Abstract
Background
Late-Onset Alzheimer’s Disease (LOAD) is a leading form of dementia. There is no effective cure for LOAD, leaving the treatment efforts to depend on preventive cognitive therapies, which stand to benefit from the timely estimation of the risk of developing the disease. Fortunately, a growing number of Machine Learning methods that are well positioned to address this challenge are becoming available.
Results
We conducted systematic comparisons of representative Machine Learning models for predicting LOAD from genetic variation data provided by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Our experimental results demonstrate that the classification performance of the best models tested yielded ∼72% of area under the ROC curve.
Conclusions
Machine learning models are promising alternatives for estimating the genetic risk of LOAD. Systematic machine learning model selection also provides the opportunity to identify new genetic markers potentially associated with the disease.
Keywords: Alzheimer’s disease, Machine learning, Benchmarking, Genome-wide association studies
Background
Alzheimer Disease (AD) is a neurodegenerative disorder that gradually destroys brain function. It is characterized by the loss of cognitive abilities such as memory, reasoning, language, and behavior. The disease leads to dementia and ultimately to death. AD is the most common form of dementia (60% – 80% cases) and occurs more often in people aged 65 and older[1]. Age is not the only risk factor for developing AD, it has been observed that there are specific inherited genetic traits that increase the risk of Early-Onset AD (EOAD) at an early age (<60). Apart from the age differences, the clinical presentation of EOAD is very similar to the presentation of late-onset AD (LOAD) and many aspects of the disease overlap with normal again in many clinical and pathological aspects. The EOAD by family inheritance is characterized by genetic mutations in the APP, PSEN1, and PSEN2, related to amyloids but only accounts for 5% of total AD [2].
The high prevalence of LOAD among the elderly is caused by the increasing life expectancy coupled with the lack of an effective treatment to either stop the advance of the sickness or reverse the damage caused by it; and up to this date, there are only two FDA-approved drugs to treat AD cognitive symptoms. An estimate from Ballard [3] shows that Alzheimer’s Disease affects between 4 and 6 percent of the population around 65 years old, that the incidence of the disease doubles every five years after 65 years of age, and by age of 85 between 30%-50% is affected by some form of AD. Therefore, there are a lot of efforts aimed at developing effective AD therapies, and it is expected that preventive ones have a greater impact before the development of the disease [4]. To apply these preventive treatments, a key component is detecting those individuals at risk at an early stage of the disease. There are multiple existing methods such as cognitive tests, magnetic resonance imaging (MRI), positron emission tomography (PET) images, cerebrospinal and blood biomarkers that can determine the development of AD [5]. But these methods do not detect the formation or propensity of the disease at a sufficiently early stage to be highly effective. Additionally, pathological postmortem examination is required for confirmatory diagnosis [6]. To complicate matters further, these biomarkers and MRI features develop in a correlated manner with the development of the disease and are at their most usefulness for prediction when the disease has progressed to the final stages.
A promising method for improving the prediction of LOAD is through the study of risk factors, and genetic testing has become an important source of information that can profile the genetic component of LOAD risk. One specific case is the gene Apolipoprotein E(APOE) and its different alleles, which have been implicated as the largest genetic risk factors for LOAD. Late-Onset Alzheimer’s Disease is a complex multifactorial disease; thus, the APOE variants do not give a definite prediction of the disease by themselves.
Multiple other genes such as CLU, PICALM, CR1 [7] have been shown to be statistically correlated and biochemically plausible. These common variants found using multiple genome-wide association studies (GWAS) have been shown to explain only 33% of the phenotypic variance of LOAD, while the expected heritability component of LOAD is around 79%, thus leaving over 40% unexplained [8]. LOAD is expected to have a known genetic component, a missing (so far) genetic component, and multiple environmental factors that contribute to the complexity of the disease [9].
The complexity of LOAD can be studied using modern machine learning (ML) strategies that leverage well-planned AD studies. With the aim to discern and discover the multiple factors that affect the onset of AD, the Alzheimer’s Disease Neuroimaging Initiative (ADNI) launched a longitudinal study to: “develop clinical, imaging, genetic, and biochemical biomarkers for the early detection and tracking of Alzheimer’s disease (AD)”. The first goal of the study is: “To detect AD at the earliest possible stage (pre-dementia) and identify ways to track the disease’s progression with biomarkers” [10]. Therefore, ADNI is a well-planned study that produces the required data to be data mined by ML. There have been several machine learning strategies that have been used to explore early stages of AD [11–13]. Most of the ML approaches are based on exploring univariate associations with MCI to AD conversions [13], and some efforts have been made in building predictive multivariate models based on merging clinical, MRI, laboratory and PET imaging [14]. These efforts have been very successful, and there are several alternatives to predict the early stages of LOAD [15]. On the other hand, similar ML approaches can be used to predict AD risk based on gene variants; but most of the efforts have been constrained to the use of advanced statistical approaches [16]. To fully explore the potential of gene biomarkers in the prediction of LOAD, multivariate ML is required. The number of approaches to be explored is very large, and their validation requires complex exploration of prediction performance and evaluation of the internal structure, i.e., what are the Single Nucleotide Polymorphisms (SNP) involved in the successful prediction of LOAD? Hence, the aim of this work was to explore the performance of genetic-based ML multivariate strategies in predicting LOAD and to describe the main genetic features associated with the risk of developing LOAD.
To achieve this goal, we used the benchmark tool implemented in FRESA.CAD (Feature Selection Algorithms for Computer Aided Diagnosis) [17, 18]. The benchmark tool evaluates statistical feature selection methods, wrapper/filter ML methods, and the ensemble of models in a coherent cross-validation and repetition method yielding a high degree of statistical confidence of the test performance. FRESA.CAD additionally has the advantage of returning the features most selected across the models and can extrapolate to a valid analysis of the gene variants which allows a more direct interpretation. We propose the hypothesis that the FRESA.CAD Benchmarking tool can achieve high predictive results by comparing and analyzing multiple Machine Learning models applied to predict the genetic risk a person has of developing Alzheimer’s Disease from genetic information only. We expect these models to explain more of the missing heritability than simpler models as the methods can represent nonlinearities from gene interactions and use a broader amount of SNPs in contrast to single markers from GWAS.
Results
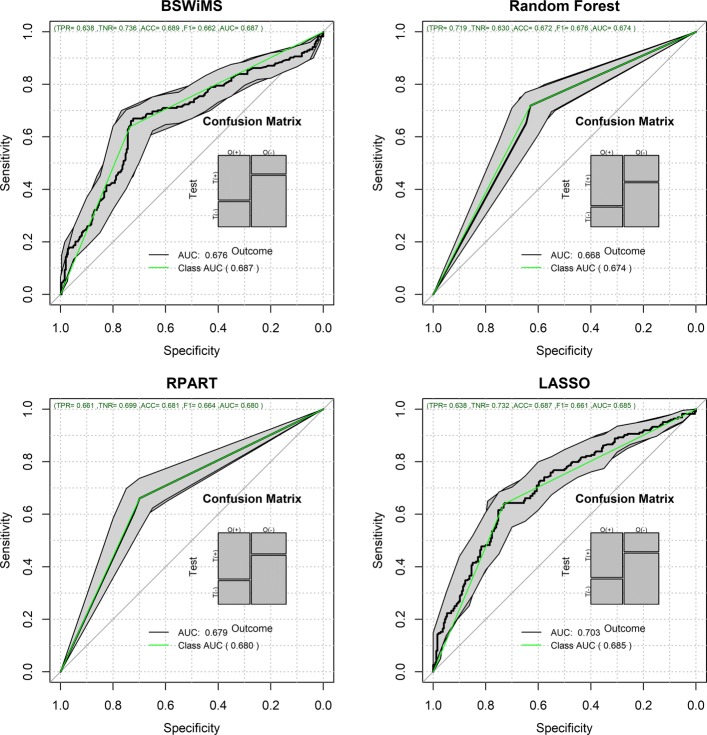
Figures 1 and 2 show the Receiver Operating Characteristic Area Under the Curve (ROC AUC) of the ML methods on the ADNI dataset. The ROC AUC ranged from 0.60 to 0.70. The BSWiMS, LASSO, and RPART had equivalent performance, and the ensemble of the methods had the best performance with a ROC score of 0.719. Figures 3, 4, 5, 6, 7 and 8 show the detailed performance analysis of the ML methods. The balanced error, the ROC AUC, the accuracy as well as specificity and sensitivity for both classifiers and the combinations with filters are depicted as bar plots. These plots indicate that the support vector machine (SVM) engine with minimum redundancy maximum relevance (mRMR) filter had the lowest performance. On the other hand, the Least Absolute Shrinkage and Selection Operator (LASSO) method gave the best results among ML methods, which was further improved by using the Ensemble of methods and achieving a ROC AUC of 0.719.
Fig. 1.
ROC Curves for the FRESA.CAD Benchmarking Classifiers ROC Curves obtained using BSWiMS, Random Forest, RPART and LASSO of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-Validation and the top 2,500 SNPs as inputs
Fig. 2.
ROC Curves for the FRESA.CAD Benchmarking Classifiers (Continued) ROC Curves obtained using SVM, KNN and the Ensemble of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-Validation and the top 2,500 SNPs as inputs
Fig. 3.
Balanced Error of the FRESA.CAD Benchmark classifiers Comparison of the Balanced Error obtained using the different classification methods of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 4.
Accuracy of the FRESA.CAD Benchmark classifiers Comparison of the Accuracy obtained using the different classification methods of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 5.
ROC AUC of the FRESA.CAD Benchmark classifiers Comparison of the ROC AUC Score obtained using the different classification methods of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 6.
ROC AUC of the FRESA.CAD Filter combinations Comparison of the ROC AUC Score obtained using the different combinations of classification methods plus filters of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 7.
Sensitivity of the FRESA.CAD Filter combinations Comparison of the Sensitivity Score obtained using the different combinations of classification methods plus filters of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 8.
Specificity of the FRESA.CAD Filter combinations Comparison of the Specificity Score obtained using the different combinations of classification methods plus filters of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
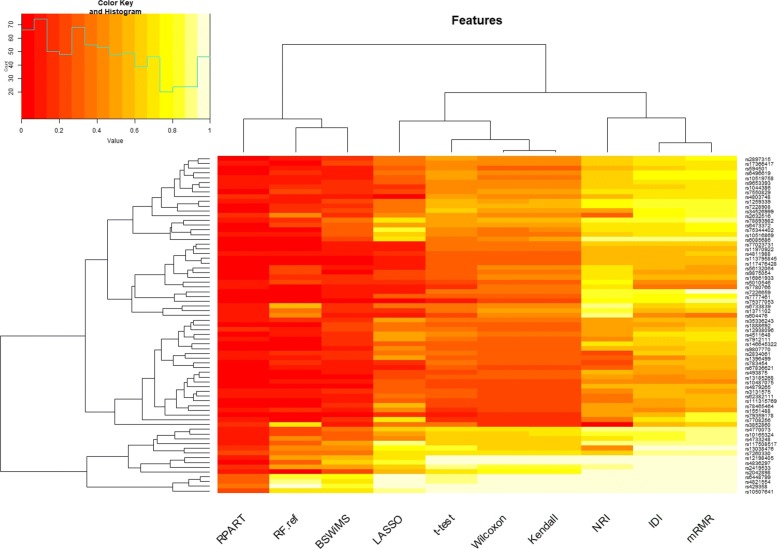
Regarding feature selection: Fig. 9 shows the Jaccard index of the different methods, while Fig. 10 shows the average number of selected features. Finally, Fig. 11 shows the top selected features by the ML method and their selection frequency. These figures show that multivariate ML methods selected different features to construct their predictive models and that those features were not constantly selected at each one of the cross-validation repetitions. The method that constantly selected the same features was BSWiMS, but it was, on average, based on a single feature. On the other extreme, the mRMR filter selected on average over 200 features at each interaction; and 50% of the selected features were common between selection sets.
Fig. 9.
Jaccard Index Jaccard Index metric of the different classifiers between features selected by each classifier of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 10.
Number of Features The number of features selected by each classifier of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
Fig. 11.
SNPs chosen more than 10% of the time as features of the FRESA.CAD Benchmark Heatmap of the main SNPs being chosen across all the classifiers. The Y axis are the main SNPs being selected while the X axis represents the different classifiers of the FRESA.CAD Benchmarking with the ADNI-Discovery dataset for the Cross-validation and using the top 2500 SNPs as input
A detailed analysis of the results presented in Fig. 11 indicates that APOE ε4 (rs429358) was chosen by all the feature selection methods. LASSO is consistently using more SNPs than net reclassification improvement (NRI) filter and NRI selected more than the other filter methods. On the other hand, the classic mRMR filter selects many markers, but the cross validation (CV) performance results were not the best. The selection frequency analysis reported by the benchmark function shows that rs67636621, rs76566842, and rs16905109 deserve further exploration. Table 1 presents the results of the eight most important SNPs that were consistently selected by the ML methods (more than 10% across feature selection methods). Most of them had a significant association with the presence of AD according to the univariate Wilcoxon test (p <0.05). The APOE ε4 variant gives a very strong predictive power, and the remaining variants are then used to further improve the models. Table 1 also shows the location and the related genes of the top SNPs. One of the notable results is SNP rs6448799 which is a variant of LOC107986178 of the HS3ST1 gene. This gene has been shown to have a near study-wide association with the “backward digits” working memory, supporting association of these variants with AD and Mild Cognitive Disorder (MCI) [24].
Table 1.
Characteristics of the top SNPs being selected as important features for the ADNI-Discovery Dataset
| SNP | Location | Function | Gene | Gene summary | WILCOX | FREQ |
|---|---|---|---|---|---|---|
| rs429358 | 19:44908684 | Missense Variant | APOE | APOE is a protein coding gene which generates alipoprotein E, a fat-binding protein crucial in many mechanisms of the body. This gene is related to Alzheimer’s Disease and Lipopoprotein Glomerulopathy among others. | 0 | 1.000 |
| rs67836621 | 19:51186703 | Noncoding (Intergenic) | Adjacent: SIGLEC20P, LOC100133225 (Pseudogene) | Unknown | 8e-04 | 0.298 |
| rs9928727 | 16:9018042 | Noncoding (Intergenic) | Adjacent: LOC105371074 (Uncharacterized), C16orf72 | Unknown | 9e-04 | 0.269 |
| rs11602604 | 11:62231065 | Noncoding (Intergenic) | Adjacent: SCGB2A1, SCGB1D2 | Unknown | 3e-04 | 0.321 |
| rs6448799 | 4:11628425 | Intron Variant | HS3ST1 (LOC107986178) | HS3ST1 is a protein coding gene which is crucial to create heparan sulfate structures that participate in sulfotransferase activity. This gene is related to Arteriosclerosis and Coronary Heart Disease. | 6e-04 | 0.288 |
| rs16905109 | 8:134194872 | Noncoding (Intergenic) | Adjacent: LOC100419617 (Pseudogene), ZFAT | Unknown | 0.0011 | 0.383 |
| rs76566842 | 9:28296478 | Intron Variant | LINGO2 | LINGO2 is a protein coding gene for the Leicine-rich Repeat Neuronal Protein. This gene is related to the Essential Tremor disease. | 0.1619 | 0.327 |
| rs9863257 | 3:27586911 | Noncoding (Intergenic) | Adjacent: RNU1-96P, RPS27P11 | Unknown | 0.1955 | 0.323 |
Figures 12 and 13 show the validation performance results of the benchmarked ML methods based on the top 1000 SNP obtained from the IGAP-independent data set. The ROC AUC ranged from 0.50 to 0.65, and the balanced error rate (BER) ranged from 0.5 to 0.39. Filtered Naive Bayes (AUC= 0.65, BER=0.42) was the top ML method, followed by RPART (AUC=0.63, BER=0.39).
Fig. 12.
Validation ROC Curves for the FRESA.CAD Benchmarking Classifiers ROC Curves obtained using BSWiMS, Random Forest, RPART and LASSO of the FRESA.CAD Benchmarking with the ADNI-Validation dataset for the Cross-validation and using the top 1000 SNPs as input
Fig. 13.
Validation ROC Curves for the FRESA.CAD Benchmarking Classifiers (Continued) ROC Curves obtained using SVM, KNN and the Ensemble of the FRESA.CAD Benchmarking with the ADNI-Validation dataset for the Cross-validation and using the top 1000 SNPs as inputs
The feature selection analysis of the validation returned a larger set of SNPs candidates. Figure 14 and Table 2 show the set of SNPs that were selected at least 10% of the time. Despite the large number of SNPs only APOE ε4 and rs6448799 appeared on both the full ADNI and IGAP-independent validation set.
Fig. 14.
Validation SNPs chosen more than 10% of the time as features of the FRESA.CAD Benchmark Heatmap of the main SNPs being chosen across all the classifiers. The Y axis are the main SNPs being selected while the X axis represents the different classifiers of the FRESA.CAD Benchmarking with the ADNI-Validation dataset for the Cross-validation and using the top 1000 SNPs as input
Table 2.
Characteristics of the top 10 SNPs being selected as important features for the ADNI-Validation Dataset
| SNP | Location | Function | Gene | Gene summary | WILCOX | FREQ |
|---|---|---|---|---|---|---|
| rs429358 | 19:44908684 | Missense Variant | APOE | APOE is a protein coding gene which generates alipoprotein E, a fat-binding protein crucial in many mechanisms of the body. This gene is related to Alzheimer’s Disease and Lipoprotein Glomerulopathy among others. | 0 | 1.000 |
| rs6448799 | 4:11628425 | Intron Variant | HS3ST1 / LOC107986178 | HS3ST1 is a protein coding gene which is crucial to create heparan sulfate structures that participate in sulfotransferase activity. This gene is related to Arteriosclerosis and Coronary Heart Disease. | 6e-04 | 0.288 |
| rs4821554 | 22:36880042 | Noncoding (Intergenic) | Adjacent: NCF4, LOC105373022 (Uncharacterized) | Unknown | 1e-04 | 0.874 |
| rs7260330 | 19:44932959 | Noncoding (Intergenic) | Adjacent: APOC1P1, APOC4-APOC2 | Unknown | 0.0027 | 0.667 |
| rs10507641 | 13:59857910 | Intron Variant | DIAPH3, | DIAPH3 is a protein coding gene that generates a Diaphanous forming protein, which regulates cell movement and adhesion. It is related to Auditory Neuropathy and Neuropathy | 0 | 0.797 |
| rs4733248 | 8:31302383 | Intron Variant | LOC101929492 (Uncharacterized) | Unknown | 0.0052 | 0.581 |
| rs13038476 | 20:4158146 | Intron Variant | SMOX | SMOX is a protein coding gene that generates the Spermine Oxidase which helps as neurotransmitters and cell regulators. It is related to Short-Rib Thoracic Dysplasia and Acute Hemorrhagic Leukoencephalitis | 0 | 0.627 |
| rs2419533 | 4:132668359 | Intron Variant | LINC01256 | LINC01256 is a non-coding RNA gene | 0.0013 | 0.716 |
| rs34526999 | 5:33728435 | Intron Variant | ADAMTS12 | ADAMTS12 is a protein coding gene that generates ADAMTS which works in pulmonary cell development or tumor processes. It is related to Brachydactyly and Intrahepatic Cholestasis of Pregnancy | 0.025 | 0.445 |
| rs2632516 | 17:58331728 | Intron Variant | TSPOAP1-AS1 | TSPOAP1-AS1 is a non-coding RNA gene | 0.02 | 0.387 |
Discussion
Most of the experimental treatments in development for LOAD require implementation at the very early stages of the disease to be effective [25]. Genetic approaches to predicting the risk of LOAD are a powerful and viable alternative to traditional biomarker-based disease prediction methods [26]. Traditional GWAS have only found SNPs that so far can only explain 33% of the estimated 79% [8] fraction of genetic risk associated with Alzheimer’s disease. While this value is low for a reliable clinical prediction, Machine learning methods have been proven to perform better in detecting candidate SNPs and predicting complex genetic diseases such as Type-2 Diabetes [27], Inflammatory Bowel Syndrome [28] and Obesity [29]. The use of machine learning-based approaches for Genetic-based Precision Medicine has increased in the current decade and shows signs of increasing [30].
This study presented the hypothesis that Benchmarking ML methods on SNP dataset can aid in discovering novel SNPs associated with the late onset of AD. Specifically, we studied the capability of the FRESA.CAD benchmarking method to discover and model the genetic risk factor. Benchmarking allowed us to gain insight in the degree of genetic risk associated with LOAD by comparing and analyzing multiple Machine Learning models applied to predict the risk a person of developing Alzheimer’s Disease from genetic information only. The Machine Learning models were expected to find linear and nonlinear relationships between genes that could explain more of the missing heritability of Alzheimer’s disease. Constructing models with the capability to detect epistasic relationships would be an important advancement compared to traditional single-variant GWAS studies. The results show that some models obtained promising results in predicting the development of the disease, namely BSWiMS, LASSO, RPART, and the Ensemble. The best ROC AUC score achieved with the ADNI-Discovery was ∼0.719 and 0.61 in the IGAP-independent subset. This result is promising considering the upper boundary set by the calculated heritability from purely genetic components (79% as described in [8]). Furthermore, the model outperforms those methods which only use the APOE4 gene, which achieve around 63 65%, and simple deep learning models, which achieve 62%.. It is noteworthy that this study showed marked differences between the ML methods in modeling LOAD. On the other hand, the ADNI results indicated a small subset of SNPs that can be used in multivariate models, while the independent IGAP study returned hundreds of possible candidates.
The models tested with the FRESA.CAD Benchmark indicated that the ensemble method had a sensitivity of 70% with a specificity of 65%, implying a strong genetic risk component in the ADNI cohort.. We also found that different feature selection methods selected common SNPs that have been already associated with Alzheimer. Thus, SNP selection based on set overlap may be a powerful method to discover clinically significant risk factors. The reduced cohort for the confirmatory validation indicated that the Naive Bayes classifier had a sensitivity of 33% with a strong specificity of 95%. The contradictory findings between the full dataset and the validation subset may be a class imbalance problem coupled with limitations regarding the size of the dataset. Regardless of the differences between cohorts, the presented results support the previous SNP finding that the APOE ε4 gene is the main risk factor for Late Onset Alzheimer’s disease [31]. Furthermore, we were able to confirm a new possible variant associated with the disease: rs6448799. According to recent GWAS studies, this last genetic variant may have a true correlation with Alzheimer’s Disease [24, 32]. Hence, FRESA.CAD Benchmark seems to be a promising tool for Genomics analysis and finding candidate clinical markers. This study is limited by the small sample size; we expect that the predictive capability of the machine learning models can be improved by increasing the sample size. Therefore, we believe that these models hold much promise for the clinical diagnosis of Late-Onset Alzheimer’s Disease and other complex diseases.
The upper limit of the genetic component alone presents a challenge for the highly precise accuracy required for a clinical diagnostic. One of the possible solutions for this problem would be to complement the genetic-based methods with imaging or clinical data. The genetic analysis could be used to detect those individuals with a higher risk of developing Alzheimer’s Disease, and then those individuals could be monitored on a yearly basis with imaging technologies to detect the development of the disease at the earliest possible moment.
LOAD polygenic scores currently available are not capable to predict mild cognitive impairment to LOAD progression [33]. Therefore, alternative models are also required for the accurate prediction of disease progression. Additionally, alternative hypothesis such as Pritchard’s Omnigenetics [34] could also be explored efficiently using ML methods to model and identify cellular networks and the respective flow of regulatory information, finding a more comprehensive and general solution.
Conclusions
This research study has shown the results of applying the FRESA.CAD Binary Classification Benchmarking algorithms to predict the risk of developing Late-Onset Alzheimer’s Disease from genetic variation data exclusively. Conducting systematic comparisons on the classification performance of machine learning algorithms is a crucial task for achieving the predictive potential of these models. Model selection methodologies used to optimize machine learning models also hold the potential for the discovery of new genetic markers associated with the disease. Given that the preliminary results show promise, we believe that a refined model could be a powerful tool for the prediction and early detection of this disease. The current models show limitations due to the complexity of the disease and the size of the datasets, both of which stand to benefit from the increasing availability of data. This paper also demonstrates that Machine Learning methods are powerful tools suited to analyze and leverage a multitude of genes that could be used in a variety of complex diseases similar to Alzheimer’s Disease. The current technological trend points toward the large-scale application of these methods with the ever-increasing demand for individual genome sequencing and the availability of much larger datasets.
Methods
Data used in the preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
We selected individuals who have either a Cognitively Normal or Alzheimer’s Disease. PLINK [19, 20] was used to read the Variant Call Format data of the WGS and to convert it to the more compact format of Binary Pedigree Files (BED). After that, we used Python 3.5 and the library PyPlink [21] to perform quality control procedures in a similar pipeline to the one described by Turner [22].
We began by performing pre-quality controls on the samples, using marker call rate, sample call rates and Minor allele frequency (MAF) filtering. Once this is done Identity-By-Descent (IBD) is performed with a value of 0.25 to find those individuals related to each other to be removed. After the binary classification filter and the IBD filter the samples are reduced from 808 individuals to 471 individuals. We named this the ADNI-Discovery dataset, it is balanced in terms of cases/controls, has a mean age of 75.5 and it is slightly skewed towards males, as is shown in Table 3.
Table 3.
Dataset and validation subset demographic metrics
| Dataset | Size | Male | Female | Mean age | Controls | Alzheimer’s cases |
|---|---|---|---|---|---|---|
| ADNI-Discovery | 471 | 252 | 219 | 75.57 | 241 | 230 |
| ADNI-Validation | 167 | 92 | 75 | 72.17 | 130 | 37 |
Afterwards, marker call rate (≤99%) and MAF filtering (≤0.01) are used to reduce the number of SNPs to only those that are useful. Then, the Hardy-Weinberg Equilibrium test is done (≤0.05) to further clean SNPs. Finally LD-Based clumping (p-value ≤0.01, r2≤0.05) is used to find those SNPs which are in Linkage Equilibrium and are statistically relevant. For a correct LD-based clumping the statistical data used as reference should be obtained from a different data set which is sufficiently large. In our case we used the statistical summary results from the International Genomics of Alzheimer’s Project (IGAP) [23] to guide the clumping algorithm and find the statistically relevant and independent candidate SNPs. These summary statistics are generated from 74,046 individuals. The Quality Control Pipeline returned 8,239 SNPs in Linkage Equilibrium after performing the LD-clump based on the IGAP Summary Statistics. Finally, for performance reasons, we reduced these 8,239 SNPs to only the top 2,500 SNPs based on their p-value (ascending) as an input to the benchmarking tool. The ADNI dataset was selected as the base of the analysis even though it has a much smaller sample size as it has the full WGS data available for each subject, while the IGAP only makes the summary statistics openly available.
For further validation, we also generated a second validation subset from the dataset where we took only those individuals in the ADNI which did not take part in the IGAP study for validation as there were some existing individuals present in both datasets. Due to the reduced data set size we further reduced the SNPs used as input to just the top 1,000 SNPs (Also based on their ascending p-value). In contrast with the full dataset, the validation set is highly unbalanced, with 78% of the samples being controls, the mean age is slightly lower as shown in Table 3.
Multivariate model-building and validation were done using the FRESA.CAD Benchmarking tool that runs the following ML methods:
Bootstrap Stage-Wise Model Selection (BSWiMS), or user-supplied cross-validated (CV) method.
Least Absolute Shrinkage and Selection Operator (LASSO)
Random Forest (RF)
Recursive Partitioning and Regression Trees (RPART)
K Nearest Neighbors (KNN) with BSWiMS features
Support Vector Machine (SVM) with minimum-Redundancy-Maximum-Relevance (mRMR) feature selection filter
The ensemble of all the above methods
The CV performance of these classification algorithms is also complemented with the following feature selection algorithms and different filters: BSWiMS, LASSO, RPART, RF, integrated discrimination improvement (IDI), net reclassification improvement (NRI), t student test, Wilcoxon test, Kendall correlation, and mRMR as filters on the following classifiers: KNN, naive Bayes, nearest centroid (NC) with normalized root sum square distance and Spearman correlation distance, RF and SVM.
The results of the CV instances executed by the binary benchmark were compared using the performance statistics and ranked by their 95% confidence interval (CI). The ranking method accumulates a positive score each time the lower CI of a performance metric is superior to the mean of the other methods and loses a point each time the mean is inferior to the top 95% CI of the other methods. The package returns the accuracy, precision, sensitivity, the balanced error rate and the ROC AUC with their corresponding 95% confidence intervals (95% CI). We used the ranking results to infer the suitability of ML methods to predict AD in the ADNI dataset.
Finally, we independently analyzed the validation subset (IGAP-independent) using the FRESA.CAD benchmarking procedure.
Acknowledgements
We wish to thank coauthor and friend Edgar Vallejo, who passed away suddenly, for his complete dedication to Bioinformatics research in Mexico as well as for being an extraordinary teacher dedicated to sharing knowledge and creating excellent researchers. We dedicate this article to his memory. We thank our colleagues from the Bioinformatics for Clinical Diagnosis Research Program, School of Medicine and Health Sciences, Tecnologico de Monterrey, for their valuable comments on this work. This work was supported by Tecnologico de Monterrey. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative(ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. We thank the International Genomics of Alzheimer’s Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer’s disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n 503480), Alzheimer’s Research UK (Grant n 503176), the Wellcome Trust (Grant n 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer’s Association grant ADGC–10–196728.
Abbreviations
- AD
Alzheimer disease
- ADNI
Alzheimer’s disease neuroimaging initiative
- APOE
Apolipoprotein E
- BED
Binary pedigree files
- BER
Balanced error rate
- BSWiMS
Bootstrap stage-wise model selection
- CI
Confidence interval
- CV
Cross validation
- EOAD
Early-onset alzheimer’s disease
- FRESA.CAD
Feature selection algorithms for computer aided diagnosis
- GWAS
Genome-wide association studies
- IBD
Identity by descent
- IDI
Integrated discrimination improvement
- IGAP
International genomics of alzheimer’s project
- KNN
K nearest neighbours
- LASSO
Least absolute shrinkage and selection operator
- LOAD
Late-onset alzheimer’s disease
- MAF
Minor alelle frequency
- MCI
Mild cognitive impairment
- ML
Machine learning
- MRI
Magnetic resonance imaging
- mRMR
Minimum redundancy maximum relevance
- NC
Nearest centroid
- NRI
Net reclassification improvement
- PET
Positron emission tomography
- RF
Random forest
- ROC
AUC Receiver operating characteristic area under the curve
- RPART
Recursive partitioning and regression trees
- SNP
Single nucleotide polymorphism
- SVM
Support vector machine
Authors’ contributions
JV processed the genetic data, converted them to a suitable format, ran the FRESA.CAD Benchmarking on the ADNI dataset and analyzed the results. EV contributed to the preparation of the manuscript and provided suggestions for the assessment of the biological relevance of the SNPs yielded by Machine Learning feature selection procedures. KE designed the quality control pipeline for preparing GWAS and provided a qualitative assessment on the predictive performance of the Machine Learning models. JT coded the FRESA.CAD R package, including the Binary Classification Benchmarking module and contributed to the preparation of the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by Escuela de Medicina y Ciencias de la Salud, Tecnologico de Monterrey and by a CONACYT scholarship for conducting graduate studies. The funding bodies did not play any role in the design of the study, in the collection, analysis and interpretation of data and in the writing of the manuscript.
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the ADNI LONI repository, http://adni.loni.usc.edu/
Ethics approval and consent to participate
Not Applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Data used in preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database (http://adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Javier De Velasco Oriol, Email: javierdevelascooriol@gmail.com.
Karol Estrada, Email: kestrada@brandeis.edu.
References
- 1.Sosa-Ortiz Ana Luisa, Acosta-Castillo Isaac, Prince Martin J. Epidemiology of Dementias and Alzheimer’s Disease. Archives of Medical Research. 2012;43(8):600–608. doi: 10.1016/j.arcmed.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Lanoiselée Hélène-Marie, Nicolas Gaël, Wallon David, Rovelet-Lecrux Anne, Lacour Morgane, Rousseau Stéphane, Richard Anne-Claire, Pasquier Florence, Rollin-Sillaire Adeline, Martinaud Olivier, Quillard-Muraine Muriel, de la Sayette Vincent, Boutoleau-Bretonniere Claire, Etcharry-Bouyx Frédérique, Chauviré Valérie, Sarazin Marie, le Ber Isabelle, Epelbaum Stéphane, Jonveaux Thérèse, Rouaud Olivier, Ceccaldi Mathieu, Félician Olivier, Godefroy Olivier, Formaglio Maite, Croisile Bernard, Auriacombe Sophie, Chamard Ludivine, Vincent Jean-Louis, Sauvée Mathilde, Marelli-Tosi Cecilia, Gabelle Audrey, Ozsancak Canan, Pariente Jérémie, Paquet Claire, Hannequin Didier, Campion Dominique. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLOS Medicine. 2017;14(3):e1002270. doi: 10.1371/journal.pmed.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballard Clive, Gauthier Serge, Corbett Anne, Brayne Carol, Aarsland Dag, Jones Emma. Alzheimer's disease. The Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 4.Sevigny J, Chiao P, Bussière T, Weinreb PH, Williams L, Maier M, et al. The antibody aducanumab reduces ab plaques in alzheimer’s disease. Nature. 2016;537:50. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 5.Li Jin, Zhang Qiushi, Chen Feng, Meng Xianglian, Liu Wenjie, Chen Dandan, Yan Jingwen, Kim Sungeun, Wang Lei, Feng Weixing, Saykin Andrew J., Liang Hong, Shen Li. Genome-wide association and interaction studies of CSF T-tau/Aβ42 ratio in ADNI cohort. Neurobiology of Aging. 2017;57:247.e1-247.e8. doi: 10.1016/j.neurobiolaging.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shao Wen, Peng Dantao, Wang Xiaomin. Genetics of Alzheimer’s disease: From pathogenesis to clinical usage. Journal of Clinical Neuroscience. 2017;45:1–8. doi: 10.1016/j.jocn.2017.06.074. [DOI] [PubMed] [Google Scholar]
- 7.Seshadri Sudha. Genome-wide Analysis of Genetic Loci Associated With Alzheimer Disease. JAMA. 2010;303(18):1832. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghavan N, Tosto G. Genetics of alzheimer’s disease: the importance of polygenic and epistatic components. Curr Neurol Neurosci Rep. 2017; 17(10):78. 10.1007/s11910-017-0787-1. [DOI] [PMC free article] [PubMed]
- 9.Panpalli Ates M., Karaman Y., Guntekin S., Ergun M.A. Analysis of genetics and risk factors of Alzheimer’s Disease. Neuroscience. 2016;325:124–131. doi: 10.1016/j.neuroscience.2016.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Saykin Andrew J., Shen Li, Foroud Tatiana M., Potkin Steven G., Swaminathan Shanker, Kim Sungeun, Risacher Shannon L., Nho Kwangsik, Huentelman Matthew J., Craig David W., Thompson Paul M., Stein Jason L., Moore Jason H., Farrer Lindsay A., Green Robert C., Bertram Lars, Jack Clifford R., Weiner Michael W. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimer's & Dementia. 2010;6(3):265–273. doi: 10.1016/j.jalz.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Torteya Antonio, Gomez-Rueda Hugo, Trevino Victor, Farber Joshua, Tamez-Pena Jose. Identification and Temporal Characterization of Features Associated with the Conversion from Mild Cognitive Impairment to Alzheimer's Disease. Current Alzheimer Research. 2018;15(8):751–763. doi: 10.2174/1567205015666180202095616. [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Torteya A, Trevino V, Tamez-Pena J. Improved multimodal biomarkers for alzheimer’s disease and mild cognitive impairment diagnosis - data from adni: 2013. p. 86700. 10.1117/12.2008100.
- 13.Martinez-Torteya Antonio, Rodriguez-Rojas Juan, Celaya-Padilla José M., Galván-Tejada Jorge I., Treviño Victor, Tamez-Peña Jose. Magnetization-prepared rapid acquisition with gradient echo magnetic resonance imaging signal and texture features for the prediction of mild cognitive impairment to Alzheimer’s disease progression. Journal of Medical Imaging. 2014;1(3):031005. doi: 10.1117/1.JMI.1.3.031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walhovd K.B., Fjell A.M., Brewer J., McEvoy L.K., Fennema-Notestine C., Hagler D.J., Jennings R.G., Karow D., Dale A.M. Combining MR Imaging, Positron-Emission Tomography, and CSF Biomarkers in the Diagnosis and Prognosis of Alzheimer Disease. American Journal of Neuroradiology. 2010;31(2):347–354. doi: 10.3174/ajnr.A1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee G, Nho K, Kang B, Sohn K-A, Kim D, Weiner MW, et al. Predicting alzheimer’s disease progression using multi-modal deep learning approach. Sci Rep. 2019; 9(1):1952. 10.1038/s41598-018-37769-z. [DOI] [PMC free article] [PubMed]
- 16.Saykin Andrew J., Shen Li, Yao Xiaohui, Kim Sungeun, Nho Kwangsik, Risacher Shannon L., Ramanan Vijay K., Foroud Tatiana M., Faber Kelley M., Sarwar Nadeem, Munsie Leanne M., Hu Xiaolan, Soares Holly D., Potkin Steven G., Thompson Paul M., Kauwe John S.K., Kaddurah-Daouk Rima, Green Robert C., Toga Arthur W., Weiner Michael W. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimer's & Dementia. 2015;11(7):792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taméz Peña JG, Martinez-Torteya A, Alanis I. Package FRESA.CAD. 2018. https://cran.r-project.org/web/packages/FRESA.CAD/index.html. Accessed 8 Aug.
- 18.Taméz Peña JG, Martinez-Torteya A, Alanis I. FRESA.CAD. 2018. http://cran.utstat.utoronto.ca/web/packages/FRESA.CAD/FRESA.CAD.pdf. Accessed 8 Aug.
- 19.Purcell Shaun, Neale Benjamin, Todd-Brown Kathe, Thomas Lori, Ferreira Manuel A.R., Bender David, Maller Julian, Sklar Pamela, de Bakker Paul I.W., Daly Mark J., Sham Pak C. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaun Purcell CC. PLINK 1.9. 2015. https://cog-genomics.org/plink/1.9/. Accessed 8 Aug.
- 21.Lemieux Perreauls L-P. PyPlink. 2015. https://lemieuxl.github.io/pyplink/pyplink.html. Accessed 8 Aug.
- 22.Turner Stephen, Armstrong Loren L., Bradford Yuki, Carlson Christopher S., Crawford Dana C., Crenshaw Andrew T., de Andrade Mariza, Doheny Kimberly F., Haines Jonathan L., Hayes Geoffrey, Jarvik Gail, Jiang Lan, Kullo Iftikhar J., Li Rongling, Ling Hua, Manolio Teri A., Matsumoto Martha, McCarty Catherine A., McDavid Andrew N., Mirel Daniel B., Paschall Justin E., Pugh Elizabeth W., Rasmussen Luke V., Wilke Russell A., Zuvich Rebecca L., Ritchie Marylyn D. Quality Control Procedures for Genome-Wide Association Studies. Current Protocols in Human Genetics. 2011;68(1):1.19.1-1.19.18. doi: 10.1002/0471142905.hg0119s68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambert J-C, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for alzheimer’s disease. Nat Genet. 2013;45:1452. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinosa A, Hernández-Olasagarre B, Moreno-Grau S, Kleineidam L, Heilmann-Heimbach S, Hernández I, et al. Exploring genetic associations of alzheimer’s disease loci with mild cognitive impairment neurocognitive endophenotypes. Front Aging Neurosci. 2018; 10:340. 10.3389/fnagi.2018.00340. 30425636[pmid]. [DOI] [PMC free article] [PubMed]
- 25.Dufouil Carole, Glymour M Maria. Prediction to prevention in Alzheimer's disease and dementia. The Lancet Neurology. 2018;17(5):388–389. doi: 10.1016/S1474-4422(18)30123-6. [DOI] [PubMed] [Google Scholar]
- 26.Alexiou A, Mantzavinos VD, Greig NH, Kamal MA. A bayesian model for the prediction and early diagnosis of alzheimer’s disease. Front Aging Neurosci. 2017; 9:77. 10.3389/fnagi.2017.00077. [DOI] [PMC free article] [PubMed]
- 27.López Beatriz, Torrent-Fontbona Ferran, Viñas Ramón, Fernández-Real José Manuel. Single Nucleotide Polymorphism relevance learning with Random Forests for Type 2 diabetes risk prediction. Artificial Intelligence in Medicine. 2018;85:43–49. doi: 10.1016/j.artmed.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Wei Zhi, Wang Wei, Bradfield Jonathan, Li Jin, Cardinale Christopher, Frackelton Edward, Kim Cecilia, Mentch Frank, Van Steen Kristel, Visscher Peter M., Baldassano Robert N., Hakonarson Hakon. Large Sample Size, Wide Variant Spectrum, and Advanced Machine-Learning Technique Boost Risk Prediction for Inflammatory Bowel Disease. The American Journal of Human Genetics. 2013;92(6):1008–1012. doi: 10.1016/j.ajhg.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montaez CAC, Fergus P, Montaez AC, Hussain A, Al-Jumeily D, Chalmers C. Deep learning classification of polygenic obesity using genome wide association study snps. In: 2018 International Joint Conference on Neural Networks (IJCNN): 2018. p. 1–8. 10.1109/IJCNN.2018.8489048.
- 30.Ho DSW, Schierding W, Wake M, Saffery R, O’Sullivan J. Machine learning snp based prediction for precision medicine. Front Genet. 2019; 10:267. 10.3389/fgene.2019.00267. [DOI] [PMC free article] [PubMed]
- 31.Wolfe Cody, Fitz Nicholas, Nam Kyong, Lefterov Iliya, Koldamova Radosveta. The Role of APOE and TREM2 in Alzheimer′s Disease—Current Understanding and Perspectives. International Journal of Molecular Sciences. 2018;20(1):81. doi: 10.3390/ijms20010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Witoelar A, Rongve A, Almdahl IS, Ulstein ID, Engvig A, White LR, et al. Meta-analysis of alzheimer’s disease on 9,751 samples from norway and igap study identifies four risk loci. Sci Rep. 2018; 8(1):18088. 10.1038/s41598-018-36429-6. 30591712[pmid]. [DOI] [PMC free article] [PubMed]
- 33.Lacour A, Espinosa A, Louwersheimer E, Heilmann S, Hernández I, Wolfsgruber S, et al. Genome-wide significant risk factors for alzheimer’s disease: role in progression to dementia due to alzheimer’s disease among subjects with mild cognitive impairment. Mole Psych. 2016;22:153. doi: 10.1038/mp.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle Evan A., Li Yang I., Pritchard Jonathan K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell. 2017;169(7):1177–1186. doi: 10.1016/j.cell.2017.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the ADNI LONI repository, http://adni.loni.usc.edu/