Summary
A major limitation in anti-tuberculosis drug screening is the lack of reliable and scalable models for homogeneous human primary macrophage cells of non-cancer origin. Here we report a modified protocol for generating homogeneous populations of macrophage-like cells from human embryonic stem cells. The induced macrophages, referred to as iMACs, presented similar transcriptomic profiles and characteristic immunological features of classical macrophages and were permissive to viral and bacterial infection, in particular Mycobacterium tuberculosis (Mtb). More importantly, iMAC production was amenable to scale up. To evaluate iMAC efficiency in high-throughput anti-tuberculosis drug screening, we performed a phenotypic screening against intracellular Mtb, involving a library of 3,716 compounds that included FDA-approved drugs and other bioactive compounds. Our primary screen identified 120 hits, which were validated in a secondary screen by dose-intracellular and -extracellular Mtb assays. Our confirmatory studies identified a novel anti-Mtb compound, 10-DEBC, also showing activity against drug-resistant strains.
Keywords: human embryonic stem cell-derived induced macrophage-like cells (iMACs), Mycobacterium tuberculosis (Mtb), drug discovery platform, anti-tuberculosis compound
Graphical Abstract
Highlights
-
•
Methods for large-scale production of hPSC-derived macrophage-like cells (iMACs)
-
•
iMACs recapitulate immune response of human monocyte in M. tuberculosis infection
-
•
High-throughput screening of 3,716 compounds using iMACs identified novel anti-tuberculosis compounds
-
•
10-DEBC inhibited intra- and extracellular growth of drug-resistant M. tuberculosis
In this article, Jung-Hyun Kim and colleagues show a protocol for generating macrophage-like cells from PSCs that exhibit features of classical phagocytes, are amenable to scale up, permissive to Mycobacterium tuberculosis infection, and suitable for intracellular high-throughput screening. By performing screening of 3,716 compounds, they found a new, potent anti-tuberculosis drug against drug-resistant strains of M. tuberculosis.
Introduction
Tuberculosis (TB) is caused by Mycobacterium tuberculosis (Mtb) and continues to be a major infectious disease around the world (World Health Organization, 2018). Standard therapy for drug-sensitive TB includes core antibiotics prescribed over a minimum period of 6 months. However, the major challenge for TB treatment is to counter the rise of multidrug-resistant (MDR) and extensively resistant (XDR) Mtb strains that are refractory to standard therapy.
On inhalation, Mtb infects resident alveolar macrophages in the distal airway that then elicit an inflammatory response, releasing chemokines and cytokines that recruit blood monocytes and neutrophils to the lungs (Cohen et al., 2018, Huang et al., 2018). The former are more permissive toward Mtb replication and harbor rather than eliminate Mtb. Mtb survives and replicates within phagosomes of macrophages by inhibiting bactericidal responses, mainly by disrupting the fusion of phagosomes with lysosomes and diminishing generation of reactive oxygen species (ROS). Therefore, targeting Mtb residing in macrophages can, in theory, lead to the discovery of new drugs.
THP-1 and U937, commonly used immortalized human macrophage-like cell lines, originate from cancer tissue and may harbor mutations that influence surface receptor expression, phagocyte function, ROS generation, metabolic state, cytokine expression, and drug detoxification capacity, which together influence Mtb replication, anti-Mtb responses, and host cell cytotoxicity (Mendoza-Coronel and Castanon-Arreola, 2016). Moreover, a comparative genomic analysis of immortalized mouse macrophage cell lines (RAW264.7 and J774.1) and mouse primary macrophages infected with Mtb showed a significant difference in the magnitude of early immune responses and induction of key effector genes of innate immunity (Chamberlain et al., 2009). Primary human blood monocyte-derived macrophages are a physiologically relevant host cell model for phenotypic screening. However, their use is greatly limited by inadequate supply, high cost, as well as reproducibility and ethical considerations.
Pluripotent stem cells (PSCs) have the capacity to self-renew and differentiate into cell types found in the adult body (Avior et al., 2016). Numerous studies have described advances in knowledge for differentiation of PSCs into diverse human primary-like cells, and their possible application to cell-based therapy, disease modeling, and toxicity testing, as well as drug screening. Recently, PSC-derived macrophage differentiation methodologies have been described, with high impact for the study of macrophage biology, as well as host-pathogen interactions and their roles in infectious diseases (Shi et al., 2017, Takata et al., 2017). Importantly, PSC-derived human primary macrophages could be of high value for drug discovery screening against intracellular Mtb. However, there is limited information for efficient scale out of homogeneous macrophages in 2D culture system using PSCs for this specific platform.
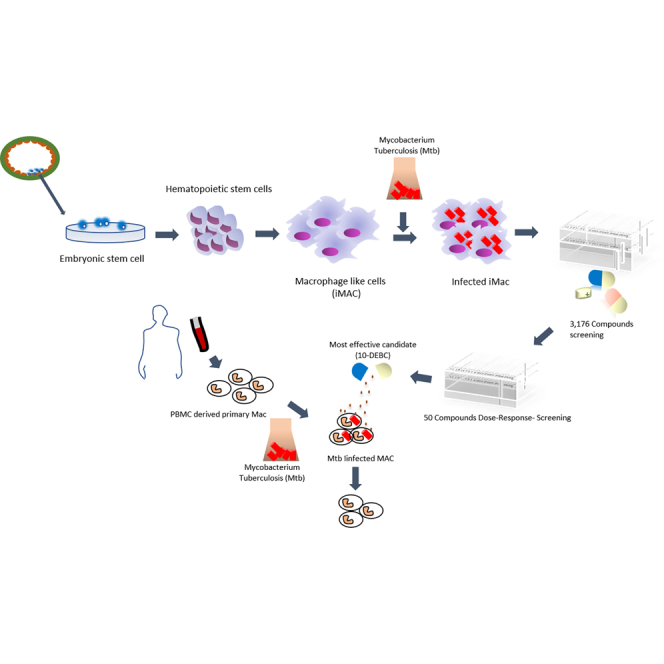
Here we report a modified protocol for generating homogeneous populations of macrophage-like cells from PSCs, which exhibit features of classical phagocytes, are amenable to scale up, permissive to Mtb infection, and suitable for high-throughput screening. Using this macrophage population as a platform, we screened a 3,716-compound library comprising Food and Drug Administration (FDA)-approved drugs and other bioactive compounds. The 120 hits were founded, and 50 hits were further validated by dose-response studies using both phenotypic and Mtb whole-cell screening methods. We ultimately identified 10-4'-(N,N-diethylamino)butyl-2-chlorophenoxazine hydrochloride (10-DEBC) as a novel anti-TB compound active against both intracellular and extracellular Mtb, as well as drug-resistant Mtb.
Results
Generation of Macrophages from Human PSCs
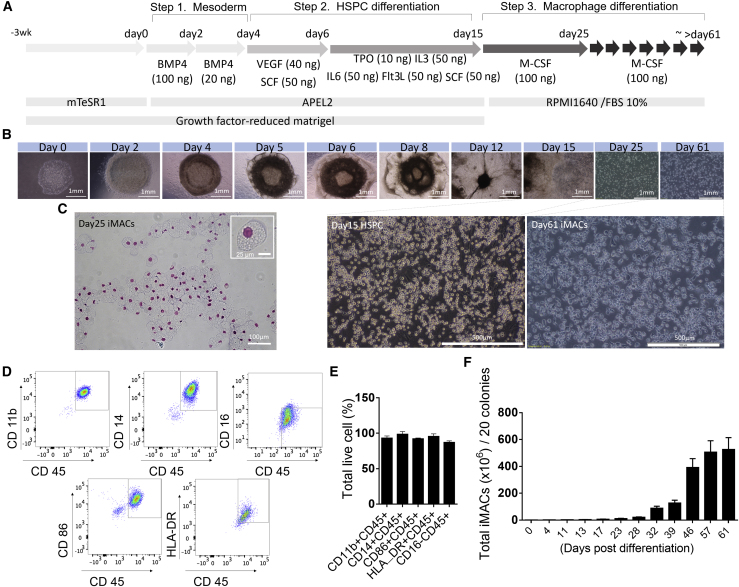
We have developed a three-step method for large-scale production of homogeneous functional macrophages from human embryonic stem cells (hESCs), by modification of previously reported methods (Figure 1A) (Yanagimachi et al., 2013). During the first step of differentiation, we noticed that the hESCs changed morphology (Figure 1B) and induced the expression of mesodermal markers (Brachyury and Mixl1) (Figure S1A) with decreased expression of pluripotent markers (Oct4 and Nanog) (Figure S1B). In the next step, we observed continuous production of CD34+CD45+ hematopoietic stem and progenitor cells (HSPCs) (Video S1) as approximately 20% of the total cells (Figures S1C and S1D). Finally, in the last step of differentiation, we obtained ∼92%–98% of induced macrophages (hence forth referred to as iMACs) that had a typical macrophage morphology (Figure 1C) and a CD11b+CD14−CD16−CD86+HLA_DR+ surface marker expression profile (Figures 1D and 1E). More importantly, the method produced about 500 million iMACs from only 20 colonies of hESCs (Figure 1F; Video S2). In addition, iMACs were generated from an iPSC cell line (CMC003) with a similar pattern (Figure S2).
Figure 1.
Differentiation and Phenotypic Characterization of Macrophages from hESCs
(A) Schematic illustration of the stepwise differentiation protocol for deriving macrophages from hESCs from day 0 to 61. Cytokine concentration indicates ng/mL.
(B) Representative bright-field image of cells at each step during differentiation from day 0 to 25. Scale bars, 1 mm (upper image) and 500 μm (lower image).
(C) Representative image of Wright-Giemsa-stained macrophages 25 days after differentiation from hESCs (400× magnification).
(D and E) Flow cytometry analysis of iMACs expressing CD11b, CD14, CD86, HLA-DR, and not expressing CD16. (D) Representative images of a dot blot are shown. (E) Percentage of positive cell populations in live cells (n = 3).
(F) Accumulated cell number of CD45+CD14+ iMACs from 20 colonies of hESCs in indicated date (n = 3). Results are shown as mean ± SD.
Production of CD34+CD45+ HSPCs. Scale bar, 200 μm. The time lapse covers a period of about 24 h, with a 30-min time interval between images. Recordings were made with a Lionheart FX system (BioTek Instruments, Winooski, VT, USA) using a 4× objective lens and data were processed with Gen5 Image+ software (BioTek Instruments).
The video shows macrophages produced from HSPCs. Scale bar, 200 μm. The time lapse covers a period of 48 h, with a 30-min time interval between images. Recordings were made with a Lionheart FX system (BioTek Instruments) using a 4× objective lens and data were processed with Gen5 Image+ software (BioTek Instruments).
While optimizing the differentiation protocol, we noticed that the base medium in steps 1–2 greatly affected the yield of HSPC and macrophage production. Among the available base media that we tested, APEL2 produced a higher percentage (Figures S3A–S3C) and number (Figure S3D) of CD34+CD45+ HSPCs. We also noticed that the efficiency of HSPCs to form granulocyte (colony-forming unit [CFU])-macrophage progenitors was higher in APEL2 cultured HSPCs compared with other media (Figure S3E). Furthermore, they generated approximately 2-fold higher number of CD11b+CD45+, CD14+CD45+, CD86+CD45+ iMACs than Stemline II-cultured HSPCs (Figure S3F).
Global Transcriptional Profile of iMACs, Human Monocyte-Derived Macrophages, and THP-1 Macrophages
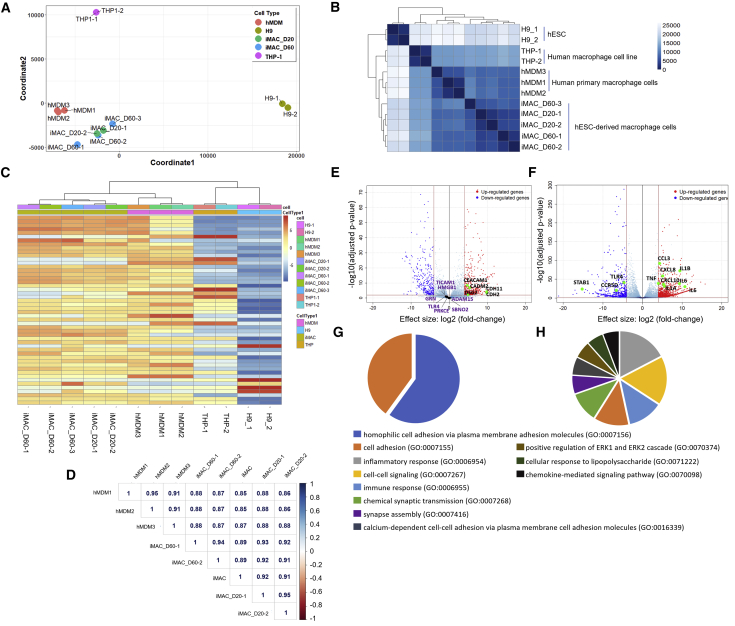
For genomic characterization of iMACs, we performed RNA sequencing (RNA-seq) of 20- and 60-day iMACs and compared their profiles with that of hESCs, human monocyte-derived macrophages (hMDMs), and THP-1 macrophages. To minimize the variations induced by the culture conditions, hMDMs and late-stage iMACs were cultured in the same media (RPMI 1640 + macrophage colony-stimulating factor+10%FBS). Multidimensional scaling provided a genome-wide overview of similarities between iMACs and hMDMs, illustrating that iMACs are transcriptionally closer to hMDMs than to hESCs. However, THP-1 macrophages showed lower transcriptional similarity to hMDMs and iMACs (Figure 2A). This inference was further supported by hierarchical and non-hierarchical clustering analysis that expression features between iMACs and hMDMs were more similar than between THP-1 cells and hMDMs (Figures 2B and 2C). Importantly, we found that the correlation between “young” (20-day iMACs) and “aged” (60-day iMACs) cultures was high (0.89–0.95, Pearson correlation coefficient Figure 2D). Although 60-day iMACs were still distinct from 20-day iMACs, we noted very close clustering of the two groups together, indicating homogeneous growth of iMACs (Figures 2A and 2D). When defining differentially expressed genes (DEGs) as having a |log2(fold change)| > log 4 and false discovery rate of 0.01, we identified 802 genes that were differentially expressed between iMACs and hMDMs, whereas 1,613 genes were differentially expressed between THP-1 macrophages and hMDMs (Table S1(1,2)). The top 5 genes highly expressed in iMACs were CEACAM1, CADM2, CDH2, CDH22, and DSG2, which were related to the cellular adhesion pathway (Figure 2E). However, the expressions of immune-related genes, such as TICAM1, HMGB1, GRN, ADAM15, TLR4, PRKCE, and SBNO2 were not significantly different (Figure 2E) between iMACs and hMDMs unlike THP-1 and hMDMs (Figure 2F) A gene ontology (GO) analysis also revealed that the DEGs in iMACs were mostly related to cell adhesion molecular pathways, as compared with hMDMs (Figure 2G). In contrast, DEGs in THP-1 cells were related to inflammatory, cell-cell signaling and immune response pathways as compared with hMDMs (Figures 2F and 2H).
Figure 2.
Transcriptomic Characterization of iMACs Compared with Human Primary Macrophage
(A) Multidimensional scaling (MDS) analyses of the transcriptome of hESC-derived macrophages (iMAC-D20 and -D60) versus hESCs (H9), hMDMs, and THP-1 monocyte-differentiated macrophages. iMAC-D20 and -D60 are iMACs obtained on day 20 and 60 after differentiation, respectively. hMDMs are human peripheral blood monocyte (CD14+)-derived macrophages. Each circle represents an independent sample.
(B) Heatmap of sample-to-sample distance matrix using Poisson distance with hierarchical clustering, depicting overall similarity of transcriptome profiles of iMACs, H9, hMDMs, and THP-1 monocyte-differentiated macrophages. Color scale indicates Poisson distance values between samples.
(C) Unsupervised non-hierarchical clustering of samples and heatmap showing variance stabilizing transformation-normalized values of the top 50 variably expressed genes. The color key from blue to red indicates low to high expression values, respectively.
(D) Spearman coefficients of log2 normalized read counts. Left color bar indicates correlation scale (blue to red, 1 to −1).
(E and F) Differentially expressed genes (DEGs) of hMDMs versus iMACs (E) and hMDMs versus THP-1 (F), |log2(fold change)| > log 4, false discovery rate (FDR) < 0.01. Representative genes related to cell adhesion molecular pathway (E) and inflammatory responses (F) were indicated in the volcano plot. Purple gene names indicated immune-related genes (GO: 0002281) in (D).
(G and H) Functional categorization of DEGs based on gene ontology (GO) annotations. iMAC vs hMDM (G), Thp1 vs hMDM (H). GO terms are shown with FDR <0.0001. FDR is shown as −log10(FDR) < 0.0001.
Phagocytosis Capacity and Immune Responses of iMACs
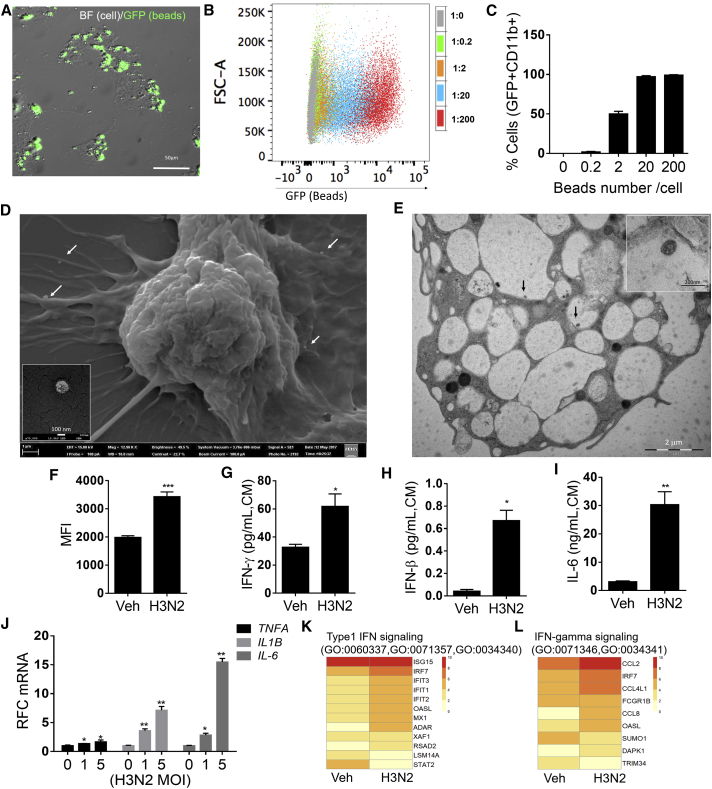
To characterize the functional properties of iMACs, we assessed their phagocytic capacity after incubation with opsonized fluorescent microbeads. iMACs actively recognized and internalized fluorescent microbeads as shown by merged bright-field images of iMACs and fluorescein isothiocyanate fluorescent beads (Figure 3A). Fluorescence-activated cell sorting (FACS) analysis showed that iMACs were able to engulf beads within 1.5 h incubation, in a dose-dependent manner (Figures 3B and 3C). Next, we infected the cells with the influenza virus H3N2, which is able to infect and replicate within alveolar macrophages (Hoeve et al., 2012, Ismail and McBride, 2017, Kim et al., 2017, Van Reeth and Adair, 1997). Scanning electron microscopy (SEM) and transmission electron microscopy (TEM) images indicated typical morphology of H3N2 viral particles in contact or internalized by the iMACs (Figures 3D and 3E). These results showed that the iMACs obtained from hESCs were functional phagocytes. Next, to study the innate immune responses of iMACs, we assessed intracellular ROS production and secretory cytokines levels. FACS analysis showed higher levels of superoxide radicals in H3N2-infected iMACs, as compared with uninfected iMACs (Figure 3F). H3N2-infected iMACs secreted significantly greater levels of interferon-γ (IFN-γ), IFN-β, and interleukin-6 (IL-6) when compared with uninfected cells (Figures 3G–3I). Consistently, mRNA levels of inflammatory cytokines and type I and II interferon signaling were elevated by H3N2 infection (Figures 3J–3L). Next, we assessed iMAC responses toward bacterial infection. Anaplasma phagocytophilum (a Gram-negative bacteria) formed morulae (Figure S4A) and the induced ROS production and inflammatory-immune response in iMACs (Figures S4B–S4D). Together, these data suggest that iMACs exhibit characteristic features of macrophages, including phagocytosis and inflammatory responses.
Figure 3.
Characterization of Phagocytosis and Innate Immune Responses of iMACs by H3N2 Viral Infection
(A) Representative micrographs of iMACs engulfing fluorescein isothiocyanate (FITC)-labeled latex beads. Bright-field (BF), GFP, and merged images (1,000× magnification) are shown.
(B and C) Percentage of CD11b+ iMACs containing FITC-labeled latex beads as determined by flow cytometry (n = 3). (B) Opsonized beads were incubated with iMACs as 1:0.2–1:200 ratio (cells:beads). CD11b+ cells were gated and GFP+ cells were presented as dot plot. (C) Percentage of GFP+ cells, mean ± SD, n = 3.
(D and E) Scanning electron micrograph (D) and transmission electron micrograph (E) of iMACs 1 day after H3N2 infection. Scale bars, 1 μm (100 nm enlarged image; left) and 2 μm (200 nm enlarged image; right). Arrows indicate H3N2 viral particles inside cells.
(F) Flow cytometry analysis of superoxide radicals in MitoSOX-stained iMACs 1 day after H3N2 infection. Bar graph represents mean fluorescence intensity (MFI) in vehicle-treated and H3N2-infected cells (n = 3).
(G–I) IFN-γ (G), IFN-β (H), and IL-6 (I) levels in culture medium (CM) of iMACs, 1 day after H3N2 infection.
(J) Relative fold change (RFC) of mRNA levels of immune response genes, after 4 h H3N2 viral infection in iMACs. (A–I) ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
(K and L) Type 1 IFN signaling (K) and IFN-γ signaling (L) in H2N3 infected iMACs. Heatmap of differentially expressed genes (p < 0.05) of iMACs on day 1 after H3N2 infection (MOI 5) detected by RNA-seq. Annotated GO terms are indicated.
iMACs Are Permissive to Mtb Infection
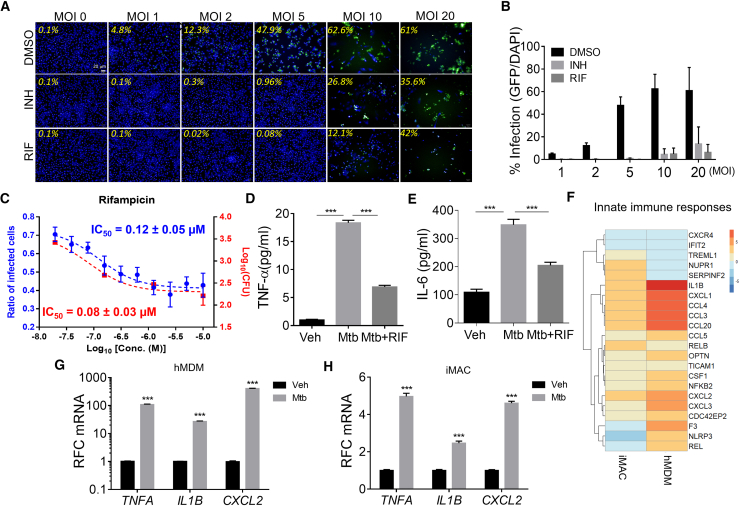
We examined whether iMACs were permissive for Mtb intracellular replication and elicited inflammatory responses after infection. Using a mycobacterial laboratory strain (Mtb H37Rv) modified to constitutively express a GFP, we found that iMACs could be infected with Mtb, from MOI ranging from 1 to 20 bacteria per cell, in a dose-dependent manner (Figure 4A). Highest infection doses (MOI 10 and 20) led to an advanced loss of cells after 5 days of infection, confirming effective intracellular replication. At lower MOIs, treatment with anti-tubercular drugs isoniazid (INH) or rifampicin (RIF) successfully rescued the cells from the infection challenge and prevented bacterial replication, leading to a sharp decrease in the infection ratio (Figure 4B), correlated with the decrease in CFU numbers (Figure 4C). Furthermore, iMACs secreted the pro-inflammatory cytokines IL-6 and tumor necrosis factor alpha after infection, and treatment with RIF was found to dampen this inflammatory response (Figures 4D and 4E).
Figure 4.
Characterization of Mtb Infection in iMACs and Anti-TB Drug Responsiveness
(A) Confocal micrographs showing the percentage of iMACs infected with GFP-labeled Mtb at various initial MOIs in the presence and absence of the anti-TB drugs INH (10 μM) or RIF (10 μM) after 5 days of infection. Infected cells harbor H37Rv-GFP bacteria (green) and are stained with DAPI (blue).
(B) Percentage of Mtb-infected iMACs obtained at various MOIs after 5 days infection, in the presence or absence of anti-TB drugs (INH or RIF). Data represent mean ± SD of three independent experiments.
(C) The correlation between the imaging method and the CFU counting method for iMACs infected with H37Rv-GFP strain. Average ± SD values are shown; n = 4 wells for the ratio of infected cells, n = 2 wells for the CFUs.
(D and E) Quantification by ELISA of secreted cytokines in culture medium of iMACs 5 days after Mtb infection (MOI 5). TNF-α (D) and IL-6 (E) concentrations are shown in pg/mL. Eight-fold diluted culture medium were used.
(F–H) Heatmap of differentially expressed genes (p < 0.05) of hMDMs on day 5 after Mtb infection (MOI 5) detected by RNA-seq, respectively (F). Annotated in GO (GO terms; innate immune and inflammatory responses genes [GO: 0045087, 0006954]). Color scale indicates log2 fold change. qRT-PCR analysis represents relative fold change (RFC) of genes expression after Mtb infection in hMDMs (G) or iMACs (H). p < 0.005 (n = 3).
To address whether iMACs showed immune responses similar to hMDMs following Mtb infection, we performed RNA-seq analysis of iMACs and hMDMs infected with Mtb. A total of 206 and 388 DEGs were identified in hMDMs and iMACs (fold change >2 fold, p < 0.05). Non-hierarchical cluster analysis and the GO analysis revealed that overall gene expressions patterns and the top signaling pathways in hMDMs and iMACs were not similar following Mtb infection (Figure S5). However, we noted a similar pattern of changes in genes associated with innate immunity and inflammation between iMACs and hMDMs, 5 days after infection at an MOI of 5 (Figure 4F; Table S1(3)). qRT-PCR confirmed upregulation of the immune-related genes TNFA, IL1B, and CXCL2 by Mtb infection, in both iMACs and hMDMs (Figures 4G and 4H). Taken together, these results suggest that the iMACs recapitulates human primary macrophages toward Mtb infection and underscore the suitability of iMACs for screening anti-tubercular compounds.
Use of iMACs for Anti-TB Drug Screening
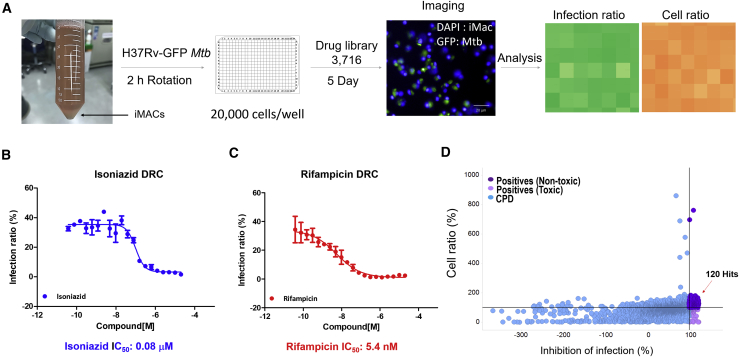
A library of 3,716 compounds was assembled from diverse libraries by gathering FDA-approved drugs for repositioning as well as bioactive compounds with known biological activities. The iMACs infected with Mtb H37Rv-GFP at an MOI of 5 were treated with compounds at a single concentration of 10 μM and analyzed after 5 days of incubation (Figure 5A). Negative controls (0.5% DMSO) as well as reference drug (RIF) were included in every plate to validate the assay performance (Figures S6A–S6C). INH and RIF were also tested separately at multiple doses and showed a dose-dependent inhibitory effect on intracellular Mtb growth, with concentrations required to reach 50% inhibition (IC50) of 0.08 μM and 5.4 nM, respectively (Figures 5B and 5C). From the primary screening, we identified 120 hits that showed more than 100% inhibition of Mtb growth without host cell cytotoxicity (Figure 5D; Table S2). Notably, among the 120 hit candidates, several WHO-recommended anti-TB drugs were present (Table S3).
Figure 5.
Compound Library Screening Using Mtb-Infected iMAC
(A) Schematic illustration of individual steps in the compound library screening using H37Rv-GFP Mtb-infected iMACs.
(B and C) Dose-response curve for INH (B) and RIF (C) in H37Rv-GFP-infected iMACs (n = 3).
(D) Primary screening results. Compounds (CPD) showing over 100% cell survival and 100% inhibition of infection are indicated as purple dots.
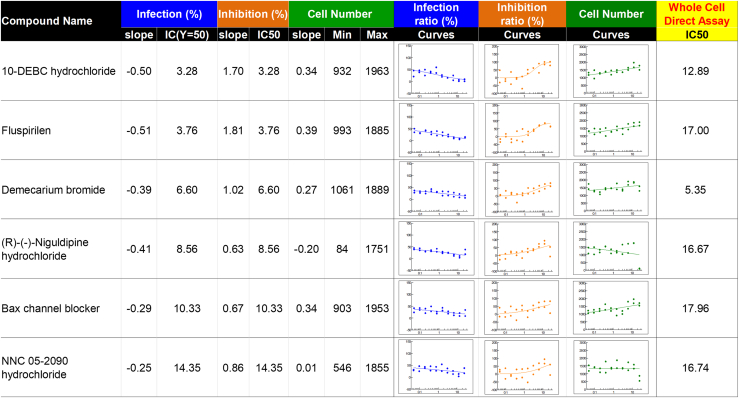
Next, we carried out a secondary screening using only the 50 “non-antibiotic” candidates, based on literature reporting from the list of 120 hits identified in the primary screening. Compound activities were validated by conducting a 10-point dose-response in both Mtb whole-cell assay and Mtb-infected intracellular assay. Of the 50 compounds tested, only 8 had an IC50 < 20 μM in the bacteria whole-cell assay, while 39 showed IC50 < 20 μM in an Mtb-infected intracellular cellular assay, with 6 compounds belonging to both categories (Figure 6). Among these, 10-DEBC was selected for further confirmation (Figure 7A). Indeed, 10-DEBC shares close structure with chlorpromazine, a phenothiazine compound clinically used to treat mental disorders and reported to have anti-TB activity, including against drug-resistant strains (Amaral and Viveiros, 2017, Kristiansen et al., 2015). In addition, 10-DEBC showed the highest activity in Mtb-infected iMAC DRC assay. It was then of interest to investigate this compound further.
Figure 6.
Hit Compounds
Dose-response curve with IC50 value of 6-hit compounds (IC50 <20 μM) in intracellular assay and whole-bacteria cell-based extracellular assay.
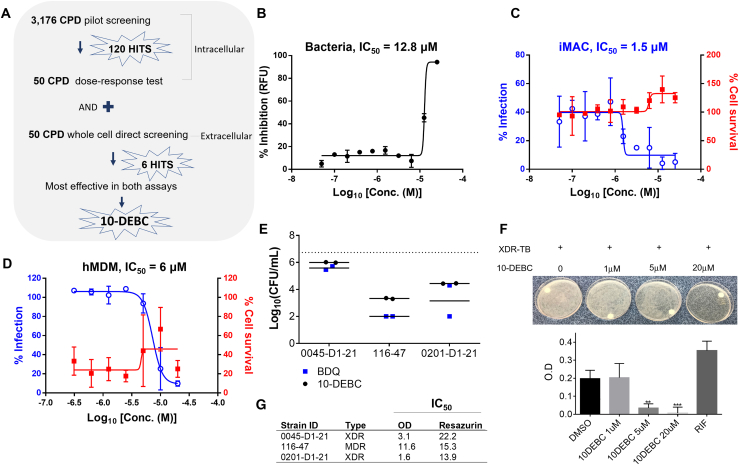
Figure 7.
Anti-TB Activity of 10-DEBC
(A) Schematic illustration of screening assay to narrow down the effective compounds.
(B) Dose-response curve of 10-DEBC against H37Rv-GFP replicating in culture medium. Data represent percent growth inhibition relative to the DMSO-treated control.
(C) Dose-response curve of 10-DEBC against H37Rv-GFP replicating inside iMACs. Data represent percentages of Mtb-infected iMACs (blue line) and surviving iMACs (red line) at 5 days after infection.
(D) Dose-response curve of 10-DEBC against H37Rv-GFP replicating in infected hMDMs treated with 10-DEBC for 5 days.
(E) Anti-mycobacterial activity of 10-DEBC against XDR Mtb replicating inside Raw264.7cells. TB-infected cells were treated with vehicle or 10-DEBC at indicated concentrations for 5 days. Cell lysates were serially diluted and 10 μL of each dilution plated in 7H11 Agar in duplicates. Colonies were counted after 21 days incubation; bedaquiline (BDQ) was used as a positive control. (B–E) Average of two independent experiments.
(F) XDR-TB-infected iMACs were treated with vehicle or 10-DEBC for 5 days. Upper images of intracellular bacterial growth on 7H10 Agar plates. The colonies were suspended in liquid media and the bacterial load was monitored by optical density at 600 nm (OD600) measurement. 10 μM of rifampicin (RIF).
(G) Extracellular anti-mycobacterial activity of 10-DEBC against a panel of three Mtb drug-resistant clinical isolates grown in 7H9 complete medium (~105/well) and treated with test compound for 5 days. Bacterial viability was also assessed with the resazurin reduction assay (n = 3).
10-DEBC Is Effective against XDR and MDR-Mtb
10-DEBC was found to inhibit the growth of Mtb in whole-cell assay with an IC50 of 12.8 μM (Figure 7B), while inhibiting replication of Mtb inside iMACs with an IC50 of 1.5 μM, without host cell toxicity even at the highest concentration of 25 μM (Figure 7C). We also validated the anti-tubercular activity of 10-DEBC in the human primary hMDM model system. Consistently with iMAC model, 10-DEBC showed similar inhibitory effect on Mtb H37Rv-GFP growth in hMDMs, with an IC50 of 6 μM (Figures 7D and S7). These data confirmed that the iMAC-based drug screening platform successfully identified a drug effective in human primary cells. Next, we evaluated 10-DEBC activity against three drug-resistant strains, including two XDR strains. As all three strains were resistant to both INH and RIF, bedaquiline (FDA-approved drug for MDR TB) was introduced as positive control for these experiments. iMACs infected at MOI 20 were treated with 10-DEBC or bedaquiline for 5 days before cell lysis and CFU enumeration. For all three strains, treatment with 10-DEBC significantly inhibited intracellular growth at a concentration of 20 μM (Figures 7E and 7F). In addition, treatment with 10-DEBC directly inhibited the growth of all three strains, with IC50 ranging from 1.6 to 11.6 μM, as determined after measuring the OD600 of the culture 5 days after treatment (Figure 7G).
Discussion
Mtb can remain within human lungs for decades (Flynn et al., 2011, Hussell and Bell, 2014). Macrophages of different lineages exhibit a variety of responses to Mtb infection and can positively or negatively influence Mtb replication (Rohde et al., 2007, Russell et al., 2009). Lungs harbor two major macrophage populations—namely, alveolar and interstitial. The former is derived from fetal liver, whereas the latter is thought to arise from blood monocytes (Guilliams et al., 2013). Macrophages derived from monocyte cell lines, such as THP-1 cells, are genomically, phenotypically, and functionally distinct from hMDMs, and their phenotype is significantly influenced by culture conditions (Aldo et al., 2013, Daigneault et al., 2010), which affect the MOI as well as the reproducibility and efficiency of intracellular anti-TB screening (McClean and Tobin, 2016). Although we examined the responses of iMACs only to a small selection of pathogens, we expect that this model system can be exploited to investigate other pathogen-induced infectious diseases.
Previous studies describing methods for producing PSC-derived macrophages are based on two major methods, achieving the differentiation of precursors into macrophages either directly, or through the formation of embryoid bodies (EB). The major challenge associated with the EB method is to control the number and size of EBs and prevent their agglomeration, which significantly affects the quality of the final product and reproducibility. Alternatively, a cytokine-based 2D direct differentiation culture method allowed to produce phenotypically well-characterized CD14+ macrophages (Yanagimachi et al., 2013), but that were not amenable to scale up. More recently, bio-reactor-based production of macrophages has been reported, and showed the possibility of scaling up of macrophage production (Ackermann et al., 2018). In our study, we found that 2D methods of differentiation can be modified for scale up to produce the desired number of macrophages. In this context, it appeared that maintaining the cells in culture for longer periods of time in wider vessels was critical to obtain larger-scale production. Importantly, we confirmed using RNA-seq that the iMACs remained relatively homogeneous after longer incubation periods, up to 60 days. We also noted that the base medium in direct differentiation protocol largely influenced HSPC yield. We tested several of the commercially available HSPC differentiation culture media and found that differentiation capacity varied according to the base medium.
It is well established now that different types of macrophages are present in human organs (Perdiguero and Geissmann, 2016), and that the function of these cells is largely influence by their environment (Amit et al., 2016). In this study, we tried to understand the similarities and differences between iMACs and primary hMDMs. The GO analysis revealed differences and similarities between iMACs and hMDMs. Approximately 800 DEGs were identified between these two cell types, and GO analysis suggested that iMACs respond differently compared with hMDMs. The nature and intensity of their responses to Mtb infection was different, with hMDMs showing differential expression several orders of magnitude higher than that of iMACs. Nevertheless, both cell types showed consistent immune activation toward bacterial and viral pathogens.
From our screening for anti-TB compounds, we identified 10-DEBC as a potential hit. The risks of finding false-positive compounds are always present due to the nature of the assay (GFP expression and image-based detection). It is thus of importance to validate the activity of hit compounds using CFU counting methods and various macrophage lines. Here we showed that 10-DEBC was effective in eradicating Mtb H37Rv-GFP replicating within several macrophage lineages, but also MDR and XDR Mtb strains using a CFU approach, thus comforting the relevance of this compound as a hit. We noted that the IC50 of 10-DEBC was significantly lowered for intracellular as compared with extracellular Mtb (1.5 versus 12.8 μM). These data suggest that 10-DEBC may target different biochemical pathways in host cells and Mtb. 10-DEBC first reported as a cell-permeable inhibitor of AKT (Thimmaiah et al., 2005). Activation of AKT1 signaling promotes intracellular Mtb survival in macrophages by multiple mechanisms including inhibition of phagosome-lysosome fusion (Kuijl et al., 2007). Genetic or pharmacological inhibition of AKT1 enhanced intracellular killing of Mtb in macrophages by promoting autophagy. The structure of 10-DEBC presents a phenoxazine core, and phenoxazine-derived compounds were shown to exhibit anti-microbial activity, including anti-mycobacterial activity by inhibiting topoisomerase I (Hayashi et al., 2010, Yu et al., 2017). Phenoxazine derivatives were also reported to inhibit mycobacterial efflux pumps involved in resistance to anti-TB drugs (Rodrigues et al., 2011). Therefore, we speculate that 10-DEBC may inhibit Mtb growth by interfering with various mechanisms, both in the bacteria and the host cell. Further studies are needed to characterize the mechanisms by which 10-DEBC mediates intracellular and extracellular killing of this pathogen, and determine whether this compound has potential for anti-TB drug development.
In conclusion, our study describes a reproducible method for large-scale production of macrophages from hESCs and demonstrates its robustness in host-pathogen phenotypic screening for drug discovery. Our iMAC-based Mtb screening platform recapitulates human primary macrophage responses to Mtb. In addition, these iMACs can provide insight into host-pathogen interactions and be used to identify candidate drugs targeting other intracellular replicating pathogens, such as Brucella abortus, Chlamydia pneumoniae, Coxiella burnetiid, and Salmonella typhimurium, among others. Finally, we report 10-DEBC as a new anti-TB compound that is effective in killing XDR and MDR-Mtb and merits further validation in a pulmonary TB disease model.
Experimental Procedures
Details are provided in Supplemental Experimental Procedures.
Ethics Approval
All experiments using hESCs and human peripheral blood mononuclear cells were approved by the institutional review board of the Korea Centers for Disease Control and Prevention (2017-03-07-C-A, 2018-06-01-P-A).
Statistical Analysis
Statistical analyses were performed using Prism v.6 software (GraphPad). Means were compared with the Student's t test, and p values <0.05 were considered significant. The results represent the mean ± SD of three independent experiments.
Author Contributions
H.H., H.S. V.D., D.S., and J.K. designed the study and performed the experiments, analyzed the data, and wrote the manuscript. H.J. H.H, V.C., V.D. J.L, J.H, J.C. and S.L. performed the experiments. H.H. S.H, M.P. designed the study, discussed the results, and performed analyses. R.K.T. and V.D. provided conceptual advice and critically revised the manuscript for intellectual content.
Acknowledgments
This study was supported by grants from the Korea Centers for Disease Control and Prevention (KCDC, 2017-NC61001–00 and 2017-NG61004–00). V.D. was supported by the French Ministry of Foreign Affairs and through grants from the National Research Foundation of Korea (NRF), funded by the Korean Ministry of Science (MSIT, grant NRF-2017M3A9G6068246). We thank to Dr. Kim Jee-Woong at KNIH, Division of biosafety Evaluation and Control department for SEM and TEM.
Published: October 31, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.10.002.
Accession Numbers
The accession number for the RNAseq data reported in this paper is GEO: GSE138398.
Supplemental Information
References
- Ackermann M., Kempf H., Hetzel M., Hesse C., Hashtchin A.R., Brinkert K., Schott J.W., Haake K., Kuhnel M.P., Glage S. Bioreactor-based mass production of human iPSC-derived macrophages enables immunotherapies against bacterial airway infections. Nat. Commun. 2018;9:5088. doi: 10.1038/s41467-018-07570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldo P.B., Craveiro V., Guller S., Mor G. Effect of culture conditions on the phenotype of THP-1 monocyte cell line. Am. J. Reprod. Immunol. 2013;70:80–86. doi: 10.1111/aji.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral L., Viveiros M. Thioridazine: a non-antibiotic drug highly effective, in combination with first line anti-tuberculosis drugs, against any form of antibiotic resistance of mycobacterium tuberculosis due to its multi-mechanisms of action. Antibiotics (Basel) 2017;6 doi: 10.3390/antibiotics6010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I., Winter D.R., Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat. Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Chamberlain L.M., Godek M.L., Gonzalez-Juarrero M., Grainger D.W. Phenotypic non-equivalence of murine (monocyte-) macrophage cells in biomaterial and inflammatory models. J. Biomed. Mater. Res. A. 2009;88:858–871. doi: 10.1002/jbm.a.31930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S.B., Gern B.H., Delahaye J.L., Adams K.N., Plumlee C.R., Winkler J.K., Sherman D.R., Gerner M.Y., Urdahl K.B. Alveolar macrophages provide an early mycobacterium tuberculosis niche and initiate dissemination. Cell Host Microbe. 2018;24:439–446.e4. doi: 10.1016/j.chom.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigneault M., Preston J.A., Marriott H.M., Whyte M.K., Dockrell D.H. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010;5:e8668. doi: 10.1371/journal.pone.0008668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.L., Chan J., Lin P.L. Macrophages and control of granulomatous inflammation in tuberculosis. Mucosal Immunol. 2011;4:271–278. doi: 10.1038/mi.2011.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., De Kleer I., Henri S., Post S., Vanhoutte L., De Prijck S., Deswarte K., Malissen B., Hammad H., Lambrecht B.N. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J. Exp. Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K., Hayashi T., Miyazawa K., Tomoda A. Phenoxazine derivatives suppress the infections caused by herpes simplex virus type-1 and herpes simplex virus type-2 intravaginally inoculated into mice. J. Pharmacol. Sci. 2010;114:85–91. doi: 10.1254/jphs.10027fp. [DOI] [PubMed] [Google Scholar]
- Hoeve M.A., Nash A.A., Jackson D., Randall R.E., Dransfield I. Influenza virus A infection of human monocyte and macrophage subpopulations reveals increased susceptibility associated with cell differentiation. PLoS One. 2012;7:e29443. doi: 10.1371/journal.pone.0029443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Nazarova E.V., Tan S., Liu Y., Russell D.G. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med. 2018;215:1135–1152. doi: 10.1084/jem.20172020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T., Bell T.J. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- Ismail N., McBride J.W. Tick-borne emerging infections: ehrlichiosis and anaplasmosis. Clin. Lab. Med. 2017;37:317–340. doi: 10.1016/j.cll.2017.01.006. [DOI] [PubMed] [Google Scholar]
- Kim C.M., Kim S.W., Kim D.M., Yoon N.R., Jha P., Jang S.J., Ahn Y.J., Lim D., Lee S.H., Hwang S.D. Case report: polymerase chain reaction testing of tick bite site samples for the diagnosis of human granulocytic anaplasmosis. Am. J. Trop. Med. Hyg. 2017;97:403–406. doi: 10.4269/ajtmh.16-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristiansen J.E., Dastidar S.G., Palchoudhuri S., Roy D.S., Das S., Hendricks O., Christensen J.B. Phenothiazines as a solution for multidrug resistant tuberculosis: from the origin to present. Int. Microbiol. 2015;18:1–12. doi: 10.2436/20.1501.01.229. [DOI] [PubMed] [Google Scholar]
- Kuijl C., Savage N.D., Marsman M., Tuin A.W., Janssen L., Egan D.A., Ketema M., van den Nieuwendijk R., van den Eeden S.J., Geluk A. Intracellular bacterial growth is controlled by a kinase network around PKB/AKT1. Nature. 2007;450:725–730. doi: 10.1038/nature06345. [DOI] [PubMed] [Google Scholar]
- McClean C.M., Tobin D.M. Macrophage form, function, and phenotype in mycobacterial infection: lessons from tuberculosis and other diseases. Pathog. Dis. 2016;74 doi: 10.1093/femspd/ftw068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Coronel E., Castanon-Arreola M. Comparative evaluation of in vitro human macrophage models for mycobacterial infection study. Pathog. Dis. 2016;74 doi: 10.1093/femspd/ftw052. [DOI] [PubMed] [Google Scholar]
- Perdiguero E.G., Geissmann F. The development and maintenance of resident macrophages. Nat. Immunol. 2016;17:2–8. doi: 10.1038/ni.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L., Ainsa J.A., Amaral L., Viveiros M. Inhibition of drug efflux in mycobacteria with phenothiazines and other putative efflux inhibitors. Recent Pat. Antiinfect. Drug Discov. 2011;6:118–127. doi: 10.2174/157489111796064579. [DOI] [PubMed] [Google Scholar]
- Rohde K.H., Abramovitch R.B., Russell D.G. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–364. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Russell D.G., Cardona P.J., Kim M.J., Allain S., Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata K., Kozaki T., Lee C.Z.W., Thion M.S., Otsuka M., Lim S., Utami K.H., Fidan K., Park D.S., Malleret B. Induced-pluripotent-stem-cell-derived primitive macrophages provide a platform for modeling tissue-resident macrophage differentiation and function. Immunity. 2017;47:183–198.e6. doi: 10.1016/j.immuni.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Thimmaiah K.N., Easton J.B., Germain G.S., Morton C.L., Kamath S., Buolamwini J.K., Houghton P.J. Identification of N10-substituted phenoxazines as potent and specific inhibitors of Akt signaling. J. Biol. Chem. 2005;280:31924–31935. doi: 10.1074/jbc.M507057200. [DOI] [PubMed] [Google Scholar]
- Van Reeth K., Adair B. Macrophages and respiratory viruses. Pathol. Biol. (Paris) 1997;45:184–192. [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; 2018. Global tuberculosis report 2018. [Google Scholar]
- Yanagimachi M.D., Niwa A., Tanaka T., Honda-Ozaki F., Nishimoto S., Murata Y., Yasumi T., Ito J., Tomida S., Oshima K. Robust and highly-efficient differentiation of functional monocytic cells from human pluripotent stem cells under serum- and feeder cell-free conditions. PLoS One. 2013;8:e59243. doi: 10.1371/journal.pone.0059243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang M., Annamalai T., Bansod P., Narula G., Tse-Dinh Y.C., Sun D. Synthesis, evaluation, and CoMFA study of fluoroquinophenoxazine derivatives as bacterial topoisomerase IA inhibitors. Eur. J. Med. Chem. 2017;125:515–527. doi: 10.1016/j.ejmech.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Production of CD34+CD45+ HSPCs. Scale bar, 200 μm. The time lapse covers a period of about 24 h, with a 30-min time interval between images. Recordings were made with a Lionheart FX system (BioTek Instruments, Winooski, VT, USA) using a 4× objective lens and data were processed with Gen5 Image+ software (BioTek Instruments).
The video shows macrophages produced from HSPCs. Scale bar, 200 μm. The time lapse covers a period of 48 h, with a 30-min time interval between images. Recordings were made with a Lionheart FX system (BioTek Instruments) using a 4× objective lens and data were processed with Gen5 Image+ software (BioTek Instruments).