression and coronary vessel formation during development as well as maintenance in disease.60, 64 In addition, hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development.65 Cardiomyoblast hedgehog signaling regulates the development of coronary veins, while perivascular cell hedgehog signaling is required for coronary arterial growth.66
Second, the secreted myocardial factor thymosin β4 is necessary for migration of EPDCs into the myocardium to form the coronary vascular plexus.67–69 In thymosin β4 knockdown hearts, the epicardium was detached and contained nodules of cells expressing endothelial and smooth muscle markers. These cells were deficient within the myocardium. In both embryonic and postnatal heart, thymosin β4 promoted endothelial cell migration and proliferation70 and initiated the embryonic coronary developmental program and epicardial progenitor cell activation.71–73 These studies suggest that thymosin β4 is a facilitator of coronary neovascularization and highlight epicardial cells as resident progenitors which, when instructed by thymosin β4, have the capacity to sustain the myocardium after ischemic damage.72, 74 However, two recent studies showed that thymosin β4 inactivation is dispensable for embryonic viability and coronary vessel development.75, 76 Further studies are required to further delineate the role of thymosin β4 in coronary vessel development and postnatal angiogenesis.
Third, normal formation of the coronary vascular plexus requires interaction between GATA4, a transcription factor that is critical for cardiac development and function77–80, and its cofactor FOG2. FOG2 binds to GATA4 to both positively and negatively regulate transcriptional activity in a context-specific manner.81, 82 Germline mutation of Fog2 resulted in severe impairment of coronary vessel development and hypoplasia of the compact myocardium, as well as structural heart defects.82 Introduction of a point mutation into Gata4 that blocks interaction with Fog2 resulted in a similar phenotype.83 Cardiomyocyte-specific deletion of Fog2 or disruption of Fog2-Gata4 interaction recapitulated the defected coronary vessel formation during embryonic development.84 Fog2-Gata4 interaction promoted expression of proangiogenesis factors and suppressed expression of angiogenesis inhibitors.84, 85
Besides these major players, there are additional molecular signals that control epicardial integrity, coronary angiogenesis, and compact myocardium formation. It is becoming increasingly clear that coronary angiogenesis in both development and regeneration depends on an orchestrated crosstalk between the epicardium, coronary VECs and the myocardium in part through secreted growth factors. This complex epicardium-myocardium communication and its impact on the coronary angiogenesis has been described in detail in other previous reviewers.22, 48, 86
Coronary VECs from SV: vein-to-artery reprogramming
If epicardial cells contribute to at most a small fraction of coronary VECs in mice, then what is the ontogeny of the majority of coronary VECs? For the initial coronary vessels emerging at atrioventricular groove, the closest connecting structures are the epicardium and the sinus venosus (Figure 2A). The SV is a transient stucture in cardiovascular development that receives venous blood from the vitelline vein, umbilical vein and common cardinal vein and returns it to the atrium.87 As the heart matures, the SV becomes integrated into the right atrium as the coronary sinus. Over a century ago, the coronary vascular network was reported to develop in the human embryo as a circulatory system with connections to the SV.88 Later, Bennett studied the development of coronary vessels in swine embryos and found that coronary vessels arose from “the descending pocket of the SV”.89 His visionary analysis showed that venous outpockets of the SV penetrated the connective tissue between atrium and SV, and these venous sprouts formed capillary networks in the subepicardial space that shared a continuous endothelial lining with the SV.89 Poelmann and colleagues investigated the origin of coronary VECs by detailed description of quail coronary development, and through the study of chimeras formed by quail tissue transplantation into chick embryos.87, 90 These studies showed that pure proepicardial transplants did not contribute to coronary VECs formation, whereas transplants containing or exclusively composed of liver sinusoids did. More importantly, these studies showed that the first coronary VECs communicated with the heart at the SV. The data were interpreted to suggest ingrowth of coronary VECs into the SV.87, 90 However, this early work on the contributions of SV to coronary vessel development remained peripheral while most attention focused on the role of proepicardium.
Recent seminal work from Red-Horse, Krasnow, and colleagues has brought to the fore the importance of vascular sprouting from the SV in coronary vessel development.29 Single cell labeling and clonal analysis were used to show a lineage relationship between SV endothelium and coronary VECs, including those of intramyocardial coronary arteries. Since SV endothelium is initially venous, the study proposed a novel process in which sprouting angiogenesis is accompanied by endothelial de-differentiation into progenitors capable of forming an entirely new vascular bed containing arteries, capillaries, and veins (Figure 4). Understanding the mechanisms that underlie vein-to-artery reprogramming might unlock numerous therapeutic opportunities by improving the durability of revascularization performed using venous grafts and by informing regenerative approaches to revascularization.91
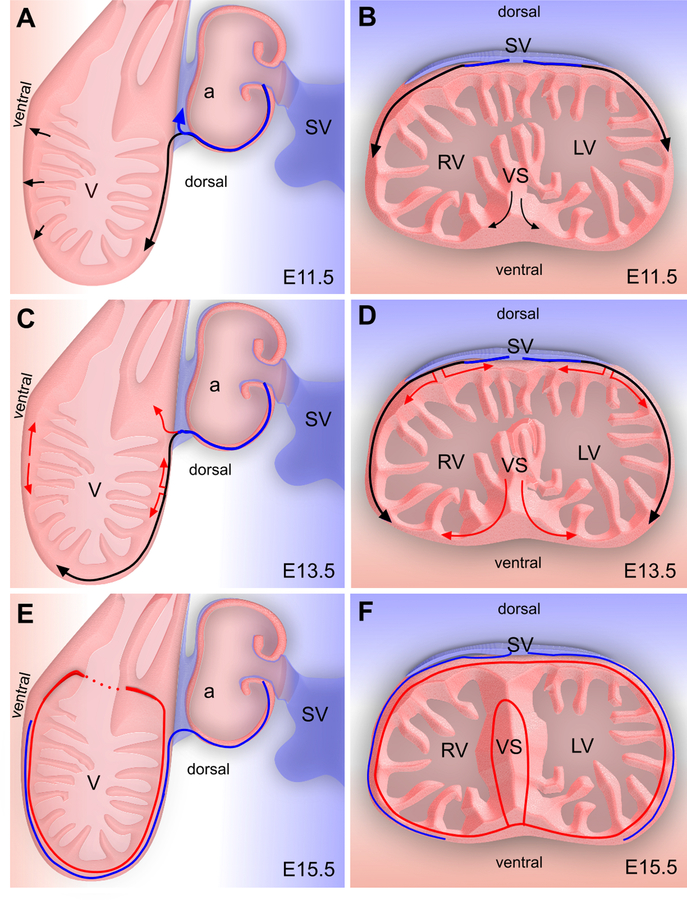
Figure 4. Coronary vessel formation in the ventricle wall and ventricular septum.
A, C, E, Sagittal view of developing heart; B, D, F, Cross-sectional view of the developing heart. Venous cells sprout from SV and dedifferentiate into undifferentiated subepicardial ECs (black) in the dorsal side of heart. As the heart continues to develop, these subepicardial ECs penetrate the myocardial wall and differentiate into arterial ECs (red), while the remaining subepicardial ECs redifferentiate into coronary veins (blue). On the ventral side of heart and in the ventricular septum, coronary ECs arise from endocardium during trabecular compaction (black) and sprout to form a coronary plexus that connects to the coronary vessels in the ventricle wall. V, ventricle; a, atrium; SV, sinus venosus; VS, ventricular septum.
The model of venous cell “reprogramming” into arteries received different views afterwards.92 Because there are no tools currently available to selectively label sinus venosus endothelial cells, the Red-Horse et al. study used a vascular endothelial cell specific Cre (VE-Cadherin-CreERT2)93 in combination with clonal analysis to establish the lineage relationship between sinus venosus ECs and coronary VECs.29 However, this Cre labels many different cell types, including endocardial cells, angioblasts, and hematopoietic cells.92 In addition, SV endothelial cell dedifferentiation and subsequent redifferentiation into coronary artery VECs was challenged by the notion that a subset of SV endothelial cells already express the arterial marker Notch1, underlining the plasticity of the embryonic microvascular endothelial cells rather than a dedifferentiation/redifferentiation program.94, 95 SV endothelial cell heterogeneity was reinforced by a recent study showing that SV contains both apelin recepetor (APJ)-positive and -negative ECs.96 The immature APJ-negative subset constituted the population that emigrated from the sinus venosus, and under the influence of angiopoietin-1 differentiated into coronary vein endothelium. Thus this study proposed selective emigration and differentiation of an immature, APJ-negative SV EC population, rather than dedifferentiation/redifferentiation of mature venous endothelium.96
We developed a novel reagent, Apln-CreER, that selectively labels subepicardial endothelial precursors but not endocardial cells. Using this reagent, we demonstrated that subepicardial endothelial precursors form most coronary VECs, including most of the intramyocardial coronary arteries.97 This conclusion was functionally reinforced by a recent study that showed that GATA factors (Gata4 and 6) regulate coronary plexus development by recruiting endothelial cells to the sub-epicardium, and that loss of subepicardial vessels in hearts lacking epicardial GATA4 and GATA6 disrupts intramyocardial endothelial cell formation.39 A subset of subepicardial vessels remained in the subepicardial space and became coronary veins.
What is the lineage relationship between subepicardial endothelial cell precursors labeled by Apln-CreER and sinus venosus endothelial cells? We used in vitro explant assays to develop supportive data that subepicardial endothelial precursors derive from sinus venosus endothelial cells by vascular sprouting,97 as suggested by Red-Horse, Krasnow, and colleagues. However, as pointed out previously, direct, quantitative in vivo data linking subepicardial endothelial cells labeled by Apln-CreER to sinus venosus is currently lacking.98 A suitable SV-specific promoter driven Cre line is needed to more firmly establish this lineage relationship.
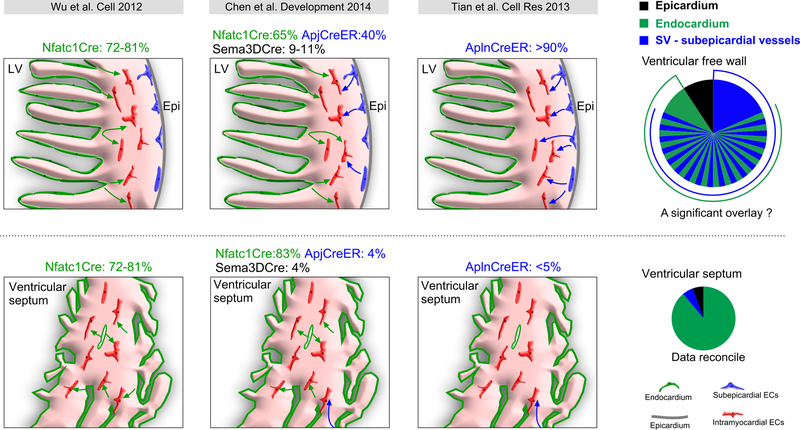
An unexpected result of the Tian et al. study was that Apln-CreER did not efficiently label coronary VECs within the ventricular septum, when it did efficiently label those in the ventricular free walls.97 This result indicates that coronary VECs in ventricular septum are not descendants from subepicardial endothelial cell precursors. Clonal analysis and direct lineage tracing of endocardial cells indicated that endocardial cells contribute to these septum vessels (Figure 4B,D,F).29, 99, 100 Blood islands budding from endocardium are found most abundantly at E12.5-E13.5 on the ventral surface of the developing heart, overlying the ventricular septum. This location coincides with a point where subepicardial vessels extending from the dorsal surface of the heart are least likely to reach by angiogenic sprouting. These blood islands have been suggested to be sites of endocardium-to-vessel formation,29, 97 and potentially this process may happen more frequently within the ventricular septum and lead to the formation of most coronary VECs within the ventricular septum. Thus the embryonic coronary vessels seem to be patterned in two distinctive compartments that irrigate the ventricular free wall and ventricular septum, respectively (Figure 4).
Taken together, recent mouse embryonic lineage tracing studies suggest that, instead of epicardium, SV is the origin for the majority of coronary vessels, including the intramyocardial coronary arteries. However, this conclusion is inferred from a combination of clonal lineage tracing and in vitro transplantation studies, and direct evidence or a quantitative estimate of the fraction of coronary VECs dervied by this mechanism, is currently lacking. It should be kept in mind that the SV is a transient structure named by its anatomic location, and it receives endothelial cells from other organs or tissues, including proepicardium43 and liver sinusoid VECs.90 SV is also partly covered by myocardium and the endothelium of sinsus venosus closely connected with and in continuity with atrium endocardium, so it may be technically challenging to dissect clearly the boundary between migrating endocardial cells of SV and atrium endocardium (discussed in the following section). Future studies are needed to find a more specific marker that could be used to label SV endocardium specifically and perform direct lineage tracing in vivo.
Currently, the molecular regulation of the coronary vessel formation by SV sprouting, dedifferentiation, and redifferentiation are incompletely understood. Recent studies identified VEGFC and angiopoietin-1 (Ang1) as key paracrine signals that promote SV sprouting. VEGFC, expressed from epicardium, regulates the formation and growth of coronary sprouts from sinus venosus.101 Similarly, cardiomyocyte-secreted Ang1 promoted the sprouting of APJ-negative ECs from SV.96 Interestingly, Ang1 stimulated the differentiation of immature endothelial sprouts into coronary vein endothelia, and cardiomyocyte-specific Ang1 deletion selectively impaired formation of subepicardial veins but not intramyocardial coronary arteries. These findings demonstrate that distinct signaling mechnisms promote formation of coronary arteries and coronary veins. Improved understanding of the process of SV sprouting and coronary artery and vein formation could significantly advance efforts in myocardial regeneration and revascularization.
Coronary VECs from endocardium
Endocardium is the specialized endothelium that lines the inner surfaces of the heart. During early heart development, endocardial cells overlying the cardiac cushions delaminate and form mesenchymal cells through epithelial to mesenchymal transition (EMT). These mesenchymal cells develop into the interstitial cells of the heart valves.22 This endocardial EMT is critical to cushion formation, and disruption of key molecular signaling impairs EMT and results in cardiac valve malformation.102–106 Since endocardium represents a large reservoir of endothelium distributed along the luminal surface of the ventricular myocardium, it would be reasonable to hypothesize that a similar delamination and migration process could yield coronary VECs.
Recently, Cre alleles based on Nfatc1 regulatory elements have permitted lineage tracing of endocardial cells. Wu et al. used constitutive Nfatc1-Cre to assess endocardial contributions to the coronary vasculature.99 This study was interpreted to show that endocardial cells overlying the ventricular trabeculae contribute to the majority of the intramyocardial coronary arteries of the embryonic ventricular wall. Sub-epicardial vessels, primarily coronary veins, were infrequently labeled by Nfatc1-Cre, which was interpreted to indicate that these arise from a different developmental mechanism (e.g., from SV endothelial cells). The Nfatc1-Cre lineage tracing data was supported by functional data obtained through manipulation of VEGFA signaling. Myocardium-specific VEGFA knockout or Nfatc1-Cre-driven endocardial VEGFR2 knockout showed significant reduction in intramyocardial coronary arteries but had less effect on subepicardial coronary veins.99 Based on these data, this study proposed a new model of coronary vascular development in which a subset of ventricular endocardial cells invade the myocardial wall from the luminal (endocardial) surface, migrate towards the outer (epicardial) surface, and differentiate into intramyocardial coronary artery endothelial cells (Figure 5 left panel). One important concept advanced by this model is that endocardial cells are not only an endothelial sheet that covers the ventricular myocardium; in addition, endocardium also contains progenitor cells that migrate into compact myocardium and trans-differentiate into coronary VECs.99 A second important possibility raised by the study is that coronary vessels may be developmentally heterogeneous, with intramyocardial coronary arteries formed primarily from endocardial sources and subepicardial veins originating from alternative precursor populations (Figure 5 left panel).99
Figure 5. Contribution of SV, endocardium and epicardium to coronary vessels in the developing hearts.
Comparison of quantitative measurements from the indicated references of the contribution of different sources to coronary VECs. Upper panels show the ventricular free walls, and the lower panels show the ventricular septum. Endocardium and SV-subepicardial endothelial progenitors were the two major sources, with endocardium making the predominant contribution to VECs in the ventricular septum. The relative contribution of SV/subepicardial endothelial progenitors and endocardium to coronary vessels in the ventricular free walls is currently a matter of debate. Resolution of this uncertainty requires new genetic tools that will distinguish endcoardium from SV/subepicardidal vessels.
The proposed model conflicts with results from other labs using different experimental approaches. First, retroviral analysis of chicken embryonic heart development showed that VECs have a different clonal origin than endocardial cells.27 Second, analysis of single cell labeling by VE-cadherin-CreERT2 showed that many VEC clones were related to SV endothelial cells, whereas only rare VEC clones were related to endocardial cells.29, 97 There is indeed a big gap between these studies, as Wu et al. found that the majority of coronary arteries (as high as 81%) originated from ventricular endocardium, but Red-Horse et al. found that very few coronary VECs were likely to arise by this mechanism.29 We showed that Apln-CreER initially labels sub-epicardial endothelial precursors, and that these give rise to over 90% of coronary VECs, including the intramyocardial subset.97, 100 These differences can perhaps be reconciled by reviewing the caveats of Cre-loxP lineage tracing, namely that interpretation hinges upon reliably excluding Cre activity in the target cell type or its other potential precursors. Wu et al. used a constitutive Nfatc1-Cre, which makes careful exclusion of Cre expression in all potential VEC sources other than endocardial cells difficult. Of particular interest is the SV, which is adjacent to atrial endocardium. Wu et al. present expression data suggesting that Nfatc1-Cre would not label SV, but this is difficult to exclude particularly using a constitutive Cre allele, and it remains possible that a portion of SV was labeled by Nfatc1-Cre (Figure 2A, B). Additionally, Nfatc1 was reported to be expressed in coronary VECs,107 which would prevent accurate assessment of ventricle endocardial contribution to coronary VECs.
Recently, Chen et al. re-investigated this controversial issue by comparing lineage tracing results from Nfatc1-Cre, Sema3d-Cre, and Apj-CreER, the latter a bacterial artificial chromosome transgenic line in which regulatory elements of Apj drive CreERT2.108 Apj is selectively expressed in sinus venosus but not epicardium nor endocardium. Nfatc1-Cre labeled most coronary vessels in ventricular septum (>80%) and a significant portion of coronary vessels in ventricle wall (eg. 65% in left ventricle), consistent with the results of Wu et al.99 Sema3d-Cre, expressed in epicardium, labeled a minority of coronary vessels (9–11%), as previously reported.43 Apj-CreER significantly labeled SV and contributed to a substantial fraction (40%) of coronary vessels in left and right ventricles, but very few (<5%) of the vessels within the ventricular septum (Figure 5 middle panel).108 Unfortunately, this study was unable to assess overlapping labeling between the Cre alleles and so was unable to resolve the relative contributions of endocardium versus SV to coronary VECs (Figure 5 middle panel). Nfatc1-Cre labeling of 65–81% of VECs108,99 may overestimate the endocardial contribution as a result of partial labeling of SV using constitutive Nfatc1-Cre or direct Cre activation in coronary VECs.107 The 40% coronary VECs labeled by Apj-CreER in the ventricle wall96 may under-estimate the contribution of SV to coronary vessels, due to incomplete Cre activation by tamoxifen or failure of the transgenic line to fully recapitulate endogenous APJ expression in SV. High labeling (>90%) by Apln-CreER in the ventricle wall may over-estimate the contribution of SV to coronary vessels97 (Figure 5 right panel), as Apln-CreER might label a subset of endocardium-derived VECs in addition to subepicardial VECs. Taken together, endocardium appears to be the main source of coronary VECs in the ventricular septum, and endocardium and SV are likely to be the main sources for coronary VECs in the embryonic ventricle wall. Current tools and analyses have been unable to definitively determine the relative contribution of each source to ventricular wall VECs, and resolution of this important question awaits development of improved lineage tracing tools and potentially the use of dual labeling strategies. At least some developing coronary vessels arise from endocardial progenitors, raising the tantalizing possibility that endocardium could be a source for coronary VECs during postnatal and adult heart growth, and could be recruited for therapeutic cardiac regeneration.
Postnatal coronary vessels: endocardium revisited
Until recently, most studies on the origins of the coronary vessels focused on the initial stages of embryonic coronary vascular development, based on the assumption that later angiogenic expansion of the coronary vascular tree occurred by outgrowth of these initial coronary vessels. Technical limitations, namely the lack of tools to selectively label either endocardial or coronary vascular endothelial cells at a chosen developmental stage, precluded rigorous testing of this assumption. We took advantage of the genetic regulatory elements of genes selectively expressed in endocardium but not VECs (Nfatc1) or in VECs but not endocardium (Apln), to achieve selective genetic labeling using the inducible Cre/loxP strategy.100 To achieve temporally regulated pulse labeling, tamoxifen-activated CreERT2 fusion protein109 was used instead of constitutive Cre. Combining these technologies yielded new genetic reagents to track the fate of endocardial or vascular endothelial cells labeled at a specific developmental stage.
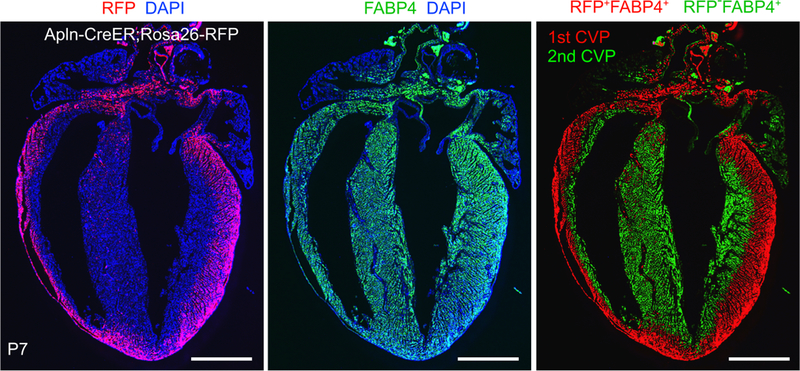
Apln-CreER, induced by a single dose of tamoxifen at the earliest stages of coronary VEC formation (E10.5), labeled nearly all coronary VECs at late gestation (E15.5) in the ventricular walls but did not label endocardium.97 Building on this observation, we tested the assumption that the adult coronary vascular tree forms by outgrowth of embryonic coronary vessels.100 In postnatal heart, most of the coronary VECs of the outer myocardial wall expressed the lineage mark made by Apln-CreER, indicating that these VECs originate from the fetal coronary tree (red staining, Figure 6, left panel). Surprisingly, a large portion of the coronary VECs in the inner myocardial wall of the postnatal heart did not express this lineage mark (green pseudo-color, Figure 6, right panel), suggesting that a substantial fraction of postnatal coronary VECs arise de novo, rather than by angiogenic expansion of pre-existing fetal coronary vessels. This result was independently verified by a different inducible Cre line (Fabp4-CreER), which like Apln-CreER selectively labeled fetal coronary vessels but not endocardium.110 To test the hypothesis that these de novo postnatal coronary VECs of the inner myocardial wall originated from endocardial progenitors, we used Nfatc1-CreERT2 (activated at E8.5) to track the fate of fetal endocardial cells in the postnatal heart. We found that endocardium formed coronary VECs de novo in the postnatal heart.100 Overall, these experiments show that coronary VECs continue to be formed de novo in the postnatal heart. Furthermore, the postnatal endocardium is not a static lining of the myocardium, but rather a dynamic source of these de novo formed postnatal coronary VECs.
Figure 6. 1st and 2nd coronary vascular population in neonatal heart.
Apln-CreER, induced at E10.5, labeled fetal coronary VECs with Cre-activated RFP expression. In the P7 postnatal heart, VECs were present throughout the myocardial wall, as demonstrated by immunostaining for the VEC-selective marker FABP4 (middle panel), but the RFP lineage tracer of fetal VECs was only observed in the outer myocardial wall (left panel). The FABP4+RFP+ VECs, descended from fetal VECs, are designated the 1st coronary vascular population (CVP; red pseudocolor, right panel) while the remaining FABP4+RFP− VECs, formed de novo in the postnatal heart, are designated the 2nd CVP (green pseudocolor, right panel). Bar = 1 mm.
How could a single layer of endocardium form such a large amount of vascular plexus at the core of a heart? Previous work in the embryonic heart suggested that endocardial cells invade the myocardium in response to high VEGFA expression in the ventricle wall, proliferate, and undergo angiogenic expansion to form the intramyocardial vascular plexus (Figure 5 left panel).99 Our recent study100 supported an alternative vessel formation model in the neonatal heart ventricular free walls. To accommodate the acute increase in wall stress that occurs due to the switch from fetal to postnatal circulations, trabecular myocardium rapidly coalesces with the inner portion of the compact myocardium, resulting in rapid expansion of compact myocardium. During this process of trabecular “compaction”, the endocardial cells on the surface of trabeculae become trapped within the compacting myocardium. In response to hypoxia and their new intramyocardial environment, the trapped endocardial progenitors rapidly form the vessels that supply the newly established compact myocardium of the inner myocardial wall.100 Similar trabecular compaction also occurs earlier in development during formation of the ventricular septum. Like the inner myocardial wall, the coronary VECs of the ventricular septum are also largely derived from endocardium (Figure 4B,D,F, VS). We describe the VECs that originate from the endocardium as the 2nd coronary vascular population, which supplies blood to the core of the heart (the inner myocardial wall and the ventricular septum). In this model, trabeculae are reservoirs of myocardium that are rapidly deployed to increase compact myocardial mass. Intrinsic to these myocardial reservoirs are endocardial progenitors which will expand to form the vasculature that will supply the newly formed myocardium. Defining the molecular mechanisms that regulate trabecular compaction and the conversion of endocardial cells to coronary VECs will be important areas for future study.111
This trapping and trans-differentiation model not only represents a new mechanism for vessel formation, but also provide critical information for the pathogenesis of congenital heart diseases such as non-compaction cardiomyopathy. The model may also provide insights on the development of coronary artery anomalies in pulmonary atresia with intact septum, where fistulae between the ventricular chamber and the coronary circulation are frequently observed. There is evidence of Thebesian vessels in developing heart: the vessels directly connect between coronary arteries and chambers of the heart.112 Interestingly, in the event of gradual closure of the orifices of the coronary arteries, the Thebesian vessels can supply the heart muscle with sufficient blood to enable it to maintain an efficient circulation.113 It will be important to understand the genetic, epigenetic, and signaling mechanisms that links coronary vessel formation with trabecular coalescence in normal development and in congenital heart disease. Unveiling the endogenous mechanisms that permit rapid development of a functional vascular supply from endocardial progenitors likewise has direct implications for both congenital heart diseases and therapeutic cardiac regenerative approaches to coronary artery disease.
Coronary orifice and truncal/stem origin
In some congenital heart diseases, the coronary stems may arise at abnormal sites, which could cause serious consequences from myocardial ischemia to sudden death.114, 115 In an extreme form, anomalous origin of the left coronary artery from the pulmonary artery (ALCAPA), infants present with myocardial ischemia, left ventricular dysfunction, mitral regurgitation,116 and the mortality rate is high if left untreated.117 More clear understanding of the developmental mechanisms that govern how coronary artery stems form and anastomose to the nascent coronary vascular plexus is a fascinating question in developmental biology with significant ramifications for congenital heart disease. Moreover, knowledge of the molecular pathways that guide coronary stem connection would also have implications for regenerative approaches to myocardial revascularization in the setting of coronary artery disease.
The nascent coronary plexus formed during early heart development initially develops in the absence of effective perfusion. At about E13.5-E14.5 of murine gestation, the nascent plexus connects to the aorta and subsequently provides nutrients and oxygen to the heart. It was once believed that coronary arteries grow out of the aorta by sprouting angiogenesis.89, 118 Elegant studies on avian embryos questioned this long-held belief and provided a reverse model in which the coronary stem and orifice forms by coalescence and remodeling of a pre-existing peritruncal coronary plexus. By staining of endothelial cells in quail embryos or ink injection of chick embryos, several groups reached the novel conclusion that the proximal coronary arteries did not grow outward from the aorta as commonly believed. Instead peritruncal capillaries encircle the outflow tract and grow into the aorta.119–121 This impression was reinforced by quail-to-chicken tissue transplantation assays, in which quail rather than chick endothelial cells formed the endothelial cells of the coronary stems.90 Mouse genetic lineage tracing studies verified this in-growth model.122 In spite of these advances, the origin remain largely undefined. Recent lineage tracing studies using Apj-CreER and Nfatc1-CreER identified at least two different sources of these peritruncal coronary VECs: SV and endocardium.123 The possibility of angiogenic sprouting from the aorta has also been re-visited,123 although direct evidence by specific lineage tracing of aortic endothelium is needed to provide more definitive evidence.
The signals that guide peritruncal capillaries to penetrate the aorta and form stable coronary stems are beginning to be revealed. At least three different cell types have been implicated. Since EPDCs act as a source of secreted mitogens and exhibit proangiogenic properties,46, 49, 124 EPDCs were among the first cell lineage investigated. Studies in chicken embryos showed that invasion of peritruncal vessels into the aorta and the formation of coronary orifices and coronary artery stems were spatially and temporally closely associated with apoptosis in the surrounding myocardium.125 When proepicardial outgrowth was delayed, apoptosis in the outflow tract diminished. Further studies revealed that EPDCs produce Fas ligand, which induces apoptosis at sites of coronary ingrowth. These EPDCs not only influence myocardial development and vascularization of compact myocardium, but also induce apoptosis in the peritruncal region during the time window when peritruncal coronary vessels connect to the aorta.126, 127 Indeed, delay of proepicardial outgrowth malfunction caused defective myocardial vascularization and absent coronary orifices. Although EPDCs contribute to few coronary VECs, the majority of coronary smooth muscle cells arise from EPDCs.14, 69, 128 The presence of EPDCs in the orifice region may provide possible cues that guide ingrowth and subsequent persistence of the proximal coronary artery.
Preotic neural crest cells are a second lineage that influences coronary stem and orifice formation. These cells preferentially distribute in the conotruncal region and differentiate into coronary smooth muscle cells. Ablation of preotic neural crest or disruption of endothelial signalling caused abnormal coronary orifice formation.129
A third cell lineage that regulates coronary stem and orifice formation was recently discovered.123 With the aorta, patches of cardiomyocytes were identified at stem sites. These intra-aortic cardiomyocytes were critical for normal formation of the coronary stems and orifices. Interestingly, in heart malformations involving outflow tract rotation defects, abnormal coronary stem implantation correlated with misplacement of these intra-aortic cardiomyocytes.
Understanding the regional signaling that governs the establishment, penetration, maintenance and growth of these two coronary artery stems would shed new light on the molecular defects that cause congenitally misplaced coronary artery stems, and may also aid in guiding or generating coronary arteries in therapies for coronary artery diseases.
Remaining challenges and future directions
Recent work has significantly advanced our understanding of the cellular origins and developmental program that regulates development of the coronary vessels. Nevertheless, major questions remain that need to be answered. What is the relative contribution of SV and ventricular endocardium to the free wall coronary vasculature, particularly the intramyocardial coronary arteries? What signals regulate SV endothelial cell outgrowth and lineage conversion to arterial VECs? What signals stimulate endocardial cells to transdifferentiate into VECs in the fetal interventricular septum or in the postnatal inner myocardial wall? What is the role of endocardium-myocardium signaling in coronary vessel formation? Does formation of vessels during coalescence of trabecular myocardium itself initiate or facilitate the compaction process? Does endothelial or endocardial plasticity exhibited in fetal life extent postnatally, during physiological growth or pathophysiological conditions? Can they be re-awakened by therapeutic stimulation? Answering these questions require new genetic tools as well as alternative methods and experiments that do not hinge on current Cre-loxP systems. Ultimately, a more complete understanding of the developmental program of coronary arteries will provide new opportunities for therapeutic cardiac revascularization and regeneration.
Acknowledgments
Funding Sources
B.Z. and X.T. was supported by National Natural Science Foundation of China (91339104, 31271552, 31222038, 31301188), National Basic Research Program of China (2013CB945302 and 2012CB945102), Shanghai Basic Research Key Project (14JC1407400), Sanofi-SIBS fellowship, SIBS Postdoc Fund 2013KIP311, China Postdoc Fund 2013M541561. W.T.P. was supported by NIH (2 R01 HL094683) and an American Heart Association Established Investigator Award.
Non-standard Abbreviations and Acronyms
- CA
coronary artery
- EC
endothelial cell
- VEC
vascular endothelial cell
- SV
sinus venosus
- PE
proepicardium
- EMT
epithelial to mesenchymal transition
- EPDC
epicardium-derived cell
- MI
myocardial infarction
Footnotes
Disclosures
None
In November, 2014, the average time from submission to first decision for all original research papers submitted to Circulation Research was 13.96 days.
References
- 1.Wu SM, Chien KR, Mummery C. Origins and fates of cardiovascular progenitor cells. Cell 2008;132:537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansson EM, Lindsay ME, Chien KR. Regeneration next: toward heart stem cell therapeutics. Cell Stem Cell 2009;5:364–377. [DOI] [PubMed] [Google Scholar]
- 3.Martin-Puig S, Wang Z, Chien KR. Lives of a heart cell: tracing the origins of cardiac progenitors. Cell Stem Cell 2008;2:320–331. [DOI] [PubMed] [Google Scholar]
- 4.Zhou B, Lin Z, Pu W. Mammalian Myocardial Regeneration. Muscle 2012;First Edition, Chapter 39:555–569. [Google Scholar]
- 5.Passier R, van Laake LW, Mummery CL. Stem-cell-based therapy and lessons from the heart. Nature 2008;453:322–329. [DOI] [PubMed] [Google Scholar]
- 6.Laflamme MA, Murry CE. Regenerating the heart. Nat Biotechnol 2005;23:845–856. [DOI] [PubMed] [Google Scholar]
- 7.Potluri R, Baig M, Mavi JS, Ali N, Aziz A, Uppal H, Chandran S. The role of angioplasty in patients with acute coronary syndrome and previous coronary artery bypass grafting. Int J Cardiol 2014 [DOI] [PubMed]
- 8.Zangi L, Lui KO, von Gise A, Ma Q, Ebina W, Ptaszek LM, Spater D, Xu H, Tabebordbar M, Gorbatov R, Sena B, Nahrendorf M, Briscoe DM, Li RA, Wagers AJ, Rossi DJ, Pu WT, Chien KR. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat Biotechnol 2013 [DOI] [PMC free article] [PubMed]
- 9.Carmeliet P. Angiogenesis in health and disease. Nat Med 2003;9:653–660. [DOI] [PubMed] [Google Scholar]
- 10.Lin Z, Pu WT. Strategies for Cardiac Regeneration and Repair. Sci Transl Med 2014;6(239):239rv1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dettman RW, Denetclaw WJ, Ordahl CP, Bristow J. Common epicardial origin of coronary vascular smooth muscle, perivascular fibroblasts, and intermyocardial fibroblasts in the avian heart. Dev Biol 1998;193:169–181. [DOI] [PubMed] [Google Scholar]
- 12.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs 2001;169:89–103. [DOI] [PubMed] [Google Scholar]
- 13.Gittenberger-de Groot AC, Vrancken Peeters MP, Mentink MM, Gourdie RG, Poelmann RE. Epicardium-derived cells contribute a novel population to the myocardial wall and the atrioventricular cushions. Circ Res 1998;82:1043–1052. [DOI] [PubMed] [Google Scholar]
- 14.Wilm B, Ipenberg A, Hastie ND, Burch JB, Bader DM. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development 2005;132:5317–5328. [DOI] [PubMed] [Google Scholar]
- 15.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A 1988;85:5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer B, McDermott J. DNA recombination with a heterospecific Cre homolog identified from comparison of the pac-c1 regions of P1-related phages. Nucleic Acids Res 2004;32:6086–6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma Q, Zhou B, Pu WT. Reassessment of Isl1 and Nkx2–5 cardiac fate maps using a Gata4-based reporter of Cre activity. Dev Biol 2008;323:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B, Pu WT. Genetic Cre-loxP Assessment of Epicardial Cell Fate Using Wt1-Driven Cre Alleles. Circ Res 2012;111:e276–e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudat C, Kispert A. Wt1 and Epicardial Fate Mapping. Circ Res 2012 [DOI] [PubMed]
- 21.Ishii Y, Garriock RJ, Navetta AM, Coughlin LE, Mikawa T. BMP signals promote proepicardial protrusion necessary for recruitment of coronary vessel and epicardial progenitors to the heart. Dev Cell 2010;19:307–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 2012;110:1628–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lie-Venema H, van den Akker NM, Bax NA, Winter EM, Maas S, Kekarainen T, Hoeben RC, deRuiter MC, Poelmann RE, Gittenberger-de Groot AC. Origin, fate, and function of epicardium-derived cells (EPDCs) in normal and abnormal cardiac development. ScientificWorldJournal 2007;7:1777–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Tuyn J, Atsma DE, Winter EM, van der Velde-van Dijke I, Pijnappels DA, Bax NA, Knaan-Shanzer S, Gittenberger-de Groot AC, Poelmann RE, van der Laarse A, van der Wall EE, Schalij MJ, de Vries AA. Epicardial cells of human adults can undergo an epithelial-to-mesenchymal transition and obtain characteristics of smooth muscle cells in vitro. Stem Cells 2007;25:271–278. [DOI] [PubMed] [Google Scholar]
- 25.Majesky MW. Development of coronary vessels. Curr Top Dev Biol 2004;62:225–259. [DOI] [PubMed] [Google Scholar]
- 26.K L, D O The epicardial signaling center in development and disease. Heart Development and Regeneration 2010
- 27.Mikawa T, Fischman DA. Retroviral analysis of cardiac morphogenesis: discontinuous formation of coronary vessels. Proc Natl Acad Sci U S A 1992;89:9504–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs). Dev Biol 2002;247:307–326. [DOI] [PubMed] [Google Scholar]
- 29.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature 2010;464:549–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature 2011;479:189–193. [DOI] [PubMed] [Google Scholar]
- 31.Rios AC, Fu NY, Lindeman GJ, Visvader JE. In situ identification of bipotent stem cells in the mammary gland. Nature 2014;506:322–327. [DOI] [PubMed] [Google Scholar]
- 32.Nahirney PC, Mikawa T, Fischman DA. Evidence for an extracellular matrix bridge guiding proepicardial cell migration to the myocardium of chick embryos. Dev Dyn 2003;227:511–523. [DOI] [PubMed] [Google Scholar]
- 33.Kattan J, Dettman RW, Bristow J. Formation and remodeling of the coronary vascular bed in the embryonic avian heart. Dev Dyn 2004;230:34–43. [DOI] [PubMed] [Google Scholar]
- 34.Vrancken Peeters MP, Gittenberger-de Groot AC, Mentink MM, Hungerford JE, Little CD, Poelmann RE. The development of the coronary vessels and their differentiation into arteries and veins in the embryonic quail heart. Dev Dyn 1997;208:338–348. [DOI] [PubMed] [Google Scholar]
- 35.Cossette S, Misra R. The identification of different endothelial cell populations within the mouse proepicardium. Dev Dyn 2011;240:2344–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Del Monte G, Casanova JC, Guadix JA, Macgrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL. Differential Notch Signaling in the Epicardium Is Required for Cardiac Inflow Development and Coronary Vessel Morphogenesis. Circ Res 2011 [DOI] [PubMed]
- 37.Wessels A, van den Hoff MJ, Adamo RF, Phelps AL, Lockhart MM, Sauls K, Briggs LE, Norris RA, van Wijk B, Perez-Pomares JM, Dettman RW, Burch JB. Epicardially derived fibroblasts preferentially contribute to the parietal leaflets of the atrioventricular valves in the murine heart. Dev Biol 2012;366:111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Norden J, Grieskamp T, Lausch E, van Wijk B, van den Hoff MJ, Englert C, Petry M, Mommersteeg MT, Christoffels VM, Niederreither K, Kispert A. Wt1 and Retinoic Acid Signaling in the Subcoelomic Mesenchyme Control the Development of the Pleuropericardial Membranes and the Sinus Horns. Circ Res 2010 [DOI] [PMC free article] [PubMed]
- 39.Kolander KD, Holtz ML, Cossette SM, Duncan SA, Misra RP. Epicardial GATA factors regulate early coronary vascular plexus formation. Dev Biol 2014;386:204–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid X receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A 2005;102:18455–18460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai CL, Martin JC, Sun Y, Cui L, Wang L, Ouyang K, Yang L, Bu L, Liang X, Zhang X, Stallcup WB, Denton CP, McCulloch A, Chen J, Evans SM. A myocardial lineage derives from Tbx18 epicardial cells. Nature 2008;454:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acharya A, Baek ST, Banfi S, Eskiocak B, Tallquist MD. Efficient inducible Cre-mediated recombination in Tcf21 cell lineages in the heart and kidney. Genesis 2011;49:870–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katz T, Singh M, Degenhardt K, Rivera-Feliciano J, Johnson R, Epstein J, Tabin C. Distinct Compartments of the Proepicardial Organ Give Rise to Coronary Vascular Endothelial Cells. Developmental Cell 2012;22:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 2008;454:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner KD, Wagner N, Bondke A, Nafz B, Flemming B, Theres H, Scholz H. The Wilms’ tumor suppressor Wt1 is expressed in the coronary vasculature after myocardial infarction. FASEB J 2002;16:1117–1119. [DOI] [PubMed] [Google Scholar]
- 46.Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J Clin Invest 2011;121:1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Wijk B, Gunst QD, Moorman AF, van den Hoff MJ. Cardiac regeneration from activated epicardium. PLoS One 2012;7:e44692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res 2010;106:818–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sucov HM, Gu Y, Thomas S, Li P, Pashmforoush M. Epicardial Control of Myocardial Proliferation and Morphogenesis. Pediatr Cardiol 2009 [DOI] [PubMed]
- 50.Morabito CJ, Kattan J, Bristow J. Mechanisms of embryonic coronary artery development. Curr Opin Cardiol 2002;17:235–241. [DOI] [PubMed] [Google Scholar]
- 51.Yang JT, Rayburn H, Hynes RO. Cell adhesion events mediated by alpha 4 integrins are essential in placental and cardiac development. Development 1995;121:549–560. [DOI] [PubMed] [Google Scholar]
- 52.Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development 1995;121:489–503. [DOI] [PubMed] [Google Scholar]
- 53.Wu H, Lee SH, Gao J, Liu X, Iruela-Arispe ML. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development 1999;126:3597–3605. [DOI] [PubMed] [Google Scholar]
- 54.Li WE, Waldo K, Linask KL, Chen T, Wessels A, Parmacek MS, Kirby ML, Lo CW. An essential role for connexin43 gap junctions in mouse coronary artery development. Development 2002;129:2031–2042. [DOI] [PubMed] [Google Scholar]
- 55.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. YAC complementation shows a requirement for Wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development 1999;126:1845–1857. [DOI] [PubMed] [Google Scholar]
- 56.von Gise A, Zhou B, Honor LB, Ma Q, Petryk A, Pu WT. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev Biol 2011;356:421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner N, Wagner KD, Theres H, Englert C, Schedl A, Scholz H. Coronary vessel development requires activation of the TrkB neurotrophin receptor by the Wilms’ tumor transcription factor Wt1. Genes Dev 2005;19:2631–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu SP, Dong XR, Regan JN, Su C, Majesky MW. Tbx18 Regulates Development of the Epicardium and Coronary Vessels. Dev Biol 2013 [DOI] [PMC free article] [PubMed]
- 59.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell 2005;8:85–95. [DOI] [PubMed] [Google Scholar]
- 60.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev 2006;20:1651–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomanek RJ, Ratajska A, Kitten GT, Yue X, Sandra A. Vascular endothelial growth factor expression coincides with coronary vasculogenesis and angiogenesis. Dev Dyn 1999;215:54–61. [DOI] [PubMed] [Google Scholar]
- 62.Tomanek RJ, Ishii Y, Holifield JS, Sjogren CL, Hansen HK, Mikawa T. VEGF family members regulate myocardial tubulogenesis and coronary artery formation in the embryo. Circ Res 2006;98:947–953. [DOI] [PubMed] [Google Scholar]
- 63.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol 2001;234:204–215. [DOI] [PubMed] [Google Scholar]
- 64.Lavine KJ, Kovacs A, Ornitz DM. Hedgehog signaling is critical for maintenance of the adult coronary vasculature in mice. J Clin Invest 2008;118:2404–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lavine KJ, Ornitz DM. Shared circuitry: developmental signaling cascades regulate both embryonic and adult coronary vasculature. Circ Res 2009;104:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavine KJ, Long F, Choi K, Smith C, Ornitz DM. Hedgehog signaling to distinct cell types differentially regulates coronary artery and vein development. Development 2008;135:3161–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smart N, Risebro CA, Melville AA, Moses K, Schwartz RJ, Chien KR, Riley PR. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature 2007;445:177–182. [DOI] [PubMed] [Google Scholar]
- 68.Masters M, Riley PR. The epicardium signals the way towards heart regeneration. Stem Cell Res 2014 [DOI] [PMC free article] [PubMed]
- 69.Smart N, Dube KN, Riley PR. Epicardial progenitor cells in cardiac regeneration and neovascularisation. Vascul Pharmacol 2013;58:164–173. [DOI] [PubMed] [Google Scholar]
- 70.Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004;432:466–472. [DOI] [PubMed] [Google Scholar]
- 71.Bock-Marquette I, Shrivastava S, Pipes GC, Thatcher JE, Blystone A, Shelton JM, Galindo CL, Melegh B, Srivastava D, Olson EN, DiMaio JM. Thymosin beta4 mediated PKC activation is essential to initiate the embryonic coronary developmental program and epicardial progenitor cell activation in adult mice in vivo. J Mol Cell Cardiol 2009;46:728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011;474:640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou B, Honor LB, Ma Q, Oh JH, Lin RZ, Melero-Martin JM, von Gise A, Zhou P, Hu T, He L, Wu KH, Zhang H, Zhang Y, Pu WT. Thymosin beta 4 treatment after myocardial infarction does not reprogram epicardial cells into cardiomyocytes. J Mol Cell Cardiol 2011;52:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smart N, Risebro CA, Clark JE, Ehler E, Miquerol L, Rossdeutsch A, Marber MS, Riley PR. Thymosin beta4 facilitates epicardial neovascularization of the injured adult heart. Ann N Y Acad Sci 2010;1194:97–104. [DOI] [PubMed] [Google Scholar]
- 75.Banerjee I, Zhang J, Moore-Morris T, Lange S, Shen T, Dalton ND, Gu Y, Peterson KL, Evans SM, Chen J. Thymosin Beta 4 is dispensable for murine cardiac development and function. Circ Res 2012;110:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Banerjee I, Moore Morris T, Evans SM, Chen J. Thymosin beta4 is not required for embryonic viability or vascular development. Circ Res 2013;112:e25–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 1997;11:1048–1060. [DOI] [PubMed] [Google Scholar]
- 78.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 1997;11:1061–1072. [DOI] [PubMed] [Google Scholar]
- 79.Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol 2004;275:235–244. [DOI] [PubMed] [Google Scholar]
- 80.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest 2009;119:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Svensson EC, Huggins GS, Lin H, Clendenin C, Jiang F, Tufts R, Dardik FB, Leiden JM. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog-2. Nat Genet 2000;25:353–356. [DOI] [PubMed] [Google Scholar]
- 82.Tevosian SG, Deconinck AE, Tanaka M, Schinke M, Litovsky SH, Izumo S, Fujiwara Y, Orkin SH. FOG-2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell 2000;101:729–739. [DOI] [PubMed] [Google Scholar]
- 83.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev 2001;15:839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou B, Ma Q, Kong SW, Hu Y, Campbell PH, McGowan FX, Ackerman KG, Wu B, Zhou B, Tevosian SG, Pu WT. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J Clin Invest 2009;119:1462–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Heineke J, Auger-Messier M, Xu J, Oka T, Sargent MA, York A, Klevitsky R, Vaikunth S, Duncan SA, Aronow BJ, Robbins J, Crombleholme TM, Molkentin JD. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest 2007;117:3198–3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tomanek RJ. Formation of the coronary vasculature during development. Angiogenesis 2005;8:273–284. [DOI] [PubMed] [Google Scholar]
- 87.Vrancken Peeters MP, Gittenberger-de Groot AC, Mentink MM, Hungerford JE, Little CD, Poelmann RE. Differences in development of coronary arteries and veins. Cardiovasc Res 1997;36:101–110. [DOI] [PubMed] [Google Scholar]
- 88.Lewis FT. The question of sinusoids. Ant Anz 1904;25:261–279. [Google Scholar]
- 89.Bennett HS. The development of the blood supply to the heart in the embryo pig. Am J Anat 1936;60:27–53 [Google Scholar]
- 90.Poelmann RE, Gittenberger-de Groot AC, Mentink MM, Bokenkamp R, Hogers B. Development of the cardiac coronary vascular endothelium, studied with antiendothelial antibodies, in chicken-quail chimeras. Circ Res 1993;73:559–568. [DOI] [PubMed] [Google Scholar]
- 91.Riley P. Developmental biology: Plumbing the heart. Nature 2010;464:498–499. [DOI] [PubMed] [Google Scholar]
- 92.Perez-Pomares JM, de la Pompa JL. Signaling during epicardium and coronary vessel development. Circ Res 2011;109:1429–1442. [DOI] [PubMed] [Google Scholar]
- 93.Monvoisin A, Alva JA, Hofmann JJ, Zovein AC, Lane TF, Iruela-Arispe ML. VE-cadherin-CreERT2 transgenic mouse: a model for inducible recombination in the endothelium. Dev Dyn 2006;235:3413–3422. [DOI] [PubMed] [Google Scholar]
- 94.Lie-Venema H, Eralp I, Markwald RR, van den Akker NM, Wijffels MC, Kolditz DP, van der Laarse A, Schalij MJ, Poelmann RE, Bogers AJ, Gittenberger-de Groot AC. Periostin expression by epicardium-derived cells is involved in the development of the atrioventricular valves and fibrous heart skeleton. Differentiation 2008;76:809–819. [DOI] [PubMed] [Google Scholar]
- 95.van den Akker NM, Caolo V, Molin DG. Cellular decisions in cardiac outflow tract and coronary development: an act by VEGF and NOTCH. Differentiation 2012;84:62–78. [DOI] [PubMed] [Google Scholar]
- 96.Arita Y, Nakaoka Y, Matsunaga T, Kidoya H, Yamamizu K, Arima Y, Kataoka-Hashimoto T, Ikeoka K, Yasui T, Masaki T, Yamamoto K, Higuchi K, Park JS, Shirai M, Nishiyama K, Yamagishi H, Otsu K, Kurihara H, Minami T, Yamauchi-Takihara K, Koh GY, Mochizuki N, Takakura N, Sakata Y, Yamashita JK, Komuro I. Myocardium-derived angiopoietin-1 is essential for coronary vein formation in the developing heart. Nat Commun 2014;5:4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Zhang Z, Zhong TP, Yang X, Yang Z, Yan Y, Baldini A, Sun Y, Lu J, Schwartz RJ, Evans SM, Gittenberger-de Groot AC, Red-Horse K, Zhou B. Subepicardial endothelial cells invade the embryonic ventricle wall to form coronary arteries. Cell Res 2013;23:1075–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang B, Pu WT. The mysterious origins of coronary vessels. Cell Res 2013;23:1063–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu B, Zhang Z, Lui W, Chen X, Wang Y, Chamberlain AA, Moreno-Rodriguez R, Markwald R, O?Rourke B, Sharp D, Zheng D, Lenz, Baldwin HS, Chang C-P, Zhou B. Endocardial Cells Form the Coronary Arteries by Angiogenesis through Myocardial-Endocardial VEGF Signaling. Cell 2012;151:1083–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tian X, Hu T, Zhang H, He L, Huang X, Liu Q, Yu W, He L, Yang Z, Yan Y, Yang X, Zhong TP, Pu WT, Zhou B. De novo formation of a distinct coronary vascular population in neonatal heart. Science 2014;345:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen HI, Sharma B, Akerberg BN, Numi HJ, Kivela R, Saharinen P, Aghajanian H, McKay AS, Bogard PE, Chang AH, Jacobs AH, Epstein JA, Stankunas K, Alitalo K, Red-Horse K. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development 2014 [DOI] [PMC free article] [PubMed]
- 102.Camenisch TD, Schroeder JA, Bradley J, Klewer SE, McDonald JA. Heart-valve mesenchyme formation is dependent on hyaluronan-augmented activation of ErbB2-ErbB3 receptors. Nat Med 2002;8:850–855. [DOI] [PubMed] [Google Scholar]
- 103.Hurlstone AF, Haramis AP, Wienholds E, Begthel H, Korving J, Van Eeden F, Cuppen E, Zivkovic D, Plasterk RH, Clevers H. The Wnt/beta-catenin pathway regulates cardiac valve formation. Nature 2003;425:633–637. [DOI] [PubMed] [Google Scholar]
- 104.High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet 2008;9:49–61. [DOI] [PubMed] [Google Scholar]
- 105.Zhang H, von Gise A, Liu Q, Hu T, Tian X, He L, Pu W, Huang X, He L, Cai CL, Camargo FD, Pu WT, Zhou B. Yap1 Is Required for Endothelial to Mesenchymal Transition of the Atrioventricular Cushion. J Biol Chem 2014;289:18681–18692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Combs MD, Braitsch CM, Lange AW, James JF, Yutzey KE. NFATC1 promotes epicardium-derived cell invasion into myocardium. Development 2011;138:1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chen HI, Sharma B, Akerberg B. The sinus venosus contributes to coronary vasculature through VEGF-C stimulated angiogenesis. Development 2014;in press [DOI] [PMC free article] [PubMed]
- 109.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun 1997;237:752–757. [DOI] [PubMed] [Google Scholar]
- 110.He L, Tian X, Zhang H, Wythe JD, Zhou B. Fabp4-CreER lineage tracing revealstwo distinctive coronary vascular populations. J Cell Mol Med 2014 [DOI] [PMC free article] [PubMed]
- 111.Burns CG, Burns CE. A crowning achievement for deciphering coronary origins. Science 2014;345:28–29. [DOI] [PubMed] [Google Scholar]
- 112.Thebesius AC. Dissertatio medica de circulo sanguinis in corde. Lugduni Batavorum 1708
- 113.Wearn JT. THE ROLE OF THE THEBESIAN VESSELS IN THE CIRCULATION OF THE HEART. J Exp Med 1928;47:293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Angelini P. Coronary artery anomalies: an entity in search of an identity. Circulation 2007;115:1296–1305. [DOI] [PubMed] [Google Scholar]
- 115.Zeina AR, Blinder J, Sharif D, Rosenschein U, Barmeir E. Congenital coronary artery anomalies in adults: non-invasive assessment with multidetector CT. Br J Radiol 2009;82:254–261. [DOI] [PubMed] [Google Scholar]
- 116.Ginde S, Earing MG, Bartz PJ, Cava JR, Tweddell JS. Late complications after Takeuchi repair of anomalous left coronary artery from the pulmonary artery: case series and review of literature. Pediatr Cardiol 2012;33:1115–1123. [DOI] [PubMed] [Google Scholar]
- 117.Arciniegas E, Farooki ZQ, Hakimi M, Green EW. Management of anomalous left coronary artery from the pulmonary artery. Circulation 1980;62:I180–9. [PubMed] [Google Scholar]
- 118.Hutchins GM, Kessler-Hanna A, Moore GW. Development of the coronary arteries in the embryonic human heart. Circulation 1988;77:1250–1257. [DOI] [PubMed] [Google Scholar]
- 119.Bogers AJ, Gittenberger-de Groot AC, Poelmann RE, Peault BM, Huysmans HA. Development of the origin of the coronary arteries, a matter of ingrowth or outgrowth? Anat Embryol (Berl) 1989;180:437–441. [DOI] [PubMed] [Google Scholar]
- 120.Waldo KL, Willner W, Kirby ML. Origin of the proximal coronary artery stems and a review of ventricular vascularization in the chick embryo. Am J Anat 1990;188:109–120. [DOI] [PubMed] [Google Scholar]
- 121.Ando K, Nakajima Y, Yamagishi T, Yamamoto S, Nakamura H. Development of proximal coronary arteries in quail embryonic heart: multiple capillaries penetrating the aortic sinus fuse to form main coronary trunk. Circ Res 2004;94:346–352. [DOI] [PubMed] [Google Scholar]
- 122.Tian X, Hu T, He L, Zhang H, Huang X, Poelmann RE, Liu W, Yang Z, Yan Y, Pu WT, Zhou B. Peritruncal Coronary Endothelial Cells Contribute to Proximal Coronary Artery Stems and Their Aortic Orifices in the Mouse Heart. PLoS ONE 2013;8:e80857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen HI, Poduri A, Numi H, Kivela R, Saharinen P, McKay AS, Raftrey B, Churko J, Tian X, Zhou B, Wu JC, Alitalo K, Red-Horse K. VEGF-C and aortic cardiomyocytes guide coronary artery stem development. J Clin Invest 2014 [DOI] [PMC free article] [PubMed]
- 124.Chen TH, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev Biol 2002;250:198–207. [DOI] [PubMed] [Google Scholar]
- 125.Velkey JM, Bernanke DH. Apoptosis during coronary artery orifice development in the chick embryo. Anat Rec 2001;262:310–317. [DOI] [PubMed] [Google Scholar]
- 126.Rothenberg F, Hitomi M, Fisher SA, Watanabe M. Initiation of apoptosis in the developing avian outflow tract myocardium. Dev Dyn 2002;223:469–482. [DOI] [PubMed] [Google Scholar]
- 127.Eralp I, Lie-Venema H, DeRuiter MC, van den Akker NM, Bogers AJ, Mentink MM, Poelmann RE, Gittenberger-de Groot AC. Coronary artery and orifice development is associated with proper timing of epicardial outgrowth and correlated Fas-ligand-associated apoptosis patterns. Circ Res 2005;96:526–534. [DOI] [PubMed] [Google Scholar]
- 128.Munoz-Chapuli R, Gonzalez-Iriarte M, Carmona R, Atencia G, Macias D, Perez-Pomares JM. Cellular precursors of the coronary arteries. Tex Heart Inst J 2002;29:243–249. [PMC free article] [PubMed] [Google Scholar]
- 129.Arima Y, Miyagawa-Tomita S, Maeda K, Asai R, Seya D, Minoux M, Rijli FM, Nishiyama K, Kim KS, Uchijima Y, Ogawa H, Kurihara Y, Kurihara H. Preotic neural crest cells contribute to coronary artery smooth muscle involving endothelin signalling. Nat Commun 2012;3:1267. [DOI] [PubMed] [Google Scholar]