Abstract
Antisense oligonucleotide (ASO) has the potential to induce off‐target effects due to complementary binding between the ASO and unintended RNA with a sequence similar to the target RNA. Conventional animal studies cannot be used to assess toxicity induced by off‐target effects because of differences in the genome sequence between humans and other animals. Consequently, the assessment of off‐target effects with in silico analysis using a human RNA database and/or in vitro expression analysis using human cells has been proposed.
Our previous study showed that the number of complementary regions of ASOs with mismatches in the human RNA sequences increases dramatically as the number of tolerated mismatches increases. However, to what extent the expression of genes with mismatches is affected by off‐target effects at the cellular level is not clear. In this study, we evaluated off‐target effects of gapmer ASOs, which cleave the target RNA in an RNase H‐dependent manner, by introducing the ASO into human cells and performing microarray analysis. Our data indicate that gapmer ASOs induce off‐target effects depending on the degree of complementarity between the ASO and off‐target candidate genes. Based on our results, we also propose a scheme for the assessment of off‐target effects of gapmer ASOs.
Keywords: antisense, Gapmer, microarray analysis, off‐target effects, pre‐mRNA
We showed that gapmer oligonucleotide (ASO) induce off‐target effects depending on the degree of complementarity between the ASO and off‐target candidate genes. Based on our results, we also proposed a scheme for the assessment of off‐target effects of gapmer ASOs.

1. INTRODUCTION
Antisense oligonucleotide (ASO) therapeutics have been developed extensively over the past several decades and are emerging as the third platform (after small molecules and biologics) for drug development. ASOs are synthetic DNA/RNA‐like single‐stranded oligonucleotides that bind to RNA through sequence‐specific Watson‐Crick base pairing. Two types of standard ASOs exist and act by different mechanisms (Kole, Krainer, & Altman, 2012). The first, gapmer ASOs, contains high‐affinity ribose modifications such as 2′‐methoxyethyl or 2′‐O, 4′‐C‐methylene‐bridged nucleic acid (also known as LNA) (Obika et al., 1997; Singh, Nielsen, Koshkin, & Wengel, 1998) on the wings and DNA in the central gap region, allowing their target to be cleaved by RNase H. So far, two 2′‐methoxyethyl gapmer ASO therapeutics, mipomersen (Kynamro; IONIS Pharmaceuticals and Genzyme) (Thomas et al., 2013) and inotersen (Tegsedi; IONIS Pharmaceuticals and Akcea Therapeutics) (Benson et al., 2018), have been approved as treatments for homozygous familial hypercholesterolemia and polyneuropathy of hereditary transthyretin‐mediated amyloidosis, respectively. The second type of standard ASOs, splice‐switching oligonucleotides, does not induce RNase H‐mediated cleavage of mRNA but instead act by sterically blocking targeted RNA without inducing its degradation. They modulate pre‐mRNA splicing and repair defective RNA to restore the production of functional proteins. Two splice‐switching oligonucleotides, eteplirsen (Exondys 51; Sarepta Therapeutics) (Aartsma‐Rus & Krieg, 2017) and nusinersen (Spinraza; Biogen and IONIS Pharmaceuticals) (Corey, 2017), have been approved as treatments for Duchenne muscular dystrophy and spinal muscular atrophy, respectively.
As with all drugs, ASOs carry the risk of causing unintended toxicity. Several likely mechanisms of ASO toxicity have been proposed (Lindow et al., 2012). One mechanism is toxicity due to hybridization‐dependent off‐target effects. Off‐target effects are the result of complementary binding between the ASO and unintended RNA with a sequence similar to the target RNA, which can cause adverse effects by affecting the expression of unintended genes. Conventional animal studies cannot be used to assess toxicity induced by off‐target effects because of differences in the genome sequence between humans and other animals. Consequently, to predict toxicity caused by off‐target effects, the assessment of off‐target effects with in silico analysis using a human RNA database and/or in vitro expression analysis using human cells has been proposed (Lindow et al., 2012).
We previously used mathematical calculations and in silico analysis to estimate the general number of complementary regions of ASOs with mismatches in human mRNA sequences using several thousand hypothetical ASOs. Our analysis showed that the number of complementary regions increases dramatically as the number of tolerated mismatches increases (Yoshida et al., 2018). However, to what extent the expression of genes with mismatches is affected by the mechanism of off‐target effects when ASOs actually act on human cells is unclear. In the present study, to clarify this point, we evaluated off‐target effects of LNA gapmer ASOs using human cells. We first performed in silico analysis of a human RNA database to identify off‐target candidate genes with complementarity to the gapmer ASOs. We then investigated the relationship between complementarity and off‐target effects by introducing the gapmer ASO into human cells and performing an exhaustive analysis of changes in gene expression using microarray profiling.
2. RESULTS
2.1. In silico analysis using a human pre‐mRNA database
Gapmer ASOs bind not just to mRNA, but also to pre‐mRNA introns to cleave and degrade pre‐mRNA (Kamola et al., 2015; Kasuya et al., 2016). Thus, when searching for off‐target candidate genes of gapmer ASOs, an in silico analysis of a database that fully includes human pre‐mRNA should be conducted. In a prior study, we showed the theoretical number of complementary regions of ASOs with mismatches in human mRNA (Yoshida et al., 2018). As the total size of human pre‐mRNA coding for proteins (1.17 Gb) is approximately 17‐fold higher than that of human mRNA (68.1 Mb) (see Section 4), the number of complementary regions in human pre‐mRNA is also correspondingly greater (Table 1, see Section 4). In the case of 18‐mer ASOs, the theoretical number of sites with perfect matches is < 1, that with one mismatch is approximately one (0.92), that with two mismatches is 23, that with three mismatches is 375, and that with four mismatches is 4,216 in the human pre‐mRNA sequences (Table 1, see the row “Length of ASO = 18”).
Figure 4.
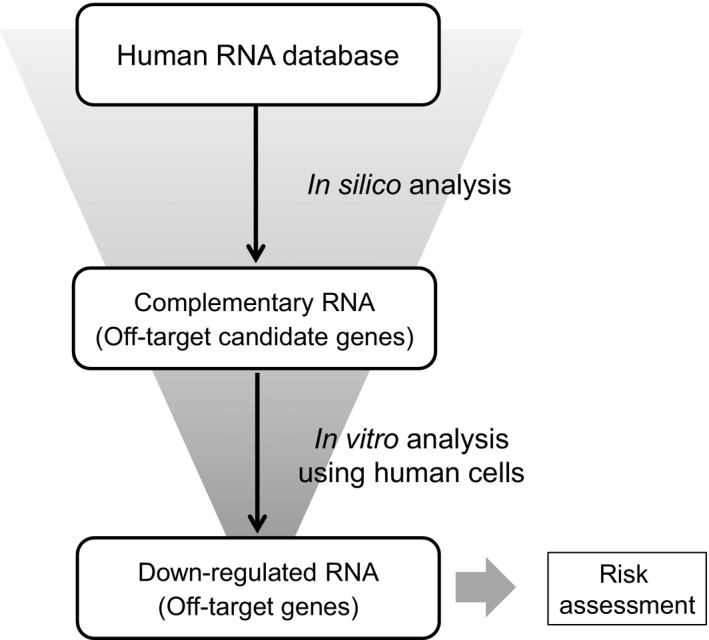
Scheme for the assessment of hybridization‐dependent off‐target effects of gapmer ASOs. In silico analysis: Off‐target candidate genes with complementary RNA sequences are selected from a human RNA database (e.g., D3G) using an appropriate search algorithm (e.g., GGGenome). In vitro analysis using human cells: The ASO is introduced into cultured human cells, and the changes in gene expression of off‐target candidate genes are analyzed. Off‐target candidate genes are narrowed down by considering those with gene expression down‐regulated to <50% as off‐target genes. Risk assessment: The risk of adverse effects emerging from the off‐target genes is investigated by comprehensive consideration of the function, etc., of the gene
Table 1.
Theoretical number of complementary regions of ASOs in the total size of the human pre‐mRNA sequences
| Length of ASO (mer) | Number of mismatches | |||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | |
| 27 | <10−1 | <10−1 | <10−1 | <10−1 | <10−1 | 1.3 |
| 26 | <10−1 | <10−1 | <10−1 | <10−1 | 0.31 | 4.1 |
| 25 | <10−1 | <10−1 | <10−1 | <10−1 | 1.1 | 13 |
| 24 | <10−1 | <10−1 | <10−1 | 0.23 | 3.6 | 43 |
| 23 | <10−1 | <10−1 | <10−1 | 0.79 | 12 | 136 |
| 22 | <10−1 | <10−1 | 0.14 | 2.8 | 39 | 425 |
| 21 | <10−1 | <10−1 | 0.50 | 9.5 | 129 | 1,314 |
| 20 | <10−1 | <10−1 | 1.8 | 33 | 417 | 4,005 |
| 19 | <10−1 | 0.24 | 6.5 | 111 | 1,335 | >104 |
| 18 | <10−1 | 0.92 | 23 | 375 | 4,216 | >104 |
| 17 | <10−1 | 3.5 | 83 | 1,249 | >104 | >104 |
| 16 | 0.27 | 13 | 294 | 4,115 | >104 | >104 |
| 15 | 1.1 | 49 | 1,029 | >104 | >104 | >104 |
| 14 | 4.4 | 183 | 3,566 | >104 | >104 | >104 |
| 13 | 17 | 679 | >104 | >104 | >104 | >104 |
| 12 | 70 | 2,508 | >104 | >104 | >104 | >104 |
| 11 | 279 | 9,196 | >104 | >104 | >104 | >104 |
| 10 | 1,114 | >104 | >104 | >104 | >104 | >104 |
In the present study, a 13‐mer LNA gapmer targeting human APOB, named gap‐A13 (Straarup et al., 2010), was used as an ASO therapeutic model. We first performed in silico analysis to identify human pre‐mRNAs with regions that are complementary to gap‐A13, or off‐target candidate genes of gap‐A13. Here, we used a complementarity measure called the “distance” (d), which is defined as the total number of mismatches, insertions or deletions between ASO and the complementary RNA sequences (Figure 1, see Section 4). The numbers of off‐target candidate genes with respect to gap‐A13 that were found were d = 0 (i.e., perfect match), 16; d = 1,849; d = 2,8526; and d = 3,8276(Figure 3a: see the row “Complementary genes”).
Figure 1.
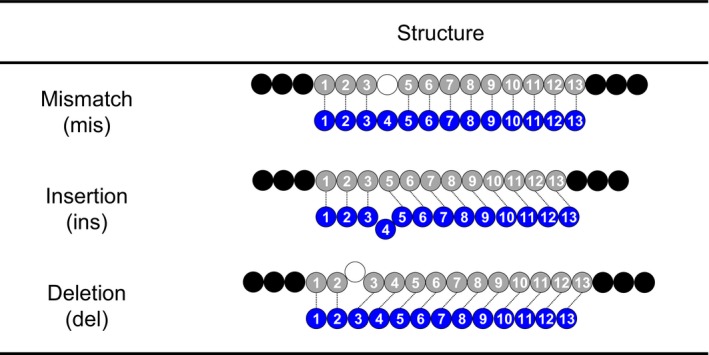
Types of alignment between ASO and complementary RNA. Gray/black: complementary RNA; blue: ASO
Figure 3.
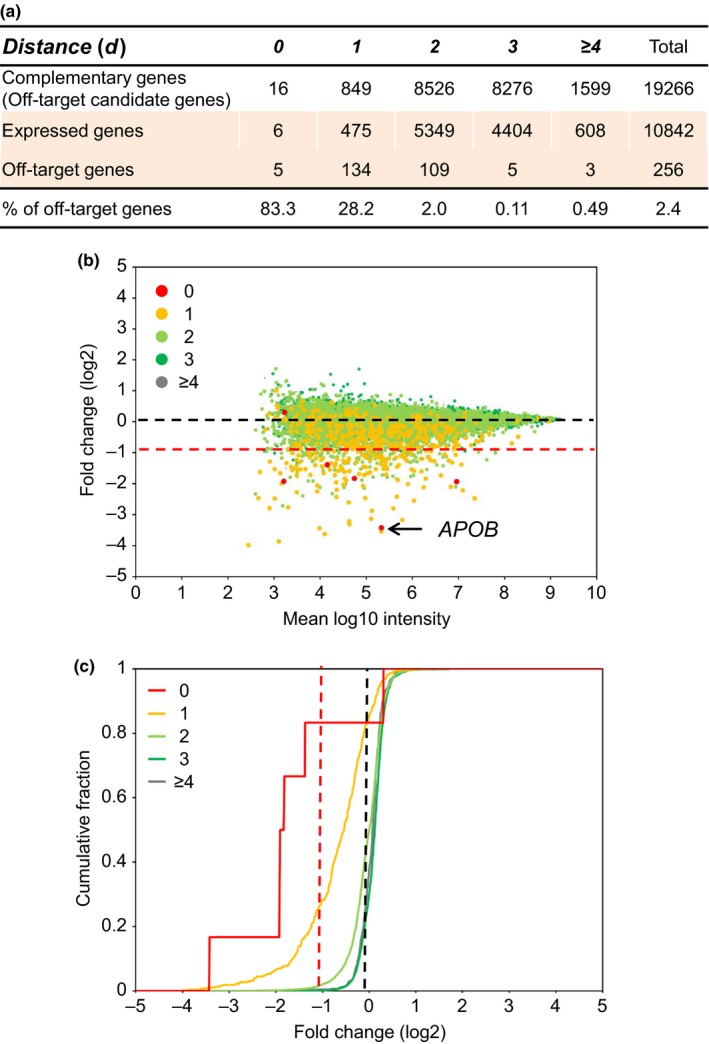
In silico and in vitro analysis of off‐target effects induced by gap‐A13. (a) Statistics from microarray analysis. Complementary genes: The number of off‐target candidate genes identified by in silico analysis. Expressed genes: The number of genes among those identified in “Complementary genes” that are on a microarray and were expressed in Huh‐7 cells. Off‐target genes: The number of genes among those in “Expressed genes” in which gene expression was down‐regulated to less than 50% of the level in the control group. % of off‐target genes: The proportion of genes in “Off‐target genes” that were in “Expressed genes.” (b) Scatter plot. The amount of gene expression in the control cells is shown on the horizontal axis, and the proportion of change in gene expression as a result of introduction of the ASO (expressed logarithmically) is shown on the vertical axis. (c) Cumulative plot. The cumulative number of genes is shown on the vertical axis, and the proportion of change in gene expression as a result of introduction of the ASO (expressed logarithmically) is shown on the horizontal axis
2.2. In vitro analysis using human cells
gap‐A13 was then introduced into the human hepatocyte cell line Huh‐7, and the changes in gene expression were comprehensively analyzed using a microarray. We first examined the expression of the on‐target gene APOB by qRT‐PCR analysis and found that was down‐regulated to 14% of the level in the control (Figure 2a). Microarray analysis was carried out in the same conditions, and APOB expression was down‐regulated to 9% of the level in the control (Table 2 and Figure 3b: red dot indicated by the arrow). To analyze off‐target effects, changes in gene expression were analyzed in each group classified by d, as described above. “Off‐target genes” were defined as genes in which gene expression was down‐regulated to <50% of the level in the control group. This is because heterozygous knockout animals, in which 50% of gene expression is retained, do not show abnormalities in most cases. The degree of change in expression is shown as a scatter plot in Figure 3b, which shows that down‐regulation is highest with d = 0 genes, shown as red dots, which have perfect complementarity. We also observed an overall trend toward down‐regulation among the d = 1 genes, shown as orange dots (Figure 3b). The expression of d = 3 and d ≥ 4 genes did not appear to be affected (Figure [Link], [Link]a,b). Analysis using a cumulative curve also showed that the expression was down‐regulated in a greater proportion of genes among those with higher complementarity (Figure 3c). Quantitative analysis showed that the numbers of off‐target genes, that is, genes down‐regulated to less than 50%, by group were d = 0, 5; d = 1, 134; d = 2, 109; d = 3, 5; and d ≥ 4, 3 (Figure 3a: Off‐target genes, Figure 3b: number of genes shown below the red dotted line). These numbers were expressed as ratios to the number of genes that could be analyzed by microarray and were expressed in Huh‐7 cells (Figure 3a: Expressed genes): d = 0, 83.3%; d = 1, 28.2%; d = 2, 2.0%; d = 3, 0.11%; and d ≥ 4, 0.49% (Figure 3a: % of off‐target genes, Figure 3c: intersection with the red dotted line).
Figure 2.
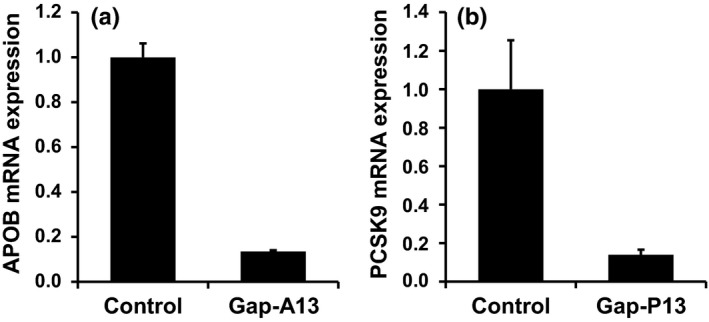
The expression level of the target genes analyzed with qRT‐PCR. (a) The mRNA expression level of the target gene, APOB, in gap‐A13‐treated cells. (b) The mRNA expression level of the target gene, PCSK9, in gap‐P13‐treated cells. The bar graph shows the mean ± standard deviation (n = 4)
Table 2.
Examples of off‐target candidate genes of gap‐A13 selected by in silico analysis
| Gene symbol | Alignment | Position | Distance (d) | mis | ins | del | mRNA level (Microarray), % | mRNA level (qRT‐PCR), % | Off‐target Effect |
|---|---|---|---|---|---|---|---|---|---|
| APOB |

|
Exon 26 | 0 | 0 | 0 | 0 | 9 | 14 | + (on‐target) |
| ASAH1 |

|
Intron 2 | 0 | 0 | 0 | 0 | 26 | 21 | + |
| LRP1B |

|
Intron 2 | 1 | 1 | 0 | 0 | 6 | 9 | + |
| KMT2E |

|
Intron1 | 1 | 1 | 0 | 0 | 9 | 12 | + |
| SCFD2 |

|
Intron5 | 1 | 0 | 1 | 0 | 11 | 10 | + |
| DAAM1 |

|
Intron1 | 1 | 0 | 1 | 0 | 21 | 25 | + |
| GPHN |

|
Intron 3 | 1 | 0 | 0 | 1 | 8 | 11 | + |
| PLCB1 |

|
Intron 3 | 1 | 0 | 0 | 1 | 28 | 24 | + |
| SULT1C2 |

|
Intron 3 | 2 | 1 | 1 | 0 | 23 | 25 | + |
| SLC5A12 |

|
Intron 4 | 2 | 0 | 2 | 0 | 29 | 23 | + |
| KCNJ16 |

|
Intron 1 | 2 | 0 | 0 | 2 | 48 | 42 | + |
| MDGA2 |

|
Intron 2 | 1 | 0 | 1 | 0 | 14 | 56 | − |
| SNX2 |

|
Exon 15 | 1 | 0 | 0 | 1 | 47 | 72 | − |
Next, the same analysis was performed using the other 13‐mer LNA gapmer, gap‐P13, which targets human PCSK9 (Gupta et al., 2010). With gap‐P13, the only d = 0 gene present that was analyzable by microarray and was expressed in Huh‐7 cells was PCSK9 itself (Figure [Link], [Link]b: red dot indicated by the arrow), with gene expression down‐regulated to 14% (Figure 2b: qRT‐PCR analysis) and 17% (Figure [Link], [Link]b: microarray analysis). In the scatter plot analysis, a tendency toward down‐regulation of d = 1 off‐target candidate genes (the orange dots), but not d = 3 and d ≥ 4 genes, was seen (Figure [Link], [Link]b–d). Analysis using a cumulative curve also showed that the expression of d = 1 and d = 2 off‐target candidate genes was down‐regulated significantly (Figure [Link], [Link]e). The ratios of off‐target genes in which gene expression was down‐regulated to less than 50% of control to expressed genes were d = 0, 100%; d = 1, 54.5%; d = 2, 15.9%; d = 3, 1.8%; and d ≥ 4, 2.8% (Figure [Link], [Link]a: % of off‐target genes, Figure [Link], [Link]e: intersection with the red dotted line).
These results indicate that for both gap‐A13 and gap‐P13, the proportion of off‐target genes was higher at a higher level of complementarity. This clearly indicates that gapmer ASOs induced off‐target effects depending on the degree of complementarity between the ASO and off‐target candidate genes. In addition, the results showed that with the 13‐mer LNA gapmers used for analysis in the present study, the expression of off‐target candidate genes up to d = 2 could actually be down‐regulated to less than 50% at the cellular level.
3. DISCUSSION
In the present study, we demonstrated that gapmer ASOs induced off‐target effects depending on the degree of complementarity that was classified by the total number of mismatches (mis), insertions (ins) or deletions (del) on the complementary binding regions, named the distance (d). Our analysis indicated that not only the expression of genes with mis, but also those with ins or del, has the potential to be affected by off‐target effects induced by gapmer ASOs (Table 2). This finding is agreement with previous reports showing off‐target effects of LNA gapmers (Hagedorn et al., 2018; Kamola et al., 2015). We further analyzed the proportion of off‐target genes to the off‐target candidate genes in mis = 1, ins = 1 and del = 1 groups, and found that a significant proportion of ins = 1 and del = 1 genes was down‐regulated to less than 50% of the control in a similar manner to that of mis = 1 genes (Table S1). These data emphasize the need to identify genes with ins and/or del as off‐target candidate genes of gapmer ASOs and support the usefulness of classification of off‐target candidate genes by the distance (d). Note that searching off‐target candidate genes based on the distance (d) can be easily performed using GGGenome by giving d as the parameter. This type of search cannot be handled easily by popular sequence search software such as BLAST.
In vitro analysis using human cells with gapmer ASO, gap‐A13, showed that 134 of 475 d = 1 expressed genes were the off‐target genes showing down‐regulation to less than 50% with respect to the control (Figure 3a: Expressed genes, Off‐target genes). Examples of these d = 1 off‐target genes were shown in Table 2 (LRP1B, KMT2E, SCFD2, DAAM1, GPHN and PLCB1). Another 341 genes were the non‐off‐target genes that were not down‐regulated to less than 50%. Examples included MDGA2 and SNX2 in Table 2, and CPNE1, CHML and CENPC in Table S2. We compared the 13‐mer sequences within the complementary region of 134 off‐target genes with those of 341 non‐off‐target genes; however, we could not identify definitive sequence rules that define off‐target genes so far. A similar analysis was performed with d = 2 off‐target genes and d = 2 non‐off‐target genes, and again we could not find any regularity. This may be partially because the cleaving activity of gapmer ASOs depends not just on the sequence of the complementary binding site, but also on the higher order structure of the RNA to which it binds complementarily. The foregoing analyses show that definitive prediction of which off‐target candidate genes will actually be subject to down‐regulation of gene expression to less than 50% of the control from an in silico study alone is difficult. Thus, also performing in vitro analysis using human cells to narrow down a list of off‐target candidate genes is useful.
The possible methods for in vitro analysis include qRT‐PCR and microarray. The number of off‐target candidate genes identified by in silico analysis, even when these were limited to d = 0 and d = 1, was 865 for gap‐A13 [16 (d = 0) + 849 (d = 1) = 865] and 475 for gap‐P13 [3 (d = 0) + 472 (d = 1) = 475]. Realistically, such numbers of genes cannot be analyzed with qRT‐PCR. With ASOs longer than 13‐mer, the number of d = 0 and d = 1 genes is expected to decrease, but longer ASOs have greater binding affinity with the complimentary RNA, so that off‐target effects may occur even with higher d numbers. Thus, if the criteria for selecting off‐target candidate genes are expanded to include d = 3, d = 4, etc., the number of off‐target candidate genes will range from several dozen to several thousand (Table 1). Taking the above into account, the most efficient method for assessing the off‐target effects of ASOs using human cells is a comprehensive method such as microarray analysis. To evaluate the validity of identifying off‐target genes based on microarray analysis, we performed qPCR analysis for 36 off‐target genes identified in microarray analysis of gapA‐13. As a result, 30 out of 36 genes were down‐regulated to less than 50% by qPCR analysis in a similar manner in microarray analysis. Other 6 genes were also down‐regulated, though the extent of down‐regulation was less in qPCR analysis than that observed in microarray analysis (examples were shown in Table 2). From these data, we suggest that the combination of comprehensive microarray analysis and specific qPCR analysis is more desirable way to determine off‐target genes.
Based on the above results, we propose a scheme for the assessment of off‐target effects of gapmer ASOs (Figure 4). When assessing the off‐target effects of gapmer ASOs, in vitro analysis using human cells is useful for narrowing down the off‐target candidate genes identified by in silico analysis to those in which gene expression is actually down‐regulated. Some of the off‐target genes identified by this scheme could be downstream targets of the on‐target gene (not real off‐target genes). These downstream targets could be identified as commonly down‐regulated genes by microarray analysis using another ASO against the same on‐target gene as described previously (Hagedorn et al., 2018); however, in the light of prediction of gene expression change in human, we consider that the downstream targets are not needed to be excluded purposely from off‐target genes because they are also predicted to be down‐regulated in humans in a similar manner to the real off‐target genes.
The present study indicates that with 13‐mer LNA gapmers, genes with complementarity up to d = 2 may be subject to off‐target effects (Figures 3 and [Link], [Link]). Therefore, in the scenario of a 13‐mer LNA gapmer being developed as an oligonucleotide therapeutic, off‐target candidate genes with complementarity up to d = 2 would be selected in the in silico analysis, which is the first stage in the assessment of off‐target effects. The induction of off‐target effects is assumed to be related to the strength of binding between the ASO and complementary RNA, and is therefore likely to vary depending not only on the length of the gapmer ASO but also on the type of chemically modified nucleic acids introduced into the ASO. In general, the strength of binding between the ASO and complementary RNA increases as the oligonucleotide becomes longer, so that LNA gapmer ASOs longer than 13‐mer may induce off‐target effects on d = 3 genes in addition to d = 0, 1 and 2 genes. Therefore, we consider that a criterion for selection of off‐target candidate genes, which is defined by d, varies depending on the length of the ASO and/or chemically modified nucleic acids used in the ASO.
In in vitro analysis using human cells, expression analysis should be investigated in conditions in which the expression of the target gene is down‐regulated to less than 50%. As for off‐target genes in which gene expression was down‐regulated to less than 50% of the control, evaluation of the possible functional consequences of altering the expression of the off‐target gene product should be done as proposed previously (Lindow et al., 2012).
4. EXPERIMENTAL PROCEDURES
4.1. Estimation of the general number of complementary regions of ASOs in human pre‐mRNA sequences
We mathematically calculated the theoretical number of complementary regions of ASOs with perfect matches or mismatches in the total size of human pre‐mRNA sequences in a similar manner as described previously (Yoshida et al., 2018). We hypothesized that the four bases (A, G, C and T) are used randomly in human pre‐mRNA sequences. The human mRNAs and pre‐mRNAs span 68 Mb and 1.17 Gb respectively, according to D3G database (release 18.04; https ://d3g.riken.jp/). D3G consists of RefSeq mRNA transcripts (NM_ and YP_ entries; O'Leary et al., 2016) and their genomic coordinates on the reference human genome (GRch38) provided in the UCSC Genome Browser (Casper et al., 2018). Human pre‐mRNA sequences are approximately 17‐fold longer than human mRNAs owing to their intronic regions.
4.2. In silico analysis
Sequence searches allowing mismatches, insertions or deletions were performed using GGGenome (https://GGGenome.dbcls.jp/), rather than the widely used BLAST software (Altschul, Gish, Miller, Myers, & Lipman, 1990). BLAST may overlook potential complementary regions, as mentioned in our previous work describing siDirect (Naito, Yamada, Ui‐Tei, Morishita, & Saigo, 2004) and CRISPRdirect (Naito, Hino, Bono, & Ui‐Tei, 2015), which are web servers for designing off‐target‐minimized siRNA and CRISPR guide RNA, respectively. GGGenome quickly searches short nucleotide sequences utilizing suffix array and FM‐index stored on solid‐state drives, and we used it to query the human pre‐mRNA sequences described above that were retrieved from D3G. Here, we used a complementarity measure called the “distance” (d), which is defined as the total number of mismatches, insertions or deletions between ASO and the complementary RNA sequences. Pre‐mRNA sequences were grouped by d according to the highest complementary site (i.e., with minimal d). For example, if two complementary sites with d = 1 and d = 3 are present in one pre‐mRNA sequence, such as one d = 1 site in exon 2 and one d = 3 site in exon 4 in one pre‐mRNA sequence, the pre‐mRNA was classified into the d = 1 group because the d = 1 site is more likely to be affected by the ASO compared to the d = 3 site. Pre‐mRNAs with complementary RNA sequence to the gapmer ASO are called “Complementary genes (Off‐target candidate genes)” in in silico analysis (Figure 3a and [Link], [Link]a).
4.3. ASOs
An LNA gapmer ASO targeting human APOB pre‐mRNA (gap‐A13) and one targeting human PCSK9 pre‐mRNA (gap‐P13) were synthesized and purified by Gene Design, Inc. Both ASOs were 13‐mer phosphorothioated oligonucleotides with a gapmer design (2‐mer LNA + 8‐mer DNA + 3‐mer LNA). Detailed sequence information is shown in Table S3.
4.4. Cell culture and transfection with ASOs
The human hepatoma cell line Huh‐7 was obtained from the Japanese Collection of Research Bioresources (JCRB). The cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's Medium (Sigma‐Aldrich) supplemented with 10% heat‐inactivated fetal bovine serum and antibiotics. Huh‐7 cells were seeded into 12‐well plates (Corning) at 1.5 × 104 cells/well (n = 4/group) and transfected with 20 nM gapmer ASO (gap‐A13 or gap‐P13) using Lipofectamine 2000 (Invitrogen, Gaithersburg, MD, USA) according to the manufacturer's protocol. Cells treated with transfection reagent in the absence of ASO were used as a control. After a further 24 hr, total RNA was isolated using a Qiagen RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. Cell viability was determined by WST‐8 assay according to the manufacturer's protocol. For analysis of off‐target effects, we determined ASO concentrations at which on‐target gene is down‐regulated to less than 25% of the control and cell viability is more than 80%.
4.5. Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR)
qRT‐PCR was performed using a One Step SYBR PrimeScript PLUS RT‐PCR Kit (Takara Bio, Inc.) and analyzed with a 7,500 Fast Real‐Time PCR System (Applied Biosystems). The primers used in this study are shown in Table S4. The level of target gene expression was normalized to that of human GAPDH.
4.6. Microarray analysis
Genome‐wide expression analysis was performed using Human Genome U133 Plus 2.0 GeneChip (Affymetrix, Inc.) according to the manufacturer's instructions. The scanned data were processed for signal values using Microarray Suite 5.0 software (Affymetrix). Genes with sufficient hybridization signals to be called “present” in at least three out of four trials were used in this study (called “Expressed genes”in Figure 3a and [Link], [Link]a). We defined “off‐target genes” as genes down‐regulated to less than 50% of the level in the control group (“Off‐target genes” in Figure 3a and [Link], [Link]a).
Supporting information
ACKNOWLEDGMENTS
This study was supported by AMED under Grant numbers JP19mk0101119, JP19ak0101073, JP19kk0305008 and JP18am0301004. We thank Mr Isamu Muto, BioInformation Technology & Science (BITS) Co., Ltd., for technical assistance with the in silico analyses.
Yoshida T, Naito Y, Yasuhara H, et al. Evaluation of off‐target effects of gapmer antisense oligonucleotides using human cells. Genes Cells. 2019;24:827–835. 10.1111/gtc.12730
Communicated by: Yoshihiro Yoneda
REFERENCES
- Aartsma‐Rus, A. , & Krieg, A. M. (2017). FDA Approves eteplirsen for duchenne muscular dystrophy: The next chapter in the eteplirsen saga. Nucleic Acid Therapeutics, 27, 1–3. 10.1089/nat.2016.0657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F. , Gish, W. , Miller, W. , Myers, E. W. , & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Benson, M. D. , Waddington‐Cruz, M. , Berk, J. L. , Polydefkis, M. , Dyck, P. J. , Wang, A. K. , … Coelho, T. (2018). Inotersen treatment for patients with hereditary transthyretin amyloidosis. The New England Journal of Medicine, 379, 22–31. 10.1056/NEJMoa1716793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, J. , Zweig, A. S. , Villarreal, C. , Tyner, C. , Speir, M. L. , Rosenbloom, K. R. , … Kent, W. J. (2018). The UCSC genome browser database: 2018 update. Nucleic Acids Research, 46, D762–D769. 10.1093/nar/gkx1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey, D. R. (2017). Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy. Nature Neuroscience, 20, 497–499. 10.1038/nn.4508 [DOI] [PubMed] [Google Scholar]
- Gupta, N. , Fisker, N. , Asselin, M. C. , Lindholm, M. , Rosenbohm, C. , Orum, H. , … Straarup, E. M. (2010). A locked nucleic acid antisense oligonucleotide (LNA) silences PCSK9 and enhances LDLR expression in vitro and in vivo. PLoS ONE, 5, e10682 10.1371/journal.pone.0010682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn, P. H. , Pontoppidan, M. , Bisgaard, T. S. , Berrera, M. , Dieckmann, A. , Ebeling, M. , … Lindow, M. (2018). Identifying and avoiding off‐target effects of RNase H‐dependent antisense oligonucleotides in mice. Nucleic Acids Research, 46, 5366–5380. 10.1093/nar/gky397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamola, P. J. , Kitson, J. D. , Turner, G. , Maratou, K. , Eriksson, S. , Panjwani, A. , … Parry, J. D. (2015). In silico and in vitro evaluation of exonic and intronic off‐target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Research, 43, 8638–8650. 10.1093/nar/gkv857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuya, T. , Hori, S.‐I. , Watanabe, A. , Nakajima, M. , Gahara, Y. , Rokushima, M. , … Kugimiya, A. (2016). Ribonuclease H1‐dependent hepatotoxicity caused by locked nucleic acid‐modified gapmer antisense oligonucleotides. Scientific Reports, 6, 30377 10.1038/srep30377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole, R. , Krainer, A. R. , & Altman, S. (2012). RNA therapeutics: Beyond RNA interference and antisense oligonucleotides. Nature Reviews Drug Discovery, 11, 125–140. 10.1038/nrd3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow, M. , Vornlocher, H.‐P. , Riley, D. , Kornbrust, D. J. , Burchard, J. , Whiteley, L. O. , … Levin, A. A. (2012). Assessing unintended hybridization‐induced biological effects of oligonucleotides. Nature Biotechnology, 30, 920–923. 10.1038/nbt.2376 [DOI] [PubMed] [Google Scholar]
- Naito, Y. , Hino, K. , Bono, H. , & Ui‐Tei, K. (2015). CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off‐target sites. Bioinformatics, 31, 1120–1123. 10.1093/bioinformatics/btu743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito, Y. , Yamada, T. , Ui‐Tei, K. , Morishita, S. , & Saigo, K. (2004). siDirect: Highly effective, target‐specific siRNA design software for mammalian RNA interference. Nucleic Acids Research, 32, W124–129. 10.1093/nar/gkh442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obika, S. , Nanbu, D. , Hari, Y. , Morio, K. , In, Y. , Ishida, T. , & Imanishi, T. (1997). Synthesis of 2'‐O,4'‐C‐methyleneuridine and ‐cytidine. Novel bicyclic nucleosides having a fixed C3'‐endo sugar puckering. Tetrahedron Letters, 38, 8735–8738. [Google Scholar]
- O'Leary, N. A. , Wright, M. W. , Brister, J. R. , Ciufo, S. , Haddad, D. , McVeigh, R. , … Pruitt, K. D. (2016). Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Research, 44, D733–745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. K. , Nielsen, P. , Koshkin, A. A. , & Wengel, J. (1998). LNA (locked nucleic acids): Synthesis and high‐affinity nucleic acid recognition. Chemical Communications, 4, 455–456. 10.1039/a708608c [DOI] [Google Scholar]
- Straarup, E. M. , Fisker, N. , Hedtjärn, M. , Lindholm, M. W. , Rosenbohm, C. , Aarup, V. , … Koch, T. (2010). Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non‐human primates. Nucleic Acids Research, 38, 7100–7111. 10.1093/nar/gkq457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, G. S. , Cromwell, W. C. , Ali, S. , Chin, W. , Flaim, J. D. , & Davidson, M. (2013). Mipomersen, an apolipoprotein B synthesis inhibitor, reduces atherogenic lipoproteins in patients with severe hypercholesterolemia at high cardiovascular risk: A randomized, double‐blind, placebo‐controlled trial. Journal of the American College of Cardiology, 62, 2178–2184. 10.1016/j.jacc.2013.07.081 [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Naito, Y. , Sasaki, K. , Uchida, E. , Sato, Y. , Naito, M. , … Inoue, T. (2018). Estimated number of off‐target candidate sites for antisense oligonucleotides in human mRNA sequences. Genes to Cells, 23, 448–455. 10.1111/gtc.12587 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


