Abstract
The placenta is a short-lived tissue required for embryonic growth and survival, and it is fetal derived. Fetal sex influences gestation, and many sexual dimorphic diseases have origins in utero. There is sex-biased gene expression in third-trimester human placentas, yet the origin of sex-specific expression is unknown. Here, we used an in vitro differentiation model to convert human embryonic stem cells (hESCs) into trophoblastic progenitor cells of the first-trimester placenta, which will eventually become mature extravillous trophoblasts and syncytiotrophoblasts. We observed significant sex differences in transcriptomic profiles of hESCs and trophoblastic progenitors, and also with the differentiation process itself. Male cells had higher dosage of X/Y gene pairs relative to female samples, supporting functions for Y-linked genes beyond spermatogenesis in the hESCs and in the early placenta. Female-specific differentiation altered the expression of several thousand genes compared with male cells, and female cells specifically upregulated numerous autosomal genes with known roles in trophoblast function. Sex-biased upregulation of cellular pathways during trophoblast differentiation was also evident. This study is the first to identify sex differences in trophoblastic progenitor cells of the first-trimester human placenta, and reveal early origins for sexual dimorphism.
Keywords: : sex differences, sex-biased gene expression, in vitro differentiation of human embryonic stem cells, XIST RNA, human trophoblastic progenitor cells, human placental development
Introduction
The placenta is required for the growth and survival of the fetus during pregnancy, and facilitates the exchange of gases, nutrients, waste products, and hormones between maternal and fetal circulation. The placenta is formed 6–7 days postconception, and is composed of specialized epithelial cells called trophoblasts [1]. Trophoblasts are one of the earliest lineage differentiation events of the mammalian embryo, arising from the outer extraembryonic trophectoderm cells of the preimplantation blastocyst. During implantation, trophoblasts invade the maternal epithelium and differentiate into two types of specialized progenitor cells [2]. Syncytiotrophoblasts (SYNs) are multinucleated terminally differentiated cells that synthesize hormones for sustaining pregnancy. Cytotrophoblasts are mononucleated undifferentiated progenitors that can differentiate into SYNs and extravillous invasive trophoblasts (EVTs), which anchor the placenta to the uterus. These early differentiation events are essential for placental formation, as impairments with trophoblast cells result in miscarriage, pre-eclampsia, and intrauterine growth restriction [1].
Our understanding of the early events of human trophoblast formation is limited due to ethical constraints of working with human embryos. Animal models have been useful for identifying trophoblast-specific lineage factors [3], yet there are significant differences between mouse and human placentation [4]. Human-specific model systems of early development are necessary to define the genetic and epigenetic mechanisms involved in this lineage. One model system for the early human placenta utilizes human embryonic stem cells (hESCs) for directed differentiation into SYN and EVT cells. Both human-induced pluripotent stem cells (hiPSCs) and hESCs, unlike their mouse counterparts, can be cultured in vitro with bone morphogenic protein 4 (BMP4) and basic fibroblast growth factor (B-FGF) withdrawal, which generates a mixture of EVTs and SYNs that are referred to as “trophoblastic cells” [5,6]. This model system is widely used to study the development of the early human placenta because it does not require access to early human embryos [6,7]. The addition of BMP4 together with the inhibitors A83-01 (A) and PD173074 (P), which block the SMAD2/3 and MEK1/2 signaling pathways, enhances trophoblastic cell differentiation without extensive generation of mesoderm, endoderm, or ectoderm cells [6,8]. Importantly, these culture conditions result in the expression of various trophoblast markers and placental hormones [6,9]. The hESCs differentiated for 12 days have similar gene expression profiles to trophectoderm cells isolated from human blastocyst-stage embryos, supporting the validity of this in vitro model system [6,9,10]. Unfortunately, two of these studies only examined hESCs from one cell line, the male H1 hESCs [6] or female H7 hESCs [6,9]. A more recent publication used two distinct male hESC lines (H1 and CA1) [10] to compare trophoblast marker expression between in vitro-derived trophoblast-like cells and primary placental cells, yet did not consider sex as a variable for gene expression.
The placenta, similar to the ovary and testis, exhibits tissue-specific expression of sex-biased genes. Human males and females exhibit different growth rates in utero, which has been attributed to sex-specific differences with placental function presumably due to gene expression differences [11,12]. Full-term placenta samples have sex-biased gene expression profiles, and the majority of differentially expressed genes are autosomal [13,14]. It is unknown whether sex influences gene expression during the formation of trophoblasts, which are critical for the development of the human placenta.
One source of gene expression variation between male and female cells comes from differences with the sex chromosomes. The Y-chromosome contains the male sex determination gene SRY and about 70 additional genes important for spermatogenesis. Y-linked genes also function beyond reproduction because they are abundantly expressed in multiple adult tissues and during development [15]. The presence of a Y-chromosome may also influence disease risk of cancer [16,17], coronary artery disease [18], autism [19], and primary biliary cirrhosis [20], yet the mechanisms whereby Y-linked genes contribute to these disease phenotypes are not known. To maintain dosage compensation of X-linked genes between the sexes, female mammals randomly silence one of their two X-chromosomes in early preimplantation development during the process of X-chromosome inactivation (XCI) [21,22]. The long noncoding RNA XIST is indispensable for the initiation of X-linked gene silencing [23,24], as XIST RNA recruits factors responsible for heterochromatin formation of the inactive X and XCI maintenance [25,26]. Nearly all female mammalian somatic cells express XIST RNA from the inactive X, and this chromosome is enriched with heterochromatic marks and XIST RNA. Unlike their mouse counterparts, human female ESCs and iPSCs are heterogeneous for XIST expression and XCI status, and the majority of the commonly used cell lines have irreversibly silenced the XIST gene [27]. XIST-negative human pluripotent stem cells (hPSCs) have a partially reactivated inactive X, yet many of the X-linked genes subject to XCI remain silenced in XIST-negative hPSCs [28–32]. Some studies have reported impaired differentiation of XIST-negative hiPSCs and hESCs [28], while other studies indicate that XIST status has no effect on differentiation capacity of these cells [32,33]. Because of this epigenetic instability, human female pluripotent stem cells are often excluded from directed differentiation experiments, and male cells are routinely used, preventing investigation of sex differences involving hESC/hiPSC-derived cells.
For this study, we sought to determine the sex-specific gene expression changes associated with the formation of human trophoblast cells using the BMP4/A/P in vitro model system. The advantage of this system is that differentiated trophoblastic progenitor cells are directly compared with the hESCs from which they originated, thus reducing the differences in genetic “background” that can occur when EVTs and SYNs from first-term pregnancies are compared with embryonic samples that may not be harvested from the same pregnancy due to ethical concerns. Here, we profiled early-stage in vitro differentiated progenitors of trophoblasts, which revealed that XIST expression status does not significantly impact global gene expression profiles, and that XIST-negative hESCs can be used for gene expression profiling when investigating sex differences for hESC/hiPSC-derived cell populations. Importantly, we demonstrate sex-specific gene expression differences in both trophoblastic progenitors and hESCs, revealing that sex-biased gene expression is present in the early human placental progenitor cells.
Materials and Methods
Cell culture of male and female hESCs and in vitro differentiation into trophoblastic cells
The hESCs (H1 p.49, H9 p.60, CHB8, p.39, HUES7 p.36, HUES8 p.22, and HUES9 p.39) and hiPSCs (hiPS-2 p.31, hiPS-12 p.31) [28] were cultured on mouse embryonic fibroblasts (MEFs) at 4% oxygen, and passaged manually to expand cells (for 70%–90% in six wells of a six-well plate) in preparation for RNA isolation (for day 0 samples) and for trophoblastic cell differentiation [28,34]. After collecting day 0 hESCs for RNA (harvesting 2 × wells of a six-well plate), we passaged cells once to remove MEFs in preparation for differentiation. For BMP4/A/P differentiation to trophoblastic cells (also performed at 4% oxygen), we used mechanical dissociation to passage the hPSC colonies, and transferred hESCs and hiPSCs onto Matrigel (R&D Biosystems)-coated plates, then the cells were grown using MEF-conditioned medium containing B-FGF [9]. After 5–7 days, we dissociated cells (passaging at 1:3) using Dispase (Life Technologies), and then transferred onto Matrigel-coated six-well plates, and ROCK inhibitor was added for 1 day. Colonies of various sizes were visible the next day. Twenty-four hours after plating, we added differentiation medium lacking B-FGF and containing 50 ng/mL BMP4, 1 μM A83-01 (Tocris), and 0.1 μM PD173074 (Tocris) (BMP4/A/P), and refreshed every 48 h during the 5-day course of differentiation. At differentiation day 5, the differentiated cells were confluent in the well, and individual colonies were no longer apparent. This study involved the use of hESCs, and was approved by the University of Pennsylvania Embryonic Stem Cell Research Oversight Committee.
Flow cytometric analysis of cellular differentiation
Before flow cytometric analysis, hESCs (HUES7 male passage 38 and HUES9 female passage 41) were differentiated into trophoblastic progenitor cells as described above. As a control, hESCs were split to generate embryoid bodies (EBs), which were formed using low-retention culture plates and placed in MEF media (without B-FGF) to allow for random differentiation. Trophoblastic progenitors and control cells were collected on differentiation day 6, then stained with Live/Dead Aqua (1:200; Invitrogen). Stained cells were fixed and permeabilized (Foxp3/Transcription Factor Staining Buffer Set; Thermo Fisher), then incubated overnight with GATA-3 (1:20, Clone 16E10A23; Biolegend) or KRT7 (1:150, KRT7/760; Abcam). The following day, KRT7 stained cells were incubated with secondary antibody (1:2,000, goat antimouse IgG H&L FITC, ab6785; Abcam) for 1 h. Flow cytometry was performed using a BD LSR Fortessa (BD Biosciences) with DiVa 6.1.3 software. Cell populations were analyzed using FlowJo v9.9.6 with the following gating scheme: FSC-A, SSC-A, Live/Dead, FSC-H, FSC-W (single cells). Single cells were analyzed for the percentage of positive events recorded for each antibody.
Fluorescence in situ hybridization and immunofluorescence
We used XIST RNA fluorescence in situ hybridization (RNA FISH) with Cy3-fluorophore-conjugated oligo probes to classify the XIST status of female hESCs and hiPSCs, as previously described [35]. For immunofluorescence (IF) analyses, cells were grown in 6-well or 12-well plates, then washed before fixation using 4% paraformaldehyde. CDX2 antibody (EPR2764Y; Abcam) and KRT7 antibody (ab215855; Abcam) were used at 1:100 dilution for IF (detection with fluorophore-conjugated secondary antibodies) and flow cytometry.
Transcriptional profiling and bioinformatic analyses
For sex-specific mRNA transcriptome analysis, we harvested cells at day 0 (undifferentiated) and at day 5 (differentiated trophoblastic progenitor cells). Total RNA was isolated for all samples at the same time using TRIzol (Invitrogen). RNA quality was assessed using an Agilent BioAnalyzer. We profiled mRNA for male and female hESCs and trophoblastic cells, and constructed libraries for all the samples at the same time, using a TruSeq Stranded mRNA Library Preparation Kit (Illumina). Pooled libraries were run on an Illumina NextSeq 500 benchtop sequencer (150-bp single-end reads) with an average depth of 30-million fragments per sample. For XIST-status transcriptome profiling (undifferentiated, HUES9 cII, hiPS2 cII and cIII, hiPS12 cII and cIII), we constructed libraries using TruSeq stranded Total RNA Library Prep Kit with Ribo-Zero (Illumina). Paired-end 100-bp fragment libraries were pooled and run on the same flow cell, and were run by the Children's Hospital of Philadelphia and Beijing Genomics Institute (BGI) Joint Genome Center using an Illumina HiSeq 4000, and sequencing was performed with an average depth of 30-million fragments per sample.
RNA-sequencing methods
Analysis of RNA-sequencing (RNA-seq) data was carried out as previously described [36]. In brief, data analyses were performed using the statistical computing environment R (v3.2.1), RStudio (v0.99), and the Bioconductor suite of packages for R [37]. Raw reads were mapped to the human reference transcriptome (Ensembl, release 92) using Kallisto [38]. Tximport [39] was used to import transcript-level count data into R and summarize to gene level. Data were normalized using the DESeq2 model to internally correct for library size [40], and filtering was carried out to remove unexpressed and lowly expressed genes [(cpm >1)> = 3]. Differentially expressed genes (false discovery rate [FDR]) <0.05 and log2-fold change ≥ 0.59) were identified using DESeq2 [40]. For plots and bar graphs comparing gene expression, estimated counts were adjusted for library size and log2 transformed. Heatmaps were created and visualized using the heatmap.2 and gplots packages [41]. Bar graphs were created in GraphPad Prism version 7.0. Data have been deposited in the Gene Expression Omnibus (GEO) database (Accession No. GSE115941).
Functional enrichment analysis
Gene Ontology (GO) term enrichment was performed using the Database for Visualization and Integrative Discovery (DAVID) v6.7 with differentially expressed genes from expression clusters as input [42]. Functional Annotation Clustering tools and GO BP “fat” terms with an enrichment score ≥1.3 (P < 0.05) were selected. Graphs were created using DataGraph. Gene Set Enrichment Analysis (GSEA, Broad Institute) was performed using MSigDB (v5.1) to query the “C2: Canonical pathways” collection, which contains 1,329 gene sets, and the “C1: Positional gene sets” collection containing 326 gene sets [43].
Results
The expression of XIST does not impact sex-specific gene expression profiling in hESCs and hiPSCs
Female hPSCs are epigenetically unstable for XCI maintenance, and the majority of available female hPSCs have irreversibly silenced the XIST gene during routine culture [29–31,34,44]. There is limited availability of National Institutes of Health (NIH)-approved XIST-positive female hESCs, and the generation and maintenance of XIST-positive hiPSCs are restricted to research groups working on cellular reprogramming that routinely check for markers of XCI. Because of limited XIST-positive female hPSC availability and the importance of sex as a biological variable in research, we asked whether XIST-negative hPSCs could be used to determine sex differences in gene expression. To address this, we performed RNA-seq to compare the genome-wide expression profiles of six samples of XIST-positive hESCs and hiPSCs with four samples of XIST-negative cell lines [28,29,32,34] and three male hESC samples. We used RNA FISH to confirm the XIST expression status for each female hPSC line, and we observed a loss of XIST RNA “clouds” at the inactive X-chromosome for XIST-negative cells (Fig. 1A). XIST-positive hPSCs contained a heterogeneous mixture of cells with different XIST statuses, where ∼70%–85% of cells retained XIST RNA signal, similar to previous observations (Fig. 1A) [28,29]. Unsupervised hierarchical clustering of all samples showed that both XIST-positive and XIST-negative cells grouped together, and grouped apart from male hESCs (Fig. 1B). Importantly, the XIST-positive and XIST-negative samples were very similar, and did not cluster together based on XIST expression (Fig. 1B). Principal component analysis (PCA) confirmed the similarities of global gene expression profiles between XIST-positive and XIST-negative hPSCs, and that male hESCs were distinct from female lines (Fig. 1C). Next, we used scatterplots to compare the average transcripts per kilobase million (TPM) for XIST-positive (n = 6) with the average for the XIST-negative hPSCs (n = 4) at the individual gene level. The global gene expression profiles of XIST-positive and XIST-negative hPSCs, representing different genetic backgrounds, were nearly identical (R2 = 0.9653; Fig. 1D, top). We compared the average of XIST-positive samples with the average for the male hESCs (n = 3), and observed many genes exhibiting a sex bias (R2 = 0.9055; Fig. 1D, middle). When we repeated this comparison using XIST-negative samples, we observed a remarkably similar distribution to XIST-positive samples (R2 = 0.8833; Fig. 1D, bottom). We conclude that XIST expression and XCI status does not impact global gene expression profiles for identifying sex-biased gene expression, and that XIST-negative hESCs, which represent the majority of female hPSCs, are a suitable model system for determining sex differences.
FIG. 1.
XIST-positive and XIST-negative female hPSCs are transcriptionally similar, and distinct from male hESCs, regardless of XIST status. (A) XIST RNA FISH (red) in single class II XIST-positive (left) and class III XIST-negative (right) nuclei counterstained with DAPI (blue). Percentage of nuclei with an XIST RNA cloud is shown. (B) Unsupervised hierarchical clustering based on Minkowski distance of male hESCs (blue), female XIST-positive hPSCs (orange), and female XIST-negative hPSCs (pink). (C) PCA for all female and male undifferentiated hPSCs, with samples colored by cell type (left) and by XIST status (right). The percentage contribution of each PC is displayed in each axis. (D) Scatterplots of TPMs from XIST-positive compared with XIST-negative female hPSCs (top), XIST-positive hPSCs compared with male hESCs (middle), and XIST-negative hPSCs compared with male hESCs (bottom). hESCs, human embryonic stem cells; hPSCs, human pluripotent stem cells; PC, principal component; PCA, principal component analysis; RNA FISH, RNA fluorescence in situ hybridization; TPMs, Transcripts Per kilobase Million. Color images available online at www.liebertpub.com/scd
hESCs exhibit sex-specific gene expression differences
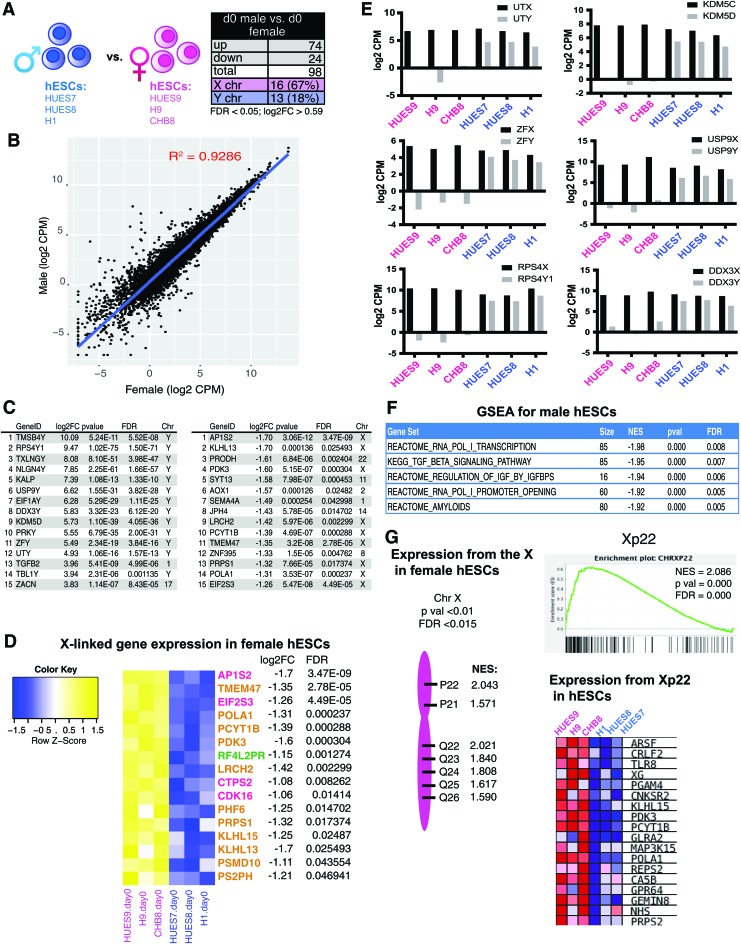
To determine the sex-specific gene expression profiles of female and male hESCs, we performed RNA-Seq using RNA isolated from distinct female (XIST negative; confirmed using RNA FISH) and male hESC lines (Fig. 2A). We selected commonly used and NIH-approved hESC lines: three female hESC lines (HUES9, H9, and CHB8) and three male hESC lines (HUES7, HUES8, and H1). We grouped the three biological replicates (from three distinct genetic backgrounds) from each sex together, and interrogated the genes that are significantly different between males and females in the averaged gene expression profiles after adjusting for multiple testing by Benjamini–Hochberg false discovery rate. We specifically included expression from the sex chromosomes in our analyses, which we hypothesized should exhibit the greatest sex differences. We found that male hESCs had significantly increased expression for 74 unique genes, and 24 genes were significantly upregulated in female hESCs (Fig. 2A; log2FC >0.59, FDR <0.05). Using a scatterplot to compare the expression on a gene-by-gene basis for male and female hESCs, we found that the overall global gene expression of hESCs was similar between the sexes (R2 = 0.9286), with a small number of genes over-represented in each group (Fig. 2B). The majority of genes significantly upregulated in females were X-linked (67%) compared with males (18% Y-linked), which had more autosomal genes upregulated (Fig. 2A, C and Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/scd). We noticed that Y-linked genes were the most abundantly expressed in male hESCs, as expected, and this list included Y-linked genes with an X-linked counterpart known to escape XCI, known as X/Y gene pairs (Fig. 2C). Male hESCs also expressed higher levels of autosomal genes important for stem cell maintenance and lineage-specific differentiation: NODAL, TGFB2, and IGFBP3 [45,46]. Female hESCs predominantly overexpressed X-linked genes (Fig. 2C), some of which are known to escape XCI (listed in pink and green; Fig. 2D) [33]. Most of the X-linked genes in this list are subject to XCI in other tissues and cell types (Fig. 2D, gold) [47]; thus, it is possible that these genes specifically escape XCI in hPSCs, or that the increased expression is from the partially eroded inactive X in XIST-negative cell lines. We found that more than half of the genes in gold were also upregulated in XIST-positive hPSCs compared with male hESCs (data not shown), indicative of hESC-specific XCI escape. In conclusion, we observed gene expression differences predominantly from the sex chromosomes in hESCs, suggesting that the inner cell mass of the human preimplantation embryo exhibits sex-biased gene expression.
FIG. 2.
Pluripotent hESCs exhibit sex-specific differences with gene expression. (A) Schematic depicting the three male and female hESCs transcriptionally profiled (left) and table showing the total number of significant DEGs between sexes (right; FDR <0.05, log2FC >0.59). (B) Scatterplot of log2 transformed reads (CPMs) of male compared with female hESCs. (C) List of the top 15 significant (P < 0.05, log2FC >0.59) most highly expressed DEGs comparing male (left) with female (right) hESCs. Genes expressed in male hESCs (left); genes expressed in female hESCs (right). (D) Heatmap for X-linked genes overexpressed in female hESCs. Genes in pink denote known XCI escape; green represents variable escape from XCI; yellow represents genes normally subject to XCI. (E) Log2 CPMs for X/Y gene pairs (black bars: X-linked; gray bars: Y-linked) in female (pink) and male (blue) hESCs. (F) GSEA of significantly enriched canonical pathways (C2CP) in male hESCs compared with female hESCs (P < 0.0005). (G) GSEA showing chromosomal locations for X-linked gene sets enriched in female hESCs (P < 0.01, FDR <0.015). CPM, counts per million; DEGs, differentially expressed genes; FDR, false discovery rate; GSEA, Gene Set Enrichment Analysis; XCI, X-chromosome inactivation. Color images available online at www.liebertpub.com/scd
Male hESCs have increased dosage of X/Y gene pairs
Because sex-linked genes exhibited the greatest expression differences among male and female hESCs, we investigated the transcript abundance for X/Y gene pairs in male and female hESCs (Fig. 2E). X/Y gene pairs are widely expressed regulators of transcription, nucleic acid binding, and translation [48]. We observed that the levels of the X-linked genes of X/Y gene pairs were similar across male and female hESC samples, indicative of dosage compensation for X-linked genes between sexes [21,22]. When considering combined dosage of the X-linked and Y-linked genes for these X/Y gene pairs, male hESCs had higher dosage for these genes (Fig. 2E). We found that male hESCs had more expression of two sets of histone demethylases (1) UTX (KDM6A) and UTY (KDM6C), and (2) KDM5C and KDM5D; zinc finger proteins ZFX and ZFY; deubiquitinases USP9X and USP9Y; and the ribosomal S4 proteins RPS4X and RPS4Y1. GSEA of the canonical pathways curated gene sets (C2:CP) confirmed enrichment for pathways of X/Y gene pairs in male cells: transcription (RNA Polymerase I transcription; FDR = 0.008), transcriptional regulation (RNA Pol I promoter opening; FDR = 0.005), and IGF pathways (regulation of IGF by IGFBPS; FDR = 0.006) (Fig. 2F). Next, we used GSEA to determine which regions across the X-chromosome were preferentially expressed in female hESCs compared with male hESCs. Seven regions on the human X-chromosome, from both the p and q arms, were significantly over-represented in female hESCs (Fig. 2G). The p22 region contained the most genes exhibiting greater expression in female hESCs (normalized enrichment score = 2.04), and the p arm is enriched for XCI escape genes [49,50]. In summary, we observed sex-specific gene expression differences in male and female hESCs predominantly from the X- and Y-chromosomes, and male hESCs exhibit increased dosage of X/Y gene pairs compared with female hESCs.
Male and female hESCs exhibit similar efficiency for in vitro differentiation to progenitors of human trophoblastic cells
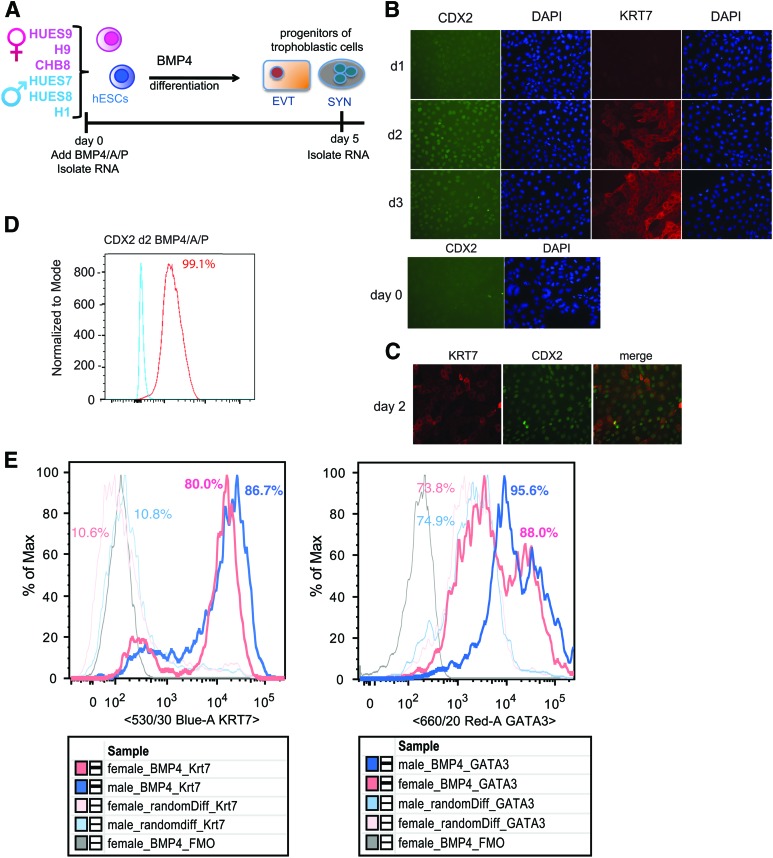
To study sex differences in gene expression during the formation of the human placenta, we differentiated male and female hESCs into progenitors of trophoblastic cells by removing B-FGF from the culture medium, and adding BMP4, A83-01 (A), and PD173074 (P) (referred to as BMP4/A/P medium) to the culture medium for 2–5 days (Fig. 3A) [6]. Complete differentiation of hESCs into trophoblast-like cells (EVTs, SYNs) requires 9–14 days [9]; here, we limited differentiation to days 2–5 to investigate sex-specific differences with the early commitment for differentiation into trophoblastic cells. We observed morphological changes after 1 day of BMP4/A/P differentiation, with cells appearing flatter and larger with enlarged nuclei, as previously observed [6,51]. Cellular differentiation was confirmed using quantitative PCR for the pluripotency marker NANOG, which decreased by day 2, and expression of trophectoderm markers KRT7 and CDX2, whose expression peaked at days 2–4 (data not shown), as described previously [9,51]. We verified that CDX2 and KRT7 proteins were expressed during BMP4/A/P differentiation using IF, and these markers appeared between days 2 and 3 of differentiation (Fig. 3B). We observed CDX2 and KRT7 protein colocalization in day 2 differentiated cells (Fig. 3C), where 99% of differentiating cells expressed CDX2 protein using flow cytometry (Fig. 3D). We did not observe any expression of CDX2 protein in undifferentiated hESCs consistent with its expression in differentiated cells and not the pluripotent state [52]. To further confirm that the in vitro differentiation protocol produced progenitors of trophoblastic cells irrespective of sex, we quantified KRT7 and GATA3 protein levels in male and female cells using flow cytometry. KRT7, a pan-trophoblast marker, was expressed at similar levels in male and female differentiation day 5 cells (Fig. 3E, left). To control for BMP4/A/P differentiation efficiency and specificity, we differentiated male and female hESCs into EBs, which generates mixed progenitors of the ectoderm, endoderm, and mesoderm lineages, and quantified the protein levels of KRT7. BMP4/A/P differentiation yielded higher levels of KRT7 protein for male and female hESCs compared with EB differentiation (Fig. 3E). BMP4/A/P Differentiation day 5 male and female cells had similar levels of GATA3 (Fig. 3E, right), which is abundantly expressed in mononuclear trophoblasts [10] and endodermal cells [53], which are present in EBs. Taken together, we conclude that the BMP4/A/P in vitro differentiation conditions induce the differentiation of both male and female hESCs with equal efficiency into early-stage progenitors of human trophoblastic cells.
FIG. 3.
Equal efficiency of BMP4/A/P differentiation of hESCs into trophoblastic progenitors for male and female cells. (A) Schematic showing the differentiation strategy of hESCs into progenitors of trophoblastic cells, and time points for RNA collection. (B) IF for CDX2 and KRT7 expression during days 1–3 of BMP4/A/P differentiation of HUES9 (top) and in undifferentiated HUES9 cells (bottom). (C) Co-IF for CDX2 and KRT7 protein in day 2 differentiated BMP4/A/P cells (HUES9). (D) Flow cytometry analysis of CDX2 protein levels on day 2 of BMP4/A/P-differentiated HUES9 (red line) compared with an undifferentiated control (blue line). (E) Representative flow plots (n = 3) for markers of trophoblastic cells (KRT7, left; GATA3, right) in male (HUES7, thick blue line) and female (HUES9, thick pink line) cells differentiated for 5 days with BMP4/A/P. Random differentiation for female and male (thin pink and blue lines, respectively) is included along with a negative control FMO (fluorescence minus one) (gray line). Gating strategy is described in the Materials and Methods section. BMP4, bone morphogenic protein 4; IF, immunofluorescence. Color images available online at www.liebertpub.com/scd
Sex distinguishes gene expression profiles of early progenitors of trophoblastic cells
To determine sex-specific gene expression profiles during the early commitment and differentiation into trophoblastic cells, we isolated total RNA from BMP4/A/P differentiation day 5 cells, derived from male and female hESCs for RNA-seq. Unbiased hierarchical clustering of hESCs and differentiated samples distinguished the two groups (Fig. 4A), and PCAs confirmed that the samples were most distinguished by differentiation, and that the trophoblastic progenitor cells (differentiation day 5) were more similar to each other than the pluripotent cells (Fig. 4B). From the hierarchical clustering and PCAs, the hESCs and trophoblastic progenitor cells cluster according to sex (Fig. 4A, B). Next, we asked whether pluripotency markers (in hESCs) or trophoblastic markers (in BMP4/A/P day 5 cells) exhibited expression differences among male and female samples. We assessed normalized counts per million (CPM) values for six pluripotency-related genes, observed similar values for male and female hESCs, and these genes were significantly downregulated in differentiation day 5 samples (Fig. 4C). We also selected six markers of human trophoblasts from first-trimester embryos and day 10–14 in vitro BMP-differentiated hESCs [9,10], and observed significant upregulation of all genes in day 5 samples compared with hESCs (Fig. 4D). Trophoblasts are the only cell type that normally expresses human leukocyte antigen G (HLA-G) [10], and we observed elevated HLA-G expression among male and female day 5 trophoblastic cells (Fig. 4D). In sum, male and female hESCs and progenitors of trophoblastic cells have distinguishable global gene expression profiles.
FIG. 4.
Transcriptomic analysis of male and female hESCs and progenitors of trophoblastic cells. (A) Cluster dendrogram of all RNA-Seq samples (females, pink; males, blue) based on Minkowski distance (ggplot). (B) PC analysis for all hESC (d0) and progenitors of trophoblastic cells (d5) samples, with female samples in pink/green and male samples in blue/purple. The percentage contribution of each principal component is displayed in each axis. (C) Log2 reads (CPM) for pluripotency markers among all samples. Two-tailed t-tests were performed to calculate significance between undifferentiated and trophoblastic cells (**P < 0.005; ****P < 0.0001). (D) Log2 reads (CPM) for markers of the early human trophoblast for male and female samples. Two-tailed t-tests were performed to calculate significance between undifferentiated and differentiation day 5 cells (**P < 0.005; ****P < 0.0001). RNA-Seq, RNA-sequencing. Color images available online at www.liebertpub.com/scd
Abundant sex differences with numbers of differentially expressed genes and gene pathways with in vitro formation of trophoblastic progenitors
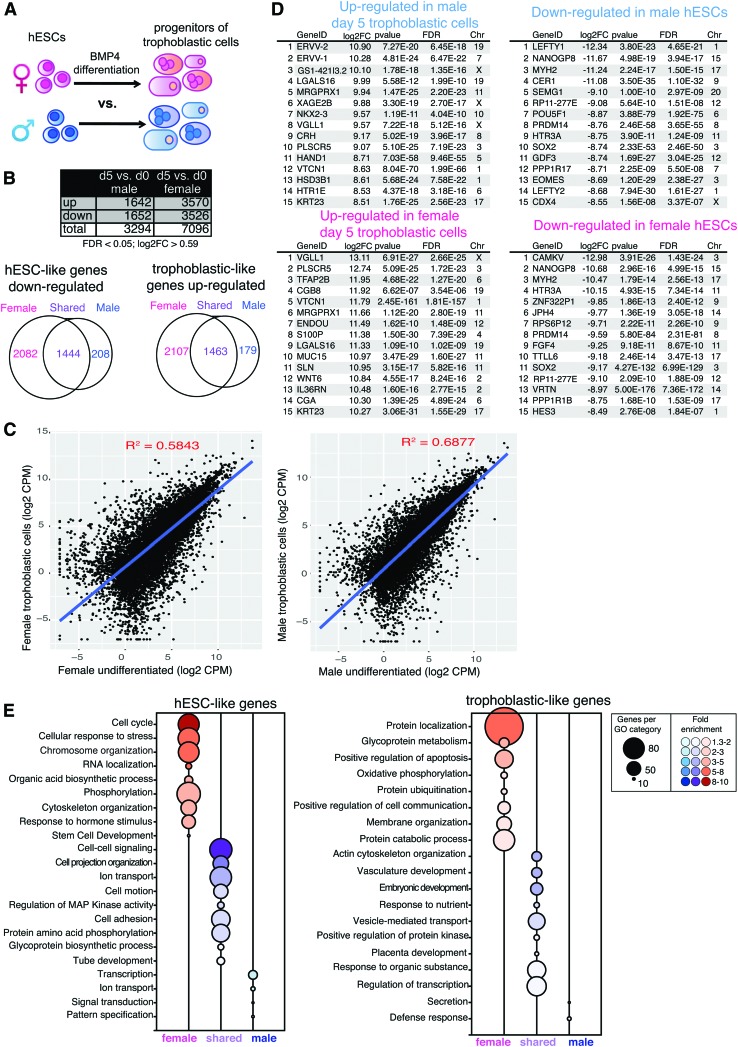
The hESCs can be differentiated into progenitors of the three germ lineages and the trophectoderm, and this conversion requires coordinated gene expression changes, which are usually characterized without regard to sex. We wondered whether the in vitro trophoblastic cell differentiation process itself might exhibit sex-biased expression differences. For these analyses, we compared male and female in vitro differentiation from hESCs to trophoblastic progenitor cells to determine sex-specific transcriptomic changes (Fig. 5A). We observed that female-specific differentiation had more differentially expressed genes compared with male differentiation (7,096 genes compared with 3,294 genes; FDR <0.05) (Fig. 5B, top). Next, we determined the number of unique and shared genes that are significantly upregulated or downregulated during male and female in vitro differentiation. For downregulated genes, 2,082 were unique to female differentiation, 208 were unique to male differentiation, and 1,444 were shared between both (Fig. 5B, bottom left). Females also displayed more upregulated genes during trophoblastic cell differentiation: 2,107 genes were uniquely female, 179 genes were unique to males, and 1,463 genes were shared between the sexes (Fig. 5B, bottom right). Scatterplots of total gene expression comparing hESCs to differentiation day 5 trophoblastic progenitors, for male and female samples, showed dramatic gene expression changes [Fig. 5C, R2 = 0.5843 (females), R2 = 0.6877 (males)], reflecting efficient differentiation with BMP4/A/P. As expected, hESCs were distinguished by elevated expression of pluripotency factors, including SOX2 and POU5F1, while progenitors of trophoblastic cells had high levels of placental markers, including IGFBP3 and KRT7, irrespective of sex (Supplementary Table S1).
FIG. 5.
Female-biased sex differences with in vitro differentiation of hESCs to trophoblastic cells. (A) Schematic showing the comparison of female BMP4/A/P differentiation (pink) with male BMP4/A/P differentiation (blue). Cells were differentiated for 5 days. (B) Top: table showing significant DEGs for female and male cells during trophoblastic differentiation (FDR <0.05, log2FC >0.59). Bottom: Venn diagrams of downregulated (left, hESC) and upregulated (right, trophoblastic cells) genes during the differentiation of trophoblastic progenitor cells in male and female cells. Numbers of genes unique to females (pink), genes unique to males (blue), and genes in shared in both males and females (purple) are shown. (C) Scatterplots (Log2 CPMs) of genes over-represented in hESCs compared with progenitors of trophoblastic cells (females, left; males, right). (D) Top: lists of the top 15 genes differentially expressed during the generation of trophoblastic progenitors (upregulated, left) from hESCs (downregulated, right) in male cells. Bottom: lists of the top 15 genes differentially expressed during the generation of trophoblastic progenitors (upregulated, left) from hESCs (downregulated, right) in female cells. (E) GO enrichment of genes significantly downregulated (left, hESCs) and upregulated (right, trophoblastic cells) during the differentiation of trophoblastic progenitor cells (GO BP “fat” terms, enrichment score ≥1.3; P < 0.05). GO, Gene Ontology. Color images available online at www.liebertpub.com/scd
Averaging all three male and three female expression sets, we generated lists for the top 15 genes overexpressed during BMP4/A/P differentiation by comparing hESCs to differentiation day 5 samples (Fig. 5D). Remarkably, more than half of the genes in these lists were unique to each sex, and nearly all of these genes were autosomal. For example, the X-linked gene CDX4 exhibited male-specific downregulation during the generation of trophoblastic progenitor cells (log2FC = −8.55, FDR = 3.37E-7; Fig. 5D and Supplementary Table S1). Female differentiation exhibited statistically significant downregulation of HES3 and FGF4, proteins important for pluripotency, survival, and growth of the inner cell mass during postimplantation development [54,55] (Fig. 5D). The most significantly downregulated genes present in both male and female cells included some known pluripotency markers: NANOGP8, PRDM14, SOX2, MYH2, HTR3A, and RP11-277E18.2 (Fig. 5D, right).
Next, we determined the genes with greatest expression in trophoblastic progenitor cells relative to hESCs for averaged male and female samples (Fig. 5D, left). Interestingly, the majority of the genes overexpressed in these cells were autosomal and also sex specific (Fig. 5D, left). Female trophoblastic progenitor cells significantly upregulated the expression of placental genes with known roles in trophoblast function and differentiation: CGA and CGB8, the alpha and beta-8 subunit of chorionic gonadotropin produced by trophoblastic cells; ENDOU, an endoribonuclease expressed in SYN [56]; SP100, a calcium-binding protein that regulates trophoblast differentiation [57]; and MUC15, mucin protein involved in trophoblast invasion [58]. Male trophoblastic progenitor cells specifically upregulated two endogenous placental retroviral genes: ERVV-1 and its paralog ERVV-2 [59], and the trophoblast-specific gene HSD3B1, involved in steroid hormone synthesis [60] (Fig. 5D). There were six genes that were consistently upregulated in both male and female samples: KRT23, VTCN1, LGALS16, MRGPRX1, PLSCR5, and the X-linked gene VGLL1 (Fig. 5D, left), which have roles in cellular differentiation, placental cells, and extracellular signaling in trophoblastic cells.
We also used GO analyses to determine enrichment of gene categories for sex-specific gene lists of upregulated and downregulated genes during in vitro differentiation of trophoblastic progenitor cells (Fig. 5E). Female samples had more GO terms for both downregulated (hESC like, Fig. 5E left) and upregulated (trophoblastic progenitor cell like, Fig. 5E right) genes during trophoblastic cell differentiation. Female hESCs downregulated pathways involved in cell cycle, chromosome organization, cellular responses to stress, phosphorylation, and cytoskeleton organization, whereas male cells downregulated transcription, ion transport, and signal transduction pathways (Fig. 5E left). Female-specific pathways upregulated during trophoblastic differentiation included protein localization, glycoprotein metabolism, membrane organization, and protein catabolism, and male cells upregulated defense responses and secretion pathways (Fig. 5E, right). The pathways upregulated in both male and female trophoblastic cells included embryonic development, vasculature development, actin cytoskeleton organization, consistent with differentiation to placental cells (Fig. 5E). In sum, we observed significant sex differences with in vitro differentiation of hESCs into trophoblastic cells, where female cells had higher numbers of differentially expressed genes and upregulated specific genes and pathways important for trophoblast cells.
Sex-biased expression of both XY-linked and autosomal genes in human trophoblastic progenitor cells
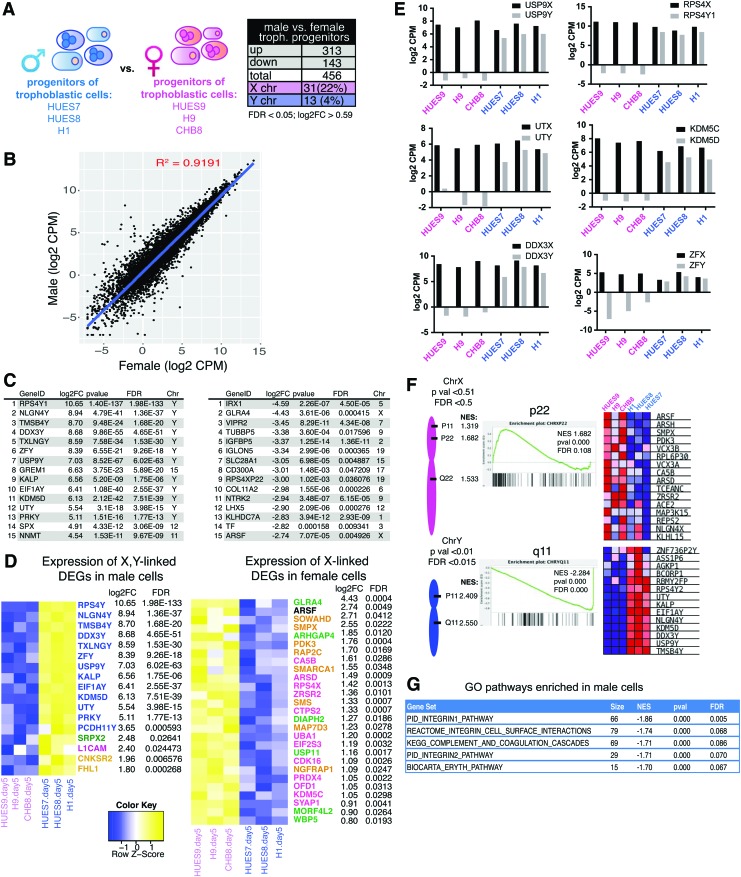
We investigated the sex-specific gene expression profiles for day 5 differentiated male and female trophoblastic progenitor cells (Fig. 6A). There were more differentially expressed genes (n = 456 genes; Fig. 6A) between male and female trophoblastic progenitors compared with hESCs (n = 98; Fig. 2A), and male samples had more upregulated genes (n = 313). We created a scatterplot to compare the average global gene expression in male and female day 5 differentiated trophoblastic progenitors, and observed similar variability compared with sex-specific hESC expression (Fig. 6B, R2 = 0.9191 compared with R2 = 0.9286). The top 15 genes exhibiting the greatest expression difference in male trophoblastic progenitor cells compared with female cells were mostly Y-linked, with the exception of the autosomal GREM1, SPX, and NNMT (Fig. 6C, left). Interestingly, of the 313 genes significantly upregulated in male trophoblastic progenitor cells, 4 were X-linked and 13 were Y-linked (Fig. 6D, left). The Y-linked genes were predominantly members of X/Y gene pairs (blue genes, Fig. 6D), and similar to our observations in male hESCs, these genes also exhibited higher dosage in male trophoblastic progenitor samples compared with female cells (Fig. 6E). Using GSEA, we found that male-biased Y-linked genes expressed in day 5 trophoblastic cells are located on p11 and q11 regions of the Y-chromosome (Fig. 6F, bottom). Although 17 of the genes upregulated in male trophoblastic progenitor cells were sex linked, the majority of genes are autosomal and from nearly every chromosome (Fig. 6A, D and Supplementary Table S1). Using GO analyses, these genes participate in integrin 1 and integrin 2 pathways (FDR = 0.005; 0.070), the integrin cell surface interaction pathway (FDR = 0.068), and the complement and coagulation cascades (FDR = 0.086) (Fig. 6G).
FIG. 6.
Sex differences in transcriptomic profiles of human trophoblastic progenitor cells. (A) Schematic depicting the three male and female trophoblastic progenitor cell samples for transcriptional profiling (left); table showing significant DEGs between sexes (right; FDR <0.05, log2FC >0.59). (B) Scatterplot of Log2 CPMs from male compared with female trophoblastic cells. (C) Lists of the top 15 significant (FDR <0.05, log2FC >0.59) DEGs comparing male with female trophoblastic cells (genes enriched in males, left; genes enriched in females, right). (D) Heatmaps for male sex-linked DEGs (left) and female sex-linked DEGs (right) in male versus female trophoblastic cells. Colored gene names in blue are from the Y-chromosome; pink denotes known escape from XCI; green denotes variable escape from XCI; yellow denotes genes subject to XCI. (E) Log2 reads (CPMs) for X-Y gene pairs (black bars: X-linked; gray bars: Y-linked) in female (pink) and male (blue) trophoblastic cells. (F) GSEA showing chromosomal locations for X-linked gene sets enriched in females (top; P < 0.05, FDR <0.05), and for Y-linked gene sets enriched in males (bottom; P < 0.01, FDR <0.015). (G) GSEA of significantly enriched canonical pathways (C2CP) in male trophoblastic cells compared with females (P < 0.0005). Color images available online at www.liebertpub.com/scd
The top 15 most differentially expressed genes in female trophoblastic progenitor cells included only 2 X-linked genes: GLRA4 and ARSF (Fig. 6C). The majority of female-specific upregulated genes were autosomal, and included the transferrin receptor (TF), placenta-expressed genes (IGFBP5, IGLON5, SLC28A1, and NTRK2), pseudogenes (TUBBP5, RPS4XP22), and genes lacking defined roles/functions in placental development (CD300A, COL11A2, LHX5, and KLHDC7A). A total of 27 X-linked genes were upregulated in female trophoblastic progenitor cells, and included X-linked genes that escape XCI (genes in pink and green; Fig. 6D right) and genes known to undergo XCI (8 genes in gold, Fig. 6D right). The eight X-linked genes (gold) overexpressed in female cells likely represent novel cell-specific XCI escape genes, as these genes were not overexpressed in female hESCs compared with males (Fig. 2). Using GSEA, we observed that X-linked genes preferentially expressed in female trophoblastic progenitors were expressed from both the p and q arms of this chromosome, and the p22 region was the most significantly enriched for female-biased gene expression (Fig. 6F). In conclusion, we found more male-specific upregulated genes in early trophoblastic progenitor cells, male-specific increased expression of X/Y pairs, and female-specific upregulation of autosomal genes.
Discussion
Fetal sex influences in utero development in healthy and complicated pregnancies, where male fetuses have increased risk of peri- and postnatal mortality, and females are smaller during gestation [61–63]. Because predisposition to many adult diseases that are sexually dimorphic (including type 2 diabetes, depression, and coronary heart disease) originate during this period [64], it is important to understand the origin of these sex differences. A number of reports profiled the placental transcriptome using full-term placentas and found sex differences [65–67], yet there are no studies examining the initial stages of trophoblast formation, which occurs early during the first trimester. Here, we used an in vitro differentiation model system, where hESCs are converted to trophoblastic cells using BMP4/A/P culture conditions, to identify sex-specific expression profiles for the progenitor cells of human placental trophoblasts. We found that there are robust sex differences in hESCs, the differentiation pathway of hESCs to trophoblastic progenitors, and in differentiation day 5 trophoblastic progenitors.
For our study, we investigated whether XIST expression status would influence gene expression comparisons between male and female hESCs. This is an important consideration because nearly all NIH-approved female hESCs are XIST negative [27]. XIST-positive hiPSCs can be generated using four-factor reprogramming, but these cells will irreversibly silence XIST during routine passage and will not recover XIST expression with differentiation [31]. Thus, XIST-positive lines are somewhat restrictive for use by the stem cell community. Here, we found that XIST expression status does not significantly affect global gene expression comparisons between male and female hPSCs. This is in agreement with previous studies demonstrating subtle gene expression differences between isogenic XIST-positive and XIST-negative hiPSCs [28]. We used unsupervised hierarchical clustering, PCA, and scatterplots, and found that the transcriptional profiles of XIST-positive and XIST-negative hPSCs were quite similar, and that sex was the most significantly distinguishing factor when comparing these profiles with male hPSCs (Fig. 1). Female hESCs and hiPSCs are often avoided by the stem cell community because of their epigenetic instability of the X and imprinted genes [32]. We do not recommend using female XIST-negative hPSCs to model X-linked diseases or for in vivo human therapies, in agreement with previous studies [28,32]. However, our findings provide strong support for using XIST-negative hPSCs for investigating sex differences in gene expression of hPSC-derived cell populations.
In our study, we compared male with female hESCs and trophoblastic progenitor cells, and discovered that male cells have greater dosage of X/Y gene pairs compared with female cells (Figs. 2E and 6E). Y-linked genes have roles beyond testis determination and spermatogenesis, exemplified by X/Y gene pairs that regulate transcription, translation, and protein stability [48], and our study demonstrates a role of X/Y genes in early placental development. We propose that the increased dosage of X/Y gene pairs in male hESCs and trophoblastic cells likely affects gene expression genome-wide. For example, male hESCs were enriched for gene pathways of transcription, RNA Pol I promoter opening, TGF-β signaling (Fig. 2F), which may contribute to the male-specific enrichment of integrin 1, integrin 2, and integrin surface interaction pathways in trophoblastic progenitors (Fig. 6G). During the preparation of this article, a study profiling human first-trimester chorionic villi samples also found significant expression of X/Y pairs with increased dosage in male samples [68]. Our study demonstrates that male-specific increased dosage of X/Y genes originates from hESCs and trophoblastic progenitors, and persists into first-trimester placental cells. It is likely that other progenitor cell populations will also exhibit male-specific increased dosage of X/Y gene pairs, and we propose that these Y-linked genes will contribute to male-specific sex-biased expression during development.
We were surprised to find strong female-biased expression with the differentiation of hESCs to trophoblastic progenitors. Female-specific differentiation exhibited about twice as many differentially expressed genes as male-specific differentiation (7,096 vs. 3,294 genes; Fig. 5B). Sex differences with the differentiation process were evident in scatterplots comparing transcriptomic profiles of hESCs with trophoblastic progenitor cells, where the female differentiation profile was more distinct (R2 = 0.5843) than male profile (R2 = 0.6877). Female hESCs upregulated 2,107 genes, whereas male samples upregulated just 179 genes during BMP4/A/P differentiation (Fig. 6B). Among these genes, we found that TFAP2B, which is a member of the AP-2 transcription factor family that regulates cellular differentiation during embryonic development [69], was dramatically upregulated (log2FC = 11.95). One possibility is that TFAP2B may function as a transcriptional activator responsible for the female-biased increased gene expression during trophoblastic cell differentiation. Additional work is necessary to investigate the targets of TFAP2B during human placental formation, and whether this transcription factor is responsible for female-biased expression in other cell types. Our study found that the female-specific genes exhibiting the most upregulation during differentiation were nearly all autosomal, and the list of genes included factors necessary for trophoblast formation and function. Because these placental/trophoblastic genes are preferentially overexpressed by differentiation day 5, it is possible that the trophoblastic cells of female placenta may develop more efficiently than its male counterpart. Also, we cannot exclude the possibility that female and male cells differentiate at different rates in vitro, which would not be evident in our study because we only examined one time point (differentiation day 5). Female-specific differentiation to trophoblastic progenitor cells was distinguished in our study by enrichment of specific pathways that promote placentation and growth, including protein localization and membrane organization. Interestingly, male-specific differentiation was enriched for defense response pathways, which suggests a sex-specific protective mechanism to enhance male placental survival from pathogens.
In our study, we found that trophoblastic progenitors, at BMP4/A/P differentiation day 5, also exhibit robust sex differences with gene expression (n = 456 genes). In male cells, we observed significant overexpression of Y-linked genes, especially those with an X-counterpart (X/Y pairs). Yet, we were surprised that female trophoblastic progenitors predominantly overexpressed autosomal genes, including the TF and genes expressed in placenta that lack functional characterization in placentation (Fig. 6C). It is possible that these autosomal genes (IGFBP5, IGLON5, SLC28A1, and NTRK2) may have female-specific roles in trophoblast cell differentiation during early pregnancy. We examined X-linked gene expression in female trophoblastic progenitors, and found 12 genes overexpressed that are known to escape XCI [47] and 8 putative XCI escape genes (Fig. 6D, pink). The X-linked gene ARSF, which encodes a sulfatase enzyme, was overexpressed in female trophoblastic progenitors (log2FC = 2.74). While it is unknown whether this gene is subject to XCI or if it escapes [47], it is possible that ARSF escapes XCI in trophoblastic progenitors. Similarly, the eight putative XCI escape genes X, which do not have known Y-chromosome paralogs, may be examples of cell-specific genes that escape XCI in the early human placenta. Additional experiments to demonstrate whether these genes (ARSF, SOWAHD, SMPX, PDK3, RAP2C, SMARCA1, SMS, MAP7D3, and NGFRAP1) are biallelically expressed in trophoblastic progenitor cells are necessary.
Our study is the first to identify sex differences in the transcriptional profiles of human trophoblastic progenitor cells, which form the placenta during early first-trimester pregnancies. We used an hESC-derived in vitro differentiation model to examine sex-biased gene expression differences in pluripotent cells, differentiation day 5 trophoblastic progenitors, and also with the differentiation process itself. Our results provide critical insight into the earliest stages of normal placental development, and identify sex-biased gene expression originating from utero. Future experiments that follow expression changes of these sexually dimorphic trophoblastic progenitor genes into later stages of placentation may shed light on the development of sex differences in adult diseases. This study demonstrates that hPSC-derived cell populations can be used to investigate sex-biased processes, and underscore the importance of sex as a variable for the design and analysis of experiments involving hPSCs.
Supplementary Material
Acknowledgments
We thank A. Delaney for assistance with flow cytometry experiments and L. King for comments on the article.
This work was supported by a Pennsylvania Health Research Formula Fund (to M.C.A.), an NIH NICHD BIRCWH (K12 HD085848) to M.C.A., an NIH T32-GM007229 (to C.M.S.), an NIH T32-HD083185 (to C.M.S.), and an NIH F31-GM123604 (to C.M.S.).
Availability of Data and Material
RNA-Seq data sets have been deposited in the Gene Expression Omnibus (GEO) database (GSE115941).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rugg-Gunn PJ. (2012). Epigenetic features of the mouse trophoblast. Reprod Biomed Online 25:21–30 [DOI] [PubMed] [Google Scholar]
- 2.Delorme-Axford E, Sadovsky Y. and Coyne CB. (2014). The placenta as a barrier to viral infections. Annu Rev Virol 1:133–146 [DOI] [PubMed] [Google Scholar]
- 3.Rossant J. and Cross JC. (2001). Placental development: lessons from mouse mutants. Nat Rev Genet 2:538–548 [DOI] [PubMed] [Google Scholar]
- 4.Carter AM. and Mess A. (2007). Evolution of the placenta in eutherian mammals. Placenta 28:259–262 [DOI] [PubMed] [Google Scholar]
- 5.Xu RH, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP. and Thomson JA. (2002). BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20:1261–1264 [DOI] [PubMed] [Google Scholar]
- 6.Amita M, Adachi K, Alexenko AP, Sinha S, Schust DJ, Schulz LC, Roberts RM. and Ezashi T. (2013). Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4. Proc Natl Acad Sci U S A 110:E1212–E1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts RM, Loh KM, Amita M, Bernardo AS, Adachi K, Alexenko AP, Schust DJ, Schulz LC, Telugu BP, Ezashi T. and Pedersen RA. (2014). Differentiation of trophoblast cells from human embryonic stem cells: to be or not to be? Reproduction 147:D1–D12 [DOI] [PubMed] [Google Scholar]
- 8.Sarkar P, Randall SM, Collier TS, Nero A, Russell TA, Muddiman DC. and Rao BM. (2015). Activin/nodal signaling switches the terminal fate of human embryonic stem cell-derived trophoblasts. J Biol Chem 290:8834–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchand M, Horcajadas JA, Esteban FJ, McElroy SL, Fisher SJ. and Giudice LC. (2011). Transcriptomic signature of trophoblast differentiation in a human embryonic stem cell model. Biol Reprod 84:1258–1271 [DOI] [PubMed] [Google Scholar]
- 10.Lee CQ, Gardner L, Turco M, Zhao N, Murray MJ, Coleman N, Rossant J, Hemberger M. and Moffett A. (2016). What is trophoblast? A combination of criteria define human first-trimester trophoblast. Stem Cell Rep 6:257–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misra DP, Salafia CM, Miller RK. and Charles AK. (2009). Non-linear and gender-specific relationships among placental growth measures and the fetoplacental weight ratio. Placenta 30:1052–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsén T, Eriksson JG, Tuomilehto J, Osmond C. and Barker DJ. (1999). Growth in utero and during childhood among women who develop coronary heart disease: longitudinal study. BMJ 319:1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buckberry S, Bianco-Miotto T, Bent SJ, Dekker GA. and Roberts CT. (2014). Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol Hum Reprod 20:810–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verburg PE, Tucker G, Scheil W, Erwich JJ, Dekker GA. and Roberts CT. (2016). Sexual dimorphism in adverse pregnancy outcomes—a retrospective Australian population study 1981–2011. PLoS One 11:e0158807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes JF. and Page DC. (2015). The biology and evolution of mammalian Y chromosomes. Annu Rev Genet 49:507–527 [DOI] [PubMed] [Google Scholar]
- 16.Cannon-Albright LA, Farnham JM, Bailey M, Albright FS, Teerlink CC, Agarwal N, Stephenson RA. and Thomas A. (2014). Identification of specific Y chromosomes associated with increased prostate cancer risk. Prostate 74:991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumanski JP, Rasi C, Lönn M, Davies H, Ingelsson M, Giedraitis V, Lannfelt L, Magnusson PK, Lindgren CM, et al. (2015). Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science 347:81–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charchar FJ, Bloomer LD, Barnes TA, Cowley MJ, Nelson CP, Wang Y, Denniff M, Debiec R, Christofidou P, et al. (2012). Inheritance of coronary artery disease in men: an analysis of the role of the Y chromosome. Lancet 379:915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee NR, Wallace GL, Adeyemi EI, Lopez KC, Blumenthal JD, Clasen LS. and Giedd JN. (2012). Dosage effects of X and Y chromosomes on language and social functioning in children with supernumerary sex chromosome aneuploidies: implications for idiopathic language impairment and autism spectrum disorders. J Child Psychol Psychiatry 53:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lleo A, Oertelt-Prigione S, Bianchi I, Caliari L, Finelli P, Miozzo M, Lazzari R, Floreani A, Donato F, et al. (2013). Y chromosome loss in male patients with primary biliary cirrhosis. J Autoimmun 41:87–91 [DOI] [PubMed] [Google Scholar]
- 21.Payer B. and Lee JT. (2008). X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42:733–772 [DOI] [PubMed] [Google Scholar]
- 22.Lyon MF. (1961). Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373 [DOI] [PubMed] [Google Scholar]
- 23.Marahrens Y, Panning B, Dausman J, Strauss W. and Jaenisch R. (1997). Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11:156–166 [DOI] [PubMed] [Google Scholar]
- 24.Penny GD, Kay GF, Sheardown SA, Rastan S. and Brockdorff N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379:131–137 [DOI] [PubMed] [Google Scholar]
- 25.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B. and Zhang Y. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135 [DOI] [PubMed] [Google Scholar]
- 26.Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T. and Wutz A. (2004). A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2:E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lessing D, Anguera MC. and Lee JT. (2013). X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 14:85–110 [DOI] [PubMed] [Google Scholar]
- 28.Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, et al. (2012). Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell 11:75–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva SS, Rowntree RK, Mekhoubad S. and Lee JT. (2008). X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A 105:4820–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD. and Fan G. (2008). X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc Natl Acad Sci U S A 105:4709–4714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tchieu J, Kuoy E, Chin MH, Trinh H, Patterson M, Sherman SP, Aimiuwu O, Lindgren A, Hakimian S, et al. (2010). Female human iPSCs retain an inactive X chromosome. Cell Stem Cell 7:329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A. and Eggan K. (2012). Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 10:595–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel S, Bonora G, Sahakyan A, Kim R, Chronis C, Langerman J, Fitz-Gibbon S, Rubbi L, Skelton RJP, et al. (2017). Human embryonic stem cells do not change their X inactivation status during differentiation. Cell Rep 18:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, Guenther MG, Welstead GG, Alagappan R, Frampton GM, Xu P, et al. (2010). Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 141:872–883 [DOI] [PubMed] [Google Scholar]
- 35.Erwin JA. and Lee JT. (2010). Characterization of X-chromosome inactivation status in human pluripotent stem cells. Curr Protoc Stem Cell Biol Chapter 1: Unit 1B.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beiting DP, Peixoto L, Akopyants NS, Beverley SM, Wherry EJ, Christian DA, Hunter CA, Brodsky IE. and Roos DS. (2014). Differential induction of TLR3-dependent innate immune signaling by closely related parasite species. PLoS One 9:e88398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 12:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bray NL, Pimentel H, Melsted P. and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527 [DOI] [PubMed] [Google Scholar]
- 39.Soneson C, Love MI. and Robinson MD. (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Love MI, Huber W. and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregory R, Warnes BB, Bonebakker L, Gentleman R, Huber TLW, Liaw A, Maechler M, Magnusson A. and Steffen Moeller M. (2016). gplots: various R programming tools for plotting data. R package version 3.0.1 [Google Scholar]
- 42.Huang da W, Sherman BT. and Lempicki RA. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- 43.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. and Mesirov JP. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dvash T, Lavon N. and Fan G. (2010). Variations of X chromosome inactivation occur in early passages of female human embryonic stem cells. PLoS One 5:e11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang KH, Chan-Ling T, McFarland EL, Afzal A, Pan H, Baxter LC, Shaw LC, Caballero S, Sengupta N, et al. (2007). IGF binding protein-3 regulates hematopoietic stem cell and endothelial precursor cell function during vascular development. Proc Natl Acad Sci U S A 104:10595–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schier AF. and Shen MM. (2000). Nodal signalling in vertebrate development. Nature 403:385–389 [DOI] [PubMed] [Google Scholar]
- 47.Balaton BP, Cotton AM. and Brown CJ. (2015). Derivation of consensus inactivation status for X-linked genes from genome-wide studies. Biol Sex Differ 6:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, et al. (2014). Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508:494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carrel L. and Willard HF. (2005). X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404 [DOI] [PubMed] [Google Scholar]
- 50.Yang F, Babak T, Shendure J. and Disteche CM. (2010). Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res 20:614–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang J. and Anguera MC. (2017). In vitro differentiation of human pluripotent stem cells into trophoblastic cells. J Vis Exp Mar 16 DOI: 10.379/55268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niakan KK. and Eggan K. (2013). Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol 375:54–64 [DOI] [PubMed] [Google Scholar]
- 53.Debacker C, Catala M. and Labastie MC. (1999). Embryonic expression of the human GATA-3 gene. Mech Dev 85:183–187 [DOI] [PubMed] [Google Scholar]
- 54.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM. and Goldfarb M. (1995). Requirement of FGF-4 for postimplantation mouse development. Science 267:246–249 [DOI] [PubMed] [Google Scholar]
- 55.van de Leemput J, Boles NC, Kiehl TR, Corneo B, Lederman P, Menon V, Lee C, Martinez RA, Levi BP, et al. (2014). CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 83:51–68 [DOI] [PubMed] [Google Scholar]
- 56.Laneve P, Gioia U, Ragno R, Altieri F, Di Franco C, Santini T, Arceci M, Bozzoni I. and Caffarelli E. (2008). The tumor marker human placental protein 11 is an endoribonuclease. J Biol Chem 283:34712–34719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu HY, Wang JX, Tong XM, Xue YM. and Zhang SY. (2015). S100P regulates trophoblast-like cell proliferation via P38 MAPK pathway. Gynecol Endocrinol 31:796–800 [DOI] [PubMed] [Google Scholar]
- 58.Shyu MK, Lin MC, Shih JC, Lee CN, Huang J, Liao CH, Huang IF, Chen HY, Huang MC. and Hsieh FJ. (2007). Mucin 15 is expressed in human placenta and suppresses invasion of trophoblast-like cells in vitro. Hum Reprod 22:2723–2732 [DOI] [PubMed] [Google Scholar]
- 59.Esnault C, Cornelis G, Heidmann O. and Heidmann T. (2013). Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet 9:e1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao TL, Kurman RJ, Jeng YM, Huang W. and Shih IeM. (2008). HSD3B1 as a novel trophoblast-associated marker that assists in the differential diagnosis of trophoblastic tumors and tumorlike lesions. Am J Surg Pathol 32:236–242 [DOI] [PubMed] [Google Scholar]
- 61.Bukowski R, Smith GC, Malone FD, Ball RH, Nyberg DA, Comstock CH, Hankins GD, Berkowitz RL, Gross SJ, et al. (2007). Human sexual size dimorphism in early pregnancy. Am J Epidemiol 165:1216–1218 [DOI] [PubMed] [Google Scholar]
- 62.Peacock JL, Marston L, Marlow N, Calvert SA. and Greenough A. (2012). Neonatal and infant outcome in boys and girls born very prematurely. Pediatr Res 71:305–310 [DOI] [PubMed] [Google Scholar]
- 63.Byrne J. and Warburton D. (1987). Male excess among anatomically normal fetuses in spontaneous abortions. Am J Med Genet 26:605–611 [DOI] [PubMed] [Google Scholar]
- 64.Barker DJ, Eriksson JG, Forsén T. and Osmond C. (2002). Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 31:1235–1239 [DOI] [PubMed] [Google Scholar]
- 65.Sood R, Zehnder JL, Druzin ML. and Brown PO. (2006). Gene expression patterns in human placenta. Proc Natl Acad Sci U S A 103:5478–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cvitic S, Longtine MS, Hackl H, Wagner K, Nelson MD, Desoye G. and Hiden U. (2013). The human placental sexome differs between trophoblast epithelium and villous vessel endothelium. PLoS One 8:e79233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Buckberry S, Bianco-Miotto T. and Roberts CT. (2014). Imprinted and X-linked non-coding RNAs as potential regulators of human placental function. Epigenetics 9:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez TL, Sun T, Koeppel AF, Lee B, Wang ET, Farber CR, Rich SS, Sundheimer LW, Buttle RA, et al. (2018). Sex differences in the late first trimester human placenta transcriptome. Biol Sex Differ 9:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Satoda M, Zhao F, Diaz GA, Burn J, Goodship J, Davidson HR, Pierpont ME. and Gelb BD. (2000). Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet 25:42–46 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-Seq data sets have been deposited in the Gene Expression Omnibus (GEO) database (GSE115941).