Abstract
Hybridization between heterospecific individuals has been documented as playing a direct role in promoting paternal leakage and mitochondrial heteroplasmy in both natural populations and laboratory conditions, by relaxing the egg-sperm recognition mechanisms. Here, we tested the hypothesis that hybridization can lead to mtDNA heteroplasmy also indirectly via mtDNA introgression. By using a phylogenetic approach, we showed in two reproductively isolated beetle species, Ochthebius quadricollis and O. urbanelliae, that past mtDNA introgression occurred between them in sympatric populations. Then, by developing a multiplex allele-specific PCR assay, we showed the presence of heteroplasmic individuals and argue that their origin was through paternal leakage following mating between mtDNA-introgressed and pure conspecific individuals. Our results highlight that mtDNA introgression can contribute to promote paternal leakage, generating genetic novelty in a way that has been overlooked to date. Furthermore, they highlight that the frequency and distribution of mtDNA heteroplasmy can be deeply underestimated in natural populations, as i) the commonly used PCR-Sanger sequencing approach can fail to detect mitochondrial heteroplasmy, and ii) specific studies aimed at searching for it in populations where mtDNA-introgressed and pure individuals co-occur remain scarce, despite the fact that mtDNA introgression has been widely documented in several taxa and populations.
Subject terms: Evolutionary ecology, Evolution
Introduction
Mitochondrial DNA (mtDNA) has been one of the most popular genetic markers for studies concerning species ecology and evolution. In animals, mtDNA has been widely used to investigate population demography, to reconstruct species phylogenies and phylogeography, and, more recently, it is even being used to identify cryptic genetic diversity and classify living organisms1–4.
Strict maternal inheritance has been undoubtedly one of the most important features underlying the mtDNA success5–8. As a consequence of this way of inheritance, a condition of homoplasmy is set up in the offspring and maintained across generations. Different mechanisms during gametogenesis and after fertilization have been described, which ensure offspring homoplasmy by selectively destroying sperm mitochondria8–10. In mammals, after fertilization, paternal mitochondria are ubiquitinated and then degraded by enzymes11; in the rice fish Oryzias latipes, paternal mtDNA is degraded12; in the nematode Caenorhabditis elegans and the fruit fly Drosophila melanogaster, paternal mitochondria are destroyed by autophagy9,13. Basically, the above selective mechanisms are based on the egg-sperm recognition, where a factor coded by the nuclear genome occurring in the eggs is able to recognize a signal produced by the paternal genome located in the sperm mitochondria, promoting their destruction just after fertilization8. However, increasing evidence is showing that maternal inheritance is often not strict, but that male mtDNA transmission can occur (a process termed paternal leakage), leading to a condition of mitochondrial heteroplasmy, where mitochondrial genomes of both parents occur within an individual (paternal leakage-driven heteroplasmy)8,10,14.
In recent years, hybridization has been shown to play a direct role in promoting paternal leakage and heteroplasmy. Indeed, given that the efficiency of egg-sperm recognition mechanisms decreases when genetic divergence between parents increases, hybridization between individuals belonging to genetically divergent lineages favours paternal leakage8. Documented cases have been observed in a wide range of vertebrate and invertebrate taxa, including insects, birds, amphibians and mammals, and most of them have been indeed shown to involve hybrid individuals from experimental crosses or natural hybrid zones8,10,15–19.
In this paper, we argue that hybridization can promote mtDNA paternal leakage and heteroplasmy also indirectly, via mtDNA introgression. Following hybridization, mtDNA introgression, defined as the permanent incorporation of genes from one species into another, has been frequently reported to such an extent that complete mtDNA replacement can also occur20–26. In many cases, however, mtDNA-introgressed and pure conspecific individuals co-exist. For example, mtDNA introgression has been observed between the sea-rock pool mosquitoes Aedes mariae and Ae. zammitii in a recently established hybrid zones along the Italian coasts of the Adriatic Sea (e.g. as a consequence of human-mediated introduction). After its introduction, Ae. mariae diffused along a transect of about 20 km, coexisting in syntopy with Ae. zammitii. Reproductive isolation between the two species is not completed and ongoing hybridization occurs which led to bidirectional mtDNA introgression (mtDNA-introgressed individuals reached 25% and 14% in Ae. mariae and Ae. zammitii, respectively)25.
The co-occurrence of mtDNA-introgressed and pure conspecific individuals has been observed not only when hybridization is ongoing, but also when ancient introgression occurred between taxa that are currently reproductively isolated27–29. For example, ancient introgression following hybridization has been documented in a zone of parapatry between the two European treefrogs Hyla arborea and H. intermedia27. Genetic analyses using both nuclear and mitochondrial diagnostic markers showed the lack of current gene exchange between the two species, while introgressed alleles were observed in both species and in all markers analysed. In particular, mtDNA of H. arborea was observed in three individuals of H. intermedia27.
Here, we suggest that in populations where conspecific mtDNA-introgressed and pure conspecific individuals co-exist, a mating between pure and introgressed individuals could favour paternal leakage, by relaxing the egg-sperm recognition mechanisms that prevent it, like a mating between two heterospecific individuals. If so, it can be expected that paternal leakage-driven heteroplasmy could also involve species that hybridized and introgressed in the past and now are fully reproductively isolated. We tested this expectation, using as study-system two hydrenid beetle species belonging to the Ochthebius genus.
Ochthebius quadricollis and O. urbanelliae are distributed along the Mediterranean Sea coasts and occur syntopically in the same rock pools in a sympatric area along the coasts of the Italian peninsula30,31. They diverged during the Plio-Pleistocene30,32 as consequence of the climatic oscillations in the western Mediterranean region, that likely led to repeated isolation and secondary contact events between the two species32. Genetic analyses of sympatric populations using nuclear markers showed that hybridization and introgression occurred in the past, and then ceased due to the action of natural selection against maladaptive hybridization, which led to full reproductive isolation (speciation by reinforcement). No F1 hybrids between O. quadricollis and O. urbanelliae occur today in sympatric populations and the analysis of mating couples showed the occurrence of assortative mating. Furthermore, mating trials under laboratory conditions showed a pattern of higher premating isolation in sympatric versus allopatric populations33, and reproductive character displacement was found in sympatric populations of O. urbanelliae34.
Here, to furnish evidences to our hypothesis, we first searched for past mtDNA introgression, by using a phylogenetic approach; then, we screened O. quadricollis and O. urbanelliae individuals from sympatric and allopatric populations for the occurrence of mtDNA heteroplasmy, by developing a multiplex allele-specific PCR assay.
Results
mtDNA diversity and phylogenetic relationships
A total of 189 individuals (84 O. quadricollis and 105 O. urbanelliae) collected along the Italian coasts (Fig. 1, Table 1) were sequenced for a 537 base pair fragment in the 5′ half of the mitochondrial COI gene (Supplementary Fig. S1). No double peaks were observed in the sequence chromatograms of any individuals, with the exception of the O. urbanelliae Cirella-3 individual (Fig. 2). The obtained sequences showed a similarity of 99–100% with the sequences of O. quadricollis or O. urbanelliae available in GenBank. Two out of the 189 individuals were excluded from the following analyses because they were heteroplasmic (see below). The analysis of nucleotide and amino-acidic polymorphism detected 48 haplotypes (Supplementary Fig. S7), defined by 97 polymorphic sites and 111 mutations (106 synonymous) (Genbank accession number MH285878-MH285928). Average uncorrected p-distance between the two clades was 7.9%, while among populations it ranged from 0.006 to 0.032 in O. quadricollis and from 0 to 0.06 in O. urbanelliae (Supplementary Table S1).
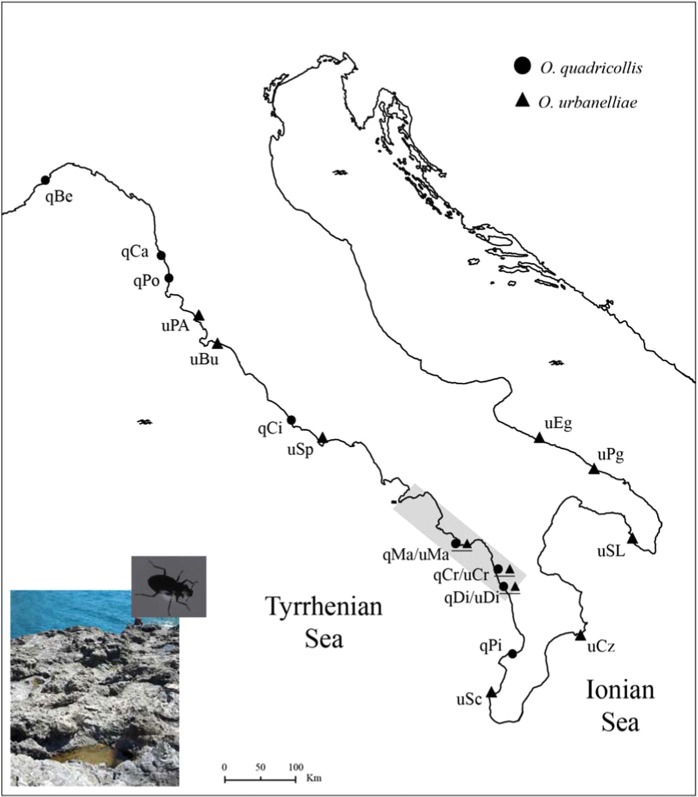
Figure 1.
Map showing sampling sites of Ochthebius quadricollis and O. urbanelliae individuals. Light grey area: sympatric area between the two species (Urbanelli 2002). Photos show a specimen of O. quadricollis and a typical sea rock pool (Photo by Alessandra Spanò).
Table 1.
Sampled populations of Ochthebius quadricollis and O. urbanelliae.
| Species | Locality | Latitude | Longitude | N | Haplotype | mtDNA introgressed individuals | Heteroplasmic individuals |
|---|---|---|---|---|---|---|---|
| O. quadricollis | Bergeggi | 44.24° | 8.42° | 11 | h1, h2, h3(2), h4(5), h5, h6 | — | — |
| Castiglioncello | 43.40° | 10.41° | 11 | h3(7), h7, h8, h9, h10 | — | — | |
| Populonia | 42.98° | 10.98° | 12 | h3, h11(7), h12(2), h13(2) | — | — | |
| Circeo | 41.22° | 13.04° | 10 | h3, h14(7), h15, h16 | — | — | |
| Cirella | 39.71° | 15.81° | 12 | h3, h11, h17(8), h34(2) | 2 | — | |
| Maratea | 39.98° | 15.70° | 7 | h17(4); h11, h13, h34 | 1 | — | |
| Diamante | 39.67° | 15.81° | 11 | h6, h13(5), h17(4), h18 | — | — | |
| Pizzo | 38.73° | 16.16° | 10 | h3, h5, h13(4), h19, h20(3) | — | — | |
| O. urbanelliae | Punta Ala | 42.80° | 10.74° | 10 | h21(10) | — | — |
| Burano | 42.40° | 11.38° | 11 | h21(10), h22 | — | — | |
| Sperlonga | 41.26° | 13.42° | 10 | h23(6), h24(4) | — | — | |
| Maratea | 39.98° | 15.70° | 12 | h17, h25(2), h26, h27, h28, h29, h30, h31, h32, h33(2) | 1 | — | |
| Cirella | 39.71° | 15.81° | 10 | h34(5), h17(2), h35 | 3 | 2 | |
| Diamante | 39.67° | 15.81° | — | — | — | — | |
| Scilla | 38.24° | 15.71° | 11 | h36(3), h37(7), h38 | — | — | |
| Capo Rizzuto | 38.90° | 17.10° | 10 | h39(4), h40(2), h41(4) | — | — | |
| S. Maria Leuca | 39.79° | 18.75° | 10 | h42(7), h43(3) | — | — | |
| Pantanagianni | 40.70° | 17.84° | 11 | h44(6), h45(4), h46 | — | — | |
| Egnazia | 40.89° | 17.37° | 10 | h44(5), h47(4), h48 | — | — |
The number of individuals analysed in each population and the haplotypes observed at mitochondrial CO I gene fragment are shown. In brackets is shown how many times each haplotype was found.
Figure 2.
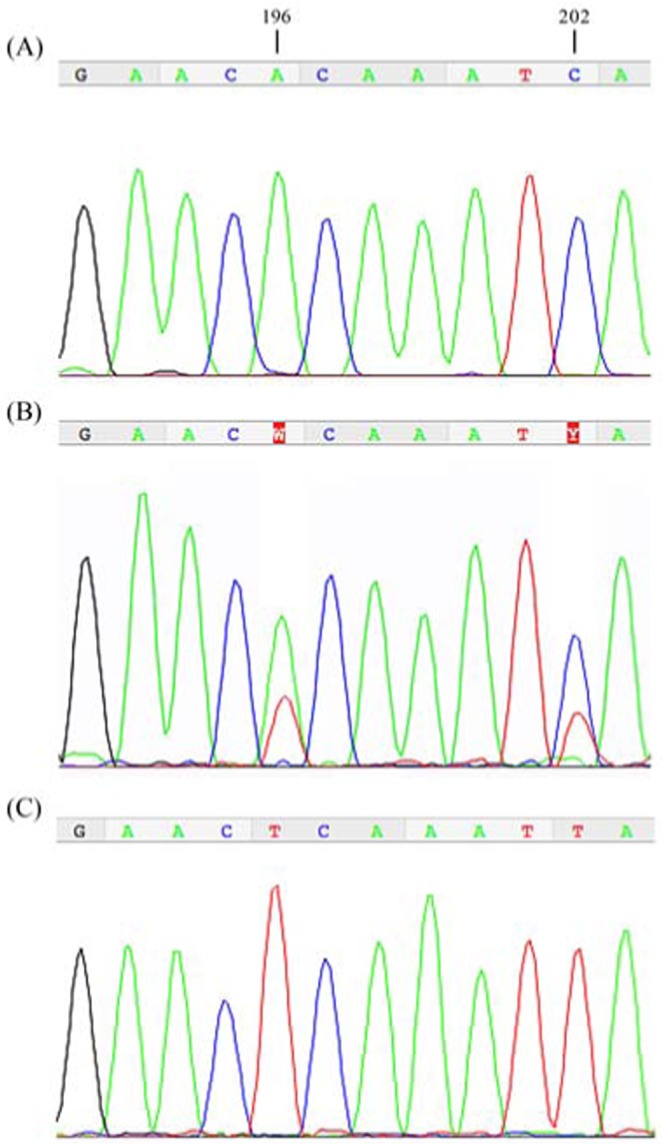
Chromatograms of the COI gene fragment showing two heteroplasmic positions. (A) O. quadricollis variant, (B) the sequence of the O. urbanelliae Cirella 3 heteroplasmic individual, (C) O. urbanelliae variant.
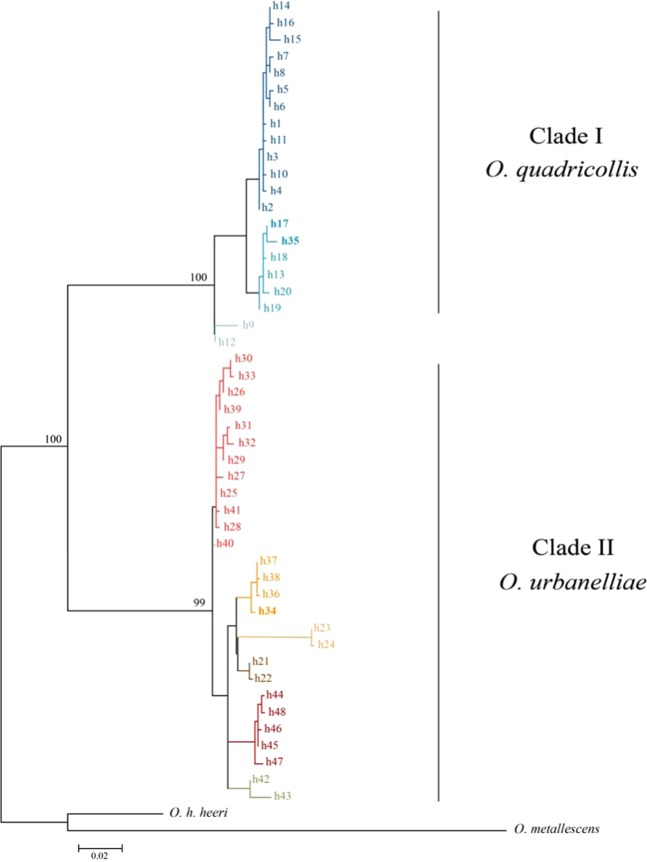
Phylogenetic analyses using the Maximum Likelihood (ML) approach was performed to reconstruct the relationships among haplotypes (Fig. 3). All haplotypes of the clade I were found in O. quadricollis individuals with the exception of the haplotypes h17 that was found in both O. quadricollis and O. urbanelliae individuals, and with the exception of the haplotype h35 that was found in one O. urbanelliae individual (Fig. 3, Table 1). All haplotypes of the clade II were found in O. urbanelliae individuals with the exception of the h34 haplotype that was found in both O. quadricollis and O. urbanelliae individuals (Fig. 3, Table 1).
Figure 3.
Phylogenetic relationships of the mtDNA haplotypes found in Ochthebius quadricollis and O. urbanelliae. Maximum likelihood (ML) tree is shown. Bootstrap values are shown above the main nodes. The haplotypes h17 and h34 (in bold) are shared between O. quadricollis and O. urbanelliae (Table 1); the haplotype h35 (in bold) was found in one O. urbanelliae individual (Table 1), but is more closely related to the O. quadricollis haplotypes.
Heteroplasmy
A multiplex allele-specific polymerase chain reaction (MAS-PCR) was used to investigate the occurrence of mtDNA heteroplasmy in O. quadricollis and O. urbanelliae (Supplementary Fig. S1). Both the MAS-PCR banding patterns and the sequencing results supported the specificity of the assay. Indeed, one band of the expected size was observed when the DNA of only O. quadricollis and O. urbanelliae was used, and two bands when DNA was mixed (Supplementary Fig. S2). Likewise, the sequences of the amplicons showed 100% identity with O. quadricollis and O. urbanelliae sequences. The MAS-PCR assay also proved to be sensitive, being able to detect an amount as low as 0.01 ng of O. quadricollis mtDNA (ratio O. urbanelliae/O. quadricollis mtDNA, 1:500), and an amount as low as 0.005 ng of O. urbanelliae mtDNA (ratio O. quadricollis/O. urbanelliae mtDNA, 1:1000) (Supplementary Fig. S2).
The MAS-PCR assay was then used to analyze all individuals that were sequenced for the COI mtDNA gene fragment to check for mitochondrial heteroplasmy. In O. urbanelliae, two heteroplasmic individuals were found (i.e. one male and one female) (Table 1). They showed two electrophoretic bands following MAS-PCR (Supplementary Fig. S3), which sequences were identical to a fragment of the O. quadricollis and O. urbanelliae haplotypes. In O. quadricollis, no heteroplasmic individuals were found.
Discussion
Paternal leakage and mtDNA heteroplasmy have been documented in several animal species, including insects, fishes, reptiles, birds and mammals8,10. Being more difficult to detect when mtDNA variation within or among populations is low, these phenomena have been often observed in secondary contact zones or laboratory crosses, as a consequence of hybridization between heterospecific individuals or between homospecific individuals belonging to highly divergent genetic lineages8,10,35. Here, we hypothesized that hybridization can lead to mtDNA paternal leakage and heteroplasmy also via mtDNA introgression. By using genetic data, we showed that: i) mtDNA introgression occurred between the two species and that mtDNA-introgressed and pure individuals co-occur in natural populations; ii) heteroplasmic individuals are present in O. urbanelliae; iii) mtDNA heteroplasmy originated through paternal leakage.
First, the phylogenetic analyses showed that O. quadricollis and O. urbanelliae share mtDNA haplotypes, as well as having some haplotypes more closely related to those of the other species than to those of its own haplogroup (Fig. 3, Table 1). This pattern is consistent with a process of mtDNA introgression and it is concordant with the signatures in the nuclear genomes of introgressive hybridization between O. quadricollis and O. urbanelliae, before the completion of the speciation process by reinforcement, shown by nuclear markers31,33. Furthermore, alternative hypotheses, such as incomplete lineage sorting can be excluded20. Contrary to what would be expected under this hypothesis, the introgressed haplotypes between O. quadricollis and O. urbanelliae are indeed not randomly distributed across the species’ range, but rather they are geographically localized within the sympatric area between the two species, where both mtDNA-introgressed and pure individuals co-occur (Fig. 1, Table 1).
Second, the screening of O. quadricollis and O. urbanelliae individuals by MAS-PCR and sequence analyses showed the occurrence of heteroplasmic individuals in O. urbanelliae (Table 1, Supplementary Fig. S3). Misleading results due to unspecific PCR amplification of the mtDNA of the two species can be confidently ruled out, as the MAS-PCR assay proved to be highly specific. Furthermore, the sequences obtained from the amplicons were identical to the COI sequences of O. quadricollis and O. urbanelliae found in the sympatric area, which also exclude the possibility that non-functional nuclear copies of mitochondrial genes (NUMTs) have been analysed.
Third, mtDNA heteroplasmy originated by paternal leakage. Empirical evidence showed that different routes can lead to mtDNA heteroplasmic individuals8,10. When mtDNA heteroplasmy is due to the leakage of paternal mitochondrial genome, it is expected that the mtDNA haplotypes within the heteroplasmic individuals derive from both maternal and paternal mtDNA lineages8,10,14. The heteroplasmic individuals found in O. urbanelliae are concordant with this expectation, as they showed one haplotype identical to the haplotypes of O. quadricollis, and the other identical to those of O. urbanelliae, which supports the hypothesis that they originated by paternal leakage. In O. quadricollis, no heteroplasmic individuals were found by the MAS-PCR screening (Table 1). Genetic or selective factors preventing paternal leakage in this species could be invoked to explain this result, or alternatively, heteroplasmic individuals may have escaped our screening by chance, if O. urbanelliae DNA was below our detection limit.
Taking into account the above results and the reproductive isolation among the two species, we argue that mtDNA introgression promoted paternal leakage in the Ochthebius beetles. Events of paternal leakage and heteroplasmy have often been ascribed to a failure of the recognition mechanisms between nuclear and mitochondrial genomes located in the eggs and sperms, and often they have been shown to involve hybridization between divergent lineages8,36. However, hybridization between O. quadricollis and O. urbanelliae cannot be invoked to explain paternal leakage and mtDNA heteroplasmy in these species, because reproductive isolation between them is completed31,33. Likewise, the hypothesis that paternal leakage originated from past hybridization events between O. quadricollis and O. urbanelliae and then was followed by maternal transmission of heteroplasmy can be excluded as well, because the heteroplasmic variants would have disappeared over time since the completion of reproductive isolation between the two species30,37–39. Paternal leakage and mtDNA heteroplasmy in the O. quadricollis and O. urbanelliae beetles would therefore be due to mates between mtDNA-introgressed and pure conspecific individuals.
The role of the mtDNA introgression as a source of evolutionary novelty has become increasingly recognized in recent years40–43. Our results, by showing that mtDNA introgression can promote paternal leakage, highlight that introgression would be able to generate genetic novelty in a way that has been neglected to date. Furthermore, they support that the frequency and distribution of paternal leakage and mtDNA heteroplasmy can be deeply underestimated in natural populations, as the commonly used PCR-Sanger sequencing approach can fail to detect mitochondrial heteroplasmy, and specific studies aimed at searching for heteroplasmy in populations where mtDNA-introgressed and pure individuals co-occur remain scarce.
In animals, the mtDNA genome has recently been shown also to recombine, both at intra- and intermolecular level8,14,44, and recombination has been suggested as an important evolutionary mechanism avoiding deleterious mutation meltdown in mtDNA (i.e. the Muller’s ratchet), as well as for the origin of new genetic combinations. Paternal leakage and mtDNA heteroplasmy, allowing two different genomes to meet, represent an intermediate but critical step for recombination. An understanding of how frequently they occur in nature, as well as the processes underlying them, represent therefore outstanding questions to be addressed.
Materials and Methods
Samples and species recognition
Ochthebius quadricollis and O. urbanelliae adult individuals were collected from 16 localities along the Italian coasts (Table 1, Fig. 1). All individuals were identified using the morphological and biochemical keys of Audisio et al.45 and Urbanelli et al.30. Standard horizontal starch gel electrophoresis was performed following the protocols described in Urbanelli31.
mtDNA sequencing and phylogenetic inferences
DNA from single adults was extracted from the tissue homogenate used for allozymic analyses by standard CTAB (cetyltrimethyl ammonium bromide) protocol46. Partial sequences of the Cytochrome Oxidase I (COI) mitochondrial gene were obtained through PCR-amplification. The universal primers pair C1-J-2183 and TL2-N-3014 was used to amplify and sequencing a COI fragment of ~900 base pairs47,48. The following specific primers were then designed and used for further analyses: OchtCOI-f 5′-accaggatttggaataattt-3′ and OchtCOI-r 5′-tccaatagaagaaataatatttc-3′. The PCR was carried out in a 25 µl volume containing 5 ng of DNA, 10 mMTris-HCl, pH 8.3, 2.0 mM MgCl2, 0.2 μM of the forward and reverse primers, 0.4 mM dNTPs, 0.3 units of NZYTaq polymerase (NZYtech, Lisbon, Portugal) and water. Negative controls including all reagents but DNA were also included in the reaction. PCR cycling procedure was: 95 °C for 5 min followed by 34 cycles of 93 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min 30 s, and a single final step at 72 °C for 10 min. PCR sequences were obtained using an ABI PRISM 3700 DNA sequencer by GATC Inc. (www.GATC.com). All individuals were double strand sequenced. The individuals found introgressed were amplified multiple times and two independent PCR products were sequenced.
Sequences were edited and aligned using the software Chromas 2.6.5 (Technelysium, Helensvale, Australia) and ClustalX 2.149, respectively, and compared with the O. quadricollis and O. urbanelliae sequences available in GenBank using BLASTN algorithm. Nucleotide and amino-acidic polymorphisms of the COI gene fragments were estimated using the software DNAsp 650. The average uncorrected p-distance between groups of haplotypes and populations was computed using Mega 7.051.
Phylogenetic relationships among haplotypes of O. quadricollis and O. urbanelliae were inferred using the Maximum Likelihood (ML) method, as implemented in PAUP 4.052. ML analysis was performed using heuristic searches with 100 rounds of random sequence addition and tree bisection-reconnection (TBR) branch swapping algorithm. The GTR + G + I substitution model was used (gamma shape parameter G = 1.480; proportion of invariable sites I = 0.669), following the substitution model inferred by jModelTest 2.053 using the Akaike Information Criterion. The robustness of the inferred ML tree topology was assessed by the non-parametric bootstrap method with 1000 replicates. Ochthebius heeri heeri (KT804242.1) and Ochthebius metallescens (HF931191.1) were used as outgroups in the phylogenetic analyses54.
Heteroplasmy screening
To screen the Ochthebius individuals for mtDNA heteroplasmy, a multiplex allele-specific polymerase chain reaction (MAS-PCR) was designed. On the basis of the CO I sequences obtained from the O. quadricollis and O. urbanelliae individuals, a common forward primer for the two species (OchCom_F, 5′-gcggtattttaagcttttca-3′) and a specific reverse primer for O. quadricollis (OchQua_R, 5′-cttgattcaaattgacatc-3′) and O. urbanelliae (OchUrb_R, 5′-actgcaccttgacttaatataa-3′) were designed. The common primer and the two species-specific primers were combined into a single multiplex PCR reaction to yield distinct PCR banding patterns that can accurately detect the mtDNA of the two species (Supplementary Fig. S1).
The specificity of the assay (i.e. the ability to correctly discriminate the mtDNA of the two species), was tested by amplifying and sequencing the DNA of known O. quadricollis and O. urbanelliae individuals and a mixture of the two species. PCR reactions were carried out in a 25 µl volume containing 5 ng of DNA, 10 mMTris-HCl, pH 8.3, 2.0 mM MgCl2, 0.4 mM dNTPs, 0.4 µM of the common forward primer, 0.4 µM of each species-specific reverse primer, and 2.5 units of NZYTaq polymerase (NZYtech, Lisbon, Portugal). In each reaction, negative controls containing all reagents but DNA were also included. PCR cycling procedure was: 95 °C for 5 min followed by 34 cycles at 93 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min 30 s, and a single final step at 72 °C for 10 min. The PCR products were then separated by electrophoresis run on 2% agarose, 0.5X TAE gel, and visualized by staining with Gelred (Sigma-Aldrich, Milan, Italy). The sizes of the DNA fragments were assessed using the 100 bp DNA ladder (Promega, Milan, Italy) run on the same gel. Three technical replicates of each reaction were performed to check for the reproducibility of the MAS-PCR assay. PCR products obtained from the amplification of O. quadricollis and O. urbanelliae individuals and mixed DNA samples were double strand sequenced (https://www.gatc-biotech.com). The bands of the mixed DNA samples were excised from the gel, purified using the NucleoSpin gel and the PCR Clean-up purification kits (Macherey-Nagel, Düren, Germany) and then double strand sequenced.
The sensitivity of the MAS-PCR assay (i.e. the smallest amount of mtDNA of both species in a sample that can be accurately detected), was tested by amplifying the DNA of O. quadricollis and O. urbanelliae at different ratios. The amount of O. quadricollis (or O. urbanelliae) DNA was gradually reduced to 0.5, 0.05, 0.01 and 0.005 ng to obtain dilutions of O. quadricollis to O. urbanelliae DNA at 1:10, 1:100, 1:500 and 1:1000. Multiplex PCR amplifications were performed and PCR products were separated by electrophoresis as described above.
After the specificity and sensitivity tests, the MAS-PCR assay was used to screen for heteroplasmy the sampled O. quadricollis and O. urbanelliae individuals. The heteroplasmic individuals were amplified three times to check for consistency, and the amplicons were excised from the gel, purified and double strand sequenced, as described above.
Supplementary information
Acknowledgements
We thank Sara Maggini for her help in sampling and laboratory analyses; Alessandra Spanò for the technical assistance; Mark Eltenton for the linguistic revision.
Author contributions
V.M., D.P. and S.U. conceived the study; V.M. and D.P. performed the laboratory work and data analyses; V.M., and D.P. drafted the manuscript; all authors read, discussed and approved the final version of the paper.
Data availability
All data generated or analysed during this study are included in this published article
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55764-w.
References
- 1.Avise JC, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematic. Ann Rev Ecol Syst. 1987;18:489–522. doi: 10.1146/annurev.es.18.110187.002421. [DOI] [Google Scholar]
- 2.Avise, J. C. Molecular markers, natural history, and evolution (2nd ed. Sunderland) (Sinauer Associates, 2004).
- 3.Ratnasingham, S. & P. D. Hebert. bold: The Barcode of Life Data System (, http://www.barcodinglife.org). Mol Ecol Notes7, 355–364 (2007). [DOI] [PMC free article] [PubMed]
- 4.Hickerson MJ, et al. Phylogeography’s past, present, and future: 10 years after Avise 2000. Mol Phyl Evol. 2010;54:291–301. doi: 10.1016/j.ympev.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13:729–744. doi: 10.1046/j.1365-294X.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 6.Ballard JWO, Rand DM. The population biology of mitochondrial DNA and its phylogenetic implications. Ann Rev Ecol Evol Syst. 2005;36:621–642. doi: 10.1146/annurev.ecolsys.36.091704.175513. [DOI] [Google Scholar]
- 7.Galtier N, Nabholz B, Glémin S, Hurst GD. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 2009;18:4541–50. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- 8.Ladoukakis D, Zouros E. Evolution and inheritance of animal mitochondrial DNA: rules and exceptions. J Biol Res Thessaloniki. 2017;24:2. doi: 10.1186/s40709-017-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato M, Sato K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim Biophys Acta. 2013;1833:1979–1984. doi: 10.1016/j.bbamcr.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Breton S, Stewart DT. Atypical mitochondrial inheritance patterns in eukaryotes. Genome. 2015;58:423–431. doi: 10.1139/gen-2015-0090. [DOI] [PubMed] [Google Scholar]
- 11.Sutovsky P, et al. Ubiquitin tag for sperm mitochondria. Nature. 1999;402:371–2. doi: 10.1038/46466. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura Y, et al. Active digestion of sperm mitochondrial DNA in single living sperm revealed by optical tweezers. Proc Natl Acad Sci USA. 2006;103:1382–7. doi: 10.1073/pnas.0506911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al Rawi S, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–7. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 14.White DJ, Wolff JN, Pierson M, Gemmell NJ. Revealing the hidden complexities of mtDNA inheritance. Mol Ecol. 2008;17:4925–4942. doi: 10.1111/j.1365-294X.2008.03982.x. [DOI] [PubMed] [Google Scholar]
- 15.Fontaine KM, Cooley JR, Simon C. Evidence for Paternal Leakage in Hybrid Periodical Cicadas (Hemiptera: Magicicada spp.) PlosOne. 2007;9:e892. doi: 10.1371/journal.pone.0000892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff JN, Nafisinia M, Sutovsky P, Ballard JWO. Paternal transmission of mitochondrial DNA as an integral part of mitochondrial inheritance in metapopulations of Drosophila simulans. Heredity. 2013;10:57–62. doi: 10.1038/hdy.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radojičic JM, Krizmanić I, Kasapidis P, Zouros E. Extensive mitochondrial heteroplasmy in hybrid water frog (Pelophylax spp.) populations from Southeast Europe. Ecol Evol. 2015;5:4529–41. doi: 10.1002/ece3.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandolfi A, et al. New evidences of mitochondrial DNA heteroplasmy by putative paternal leakage between the rock partridge (Alectoris graeca) and the chukar partridge (Alectoris chukar) PlosOne. 2017;12:e0170507. doi: 10.1371/journal.pone.0170507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mastrantonio V, et al. Paternal leakage and mtDNA heteroplasmy in Rhipicephalus spp. ticks. Sci Rep. 2019;9:1460. doi: 10.1038/s41598-018-38001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol. 2012;21:3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 21.Pons JM, Sonsthagen S, Dove C, Crochet. PA. Extensive mitochondrial introgression in North American Great Black-backed Gulls (Larus marinus) from the American Herring Gull (Larus smithsonianus) with little nuclear DNA impact. Heredity. 2013;112:226–239. doi: 10.1038/hdy.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zieliński P, et al. No evidence for nuclear introgression despite complete mtDNA replacement in the Carpathian newt (Lissotriton montandoni) Mol Ecol. 2013;22:1884–1903. doi: 10.1111/mec.12225. [DOI] [PubMed] [Google Scholar]
- 23.Canestrelli D, Bisconti R, Nascetti G. Extensive unidirectional introgression between two salamander lineages of ancient divergence and its evolutionary implications. Sci Rep. 2014;4:6516. doi: 10.1038/srep06516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halas D, Simons AM. Cryptic speciation reversal in the Etheostoma zonale (Teleostei: Percidae) species group, with an examination of the effect of recombination and introgression on species tree inference. Mol Phyl Evol. 2014;70:13–28. doi: 10.1016/j.ympev.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Mastrantonio V, Porretta D, Urbanelli S, Crasta G, Nascetti G. Dynamics of mtDNA introgression during species range expansion: insights from an experimental longitudinal study. Sci Rep. 2016;6:30355. doi: 10.1038/srep30355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bisconti R, Porretta D, Arduino P, Nascetti G, Canestrelli D. Hybridization and extensive mitochondrial introgression among fire salamanders in peninsular Italy. Sci Rep. 2018;8:13187. doi: 10.1038/s41598-018-31535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verardi A, Canestrelli D, Nascetti G. Nuclear and mitochondrial patterns of introgression between the parapatric European treefrogs Hyla arborea and H. intermedia. Ann Zool Fennici. 2009;46(4):247–58. doi: 10.5735/086.046.0402. [DOI] [Google Scholar]
- 28.Colliard C, et al. Strong reproductive barriers in a narrow hybrid zone of West-Mediterranean green toads (Bufo viridis subgroup) with Plio-Pleistocene divergence. BMC Evol Biol. 2010;10:232. doi: 10.1186/1471-2148-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallinoto M, Cunha DB, Bessa-Silva A, Sodre D, Sequeira F. Deep divergence and hybridization among sympatric Neotropical toads. Zool J Linn Soc. 2017;180:647–660. doi: 10.1093/zoolinnean/zlw001. [DOI] [Google Scholar]
- 30.Urbanelli S, Sallicandro P, De Vito E, Colonnelli E, Bullini L. Molecular reexamination of the taxonomy of Ochthebius (Calobius) (Coleoptera: Hydraenidae) from the Mediterranean and Macaronesian Regions. Ann Entomol Soc Am. 1996;89:623–636. doi: 10.1093/aesa/89.5.623. [DOI] [Google Scholar]
- 31.Urbanelli S. Genetic divergence and reproductive isolation in the Ochthebius (Calobius) complex (Coleoptera: Hydraenidae) Heredity. 2002;88:333–341. doi: 10.1038/sj.hdy.6800046. [DOI] [PubMed] [Google Scholar]
- 32.Antonini G, et al. Molecular phylogeography of two Italian sibling species of Calobius (Coleoptera, Hydraenidae, Ochthebiinae) inhabiting Mediterranean marine rock-pools. Mar Biol. 2010;157:371–381. doi: 10.1007/s00227-009-1324-9. [DOI] [Google Scholar]
- 33.Urbanelli S, Porretta D. Evidence of reinforcement of premating isolation between two species of the genus Ochthebius (Coleoptera:Hydraenidae) Evolution. 2008;62:1520–1527. doi: 10.1111/j.1558-5646.2008.00381.x. [DOI] [PubMed] [Google Scholar]
- 34.Porretta D, Urbanelli S. Evolution of premating reproductive isolation among conspecific populations of the sea rock‐pool beetle Ochthebius urbanelliae driven by reinforcing natural selection. Evolution. 2012;66:1284–1295. doi: 10.1111/j.1558-5646.2011.01535.x. [DOI] [PubMed] [Google Scholar]
- 35.Sato K, Sato M. Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J Biochem. 2017;162:247–53. doi: 10.1093/jb/mvx052. [DOI] [PubMed] [Google Scholar]
- 36.Dokianakis E, Ladoukakis ED. Different degree of paternal mtDNA leakage between male and female progeny in interspecific Drosophila crosses. Ecol Evol. 2014;4:2633–41. doi: 10.1002/ece3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solignac M, Génermont J, Monnerot M, Mounolou JC. Genetics of mitochondria in Drosophila: mtDNA inheritance in heteroplasmic strains of D. mauritiana. Mol Gen Gen. 1984;197:183–188. doi: 10.1007/BF00330961. [DOI] [Google Scholar]
- 38.Rand DM, Harrison RG. Mitochondrial DNA transmission genetics in crickets. Genetics. 1986;114:955–970. doi: 10.1093/genetics/114.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nunes MDS, Dolezal M, Schlötterer C. Extensive paternal mtDNA leakage in natural populations of Drosophila melanogaster. Mol Ecol. 2013;22:2106–17. doi: 10.1111/mec.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boratyński Z, et al. Introgression of mitochondrial DNA among Myodes voles: consequences for energetics? BMC Evol Biol. 2011;11:355. doi: 10.1186/1471-2148-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries. J Her. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 42.Llopart A, et al. Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea. Mol Ecol. 2014;23:1124–1136. doi: 10.1111/mec.12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloan DB, Havird JC, Sharbrough J. The on-again, off-again relationship between mitochondrial genomes and species boundaries. Mol Ecol. 2017;26:2212–2236. doi: 10.1111/mec.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rokas A, Ladoukakis E, Zouros E. Animal mitochondrial DNA recombination revisited. Trend. Ecol Evol. 2003;18:411–417. doi: 10.1016/S0169-5347(03)00125-3. [DOI] [Google Scholar]
- 45.Audisio P, et al. Molecular and morphological evidence of a new sibling species of Calobius (Coleoptera: Hydraenidae) of the C. quadricollis complex from peninsular Italy. Ital J Zool. 2010;77:29–37. doi: 10.1080/11250000902845738. [DOI] [Google Scholar]
- 46.Navajas M, Lagnel J, Fauvel G, De Moraes G. Sequence variation of ribosomal internal transcribed spacers (ITS) in commercially important phytoseiidae mites. Exp Appl Acarol. 1999;23:851–859. doi: 10.1023/A:1006251220052. [DOI] [PubMed] [Google Scholar]
- 47.Simon C, et al. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am. 1994;87:651–701. doi: 10.1093/aesa/87.6.651. [DOI] [Google Scholar]
- 48.Porretta D, Gargani M, Bellini R, Calvitti M, Urbanelli S. Isolation of microsatellite markers in the tiger mosquito Aedes albopictus (Skuse) Mol Ecol Notes. 2006;6:880–881. doi: 10.1111/j.1471-8286.2006.01384.x. [DOI] [Google Scholar]
- 49.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 50.Rozas J, et al. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Datasets. Mol Biol Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 51.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swofford, D. L. PAUP*: phylogenetic analysis using parsimony, version 4.0b10 (2016).
- 53.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sabatelli S, et al. Molecular ecology and phylogenetics of the water beetle genus Ochthebius revealed multiple independent shifts to marine rockpools lifestyle. Zool Scripta. 2016;45:175–186. doi: 10.1111/zsc.12141. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article