Abstract
D-Apiose is a C-branched pentose sugar important for plant cell wall development. Its biosynthesis as UDP-D-apiose involves decarboxylation of the UDP-D-glucuronic acid precursor coupled to pyranosyl-to-furanosyl sugar ring contraction. This unusual multistep reaction is catalyzed within a single active site by UDP-D-apiose/UDP-D-xylose synthase (UAXS). Here, we decipher the UAXS catalytic mechanism based on crystal structures of the enzyme from Arabidopsis thaliana, molecular dynamics simulations expanded by QM/MM calculations, and mutational-mechanistic analyses. Our studies show how UAXS uniquely integrates a classical catalytic cycle of oxidation and reduction by a tightly bound nicotinamide coenzyme with retro-aldol/aldol chemistry for the sugar ring contraction. They further demonstrate that decarboxylation occurs only after the sugar ring opening and identify the thiol group of Cys100 in steering the sugar skeleton rearrangement by proton transfer to and from the C3’. The mechanistic features of UAXS highlight the evolutionary expansion of the basic catalytic apparatus of short-chain dehydrogenases/reductases for functional versatility in sugar biosynthesis.
Keywords: Enzyme catalysis, glycobiology, short-chain dehydrogenases/reductases (SDR), aldol cleavage, oxidative decarboxylation
Introduction
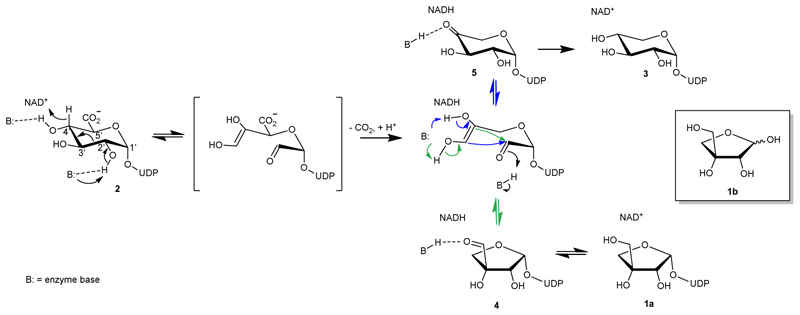
D-Apiose (3-C-(hydroxymethyl)-D-glycero-tetrose; 1b in Figure 1) is a C-branched pentofuranose widely distributed in plants1–3 and recently also found in bacteria.4 In plants, apiose is present in pectin polysaccharides and certain glycosylated natural products.1 By forming borate ester cross-links between polysaccharide chains, apiose residues are essential for cell wall development and stability.1,3,5,6 The degradation of apiose-containing polysaccharides by human-gut microorganisms7 and the biological routes of catabolism of the apiose monosaccharide3 have recently attracted considerable attention. Chemical de novo synthesis of apiose was developed.8 Naturally, however, apiose is synthesized as UDP-apiose (1a) from UDP-D-glucuronic acid (UDP-GlcA; 2) by UDP-apiose/UDP-xylose synthases (UAXS; Figure 1).2,4,6,9,10 The UAXS reaction is unusually challenging to be performed by a single enzyme. It involves decarboxylation coupled to sugar ring contraction to generate a five-membered ring furanosyl product.11–13 The enzyme also produces UDP-xylose, a six-membered ring UDP-xylopyranose (3), in consistent ratios, typically about 1:1 at neutral pH.6,13 A plausible reaction path has been proposed based on mechanistic studies spanning several decades, as shown in Figure 1.6,9–19 The relative timing of the catalytic steps has been recently elucidated from kinetic isotope effects on the enzymatic rates as well as on the distribution of UDP-apiose (1a) and UDP-xylose (3).13
Fig. 1. Proposed mechanism of UAXS.
A full representation of the steps of substrate oxidation, aldol cleavage for ring opening, and decarboxylation is given in Supplementary Figure 1. The green and blue arrows indicate the aldol reactions leading to the ring-contracted furanosyl product and to the pyranosyl product, respectively.
UDP-GlcA is first oxidized at C4' by the tightly bound NAD+. Subsequently, deprotonation of the 2'-OH facilitates a C2'-C3' aldol cleavage.11–13 Decarboxylation thus occurs on the ring-opened oxidized substrate.13 Rearrangement of the carbon skeleton yields either UDP-apiose aldehyde (4) or UDP-4-keto-xylose (5) whose reduction by NADH gives the respective furanosyl or pyranosyl products. However, despite these extensive studies, it remains obscure how the UAXS can incorporate such a complex mechanism and orchestrate the involved multiple catalytic steps in a single active site.
The mechanistic complexity of UAXS becomes even more fascinating when viewed from an evolutionary perspective. The enzyme is classified as member of the sugar nucleotide epimerase/dehydratase subclass of the short-chain dehydrogenase/reductase superfamily (SDR).20–22 Indeed, UAXS incorporates the distinctive SDR sequence signature, a highly conserved Thr/Tyr/Lys motif, for catalytic oxidoreduction by NAD+/NADH.22,23 The catalytic roles of these residues are well described from studies of other SDRs and involve Tyr as Brønsted acid/base for reduction/oxidation.23,24 Functional SDR homologs of UAXS (Supplementary Figure 2), such as UDP-xylose synthase (UXS)24–26 and UDP-GlcA dehydrogenase/decarboxylase (ArnA)27,28, catalyze decarboxylation of UDP-GlcA (2) and form UDP-xylose (3) (UXS) or UDP-4-keto-xylose (5) (ArnA).28 However, neither UXS nor ArnA promote the rearrangements leading to sugar ring contraction28, which is an activity specific of UAXS. Contrary to UAXS (Figure 1), UXS and ArnA catalyze oxidative decarboxylation of UDP-GlcA (2) without opening of the sugar ring. Core problem for the mechanistic inquiry thus becomes to understand how UAXS integrates retro-aldol/aldol rearrangement of the sugar-ring carbon skeleton with the established11–13 SDR catalytic cycle of substrate oxidation, decarboxylation, and reduction.
Here, we report crystal structures of the ternary complexes of Arabidopsis thaliana AtUAXS6 (in the following text named UAXS for convenience) with NAD+ and UDP (3.0 Å resolution), and NADH and UDP-GlcA (2) (3.5 Å resolution), respectively. Using the structural evidence of the enzyme in combination with Molecular Dynamics (MD) simulations and hybrid Quantum Mechanics/Molecular Mechanics (QM/MM) calculations, we identify the key catalytic elements of UAXS. Mutational analysis of the active site interactions substantiates their proposed roles in catalysis. In particular, we find that pyranosyl ring distortion in the UDP-GlcA (2) substrate, away from the preferred 4C1 chair conformation in solution to a relatively flexible skew-boat (1S3, 1S5) or boat (1,4B) conformation in the Michaelis complex, serves to align precisely the reactive groups of the substrate (4’-OH, C4’-H, 2’-OH) with the catalytic groups of the enzyme. Comparisons of UAXS with human UXS and ArnA reveal the remarkable catalytic subtlety involved in each enzyme to achieve conversion of UDP-GlcA (2) according to the corresponding individual reaction path (Figure 1, Supplementary Figure 1). We show that UAXS uses only minimal extensions to the SDR core catalytic machinery to establish its unique catalytic functionality.
Results
The overall structure of UAXS features a highly conserved active site architecture
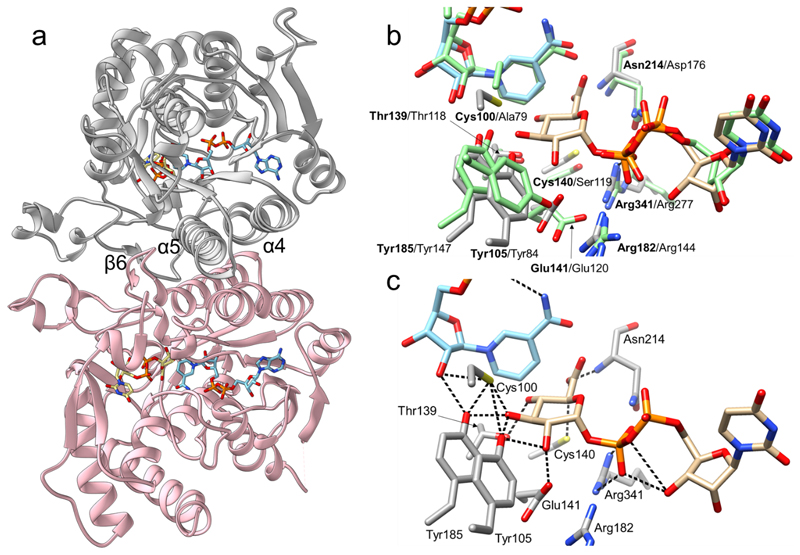
We first determined the crystal structure of wild-type UAXS in complex with the NAD+ coenzyme and UDP (3.0 Å resolution; Supplementary Table 1, Supplementary Figures 3–8). The structure consists of a single polypeptide chain (residues 8-382), with a surface loop that could not be traced during modelling (residues 61-68). The protein is composed of 10 α-helices and 12 β-strands (Figure 2a). The asymmetric unit contains two protein subunits arranged into a homodimer, as reported for other UAXS enzymes in solution.29 Like other dimeric SDRs 20–22, the inter-subunit interface is stabilized by hydrophobic interactions30 between a pair of two-fold related α-helices, forming a four-helical bundle, and two elongated loops (Figure 2a). The UAXS monomer is built of two domains: a large NAD+-binding domain (residues 8-215, 263-292, 343-358), which is a modified version of the classical Rossmann fold, and a considerably smaller UDP-GlcA-binding domain (residues 216-262, 293-342, 359-382). This two-domain folding topology is typical of SDRs and has high structural similarity with its closest functional homologs, namely human UXS and ArnA.25,27,28 Compared to human UXS (28% sequence identity; overall RMSD of 1.6 Å)25, UAXS exhibits several elongated loops (residues 59-67, 147-165, 224-234, 310-321 and 326-336) and contains an additional helix (α3, residues 80-90) which is replaced by a loop in human UXS. Noteworthy, an UAXS loop (residues 102-109) makes a larger turn around the active site than does the equivalent loop of human UXS (residues 81-88), as shown in Supplementary Figure 9.
Fig. 2. Crystallographic analysis of the three-dimensional structure of UAXS from Arabidopsis thaliana.
(a) The UAXS subunit features the typical SDR fold. The enzyme is a functional dimer with chain A shown in grey and chain B shown in pink. The dimer interface of UAXS is composed of helices α4 (residues 109-129) and α5 (residues 181-202). There are additional dimer contacts from strand β6 (residues 170-179) and from an extended loop. NAD+ is represented in cyan carbons and UDP-GlcA (2) in beige carbons. Oxygen atoms are in red, nitrogen atoms in blue, phosphorus atoms in orange, and sulfur atoms in yellow. (b) Structural comparison/overlay of the active sites of UAXS and human UXS in complex with UDP and NAD+ (PDB entry 2B69; light green carbon atoms). Most of the residues are conserved and have similar conformations. The main changes are replacements of Cys100 and Cys140 of UAXS with Ala79 and Ser119 in human UXS (Supplementary Figure 2). (c) A model for the UAXS Michaelis complex (enzyme:NAD+:UDP-GlcA) was built by combining the ligands from two different structures; the wild-type enzyme bound to NAD+ and UDP (Supplementary Figure 7), and the C100A mutant bound to NADH and UDP-GlcA (Supplementary Figure 8). Enzyme residues are displayed in light grey carbon atoms, UDP-GlcA is represented with beige carbon atoms, NAD+ with cyan carbons. Cys100 was modelled in a rotameric conformation suitable to interact with 3’-OH of the sugar substrate. Possible H-bond interactions between the labelled residues and the ligands are shown as black dashed lines.
The active site of UAXS is located in a cavity formed between the NAD+- and UDP-GlcA-binding domains and is made up by seven highly conserved residues (Figure 2a). Thr139, Tyr185 and Lys189 (note: the lysine is not shown in Figure 2 but it is shown later in Figure 3) form the characteristic signature of SDR catalytic center20–22, whereas Tyr105, Glu141, Arg182 and Arg341 are distinct features of the UAXS/UXS/ArnA group of enzymes. As in human UXS24–26,28 and ArnA27,30, interaction between Glu141 and Arg341 fastens together two long loops (residues 128-173 and 332-345), thus closing the active site (Supplementary Figure 10). Sequence changes that distinguish the UAXS active site from the active sites of human UXS (Figure 2b) and ArnA (Supplementary Figure 11) are Cys100 and Cys140, which are Ala or Ser in the two other enzymes.25,30 This sequence conservation is matched by highly similar ligand-binding modes, as revealed by the comparison of our structure of wild-type UAXS in complex with NAD+ and UDP with the structures of human UXS28 (Figure 2b) and substrate-bound ArnA30 (Supplementary Figure 11). Specifically, NAD+ is buried deeply in the protein (Figure 2a), consistent with the observation that the coenzyme is tightly bound by UAXS. Indeed, biochemical data show that UAXS retains one coenzyme/protein monomer throughout purification. Interestingly, between 30 and 80% of the bound coenzyme in UAXS was isolated as NADH (Supplementary Figure 12). However, incubation of the enzyme in the presence of NAD+ results in the complete exchange for the oxidized form of the coenzyme (Supplementary Figure 13). Binding of coenzyme partly in the reduced state is likely due to recombinant (over)production. Other SDR enzymes, which like UAXS contain tightly bound NAD+ or NADP+ (e.g., epimerases, dehydratases, decarboxylases), show similar effect, with the portion of reduced coenzyme varying strongly dependent on the enzyme and the conditions used.31,32
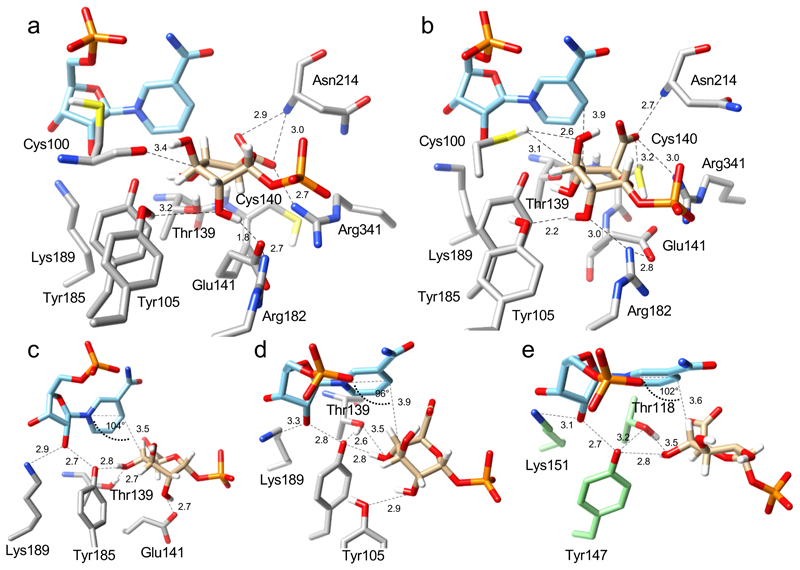
Fig. 3. Positioning of the UDP-GlcA substrate for catalytic oxidation and ring opening at the active site of UAXS, revealed by QM/MM calculations.
The QM/MM calculations were performed on selected, highly representative snapshots from the MD simulations. (a) Close-up structure and key interactions for the proposed Michaelis complex; the ternary NAD+:UDP-GlcA:enzyme complex is from Replica 1 (Supplementary Video 1). The pyranosyl ring of GlcA adopts a 1S5 conformation. (b) Close-up structure and key interactions for an alternative ternary complex, prominently present in MD simulations from Replica 2 (Supplementary Video 2). The pyranosyl ring of GlcA adopts a 1C4 conformation. (c), (d) Further close ups, taken from panels a and b, respectively, to highlight the catalytic interactions for oxidation of the C4’ alcohol group, via hydride transfer to NAD+ and proton transfer to Tyr185, and sugar ring opening, via deprotonation of the 2’-hydroxy group. Note: the orientations from panels a and b were adapted for better viewing. (e) Modeled Michaelis complex of human UXS with bound UDP-GlcA (2) and NAD+.25
Structural basis of catalysis by UAXS
Co-crystallization or soaking of wild-type UAXS with UDP-GlcA (2) proved unfruitful. Electron density in the area supposedly occupied by the substrate's α-glucuronyl moiety showed discontinuity from the density of UDP probably due to a phosphate from the crystallization solution. However, using a lower activity UAXS variant (C100A; Table 1) and co-crystallization with NADH, we obtained a structure with well-defined electron density for the complete UDP-GlcA (2) molecule bound to the enzyme (Supplementary Figure 8). In this structure, the nicotinamide ring could not be traced although clear electron density was found for the remaining part of the bound coenzyme. The Ala100 is in a position that could as well accommodate the Cys100 of wild-type UAXS (Figure 2c and Supplementary Figure 8). Residues surrounding NAD+ and UDP-GlcA (2) in the C100A variant structure have the same conformation as in the wild-type UAXS (Supplementary Figure 14). We therefore combined the two structures to obtain a model of the NAD+:UDP-GlcA:enzyme ternary complex (Figure 2c). We validated the resulting enzyme complex through comparison with ArnA for which a complex structure with UDP-GlcA (2) has been reported (PDB entry 1Z7E).30 ArnA and UAXS structures share 37% residue identity and have RMSD of atomic positions of 1.2 Å. Most importantly, the UDP-GlcA (2) accommodates almost identically in the two structures, as shown in Supplementary Figure 11.
Table 1. Activity and selectivity of wild-type UAXS and variant enzymes for conversion of UDP-GlcA (2).
| Enzyme | kcat[a] [min-1] | UDP-apiose (1a) [%] |
UDP-xylose (3) [%] |
UDP-4-keto-xylose[b] (5) [%] | Apiose cyclic 1,2-phosphate (1) [%] |
|---|---|---|---|---|---|
| Wild-type | 0.49 | 59 | 26 | 8 | 7 |
| Y185F | 0 | n.a. | n.a. | n.a. | n.a. |
| Y105F | 0.05 | 18 | 41 | 32 | 9 |
| Y105A | 0.13 | 0 | 50 | 50 | 0 |
| T139V | 3 × 10-3 | 0 | 0 | 100 | 0 |
| C100A | 0.12 | 18 | 61 | 15 | 6 |
| C100S | 0.09 | 0 | 21 | 79 | 0 |
| C140S | 0.07 | 29 | 26 | 35 | 10 |
| C140A | 0.11 | 15 | 33 | 52 | 0 |
| C100A/C140S | 0.23 | 27 | 32 | 33 | 8 |
| E141A | 3 × 10-3 | 0 | 0 | 100[b] | 0 |
Activity (kcat) was measured by HPLC-UV as depletion of substrate in H2O at pH 7.0 (Supplementary Figures 17-32). Selectivity for product formation was determined by 1H-NMR in D2O (pD 7.0) (Supplementary Figure 33-35). Reaction conditions: 50 mM potassium phosphate buffer; 30 °C; 2 mM substrate; 0.1 mM NAD+ (1.0 mM for T139V and E141A variants). An appropriate enzyme concentration between 0.5 and 14.3 mg/mL was used. Note: there was no solvent isotope effect on the kcat (see Supplementary Figure 36). n.a. = not applicable. Data are from multiple determinations have relative S.D. ≤ 10% (except Y105F, 20%; see Supplementary Figures 17-32).
22 mg/mL enzyme added for 1H-NMR measurement.
Substrate positioning for catalysis in the Michaelis complex of UAXS
With the support of the crystallographic data, we performed molecular dynamics (MD) simulations with GLYCAM0633 combined with hybrid QM/MM calculations to further examine the positioning of the UDP-GlcA (2) substrate in the UAXS active site. Of note, MD simulations have aided previously the identification of a likely substrate positioning for catalysis in human UXS.25 600-ns MD simulations in two replicas of the NAD+:UDP-GlcA:enzyme complex were initially carried out with Tyr185 in protonated or unprotonated form. The pKa of the homologous tyrosine in related SDRs (epimerase, dehydratase)32,34 is ~6.5, making it necessary to consider both protonation states of Tyr185. Only with unprotonated Tyr185, we identified conformations that were plausibly catalytic, in (a) showing proper arrangement of substrate and NAD+ for hydride transfer and (b) having the conserved SDR catalytic dyad positioned for the required deprotonation of the 4’-OH (Tyr185, Thr139; Fig. 3). A representative snapshot from each MD trajectory was further optimized using hybrid QM/MM calculations at the DFT (density functional theory) level of theory (Figure 3a-d). Thorough analysis of the MD trajectories for the Tyr185-deprotonated enzyme revealed considerable flexibility in the active-site interactions of the bound UDP-GlcA (2) (SupplementaryVideos 1, 2). This flexibility arises due to seemingly coordinated, atomic motions of the active-site residues, the NAD+, and the substrate. It involves substantial rearrangements in the sugar ring conformation, as shown in detailed maps of the puckering geometries adopted along the full MD trajectories (Supplementary Figure 15).
In Replica 1, the pyranosyl ring of UDP-GlcA is distorted from the preferred 4C1 chair conformation in solution to skew-boat (1S3, 1S5) or boat (1,4B) conformations. The ring-flipped 1C4 conformation is present to a minor extent (Supplementary Figure 15a). The nicotinamide C4 is above the substrate C4’, with distance (3.5 Å) and angle (104°; relative to the nicotinamide ring C4-N1 axis) well set for hydride transfer (Figure 3a,c; Supplementary Video 1). The 4’-OH is oriented by hydrogen bonds with Tyr185 and Thr139. The polarity of these hydrogen bonds indicates proton abstraction to the phenolate of Tyr185. Catalytic function of the tyrosine-threonine dyad in alcohol oxidation is analogous to other SDR enzymes,22,23,25,32,34 as visible from the comparison with the proposed Michaelis complex of human UXS that was obtained from a previous study (Figure 3e).25 The substrate 2’-OH is hydrogen-bonded to Glu141 whereas the 5’-carboxylate is oriented equatorially and interacts with Arg341 and Asn214 (main chain NH). Furthermore, the C5’ of UDP-GlcA (2) is close to Cys140, which is a good candidate to play a role in the obligatory proton uptake to the C5’ during decarboxylation (Supplementary Figure 1). Indeed, an alternative snapshot from Replica 1 has the Cys140 side chain oriented toward the C5’ (Supplementary Figure 16a).
The other prominent enzyme complex obtained from the MD simulations (Replica 2) features the pyranosyl ring of UDP-GlcA (2) in a 1C4chair conformation (Supplementary Figure 15b) with the carboxylate oriented axially. The sugar remains reasonably positioned for hydride transfer (C4’-C4 distance of 3.9 Å; angle of 96° relative to the nicotinamide ring C4-N1 axis) and the 4’-OH retains the characteristic hydrogen bond with the Thr139-Tyr185 dyad (Figure 3b,d and Supplementary Video 2). The substrate sugar-protein interactions are further enriched by hydrogen bonds between the 2’-OH and Tyr105 (Figure 3d) and the direct contacts of the C3’ and C5’ atoms with the side chains of Cys100 and Cys140, respectively (Supplementary Figure 16b). Moreover, in a representative portion of the simulated structures (~30%), a water molecule coordinated by Glu141 is placed close to the C5’ and, therefore, may establish a proton conduit (Supplementary Figure 16c).
Collectively, the analysis of the MD trajectories and the QM/MM optimized models highlight several features that are relevant for catalysis (Figure 1 and Supplementary Figure 1). The pyranosyl ring of UDP-GlcA can adopt multiple conformations that deviate from the 4C1 chair conformation and yet retain acceptable geometry for hydride transfer to the NAD+. The carboxylate at C5’ can be equatorially (Replica 1) or axially (Replica 2) oriented. Change in orientation from axial to equatorial can be catalytically relevant by modulating the reactivity of the C5’ carboxylate for decarboxylation. Glu141 and Tyr105 are the hydrogen bond partners for the 2’OH of the substrate and may thereby be involved in ring opening. Cys140 is consistently found in proximity to the C5’ and, possibly together with nearby waters and side chains, is positioned well to protonate the C5’ in the decarboxylation step. Cys100 adopts flexible conformations with its side chain pointing out of (Figure 3a) and into the active site (Figure 3b). In the “in” conformation, its thiol group points towards the sugar C3’ atom, in a position suited for subsequent proton transfer to and from the C3’.
Probing the catalytic roles by mutagenesis
Residues suggested from the structural and computational studies to have a role in enzymatic catalysis were first probed by site-directed mutagenesis (Table 1). Our strategy was to target residues that are (i) part of the conserved SDR catalytic core (Tyr185-Thr139 dyad)20–22, (ii) characteristic of UAXS/UXS/ArnA sub-group of enzymes (Tyr105, Glu141)25,30 and (iii) specifically present only in the UAXS active site (Cys100, Cys140) (Figure 2b,c). The C100A-C140S variant was made with the aim of creating a UXS/ArnA-like active site in UAXS (Figure 2, Supplementary Figure 2, 11). Activities of purified enzymes were initially measured with an HPLC-UV-based substrate depletion assay as summarized in Table 1 (Supplementary Figures 17-32). The Y185F protein was inactive, in agreement with a role in acid/base catalysis of Tyr185, as generally observed in the SDR enzymes.20–23 The T139V variant retained a very low level of activity, consistent with an auxiliary25 yet highly relevant function for the original threonine (Supplementary Figures 22, 25). The E141A variant also showed very low activity (Supplementary Figure 32), in agreement with the suggestion that Glu141 has a role in forming the reactive UAXS-substrate complex and stabilizing the active-site conformation (Figure 3a,b; Supplementary Figure 10). All other variants (C100A, C100S, Y105F, Y105A, C140A, C140S, C100A/C140S) exhibited 2- to 9-fold lower activities than the wild-type enzyme (Table 1).
The active variants were subjected to NMR experiments to determine the distribution of UDP-apiose (1a), UDP-xylose (3) and UDP-4-keto-xylose (5) that can be generated by UAXS upon conversion of UDP-GlcA (2) (Table 1 and Figure 1).19 Spontaneous decomposition of UDP-apiose (1a) gives the cyclic 1,2-apiosyl-phosphate (1, Supplementary Figure 1), which we also measured (Supplementary Figure 33).13,19 In addition, NMR was employed for detection of reaction intermediates accumulated prior to or after decarboxylation (Supplementary Figure 34, 35).19 Wild-type UAXS was found to have clear preference for UDP-apiose (1a) which is produced in 2.5-fold excess over UDP-xylose (3) with minimal leakage of intermediates (8% of UDP-4-keto-xylose, 5). This reactivity pattern was found to be altered in all studied variants, which were indiscriminate (C140S) regarding formation of UDP-apiose (1a) and UDP-xylose (3) or exhibited “inverted” (C100A, Y105F, C140A, C100A/C140S) preference for the xylose over the apiose product (Table 1, Supplementary Figure 34, 35). Importantly, the C100S and Y105A variants produced no UDP-apiose (1a) and are mechanistically very insightful because their reactions may no longer involve ring opening. Moreover, all variants featured an enhanced accumulation of the UDP-4-keto-xylose (5) intermediate, which was the only detectable product for the minimally active T139V and E141A variants. Accumulation of the UDP-apiose aldehyde (4) intermediate was not detected in any variant. Thus, the trend common to all the variants was a reduced or abolished capacity to produce a ring-contracted apiose product.
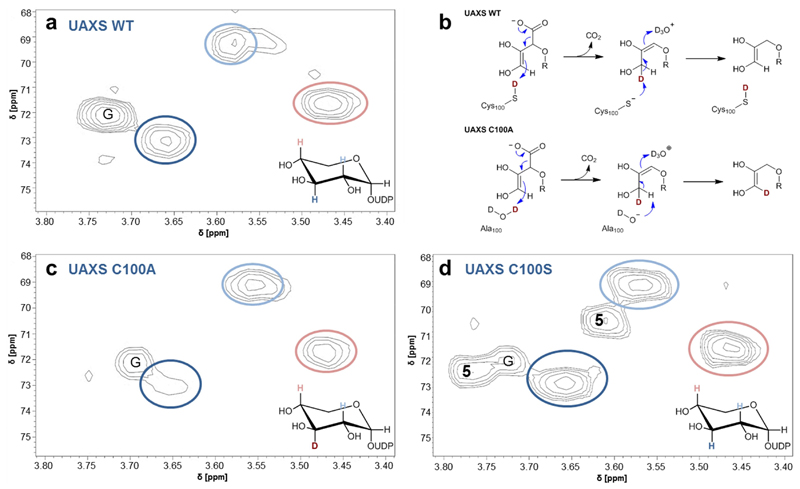
Incorporation of solvent deuterium into UDP-xylose C3’
The mechanistic proposal for UAXS (Supplementary Figure 1) entails a proton transfer to and from the C3’ of the ring-opened reaction intermediates prior to and after the decarboxylation, respectively. From its position in the enzyme structure and the outcome of the mutagenesis experiments, Cys100 was the prime candidate residue to catalyze these proton transfers (Figure 2c, 3a,b). To assess the role of Cys100, we analyzed the C100A and C100S variants along with wild-type UAXS and measured incorporation of solvent deuterium into the C3’ of UDP-xylose (3) released during conversion of UDP-GlcA (2) by these enzymes (Figure 4, Supplementary Figure 37, 38). It was known from a previous study13, and we confirmed here for different pH conditions, that wild-type UAXS does not promote deuterium incorporation at C3’ from the solvent (Figure 4a). However, as shown in Figure 4b, we considered that a C100A variant might promote deuterium incorporation in the case that water substituted for the supposed proton transfer function of the original thiol side chain of Cys100. The UDP-xylose (3) formed by the C100A variant indeed showed H/D exchange at the C3’ (Figure 4c). The degree of deuterium labeling was large, with only traces of proton remaining at the C3’ (Figure 4c). A role of Cys100 in proton transfer to the C3’ (Figure 4b) was strongly supported from these results. The C100S variant, by contrast, did not incorporate solvent deuterium into the C3’ of UDP-xylose (3) and UDP-4-keto-xylose (5), as shown in Figure 4d. This finding, together with the evidence that ring-contracted apiosyl product cannot be formed by the C100S variant (Table 1), suggests that this variant proceeds in oxidative decarboxylation like human UXS, without ring opening. The pKa (~13) of serine is much larger than that of cysteine (~8.5) and it does not allow Ser100 to catalyze proton transfer analogously as the Cys100.
Fig. 4. Deuterium incorporation at C3’ during conversion of UDP-GlcA (2) by wild-type and C100A and C100S forms of UAXS.
Panels (a), (b) and (d) show results of hetero-nuclear single quantum coherence (HSQC) experiments, analyzing the UDP-xylose (3) isolated from reactions in D2O (pD 7.0) of wild-type UAXS (a), C100A variant (c) and C100S variant (d). Relevant hydrogens at carbons 2’, 3’ and 4’ are highlighted in color and are indicated in the spectra with correspondingly colored circles. G = signal derived from traces of glycerol. 5 = signals derived from UDP-4-keto-xylose (C3-H at ∼3.78 ppm; C2-H at ∼3.61 ppm). Panel (b) shows protonation and deprotonation events during the decarboxylation, leading to deuterium incorporation at C3’ in the reaction of the C100A variant but not in the reaction of the wild-type enzyme. Note: in all enzymatic reactions, the C5’ in UDP-xylose (3) is partly deuterated (50%) from solvent, as expected from the proposed mechanism (panel b).
UAXS reaction uncoupled from the decarboxylation
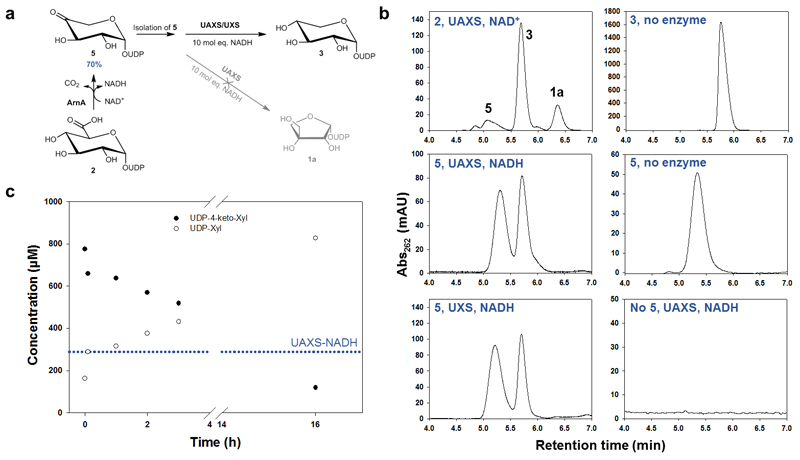
The steps post decarboxylation, leading to rearrangement of the five-carbon skeleton to yield UDP-4-keto-xylose (5) and UDP-apiose aldehyde (4), have so far eluded detailed characterization. Several studies6,12,35 including our own (data not shown) indicate that UDP-xylose (3) is not detectably converted into UDP-apiose (1a). The UDP-apiose (1a) is too unstable for isolation in relevant/required amounts and purity36, prohibiting product interconversion in the other direction (1a → 3). [Note: we considered purification of UDP-apiose (1a) in the presence of organic amines (e.g., trimethylamine) that appear to be somewhat stabilizing.36 However, our attempt failed after the last step of removing the amine by size-exclusion chromatography. The isolated UDP-apiose (1a) rapidly decomposed in water.] Liu and co-workers12 demonstrated that a chemically stable phosphonate analogue of UDP-apiose (1a) obtains the corresponding UDP-xylose analogue enzymatically. The actual turnover product is cyclic xylose 1,2-phosphonate (and UMP), which however renders the overall reaction irreversible. Using the C-terminal decarboxylase fragment of ArnA (Supplementary Figure 39)27,28, we established a new protocol for the synthesis of UDP-4-keto-xylose (5) in excellent yields (≥ 90%) as the sole product of UDP-GlcA (2) conversion in the presence of NAD+ (Figure 5a, Supplementary Figure 40, 41). By means of a highly sensitive (herein developed) HPLC-UV assay, we could thereby show that UDP-4-keto-xylose is not a substrate for ring rearrangement by UAXS (Figure 5b,c, Supplementary Figures 42-45). No UDP-apiose (1a) was formed from UDP-4-keto-xylose (5). Rather, UDP-4-keto-xylose is gradually converted into UDP-xylose (3) in the presence of externally added NADH (Figure 5b). The reduction of UDP-4-keto-xylose (5) by both wild-type UAXS and C100S variant features a kinetic behavior similar to that seen for the same reaction catalyzed by human UXS (Figure 5c, Supplementary Figure 45). The reaction time course characteristically involves an initial “burst” of UDP-xylose (3) release, with the product amount corresponding to the molar equivalent of enzyme-NADH present in the reaction (Figure 5c, Supplementary Figure 44). The burst rate (~1 – 3 min-1) is consistent with the kcat for the full enzymatic reaction starting from UDP-GlcA (2) (Table 1).
Fig. 5. Reaction of UAXS with UDP-4-keto-xylose (5) to study ring opening and reduction by the enzyme.
Panel (a) shows the different steps of the experiment. The UDP-4-keto-xylose (5) was synthesized from UDP-GlcA (2) using the enzyme ArnA. The isolated compound 5 was then subjected to conversion by UAXS in the presence of excess of NADH. As a reference, the reaction was also performed with UXS. Panel (b) shows HPLC traces from the conversions of UDP-4-keto-xylose (5) with wild-type UAXS and human UXS as well as the most relevant controls. Panel (c) shows a time course for the reduction of UDP-4-keto-xylose (5) by wild-type UAXS. The blue dotted line indicates the concentration of enzyme-bound NADH in UAXS (here ∼60% of 412 μM enzyme).
Requirement for coenzyme exchange (NAD+ → NADH) to continue the reduction of UDP-4-keto-xylose (5) explains the appearance of a second, slower phase in the reaction time course (Figure 5c). In addition, we found that reduction of UDP-4-keto-xylose (5) in D2O does not involve deuterium incorporation at C3’ (Supplementary Figure 46). These results demonstrate that UDP-4-keto-xylose (5) is a substrate for reduction, but not for aldol/retro-aldol rearrangement, by UAXS (Figure 1). Therefore, formation of UDP-4-keto-xylose (5) is a quasi-irreversible step of the enzymatic reaction path, providing strong support to the mechanistic notion of a sugar ring opening that precedes the decarboxylation.13
Discussion
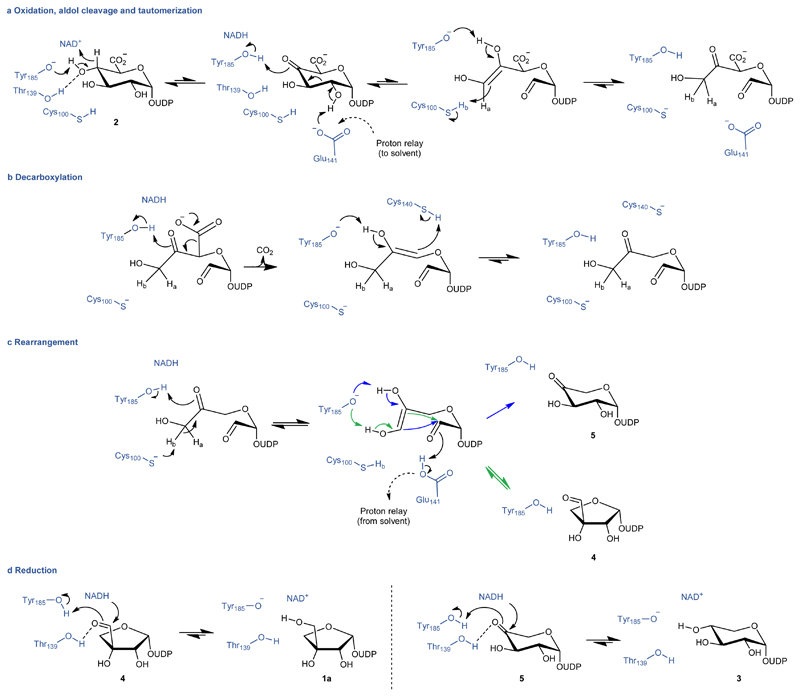
Based on the evidences presented, we propose a detailed catalytic mechanism for the enzymatic reaction of UAXS (Figure 6). We assign functions to the main active-site residues in each step of the catalytic conversion. The proposed mechanism involves a largely conserved exploitation of the enzyme's SDR catalytic machinery for NAD-dependent oxidoreduction (Figure 6a,d).20–23,25 The enzyme affords catalysis of retro-aldol/aldol rearrangement for sugar ring opening/contraction11–13 thanks to a strategic residue substitution to introduce two cysteines into the active site (Figure 2, Figure 3a-d). Following an initial “activation” of the substrate by oxidation at C4’, facilitated by Tyr185 as the catalytic base (Figure 6a), the characteristic steps of ring opening and decarboxylation would be promoted by concerted general acid-general base catalysis. The sugar ring puckering (1S3, 1,4B, 1S5,) observed in the Replica 1 represents a plausible model of the reactant complex. In this conformation, the carboxylate at C5’ is equatorially oriented to delay decarboxylation that characteristically follows aldol cleavage in UAXS. Thus, there seems to be chemical rationale for UAXS, in contrast to UXS, to favor an equatorial orientation of the carboxylate at C5’ (Figure 3a,c,e).
Fig. 6. Proposed enzymatic mechanism of UAXS.
In steps a – c, the proton transferred by Cys100 to and from the C3' is indicated as Hb. In step c, the ring closure to give UDP-4-keto-xylose (5) is not detectably reversible. In step d, the reductions are indicated to be reversible in principle. However, oxidation of UDP-xylose (3) by UAXS was not detectable.11,12
The C2'-C3' aldol cleavage requires the catalytic deprotonation of the C2'-OH group (Figure 6a).11–13 MD simulation and QM/MM calculations reveal that Glu141 and Tyr105 are well positioned to fulfil that role. Specifically, Replica 1 structure suggests that Glu141 is directly responsible for this catalytic step whereas Tyr105 might form a conduit for proton release and/or modulate the pKa of Glu141 (Figure 3a and Supplementary Video 1). Mutagenesis data support the proposed function of these residues as the E141A variant shows only trace activity and is completely devoid of UDP-apiose formation whereas the Y105F enzyme is impaired in the ability to form UDP-apiose (1a) (Table 1). Precise structural positioning for proper functional interplay between the catalytic residues promoting C4’ alcohol oxidation and ensuing aldol cleavage appears to be essential for the UAXS type of reactivity. This is illustrated by the Y105A variant, which retains ~27% of wild-type activity for the conversion of UDP-GlcA (2) but shows complete disruption of the UDP-apiose (1a) formation (Table 1). The effect likely arises from the loss of aromatic stacking of Tyr105 which seems to help the positioning of Tyr185 in the wild-type enzyme (Figure 2b,c) to enable reaction via ring opening.
In the next catalytic step, protonation of the C3’ stabilizes the ring-opened oxidized substrate by preventing re-closure and promotes its decarboxylation (Figure 6a-b). Our results identify Cys100 as the critical determinant for this step of the UAXS reaction. The C100S variant is unable to produce any apiose product and our isotope labelling experiments suggest that it reacts through the UXS path without ring opening. The apiose-producing activity of the C100A variant necessitates that proton uptake to the C3’ occurs from the solvent. The keto-enol tautomerization steps before and after the decarboxylation involve reversible proton transfers to and from the C3’ (Figure 6a-c), which can lead to the observed deuterium incorporation at C3’ in the reaction of the C100A variant. In the wild-type enzyme, where Cys100 is present, the proton transfers appear to be selective in that only the previously transferred hydrogen (or deuterium) is re-abstracted from the C3' and no isotope exchange is observed. Finally, the distinct use made by UAXS of the highly conserved Glu141 in catalysis to aldol cleavage emphasizes the requirement for an alternative residue (Cys140) managing the proton transfer to the C5’ during decarboxylation. Mechanistic explanation is thus provided for why UAXS has uniquely adopted the readily de-protonatable cysteine at position 140 where UXS and ArnA have a serine.
The space vacated by the release of the carboxylic group gives room to the rearrangement of the ring open structure leading to closure and formation of the five-membered apiose ring (Figure 6c). Evidence that UDP-4-keto-xylose (5) is not a substrate for retro-aldol/aldol reaction by the enzyme suggests that ring closure leading to the formation of UDP-4-keto-xylose (5) is a largely spontaneous reaction. Rather than catalytic facilitation, therefore, this step would seem to primarily require selectivity control from the enzyme, to promote rearrangement of the five-carbon skeleton into the UDP-apiose aldehyde (4). The prime candidate residue for steering the ring closure is Tyr185 as this residue interacts with the reactive hydroxyl/carbonyl groups in the substrate (Figure 6c). Glu141 might participate in proton uptake to the C2’ oxygen from the solvent, employing the proton conduit identified in the protein structure (Supplementary Figure 47). The final reduction of UDP-apiose aldehyde (4) to UDP-apiose (1a), and of UDP-4-keto-xylose (5) to UDP-xylose (3), likely involves Tyr185 as the catalytic proton donor (Figure 6d).
In conclusion, the mechanistic proposal builds on deepened understanding of how the core catalytic apparatus of SDR was expanded in UAXS to incorporate catalysis to sugar ring opening/contraction as a unique functionality of this enzyme. Our findings also reveal distinct catalytic strategies employed by UAXS and UXS to achieve conversion of UDP-GlcA (2) according to the corresponding individual reaction path. Interestingly, each enzyme appears to have found its own way of exploiting a specific substrate conformation to promote catalysis. While UXS utilizes distortion in sugar ring pucker to drive the oxidative decarboxylation from pyranosyl substrate25, UAXS positions the UDP-GlcA (2) to enable C2’-C3’ aldol cleavage coupled to alcohol oxidation at C4’. UAXS adds a genuinely new facet of SDR catalytic principle to extend the functional diversity of this important enzyme superfamily.
Methods
The methods used are summarized in full detail in the Supplementary Information. For enzyme production and purification see Supplementary Methods and Supplementary Figures 3-5 and 39. Protein crystallography and structure determination are reported in Supplementary Methods, Supplementary Table 1 and Supplementary Figures 6-11 and 14. Molecular dynamics simulation experiments and QM/MM calculations are available in Supplementary Methods and Supplementary Figures 15, 16 and 47. Kinetic analysis of enzymes is reported in Supplementary Methods and presented graphically in Supplementary Figures 23-32. For product analysis by HPLC and NMR see Supplementary Methods and Supplementary Figures 17-22, 33-36 and 40-46.
Supplementary Material
Supplementary information is available for this paper.
Acknowledgments
Contributions of Paola Scudieri (enzyme characterization), Alexander Lepak (UDP-apiose HPLC assay) and Joan Coines (analysis of MD trajectories) are gratefully acknowledged. This work was supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol, and the Government of Lower Austrian and Business Agency Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG. Funding from the Austrian Science Funds (FWF; I-3247 to B.N. and A.J.E.B.) and by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Program (2018–2022) – Department of Biology and Biotechnology “L. Spallanzani” University of Pavia, is acknowledged.
Abbreviations
- UDP-GlcA
UDP-glucuronic acid
- UAXS
UDP-apiose/UDP-xylose synthase
- SDR
short-chain dehydrogenase/reductase superfamily
- UXS
UDP-xylose synthase
- ArnA
UDP-GlcA dehydrogenase/decarboxylase
- MD
molecular-dynamics
Footnotes
Data availability
Coordinates and structure factors have been deposited with the Protein Data Bank with accession codes 6H0N and 6H0P. All other data is available from the authors upon reasonable request.
Author contributions
S.S. performed protein crystallization and determined the structures. K.D.D., M.P., and C.R.V. performed MD simulations and QM calculations, and analyzed the data with A.B. A.D. designed enzyme variants and performed biochemical characterization with F. de G. and A.B. A.B., A.D., and H.W. performed the NMR analyses. A.B. performed substrate synthesis and product isolation. A.B. and A.D. performed the mechanistic analyses. B.N. and A.M. designed and supervised the research. The manuscript was written with contributions from all authors. B.N., A.D., A.M. S.S., and A.B. wrote the paper.
Competing interests
The authors declare no competing interests.
References
- 1.Picmanova M, Moller BL. Apiose: One of nature's witty games. Glycobiology. 2016;26:430–442. doi: 10.1093/glycob/cww012. [DOI] [PubMed] [Google Scholar]
- 2.Smith J, et al. Functional characterization of UDP-apiose synthases from bryophytes and green algae provides insight into the appearance of apiose-containing glycans during plant evolution. J Biol Chem. 2016;291:21434–21447. doi: 10.1074/jbc.M116.749069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter MS, et al. Functional assignment of multiple catabolic pathways for D-apiose. Nat Chem Biol. 2018;14:696–705. doi: 10.1038/s41589-018-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith JA, Bar-Peled M. Synthesis of UDP-apiose in Bacteria: The marine phototroph Geminicoccus roseus and the plant pathogen Xanthomonas pisi. PLoS One. 2017;12:e0184953. doi: 10.1371/journal.pone.0184953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsunaga T, et al. Occurrence of the primary cell wall polysaccharide rhamnogalacturonan II in pteridophytes, lycophytes, and bryophytes. Implications for the evolution of vascular plants. Plant Physiol. 2004;134:339–351. doi: 10.1104/pp.103.030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molhoj M, Verma R, Reiter WD. The biosynthesis of the branched-chain sugar D-apiose in plants: functional cloning and characterization of a UDP-D-apiose/UDP-D-xylose synthase from Arabidopsis. Plant J. 2003;35:693–703. doi: 10.1046/j.1365-313x.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- 7.Ndeh D, et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 2017;544:65–70. doi: 10.1038/nature21725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim M, Kang S, Rhee YH. De Novo synthesis of furanose sugars: Catalytic asymmetric synthesis of apiose and apiose-containing oligosaccharides. Angew Chem Int Ed. 2016;55:9733–9737. doi: 10.1002/anie.201604199. [DOI] [PubMed] [Google Scholar]
- 9.Picken JM, Mendicino J. The biosynthesis of D-apiose in Lemna minor. J Biol Chem. 1967;242:1629–1634. [PubMed] [Google Scholar]
- 10.Sandermann H, Jr, Tisue GT, Grisebach H. Biosynthesis of D-apiose. IV. Formation of UDP-apiose from UDP-D-glucuronic acid in cell-free extracts of parsley (Apium petroselinum L.) and Lemna minor. Biochim Biophys Acta. 1968;165:550–552. doi: 10.1016/0304-4165(68)90239-0. [DOI] [PubMed] [Google Scholar]
- 11.Choi SH, Ruszczycky MW, Zhang H, Liu HW. A fluoro analogue of UDP-α-D-glucuronic acid is an inhibitor of UDP-α-D-apiose/UDP-α-D-xylose synthase. Chem Commun. 2011;47:10130–10132. doi: 10.1039/c1cc13140k. [DOI] [PubMed] [Google Scholar]
- 12.Choi SH, et al. Analysis of UDP-D-apiose/UDP-D-xylose synthase-catalyzed conversion of UDP-D-apiose phosphonate to UDP-D-xylose phosphonate: Implications for a retroaldol-aldol mechanism. J Am Chem Soc. 2012;134:13946–13949. doi: 10.1021/ja305322x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eixelsberger T, et al. Isotope probing of the UDP-apiose/UDP-xylose synthase reaction: Evidence of a mechanism via a coupled oxidation and aldol cleavage. Angew Chem Int Ed. 2017;56:2503–2507. doi: 10.1002/anie.201609288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendicino J, Abouissa H. Conversion of UDP-D-glucuronic acid to UDP-D-apiose and UDP-D-xylose by an enzyme isolated from Lemna minor. Biochim Biophys Acta. 1974;364:159–172. doi: 10.1016/0005-2744(74)90143-0. [DOI] [PubMed] [Google Scholar]
- 15.Kelleher WJ, Grisebach H. Hydride transfer in the biosynthesis of uridine diphospho-apiose from uridine diphospho-D-glucuronic acid with an enzyme preparation of Lemna minor. Eur J Biochem. 1971;23:136–142. doi: 10.1111/j.1432-1033.1971.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 16.Baron D, Grisebach H. Further studies on the mechanism of action of UDP-apiose/UDP-xylose synthase from cell cultures of parsley. FEBS J. 1973;38:153–159. [Google Scholar]
- 17.Gebb C, Baron D, Grisebach H. Spectroscopic evidence for the formation of a 4-keto intermediate in the UDP-apiose/UDP-xylose synthase reaction. Eur J Biochem. 1975;54:493–498. doi: 10.1111/j.1432-1033.1975.tb04161.x. [DOI] [PubMed] [Google Scholar]
- 18.Kelleher WJ, Baron D, Ortmann R, Grisebach H. Proof for the origin of the branch hydroxymethyl carbon of D-apiose from carbon 3 of D-glucuronic acid. FEBS Lett. 1972;22:203–204. doi: 10.1016/0014-5793(72)80045-0. [DOI] [PubMed] [Google Scholar]
- 19.Guyett P, Glushka J, Gu X, Bar-Peled M. Real-time NMR monitoring of intermediates and labile products of the bifunctional enzyme UDP-apiose/UDP-xylose synthase. Carbohydr Res. 2009;344:1072–1078. doi: 10.1016/j.carres.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson B, et al. The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem-Biol Interact. 2009;178:94–98. doi: 10.1016/j.cbi.2008.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavanagh KL, Jornvall H, Persson B, Oppermann U. Medium- and short-chain dehydrogenase/reductase gene and protein families: The SDR superfamily: Functional and structural diversity within a family of metabolic and regulatory enzymes. Cell Mol Life Sci. 2008;65:3895–3906. doi: 10.1007/s00018-008-8588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jornvall H, et al. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 23.Filling C, et al. Critical residues for structure and catalysis in short-chain dehydrogenases/reductases. J Biol Chem. 2002;277:25677–25684. doi: 10.1074/jbc.M202160200. [DOI] [PubMed] [Google Scholar]
- 24.Thibodeaux CJ, Melancon CE, Liu HW. Natural-product sugar biosynthesis and enzymatic glycodiversification. Angew Chem Int Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eixelsberger T, et al. Structure and mechanism of human UDP-xylose synthase: Evidence for a promoting role of sugar ring distortion in a three-step catalytic conversion of UDP-glucuronic acid. J Biol Chem. 2012;287:31349–31358. doi: 10.1074/jbc.M112.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bar-Peled M, Griffith CL, Doering TL. Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: The pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proc Natl Acad Sci U S A. 2001;98:12003–12008. doi: 10.1073/pnas.211229198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gatzeva-Topalova PZ, May AP, Sousa MC. Crystal structure of Escherichia coli ArnA (PmrI) decarboxylase domain. A key enzyme for lipid A modification with 4-amino-4-deoxy-L-arabinose and polymyxin resistance. Biochemistry. 2004;43:13370–13379. doi: 10.1021/bi048551f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polizzi SJ, et al. Human UDP-α-D-xylose synthase and Escherichia coli ArnA conserve a conformational shunt that controls whether xylose or 4-keto-xylose is produced. Biochemistry. 2012;51:8844–8855. doi: 10.1021/bi301135b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matern U, Grisebach H. UDP-apiose/UDP-xylose synthase. Subunit composition and binding studies. Eur J Biochem. 1977;74:303–312. doi: 10.1111/j.1432-1033.1977.tb11394.x. [DOI] [PubMed] [Google Scholar]
- 30.Gatzeva-Topalova PZ, May AP, Sousa MC. Structure and mechanism of ArnA: Conformational change implies ordered dehydrogenase mechanism in key enzyme for polymyxin resistance. Structure. 2005;13:929–942. doi: 10.1016/j.str.2005.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfeiffer M, et al. A parsimonious mechanism of sugar dehydration by human GDP-mannose-4,6-dehydratase. ACS Catalysis. 2019;9:2962–2968. doi: 10.1021/acscatal.9b00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gerratana B, Cleland WW, Frey PA. Mechanistic roles of Thr134, Tyr160, and Lys 164 in the reaction catalyzed by dTDP-glucose 4,6-dehydratase. Biochemistry. 2001;40:9187–9195. doi: 10.1021/bi0108249. [DOI] [PubMed] [Google Scholar]
- 33.Kirschner KN, et al. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berger E, et al. Acid-base catalysis by UDP-galactose 4-epimerase: Correlations of kinetically measured acid dissociation constants with thermodynamic values for tyrosine 149. Biochemistry. 2001;40:6699–6705. doi: 10.1021/bi0104571. [DOI] [PubMed] [Google Scholar]
- 35.Baron D, Grisebach H. Further studies on mechanism of action of UDP-apiose/UDP-xylose synthase from cell-cultures of parsley. Eur J Biochem. 1973;38:153–159. [Google Scholar]
- 36.Fujimori T, et al. Practical preparation of UDP-apiose and its applications for studying apiosyltransferase. Carbohydr Res. 2019;477:20–25. doi: 10.1016/j.carres.2019.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.