Abstract
Background
The morbidity of thyroid carcinoma has been rising worldwide and increasing faster than any other cancer type. The most common subtype with the best prognosis is papillary thyroid cancer (PTC); however, the exact molecular pathogenesis of PTC is still not completely understood.
Methods
In the current study, 3 gene expression datasets (GSE3678, GSE3467, and GSE33630) and 2 miRNA expression datasets (GSE113629 and GSE73182) of PTC were selected from the Gene Expression Omnibus (GEO) database and were further used to identify differentially expressed genes (DEGs) and deregulated miRNAs between normal thyroid tissue samples and PTC samples. Then, Gene Ontology (GO) and pathway enrichment analyses were conducted, and a protein-protein interaction (PPI) network was constructed to explore the potential mechanism of PTC carcinogenesis. The hub gene detection was performed using the CentiScaPe v2.0 plugin, and significant modules were discovered using the MCODE plugin for Cytoscape. In addition, a miRNA-gene regulatory network in PTC was constructed using common deregulated miRNAs and DEGs.
Results
A total of 263 common DEGs and 12 common deregulated miRNAs were identified. Then, 6 significant KEGG pathways (P < 0.05) and 82 significant GO terms were found to be enriched, indicating that PTC was closely related to amino acid metabolism, development, immune system, and endocrine system. In addition, by constructing a PPI network and miRNA-gene regulatory network, we found that hsa-miR-181a-5p regulated the most DEGs, while BCL2 was targeted by the most miRNAs.
Conclusions
The results of this study suggested that hsa-miR-181a-5p and BCL2 and their regulatory networks may play important roles in the pathogenesis of PTC.
Keywords: Papillary thyroid cancer (PTC), Differentially expressed genes (DEGs), Deregulated miRNAs, BCL2, hsa-miR-181a-5p
Background
The morbidity of thyroid carcinoma has been rising worldwide and increasing faster than any other type of cancer, mainly due to the increasing use of diagnostic equipment [1]. It was reported that in 2017 thyroid cancer was the fifth most common cancer among American women [2]. Remarkably, thyroid cancer is 3 times more common in women than in men [2]. Similarly, thyroid cancer was the fastest growing cancer among Chinese from 1988 to 2013. Between 1988 and 2013, the incidence of thyroid cancer increased by an average of 14.73% per year in men and 18.98% per year in women [3]. Papillary thyroid cancer (PTC) is the most common subtype of differentiated thyroid cancers (DTCs), with the best overall prognosis [4]. However, the biggest challenge in treating PTC is to identify an easy method for the early recognition of benign or malignant nodules and the detection of overtreatment.
In recent years, a number of risk genes of PTC have been identified, including Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1 (CITED1) [5], LDL receptor related protein 4 (LRP4) [6] and tektin 4 (TEKT4) [7], whose downregulation can significantly inhibit the proliferation, migration, and invasion of PTC cells. Interleukin 17 receptor A (IL17RA) polymorphisms, which play an important role in tumor development, have been found to influence both the unilateral and bilateral development of PTC [8]. In addition, increased expression of flavin-containing monooxygenase 1 (encoded by the FMO1 gene) could serve as a biomarker that independently predicts favorable recurrence-free survival in classical PTC patients [9]. These observations suggest that an increasing number of genes are crucial for the pathogenesis of PTC.
In addition, miRNAs also play an important role in the pathogenesis of various cancers, especially PTC. For example, miR-524-5p can inhibit cell migration, invasion, and apoptosis by targeting FOXE1 and ITGA3 in PTC [10]. In addition, miR-215 was found to target ARFGEF1 and inhibit the proliferation and metastasis of PTC by regulating the epithelial-mesenchymal transformation [11]. Furthermore, miR-509 [12], miR-1270 [13], miR-128 ,[14] and many other miRNAs can inhibit PTC by targeting specific genes. These studies focused on one specific gene or miRNA; however, the comprehensive view of how these miRNAs and genes affect PTC remains unknown. The aim of our study was to screen significant gene and miRNA changes through bioinformatics methods to provide guidance for the study of PTC mechanisms and clinical treatment.
In this study, 3 gene expression datasets (GSE3678, GSE3467, and GSE33630) and 2 miRNA expression datasets (GSE113629 and GSE73182) (Sample analysis was shown in Additional file 1: Figure S1) of PTC were selected from the GEO database that were further used to identify DEGs and deregulated miRNAs between normal thyroid tissue samples and PTC samples. As a result, 263 DEGs and 12 deregulated miRNAs were identified based on the criteria we set. Then, GO and pathway enrichment analyses were conducted, and a PPI network was constructed to explore the potential mechanism of PTC carcinogenesis. The hub gene detection was performed using the CentiScaPe v2.0 plugin, and significant modules were discovered using the MCODE plugin for Cytoscape. In addition, a miRNA-gene regulatory network of PTC was constructed using common deregulated miRNAs and DEGs, and we found that hsa-miR-181a-5p regulated the most DEGs, while BCL2 was targeted by the most miRNAs in this network. However, the specific mechanisms of how hsa-miR-181a-5p could regulate BCL2 need further experiments. In conclusion, hsa-miR-181a-5p and BCL2 are expected to be distinctive biomarkers of benign or malignant tumors and potential therapeutic targets of PTC.
Methods
Acquisition of gene and miRNA expression profile microarray data
The microarray data were acquired from the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo) [15]. Three gene expression datasets (GSE3678, GSE3467, and GSE33630) and 2 miRNA expression datasets (GSE113629 and GSE73182) of PTC were included in this study.
Dataset GSE3678 included 7 PTC samples and 7 paired normal thyroid tissue samples; dataset GSE3467 included 9 PTC patients with paired tumor and normal thyroid tissue; and dataset GSE33630 included 49 PTC samples and 45 normal thyroid tissue samples. These 3 gene expression datasets were all based on the platform of GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array [16–19].
The miRNA dataset GSE113629, based on the GPL24741 Agilent-070156 Human_miRNA_V21.0_Microarray 046064 platform, included matched neoplasms and normal thyroid tissues from 5 patients with PTC. The GSE73182 dataset based on the GPL20194 Agilent-035758 Human miRBASE 16.0 plus 031181 platform included 19 primary papillary thyroid carcinomas and 5 normal thyroids [20, 21].
Identification of DEGs and deregulated miRNA
The interactive web tool GEO2R (www.ncbi.nlm.nih.gov/geo/geo2r) was used to screen the DEGs and deregulated miRNAs between normal thyroid tissue samples and PTC samples [15]. The Benjamin and Hochberg false discovery rate (FDR) method was used to correct the adjusted P value and correct the occurrence of false positive results. The cutoff standard was defined as P value< 0.01, adjusted P value < 0.01 and |logFC| > 1.
GO terms and KEGG pathway enrichment analysis
Pathway data were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/) [22] to examine specific pathways. To identify the Gene Ontology (GO) annotation and pathways in which DEGs were enriched, functional annotation tools were used for GO terms and KEGG pathway enrichment analysis in Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/) [23]. The significance level of KEGG pathway enrichment was calculated using a cutoff of P value < 0.05. A GO term was considered significantly enriched if it showed a P value < 0.05.
Construction of the PPI network
The PPI network of DEGs was identified using the Search Tool for the Retrieval of Interacting Genes (STRING) (http://string-db.org/) database [24]. Subsequently, Cytoscape software (v 3.6.1) was used to visualize the PPI network.
Topological measurements of the PPI network
Several common topological measurements were investigated to reveal the basic features of the PPI network. The node degree, neighborhood connectivity, topological coefficients, and clustering coefficients were analyzed for the whole network.
Hub gene identification and module analysis of the PPI network
The hub genes in this PPI network were defined as nodes with a connective degree > 10 and identified using the CentiScaPe v2.0 plugin for Cytoscape [25]. The most significant modules in the PPI network were identified by the Molecular Complex Detection (MCODE) plugin [26] with MCODE scores ≥ 5, degree cutoff = 2, node score cutoff = 0.2, Max depth = 100 and k-core = 3.
Construction of a network of deregulated miRNAs targeting DEGs
The miRNA-gene target data were extracted from miRTarBase [27] (http://mirtarbase.mbc.nctu.edu.tw/php/index.php). When miRNA-gene pairs showed strong evidence of interaction in humans and matched the DEGs we identified, they were selected. Subsequently, Cytoscape software (v 3.6.1) was used to visualize the miRNA-gene network.
Results
Identification of DEGs and deregulated miRNAs in PTC
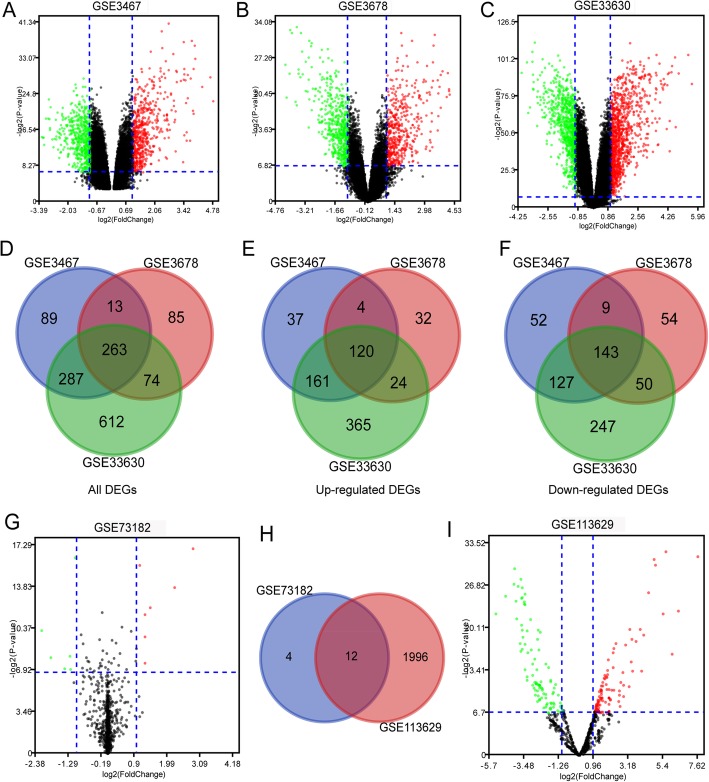
The gene expression datasets GSE3678, GSE3467, and GSE33630 were acquired from the GEO database. DEGs between normal thyroid tissue and PTC samples were screened using GEO2R. As a result, 436, 653, and 1237 DEGs were identified from the GSE3467, GSE3678, and GSE33630 datasets, respectively. Volcano plots were generated for the 3 gene expression datasets for intuitive representation of the DEGs (Fig. 1a–c). The green plots represent downregulated DEGs, the red plots represent upregulated DEGs, and the black plots are not DEGs. In addition, Venn diagrams were also drawn for comparison of the number of total DEGs, upregulated DEGs, and downregulated DEGs in the three datasets. As a result, 263 common DEGs were obtained (Fig. 1d), comprising 120 coupregulated genes and 143 codownregulated genes (Fig. 1e, f).
Fig. 1.
Volcano plot and Venn diagram of DEGs and deregulated miRNAs in gene/miRNA expression profiling datasets. a–c Volcano plots of DEGs in normal thyroid tissue and PTC samples in the GSE3467, GSE3678, and GSE33630 datasets. d–f Venn diagrams illustrating the number of all upregulated and downregulated DEGs in three gene expression datasets. The intersection in the center represents the common DEGs among the three datasets. g, i Volcano plots of deregulated miRNAs in normal thyroid tissue and PTC samples in GSE73182 and GSE113629 datasets, respectively. h Venn diagrams of all deregulated miRNAs in the two datasets. The intersection in the center represents the common deregulated miRNAs between the two datasets. DEGs and deregulated miRNAs were selected by P value < 0.01 and |logFC| > 1. The x-axis shows the fold-change in gene/miRNA expression, and the y-axis shows the statistical significance of the differences. Colors represent different genes/miRNAs: black for genes/miRNAs without significantly different expression, red for significantly upregulated genes/miRNAs and green for significantly downregulated genes/miRNAs. PTC, papillary thyroid cancer; FC, fold change
The miRNA datasets GSE113629 and GSE73182 were also analyzed to screen deregulated miRNAs. The volcano plots showed that 16 and 2008 deregulated miRNAs were identified from the GSE73182 and GSE113629 datasets, respectively (Fig. 1g, i). Among them, 8 downregulated miRNAs and 8 upregulated miRNAs in the GSE73182 dataset, 96 downregulated miRNAs and 1912 upregulated miRNAs in the GSE113629 dataset, were identified. In addition, a Venn diagram was generated for comparison of the number of deregulated miRNAs in the two miRNA datasets; thus, 12 common deregulated miRNAs were acquired (Fig. 1h).
Pathway enrichment analysis of common DEGs
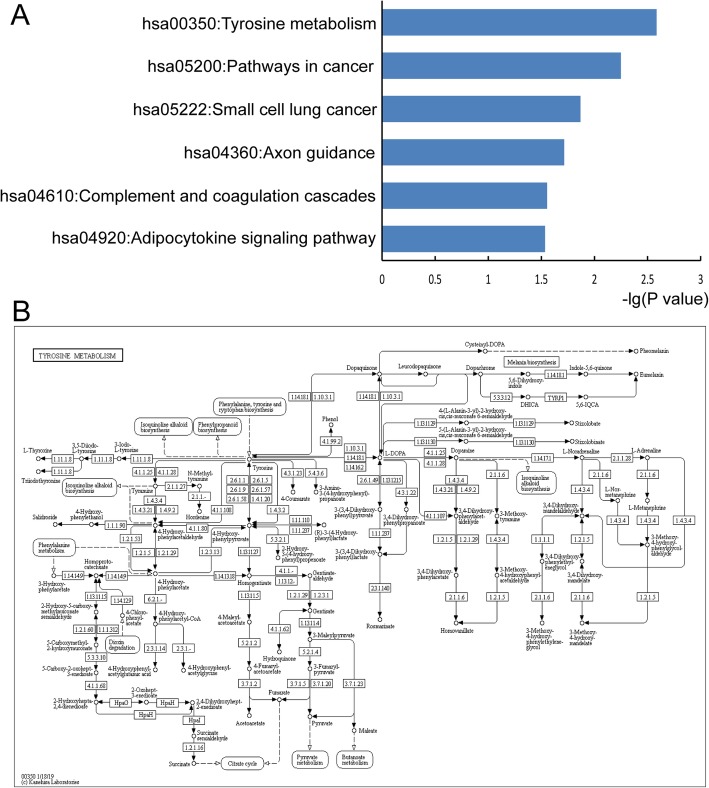
With the aid of the David database, KEGG pathway enrichment analysis was performed using common DEGs. As a result, 6 KEGG pathways (P < 0.05) were significantly enriched, and the top 6 pathways are shown in Fig. 2a. These common DEGs were identified to be enriched in the pathways of ‘Tyrosine metabolism’, ‘Pathways in cancer’, ‘Small cell lung cancer’, ‘Axon guidance’, ‘Complement and coagulation cascades’ and ‘Adipocytokine signaling pathway’. The most significant pathway ‘Tyrosine metabolism’ is shown in Fig. 2b. In addition, a classification of these pathways was also performed, which showed that the six pathways were divided into three major categories (Metabolism, Human Diseases and Organismal Systems) and five minor categories (Amino acid metabolism, Cancers, Development, Immune system and Endocrine system) (Table 1). These results indicated that PTC was closely related to amino acid metabolism, development, immune system, and endocrine system.
Fig. 2.
Pathway enrichment analysis of common DEGs. a Top six significant pathways of DEGs with KEGG enrichment (P < 0.05). b In-depth study of the top PTC-associated pathway (hsa00350: Tyrosine metabolism). DEGs, differentially expressed genes; KEGG, Kyoto encyclopedia of genes and genomes; PTC, papillary thyroid cancer
Table 1.
Six significant KEGG pathways and their classification
| Pathway | P value | Classification |
|---|---|---|
| hsa00350:Tyrosine metabolism | 0.002602 | Metabolism; Amino acid metabolism |
| hsa05200:Pathways in cancer | 0.005643 | Human Diseases; Cancers: Overview |
| hsa05222:Small cell lung cancer | 0.013589 | Human Diseases; Cancers: Specific types |
| hsa04360:Axon guidance | 0.019287 | Organismal Systems; Development |
| hsa04610:Complement and coagulation cascades | 0.027889 | Organismal Systems; Immune system |
| hsa04920:Adipocytokine signaling pathway | 0.029209 | Organismal Systems; Endocrine system |
GO enrichment analysis of common DEGs
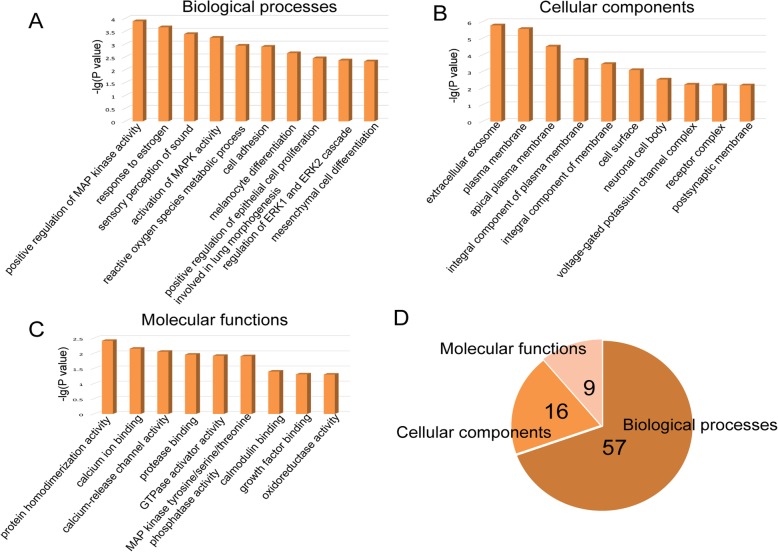
To better understand common DEGs, a GO enrichment analysis was also performed using DAVID. As a result, a total of 82 significant GO terms were identified with a cutoff of P < 0.05. The top biological processes (BPs), cellular components (CCs), and molecular functions (MFs) are shown in Fig. 3 a, b, and c, respectively. A total of 57 BPs, 16 CCs, and 9 MFs were included in the 82 significant GO terms (Fig. 3d). The top 10 BPs were ‘positive regulation of MAP kinase activity’, ‘response to estrogen’, ‘sensory perception of sound’, ‘activation of MAPK activity’, ‘reactive oxygen species metabolic process’, ‘cell adhesion’, ‘melanocyte differentiation’, ‘positive regulation of epithelial cell proliferation involved in lung morphogenesis’, ‘regulation of ERK1 and ERK2 cascade’ and ‘mesenchymal cell differentiation’. In addition, the most significant CC was the ‘extracellular exosome’, and the most significant MF was ‘protein homodimerization activity’. These results further illustrate the fundamental characteristics and functions of cell adhesion, differentiation, proliferation, and MAPK activity in PTC. Therefore, the roles of these DEGs could be better understood by analyzing these significantly enriched GO terms in PTC pathogenesis.
Fig. 3.
GO enrichment analysis of common DEGs. a The top 10 significant biological processes (P < 0.05). b The top 10 significant cellular components (P < 0.05). c The top 9 significant molecular functions (P < 0.05). d A pie chart of the proportion of the significant GO terms, including 57 biological processes, 16 cellular components and 9 molecular functions. DEGs, differentially expressed genes; GO, Gene Ontology
Construction of the PPI network and module analysis
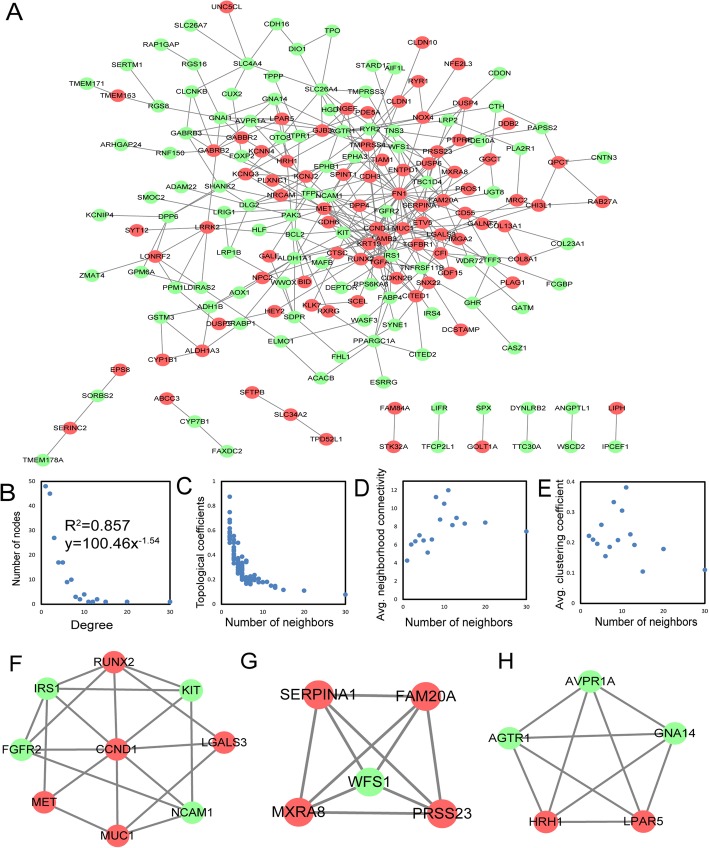
To further explore the interaction among the 263 common DEGs, a PPI network was constructed (Fig. 4a). The PPI network contained 189 nodes and 346 edges. To explore the basic characteristics of the PPI network, the topological features of the network were analyzed in terms of degrees, topological coefficients, neighborhood connectivity, and clustering coefficients (Fig. 4b–e). It was observed that this network followed a natural rule that the majority of nodes had a low degree and that only a few nodes were highly connected with the others. Similar to other biological networks, the degree distribution of this network displayed a power law distribution of f(x) = 100.46x−1.54 with an R2 of 0.857, indicating that the network followed a scale-free distribution and behaved like a small-world network [28].
Fig. 4.
PPI network constructed from the common DEGs and their topological features and significant modules. a A PPI network was constructed from STRING using the 164 common DEGs. The nodes represent proteins, the edges represent the interactions of proteins and the green circles and red circles indicate downregulated and upregulated DEGs, respectively. b–e The basic topological features of the network included degrees, topological coefficients, neighborhood connectivity, and clustering coefficients. The degree distribution of this network displayed a power law distribution of f(x) = 100.46x−1.54 and an R2 of 0.857. f–h Three significant modules in the PPI network with MCODE scores ≥ 5. Green circles and red circles indicate downregulated and upregulated DEGs, respectively. PPI, protein-protein interaction; DEGs, differentially expressed genes; STRING, Search Tool for the Retrieval of Interacting Genes; MCODE, Molecular Complex Detection
Furthermore, the hub genes in the PPI network with a connectivity degree > 10 were identified using the CentiScaPe v2.0 plugin for Cytoscape. The most significant 11 node degree genes were FN1, CCND1, MET, RUNX2, IRS1, SERPINA1, KRT19, FGFR2, MUC1, NCAM1, and WFS1. Detailed information on the 11 hub genes is summarized in Table 2. Among these 11 hub genes, IRS1 and WFS1 had not been previously validated in PTC, which prompted us to further investigate the role of IRS1 and WFS1 in PTC.
Table 2.
Detailed information on the 11 hub genes identified in the PPI network
| Gene | Full name | Overview | Expression in PTC | Reference |
|---|---|---|---|---|
| FN1 | Fibronectin 1 | FN1 is involved in cell adhesion and migration processes including host defense and metastasis. | Upregulated | [29] |
| CCND1 | Cyclin D1 | CCND1 has been demonstrated to interact with tumor suppressor protein Rb. | Upregulated | [30] |
| MET | MET proto-oncogene, receptor tyrosine kinase | MET plays a role in cellular survival, embryogenesis, and cellular migration and invasion. | Downregulated | [31] |
| RUNX2 | RUNX family transcription factor 2 | RUNX2 is essential for osteoblastic differentiation and skeletal morphogenesis. | Upregulated | [32] |
| IRS1 | Insulin receptor substrate 1 | Mutations in IRS1 are associated with type II diabetes and susceptibility to insulin resistance. | No relevant biological experiments | [33] |
| SERPINA1 | Serpin family A member 1 | The protein encoded by SERPINA1 is an inhibitor whose targets include elastase, plasmin, thrombin, trypsin, chymotrypsin, and plasminogen activator. | Upregulated | [34] |
| KRT19 | Keratin 19 | KRT19 is responsible for the structural integrity of epithelial cells. It is specifically expressed in the periderm. | Upregulated | [35] |
| FGFR2 | Fibroblast growth factor receptor 2 | FGFR-2 is involved in regulating cell proliferation, migration and differentiation, as well as in the response to injury and tissue repair. | Downregulated | [36] |
| MUC1 | Mucin 1, cell surface associated | Overexpression, aberrant intracellular localization, and changes in the glycosylation of MUC1 have been associated with carcinomas. | Downregulated | [37] |
| NCAM1 | Neural cell adhesion molecule 1, also known as CD56 | NCAM1 is involved in the development of the nervous system, the expansion of T cells and dendritic cells with a regulatory role in cell motility and migratory capacity of neoplastic cells. | Downregulated | [38] |
| WFS1 | Wolframin ER transmembrane glycoprotein | Mutations in WFS1 are associated with Wolfram syndrome, also called DIDMOAD (diabetes insipidus, diabetes mellitus, optic atrophy, and deafness), an autosomal recessive disorder. | No related references | [39] |
The MCODE plug-in was also used to identify significant clusters. As a result, 3 clusters were identified. One cluster consisted of 9 nodes and 20 edges and included RUNX2, IRS1, KIT, FGFR2, CCND1, LGALS3, MET, MUC1, and NCAM1, which exhibited the highest score (Fig. 4f). Another cluster containing 9 nodes and 20 edges, including SERPINA, FAM20A, WFS1, MXRA8, and PRSS23, also possessed a strong connection (Fig. 4g). Furthermore, the third cluster contained 5 nodes and 10 edges and included AVPR1A, AGTR1, GNA14, HRH1, and LPAR5 (Fig. 4h). These findings strongly suggested that the hub genes we identified could play critical roles in the pathogenesis of PTC.
Comprehensive analysis of common deregulated miRNA and DEGs
The miRNA-gene pairs were based on the 12 common deregulated miRNAs and 263 common DEGs. Thus, 35 miRNA-gene pairs were acquired that included 11 common deregulated miRNAs and 20 common DEGs. We constructed a miRNA-gene regulating network, and the network is shown in Fig. 5a, which illustrates that certain miRNAs play important roles in regulating DEGs. In Fig. 5a, the larger the size of the modules, the greater the degree they have. The nodes with greater degrees tend to be network hubs and are usually considered to play critical roles in maintaining the overall connectivity of the network [40]. In this network, hsa-miR-181a-5p regulated the most DEGs, while BCL2 was targeted by the most miRNAs; in other words, hsa-miR-181a-5p and BCL2 had the greatest degrees among miRNAs and DEGs, respectively, and they were considered to be network hubs. Therefore, we generated subnetworks with hsa-miR-181a-5p and BCL2 as the central nodes (Fig. 5b, c). This information may be important in establishing underlying molecular mechanisms of PTC, which may be used in the development of targets for further research and diagnosis.
Fig. 5.
miRNA-gene regulatory network. a A global view of the network. The network was constructed from common DEGs and deregulated miRNAs. The network contained 11 common deregulated miRNAs, 20 common DEGs, and 35 miRNA-gene pairs. The orange triangle represents miRNA, while the blue square represents DEG. b A subnetwork with hsa-miR-181a-5p as the central node. c A subnetwork with BCL2 as the central node. DEGs, differentially expressed genes
Discussion
Although great progress has been made in PTC research in recent decades, the pathogenesis of PTC still needs to be further clarified. The urgent need for better treatment of PTC has sparked a search for an easy method for the early recognition of benign or malignant modules. If there is a way to find a certain deregulated gene or ncRNA for the early recognition of PTC, then the treatment of PTC could be further improved.
In our present study, PTC-associated deregulated genes and miRNAs were identified based on 3 gene and 2 miRNA expression microarray datasets. A total of 263 DEGs and 12 deregulated miRNAs were selected. Then, pathway and GO enrichment analyses were performed using the 263 DEGs to elucidate the function of PTC-associated DEGs. In addition, PPI networks were constructed to discover hub genes and core clusters. In addition, a deregulated miRNA-gene network was also constructed, and hsa-miR-181a-5p was found to regulate the most DEGs, while BCL2 was targeted by the most miRNAs.
Pathway enrichment analysis using DEGs of PTC provided an insightful overview in elucidating the mechanism of PTC. Among the top 6 pathways, two were associated with Human Diseases; Cancers, while three were categorized into Organismal Systems and were predicted to attribute to development (axon guidance), immune system and endocrine system, and the last pathway was related to amino acid metabolism, suggesting that the pathogenesis of PTC may be closely related to amino acid metabolism, axon guidance, immune system, and endocrine system. For example, Li Y et al. discovered metabolic changes associated with PTC by nuclear magnetic resonance (NMR)–based metabolomic technique, including branched chain amino acid metabolism (leucine and valine), other amino acid metabolism (glycine and taurine) and other metabolisms of other substances, such as glycolysis, tricarboxylic acid cycle, choline metabolism, and lipid metabolism, among which amino acid metabolism function as an oncogenic substance, suggesting that amino acid metabolism may play an important part in the pathogenesis of PTC [41]. Slits, representative axon guidance molecules have been reported whose overexpression regulates the activity of Rho GTPase by inhibiting the transcriptional activity of beta-catenin and inhibiting the cell proliferation, migration, and invasion of thyroid cancer [42], which was consistent with our pathway enrichment results. In addition, a distinct tumor immune microenvironment exists in PTC, correlating with pathological aggressiveness [43]. All these findings indicated a complex mechanism of PTC involving the immune and endocrine system with the participation of amino acid metabolism and axon guidance.
To verify the interaction between the functions of DEGs identified, a PPI network was constructed in which 11 hub genes with the highest connective degree were selected that included FN1, CCND1, MET, RUNX2, IRS1, SERPINA1, KRT19, FGFR2, MUC1, NCAM1, and WFS1. Some of these genes have been reported to be closely associated with PTC; however, their precise roles and molecular mechanisms have not yet been fully elucidated. Fibronectin 1 (FN1), with the highest connective degree in this PPI network, encodes fibronectin, which is involved in cell adhesion and migration processes, including host defense and metastasis [44]. The overexpression of FN1 is an important determinant of PTC aggressiveness [29]. In addition, FN1 is targeted by miR-139 and functions in inhibiting tumorigenesis in PTC cells [45]. CCND1, short for cyclin D1, has been demonstrated to interact with tumor suppressor protein Rb [46]. Mutations, amplification, and overexpression of CCND1, which alters cell cycle progression, occurred frequently in a variety of tumors, including PTC, and may lead to tumorigenesis [30, 47].
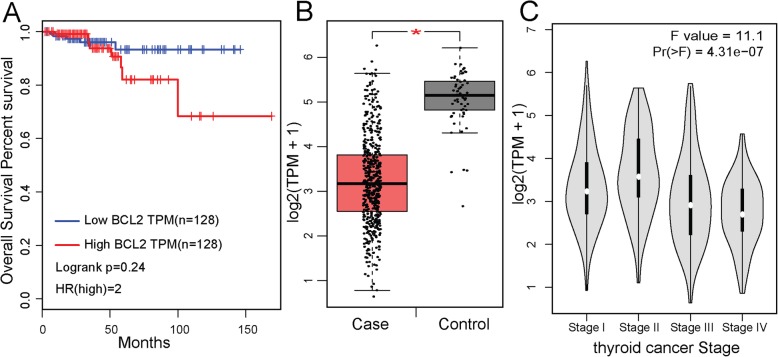
In addition, a miRNA-gene regulatory network was constructed to further explore the association between deregulated miRNAs and DEGs in PTC. The results showed that hsa-miR-181a-5p and BCL2 were the most impactful miRNA and gene of PTC. However, few biological studies have focused on the relationship between hsa-miR-181a-5p and PTC. By consulting the literature in PubMed, only one study was found to have reported the underlying mechanism that miR-181a-5p was oppositely expressed in the exosomes of both PTC and follicular thyroid cancer (FTC), which can help distinguish these two types of TC through comparison [48]. Similarly, studies on the relationship between BCL2 and PTC are also very rare, and only a few studies have verified the correlation between BCL2 and PTC. For example, the synonymous SNP rs1801018 and the G allele of the BCL2 gene were discovered by Eun et al., and this alteration may be related to the bilaterality and multifocality of PTC [49]. It has been found that loss of BCL2 is associated with dedifferentiation in thyroid tumors [50]; however, whether there is a connection that deregulation of BCL2 influences PTC remains unclear. Our study provides a novel way to discover how BCL2 and hsa-miR-181a-5p act on PTC through biological experiments. Next, we used the GEPIA database [51] to analyze the predictive power of BCL2 for the clinical stage and survival of TCGA thyroid cancer (Fig. 6). The survival analysis results showed that there was no statistically significant association between the expression level of BCL2 and the survival of patients with thyroid cancer. However, observed that high levels of BCL2 expression could be correlated with a 2-fold higher mortality rate in patients than in the low-expression group (Fig. 6a, HR (high) = 2). Therefore, the expression level of BCL2 can be used as a predictor of survival in patients with thyroid cancer. In addition, we found that in TCGA thyroid cancer data, BCL2 was differentially expressed between cases and controls. BCL2 was generally downregulated in tumor tissues, and this trend became more pronounced as the disease progressed, so BCL2 also has a potential to be a dynamic signal for estimating the progression and prognosis of thyroid cancer (Fig. 6b, c).
Fig. 6.
The predictive power of BCL2 for the clinical stage and patient survival. a The Kaplan-Meier curve of TCGA samples categorized by the expression level of BCL2. b The boxplot of the BCL2 expression level in the Case and Control groups. c Expression box diagram of BCL2 in different tumor stages
However, there are still some limitations of this study. (i) Different methods of functional analyses have different advantages and disadvantages. In this study, we applied DAVID for functional analyses. The limitations of the DAVID tool include that it only uses the number of genes, regardless of the gene expression level or differential expression value. In addition, to obtain the genes of interest or differential expression, an artificial threshold was needed. In addition, it usually uses the most significant genes and ignores the genes with no significant differences, which may result in a loss of genes with lower significance but a more critical role, resulting in decreased detection sensitivity. (ii) Microarray data were obtained from the GEO database, not generated by the authors. (iii) This study focused on bioinformatics methods to screen candidate genes and miRNAs for PTC; however, the predicted results should be confirmed by laboratory data. Further validation with larger sample sizes and in vitro and in vivo experiments are required to confirm these results. (iv) BCL2 has two main isoforms (1G5M and 1G5O/1GJH), and the structures of the two BCL2 isoforms were found to be very similar [52, 53]. Further experiments could be performed to recognize the differences in the results for different isoforms of BCL2 and may be useful for the treatment of cancers.
Conclusions
In conclusion, we systematically identified DEGs and deregulated miRNAs through microarray datasets. Pathway and GO enrichment analysis gave us an insightful view of the functions of DEGs. Furthermore, the construction of the PPI network and miRNA-gene regulatory network provided us with hsa-miR-181a-5p and BCL2 for further biological experiments to determine the regulatory relationship between them in PTC. As biomarkers, hsa-miR-181a-5p and BCL2 have enormous potential for distinguishing benign or malignant nodules and guiding clinical treatments. These findings will provide important clues for investigating the pathogenesis and therapeutic methods of PTC.
Supplementary information
Additional file 1: Figure S1. Sample analysis
Acknowledgements
Not applicable.
Abbreviations
- BPs
Biological processes
- CCND1
Cyclin D1
- CCs
Cellular components
- CITED1
Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 1
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- DEGs
Differentially expressed genes
- DTCs
Differentiated thyroid cancers
- FDR
False discovery rate
- FMO1
Flavin-containing monooxygenase 1
- FN1
Fibronectin 1
- FTC
Follicular thyroid cancer
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- IL17RA
Interleukin 17 receptor A
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LRP4
LDL receptor related protein 4
- MCODE
Molecular Complex Detection
- MFs
Molecular functions
- NMR
Nuclear magnetic resonance
- PPI
Protein-protein interaction
- PTC
Papillary thyroid cancer
- STRING
The Search Tool for the Retrieval of Interacting Genes
- TEKT4
Tektin 4
Authors’ contributions
CZ, CB, SM, and XG designed the experiment. LG and XG undertook the data acquisition. PY and SM were involved in the interpretation of data. CZ and CB analyzed and visualized the data. CZ drafted the manuscript. CZ and CB revised the manuscript. The final manuscript was read and approved by all authors.
Funding
This work was supported by the Youth project of Haiyan scientific research of Harbin Medical University Cancer Hospital (grant nos. JJQN2019-10).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cong Zhang and Chunrui Bo contributed equally to this work.
Contributor Information
Susheng Miao, Email: 38898546@qq.com.
Xin Gu, Email: Dr_guxin@outlook.com.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12957-019-1755-9.
References
- 1.Morris LG, Tuttle RM, Davies L. Changing trends in the incidence of thyroid cancer in the United States. JAMA Otolaryngol Head Neck Surg. 2016;142:709–711. doi: 10.1001/jamaoto.2016.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Jiang Y, Fang Q, Yuan H, Cai N, Suo C, Ye W, Chen X, Zhang T. Future of cancer incidence in Shanghai, China: predicting the burden upon the ageing population. Cancer Epidemiol. 2019;60:8–15. doi: 10.1016/j.canep.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388:2783–2795. doi: 10.1016/S0140-6736(16)30172-6. [DOI] [PubMed] [Google Scholar]
- 5.Xia E, Wang Y, Bhandari A, Niu J, Yang F, Yao Z, Wang O. CITED1 gene promotes proliferation, migration and invasion in papillary thyroid cancer. Oncol Lett. 2018;16:105–112. doi: 10.3892/ol.2018.8653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou X, Xia E, Bhandari A, Zheng C, Xiang J, Guan Y, Zhang X. LRP4 promotes proliferation, migration, and invasion in papillary thyroid cancer. Biochem Biophys Res Commun. 2018;503:257–263. doi: 10.1016/j.bbrc.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Zheng Z, Zhou X, Cai Y, Chen E, Zhang X, Wang O, Wang Q, Liu H. TEKT4 promotes papillary thyroid cancer cell proliferation, colony formation, and metastasis through activating PI3K/Akt pathway. Endocr Pathol. 2018;29:310–316. doi: 10.1007/s12022-018-9549-0. [DOI] [PubMed] [Google Scholar]
- 8.Lee YC, Chung JH, Kim SK, Rhee SY, Chon S, Oh SJ, Hong IK, Eun YG. Association between interleukin 17/interleukin 17 receptor gene polymorphisms and papillary thyroid cancer in Korean population. Cytokine. 2015;71:283–288. doi: 10.1016/j.cyto.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Luo J, Zhang B, Cui L, Liu T, Gu Y. FMO1 gene expression independently predicts favorable recurrence-free survival of classical papillary thyroid cancer. Future Oncol. 2019;15:1303–1311. doi: 10.2217/fon-2018-0885. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Chen X, Lin T, Chen X, Yan J, Jiang S. MicroRNA-524-5p suppresses the progression of papillary thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell autophagy and cycling pathways. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han J, Zhang M, Nie C, Jia J, Wang F, Yu J, Bi W, Liu B, Sheng R, He G, et al. miR-215 suppresses papillary thyroid cancer proliferation, migration, and invasion through the AKT/GSK-3beta/Snail signaling by targeting ARFGEF1. Cell Death Dis. 2019;10:195. doi: 10.1038/s41419-019-1444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang S, Wang Q, Li D, Huang B, Hou X, Wang D. MicroRNA509 targets PAX6 to inhibit cell proliferation and invasion in papillary thyroid carcinoma. Mol Med Rep. 2019;19:1403–1409. doi: 10.3892/mmr.2018.9750. [DOI] [PubMed] [Google Scholar]
- 13.Yi T, Zhou X, Sang K, Zhou J, Ge L. MicroRNA-1270 modulates papillary thyroid cancer cell development by regulating SCAI. Biomed Pharmacother. 2019;109:2357–2364. doi: 10.1016/j.biopha.2018.08.150. [DOI] [PubMed] [Google Scholar]
- 14.Cao XZ, Bin H, Zang ZN. MiR-128 suppresses the growth of thyroid carcinoma by negatively regulating SPHK1. Biomed Pharmacother. 2019;109:1960–1966. doi: 10.1016/j.biopha.2018.08.052. [DOI] [PubMed] [Google Scholar]
- 15.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aragon AD, Quinones GA, Thomas EV, Roy S, Werner-Washburne M. Release of extraction-resistant mRNA in stationary phase Saccharomyces cerevisiae produces a massive increase in transcript abundance in response to stress. Genome Biol. 2006;7:R9. doi: 10.1186/gb-2006-7-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomas G, Tarabichi M, Gacquer D, Hebrant A, Dom G, Dumont JE, Keutgen X, Fahey TJ, 3rd, Maenhaut C, Detours V. A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic. Oncogene. 2012;31:4490–4498. doi: 10.1038/onc.2011.626. [DOI] [PubMed] [Google Scholar]
- 19.Dom G, Tarabichi M, Unger K, Thomas G, Oczko-Wojciechowska M, Bogdanova T, Jarzab B, Dumont JE, Detours V, Maenhaut C. A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br J Cancer. 2012;107:994–1000. doi: 10.1038/bjc.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minna E, Romeo P, Dugo M, De Cecco L, Todoerti K, Pilotti S, Perrone F, Seregni E, Agnelli L, Neri A, et al. miR-451a is underexpressed and targets AKT/mTOR pathway in papillary thyroid carcinoma. Oncotarget. 2016;7:12731–12747. doi: 10.18632/oncotarget.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia F, Wang W, Jiang B, Chen Y, Li X. DNA methylation-mediated silencing of miR-204 is a potential prognostic marker for papillary thyroid carcinoma. Cancer Manag Res. 2019;11:1249–1262. doi: 10.2147/CMAR.S184566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 24.Szklarczyk Damian, Morris John H, Cook Helen, Kuhn Michael, Wyder Stefan, Simonovic Milan, Santos Alberto, Doncheva Nadezhda T, Roth Alexander, Bork Peer, Jensen Lars J., von Mering Christian. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research. 2016;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with CentiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amaral LA, Scala A, Barthelemy M, Stanley HE. Classes of small-world networks. Proc Natl Acad Sci U S A. 2000;97:11149–11152. doi: 10.1073/pnas.200327197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sponziello M, Rosignolo F, Celano M, Maggisano V, Pecce V, De Rose RF, Lombardo GE, Durante C, Filetti S, Damante G, et al. Fibronectin-1 expression is increased in aggressive thyroid cancer and favors the migration and invasion of cancer cells. Mol Cell Endocrinol. 2016;431:123–132. doi: 10.1016/j.mce.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Yin Y, Hong S. MiR-195 Inhibits Tumor Growth and Metastasis in Papillary Thyroid Carcinoma Cell Lines by Targeting CCND1 and FGF2. Int J Endocrinol. 2017;2017:6180425. [DOI] [PMC free article] [PubMed]
- 31.Fu YT, Zheng HB, Zhou L, Zhang DQ, Liu XL, Sun H. Valproic acid, targets papillary thyroid cancer through inhibition of c-Met signalling pathway. Am J Transl Res. 2017;9:3138–3147. [PMC free article] [PubMed] [Google Scholar]
- 32.Han M, Chen L, Wang Y. miR-218 overexpression suppresses tumorigenesis of papillary thyroid cancer via inactivation of PTEN/PI3K/AKT pathway by targeting Runx2. Onco Targets Ther. 2018;11:6305–6316. doi: 10.2147/OTT.S172152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahmutovic L, Bego T, Sterner M, Gremsperger G, Ahlqvist E, Velija Asimi Z, Prnjavorac B, Hamad N, Causevic A, Groop L, Semiz S. Association of IRS1 genetic variants with glucose control and insulin resistance in type 2 diabetic patients from Bosnia and Herzegovina. Drug Metab Pers Ther. 2019;34(1). [DOI] [PubMed]
- 34.Vierlinger K, Mansfeld MH, Koperek O, Nohammer C, Kaserer K, Leisch F. Identification of SERPINA1 as single marker for papillary thyroid carcinoma through microarray meta analysis and quantification of its discriminatory power in independent validation. BMC Med Genet. 2011;4:30. doi: 10.1186/1755-8794-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, et al. Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci U S A. 2001;98:15044–15049. doi: 10.1073/pnas.251547398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redler A, Di Rocco G, Giannotti D, Frezzotti F, Bernieri MG, Ceccarelli S, D'Amici S, Vescarelli E, Mitterhofer AP, Angeloni A, Marchese C. Fibroblast growth factor receptor-2 expression in thyroid tumor progression: potential diagnostic application. PLoS One. 2013;8:e72224. doi: 10.1371/journal.pone.0072224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhan XX, Zhao B, Diao C, Cao Y, Cheng RC. Expression of MUC1 and CD176 (Thomsen-Friedenreich antigen) in papillary thyroid carcinomas. Endocr Pathol. 2015;26:21–26. doi: 10.1007/s12022-015-9356-9. [DOI] [PubMed] [Google Scholar]
- 38.Abouhashem NS, Talaat SM. Diagnostic utility of CK19 and CD56 in the differentiation of thyroid papillary carcinoma from its mimics. Pathol Res Pract. 2017;213:509–517. doi: 10.1016/j.prp.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 39.Hardy C, Khanim F, Torres R, Scott-Brown M, Seller A, Poulton J, Collier D, Kirk J, Polymeropoulos M, Latif F, Barrett T. Clinical and molecular genetic analysis of 19 Wolfram syndrome kindreds demonstrating a wide spectrum of mutations in WFS1. Am J Hum Genet. 1999;65:1279–1290. doi: 10.1086/302609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Greenbaum D, Xin Lu H, Zhu X, Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;20:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Li Y, Chen M, Liu C, Xia Y, Xu B, Hu Y, Chen T, Shen M, Tang W. Metabolic changes associated with papillary thyroid carcinoma: a nuclear magnetic resonance-based metabolomics study. Int J Mol Med. 2018;41:3006–3014. doi: 10.3892/ijmm.2018.3494. [DOI] [PubMed] [Google Scholar]
- 42.Jeon MJ, Lim S, You MH, Park Y, Song DE, Sim S, Kim TY, Shong YK, Kim WB, Kim WG. The role of Slit2 as a tumor suppressor in thyroid cancer. Mol Cell Endocrinol. 2019;483:87–96. doi: 10.1016/j.mce.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Means C, Clayburgh DR. Tumor immune microenvironment characteristics of papillary thyroid carcinoma are associated with histopathological aggressiveness and BRAF mutation status. 2019. [DOI] [PubMed] [Google Scholar]
- 44.Liao YX, Zhang ZP, Zhao J, Liu JP. Effects of fibronectin 1 on cell proliferation, senescence and apoptosis of human glioma cells through the PI3K/AKT signaling pathway. Cell Physiol Biochem. 2018;48:1382–1396. doi: 10.1159/000492096. [DOI] [PubMed] [Google Scholar]
- 45.Ye Y, Zhuang J, Wang G, He S, Ni J, Xia W. MicroRNA-139 targets fibronectin 1 to inhibit papillary thyroid carcinoma progression. Oncol Lett. 2017;14:7799–7806. doi: 10.3892/ol.2017.7201. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Komori T. Regulation of Rb family proteins by Cdk6/Ccnd1 in growth plates. Cell Cycle. 2013;12:2161–2162. doi: 10.4161/cc.25515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aytekin T, Aytekin A, Maralcan G, Gokalp MA, Ozen D, Borazan E, Yilmaz L. A cyclin D1 (CCND1) gene polymorphism contributes to susceptibility to papillary thyroid cancer in the Turkish population. Asian Pac J Cancer Prev. 2014;15:7181–7185. doi: 10.7314/APJCP.2014.15.17.7181. [DOI] [PubMed] [Google Scholar]
- 48.Samsonov R, Burdakov V, Shtam T, Radzhabovа CZ, Vasilyev D, Tsyrlina E, Titov S, Ivanov M, Berstein L, Filatov M, et al. Plasma exosomal miR-21 and miR-181a differentiates follicular from papillary thyroid cancer. Tumour Biol. 2016;37:12011–12021. doi: 10.1007/s13277-016-5065-3. [DOI] [PubMed] [Google Scholar]
- 49.Eun YG, Hong IK, Kim SK, Park HK, Kwon S, Chung DH, Kwon KH. A polymorphism (rs1801018, Thr7Thr) of BCL2 is associated with papillary thyroid cancer in Korean population. Clin Exp Otorhinolaryngol. 2011;4:149–154. doi: 10.3342/ceo.2011.4.3.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A, Jain S, Khurana N, Kakar AK. Expression of p63 and Bcl-2 in malignant thyroid tumors and their correlation with other diagnostic immunocytochemical markers. J Clin Diagn Res. 2016;10:Ec04–Ec08. doi: 10.7860/JCDR/2016/13899.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–w102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Oltersdorf T, Fesik SW. Solution structure of the antiapoptotic protein bcl-2. Proc Natl Acad Sci U S A. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsujimoto Y, Croce CM. Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc Natl Acad Sci U S A. 1986;83:5214–5218. doi: 10.1073/pnas.83.14.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Sample analysis
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.