Abstract
Dissemination of metastatic precursors from primaries is the primary reason for patient death. Dissemination encompasses tumor cells invasion of stroma, followed by intravasation through the endothelium barrier into the bloodstream. Here, we describe how geminin-overexpressing tumor cells acquire dissemination ability. Acetylated HMGB1 (Ac-HMGB1) secreted by geminin-overexpressing cells activates RAGE and CXCR4 expression on mesenchymal stem cells (MSCs) located in tumor stroma. Through secreting CXCL12, geminin-overexpressing cells recruit these CXCR4+-MSCs into the tumor. Within the tumor, MSCs differentiate into S100A4-secreting cancer-associated fibroblasts (CAFs). S100A4, in a reciprocal manner, activates geminin-overexpressing cells to secrete CCL2 that recruits M0-macrophages from the stroma into the tumor. Within the tumor, CCL2 polarizes M0-macrophages into Gas6-secreting M2-tumor-associated macrophages (M2-TAMs). In concert, geminin-overexpression, S100A4/RAGE and Gas6/AXL signaling promote the invasive and intravasation abilities in geminin-overexpressing cells through exacerbating their stemness and epithelial-to-mesenchymal phenotypes and enhancing expression and functional interaction of CD151 and α3β1-integrin in geminin-overexpressing cells. Tumors formed following injection of geminin-overexpressing cells admixed with MSCs/CAFs grew faster, metastasized earlier, especially to lungs, and were extremely sensitive to anti-c-Abl, anti-RAGE, and anti-AXL drugs. These data support an intrinsic ability in geminin-overexpressing tumor cells to promote their metastatic potential through recruitment and bi-directional interactions with MSCs/CAFs and M2-TAMs.
Subject terms: Breast cancer, Breast cancer
Introduction
Geminin is a multifunctional protein that inhibits DNA-replication in S-phase1, protect against genomic instability2, and safeguard against chemically induced carcinogenesis3. Although these functions could support a tumor suppressor function for geminin2,3, ablation in mouse embryos or human cells didn′t induce endoreplication or tumorigenesis without stimulus3,4. To date, there have never been any reports describing any genetic or epigenetic alterations in the geminin gene in human breast cancer. Our sequencing efforts of the whole gene in ~150 breast tumors confirmed this lack of alterations in geminin gene4.
In contrast, geminin is overexpressed in many cancers, including breast cancers, suggesting an oncogenic function instead for geminin5–9. Mechanisms to explain this oncogenic role range from ability to maintain pluripotency in early cell lineages10,11, promotion of epithelial-to-mesenchymal transition (EMT) in embryonic stem cells12, and geminin ability to control proper cytokinesis in G2/M/early G1-phases1,4 in normal human mammary epithelial (HME) cells, while cytokinesis failure and production of aneuploid, aggressive breast cancer cells when overexpressed13,14.
In normal cells, during G2/M/early G1, geminin helps Aurora B chromosome condensation function, and TopoIIα chromosome decatenation function. In breast cancer cells, geminin overexpression blocks Aurora B histone H3-(serine 10) phosphorylation, leading to failure in chromosome condensation, aborted cytokinesis, and aneuploidy13,14. In geminin-overexpressing breast cancer cells, TopoIIα is prematurely de-sumoylated and released from chromosomal decatenation sites, also leading to aneuploidy due to elevated chromosomal breakage14.
Our recent work showed that, while during S-phase, geminin is a nuclear soluble-serine/threonine (S/T) phosphorylated protein15, during G2/M/early G1-phases, it becomes chromatin bound-tyrosine (Y) phosphorylated protein4. While overexpression of wild-type geminin triggers aneuploidy, a single Y-to-A mutant geminin (contains 3 Y-residues at position: 98, 111, and 150)4 triggers apoptosis13, suggesting geminin oncogenic function requires simultaneous phosphorylation on all 3Ys, implicating the upstream kinases in this function13. Our intense search for these kinases revealed the non-receptor tyrosine kinase, c-Abl phosphorylates geminin-Y150 in vitro, and in vivo4,13,16. Indeed, imatinib and nilotinib (two c-Abl inhibitors) promote geminin-overexpressing cell death, in vitro, and geminin-overexpressing tumor regression, in vivo16. Immunohistochemical analysis of several large human breast tumor cohorts revealed geminin overexpression in ~50% of the tumors, while c-Abl overexpression in >90% of the tumors16. These analyses also revealed geminin-negative tumors (i.e., expressing normal tissue level), overexpress cytoplasmic c-Abl, while geminin-overexpressing tumors (especially TNBCs) overexpress nuclear c-Abl16. We now refer to geminin/nuclear c-Abl co-overexpressing tumors as “GemOE” tumors.
High-mobility group box 1 (HMGB1) is a ubiquitous DNA-binding protein with essential DNA metabolism functions17. HMGB1 can be released passively from necrotic cells or actively from activated immune cells, hypoxic, or inflamed cancer cells18. HMGB1 secretion requires hyper-acetylation on the chromatin10,19. Recently, we described how geminin helps acetylate chromatic HMGB1 and release it from GemOE tumor cells10, where it through binding to receptor for advanced glycation end products (RAGE) on GemOE tumor cells activates NF-κB-induced survival, especially those exposed to the harsh condition of hypoxia and inflammation10 within the tumor core (aka “aggressiveness niche”20). Binding of extracellular Ac-HMGB1 to RAGE on naïve mesenchymal stem cells (MSCs) activates NF-κB signaling-induced CXCR4 expression. CXCR4-expressing MSCs are then recruited to CXCL12/SDF1-secreting GemOE cells, in vitro, and into the aggressiveness niche, in vivo10,20.
We expand these data, here, and show MSCs activated by Ac-HMGB1 secrete the calcium-binding protein, S100A4 (aka metastasin)21–24, a known promoter of breast cancer proliferation, invasion, and metastasis24–26. In, TNBCs, expression of a nuclear/cytoplasmic S100A4 is associated with high histological tumor grade and inferior metastasis-free and overall survival24,27. We show S100A4 entrains GemOE cells to recruit macrophages into the aggressiveness niche and polarizes them to Gas6-secreting M2-TAMs. GemOE tumor cells overexpress the tyrosine kinase receptor, AXL, that binds Gas628. AXL is overexpressed in breast cancers29–32 (especially ERα-negative tumors29,33). Activation of AXL and RAGE in GemOE tumor cells converts them into metastatic precursors capable of dissemination from primary tumors through exacerbating the stemness and EMT phenotypes31 in them, and the expression and functional interaction of the intravasation-inducing CD151 and α3β1-integrin34.
Results
GemOE cells recruit and activate MSCs into S100A4-secreting CAFs
Extracellular Ac-HMGB1 activation of RAGE on naïve MSCs triggers CXCR4 expression and recruitment towards CXCL12-secreting GemOE cells10. To expand these data, normal HME, or two of the 1° orthotopic GemOE mammary tumors; Gem240, and Gem257 cells were grown (24 h) under normoxia (20% O2) or hypoxia (1% O2) in Dox-containing media in the presence or absence of imatinib4,16. ELISA revealed that compared to CM from cells expressing low-level geminin, induced Gem240 and Gem257 cells CM contained ~3-fold higher HMGB1 (Fig. 1A, white, and compare white to blue, Suppl. Fig. 1). Hypoxia did not affect normal HME or Dox-uninduced cells (Fig. 1, red, and compare blue and black, Suppl. Fig. 1), while exacerbated HMGB1 secretion from Dox-induced cells (Fig. 1A, red, and Suppl. Fig. 1). Imatinib blocked hypoxia-induced effects (compare black to red, Fig. 1A). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 2).
Figure 1.
GemOE cells recruit and activate MSCs. (A) The level of HMGB1 secreted from the indicated cell lines under normoxic or hypoxic conditions in the absence or presence of imatinib. Assay performed 3 separate times, each in triplicates. (B) The levels of RAGE and TLR4 in MSCs exposed to MSCs [−] or indicated cell lines CM for 24 h. The blot was repeated 3 separate times. (C) Real-time RT/PCR analysis of RAGE mRNA and CXCR4 mRNA in MSCs 24 h following exposure to Ac-rHMGB1 or CM from Dox-induced Gem240 or Gem257 cells supplemented with the vehicles, HMGB1 NeuAb, imatinib, TAK-242, glycyrrhizin, BAY 11 7082 or MK-2206. Assay performed 3 separate times, each in triplicates. (D) The effect of the indicated cells CM on the migration of MSCs performed for 24 h in Boyden chambers in the presence of the vehicle, HMGB1 or CXCL12 NeuAb. Assay performed 3 separate times, each in triplicates. (E) The levels of RAGE and TLR4 in the indicated cell lines exposed 24 h to normoxic (upper) or hypoxic (lower). The blot was repeated 3 separate times. (F) The level of S100A4 secreted from MSCs exposed 24 h to indicated cell lines CM under normoxic or hypoxic conditions in the absence or presence of HMGB1 NeuAb. Assay performed 3 separate times, each in triplicates. (G) Schematic representation showing the data discussed in the Figure.
Naïve MSCs (see [−], Fig. 1B) are RAGE-negative35,36, remain negative after exposure to HME cells CM (Fig. 1B). In contrast, exposure (24 h) to Dox-induced Gem197, Gem240, or Gem257 cells CM induced RAGE expression on MSCs surface (blot for membrane proteins, Fig. 1B). In contrast, whether naïve or exposed to CM from any of these cell lines, MSCs express equally high levels of TLR4 (a second HMGB1 receptor37,38) on their surface (Fig. 1B).
RNAs were isolated from naïve MSCs exposed (24 h) to Ac-rHMGB1 (10 µg/ml), or CM from Dox-induced Gem240 or Gem257 cells in the presence of vehicles (IgG isotype + DMSO), HMGB1 neutralizing antibody (NeuAb, 10 µg/ml), imatinib, TLR4 TAK-242, glycyrrhizin, Bay 11-7082, or MK-2206. RT/qPCR showed RAGE mRNA increases ~30fold in naïve MSCs exposed to Ac-rHMGB1, or CM from Dox-induced Gem240 or Gem257 cells (compare [−] to vehicles, white, Fig. 1C). All drugs except glycyrrhizin and MK-2206 blocked this increase (white bars, Fig. 1C). Moreover, previously, we showed that CXCR4 expression increases in naïve MSCs by Ac-HMGB1 downstream of RAGE10. CXCR4 mRNA level was also increased by ~30fold in naïve MSCs exposed to Ac-rHMGB1 or CM from Dox-induced Gem240 cells or Gem257 cells (compare [−] to vehicle, black bars, Fig. 1C). All drugs except MK-2206 blocked this increase in CXCR4 mRNA expression (black bars, Fig. 1C). Together reinforce our previous published data10 and expand them to show Ac-HMGB1 activates TLR4 first to trigger RAGE expression, which then also through activation by Ac-HMGB1 triggers CXCR4 expression. Noteworthy, Ac-HMGB1 in both situations activated NF-κB rather than AKT signaling in MSCs10.
Naïve MSCs layered in inserts of Boyden chambers efficiently migrated towards Dox-induced Gem197, Gem240, or Gem257 cells CM not MSCs CM or HME cells CM added in the lower chamber within 24 h (Fig. 1D). The recruitment was almost completely blocked in the presence of HMGB1 or CXCL12 NeuAbs (Fig. 1D). Together, reinforce that a primary function for RAGE/CXCR4 activation in naïve MSCs is to promote their migration towards GemOE tumor cells (step 1, Fig. 1G).
Unlike naïve MSCs (Fig. 1B), HME and Dox-induced GemOE cells express no TLR4 while expressing equally high-levels of RAGE on their surface (Fig. 1E, upper). Hypoxia did not alter this pattern (Fig. 1E, lower). RAGE also binds the S100 family members23,24,27,39. Since RAGE is constitutively expressed on GemOE cells, we evaluated whether activated MSCs express/release HMGB1 or S100 proteins.
RNAs were isolated from naïve MSCs exposed (24 h) to naïve MSCs CM, normal HME cells CM, Dox-induced Gem240, or Gem257 cells CM. RT/qPCR showed that only exposure to Dox-induced Gem240 or Gem257 cells CM induced expression of S100A2, 4, 6, 8, and 9, not S100A1, 7, or HMGB1 mRNAs in naïve MSCs (Suppl. Fig. 3A). Because of S100A4 role in breast cancer metastasis23–26,40, we pursued it further. We found that HMGB1 NeuAb blocked the elevation of S100A4 mRNAs expression in MSCs following exposure to Dox-induced Gem240 or Gem257 cells CM (Suppl. Fig. 3B).
Moreover, ELISA showed that neither normal HME cells nor Dox-induced GemOE cells secrete S100A4 (not shown). In contrast, naïve MSCs exposed (24 h) to Dox-induced Gem240 or Gem257 cells CM only secrete high-levels of S100A4 (Fig. 1F, white bars). This secretion increased further when hypoxic Dox-induced Gem240 or Gem257 cells CM was used instead (Fig. 1F, red) and almost completely blocked by HMGB1 NeuAb (Fig. 1F, black). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 4). Together suggest that in the vicinity of GemOE cells, MSCs differentiate into S100A4-secreting cancer-associated fibroblasts10,41 (i.e., CAFs, Fig. 1G, step 2).
S100A4-activated GemOE cells attract and polarize macrophages into Gas6-secreting M2-TAMs
Recently, we showed that IRISOE cells secrete high-levels of CCL2, in vitro, and in vivo to recruit macrophages42. To define whether bidirectional interaction between GemOE cells and MSCs through S100A4 exists was studied next.
CM from normal HME cells or Dox-induced Gem240 or Gem257 cells exposed to normoxia or hypoxia (24 h) re-conditioned by exposure to MSCs (another 24 h, Fig. 2A1) were re-added to the same cell line in the absence or presence of S100A4 NeuAb or glycyrrhizin (24 h, Fig. 2A2). ELISA showed MSCs (Fig. 2B, blue), and normal HME cells exposed to their normoxic or hypoxic CM re-conditioned by MSCs (Fig. 2B, orange and red, respectively) or not (Fig. 2B, white) did not secrete CCL2. In contrast, Dox-induced Gem240 or Gem257 cells secreted low-levels of CCL2 when exposed to their CM (Fig. 2B, white), slightly increased following exposure to their normoxic CM re-conditioned by MSCs (Fig. 2B, orange), and increased even further when exposed to their hypoxic CM reconditioned by MSCs (Fig. 2B, red). Importantly, S100A4 NeuAb (Fig. 2B, green) and glycyrrhizin (Fig. 2B, black) both blocked these increases. One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 5).
Figure 2.
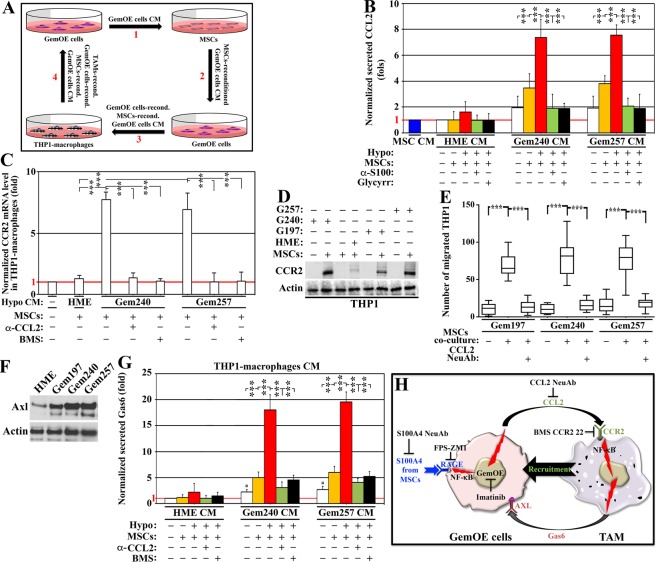
MSCs-reactivated GemOE cells recruit and activate macrophages. (A) Schematic representation of the co-culture experiments performed in this study. (B) The level of CCL2 secreted from the indicated cell lines under normoxic or hypoxic condition co-cultures with MSCs in the absence or presence of S100A4 NeuAb or glycyrrhizin. Assay performed 3 separate times, each in triplicates. (C) Real-time RT/PCR analysis of the CCR2 mRNA in THP1-macrophages exposed 24 h to the co-cultures of the indicated cell lines with MSCs supplemented with CCL2 NeuAb or BMS CCR2 22. Assay performed 3 separate times, each in triplicates. (D) The expression of CCR2 protein in the indicated cell lines alone or co-cultured with MSC for 24 h. The blot was repeated 3 separate times. (E) The effect of the CM from the indicated co-cultures with MSCs for 24 on the migration of THP1-macrophages performed for 24 h in Boyden chambers in the presence of vehicle or CCL2 NeuAb. Assay performed 3 separate times, each in triplicates. (F) The levels of AXL in the indicated cells. The blot was repeated 3 separate times. (G) The level of Gas6 secreted from THP1-macrophages exposed 24 h to the indicated co-cultures generated under normoxic or hypoxic conditions in the absence or presence of CCL2 NeuAb or BMS CC2 22. Assay performed 3 separate times, each in triplicates. (H) Schematic representation showing the data discussed in the Figure.
The receptor for CCL2, CCR2, controls the trafficking of monocytes into damaged tissues or tumors42,43. Recently, we showed CCL2 upregulates CCR2 expression on unpolarized macrophages42. PMA-treated THP1-macrophages (hereafter THP1s) were exposed (24 h) to HME cells/or Dox-induced Gem240 or Gem257 cells/MSCs co-cultures CM (Fig. 2A3). Real-time RT/qPCR and western blot analysis of these THP1s showed low-levels CCR2 mRNA in unexposed THP1s ([−], Fig. 2C). While MSCs only CM did not induce CCR2 mRNA and protein expression, a slight increase was detected in THP1s exposed to hypoxic HME cells/MSCs co-culture CM (Fig. 2C,D). In contrast, high-levels CCR2 mRNA was detected in THP1s exposed to hypoxic Dox-induced Gem240 or Gem257 cells/MSCs co-culture CM (Fig. 2C). This induction was significantly blocked by CCL2 NeuAb and BMS CCR2 22 (Fig. 2C). Importantly, high-levels CCR2 protein expression was detected in THP1s exposed to Dox-induced Gem197, Gem240, or Gem257 cells/MSCs co-culture CM only (Fig. 2D). Taken together, confirming the role of CCL2 in CCR2 expression in THP1s and primary macrophages44,45.
Many THP1s layered in inserts of Boyden chambers migrated 24 h later towards Dox-induced Gem197, Gem240, or Gem257 cells/MSCs co-cultures CM only in the lower chambers. Importantly, in the presence of CCL2 NeuAb, this migration was significantly blocked (Fig. 2E), supporting the CCL2/CCR2 role in macrophages recruitment into GemOE tumors.
Our analysis of receptors upregulated on the surface of the 1° orthotopic GemOE mammary tumors revealed many important metastasis inducing receptors. One such receptor, AXL, was chosen for further analysis in this study due to its pronounced role in intravasation46 and breast cancer metastasis30. Compared to normal HME cells, Dox-induced Gem197, Gem240, and Gem257 cells express significantly higher levels of AXL on their surfaces (western done on cell membrane extracts, Fig. 2F).
The ligand for AXL; Gas6 also has a notable role in breast cancer progression28. Interestingly, compared to normal HME cells CM, Dox-induced Gem240, or Gem257 cells alone CM induced low-levels Gas6 secretion from THP1s (see Fig. 2A4,G, white). This secretion was significantly increased when THP1s were instead exposed to normoxic Dox-induced Gem240 or Gem257 cells/MSCs co-culture CM (Fig. 2G, orange). Even higher secretion was detected when hypoxic Dox-induced Gem240 or Gem257 cells/MSCs co-culture CM (Fig. 2G, red). Importantly, CCL2 NeuAb and BMS CCR2 22 significantly blocked Gas6 secretion from THP1s exposed to the hypoxic co-cultures CM (Fig. 2G, green and black, respectively). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 6). Together suggest S100A4 re-activate GemOE cells to secrete CCL2 that attracts macrophages and polarizes them into Gas6-secreting M2-tumor-associated macrophages (M2-TAMs42,47,48, Fig. 2H).
Imatinib-sensitive MSCs and TAMs recruitment in vivo
Previously, we injected shCtrl-, sh-geminin- or shc-Abl-expressing GemOE cells into athymic female mice mammary fat pads. Geminin- or c-Abl-depleted cells developed <20% size tumors, compared to control cells16, supporting the intimate relationship between c-Abl and geminin and giving credence to using imatinib to treat GemOE mammary tumors (especially TNBC) see introduction and16.
We injected 4 × 106 Gem240 or Gem257 cells in athymic female mice (n = 24/cell line, Fig. 3A), and kept the mice on Dox-supplemented drinking water. Mice developed ~0.5 cm3 tumors within 4weeks, at which time (i.e., day -1), mice were randomized into two groups: one received vehicle (DMSO, n = 12/cell line), and the other 10 mg/kg of imatinib (orally, n = 12/cell line, Fig. 3A). We activated GFP-human MSCs (GFP-hMSCs) by culturing for a week in CM from Dox-induced Gem240 or Gem257 cells (changed daily), and GFP-THP1s by culturing in Dox-induced Gem240 or Gem257 cells/MSCs co-cultures CM (also changed daily). On day (0), 4 × 105 of activated GFP-hMSCs or GFP-THP1s were intracardiac (i.c., through the left ventricle) injected in vehicle (n = 6/cell line) or imatinib (n = 6/cell line) treated mice and treatments were administrated at that time and daily thereafter on days 1, 2, and 3 (Fig. 3A). Tumors and peripheral blood (PB) were collected from all mice on day 4 (Fig. 3A).
Figure 3.
GemOE tumor cells recruit and interact with MSCs and TAMs, in vivo. (A) Schematic representation of the in vivo assay discussed in the Figure. Representative images of GFP-MSCs (B1) or GFP-THP1-macrophages (B2) found in GemOE tumors treated in vivo with vehicle (lefts) or imatinib (rights) according to the protocol in (A). The tumors were dissociated into single cells and cultured overnight before photographing. The numbers of GFP-MSCs (C1) or GFP-THP1-macrophages (C2) found in GemOE tumors treated in vivo with vehicle (blacks) or imatinib (red and organ) according to the protocol in (A). The tumors were dissociated into single cells and cultured overnight before counting. The levels of HMGB1 (D), CXCL12 (E), and S100A4 (F) in sera isolated from PBs of naïve mice (before tumor cells injection), tumor-bearing mice before or after vehicle or imatinib treatments. (G) Kaplan Meir curve of the recurrence-free survival in indicated numbers of patients overexpressing geminin + HMGB1 + S100A4 vs. low expressing patients.
Part of each tumor was dissociated into single cells and grew in culture overnight. Next day GFP+-cells were counted in at least five high magnification fields (HF) from at least three tumors treated with vehicle or imatinib, and data for both cell lines were pooled (similar results). We detected 10.1 ± 4.2 GFP-hMSCs/HF in cultures from vehicle-treated tumors (Fig. 3B1, left, and 3C1, black) vs. 1.2 ± 1.4 in cultures from imatinib-treated tumors (Fig. 3B1, right and 3C1, red, p = 1 × 10−6). Similarly, there was 11.1 ± 6.4 GFP-THP1s/HF in cultures from vehicle-treated tumors (Fig. 3B2, left and 3C2, black) vs. 1.2 ± 1.6 in cultures from imatinib-treated tumors (Fig. 3B2, right, and 3C2, orange, p = 1 × 10−6).
ELISA performed on sera isolated from the PBs showed (here too, data for similar treated mice were pooled, similar results) that consistent with others49–51 and our previous10,16 data, HMGB1 level was 14.9 ± 7.4 ng/ml in sera from naïve mice (n = 48, Fig. 3D, yellow), increased to 154.0 ± 36.3 ng/ml in sera from tumor-bearing mice (n = 48, collected on day (−1), p < 1 × 10−6, Fig. 3D, blue), remained high at 155.2 ± 42.9 ng/ml in sera from vehicle-treated mice (n = 24, collected at euthanasia, p < 1 × 10−6, Fig. 3D, red), dropped to 24.8 ± 12.5 ng/ml in sera from imatinib-treated mice (n = 24, p < 1 × 10−6, Fig. 3D, brown). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 7).
Similarly, CXCL12 level was 1.8 ± 9.7 ng/ml in sera from naïve mice (Fig. 3E, yellow), increased to 18.4 ± 6.4 ng/ml in tumor-bearing mice (p < 1 × 10−6, Fig. 3E, blue), remained high at 17.1 ± 6.3 ng/ml in vehicle-treated mice (p < 1 × 10−6, Fig. 3E, red), dropped to 4.1 ± 2.1 ng/ml in imatinib-treated mice (p < 1 × 10−6, Fig. 3E, brown). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 8).
Finally, S100A4 level was 1.4 ± 0.8 ng/ml in sera from naïve mice (Fig. 3F, yellow), increased to 15.5 ± 3.7 ng/ml in tumor-bearing mice (p < 1 × 10−6, Fig. 3F, blue), remained high at 16.3 ± 5.1 ng/ml in vehicle-treated mice (p < 1 × 10−6, Fig. 3F, red), dropped to 6.3 ± 2.7 ng/ml in imatinib-treated mice (p < 1 × 10−6, Fig. 3F, brown). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 9). Interestingly, analysis of TCGA data confirmed that compared to low geminin + HMGB1 + S100A4-expressing patients, patients expressing high levels had an inferior recurrence-free survival (RFS, p = 0.007, compare red to black, Fig. 3G).
Further evidence for the recruitment of MSCs and TAMs into GemOE tumors
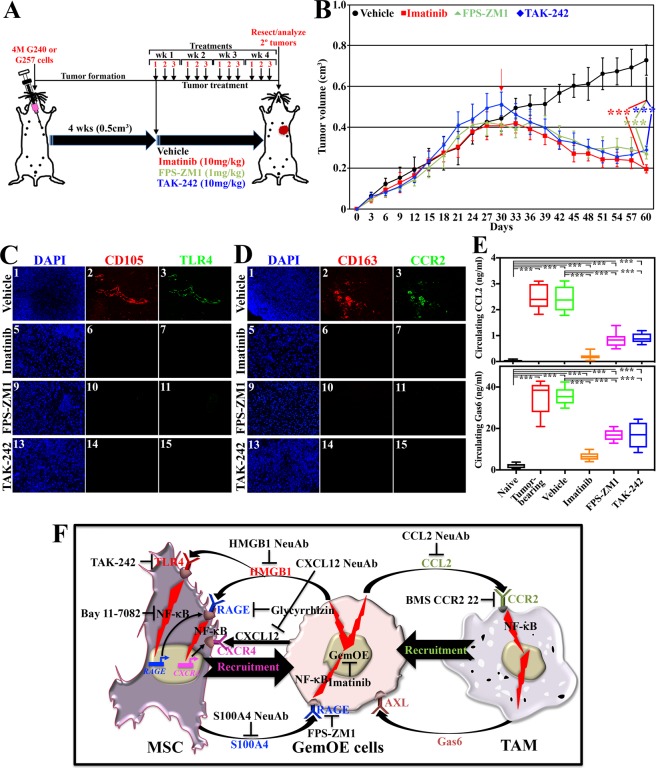
Again, based on our previous experience with imatinib16, and because our model specifically calls for macrophages recruitment as a subsequent step for the MSCs recruitment (Fig. 2H), we aimed at further analyze these interactions through inhibiting the two receptors; RAGE and TLR4 we propose mostly involved in MSCs recruitment. We injected 4 × 106 Gem240, or Gem257 cells in athymic female mice (n = 24/cell line, see Fig. 4A) kept on Dox-supplemented drinking water. The mice developed ~0.5 cm3 tumor volume within 4weeks, at which time they were randomized into 4 groups received vehicle (DMSO, n = 6), imatinib (10 mg/kg, n = 6), FPS-ZM152 (10 mg/kg, intraperitoneally [i.p.], 1 mg/kg, n = 6), or TAK-24253 (10 mg/kg, i.p. n = 6). Drugs were administered 3times/week for 4weeks (Fig. 4A).
Figure 4.
Bi-directional interaction of GemOE cells with stromal cells in GemOE tumors. (A) Schematic representation of the in vivo assay discussed in the Figure. (B) Gem240 tumor volume (Gem257 cells produced identical data) following the protocol described in (A) in the presence of the indicated inhibitors. The red arrow indicates the start of treatments. Student t-test was used to compare vehicle-treated tumors and each treatment separately. Fluorescence IHC staining of sections from the tumors above using mouse-specific anti-CD105 (MSCs specific biomarker) and anti-TLR4 (C), or mouse-specific anti-CD163 (macrophage-specific biomarker) and anti-CCR2 (D). (E) The level of CCL2 (upper) and Gas6 (lower) in naïve mice, tumor-bearing mice before or after vehicle, imatinib, FPS-ZM1, and TAK-242 treatments at tumor resection. (F) Schematic representation showing the data discussed so far.
Vehicle-treated Gem240 (identical results were obtained using Gem257) tumors grow ~200% by day 60 (Fig. 4B, black), while imatinib (Fig. 4B, red), FPS-ZM1 (Fig. 4B, green) or TAK-242 (Fig. 4B, blue) treated tumors regressed by ~50% (p < 1 × 10−6 for all drugs, Fig. 4B). All tumors were resected, paraffin-embedded, sectioned at 4 µm, and immune-stained. Vehicle-treated tumors only showed high-levels CD105 (mouse MSCs fibrogenic differentiation biomarker54)/TLR4 co-stained cells (Fig. 4C1–3), and CD163 (M2-macrophage-specific biomarker55)/CCR2 co-stained cells (Fig. 4D1–3). Imatinib (Fig. 4C5–7,D5–7), FPS-ZM1 (Fig. 4C9–11,D9–11), or TAK-242 (Fig. 4C13–15,D13–15) treated mice were negative for both cells.
Finally, sera were isolated from PBs from these mice at euthanasia. ELISA (data from both cell lines were pooled) showed 0.052 ± 0.030 ng/ml of CCL2 in naïve mice sera (n = 48, Fig. 4E, upper-black), increased to 2.358 ± 0.524 ng/ml in tumor-bearing mice sera (n = 48, p < 1 × 10−6, Fig. 4E, upper-red), remained high at 2.418 ± 0.512 ng/ml in vehicle-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, upper-green). The level dropped to 0.205 ± 0.119 ng/ml in imatinib-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, upper-orange), to 0.756 ± 0.269 ng/ml in FPS-treated mice (n = 12, p < 1 × 10−6, Fig. 4E, upper-pink), and to 0.962 ± 0.182 ng/ml in TAK-242-treated mice (n = 12, p < 1 × 10−6, Fig. 4E, upper-blue). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 10).
Similarly, 1.865 ± 0.999 ng/ml of Gas6 was detected in naïve mice sera (n = 48, Fig. 4E, lower-black), increased to 33.534 ± 6.738 ng/ml in tumor-bearing mice sera (n = 48, p < 1 × 10−6, Fig. 4E, lower-red), remained high at 35.505 ± 4.378 in the vehicle-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, lower-green). The level dropped to 7.073 ± 1.857 ng/ml in imatinib-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, lower-pink), to 9.842 ± 2.256 ng/ml in FPS-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, lower-pink) and to 10.539 ± 2.772 ng/ml in TAK-242-treated mice sera (n = 12, p < 1 × 10−6, Fig. 4E, lower-blue). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 11). Together, suggest blocking HMGB1 secretion or function prevents recruitment of MSCs and TAMs into GemOE tumors10 and their conversion into pro-tumor S100A4-secreting CAFs and Gas6-secreting M2-TAMs (Fig. 4F) leading to tumor regerssion56,57.
The bi-directional loops are involved in GemOE-induced TNBC metastasis
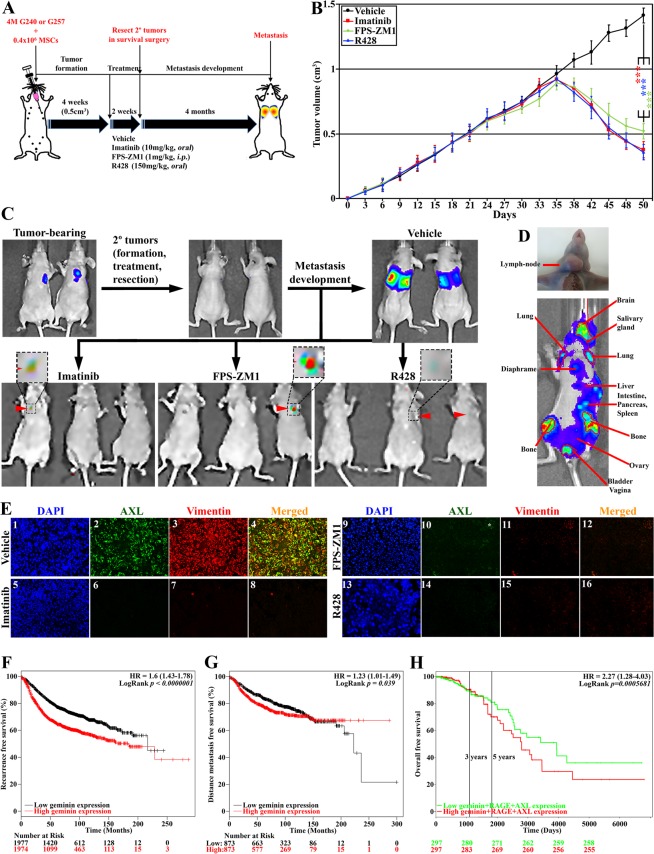
To evaluate HMGB1/S100A4 and CCL2/Gas6 bi-directional loops role in GemOE cells metastatic potential (cf. Fig. 4F), we injected 4 × 106 Gem240 or Gem257 cells admixed with 4 × 105 hMSCs in female athymic mice (48mice/cell line, Fig. 5A) kept on Dox-supplemented drinking water. The mice developed ~0.5 cm3 tumor volume within 4weeks (Fig. 5A), at which time they were randomized into 4 groups/cell line treated with vehicle (DMSO, n = 12), imatinib (n = 12, 10 mg/kg, orally), FPS-ZM1 (n = 12, 1 mg/kg, i.p.), or R428 (n = 12, 150 mg/kg, orally) daily for 2 weeks (Fig. 5A). Half of the mice were maintained for tumor growth (Fig. 5B). In the other half, tumors were resected in survival surgeries, and metastasis was followed using IVIS for a maximum of 4months (Fig. 5A).
Figure 5.
The bi-directional pathways elevate the GemOE tumor metastasis. (A) Schematic representation of the in vivo assay discussed in the Figure. (B) The volume of orthotopic mammary GemOE tumors generated using Gem240 cells. At ~0.5 cm3 around week 4, mice were randomized and divided into 4 groups treated with vehicle, imatinib, FPS-ZM1, or R428 daily for 2 weeks (weekend off). Refer to the Results section for more information. Student t-test was used to compare vehicle-treated tumors and each treatment separately. (C) Representative images of mice bearing tumors before (upper left), after (upper middle) tumors resection, and macro-metastasis developed within 4 months following vehicle (upper right), not imatinib (lower left), FPS-ZM1 (Lower middle) and R428 (lower right) treatments. Insets show micro-metastasis in the lungs in the treated mice. (D) Lymph-node (upper) and widespread (lower) metastases in GemOE tumor-bearing mice. (E) Fluorescence IHC staining of sections from the tumors above using anti-AXL and anti-vimentin. Kaplan Meir curves of the recurrence-free survival (F) or distant-metastasis-free survival (G) of geminin overexpressing patients vs. geminin low expressing patients. (H) Kaplan Meir curve of the overall survival of geminin + RAGE + AXL overexpressing patients vs. low expressing patients.
Tumors in vehicle-treated mice increased ~50% within the treatment period (Fig. 5B, black). In contrast, tumors in mice treated with imatinib (Fig. 5B, red), FPS-ZM1 (Fig. 5B, green), or R428 (Fig. 5B, blue) regressed by >50%.
Only in vehicle-treated mice, we detected macro-metastases, especially to the lungs (n = 12, i.e., 6/cell line, Fig. 5C, upper right). Noteworthy, one mouse also developed a lymph-node metastasis (Fig. 5D, upper), and another developed widespread metastasis in almost all organs (Fig. 5D, lower). In contrast, none of the mice injected with either cell line developed metastasis following imatinib (Fig. 5C, lower left), FPS-ZM1 (Fig. 5C, lower middle), or R428 (Fig. 5C, lower right) treatment, although few mice developed very small micro-metastases (arrowheads and insets in Fig. 5C, lower). Detail analysis of these metastases will be reported elsewhere. Importantly, only in vehicle-treated mice, we detected AXL+/vimentin+ cells in the tumors (compare Fig. 5E1–4 to E5–8,E9–12,E13–16).
To support these data further, we carried out Kaplan Meier analysis using several publicly available breast cancer data sets58–60. Geminin-overexpressing patients had inferior recurrence-free survival (RFS, n = 3955, HR = 1.6, 95% CI = 1.43–1.78, p < 1 × 10−6, Fig. 5F) and distance metastasis-free survival (DMFS, n = 1747, HR = 1.23, 95% CI = 1.01–1.49, p = 0.039, Fig. 5G). Geminin + RAGE + AXL-overexpressing patients had inferior overall survival (OS, n = 594, HR = 2.27, 95% CI = 1.28–4.03, p = 0.0005681, Fig. 5H), and geminin + HMGB1 + RAGE + CCL2 + AXL-overexpressing patients had inferior lung metastasis-free survival (LMFS, n = 58, HR = 7.35, 95% CI = 2.38–22.63, p = 0.0005123, Suppl. Fig. 12). Together, confirm that positive feedback loops with MSCs/CAFs and/or M2-TAMs, especially within the aggressiveness niche through RAGE and AXL, respectively (cf. Fig. 4F) worsen the outcomes for patients with GemOE/TNBC tumors.
AXL and RAGE enhance GemOE cell invasion ability
HME, Gem197, Gem240, and Gem257 cells were switched to Dox-containing serum-free (SF)-media for 24 h before they were treated in Dox-containing SF-media with none or S100A4 + Gas6 plus vehicle, FPS-ZM1, R428, or both for 30 mins or 24 h.
Following 30 min treatments, the membrane/cytoplasmic fractions were isolated. HME cells express no/low-levels of the total (T-)AXL (see below), p-AXLY779, T-AKT, p-AKTT308/S473, T-ERK, p-ERKT183/Y185, NF-κB and survivin under all conditions (Fig. 6A). In contrast, in SF-media, Dox-induced Gem197, Gem240, or Gem257 cells express >5-fold higher T-AXL (see below), T-AKT, NF-κB, and survivin, and similar levels of T-ERK compared to HME cells (Fig. 6A). Treatment with FPS-ZM1, R428, or both did not significantly affect the expression of total proteins (Fig. 6A). Although the levels of p-AXLY779, p-AKTT308/S473, and p-ERKT183/Y185 were not significantly affected by S100A4 + Gas6 plus vehicle treatment, FPS-ZM1, R428 and even more so in both treatments significantly decreased the levels of these proteins (Fig. 6A), suggesting cross-talk between AXL and RAGE that activates AXL, AKT, and to some extent, ERK signaling in GemOE cells (cf. Fig. 6H).
Figure 6.
The roles of RAGE and AXL in promoting invasion in GemOE tumor cells. The expression of the indicated proteins in the indicated cell lines following 24 h of growth in Dox-containing SF-media followed by a switch to Dox-containing SF-media supplemented with none, or S100A4 + Gas6 plus vehicle, FPS-ZM1, R428, or FPS-ZM1 + R428 for 30 mins (A) or 24 h (B). (C) Representative images of the invasion of Dox-induced Gem240 or Gem257 cells through Matrigel-coated inserts of Boyden chambers 24 h after incubation with SF-media supplemented with none, or S100A4 + Gas6 plus vehicle, plus FPS-ZM1, plus R428, or plus FPS-ZM1 + R428. (D) Quantitative analysis of the data presented in (C). Assay performed 3 separate times, each in triplicates. (E) The expression of geminin and AXL in the indicated cell lines following 24 h growth in Dox-containing SF-media followed by switching to Dox-containing SF-media supplemented with none or S100A4 with or without FPS-ZM1. (F) The expression of geminin and AXL in the indicated cell lines following 24 h growth in Dox-containing SF- media followed by switching to Dox-containing SF-media supplemented with none or Gas6 with or without R428. (G) The expression of MMP2 and MMP9 (pro and active forms) in the indicated cell lines following 24 h growth in Dox-containing SF-media followed by switching to Dox-containing SF-media supplemented with none, or S100A4 + Gas plus vehicle, plus FPS-ZM1, plus R428, or plus FPS-ZM1 + R428 for 24 h. (H) Schematic representation showing the data discussed in the Figure.
Following the 24 h treatments, cells were sonicated, and whole-cell extracts prepared. HME cells grown in SF-medium expressed low levels of geminin, CDH2, and survivin, while detectable levels of Twist1, Slug, Oct4, and Sox2 (Fig. 6B). In contrast, in SF-media, all Dox-induced GemOE cells expressed high levels of geminin, CDH2, and survivin, and even higher levels of Twist1, Slug, Oct4, and Sox2 compared to HME cells (Fig. 6B). None of the treatments significantly affected the expression of these proteins in HME cells (Fig. 6B), while in SF-media, S100A4 + Gas6 plus vehicle elevated the expression of all proteins to different degrees in the three Dox-induced GemOE cell lines (Fig. 6B). In SF-media, S100A4 + Gas6 plus FPS-ZM1 significantly decreased the expression of the majority of the proteins except for geminin to different degrees in the three Dox-induced GemOE cell lines (Fig. 6B), plus R428 treatment decreased expression of all proteins in some instances marginally, e.g., Twist and Sox2, yet in others significantly, e.g., Slug and Oct4 (Fig. 6B). In SF-media, S100A4 + Gas6 plus both drugs treatment, decreased the expression of all proteins even geminin to a level similar to that observed with R428 alone (Fig. 6B), suggesting that RAGE perhaps primes AXL effects.
Next, we layered Gem240 or Gem257 cells in Matrigel-coated inserts of Boyden chambers. In the lower well, we added SF-media containing Dox and none or S100A4 + Gas6 plus vehicle, FPS-ZM1, R428, or both. After 24 h, we counted cells invaded the Matrigel and migrated to the other side of the inserts as an in vitro confirmation of their stemness and EMT phenotype described above.
Although, many Dox-induced Gem240 and Gem257 cells invaded in SF-condition (Fig. 6C,D), a significant increase in the number of invaded cells was detected in the presence of S100A4 + Gas6 plus vehicle (Fig. 6C,D). On the other hand, in the presence of FPS-ZM1 or R428, S100A + Gas6 effect on invasion was significantly blocked (Fig. 6C,D). Again, the reduction in invasion was the same in the presence of both drugs or R428 alone (Fig. 6C,D). One-way ANOVA test followed by post hoc Bonferroni tests confirmed these data (Suppl. Figs. 13 and 14). Together, reinforces that RAGE activation perhaps primes GemOE/TNBC cells to Gas6 effects by affecting geminin and/or AXL expression and/or activity.
To test this hypothesis, we grow HME, Gem197, Gem240, or Gem257 in Dox-containing SF-media for 24 h then switched them to Dox-containing SF-media supplemented with S100A4 plus vehicle or FPS-ZM1 (Fig. 6E) or supplemented with Gas6 plus vehicle or R428 (Fig. 6F). First, in SF-media, S100A4 elevated geminin and AXL levels in all Dox-induced GemOE (not HME) cells (compare green to red in Fig. 6E). Importantly, the expression of geminin and AXL proteins were significantly decreased in the presence of S100A4 plus FPS-ZM1 (compare blue to green in Fig. 6E). It is possible that S100A4/RAGE signaling stabilizes geminin protein (exogenous geminin is expressed from a heterologous promoter) and/or induces its transcription. Geminin, then could affect AXL gene transcription and/or protein stabilization.
Second, in SF-media, Gas6 did not affect geminin expression, while increased AXL expression in all Dox-induced GemOE cells (compare green to red in Fig. 6F). R428 did not affect geminin expression in Dox-induced GemOE cells but inhibited AXL expression (compare blue to green in Fig. 6F). It is possible that S100A4/RAGE signaling separately affects geminin or AXL. Alternatively, positive feedback between geminin and AXL is also possible. We favor the latter since only in Dox-induced GemOE cells, AXL was expressed (Fig. 6E,F).
Finally, in SF-media, Dox-induced Gem240 and Gem257 cells express relatively high levels of pro-/active-MMP2 and -MMP9. Expression of both significantly increased in the presence of S100A4 + Gas6 plus vehicle (Fig. 6G) and was blocked in the presence of FPS-ZM1, R428, or both drugs (Fig. 6G). The bidirectional interactions with MSCs/CAFs and M2-TAMs through S100A4/RAGE and Gas6/AXL signaling that activated AKT, ERK, and NF-κB in GemOE cells elevate geminin and/or AXL expression triggering TNBC cells stemness, EMT, and their invasion ability (cf. Fig. 6H).
AXL and RAGE enhance GemOE cells’ intravasation ability
The integrin α3 and β1 (hereafter α3β1-integrin) complex with CD151 (expressed specifically on intravasating tumor cells) is essential to initiate and maintains a tight interaction with the extracellular matrices (ECMs); e.g., laminin I (LMN I), collagen IV (COL IV) and fibronectin (FN) on the basal side of endothelial cells of the vessel61–63. To study this in our system, we grew Gem240 or Gem257 for 24 h on uncoated wells or wells that were coated with LMN I, COL IV, or FN in SF-media containing Dox and supplemented with none or S100A4 + Gas6 plus vehicle, FPS-ZM1, R428 or both. The next day, wells were washed 3 times, and the cells remaining attached to each well were counted in HF and blotted.
The Gem240 cells (identical results were obtained with Gem257 cells, not shown) showed a high binding ability to uncoated wells under all conditions (white, Fig. 7A). In comparison, binding to LMN I, COL IV, and FN was significantly decreased under SF conditions (none, Fig. 7A). The binding to all matrices was restored in SF containing S100A4 + Gas6 plus vehicle (Fig. 7A), which was blocked by FPS-ZM1, R428, or both treatments (Fig. 7A). One-way ANOVA, followed by post hoc Bonferroni tests, confirmed these data (Suppl. Fig. 15). Together, suggest that AXL and/or RAGE positively affect the binding of intravasating tumor cells with the ECM on the vessels within GemOE tumors.
Figure 7.
The roles of RAGE and AXL in promoting intravasation in GemOE tumor cells. (A) The number of Gem240 cells (Gem257 cells showed identical results) attached to non-coated or LMN I-, COL IV-, or FN-coated wells 24 h after platting followed by washing and counting of attached cells in at least 4 HPFs. Each assay was performed 3 separate times, each in triplicates. (B) The expression of the indicated proteins in the indicated cell lines following 24 h growth in Dox-containing SF-media followed by a switch to Dox-containing SF-media supplemented with none, or Gas6 plus vehicle, or plus R428 for 24 h. Note, the actin blot is the same as in Fig. 6F because it is from the same experiment. Percentage of AXL+/β1-integrin+ populations (red), the AXL+/CD151+ populations (white), or β1-integrin+/CD151+ populations (black) derived from FACS analysis of non-permeabilized Dox-induced Gem197 (C), Gem240 (D), and Gem257 (E) treated as indicated. Each assay was performed 3 separate times, each in triplicates. (F) Schematic representation of the data in the entire study as well as future directions.
To study the contribution of CD151 and α3β1 in this binding, we again in SF-media grow normal HME cells, or Dox-induced Gem197, Gem240, or Gem257 cells in the presence of none, or Gas6 plus vehicle or plus R428. HME expressed very low levels of α3-integrin, β1-integrin, while a high level of β4-integrin (β4) and no CD151 (Fig. 7B). In contrast, in SF-media, all Dox-induced GemOE cell lines expressed high levels of α3-integrin and β1-integrin, low levels of β4-integrin, and very high levels of CD151 (red in Fig. 7B). Gas6 treatment did not affect the expression of these proteins in normal HME cells, but significantly increased expression of α3-integrin, β1-integrin and CD151 in all Dox-induced GemOE cells by >2fold (compare green to red in Fig. 7B). Importantly, inactivating AXL significantly blocked Gas6-induced expression of all four proteins in all Dox-induced GemOE cell lines (compare blue to green in Fig. 7B), suggesting a direct role for AXL in GemOE intravasation ability.
To establish this further, Gem197, Gem240, and Gem257 cells (HME cells were omitted from this analysis since they did not express CD151) grown in Dox-containing SF-media supplemented with none or A4 + Gas6 plus vehicle, FPS-ZM1, R428 or both (24 h). Non-permeabilized cells were first stained with FITC-anti-CD44, PE-anti-CD24. The stem-like (i.e., CD44+/CD24−) population within each cell line was FACS sorted out and stained with PE-Cy7-anti-AXL, 561 620_20-anti-β1-integrin, and APC-anti-CD151 for FACS analysis.
AXL+/β1+ cell populations
All cell lines contained ~40% of AXL+/β1+ cell populations in SF-media (red, Fig. 7C–E), that didn’t change in the presence of S100A4 + Gas6 plus vehicle in any of the cell lines (red, Fig. 7C–E). Although, S100A4 + Gas6 plus FPS-ZM1 or plus R428 didn’t change this population in any of the cell lines, in S100A4 + Gas6 plus both a slightly yet significantly decreased in the AXL+/β1+-populations in all cell lines was detected (red, Fig. 7C–E).
AXL+/CD151+ cell populations
All cell lines contained >60% of AXL+/CD151+ cell populations in SF-media (white, Fig. 7C–E), that didn’t change in S100A4 + Gas6 plus vehicle, plus FPS-ZM1 or plus R428 treatments (white, Fig. 7C–E). Importantly, a significant decrease in the percentage of the AXL+/CD151+ population was observed in all cell lines following S100A4 + Gas6 plus both drug treatment (Fig. 7C–E).
β1-integrin+/CD151+ cell populations
All cell lines contained 25–40% of β1+/CD151+ cell populations in SF-media (black, Fig. 7C–E). Again, this percentage was not significantly changed in S100A4 + Gas6 plus vehicle, plus FPS-ZM1 or plus R428 treatments (black, Fig. 7C–E). Importantly, a significant decrease in the percentage of this β1+/CD151+ cell populations was observed in all cell lines following treatment with S100A4 + Gas6 plus both drugs (black, Fig. 7C–E). Taken together, we propose that GemOE triggers expression and/or activation of AXL and RAGE through interactions with the microenvironment that, in turn, enhances the expression and the functional interaction between α3β1-integrin and CD151. This complex by tightly binding to the ECM on the tumor side of endothelial cells enhances the intravasation ability of GemOE/TNBC metastatic precursors through the endothelium barrier upon dissemination (cf. Fig. 7F).
Discussion
The vast majority of breast cancer deaths are due to metastatic diseases. Inflammation and hypoxia within the tumor microenvironment, especially the core (i.e., the aggressiveness niche20) exacerbate metastasis. Here, we elucidated the mechanistic role of the bi-directional interactions between GemOE tumor cells and the microenvironment in promoting GemOE metastatic precursors invasion and intravasation abilities.
MSCs and TAMs are recruited into tumors through tumor secreted factors. MSCs are recruited by Ac-HMGB1/CXCL1227,64, while TAMs by CCL231,65. We resolved an issue that was unclear in our previous study10, how Ac-HMGB1 induced CXCR4 expression through RAGE in the RAGE-negative naïve MSCs? We found that Ac-HMGB1 activates TLR4 first, which then upregulates RAGE expression in naïve MSCs66–68 (cf. Fig. 1G). Earlier reports also showed that HMGB1 promotes CXCR4 expression through RAGE and TLR410,69.
Interestingly, in our experiments, Ac-HMGB1 activated NF-κB rather than AKT signaling via TLR4. A recent study showed that lipopolysaccharide (LPS) signaling activates AKT via TLR4 to promote breast cancer metastasis66. It is possible that during breast cancer metastasis, Ac-HMGB1 and LPS each initiate a specific signaling pathway downstream of TLR470.
Another important aspect of our studies is that extracellular S100A4 instigates a tumor-supportive microenvironment in GemOE/TNBC tumors, as was recently shown71. High S100A4 expression associates with poor outcome in early-stage cancers24 and direct interaction between S100A4 and RAGE was recently shown to promote motility in colorectal cancer cells via ERK-dependent mechanism72,73, and prostate cancer cells via NF-κB-activated pathway40. Chronic inflammation enhances RAGE expression in many cancers25,72,74,75, which was directly linked to low-rate patient survival. In our current analysis of a public dataset of >1700 breast cancer samples, low RFS in patients with high geminin + HMGB1 + S100A4 (two RAGE ligands)-expressing patients was observed (Fig. 3H). This observation supports the conclusion that RAGE signaling is involved in enhancing aggressiveness in GemOE TNBC tumors (cf. Fig. 3G). We propose that Ac-HMGB1 plays a previously unappreciated role in early-stage TNBCs.
TAMs presence within TNBC tumors is recognized as a critical factor in tumor progression and could be a prognostic factor of a worse outcome49,50,56,57. Most human tumors exhibit TAMs with an M2-like phenotype involved in promoting EMT and contributing to tumor progression and drug resistance49–51. In keeping, the presence of a high number of CD163+-M2 macrophages76 in GemOE orthotopic TNBC tumors was correlated with aggressive behavior50. We propose that TAM secretome, including Gas6, contributes to this aggressiveness through activation of the AXL receptor, which is overexpressed on GemOE/TNBC tumor cells. Indeed, recently published reports49 demonstrate that mesenchymal-like breast cancer cells induce the release of Gas6 selectively from M2-type macrophages49.
It is possible that along with c-Abl, AXL is an additional therapeutic target to prevent GemOE metastatic precursors dissemination from TNBC primaries. It is possible that in vivo selective inhibition of AXL using R428 could cooperate with imatinib to impair GemOE/TNBC cell invasion and intravasation, entrapping these GemOE/TNBC metastatic precursors within primaries to be resected during surgery reducing the metastatic burden. In support of this conclusion, we found that high geminin + AXL expression was associated with reduced RFS, DMFS, OS and LMFS in TNBC patients (cf. Fig. 5F–H, and Suppl. Fig. 12)
Our study has some limitations. Although, we provide evidence for intrinsic ability in GemOE/TNBC tumor cells to recruit and activate stromal elements, such as MSCs and TAMs, the specific requirement of RAGE and AXL for the interaction with MSCs and TAMs, and the distinct biological role in different subtypes of the TNBC disease warrant further studies. Despite these limitations, our results suggest that RAGE and AXL are prognostic indicators of outcome in GemOE/TNBCs patients.
Another limitation is the fact that direct testing for S100A4 in TAMs recruitment into GemOE/TNBC tumors should be done, in vivo. Although all the drugs used in this study are specific for their cognate target, their effect in our system should be tested in future studies to provide mechanistic insight, for instance, using the same assays except in cell lines harboring knockdown/knockout of these targets. Also, in future assays, we will assess whether the recruitment of M2-macrophages only or the macrophages, in general, is affected by imatinib, for example, using FACS analysis for the CD11b/F4/80 population within GemOE/TNBC tumors.
Moreover, we demonstrated AXL-mediated elevation in CD151, α3, and β1-integrin expression and functional interaction enhance binding to ECM in vitro, which could also be the case on vessels within tumors. This observation would be consistent with previous reports suggesting AXL’s role in breast cancer cells intravasation of the endothelium barrier into the bloodstream46. Noteworthy here, in preliminary data (not shown), we found that β1-integrin is phosphorylated in GemOE. An exciting possibility is that this phosphorylation is AXL- or AXL/CD151-driven. In support of the former, R428 abolished this phosphorylation event in GemOE cells (not shown). CD151 may also be a target along with c-Abl and AXL for therapy of GemOE/TNBC tumors.
Our analysis showing the opposite effect for GemOE on β4-integrin expression is intriguing. At present, the functional significance of this observation remains unknown. It is possible since α3β4-integrin is also a receptor for ECM binding in breast cancers77, that β4 is involved in the extravasation rather than the intravasation step of GemOE/TNBC metastatic spread, as described recently48. This will be investigated further in a follow-up study. Another critical experiment that will also be reported in a soon to be published article is the importance of MSCs/CAFs in the proposed intravasation of GemOE/TNBC cells.
These findings implicate synergistic signals simultaneously mediate multiple mechanisms in GemOE/TNBC intravasation. This study provides new understanding of the signaling pathways activated in GemOE/TNBC cells through interactions with stromal cells (MSCs/CAFs and M2-TAMs) that modulate the intravasation ability of tumor cells and underlines the importance of probing the process of tumor dissemination from an essential yet hitherto under-explored bi-directional interactions between GemOE/TNBC tumor cells and their microenvironmental cells. The dependence of tumor intravasation on the microenvironment provides yet another point of intervention to prevent tumor cell dissemination and patients’ demise78.
Methods
Cell culture
Human MSCs were purchased from Texas A&M HSC COM Institute for Regenerative Medicine, expanded, and frozen down. THP-1 cells were purchased from ATCC (catalog number: TIB-202TM), expanded, and frozen down. For both cell types, one vial is usually propagated for ≤5 generations to perform experiments before a new vail is used. Parental HME cell line (i.e., normal HME, used as control through the studies performed here) was transfected with a retrovirus expressing doxycycline (Dox)-inducible geminin allele14. Several clones from these cells were generated by antibiotic-selection, expanded, and tested for geminin expression14. A clone named Gem9 overexpressing a level of geminin similar to that observed in breast cancer cell lines was selected to use further. Gem9 cells were orthotopically injected in Dox-supplemented female SCID mice mammary fat pads (n = 10) and the primary (1°)-geminin overexpressing tumors developed were collected13 and used to generate the 1° geminin-overexpressing tumor cell lines we named; Gem197, Gem240, Gem256, Gem257, and Gem270. These cell lines were described earlier10, and are usually maintained in RPMI medium supplemented with 10% FBS, 1% antibiotics, and 2 µg/ml doxycycline to induce geminin expression will be used to perform the assays in this study. All commercial and in-house cell lines were authenticated by STR profiling and tested for mycoplasma contamination.
Growth factors, cytokines, and drugs
Recombinant (r)S100A4, rGas6, and rHMGB1, were from Sino Biological. Acetylation of rHMGB1 was described earlier10. Imatinib, RAGE inhibitor; glycyrrhizin79,80, NF-κB inhibitor; Bay 11-708281, or AKT inhibitor; MK-220682,83, RAGE inhibitor; FPS-ZM1, and CCR2 inhibitor; BMS CCR2 22 (Toronto Research Chemicals Inc.), TLR4 inhibitor; TAK-24268,84 (TOCRIS), Glycyrrhizic acid ammonium salt (Sigma) and AXL-specific inhibitor, R428 (Cayman Chemical). All were dissolved in DMSO, working concentration = 10 µM.
Antibodies
Mouse [m]-Twist (ab50887), m-CD105 (ab114052), m-HMGB1 (ab77302), rabbit [r]-Geminin (ab12147), r-TLR4 (ab13556), r-AXL (ab37861), r-RAGE (ab172473), m-α3-integrin (ab8985), m-β1-integrin (ab24693), r-CCR2 (ab32144) were from abcam. R-NF-κB/p65 (sc-372), m-OCT4 (sc-5279), r-ERK1/2 (137F5), r-Slug (#9585), r-Survivin (6E4), goat [g] MMP2 (sc-6838), g-MMP9 (sc-6840), m-p-ERK (sc-7383) were from Santa Cruz. R-SDF1 (#3740), m-Sox2 (#4900), r-AKT1 (C73H1b), r-p-AKT (T308 “C31E5E/S437”/S437 “D9E”) were from Cell signaling. M-HMGB1 (#07-584), m-vimentin (IF01) were from Millipore. PE-CD163 (560933), m-N-Cadherin (610920) were from R&D Systems. M-p-AXL (MAB6965), m-CCL2 (MAB679) were from ThermoFisher. APC-AXL (DS7HAXL) was from BD Biosciences. M-Actin (#cp01) was from Calbiochem. M-CD151 (350405) was from Biolegend. M-PE-Vio770-CD151 (130-103-728) was from Miltenyi. M-β4-integrin (MCA1456T) was from BIO-RAD. R-S100A4 (GTX62977) was from GeneTax. All antibodies were tested before use.
Serum isolation from mouse blood
Serum over plasma was chosen to study because it is more representative of inflammation. Samples were collected in a covered test tube and allowed to clot undisturbed at RT for ~30 min. The clot was then removed by centrifuging at 1,000–2,000 g for 10 min at 4 °C. The isolated serum is immediately transferred into a clean polypropylene tube aliquoted into 0.5 µl aliquots and stored in −80 °C.
ELISA analysis
Wells of a PVC microtiter plate were coat with the antigen by pipetting 50 μl of the conditioned medium (CM) or Serum (or dilution thereof in PBS) in triplicates and plate covered and incubated for overnight at 4 °C. After washing 3x with PBS, blocking of none-specific sites was done using 5% BSA in PBS incubated for ~2 h at room temperature (RT) followed by 3x washing in PBS. Diluted 1° antibody was added for 2 hours at RT, followed by 3x washing in PBS. HRP-conjugated 2° antibody diluted in blocking buffer was incubated 1 hour at RT then 3x washed with PBS. Detection using OPD (o-phenylenediamine dihydrochloride) tables and detection at 492 nm was done.
Cytokine array
HME and HME/geminin cells were assessed for differential cytokine secretion using human cytokine antibody array (RayBio). Conditioned media from an equal number of HME and HME/geminin cells plated in a serum-free medium for 20 h under standard conditions were used to perform according to the manufacturer’s instructions and previously described10.
Co-culture experiment
Boyden chambers (BD biosciences) of 8 µm (for migration) or 0.4 µm (for secretome) pore size were used. Certain cells (or their CM) were layered in the lower chamber with or without neutralizing antibodies, and test cells were layered in the transwell inserts. Cells migrated to the lower compartment of the Boyden chamber were counted and plotted 24 h later. Occasionally, hypoxia was introduced.
Conditioned media transfer experiment
Conditioned medium (CM) generated for analysis of secreted cytokines and surface receptor expression under normoxia (20% O2 for 24 h), or hypoxia (1% O2 for 24 h) conditions from an equal number of cells were transferred onto MSCs in the presence or absence of drugs or NeuAb. MSCs re-CM medium was transferred onto HME or GemOE cells with or without drugs or NeuAb for 24 h to re-reconditioned. The resultant CM was then transferred to an equal number of THP1-macrophages for 24 h to be re-re-reconditioned. Finally, this CM was re-added onto HME or GemOE cells for 24 h. At every step, the receiving cells were seeded at equal numbers to avoid number variations discrepancies. At various points, ELISA on CM and western blot on membrane fractions or whole-cell extracts generated by sonication was done.
Quantitative Real-time RT/PCR
In brief, 100 ng of total RNA isolated using TRIzol was used for qRT/PCR using iScriptTM One-Step RT-PCR kit with SYBR Green (Bio-Rad) as previously described10. The primer sequences used in this study are:
RAGE: Forward: 5′-GACTCTTAGCTGGCACTTGGAT-3′ and Reverse: 5′-GGACTTCACAGGTCAGGGTTAC-3′85,
CXCR4: Forward: 5′-TTCTACCCCAATGACTTGTG-3′ and Reverse: 5′-ATGTAGTAAGGCAGCCAACA-3′86,
S100A1: Forward: 5′-CCATGGAGACCCTCATCAAT-3′ and Reverse: 5′-TTCTGGACATCCAGGAAGC-3′87,
S100A2: Forward: 5′-GAACTTCTGCACAAGGAGCTG-3′ and Reverse: 5′-GACAGTGATGAGTGCCAGGA-3′,
S100A4: Forward: 5′-CCACAAGTACTCGGGCAAAG-3′ and Reverse: 5′-GTCCCTGTTGCTGTCCAAGT-3′88,
S100A6: Forward: 5′-AAGCTGCAGGATGCTGAAAT-3′ and Reverse: 5′-CCCTTGAGGGCTTCATTGTA-3′89,
S100A7: Forward: 5′-AGACGTGATGACAAGATTGAC-3′ and Reverse: 5′-TGTCTTTTTTCTCAAAGACGTC-3′90,
S100A8: Forward: 5′-GCTAGAGACCGAGTGTCCTCAG-3′ and Reverse: 5′-GCCCATCTTTATCACCAGAATG-3′,
S100A9: Forward: 5′-TGGAGGACCTGGACACAAATG-3′ and Reverse: 5′-TCGTCACCCTCGTGCATCTT-3′91,
HMGB1: Forward: 5′-ATATGGCAAAAGCGGACAAG-3′ and Reverse: 5′-AGGCCAGGATGTTCTCCTTT-3′92,
GAPDH: Forward: 5′-TGCACCACCAACTGCTTAGC-3′ and Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′93.
In vivo tumorigenicity assay
The University of Mississippi Medical Center IACUC committees approved all animal experiments. All experiments were performed in accordance with NIH guidelines and regulations. Six- to eight-week-old anesthetized immune-compromised Nu/Nu (Harlan) mice were injected with cells re-suspended in 100 µl RPMI medium and Matrigel (1:1) using a 27-gauge needle orthotopically in the 2nd left mammary gland. Tumors were monitored weekly by i.p. injection of 100 µl of D-luciferin (15 mg/mL in PBS) and anesthetized mice using a mix of oxygen and isoflurane gas were photographed for luciferase in vivo signals with IVIS machine (Xenogen). Tumor volume was assessed twice weekly by caliper (Life Sciences) using the formula L.W/2 (L = longest and W = shortest diameter of the tumor). Tumor initiation was defined as the time when tumors were 3 mm in diameter. Mice were sacrificed when the tumors reached <1.5 cm3 in volume or at the indicated time points in Results. At the end of the experiments, mice were euthanized by compressed 100% CO2 gas, tumors and organs (e.g., lung) were resected, weighed, fixed in formalin, and later cut at 4 µm for histological and immunohistochemical analysis. Some tumor parts were flash-frozen for DNA, RNA, and protein preparation later. In some instances, from anesthetized mice (using a mix of oxygen and isoflurane gas), 200 µl of blood was drawn from the heart before they were euthanized.
In vivo drug treatments
Tumor-bearing mice were treated with the drugs, concentration, and routes indicated in Results.
Preparation and injection of GFP-MSCs or -THP1s
GFP-expressing MSCs or THP1 were generated using lentivirus GFP-expressing plasmid. Antibiotics selected clones were propagated and stored frozen. Tumor-bearing mice and treated as described in Results were intracardiac injected through the left ventricle. Following tumor resection, part of the tumor was dissociated using Collagenase I and the protocol used by94, grown in culture for 24 h to adhere, and GFP-cells were photographed, counted, and plotted using Prism 7®.
Fluorescence IHC
IHC was performed as previously described95. Briefly, 5μm thick paraffin-embedded sections of tumor tissue excised from GemOE orthotopic mammary tumors were deparaffinized, rehydrated, and washed in PBS. Antigen retrieval for all antibodies was done using Sodium citrate buffer (10 mM Sodium citrate, 0.05% Tween 20, pH 6.0), 10 min at 95 °C. Slides were blocked with 10% normal serum (2° antibody species) for 1 h at RT, washed, and probed with 1° antibodies overnight at 4 °C. Slides were exposed to Alexa Fluor 568 (red), and 488 (green) conjugated secondary antibody for 1 h at RT and counterstained and mounted with VECTASHIELD mounting medium with DAPI (Vector) and imaged under the fluorescence microscope.
Overall survival (OS) and replace-free survival (RFS) and metastasis-free survival (MFS) analysis
The association of geminin alone or combined with the other factors was investigated for stratified patient cohorts using overall (OS) and relapse/recurrent- (RFS), or distant metastasis- (MFS), or lung metastasis- (LMFS) -free survival using the PROGgeneV2 - Pan-Cancer Prognostics Database (http://watson.compbio.iupui.edu/chirayu/proggene/database) or the Kaplan-Meier survival analysis.
Statistical analysis
Comparisons of treatment outcomes were tested for statistical differences using the Student t-test for paired data. Additionally, SPSS software was used to perform the ANOVA test followed by a post hoc Bonferroni correction for comparing multiple groups. The association of mRNA transcript expression with various clinicopathological parameters was also analyzed. Statistical significance was assumed at a P-value are *≤0.05, **≤0.01 and ***≤0.001.
Supplementary information
Acknowledgements
Wael is Dr. Lawrence & Mrs. Bo Hing Chan Tseu, American Cancer Society Research Scholar. This research was funded in part by grant # RSG-09-275-01 from American Cancer Society.
Author contributions
W. e.S. performed the in vivo experiments, analyzed all the data, interpreted all the data, and wrote the manuscript, D.R. performed some in vitro experiments, and J.K. performed statistical analysis. All authors read and approved the final manuscript.
Data availability
The data that support the findings of this study are available from the corresponding author [WeS] upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-55702-w.
References
- 1.McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/S0092-8674(00)81209-X. [DOI] [PubMed] [Google Scholar]
- 2.Melixetian M, et al. Loss of Geminin induces rereplication in the presence of functional p53. The Journal of cell biology. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Champeris Tsaniras S, et al. Geminin ablation in vivo enhances tumorigenesis through increased genomic instability. The Journal of pathology. 2018;246:134–140. doi: 10.1002/path.5128. [DOI] [PubMed] [Google Scholar]
- 4.Nakuci E, Xu M, Pujana MA, Valls J, Elshamy WM. Geminin is bound to chromatin in G2/M phase to promote proper cytokinesis. The international journal of biochemistry & cell biology. 2006;38:1207–1220. doi: 10.1016/j.biocel.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Hazama Y, et al. The Analysis of Cell Cycle-related Proteins in Ovarian Clear Cell Carcinoma Versus High-grade Serous Carcinoma. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists. 2018;37:516–524. doi: 10.1097/pgp.0000000000000461. [DOI] [PubMed] [Google Scholar]
- 6.Montanari M, et al. Increased expression of geminin stimulates the growth of mammary epithelial cells and is a frequent event in human tumors. Journal of cellular physiology. 2005;202:215–222. doi: 10.1002/jcp.20120. [DOI] [PubMed] [Google Scholar]
- 7.Di Bonito M, et al. Overexpression of Cell Cycle Progression Inhibitor Geminin is Associated with Tumor Stem-Like Phenotype of Triple-Negative Breast Cancer. Journal of breast cancer. 2012;15:162–171. doi: 10.4048/jbc.2012.15.2.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Andrade BA, et al. Expression of minichromosome maintenance 2, Ki-67, and geminin in oral nevi and melanoma. Annals of diagnostic pathology. 2013;17:32–36. doi: 10.1016/j.anndiagpath.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 9.Aizawa M, et al. Geminin expression in pancreatic neuroendocrine tumors: possible new marker of malignancy. Pancreas. 2012;41:512–517. doi: 10.1097/MPA.0b013e31823ca3e0. [DOI] [PubMed] [Google Scholar]
- 10.Ananthula S, et al. Geminin overexpression-dependent recruitment and crosstalk with mesenchymal stem cells enhance aggressiveness in triple negative breast cancers. Oncotarget. 2016;7:20869–20889. doi: 10.18632/oncotarget.8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tabrizi GA, Bose K, Reimann Y, Kessel M. Geminin is required for the maintenance of pluripotency. PloS one. 2013;8:e73826. doi: 10.1371/journal.pone.0073826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slawny N, O’Shea KS. Geminin promotes an epithelial-to-mesenchymal transition in an embryonic stem cell model of gastrulation. Stem cells and development. 2013;22:1177–1189. doi: 10.1089/scd.2012.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanchard Z, et al. Geminin overexpression induces mammary tumors via suppressing cytokinesis. Oncotarget. 2011;2:1011–1027. doi: 10.18632/oncotarget.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner L, Malik R, Shimizu Y, Mullins N, ElShamy WM. Geminin overexpression prevents the completion of topoisomerase IIalpha chromosome decatenation, leading to aneuploidy in human mammary epithelial cells. Breast cancer research: BCR. 2011;13:R53. doi: 10.1186/bcr2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulartz M, et al. Expression and phosphorylation of the replication regulator protein geminin. Biochemical and biophysical research communications. 2003;305:412–420. doi: 10.1016/s0006-291x(03)00773-3. [DOI] [PubMed] [Google Scholar]
- 16.Blanchard Z, et al. Geminin overexpression promotes imatinib sensitive breast cancer: a novel treatment approach for aggressive breast cancers, including a subset of triple negative. PloS one. 2014;9:e95663. doi: 10.1371/journal.pone.0095663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochemical Society transactions. 2012;40:341–346. doi: 10.1042/bst20120014. [DOI] [PubMed] [Google Scholar]
- 18.Andersson U, Yang H, Harris H. Extracellular HMGB1 as a therapeutic target in inflammatory diseases. Expert opinion on therapeutic targets. 2018;22:263–277. doi: 10.1080/14728222.2018.1439924. [DOI] [PubMed] [Google Scholar]
- 19.Lu B, et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3068–3073. doi: 10.1073/pnas.1316925111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ElShamy WM, Sinha A, Said N. Aggressiveness Niche: Can It Be the Foster Ground for Cancer Metastasis Precursors? Stem cells international. 2016;2016:4829106. doi: 10.1155/2016/4829106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan XL, et al. Hepatocellular carcinoma-associated mesenchymal stem cells promote hepatocarcinoma progression: role of the S100A4-miR155-SOCS1-MMP9 axis. Hepatology (Baltimore, Md.) 2013;57:2274–2286. doi: 10.1002/hep.26257. [DOI] [PubMed] [Google Scholar]
- 22.Wen L, Zhang C, Nong Y, Yao Q, Song Z. Mild electrical pulse current stimulation upregulates S100A4 and promotes cardiogenesis in MSC and cardiac myocytes coculture monolayer. Cell biochemistry and biophysics. 2013;65:43–55. doi: 10.1007/s12013-012-9402-x. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Y, et al. Overexpression of S100A4 protein may be associated with the development and progression of pancreatic cancer. Journal of cancer research and therapeutics. 2018;14:S159–s166. doi: 10.4103/0973-1482.172582. [DOI] [PubMed] [Google Scholar]
- 24.Egeland EV, et al. Prognostic significance of S100A4-expression and subcellular localization in early-stage breast cancer. Breast cancer research and treatment. 2017;162:127–137. doi: 10.1007/s10549-016-4096-1. [DOI] [PubMed] [Google Scholar]
- 25.Boye K, Maelandsmo GM. S100A4 and metastasis: a small actor playing many roles. The American journal of pathology. 2010;176:528–535. doi: 10.2353/ajpath.2010.090526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherbet GV, Lakshmi MS. S100A4 (MTS1) calcium binding protein in cancer growth, invasion and metastasis. Anticancer research. 1998;18:2415–2421. [PubMed] [Google Scholar]
- 27.Mundim FG, et al. Breast Carcinoma-associated Fibroblasts Share Similar Biomarker Profiles in Matched Lymph Node Metastasis. Applied immunohistochemistry & molecular morphology: AIMM. 2016;24:712–720. doi: 10.1097/pai.0000000000000253. [DOI] [PubMed] [Google Scholar]
- 28.Mc Cormack O, et al. Growth arrest-specific gene 6 expression in human breast cancer. British journal of cancer. 2008;98:1141–1146. doi: 10.1038/sj.bjc.6604260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leconet W, et al. Therapeutic Activity of Anti-AXL Antibody against Triple-Negative Breast Cancer Patient-Derived Xenografts and Metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2017;23:2806–2816. doi: 10.1158/1078-0432.Ccr-16-1316. [DOI] [PubMed] [Google Scholar]
- 30.Park JS, et al. Suppression of the metastatic spread of breast cancer by DN10764 (AZD7762)-mediated inhibition of AXL signaling. Oncotarget. 2016;7:83308–83318. doi: 10.18632/oncotarget.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, et al. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3beta/beta-catenin Signaling. Theranostics. 2016;6:1205–1219. doi: 10.7150/thno.15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu X, et al. AXL kinase as a novel target for cancer therapy. Oncotarget. 2014;5:9546–9563. doi: 10.18632/oncotarget.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed L, et al. Increased tumor cell expression of Axl is a marker of aggressive features in breast cancer among African women. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2015;123:688–696. doi: 10.1111/apm.12403. [DOI] [PubMed] [Google Scholar]
- 34.Abdel-Rahman WM, Al-Khayyal NA, Nair VA, Aravind SR, Saber-Ayad M. Role of AXL in invasion and drug resistance of colon and breast cancer cells and its association with p53 alterations. World journal of gastroenterology. 2017;23:3440–3448. doi: 10.3748/wjg.v23.i19.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasser MW, et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer research. 2015;75:974–985. doi: 10.1158/0008-5472.Can-14-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwak T, et al. Targeting of RAGE-ligand signaling impairs breast cancer cell invasion and metastasis. Oncogene. 2017;36:1559–1572. doi: 10.1038/onc.2016.324. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Yu X, Wang Q, Lu Y, Chen H. Expression and clinical significance of SATB1 and TLR4 in breast cancer. Oncology letters. 2017;14:3611–3615. doi: 10.3892/ol.2017.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Apetoh L, et al. The interaction between HMGB1 and TLR4 dictates the outcome of anticancer chemotherapy and radiotherapy. Immunological reviews. 2007;220:47–59. doi: 10.1111/j.1600-065X.2007.00573.x. [DOI] [PubMed] [Google Scholar]
- 39.Logsdon CD, Fuentes MK, Huang EH, Arumugam T. RAGE and RAGE ligands in cancer. Current molecular medicine. 2007;7:777–789. doi: 10.2174/156652407783220697. [DOI] [PubMed] [Google Scholar]
- 40.Siddique HR, et al. The S100A4 Oncoprotein Promotes Prostate Tumorigenesis in a Transgenic Mouse Model: Regulating NFkappaB through the RAGE Receptor. Genes & cancer. 2013;4:224–234. doi: 10.1177/1947601913492420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. The Journal of experimental medicine. 2014;211:1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryan D, et al. A niche that triggers aggressiveness within BRCA1-IRIS overexpressing triple negative tumors is supported by reciprocal interactions with the microenvironment. Oncotarget. 2017;8:113294. doi: 10.18632/oncotarget.23573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao W, et al. A Natural CCR2 Antagonist Relieves Tumor-associated Macrophage-mediated Immunosuppression to Produce a Therapeutic Effect for Liver Cancer. EBioMedicine. 2017;22:58–67. doi: 10.1016/j.ebiom.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Z, et al. CCL2/CCR2 Axis Promotes the Progression of Salivary Adenoid Cystic Carcinoma via Recruiting and Reprogramming the Tumor-Associated Macrophages. Frontiers in oncology. 2019;9:231. doi: 10.3389/fonc.2019.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. Kif4A mediate the accumulation and reeducation of THP-1 derived macrophages via regulation of CCL2-CCR2 expression in crosstalking with OSCC. Scientific reports. 2017;7:2226. doi: 10.1038/s41598-017-02261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goyette MA, et al. The Receptor Tyrosine Kinase AXL Is Required at Multiple Steps of the Metastatic Cascade during HER2-Positive Breast Cancer Progression. Cell reports. 2018;23:1476–1490. doi: 10.1016/j.celrep.2018.04.019. [DOI] [PubMed] [Google Scholar]
- 47.Sierra-Filardi E, et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. Journal of immunology (Baltimore, Md.: 1950) 2014;192:3858–3867. doi: 10.4049/jimmunol.1302821. [DOI] [PubMed] [Google Scholar]
- 48.Abdel-Ghany M, Cheng HC, Elble RC, Pauli BU. The breast cancer beta 4 integrin and endothelial human CLCA2 mediate lung metastasis. The Journal of biological chemistry. 2001;276:25438–25446. doi: 10.1074/jbc.M100478200. [DOI] [PubMed] [Google Scholar]
- 49.Bottai G, et al. AXL-associated tumor inflammation as a poor prognostic signature in chemotherapy-treated triple-negative breast cancer patients. NPJ breast cancer. 2016;2:16033. doi: 10.1038/npjbcancer.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reiman JM, Knutson KL, Radisky DC. Immune promotion of epithelial-mesenchymal transition and generation of breast cancer stem cells. Cancer research. 2010;70:3005–3008. doi: 10.1158/0008-5472.Can-09-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong Y, et al. Effects of RAGE-Specific Inhibitor FPS-ZM1 on Amyloid-beta Metabolism and AGEs-Induced Inflammation and Oxidative Stress in Rat Hippocampus. Neurochemical research. 2016;41:1192–1199. doi: 10.1007/s11064-015-1814-8. [DOI] [PubMed] [Google Scholar]
- 53.Takashima K, et al. Analysis of binding site for the novel small-molecule TLR4 signal transduction inhibitor TAK-242 and its therapeutic effect on mouse sepsis model. British journal of pharmacology. 2009;157:1250–1262. doi: 10.1111/j.1476-5381.2009.00297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee SB, et al. Identification of a distinct subpopulation of fibroblasts from murine dermis: CD73(−) CD105(+) as potential marker of dermal fibroblasts subset with multipotency. Cell biology international. 2016;40:1008–1016. doi: 10.1002/cbin.10623. [DOI] [PubMed] [Google Scholar]
- 55.Zhang B, et al. Increased circulating M2-like monocytes in patients with breast cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2017;39:1010428317711571. doi: 10.1177/1010428317711571. [DOI] [PubMed] [Google Scholar]
- 56.Jinushi M, et al. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12425–12430. doi: 10.1073/pnas.1106645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su S, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer cell. 2014;25:605–620. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 58.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast cancer research and treatment. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 59.Goswami CP, Nakshatri H. PROGgene: gene expression based survival analysis web application for multiple cancers. Journal of clinical bioinformatics. 2013;3:22. doi: 10.1186/2043-9113-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goswami CP, Nakshatri H. PROGgeneV2: enhancements on the existing database. BMC cancer. 2014;14:970. doi: 10.1186/1471-2407-14-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deng X, et al. Integrin-associated CD151 drives ErbB2-evoked mammary tumor onset and metastasis. Neoplasia (New York, N.Y.) 2012;14:678–689. doi: 10.1593/neo.12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gustafson-Wagner E, Stipp CS. The CD9/CD81 tetraspanin complex and tetraspanin CD151 regulate alpha3beta1 integrin-dependent tumor cell behaviors by overlapping but distinct mechanisms. PloS one. 2013;8:e61834. doi: 10.1371/journal.pone.0061834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwon MJ, et al. Clinical significance of CD151 overexpression in subtypes of invasive breast cancer. British journal of cancer. 2012;106:923–930. doi: 10.1038/bjc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huber R, et al. Tumour hypoxia promotes melanoma growth and metastasis via High Mobility Group Box-1 and M2-like macrophages. Scientific reports. 2016;6:29914. doi: 10.1038/srep29914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han R, et al. Estrogen promotes progression of hormone-dependent breast cancer through CCL2-CCR2 axis by upregulation of Twist via PI3K/AKT/NF-kappaB signaling. Scientific reports. 2018;8:9575. doi: 10.1038/s41598-018-27810-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li J, et al. TLR4 Promotes Breast Cancer Metastasis via Akt/GSK3beta/beta-Catenin Pathway upon LPS Stimulation. Anatomical record (Hoboken, N.J.: 2007) 2017;300:1219–1229. doi: 10.1002/ar.23590. [DOI] [PubMed] [Google Scholar]
- 67.Sun NK, Huang SL, Chang TC, Chao CC. TLR4 and NFkappaB signaling is critical for taxol resistance in ovarian carcinoma cells. Journal of cellular physiology. 2018;233:2489–2501. doi: 10.1002/jcp.26125. [DOI] [PubMed] [Google Scholar]
- 68.Yu P, Cheng X, Du Y, Huang L, Dong R. TAK-242 can be the potential agents for preventing invasion and metastasis of hepatocellular carcinoma. Medical hypotheses. 2013;81:653–655. doi: 10.1016/j.mehy.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 69.Tirone M, et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. The Journal of experimental medicine. 2018;215:303–318. doi: 10.1084/jem.20160217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao SJ, et al. Triggering of Toll-like receptor 4 on metastatic breast cancer cells promotes alphavbeta3-mediated adhesion and invasive migration. Breast cancer research and treatment. 2012;133:853–863. doi: 10.1007/s10549-011-1844-0. [DOI] [PubMed] [Google Scholar]
- 71.Prasmickaite Lina, Tenstad Ellen M., Pettersen Solveig, Jabeen Shakila, Egeland Eivind V., Nord Silje, Pandya Abhilash, Haugen Mads H., Kristensen Vessela N., Børresen-Dale Anne-Lise, Engebråten Olav, Maelandsmo Gunhild M. Basal-like breast cancer engages tumor-supportive macrophages via secreted factors induced by extracellular S100A4. Molecular Oncology. 2018;12(9):1540–1558. doi: 10.1002/1878-0261.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahlmann M, et al. RAGE mediates S100A4-induced cell motility via MAPK/ERK and hypoxia signaling and is a prognostic biomarker for human colorectal cancer metastasis. Oncotarget. 2014;5:3220–3233. doi: 10.18632/oncotarget.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schneider M, et al. S100A4 is upregulated in injured myocardium and promotes growth and survival of cardiac myocytes. Cardiovascular research. 2007;75:40–50. doi: 10.1016/j.cardiores.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 74.Heijmans J, et al. Rage signalling promotes intestinal tumourigenesis. Oncogene. 2013;32:1202–1206. doi: 10.1038/onc.2012.119. [DOI] [PubMed] [Google Scholar]
- 75.Luo Y, et al. High mobility group box 1 released from necrotic cells enhances regrowth and metastasis of cancer cells that have survived chemotherapy. European journal of cancer (Oxford, England: 1990) 2013;49:741–751. doi: 10.1016/j.ejca.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 76.Tiainen S, et al. High numbers of macrophages, especially M2-like (CD163-positive), correlate with hyaluronan accumulation and poor outcome in breast cancer. Histopathology. 2015;66:873–883. doi: 10.1111/his.12607. [DOI] [PubMed] [Google Scholar]
- 77.Mercurio AM, Rabinovitz I, Shaw LM. The alpha 6 beta 4 integrin and epithelial cell migration. Current opinion in cell biology. 2001;13:541–545. doi: 10.1016/S0955-0674(00)00249-0. [DOI] [PubMed] [Google Scholar]
- 78.Khuon S, et al. Myosin light chain kinase mediates transcellular intravasation of breast cancer cells through the underlying endothelial cells: a three-dimensional FRET study. Journal of cell science. 2010;123:431–440. doi: 10.1242/jcs.053793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin SC, et al. Glycyrrhizic acid induces human MDA-MB-231 breast cancer cell death and autophagy via the ROS-mitochondrial pathway. Oncology reports. 2018;39:703–710. doi: 10.3892/or.2017.6123. [DOI] [PubMed] [Google Scholar]
- 80.Park SY, et al. Anti-carcinogenic effects of non-polar components containing licochalcone A in roasted licorice root. Nutrition research and practice. 2014;8:257–266. doi: 10.4162/nrp.2014.8.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viola K, et al. Bay11-7082 inhibits the disintegration of the lymphendothelial barrier triggered by MCF-7 breast cancer spheroids; the role of ICAM-1 and adhesion. British journal of cancer. 2013;108:564–569. doi: 10.1038/bjc.2012.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi AR, et al. Co-treatment of LY294002 or MK-2206 with AZD5363 Attenuates AZD5363-induced Increase in the Level of Phosphorylated AKT. Anticancer research. 2016;36:5849–5858. doi: 10.21873/anticanres.11170. [DOI] [PubMed] [Google Scholar]
- 83.Wisinski KB, et al. Phase I Study of an AKT Inhibitor (MK-2206) Combined with Lapatinib in Adult Solid Tumors Followed by Dose Expansion in Advanced HER2+ Breast Cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2016;22:2659–2667. doi: 10.1158/1078-0432.Ccr-15-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang X, et al. Hypoxia-inducible factor-1alpha mediates the toll-like receptor 4 signaling pathway leading to anti-tumor effects in human hepatocellular carcinoma cells under hypoxic conditions. Oncology letters. 2016;12:1034–1040. doi: 10.3892/ol.2016.4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Crabtree M, et al. RAGE mRNA expression and its correlation with nuclear factor kappa beta mRNA expression in inflamed human periradicular tissues. Journal of endodontics. 2008;34:689–692. doi: 10.1016/j.joen.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Speetjens FM, et al. Nuclear localization of CXCR4 determines prognosis for colorectal cancer patients. Cancer microenvironment: official journal of the International Cancer Microenvironment Society. 2009;2:1–7. doi: 10.1007/s12307-008-0016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lisachev Pavel D., Shtark Mark B., Sokolova Olga O., Pustylnyak Vladimir O., Salakhutdinova Mary Yu., Epstein Oleg I. A Comparison of the Dynamics of S100B, S100A1, and S100A6 mRNA Expression in Hippocampal CA1 Area of Rats during Long-Term Potentiation and after Low-Frequency Stimulation. Cardiovascular Psychiatry and Neurology. 2010;2010:1–6. doi: 10.1155/2010/720958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee OJ, et al. Expression of calcium-binding proteins S100A2 and S100A4 in Barrett’s adenocarcinomas. Neoplasia (New York, N.Y.) 2006;8:843–850. doi: 10.1593/neo.06481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang XH, et al. Increased expression of S100A6 promotes cell proliferation in gastric cancer cells. Oncology letters. 2017;13:222–230. doi: 10.3892/ol.2016.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Castro A, Minty F, Hattinger E, Wolf R, Parkinson EK. The secreted protein S100A7 (psoriasin) is induced by telomere dysfunction in human keratinocytes independently of a DNA damage response and cell cycle regulators. Longevity & healthspan. 2014;3:8. doi: 10.1186/2046-2395-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan LL, et al. Effects of silencing S100A8 and S100A9 with small interfering RNA on the migration of CNE1 nasopharyngeal carcinoma cells. Oncology letters. 2015;9:2534–2540. doi: 10.3892/ol.2015.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dong YD, et al. Expression and clinical significance of HMGB1 in human liver cancer: Knockdown inhibits tumor growth and metastasis in vitro and in vivo. Oncology reports. 2013;29:87–94. doi: 10.3892/or.2012.2070. [DOI] [PubMed] [Google Scholar]
- 93.Cicinnati VR, et al. Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC cancer. 2008;8:350. doi: 10.1186/1471-2407-8-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Leelatian N, et al. Preparing Viable Single Cells from Human Tissue and Tumors for Cytomic Analysis. Current protocols in molecular biology. 2017;118:25c.21.21–25c.21.23. doi: 10.1002/cpmb.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Paul BT, Blanchard Z, Ridgway M, ElShamy WM. BRCA1-IRIS inactivation sensitizes ovarian tumors to cisplatin. Oncogene. 2015;34:3036–3052. doi: 10.1038/onc.2014.237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author [WeS] upon reasonable request.